Significance
Many ion channels are composed of related but structurally distinct membrane protein subunits, each determining the functional properties of the channel. Although their composition is carefully regulated, the mechanisms underlying their assembly are poorly understood. We show that mRNA transcripts encoding human ether-à-go-go-related gene (hERG) 1a and 1b subunits, which assemble to form channels mediating cardiac repolarization, are physically associated and can be coimmunoprecipitated with an Ab against the nascent hERG 1a protein. Their association allows the subunits to interact during protein translation to form the heteromeric ion channel. These findings are important for understanding hERG channels, which are a target for acquired and congenital long QT syndrome, and may be generalizable to oligomeric proteins serving a wide range of biological functions.
Keywords: KCNH2, hERG, cotranslational assembly, microtranslatome, hERG 1a/1b
Abstract
Oligomers of homomeric voltage-gated potassium channels associate early in biogenesis as the nascent proteins emerge from the polysome. Less is known about how proteins emerging from different polysomes associate to form hetero-oligomeric channels. Here, we report that alternate mRNA transcripts encoding human ether-à-go-go-related gene (hERG) 1a and 1b subunits, which assemble to produce ion channels mediating cardiac repolarization, are physically associated during translation. We show that shRNA specifically targeting either hERG 1a or 1b transcripts reduced levels of both transcripts, but only when they were coexpressed heterologously. Both transcripts could be copurified with an Ab against the nascent hERG 1a N terminus. This interaction occurred even when translation of 1b was prevented, indicating the transcripts associate independent of their encoded proteins. The association was also demonstrated in cardiomyocytes, where levels of both hERG transcripts were reduced by either 1a or 1b shRNA, but native KCNE1 and ryanodine receptor 2 (RYR2) transcripts were unaffected. Changes in protein levels and membrane currents mirrored changes in transcript levels, indicating the targeted transcripts were undergoing translation. The physical association of transcripts encoding different subunits provides the spatial proximity required for nascent proteins to interact during biogenesis, and may represent a general mechanism facilitating assembly of heteromeric protein complexes involved in a range of biological processes.
Most oligomeric membrane proteins are heteromers of subunits whose respective structures confer specific properties. For example, different GABAA receptor subunits confer distinct single-channel conductances, gating kinetics and pharmacological sensitivities to benzodiazepines and other drugs (1). For voltage-gated K (Kv) channels, different subunits assemble to provide functional diversity within subfamilies, but not across subfamily boundaries (2, 3). How the right subunits find each other to assemble into multimeric proteins with the appropriate stoichiometry and functional properties is a major unanswered question.
Cardiac IKr, a repolarizing current in the heart (4), is produced by heteromeric assemblies of human ether-à-go-go-related gene (hERG) 1a and 1b subunits (5–10). Perturbations of IKr by congenital mutations in the KCNH2/hERG1 gene or by drug block can prolong the ventricular action potential and associated QT interval to cause long QT syndrome and sudden cardiac death (11–13). hERG 1a and 1b subunits are encoded by alternate transcripts that arise from distinct promoters within the hERG1 locus (7, 8). The subunits are identical except for their N termini, which differ in length and primary sequence. The hERG 1a subunits can form homomeric channels and produce membrane currents in heterologous expression systems. In contrast, the 1b subunits are largely retained in the endoplasmic reticulum (ER) unless coexpressed with hERG 1a, which masks an ER retention signal in the hERG 1b N terminus (14). Compared with 1a homomers, 1a/1b heteromers exhibit accelerated channel gating, twice the repolarizing current magnitude, and distinct pharmacology (15, 16). Both subunits are expressed in native tissue, where they colocalize to T-tubular structures of ventricular myocytes (10). In isolated cardiomyocytes, IKr channels can be functionally converted to homomeric 1a-like channels by exogenous expression of the 1a-specific Per-Arnt-Sim (PAS) domain. The result is diminished repolarizing current amplitude and cellular manifestations of proarrhythmia, including prolonged action potential duration (APD), APD variability, and early afterdepolarizations (9). These findings reinforce the importance of both hERG 1a and 1b subunits in cardiac repolarization.
Deutsch and coworkers (17–19) established that N-terminal domains of Kv1.3 homo-oligomers associate cotranslationally as the nascent proteins emerge from the polysome. The assembly of hetero-oligomeric hERG 1a/1b channels has been suggested similarly to involve cotranslational, cytosolic N-terminal interactions between subunits (20). In this case, the hERG 1a and 1b N termini are structurally distinct, but were shown to interact in a dose-dependent manner in vitro. In cells, the 1a N-terminal fragment can disrupt 1b subunit homo-oligomerization and core glycosylation. Because core glycosylation occurs cotranslationally (21, 22), these observations suggest the two subunits associate via N-terminal interactions early in biogenesis (20). Such a cotranslational interaction implies that the 1a and 1b mRNA transcripts and their respective polysomes must be located in close physical proximity to each other. How is this proximity achieved?
Previously, we conducted 1b knockdown experiments with shRNA that specifically targets hERG 1b, but not hERG 1a, expressed independently in HEK293 cells. Surprisingly, the 1b-specific shRNA reduced both 1a and 1b transcripts when they were coexpressed in HEK293 cells (9), suggesting the transcripts were associated. Here, we tested this hypothesis and the more general idea that heteromeric channel assembly is mediated by a complex comprising the encoding mRNA species. We found that hERG 1a and 1b transcripts are associated in HEK293 cells, along with the nascent 1a polypeptide. The association was also observed in cardiomyocytes derived from human induced pluripotent stem cells (iPSC-CMs) for mRNAs encoding hERG subunits, but not other cardiac proteins tested. These results identify a pool of associated transcripts, or “microtranslatome,” as a key mechanism in the biogenesis of a multimeric ion channel whose function critically relies on the contribution of distinct protein subunits.
Results
mRNA Transcript Association in HEK293 Cells.
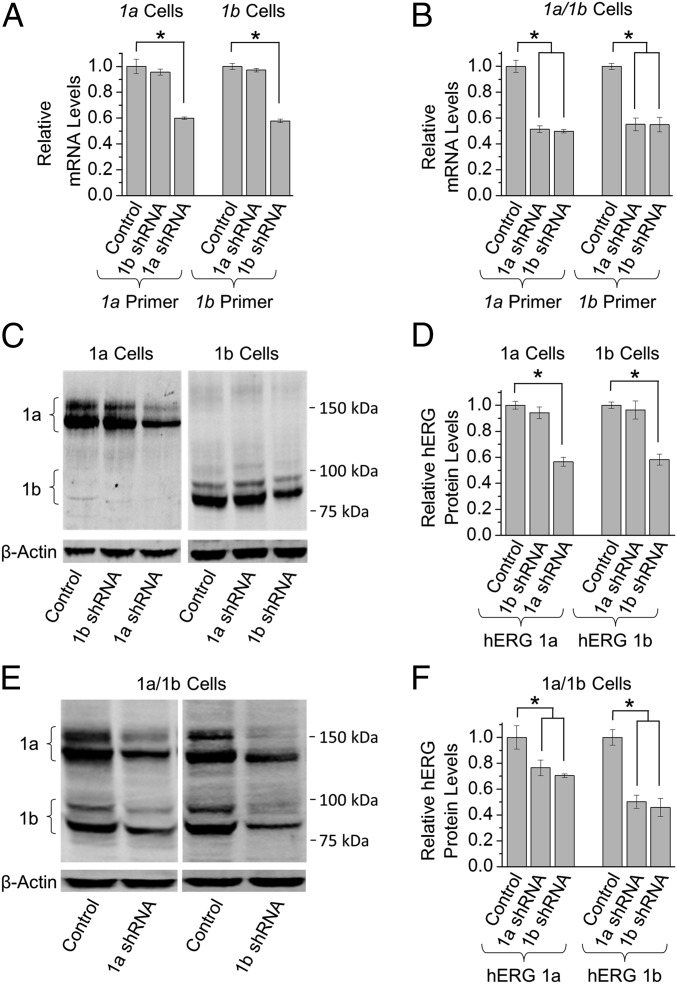
We first tested the effects of 1a- and 1b-specific shRNA on the corresponding transcript quantities using HEK293 cells transiently transfected with either the hERG 1a or hERG 1b subunit. Each shRNA, compared with a nontargeting shRNA control, reduced its corresponding mRNA levels by about half (Fig. 1A). The 1b shRNA had no effect on hERG 1a mRNA expressed alone (Fig. 1A, Left), and 1a shRNA had no effect on hERG 1b expressed alone (Fig. 1A, Right). However, when hERG 1a and 1b were coexpressed, either shRNA (compared with nontargeting shRNA control) reduced levels of both 1a and 1b mRNA by roughly half (Fig. 1B). A corresponding reduction in protein levels indicates that the shRNA targeted transcripts undergoing translation whether expressed alone (Fig. 1 C and D) or in combination (Fig. 1 E and F). Thus, selective targeting of either transcript causes a coordinate reduction of both transcript levels not attributable to off-target effects. This observation suggests an association of the coexpressed transcripts.
Fig. 1.
Cotranslational co-knockdown of hERG 1a and 1b mRNA by transcript-specific shRNA constructs. (A) Effects of transcript-specific shRNA vs. nontargeting control on mRNA levels (mean ± SEM) in HEK293 cells expressing either hERG 1a (Left) or hERG 1b (Right) (Materials and Methods). (B) Effects of transcript-specific shRNA vs. nontargeting shRNA controls on mRNA levels from HEK293 cells coexpressing hERG 1a and 1b. The mRNA levels were significantly reduced using either shRNA construct. (C) Exemplar and (D) quantification of Western blots corresponding to A showing transcript-specific reduction of hERG 1a (Left) or hERG 1b (Right) protein. (E) Exemplar and (F) quantification of Western blots showing effects of transcript-specific shRNA vs. nontargeting controls on coexpressed hERG 1a and hERG 1b protein. Both subunits were reduced, corresponding to the decrease of mRNA in B (n = 3–5). Statistical significance was assessed using ANOVA and a Bonferroni post hoc t test. *P < 0.05.
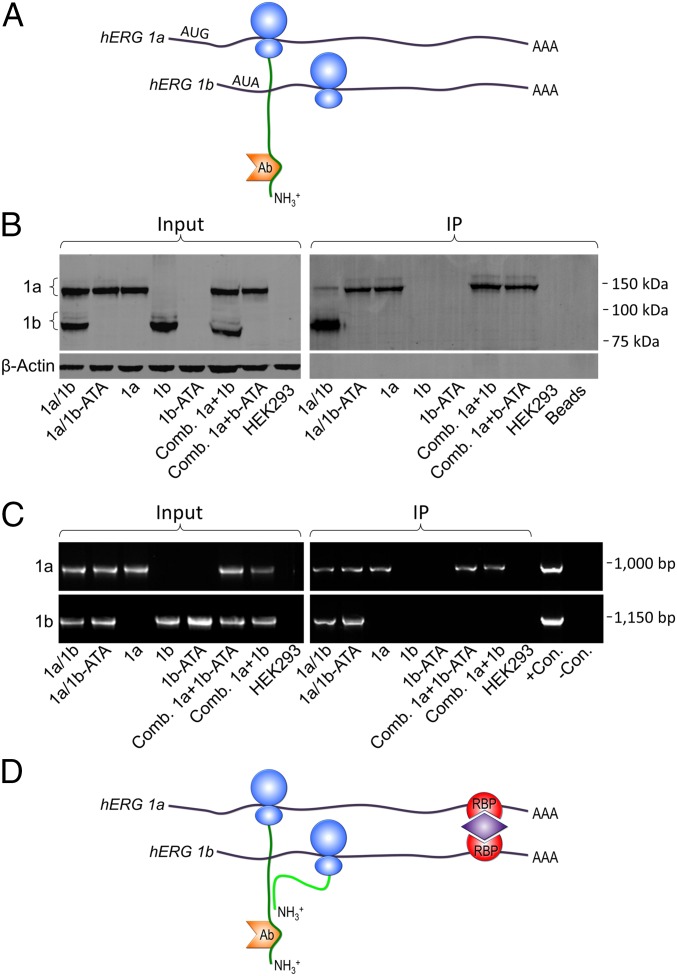
We reasoned that if alternate hERG 1a and 1b transcripts physically associate during translation, we should be able to immunoprecipitate both transcripts with an Ab to one of their nascent proteins. We tested this hypothesis in HEK293 cells coexpressing hERG 1a and 1b using a 1a-specific, N-terminal Ab to immunoprecipitate the nascent hERG 1a protein (Fig. 2A). We carried out RT-PCR to identify transcripts associated with the 1a protein (Fig. 2B). In the input lanes (Fig. 2B, Left), no signal was amplified by the 1b primers when 1a was expressed alone, or by the 1a primers when 1b was expressed alone, demonstrating the specificity of the PCR primers. No signal was amplified in the untransfected HEK293 cells. The immunoprecipitation (IP) lanes (Fig. 2B, Right) show that both 1a and 1b transcripts were immunoprecipitated by the hERG 1a-specific Ab. When expressed alone, only the 1a signal was present, but not the 1b, a control indicating that the Ab did not directly interact with the 1b mRNA. It is apparent that the two transcripts associate only when coexpressed because the 1b transcript did not coimmunoprecipitate when lysates from cells independently expressing 1a and 1b were combined. We conclude that the 1a-specific Ab immunoprecipitated not only the 1a protein but also associated 1a and 1b mRNA.
Fig. 2.
hERG 1a and 1b transcripts associate with nascent hERG 1a protein. (A) Schematic depicting cotranslational association of hERG 1a and hERG 1b polypeptides (green) as they emerge from ribosomes (blue) on neighboring hERG transcripts (black). The hERG 1a-specific Ab (orange) used to immunoprecipitate the mRNA/protein complex is shown bound to the 1a N terminus. (B) RT-PCR products showing hERG mRNA transcripts from HEK293 cells before (Input) and after (IP) using a hERG 1a-specific Ab. Input lanes show primers were specific to each transcript. IP lanes show that, when coexpressed, both species coimmunoprecipitated with the hERG 1a Ab. When expressed alone, 1a, but not 1b, copurified with the 1a Ab, indicating the Ab did not bind 1b mRNA directly. The 1b failed to copurify with the 1a Ab when lysates were mixed from cells separately expressing hERG 1a and 1b (Comb. 1a+1b). Blank HEK293 and H2O lanes show an absence of contaminating templates; control (+Con.) 1a and 1b represent signal amplified from the purified cDNA template.
We considered the possibility that the transcripts could coimmunoprecipitate based solely on the interactions of their nascent polypeptides (see Fig. 2A). To test this hypothesis, we prevented hERG 1b mRNA translation by mutating its translation initiation site in the cDNA (ATG to ATA or AUA in mRNA; Fig. 3A). The Western blot in Fig. 3B shows robust expression of WT 1b alone or in combination with 1a, but no hERG 1b protein when the translation start site was mutated (Fig. 3B, Left). No evidence of downstream initiation of protein synthesis was observed, indicating that the mutation fully abrogated 1b translation (Figs. 3B and Fig. S1). The IP lanes also failed to show any signal for the 1b-ATA mutant protein coexpressed with hERG 1a (Fig. 3B, Right). The WT 1b protein expressed alone did not immunoprecipitate with the 1a-specific Ab, further demonstrating Ab specificity. These results established that we could prevent hERG 1b translation by mutating the translation start site.
Fig. 3.
mRNA transcripts interact independent of the nascent hERG 1a and 1b protein interaction. (A) Cartoon showing experimental design, with 1a-specific Ab used to immunoprecipitate hERG 1a protein in the absence of 1b protein. (B) Western blot demonstrating loss of hERG 1b protein when the translation initiation site is mutated (1b-ATA). Lanes indicate constructs transfected in Input lanes (Left) and IP lanes (Right). Combined lysates from cells independently expressing hERG 1a and hERG 1b or hERG 1b-ATA are also shown (Comb.). β-Actin is shown as a loading control taken from the same gel. (C, Left) RT-PCR representing hERG mRNA transcripts expressed HEK293 cell lysate (Input). (C, Right) IP (as per the scheme in B) shows 1b-ATA transcript copurified with 1a-specific Ab but only when coexpressed with 1a. (D) Working model for the mechanism of hERG 1a and 1b transcript association.
Fig. S1.
ATA mutation in hERG 1b prevents hERG protein synthesis. A Western blot depicts hERG protein from blank HEK293 cells or HEK cells transfected with either WT hERG 1b or the hERG 1b ATA mutant. The hERG protein was labeled using a C-terminal Ab. These data exclude initiation of translation downstream of the ATA mutation.
We then used RT-PCR to determine whether preventing hERG 1b translation affected association of hERG 1a and 1b transcripts. It is clear from the input lanes (Fig. 3C, Left) that the mutated cDNA (1b-ATA) produced a stable transcript. With IP, the mutant 1b signal copurified with hERG 1a, but only when coexpressed in the same cell and not from combined lysates independently expressing the two transcripts (Fig. 3C, Right). Thus, the transcripts associated even when the nascent hERG 1a and 1b polypeptides did not interact, indicating that an interaction at the RNA level unites the transcripts encoding the hERG 1a/1b heteromultimers underlying cardiac IKr. Approaches such as RNA-sequencing (RNA-seq) and mass spectrometry (MS) will be required to identify candidate proteins mediating these interactions, as illustrated in a working model in Fig. 3D.
mRNA Transcript Association in Cardiomyocytes.
To determine whether the hERG 1a and 1b transcripts similarly associate under native conditions, we used iPSC-CMs in which native IKr is produced by heteromeric channels comprising hERG 1a and 1b subunits (9). In the iPSC-CMs, like HEK293 cells, either transcript-specific shRNA reduced levels of both hERG 1a and 1b by about 50% or more (Fig. 4A). The effect was specific for hERG mRNA, with no changes observed in levels of transcripts encoding KCNE1, a protein that interacts with KCNQ1 to form IKs channels (23, 24), or the type 2 ryanodine receptor (RYR2) involved in excitation–contraction coupling (25).
Fig. 4.
hERG 1a and 1b transcripts are associated in cardiomyocytes. (A) Effects of transcript-specific shRNA vs. nontargeting controls on mRNA levels (mean ± SEM) from iPSC-CMs (Materials and Methods). Either shRNA reduced hERG 1a and 1b (two panels on left), but not KCNE1 or RYR2 mRNA (two panels on right) (n = 3). (B, Left, Input) RT-PCR products from lysates of iPSC-CMs; HEK293 cells expressing 1a/1b, 1a, and 1b, respectively, and blank HEK293 cells. (B, Right, IP) Signals amplified from samples immunoprecipitated with the 1a-specific Ab show that 1a and 1b transcripts associate in iPSC-CMs. Signal from HEK293 cells expressing both transcripts (1a/1b) is shown as a control. Amplification of hERG 1a and 1b cDNA is shown as a positive control. Statistical significance was assessed using ANOVA and Bonferroni post hoc t tests. *P < 0.05.
To assay directly for transcript interaction in iPSC-CMs, we used the hERG 1a-specific Ab to immunoprecipitate the nascent hERG 1a protein and assay for associated transcripts using RT-PCR (Fig. 4B). Transcript-specific primers amplified signal corresponding to both 1a and 1b mRNA species in the IP. These findings demonstrate that in cardiomyocytes, like HEK293 cells, nascent hERG 1a polypeptides are in a complex with hERG 1a and 1b transcripts encoding heteromeric hERG subunits.
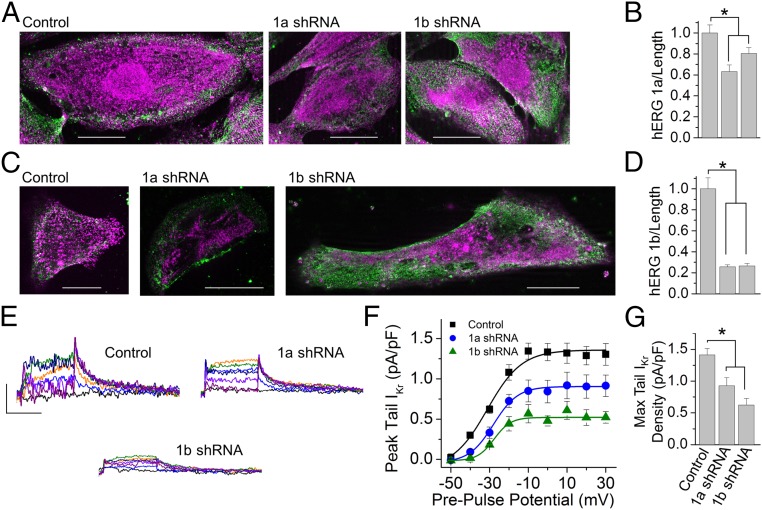
To evaluate whether the transcripts exhibiting co-knockdown in the cardiomyocytes were those transcripts actively undergoing translation, we measured hERG protein expression using two independent methods: immunocytochemistry and IKr current density (Fig. 5). Blind scoring of cells using isoform-specific Abs revealed a reduction in levels of both proteins by either 1a- or 1b-specific shRNA (Fig. 5 A–D). Peak IKr tail current density was correspondingly reduced (Fig. 5 E–G and Materials and Methods). These experiments independently confirm that the mRNA transcripts targeted by the shRNA in the iPSC-CMs were actively undergoing protein translation. The observation that either shRNA can reduce current magnitude underscores the importance of both subunits in IKr channel function (9).
Fig. 5.
shRNA knockdown in cardiomyocytes targets mRNA undergoing protein translation. (A) hERG 1a (magenta) and β-actin (green) immunofluorescence from iPSC-CMs transfected with nontargeting shRNA (Left, Control), 1a shRNA (Middle), or 1b shRNA (Right). (Scale bars: 20 μM.) (B) Quantification of data represented in A shows 1a fluorescence (mean ± SEM) was significantly reduced by either 1a- or 1b-specific shRNA compared with nontargeting control shRNA, indicating a reduction of hERG 1a protein (n = 17–21 cells). P ≤ 0.02. (C) hERG 1b (magenta) and β-actin (green) immunofluorescence from iPSC-CMs transfected with nontargeting shRNA (Left, Control), 1a shRNA (Middle), or 1b shRNA (Right). (Scale bars: 20 μM.) (D) Quantification of data represented in C shows 1b fluorescence was significantly reduced by either 1a- or 1b-specific shRNA compared with control cells, indicating a reduction of hERG 1b protein (n = 19–22 cells). P < 0.001. (E) IKr measured as E-4031–sensitive membrane currents recorded from iPSC-CMs transfected with nontargeting shRNA (Left, Control), 1a shRNA (Right), or 1b shRNA (Lower). (Scale bars: 0.5 pA/pF by 2 s.) (F) Quantification of peak tail current represented by traces in E plotted as a function of prepulse potential and fitted with a Boltzmann function for IPSC-CMs transfected with nontargeting control shRNA (black), 1a shRNA (blue), and 1b shRNA (green). (G) shRNA transfection significantly reduced peak tail IKr density (n = 4–9 cells). Max, maximum. P ≤ 0.02. Statistical significance was assessed using an ANOVA and a Bonferroni post hoc t test. *P < 0.05.
Discussion
In this study, we demonstrate that hERG 1a/1b heteromeric channels form by the cotranslational association of hERG 1a and 1b transcripts, which can be coordinately regulated. Using transcript-specific RNAi in HEK293 cells or human iPSC-CMs, levels of both transcripts were reduced when either was targeted. Moreover, both transcripts could be coimmunoprecipitated with an Ab specific to the nascent hERG 1a polypeptide. The co-knockdown effect was exerted on transcripts undergoing translation, as demonstrated by a corresponding effect on protein levels measured by Western blot, immunocytochemistry, and membrane current density. Association of the two nascent proteins was not required for interaction of their encoding transcripts, which coimmunoprecipitated even in the absence of hERG 1b when its translation was prevented. Thus, these alternate transcripts are associated during protein translation, part of a microtranslatome that supports the avid assembly of hERG 1a and 1b subunits into heteromeric channels producing cardiac IKr.
Previous studies in yeast have demonstrated that transcripts associated with specific cellular functions can be copurified with one or more of the encoded proteins (26, 27). Current models suggest the transcripts are coimmunoprecipitated as a consequence of cotranslational assembly of their encoded proteins. For example, tip1 and tea2p transcripts encoding components of a molecular motor could be copurified with the Tea2p kinesin protein, but not when the tip1 translation initiation site was mutated; thus, the association occurs via the nascent proteins and not directly through the transcripts (26). A similar model was suggested by studies on the SET1C histone methyltransferase complex, which were among the first studies demonstrating that the association of protein with a transcript was not through binding of the protein to the mRNA but rather by copurification of the polysomal complex comprising the transcript and the nascent protein emerging from the ribosome (28). These models echoed early studies describing cotranslational association of nascent cytoskeletal proteins with the extant cytoskeleton (29). Our study, in contrast, describes the association of transcript species independent of the nascent protein interaction. Such an interaction would increase the likelihood that the proteins would be synthesized together in time and space, a requirement for the biogenesis of a heteromeric channel if it is to follow the same steps of folding and assembly identified for homotetramers (17, 18, 30–32).
What is the mechanism of transcript association? Studies in other systems using RNA IP combined with DNA microarray analysis (RIP-ChIP) point to a role for RNA binding proteins (RNABPs). In yeast, different members of the Puf class of RNABPs each coimmunoprecipitate multiple transcripts destined for a given organelle, such as the mitochondria or nucleolus (33). In this regard, the RNABPs act as a zip code to ensure delivery to an appropriate cellular address for local protein synthesis. The mRNAs encoding proteins in macromolecular complexes mediating specific cellular functions, such as cell cycle control, were also copurified with mRNAs encoding associated RNABPs (26). In mammalian systems, RNABPs have been characterized in neurons, where they are associated with “biologically coherent” (34) groups of mRNA encoding proteins related to a recognized function [e.g., synaptic transmission (35–37)]. For hERG transcripts, RNABPs might oligomerize or bind to another protein to mediate 1a and 1b transcript association, a hypothesis depicted in Fig. 3D. Further work will be required to establish the full constituency of the microtranslatomes that encode the heterotetrameric IKr channel and regulate cardiac excitability.
Given the association of the transcripts encoding the two hERG subunits, it may seem surprising that no evidence for association of hERG and KCNE1 transcripts was found in cardiomyocytes. As early as 1995, it was suggested that the KCNE1 protein (also known as MiRP1 or minK) is a subunit of IKr channels based on experiments in which antisense KCNE1 oligonucleotides reduced IKr current levels in AT1 cardiomyocyte-like cells (38). A subsequent report using HEK293 cells supported this idea by showing that KCNE1 alters hERG function (39), whereas another study using CHO cells did not (40). If KCNE1/minK participates as subunits or regulators of IKr channels, it appears from our findings that it does so posttranslationally, perhaps as described for IKs channels (41); alternatively, it may assemble from nonassociating transcripts, or in cell types other than the two examined here.
The observation that shRNA selectively targeting one hERG mRNA species can coordinately knock down the alternate transcript in HEK293 cells and cardiomyocytes raises several questions. In the hERG 1a/1b mRNA complex, are the associated mRNA transcripts codegraded or are they degraded independently? Alternatively, does loss of one mRNA reduce stability of the other? For example, a miRNA binding site could be unmasked when the two transcripts are not coupled, rendering them susceptible to degradation. Off-target effects do not account for the coupled reduction in mRNA levels because each shRNA has no apparent affinity for the alternate transcript when separately expressed. This finding suggests that specific shRNA-induced reduction does not always lead to the surgical elimination of only a single transcript: Other transcripts not assayed but functionally related and physically complexed may also be affected.
It makes sense that two transcripts encoding proteins that assemble cotranslationally should be located within close proximity so that their nascent proteins can interact. Such a mechanism can also regulate stoichiometry of macromolecular constituents, as demonstrated in Escherichia coli using ribosomal profiling (42). By measuring the quantitative translation rates of nearly all cellular proteins synthesized in a single experiment, Weissman and coworkers (42) found support for the principle of “proportional synthesis” of proteins in ratios that reflect stoichiometry. This mechanism thus controls ratios of proteins by local translation and can buffer the effects of fluctuating or “bursty” transcription to ensure appropriate composition (43). For IKr, the subunit composition is critical for cardiac repolarization: Failure of a hERG 1b subunit to associate with 1a means that 1b protein will be trapped in the ER (7, 14) and the 1a homomeric channels exiting to the surface will produce half the repolarizing current owing to their different gating properties (15). Through amino terminal interactions, hERG 1a masks an ER retention signal in hERG 1b and promotes heteromerization. Thus, translational control of heteromeric channel assembly is an important mechanism contributing to cardiac excitability and a potential disease target for any system in which heteromeric subunit assembly is crucial to normal function.
Materials and Methods
Cell Culture and Transfection.
iCell-induced pluripotent iPSC-CMs (Cellular Dynamics International) were plated per the manufacturer’s instructions and maintained in culture up to 45 d postplating. HEK293 cells were cultured in 35-mm dishes in DMEM supplemented with 10% (vol/vol) FBS and grown to 80–90% confluence. Nontargeting shRNA controls and transcript-specific hERG shRNA were purchased from Sigma–Aldrich (1a shRNA: TACCGCACCATTAGCAAGATT; 1b shRNA: CCACAACCACCCTGGCTTCAT). Cells were transfected using 2.5 μL/mL Lipofectamine 2000 (Life Technologies) per well and 1 μg/mL hERG 1a-pcDNA3 or hERG 1b-pcDNA3, or a total of 2 μg when the two plasmids were cotransfected. Twenty-four hours following hERG transfection, cells were transfected with 1 μg/mL DNA encoding either the shRNA or nontargeting shRNA control. Assays were completed 48 h following shRNA transfection.
RNA Isolation and Quantitative PCR.
Cells were transferred to 1.5-mL centrifuge tubes on ice and homogenized in 400 μL of cold TriZol solution (Life Technologies). Homogenates were incubated for 5 min at room temperature. The sample was then combined with 80 μL of chloroform, mixed vigorously for 15 s, incubated at room temperature for 10 min, and centrifuged at 12,000 × g for 15 min at 4 °C to allow phase separation. The clear RNA-containing aqueous phase was harvested for DNaseI treatment and purification using an RNeasy Mini Kit (Qiagen): 85 μL of aqueous phase was added to a mixture of 12.5 μL of DNA digestion buffer (RDD) and 2.5 μL of DNaseI, and incubated at room temperature for 30 min. Following incubation, 350 μL of RNA lysis buffer (RLT) and 250 μL of 100% ethanol were added to the sample, mixed, applied to RNeasy spin columns (Qiagen), and centrifuged at 10,000 × g for 15 s. Columns were washed with 500 μL of RPE buffer by centrifugation. The RNA sample was then eluted using 50 μL of RNase-free ddH2O.
RT was performed using an iScript Reverse Transcription Supermix Kit (BIORAD) and an Eppendorf thermocycler per the manufacturer’s instructions. We carried out quantitative real-time PCR using a TaqMan Gene Expression Assay (Life Technologies) and calculated hERG mRNA transcript levels relative to GAPDH using the ΔΔCT cycle threshold method. Triplicate quantitative PCR experiments completed on the same day, and averaged, represent an individual sample. These averaged samples were normalized to nontargeting controls carried out simultaneously. TaqMan assays (Life Technologies) were transcript-specific: hERG 1a (assay no. Hs00165120_m1), hERG 1b (assay no. Hs04234675_m1), KCNE1 (assay no. Hs01093259_m1), RYR2 (assay no. Hs00892883_m1), and human GAPDH (assay no Hs99999905_m1) transcripts. Transcript quantity was assessed in the linear range of nucleotide amplification, and all data were reported relative to nontargeting shRNA control values. ddH2O was used as a negative control.
Ribonucleoprotein Complex Isolation.
Ribonucleoprotein (RNP) complexes were isolated using a RiboCluster Profiler TM RIP-Assay Kit (Medical & Biological Laboratories). Cells were centrifuged for 5 min at 300 × g. The pellet was washed using ice-cold PBS, suspended in 500 μL of lysis buffer, vortexed, and finally centrifuged at 12,000 × g for 5 min at 4 °C. Lysate was then precleared by adding protein A/G agarose beads, mixed for 1 h at 4 °C, and centrifuged at 2,000 × g for 1 min at 4 °C. (Following centrifugation, a 10-μL aliquot was combined with Laemmli’s sample buffer for quality control by Western blot.) Ab-immobilized protein A/G agarose beads were mixed with hERG 1a-specific Ab (mAb no. 12889S; Cell Signaling) for 4 h at 4 °C and then combined with 500 μL of the precleared lysate, incubated overnight at 4 °C, and centrifuged at 2,000 × g for 1 min at 4 °C. (One hundred microliters of Ab-immobilized beads/RNP complex was centrifuged, and the pellet was suspended in 20 μL of Laemmli’s buffer for quality control by Western blot.) Washed Ab-immobilized beads/RNP complex solution was centrifuged again, supernatant was discarded, and guanidine hydrochloride solution was added to dissociate beads from RNP complexes. The solution was vortexed, and supernatant was transferred to RNA coprecipitator solution-containing microcentrifuge tubes to precipitate target RNAs. The solution was incubated in ice-cold 2-propanol for 20 min at −20 °C and then centrifuged at 12,000 × g for 10 min at 4 °C. The pellet was rinsed with 70% ethanol and recentrifuged, and the pellet was dried and then resuspended in nuclease-free H2O. The target RNAs were then analyzed using RT-PCR. (For additional quality control, total RNA was isolated from 10 μL of precleared cell lysate.) RT was performed using an iScript Reverse Transcription Supermix Kit (BIORAD) and an Eppendorf thermocycler as per the manufacturer’s instructions. cDNA was then amplified using 1a-specific (forward: AGAACTGCGCCGTCATCTAC; reverse: ACATTGTGGGTTCGCTCCTT) and 1b-specific (forward: CATCTCCAGCCTCGTGG; reverse: GTGTGGTCTTGAACTTCATGGC) primers. The 1a and 1b primers produced 1,000-bp and 1,150-bp products, respectively.
Western Blot.
Cells were lysed using buffer containing 150 mM Tris⋅NaCl, 25 mM Tris⋅HCl, 10 mM NaEGTA, 20 mM NaEDTA, and 5 mM glucose, and supplemented with 1% Triton X-100, 50 μg/mL 1,10 phenanthrolene, 0.7 μg/mL pepstatin A, 1.56 μg/mL benzamidine, and 1× Complete Minicab (Roche Applied Science). Solution was sonicated, incubated on ice for 30 min, and centrifuged for 10 min at 10,000 × g at 4 °C. Protein concentration of supernatant was assessed using a BIORAD protein assay. Thirty micrograms of protein per lane was electrophoresed on a 7.5% SDS/polyacrylamide gel and then transferred onto PVDF membranes. Membranes were blocked and incubated overnight with 1:2,000 anti–β-actin (ab8226; abeam) and either C-terminal pan–anti-hERG (ALX-215049-R100; Enzol Life Sciences) or antiribosomal protein L13A (C-11) (sc-390131; Santa Cruz Biotechnology) primary Abs. Membranes were washed and then incubated with 1:1,000 dilutions of secondary Ab Alexa Fluor 647 goat anti-rabbit IgG A-21245 (Life Technologies) for 1 h and imaged using a Chelidon-MP Imaging System (BIORAD).
Immunocytochemistry.
Immunocytochemistry was completed using subunit-specific Abs according to standard laboratory protocols as previously described (9). Briefly, cells were labeled using primary Abs specific for β-actin (mAbcam 8224; Abcam) and hERG 1a (ALX-215-050-R100; Enzo Life Sciences) or hERG 1b (ALX-215-051-R100; Enzo Life Sciences). Cells were washed and then incubated with blocking solution containing 1:1,000 dilutions of the secondary Abs Alexa Fluor 488 goat anti-mouse IgG (H+L) A-11029 for β-actin and Alexa Fluor 647 goat anti-rabbit IgG A-21245 (both from Life Technologies) for hERG. Cell selection was blinded, based on a robust β-actin signal, and morphology was assessed using bright-field imaging to verify cell health and diminish bias. Fluorescence was quantified based on the integral of the profile of fluorescence across the cell, avoiding the nucleus. Application of primary and secondary Abs alone acted as negative controls.
Electrophysiology.
All recordings were done using an Axon 200A amplifier and Clampex (Molecular Devices). Data were sampled at 10 kHz and low-pass-filtered at 1 kHz. Cells were perfused at 2 mL⋅min−1 with bath solution containing 150 mM NaCl, 5.4 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 15 mM glucose, 10 mM Hepes, and 1 mM Na-pyruvate, and titrated to pH 7.4 using NaOH. Recording pipettes (2–4.5 MΩ) were pulled from borosilicate glass capillaries using a P-97 Micropipette Puller System (Sutter Instruments). Electrodes were backfilled with intracellular solution containing 5 mM NaCl, 150 mM KCl, 2 mM CaCl2, 5 mM EGTA, 10 mM Hepes, and 5 mM MgATP, and titrated to pH 7.2 using KOH. All voltage protocols were completed at 36 ± 1 °C, before and after bath perfusion of 2 μM E-4031 and an IKr-specific blocker, and the difference in current was taken to represent IKr (9, 44). To measure IKr, cells were held at −40 mV to inactivate voltage-gated sodium currents and then stepped to a 3-s conditioning prepulse (−50 mV to +30 mV in 10-mV increments). Steady-state current was taken as the 5-ms average current at the end of the prepulse. Tail currents were measured following return to the −40 mV test pulse. To describe IKr voltage dependence, peak tail current was normalized to cellular capacitance, plotted as a function of prepulse potential, and fitted with a Boltzmann equation:
where A1 and A2 represent the maximum and minimums of the fit, respectively; V is the membrane potential; and V0 is the midpoint.
Analysis was completed using Clampfit (Molecular Devices) and Origin (OriginLab). Means were compared using a Student’s t test. Where applicable, an ANOVA and Bonferroni post hoc t tests were used. Statistical significance was taken at P < 0.05.
Acknowledgments
We thank Drs. James Dahlberg and Elsebet Lund for advice and Drs. Cynthia Czajkowski, Andrew Harris, and Greg Starek for critical comments on an earlier version of the manuscript. This study was supported by NIH Grants NHLBI 5R01 HL081780 and 1R01 NS081320 (to G.A.R.), the University of Wisconsin Training Program in Translational Cardiovascular Science (Grant NHLBI 5T32HL007936 to D.K.J.), and a postdoctoral training award from the University of Wisconsin Stem Cell and Regenerative Medicine Center (to D.K.J.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1521577113/-/DCSupplemental.
References
- 1.Sigel E, Steinmann ME. Structure, function, and modulation of GABA(A) receptors. J Biol Chem. 2012;287(48):40224–40231. doi: 10.1074/jbc.R112.386664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu J, Yu W, Jan YN, Jan LY, Li M. Assembly of voltage-gated potassium channels. Conserved hydrophilic motifs determine subfamily-specific interactions between the alpha-subunits. J Biol Chem. 1995;270(42):24761–24768. doi: 10.1074/jbc.270.42.24761. [DOI] [PubMed] [Google Scholar]
- 3.Lin TF, et al. The subfamily-specific assembly of Eag and Erg K+ channels is determined by both the amino and the carboxyl recognition domains. J Biol Chem. 2014;289(33):22815–22834. doi: 10.1074/jbc.M114.574814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanguinetti MC, Jurkiewicz NK. Two components of cardiac delayed rectifier K+ current. Differential sensitivity to block by class III antiarrhythmic agents. J Gen Physiol. 1990;96(1):195–215. doi: 10.1085/jgp.96.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanguinetti MC, Jiang C, Curran ME, Keating MT. A mechanistic link between an inherited and an acquired cardiac arrhythmia: HERG encodes the IKr potassium channel. Cell. 1995;81(2):299–307. doi: 10.1016/0092-8674(95)90340-2. [DOI] [PubMed] [Google Scholar]
- 6.Trudeau MC, Warmke JW, Ganetzky B, Robertson GA. HERG, a human inward rectifier in the voltage-gated potassium channel family. Science. 1995;269(5220):92–95. doi: 10.1126/science.7604285. [DOI] [PubMed] [Google Scholar]
- 7.London B, et al. Two isoforms of the mouse ether-a-go-go-related gene coassemble to form channels with properties similar to the rapidly activating component of the cardiac delayed rectifier K+ current. Circ Res. 1997;81(5):870–878. doi: 10.1161/01.res.81.5.870. [DOI] [PubMed] [Google Scholar]
- 8.Lees-Miller JP, Kondo C, Wang L, Duff HJ. Electrophysiological characterization of an alternatively processed ERG K+ channel in mouse and human hearts. Circ Res. 1997;81(5):719–726. doi: 10.1161/01.res.81.5.719. [DOI] [PubMed] [Google Scholar]
- 9.Jones DK, et al. hERG 1b is critical for human cardiac repolarization. Proc Natl Acad Sci USA. 2014;111(50):18073–18077. doi: 10.1073/pnas.1414945111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones EM, Roti Roti EC, Wang J, Delfosse SA, Robertson GA. Cardiac IKr channels minimally comprise hERG 1a and 1b subunits. J Biol Chem. 2004;279(43):44690–44694. doi: 10.1074/jbc.M408344200. [DOI] [PubMed] [Google Scholar]
- 11.Curran ME, Landes GM, Keating MT. Molecular cloning, characterization, and genomic localization of a human potassium channel gene. Genomics. 1992;12(4):729–737. doi: 10.1016/0888-7543(92)90302-9. [DOI] [PubMed] [Google Scholar]
- 12.Furutani M, et al. Novel mechanism associated with an inherited cardiac arrhythmia: Defective protein trafficking by the mutant HERG (G601S) potassium channel. Circulation. 1999;99(17):2290–2294. doi: 10.1161/01.cir.99.17.2290. [DOI] [PubMed] [Google Scholar]
- 13.Trudeau MC, Leung LM, Roti ER, Robertson GA. hERG1a N-terminal eag domain-containing polypeptides regulate homomeric hERG1b and heteromeric hERG1a/hERG1b channels: A possible mechanism for long QT syndrome. J Gen Physiol. 2011;138(6):581–592. doi: 10.1085/jgp.201110683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phartiyal P, Sale H, Jones EM, Robertson GA. Endoplasmic reticulum retention and rescue by heteromeric assembly regulate human ERG 1a/1b surface channel composition. J Biol Chem. 2008;283(7):3702–3707. doi: 10.1074/jbc.M708999200. [DOI] [PubMed] [Google Scholar]
- 15.Sale H, et al. Physiological properties of hERG 1a/1b heteromeric currents and a hERG 1b-specific mutation associated with long-QT syndrome. Circ Res. 2008;103(7):e81–e95. doi: 10.1161/CIRCRESAHA.108.185249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abi-Gerges N, et al. hERG subunit composition determines differential drug sensitivity. Br J Pharmacol. 2011;164(2b):419–432. doi: 10.1111/j.1476-5381.2011.01378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu J, Robinson JM, Edwards D, Deutsch C. T1-T1 interactions occur in ER membranes while nascent Kv peptides are still attached to ribosomes. Biochemistry. 2001;40(37):10934–10946. doi: 10.1021/bi010763e. [DOI] [PubMed] [Google Scholar]
- 18.Robinson JM, Deutsch C. Coupled tertiary folding and oligomerization of the T1 domain of Kv channels. Neuron. 2005;45(2):223–232. doi: 10.1016/j.neuron.2004.12.043. [DOI] [PubMed] [Google Scholar]
- 19.Tu L, Wang J, Helm A, Skach WR, Deutsch C. Transmembrane biogenesis of Kv1.3. Biochemistry. 2000;39(4):824–836. doi: 10.1021/bi991740r. [DOI] [PubMed] [Google Scholar]
- 20.Phartiyal P, Jones EM, Robertson GA. Heteromeric assembly of human ether-à-go-go-related gene (hERG) 1a/1b channels occurs cotranslationally via N-terminal interactions. J Biol Chem. 2007;282(13):9874–9882. doi: 10.1074/jbc.M610875200. [DOI] [PubMed] [Google Scholar]
- 21.Rothman JE, Lodish HF. Synchronised transmembrane insertion and glycosylation of a nascent membrane protein. Nature. 1977;269(5631):775–780. doi: 10.1038/269775a0. [DOI] [PubMed] [Google Scholar]
- 22.Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- 23.Barhanin J, et al. K(V)LQT1 and lsK (minK) proteins associate to form the I(Ks) cardiac potassium current. Nature. 1996;384(6604):78–80. doi: 10.1038/384078a0. [DOI] [PubMed] [Google Scholar]
- 24.Sanguinetti MC, et al. Coassembly of K(V)LQT1 and minK (IsK) proteins to form cardiac I(Ks) potassium channel. Nature. 1996;384(6604):80–83. doi: 10.1038/384080a0. [DOI] [PubMed] [Google Scholar]
- 25.Otsu K, et al. Molecular cloning of cDNA encoding the Ca2+ release channel (ryanodine receptor) of rabbit cardiac muscle sarcoplasmic reticulum. J Biol Chem. 1990;265(23):13472–13483. [PubMed] [Google Scholar]
- 26.Duncan CD, Mata J. Widespread cotranslational formation of protein complexes. PLoS Genet. 2011;7(12):e1002398. doi: 10.1371/journal.pgen.1002398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duncan CD, Mata J. Cotranslational protein-RNA associations predict protein-protein interactions. BMC Genomics. 2014;15:298. doi: 10.1186/1471-2164-15-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halbach A, et al. Cotranslational assembly of the yeast SET1C histone methyltransferase complex. EMBO J. 2009;28(19):2959–2970. doi: 10.1038/emboj.2009.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fulton AB, L’Ecuyer T. Cotranslational assembly of some cytoskeletal proteins: implications and prospects. J Cell Sci. 1993;105(Pt 4):867–871. doi: 10.1242/jcs.105.4.867. [DOI] [PubMed] [Google Scholar]
- 30.Sheng Z, Skach W, Santarelli V, Deutsch C. Evidence for interaction between transmembrane segments in assembly of Kv1.3. Biochemistry. 1997;36(49):15501–15513. doi: 10.1021/bi971490j. [DOI] [PubMed] [Google Scholar]
- 31.Deutsch C. Potassium channel ontogeny. Annu Rev Physiol. 2002;64:19–46. doi: 10.1146/annurev.physiol.64.081501.155934. [DOI] [PubMed] [Google Scholar]
- 32.Kosolapov A, Deutsch C. Folding of the voltage-gated K+ channel T1 recognition domain. J Biol Chem. 2003;278(6):4305–4313. doi: 10.1074/jbc.M209422200. [DOI] [PubMed] [Google Scholar]
- 33.Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2(3):E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Darnell RB. RNA protein interaction in neurons. Annu Rev Neurosci. 2013;36:243–270. doi: 10.1146/annurev-neuro-062912-114322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ule J, et al. Nova regulates brain-specific splicing to shape the synapse. Nat Genet. 2005;37(8):844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- 36.Liu-Yesucevitz L, et al. Local RNA translation at the synapse and in disease. J Neurosci. 2011;31(45):16086–16093. doi: 10.1523/JNEUROSCI.4105-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amrute-Nayak M, Bullock SL. Single-molecule assays reveal that RNA localization signals regulate dynein-dynactin copy number on individual transcript cargoes. Nat Cell Biol. 2012;14(4):416–423. doi: 10.1038/ncb2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang T, Kupershmidt S, Roden DM. Anti-minK antisense decreases the amplitude of the rapidly activating cardiac delayed rectifier K+ current. Circ Res. 1995;77(6):1246–1253. doi: 10.1161/01.res.77.6.1246. [DOI] [PubMed] [Google Scholar]
- 39.McDonald TV, et al. A minK-HERG complex regulates the cardiac potassium current I(Kr) Nature. 1997;388(6639):289–292. doi: 10.1038/40882. [DOI] [PubMed] [Google Scholar]
- 40.Weerapura M, Nattel S, Chartier D, Caballero R, Hébert TE. A comparison of currents carried by HERG, with and without coexpression of MiRP1, and the native rapid delayed rectifier current. Is MiRP1 the missing link? J Physiol. 2002;540(Pt 1):15–27. doi: 10.1113/jphysiol.2001.013296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang M, et al. Dynamic partnership between KCNQ1 and KCNE1 and influence on cardiac IKs current amplitude by KCNE2. J Biol Chem. 2009;284(24):16452–16462. doi: 10.1074/jbc.M808262200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li GW, Burkhardt D, Gross C, Weissman JS. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157(3):624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123(6):1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 44.Ma J, et al. High purity human-induced pluripotent stem cell-derived cardiomyocytes: Electrophysiological properties of action potentials and ionic currents. Am J Physiol Heart Circ Physiol. 2011;301(5):H2006–H2017. doi: 10.1152/ajpheart.00694.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]