Significance
Bacterial pathogens use a variety of strategies to evade host cell innate immune responses. For some, this includes the establishment of an intracellular replicative niche. Although many intracellular pathogens remodel phagosomes to support bacterial replication, others lyse their internalization vacuole to reside within the host cell cytosol. Little is currently known regarding how bacteria, particularly Gram-negative pathogens, mediate phagosomal escape. Using complementary reductionist and functional interchangeability experimental approaches, we demonstrate that the type III secretion system machinery itself directly modulates the extent to which bacteria escape from phagosomes. Given the high prevalence of type III secretion systems among intracellular bacterial pathogens, these studies have identified a potential means by which the intracellular niche of Gram-negative bacteria is defined.
Keywords: type III secretion system, phagosomal escape, vacuole lysis, Shigella, Salmonella
Abstract
Upon entry into host cells, intracellular bacterial pathogens establish a variety of replicative niches. Although some remodel phagosomes, others rapidly escape into the cytosol of infected cells. Little is currently known regarding how professional intracytoplasmic pathogens, including Shigella, mediate phagosomal escape. Shigella, like many other Gram-negative bacterial pathogens, uses a type III secretion system to deliver multiple proteins, referred to as effectors, into host cells. Here, using an innovative reductionist-based approach, we demonstrate that the introduction of a functional Shigella type III secretion system, but none of its effectors, into a laboratory strain of Escherichia coli is sufficient to promote the efficient vacuole lysis and escape of the modified bacteria into the cytosol of epithelial cells. This establishes for the first time, to our knowledge, a direct physiologic role for the Shigella type III secretion apparatus (T3SA) in mediating phagosomal escape. Furthermore, although protein components of the T3SA share a moderate degree of structural and functional conservation across bacterial species, we show that vacuole lysis is not a common feature of T3SA, as an effectorless strain of Yersinia remains confined to phagosomes. Additionally, by exploiting the functional interchangeability of the translocator components of the T3SA of Shigella, Salmonella, and Chromobacterium, we demonstrate that a single protein component of the T3SA translocon—Shigella IpaC, Salmonella SipC, or Chromobacterium CipC—determines the fate of intracellular pathogens within both epithelial cells and macrophages. Thus, these findings have identified a likely paradigm by which the replicative niche of many intracellular bacterial pathogens is established.
Intracellular bacterial pathogens use a variety of elaborate means to survive within host cells. Postinvasion, some such as Legionella, Salmonella, and Chlamydia species modify bacteria-containing vacuoles to avoid death via phagosomal acidification or lysosomal fusion. Others, including Shigella, Listeria, Rickettsia, and Burkholderia species, rapidly escape from phagosomes into the cytosol of infected cells. Although escape from phagosomes by the classic intracytoplasmic Gram-positive bacterium Listeria monocytogenes is well understood (1), much less is known regarding how Gram-negative pathogens, including the model professional intracytoplasmic Shigella species, enter the cytosol.
During the course of an infection, many Gram-negative pathogens, including Shigella, Salmonella, enteropathogenic Escherichia coli, and Yersinia species, use type III secretion systems (T3SSs) as injection devices to deliver multiple virulence proteins, referred to as effectors, directly into the cytosol of infected cells (2). T3SSs are composed of ∼20 proteins and sense host cell contact via a tip complex at the distal end of a needle filament, which then acts as a scaffold for the formation of a translocon pore in the host cell membrane. Although components of their type III secretion apparatus (T3SA) are relatively well conserved, each pathogen delivers a unique repertoire of effectors into host cells, likely accounting for the establishment of a variety of replicative niches. For example, Salmonella and Shigella secreted effectors promote the uptake of these bacteria into nonphagocytic cells, whereas those from Yersinia inhibit phagocytosis by macrophages.
All four pathogenic Shigella species—Shigella flexneri, Shigella sonnei, Shigella boydii, and Shigella dysenteriae—deliver ∼30 effectors into host cells, the majority of which are encoded on a large virulence plasmid (VP) alongside the genes for all of the proteins needed to form a T3SA (3). These secreted proteins play major roles in Shigella pathogenesis, including host cell invasion and modulation of innate immune response. One effector, IpgD, promotes the efficiency of Shigella phagosomal escape, although it is not absolutely required for this process (4). Interestingly, IpaB and IpaC, components of the Shigella translocon, the portion of the T3SA that inserts into the host cell membrane, have been implicated to mediate phagosomal escape based on the behavior of recombinant proteins (5–7). The physiologic relevance of these findings has not yet been directly addressed, as strains that lack either of these two proteins are completely impaired in the delivery of Shigella effectors into host cells (8).
Here, using a reductionist approach, we directly tested a role for the Shigella translocon apparatus in phagosomal escape. Using an innovative reengineering approach, we introduced a functional effectorless Shigella T3SA into a nonpathogenic laboratory strain of DH10B E. coli. Remarkably, upon entry into host epithelial cells, these bacteria efficiently escape from phagosomes. This demonstrates for the first time, to our knowledge, in the context of an infection, a direct role for the Shigella T3SA in mediating vacuole lysis. Despite structural conservation across T3SS families, we further observed that, in the absence of any type III effectors, the Ysc T3SA mediates little to no Yersinia phagosomal escape, suggesting that not all injectisomes have equivalent functions. Lastly, by exploring the functional interchangeability of translocon components of the Shigella, Salmonella, and Chromobacterium T3SA, we demonstrate that one translocon protein controls the extent to which these intracellular pathogens escape into the cytosol of infected cells, thus demonstrating a major role for the T3SA in determining the site of the replicative niche of intracellular bacteria.
Results
mT3_E. coli Invade HeLa Cells at Levels Similar to WT Shigella.
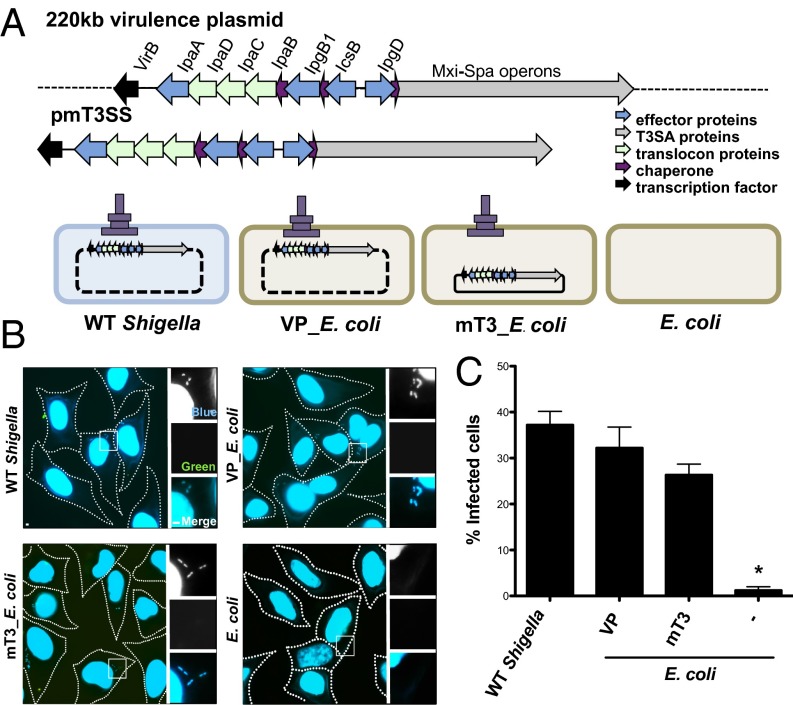
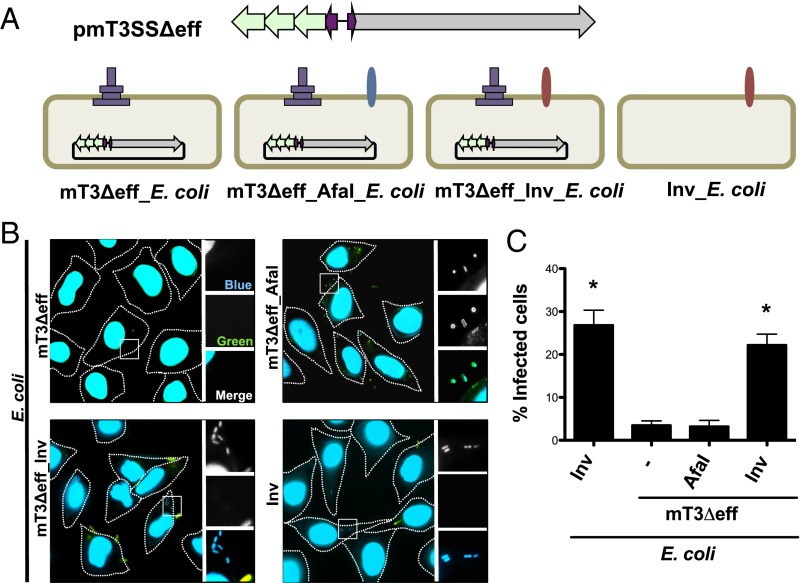
We recently reported the development of mT3 E. coli, laboratory strains of E. coli (i.e., DH10B) that express a functional Shigella T3SS (9). These strains carry a 31-kb region of the 220-kb S. flexneri VP, either on a plasmid (pmT3SS) or chromosomally integrated, plus a plasmid that encodes either the master Shigella T3SS transcriptional regulator, VirF, or its downstream target, VirB (10). All of the genes needed to form a functional T3SA, and four effectors are present within this fragment of transplanted Shigella DNA (Fig. 1A). Three of the effectors—IpaA, IpgB1, and IpgD—play roles in the invasion of Shigella into nonphagocytic epithelial cells (11), likely accounting for the ability of mT3 E. coli strains, like wild-type (WT) Shigella, to efficiently invade epithelial cells (9).
Fig. 1.
mT3_E. coli efficiently invade epithelial cells. (A) Schematic of plasmids and strains. The dashed line represents the region of VP encoded outside of that present in mT3_E. coli, which encodes ∼21 Shigella effectors. (B) Representative images of HeLa cells infected with the designated strains that have been differentially labeled at 1 h p.i. using inside/outside microscopy assay to identify intra- (blue) versus extracellular (blue/green) bacteria. The HeLa cell perimeter is denoted by a dashed line. The enlarged images are from the boxed region in the larger panel. (C) Quantification of the percentage of cells that contain intracellular bacteria. For each infection, 100 HeLa cells were counted. *P < 0.05 for strains versus WT Shigella as determined by one-way ANOVA with Dunnett’s post hoc analysis. Data are the mean ± SEM of four independent experiments.
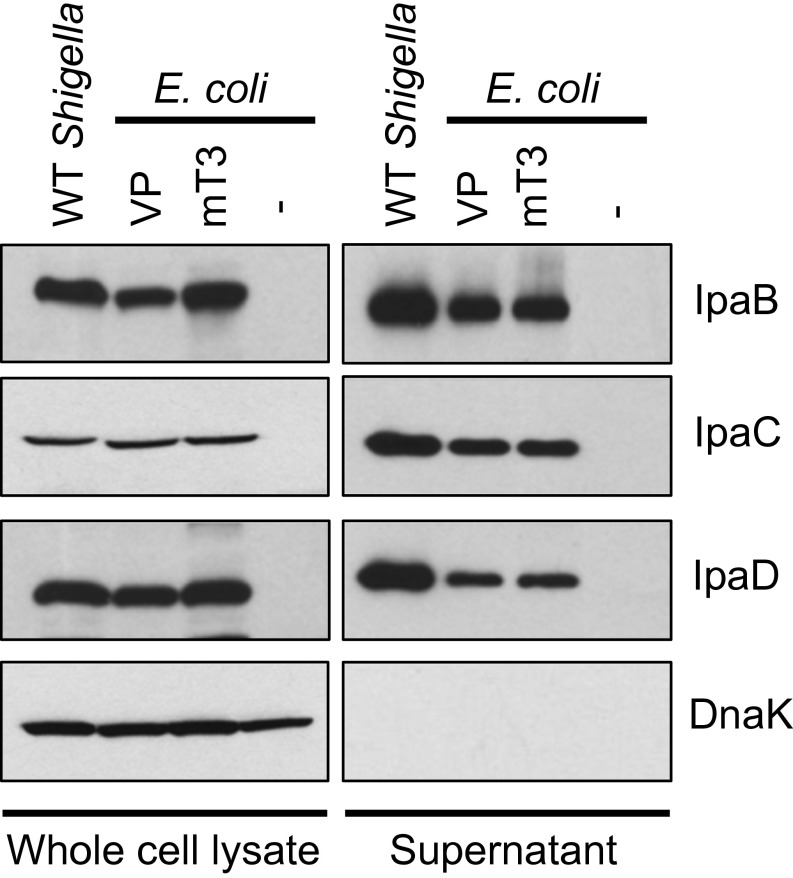
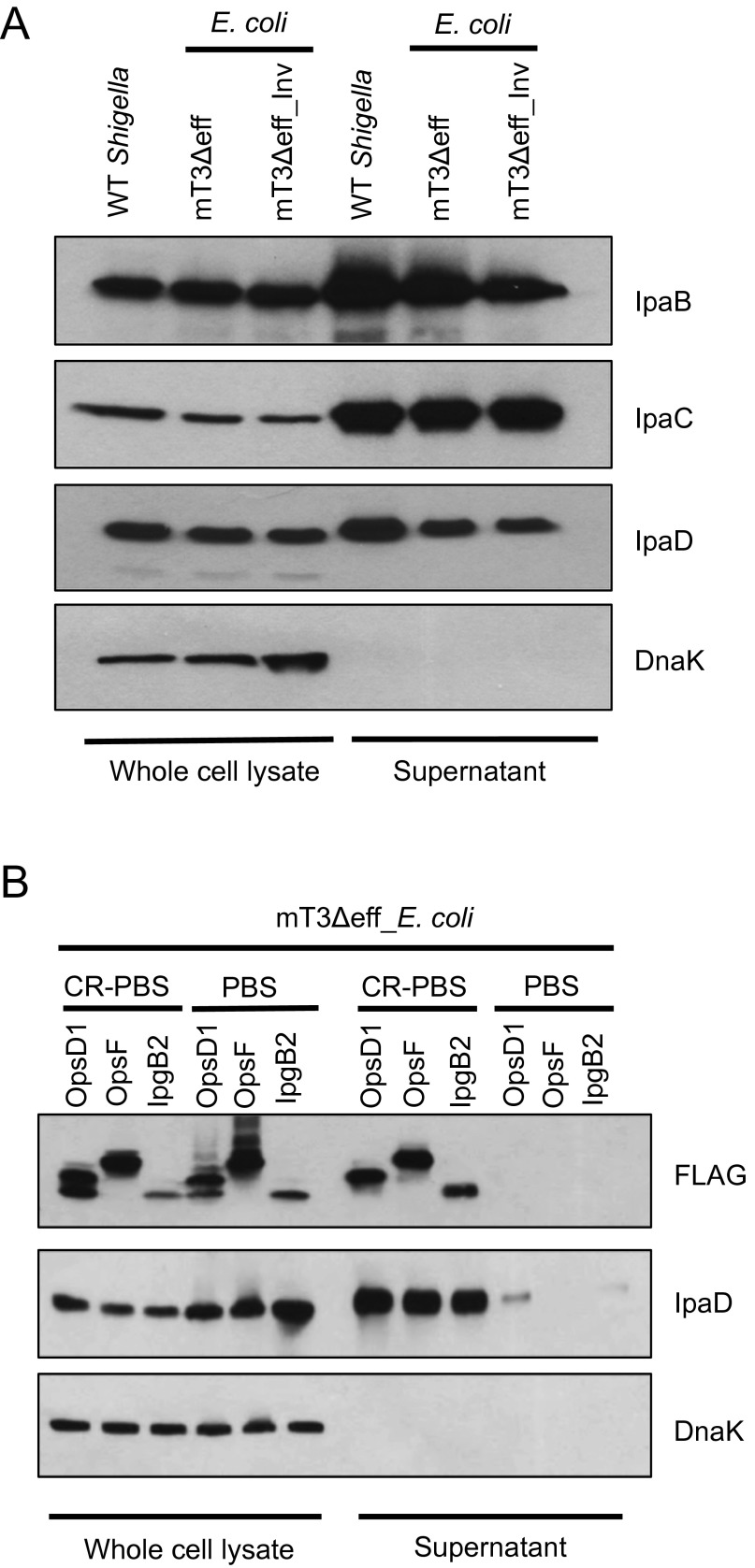
Here, we focused our efforts on further characterization of one of these strains, a variant of DH10B E. coli whereby the activity of T3SS is controlled by an isopropyl β-d-1-thiogalactopyranoside (IPTG)-inducible allele of virB, a strain hereafter referred to as mT3_E. coli (Fig. 1A). As shown in Fig. S1, this strain as well as DH10B E. coli that carry the intact 220-kb Shigella VP (VP_E. coli) synthesize and secrete the three exported components of the Shigella T3SS—IpaD, the injectisome needle tip protein, as well as IpaB and IpaC—which form a translocon pore in the host membrane, at levels equivalent to WT Shigella.
Fig. S1.
mT3_E. coli secrete components of the Shigella T3SS at levels equivalent to WT Shigella. Each of the designated strains was grown under conditions that induce secretion via the Shigella T3SS. Proteins present in the pellet (whole-cell lysate) and supernatant fractions were separated by SDS/PAGE and probed with antibodies against IpaB, IpaC, IpaD, components of the Shigella T3SA, and DnaK, a cytoplasmic protein (bacterial lysis control). The immunoblots shown are representative of at least three experiments.
We next assessed the functional activity of the Shigella T3SS in E. coli by comparing the ability of WT Shigella, VP_E. coli, and mT3_E. coli to invade epithelial cells, an early T3SS-dependent step. An inside/outside microscopy-based assay was used to differentiate between internalized and cell-associated bacteria to determine the percentage of HeLa cells that contain intracellular bacteria (Fig. 1B). At 1 h postinfection (p.i.), only strains that express a functional T3SS, mT3_E. coli, VP_E. coli, and WT Shigella, but not unmodified DH10B E. coli, were observed within epithelial cells (Fig. 1C). WT Shigella, VP_E. coli, and mT3_E. coli invaded epithelial cells at nearly equivalent levels (37% vs. 32% vs. 26%, respectively). Together, the results of the secretion and invasion assays demonstrate that the Shigella T3SS is expressed and exhibits similar levels of activity in the context of mT3_E. coli, VP_E. coli, and WT Shigella.
mT3_E. coli Efficiently Escape from Phagosomes Within Epithelial Cells.
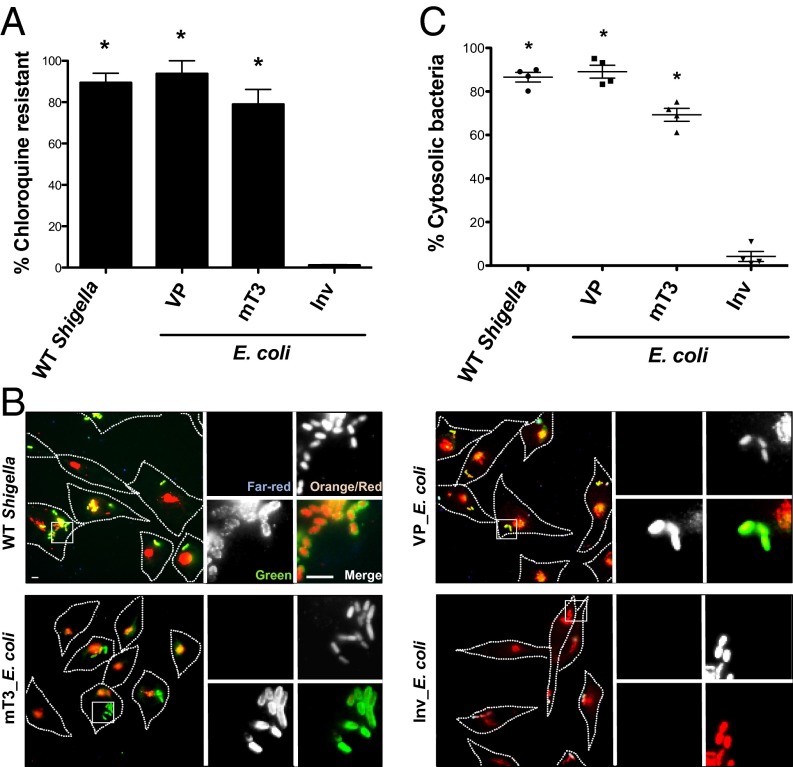
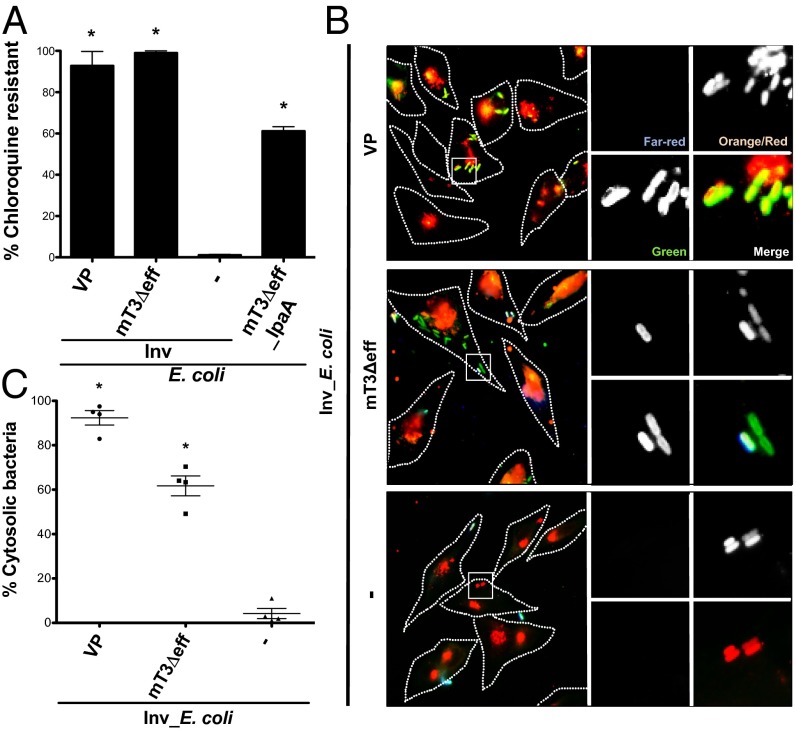
Using two complementary assays, we next investigated the intracellular fate of mT3_E. coli. Given our prior observation that these bacteria replicate poorly within host cells (9), we were interested in determining whether these bacteria, like WT Shigella, escape from phagosomes into the cytosol of epithelial cells. First, we investigated the sensitivity of intracellular WT Shigella, mT3_ E. coli, and VP_E. coli to chloroquine (CHQ), an antimicrobial agent that only reaches bactericidal levels when concentrated within phagosomes (12). When exposed to CHQ at 1 h p.i. for 1 h, almost all intracellular Shigella (90%) exhibited CHQ resistance (CHQR) (Fig. 2A), consistent with the observation that Shigella rapidly escape from phagosomes within epithelial cells (13). Similarly, 94% of VP_E. coli and 79% of mT3_E. coli exhibited CHQR, suggesting that these strains, like WT Shigella, also enter the cytosol of epithelial cells. In contrast, only 1% of DH10B E. coli that express Yersinia Invasin (Inv) (Inv_E. coli), a bacterial adhesin that promotes the uptake of E. coli into epithelial cells (14), exhibited CHQR.
Fig. 2.
mT3_E. coli efficiently escape from phagosomes in epithelial cells. HeLa cells were infected with the designated strains [multiplicity of infection (MOI) of 100 (WT Shigella, VP_E. coli, mT3_E. coli) or 10 (Inv_E. coli)] grown under conditions that induce expression of their T3SSs and assessed for phagosomal escape. (A) CHQR assay. HeLa cells were infected for 1 h and then treated with gentamicin (gent) ± CHQ for an additional 1 h before cell monolayers were lysed and bacteria enumerated. The percentage of CHQR bacteria was calculated as the ratio of (CHQR + gentR)/(gentR) bacteria. Infections were conducted in duplicate for each experiment. (B) Representative images of infected cells visualized using the digitonin permeabilization assay. HeLa cells were infected with variants of the designated strains that constitutively express mCherry. Before fixation, cells were stained with Alexa Fluor 647 (far-red) goat-conjugated antibacterial antibodies to identify extracellular bacteria. Monolayers were then treated with digitonin to selectively permeabilize the plasma membrane and labeled with Alexa Fluor 555 (orange)-conjugated anti-GM130 (peripheral Golgi matrix protein) antibodies and antibacterial primary and Alexa Fluor 488 (green) secondary antibodies to identify permeabilized cells and label intracytoplasmic and extracellular bacteria (green). The enlarged images are from the boxed region in the larger panel. (C) Quantification of the percentage of cytoplasmic bacteria as defined by the ratio of intracytoplasmic to total intracellular (green/red)/(green/red + red) bacteria associated with digitonin-permeabilized (GM130-positive) cells. For each infection, 100 intracellular bacteria were counted; extracellular bacteria (far-red channel) were excluded from calculations. (A and C) *P < 0.05 for strains versus Inv_E. coli as determined by one-way ANOVA with Dunnett’s post hoc analysis. The mean ± SEM of four independent experiments is shown.
To confirm the CHQR findings, we performed a previously established digitonin permeabilization fluorescent microscopy assay to deliver anti-LPS antibodies into the cytosol in order to directly visualize the location of intracellular bacteria within epithelial cells at 1 h p.i. (15, 16). As predicted, and consistent with our CHQR data, almost all WT Shigella (87%), as well as the majority of VP_E. coli (89%) and mT3_E. coli (69%), were detected by anti-LPS antibodies, indicating these bacteria were free in the cytosol or within phagosomes with compromised membranes (Fig. 2 B and C). However, only 4% of Inv_E. coli were labeled and thus the majority remained confined to intact phagosomes. The results from the CHQR and digitonin permeabilization assays demonstrate that all of the genetic information necessary to promote the uptake and entry of bacteria into the cytosol of host epithelial cells is present within the 31-kb fragment of Shigella DNA introduced into mT3_E. coli.
mT3Δeff_E. coli Are Unable to Attach to or Invade Epithelial Cells.
The 31-kb region of Shigella DNA present in mT3_E. coli encodes proteins needed to form a functional T3SA and four effectors, three of which—IpgB1, IpgD, and IpaA—play roles in mediating host cell invasion and one—IpgD—that promotes phagosomal escape (4). Furthermore, studies with purified IpaB and IpaC have implicated these proteins in mediating vacuole lysis (5–7); however, given the absolute requirement of the IpaB–IpaC translocon for the delivery of effectors into host cells, a direct physiologic role for either protein in phagosomal escape has not yet been possible to assess.
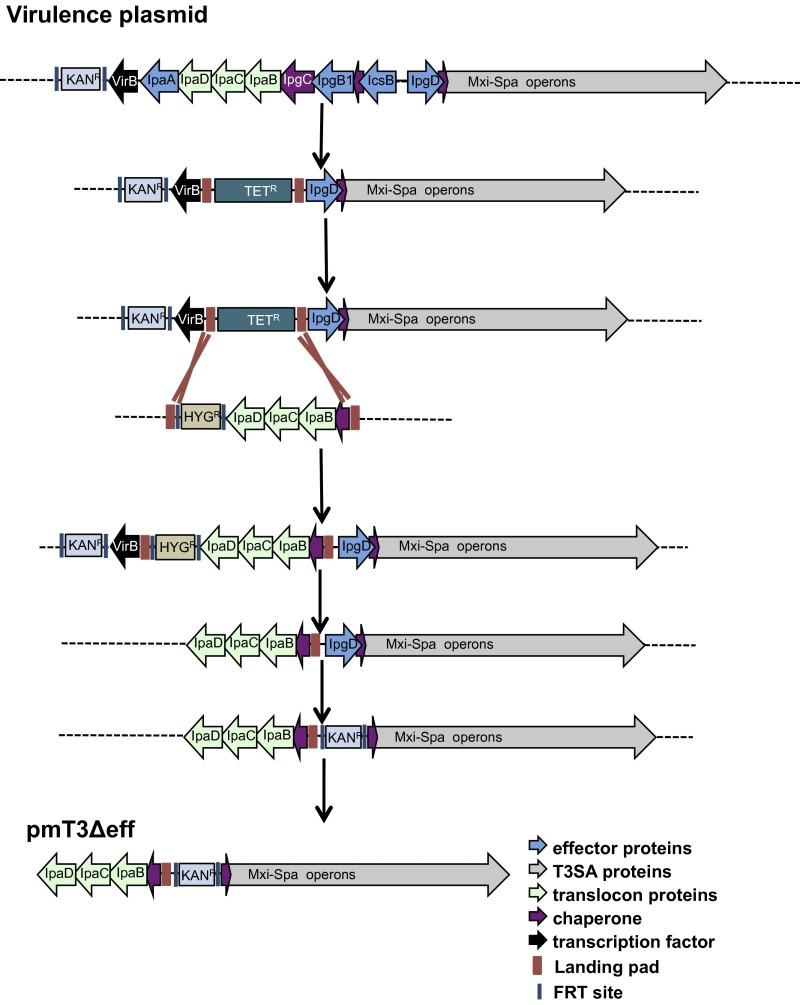
To investigate whether effectors or the T3SA mediate phagosomal escape, we conducted a series of genetic modifications (Fig. S2 and SI Materials and Methods) to remove genes encoding the four effectors to generate mT3Δeff_E. coli, a variant of DH10B E. coli whereby the activity of an effectorless T3SS is controlled by an IPTG-inducible allele of VirB (Fig. 3A). As shown in Fig. S3A, mT3Δeff_E. coli secretes IpaB, IpaC, and IpaD at levels similar to WT Shigella. This strain expresses a fully functional translocon apparatus, as exogenously supplemented effectors are not constitutively secreted (a phenotype associated with ΔipaB Shigella) but rather are released from the bacteria only in the presence of the dye Congo red (CR), an in vitro activator of the Shigella T3SS (17) (Fig. S3B).
Fig. S2.
Schematic of strategy used to generate pmT3SSΔeff. See SI Materials and Methods for details.
Fig. 3.
Only mT3Δeff_E. coli that express Yersinia Inv enter epithelial cells. (A) Schematic representation of designated plasmids and strains. (B) Inside/outside microscopy assay as described in Fig. 1 of HeLa cells infected for 1 h with the designated strains at an MOI of 100 (mT3∆eff_E. coli) or 10 (mT3∆eff_AfaI_E. coli, mT3∆eff_Inv_E. coli, Inv_E. coli). (C) Quantification of the percentage of infected cells, defined as those that contain intracellular bacteria. For each infection, 100 HeLa cells were counted. *P < 0.05 for strains versus mT3∆eff_E. coli as determined by one-way ANOVA with Dunnett’s post hoc analysis. Data are expressed as the mean ± SEM of four independent experiments.
Fig. S3.
mT3Δeff_E. coli express a functional T3SS. (A) WT Shigella and mT3Δeff_E. coli were grown under conditions that induce activity of the Shigella T3SS. (B) mT3Δeff_E. coli that carry plasmids encoding IPTG-inducible alleles of the designated Shigella effectors were grown under conditions that induce expression of the Shigella T3SS and resuspended in PBS ± CR to assess for leaky activity of Shigella T3SS in the absence of the activator (CR). (A and B) Proteins present in the pellet (whole-cell lysate) and supernatant fractions were separated by SDS/PAGE and immunoblotted with antibodies against IpaB, IpaC, IpaD, and FLAG, for detection of type III secreted effector, and DnaK. The blots shown are representative of at least three experiments.
We next investigated whether this strain, like mT3_E. coli, can invade epithelial cells. Using the inside/outside microscopy assay, we observed very few mT3Δeff_E. coli bacteria within, or associated with, epithelial cells at 1 h p.i. (Fig. 3 B and C). Given our interest in assessing a role for the Shigella translocon apparatus in mediating phagosomal escape, we tested modifications that might promote the uptake of mT3Δeff_E. coli into host cells (Fig. 3 B and C). First, we investigated the fate of mT3Δeff_E. coli that express AfaI, an afimbrial adhesin from uropathogenic E. coli that promotes attachment to the extracellular surface of mammalian cells (18). This modification resulted in the adherence to, but not uptake of, mT3Δeff_E. coli into host cells. In contrast, mT3Δeff_Inv_E. coli, mT3Δeff_E. coli that express Yersinia Inv, entered HeLa cells at levels similar to Inv_E. coli (Fig. 3 B and C) and mT3_E. coli (Fig. 1C) (22% vs. 27% vs. 26%, respectively).
The Shigella T3SA Is Sufficient to Mediate Phagosomal Escape.
Using the invasion-competent mT3Δeff_E. coli (mT3Δeff_Inv_E. coli), we next investigated the intracellular fate of bacteria expressing an effectorless Shigella T3SA using the CHQR and digitonin permeabilization assays. When infected cells were treated with CHQ at 1 h p.i. for 1 h, both VP_Inv_E. coli and mT3Δeff_Inv_E. coli exhibited high levels of CHQR, 93% and 99%, respectively, suggesting that both strains efficiently escape from their internalization vacuole into the cytosol of epithelial cells (Fig. 4A). Using the digitonin permeabilization assay, we similarly observed substantial numbers of intracytoplasmic mT3Δeff_Inv_E. coli, albeit at slightly lower levels than VP_Inv_E. coli (62% vs. 92%). In contrast, very few Inv_E. coli (4%) were detected within the cytosol of host cells, confirming that Inv alone in E. coli does not mediate phagosomal escape (14) (Fig. 4 B and C).
Fig. 4.
The Shigella T3SA itself is sufficient to mediate phagosomal escape. HeLa cells were infected with the designated strains at an MOI of 100 (mT3Δeff_IpaA_E. coli) or 10 (VP_Inv_E. coli, mT3Δeff_Inv_E. coli, Inv_E. coli) and assessed for phagosomal escape. (A) CHQR and (B and C) digitonin permeabilization assays as described in Fig. 2. *P < 0.05 for strains versus Inv_E. coli as determined by one-way ANOVA with Dunnett’s post hoc analysis. Data are expressed as the mean ± SEM of four independent experiments.
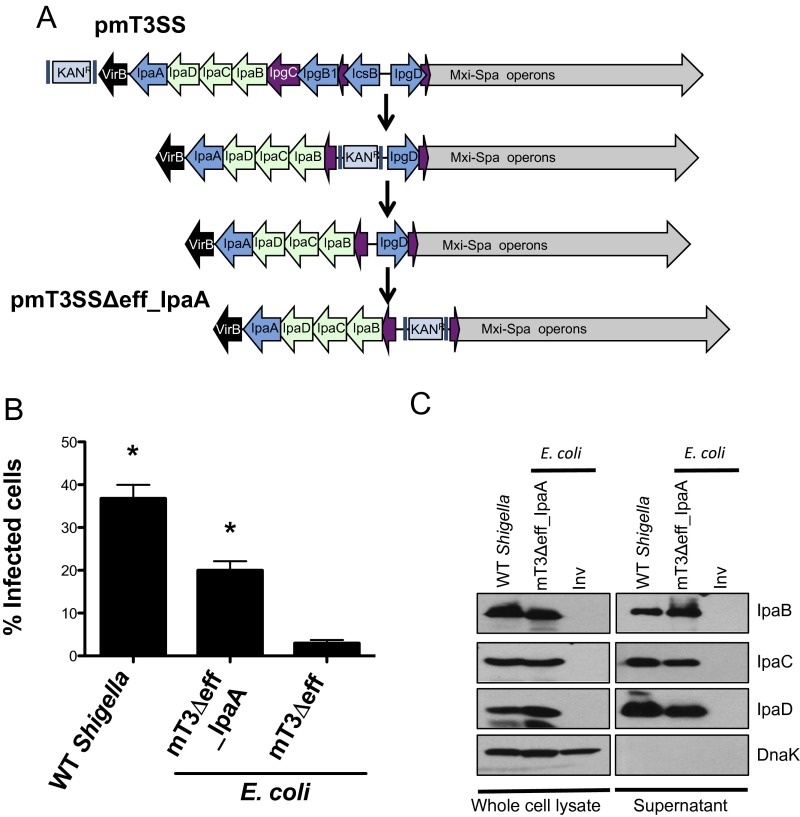
Inv mediates uptake of Yersinia into host cells via a “zipper”-type mechanism mediated by interactions with α5β1 integrins, a pathway that could potentially affect mT3Δeff_Inv_E. coli vacuolar lysis (19). To address this possibility, we developed a means to introduce mT3_E. coli into epithelial cells via a Shigella-like “trigger”-type mechanism (19). By sequentially deleting regions of DNA that encode three effectors present in mT3_E. coli, we generated mT3Δeff_IpaA_E. coli, a strain that encodes only a single effector (IpaA) (Fig. S4A). mT3Δeff_IpaA_E. coli invade epithelial cells about half as well as WT Shigella (Fig. S4B) (20% vs. 37%). The invasion defect is likely solely due to the absence of the two deleted effectors that are involved in invasion—IpgB1 and IpgD—as mT3Δeff_IpaA_E. coli and WT Shigella secrete similar levels of IpaB, IpaC, and IpaD (Fig. S4C). Intracellular mT3Δeff_IpaA_E. coli, like mT3Δeff_Inv_E. coli, exhibited high levels of CHQR (61%) (Fig. 4A). The slightly lower levels of CHQR observed for mT3Δeff_IpaA_E. coli versus mT3Δeff_Inv_E. coli might reflect differences due to the presence of IpaA or the nature of the phagosomes generated when uptake is mediated via zipper versus trigger mechanisms. Nevertheless, the complementary mT3Δeff_IpaA_E. coli and mT3Δeff_Inv_E. coli data demonstrate that the Shigella T3SA plays a major role in mediating phagosomal escape.
Fig. S4.
Characterization of mT3Δeff_IpaA_E. coli. (A) Schematic of strategy used to generate pmT3Δeff_IpaA_E. coli. See SI Materials and Methods for details. (B) Quantification of the percentage of infected cells at 1 h p.i., defined as those that contain intracellular bacteria as assayed via inside/outside staining described in Fig. 1. For each infection, 100 HeLa cells were counted. *P < 0.05 for strains versus mT3∆eff_E. coli as determined by one-way ANOVA with Dunnett’s post hoc analysis. Data are expressed as the mean ± SEM of four independent experiments. (C) Each of the designated strains was grown under conditions that induce secretion via the Shigella T3SS. Proteins present in the pellet (whole-cell lysate) and supernatant fractions were separated by SDS/PAGE and immunoblotted with antibodies against IpaB, IpaC, IpaD, and DnaK. The blots shown are representative of at least three experiments.
The Yersinia T3SA Does Not Mediate Phagosomal Escape.
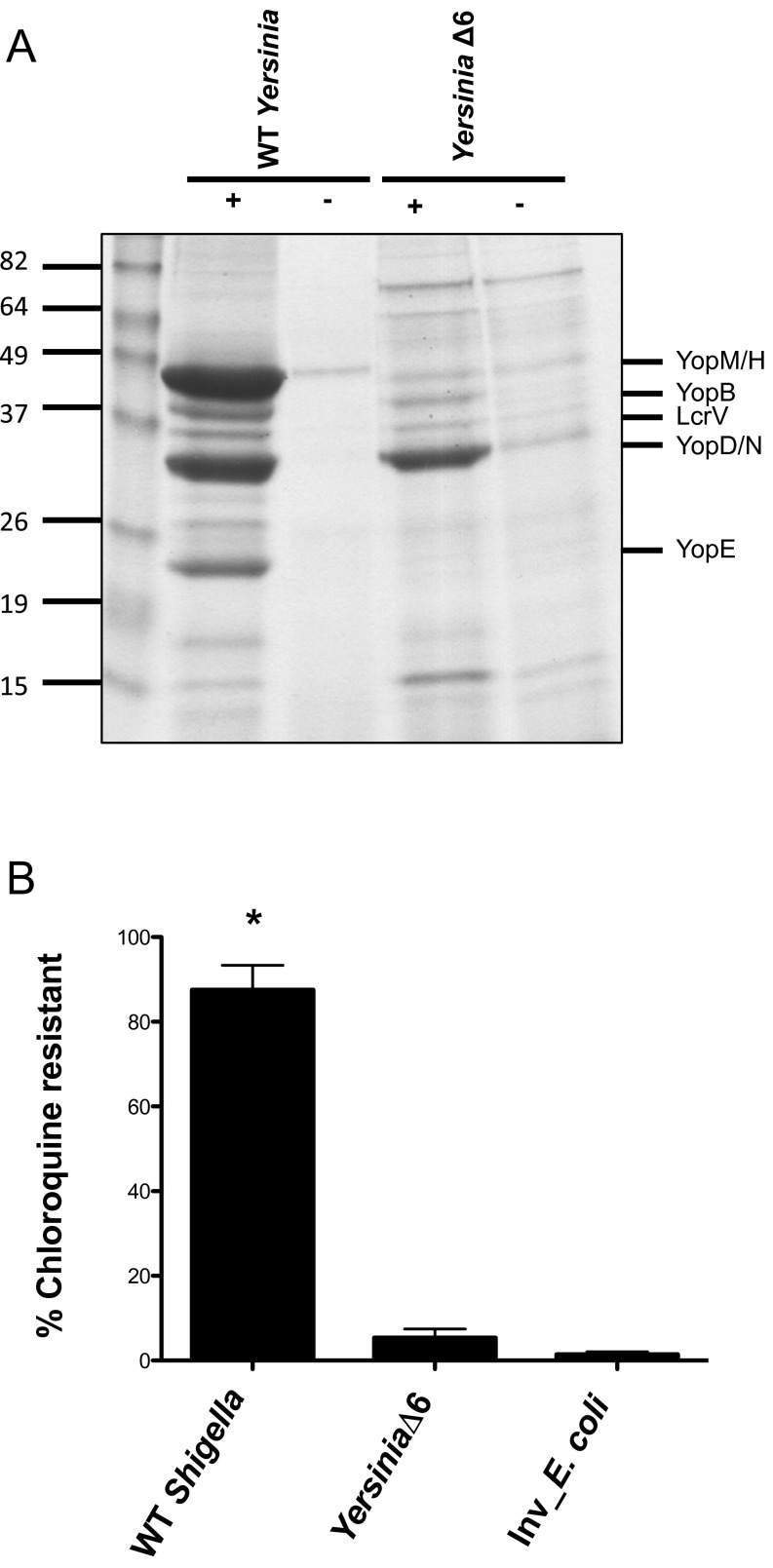
One model to account for the ability of the Shigella T3SA to mediate escape is through the formation of pores in the phagosomal membrane. If this is the case, then it seems likely that all intracellular bacteria that express a T3SS would escape from phagosomes into the cytosol, particularly in the absence of effectors that might prevent such lytic activity (i.e., Yersinia YopE) (20). To test this possibility, we investigated the behavior of YPIIIΔ6, an effectorless Yersinia pseudotuberculosis strain (21) that forms pores in host cell membranes (22). Although most Yersinia remain extracellular, some invade and establish a replicative niche within modified phagosomes in macrophages (23). When grown under conditions that activate its T3SS (Fig. S5A), we observed that, as previously reported, WT Yersinia rapidly induced the rounding and detachment of epithelial cells (24), precluding further analyses. However, YPIIIΔ6 did not and, like mT3Δeff_Inv_E. coli, efficiently invaded epithelial cells. In contrast, when exposed to CHQ at 1 h p.i. for 1 h, YPIIIΔ6 exhibited only 5% CHQR (Fig. S5B), indicating that effectorless Yersinia reside primarily within phagosomes, thus demonstrating that phagosomal escape is not a uniform property of all T3SA.
Fig. S5.
Yersinia T3SA does not mediate phagosomal escape. (A) WT and YPIIIΔ6 Yersinia secrete comparable levels of T3SA components. WT and YPIIIΔ6 Yersinia were grown overnight with aeration at 26 °C, then back-diluted, and incubated at 37 °C for 5 h in 2XYT media ± 20 mM NaOxalate and 20 mM MgCl2, the presence of which induces Yersinia T3SS. Proteins in the secreted supernatant fractions were TCA precipitated, separated by SDS/PAGE, and visualized by staining with GelCode Blue. (B) CHQR assays of HeLa cells infected with Shigella at an MOI of 100 and YPIIIΔ6, Inv_E. coli strains at an MOI of 10 were exposed to gent ± CHQ for 1 h at 1 h p.i. *P < 0.05 for strains versus Inv_E. coli as determined by one-way ANOVA with Dunnett’s post hoc analysis. The mean ± SEM of at least four independent experiments is shown.
A Single Component of the Translocon Apparatus Mediates Phagosomal Escape Efficiency.
Salmonella pathogenicity island 1 (SPI1) and Shigella T3SA belong to the Inv/Mxi-Spa family of T3SS (25), and accordingly, their proteins share a moderate degree of sequence similarity. Interestingly, although most Salmonella enterica serovar Typhimurium (Salmonella) reside within phagosomes, a significant number of Salmonella (∼15–20%) escape from nascent vacuoles to establish an intracytoplasmic replicative niche in epithelial cells in a SPI1 T3SS-dependent manner (15, 26). There is evidence to suggest that this vacuole lysis is linked to the SPI1 T3SA itself, rather than seven of its translocated effectors (15). Given the observed differences in the ability of the phylogenetically related Shigella and Salmonella T3SA to mediate phagosomal escape, we hypothesized that the degree to which these pathogens enter the cytosol reflects the action of their T3SA, specifically one or more translocator proteins, the only three T3SA proteins that directly contact host cell membranes. To test this possibility, we investigated whether swapping the translocator proteins from Salmonella with their Shigella orthologs would promote Salmonella phagosomal escape, an approach that had previously suggested a role for Shigella IpaC in this process, at least when overexpressed in Salmonella (6).
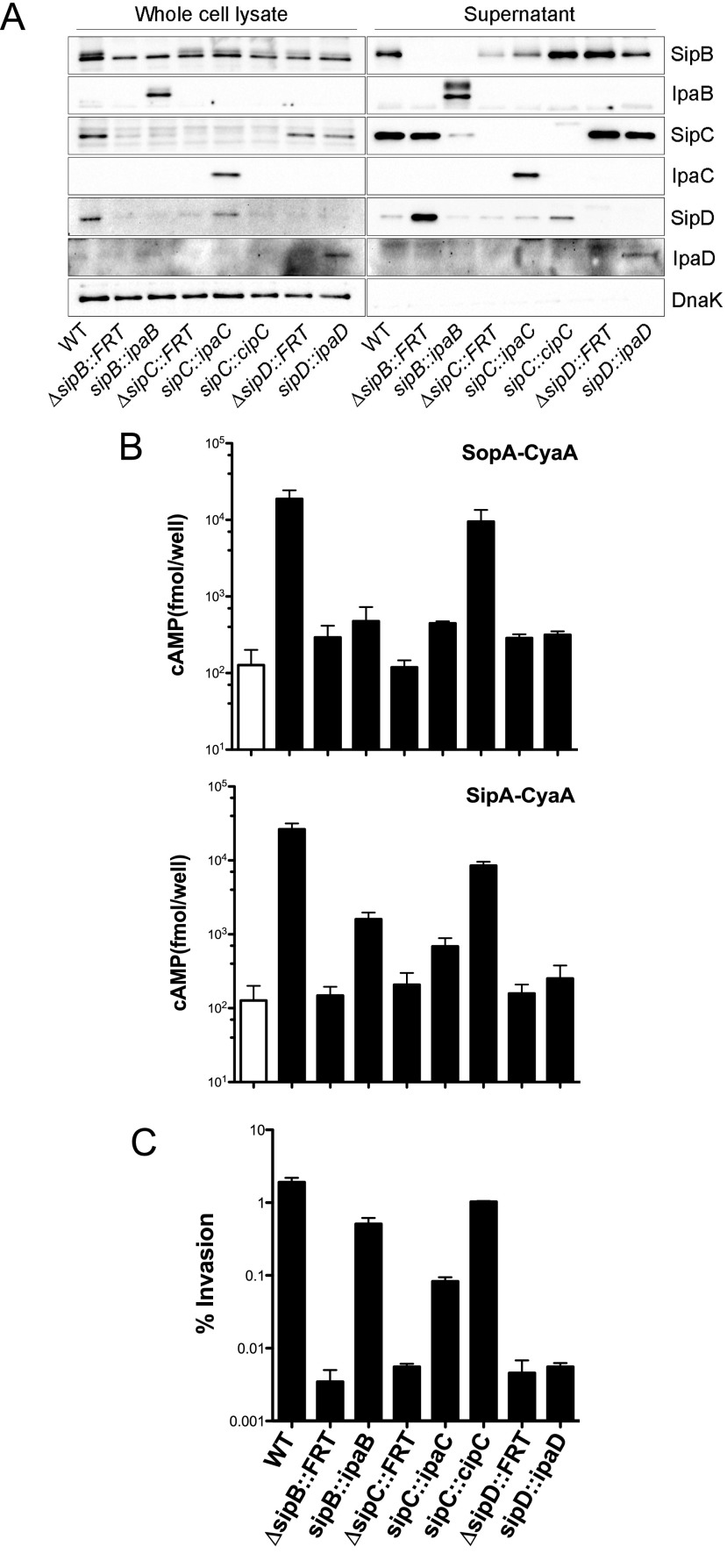
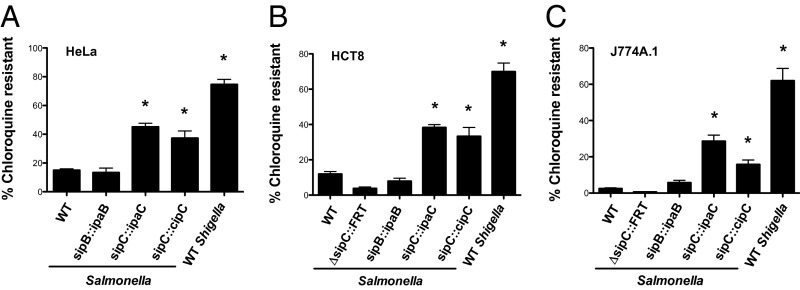
We replaced sipB, sipC, and sipD from Salmonella with their respective Shigella orthologs (ipaB, ipaC, and ipaD) using allelic exchange to generate sipB::ipaB, sipC::ipaC, and sipD::ipaD Salmonella strains. Such chromosomal replacement preserves the genetic organization of the operon and stoichiometry of the translocator proteins. Each of these orthologs was expressed and recognized as a secreted substrate by the Salmonella SPI1 T3SS (Fig. S6A). Furthermore, the translocation of type III effectors (Fig. S6B) and invasion into epithelial cells (Fig. S6C) of sipB::ipaB and sipC::ipaC Salmonella indicated that IpaB and IpaC could functionally replace SipB and SipC, respectively. IpaD did not substitute for SipD (Fig. S6 B and C). We next investigated the intracellular fate of sipB::ipaB and sipC::ipaC Salmonella using the CHQR assay; the sipD::ipaD strain was not studied further given the lack of complementation. As shown in Fig. 5, replacement of SipC with IpaC, but not SipB with IpaB, significantly altered the fate of intracellular Salmonella in three different cell lines. Compared with WT Salmonella, at 1.5 h p.i., phagosomal escape by sipC::ipaC Salmonella was increased by 3.0-, 3.2-, and 11.6-fold within HeLa epithelial cells (Fig. 5A), HCT-8 intestinal epithelial cells (Fig. 5B), and J774A.1 murine macrophages (Fig. 5C), respectively. Therefore, the presence of IpaC, but not IpaB, leads to enhanced phagosome lysis by Salmonella.
Fig. S6.
Components of the Shigella, Salmonella, and Chromobacterium translocon are functionally interchangeable. (A) The designated Salmonella strains were grown overnight shaking at 37 °C and then subcultured for 3.5 h with shaking. Proteins present in the pellet (whole-cell lysate) and supernatant fractions were separated by SDS/PAGE and probed with antibodies against SipB, IpaB, SipC, IpaC, SipD, IpaD, and DnaK. (B) J774A.1 cells were infected with the designated strains, each of which carries a plasmid that expresses SipA–CyaA or SopA–CyaA. The relative amounts of translocated CyaA fusion proteins at 1 h p.i. reflect the cAMP levels present in clarified infected cell lysates. The open bar represents WT Salmonella with no pBAD18 vector. (C) HeLa cells infected with the Salmonella strains grown as described in A were exposed to gent at 30 min p.i. The monolayers were lysed at 1 h p.i. and internalized bacteria enumerated from colony-forming units. Invasion reflects the number of gentR bacteria as a percentage of the original inoculum. Data are mean ± SEM for at least three independent experiments.
Fig. 5.
The T3SA translocon plays a major role in determining the fate of intracellular bacterial pathogens. CHQR assays of (A) HeLa epithelial, (B) HCT-8 epithelial, or (C) J774A.1 macrophage-like cells infected with the designated strains grown under conditions that induce expression of the Salmonella SPI1 or Shigella Mxi-Spa T3SS. Cells were infected with WT Shigella (MOI of 20–100) or the Salmonella strains at an MOI of 10–50 and exposed to gent ± CHQ at 30 min p.i. for 1 h. *P < 0.05 for strains versus WT Salmonella as determined by one-way ANOVA with Dunnett’s post hoc analysis. The mean ± SEM of at least three independent experiments is shown.
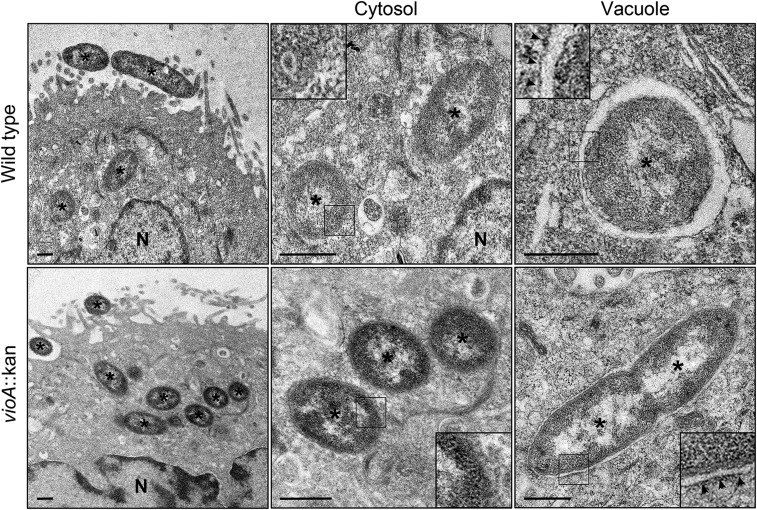
T3SSs have been divided into eight distinct families based on phylogenetic analyses (25). The Salmonella SPI1 and Shigella Mxi-Spa systems are the best characterized members of the Inv/Mxi-Spa family, which also includes T3SSs from Chromobacterium (Cpi-1), Yersinia (Ysa), and Burkholderia (Bsa). Like Salmonella and Shigella spp., entry of Chromobacterium violaceum into nonphagocytic cells is dependent upon its Inv/Mxi-Spa T3SS (27), but its intracellular niche is unknown. By transmission electron microscopy (TEM), we identified that Chromobacterium is a cytosolic bacterium in epithelial cells, with 47% (n = 30 bacteria) and 97% (n = 76) free in the cytosol at 1 h and 3 h p.i., respectively. Furthermore, vacuole lysis is independent of the violacein pigment encoded by vioA; 53% (n = 36) and 96% (n = 96) of ΔvioA bacteria were in the cytosol at 1 h and 3 h p.i., respectively (Fig. S7). To test whether the ability of Shigella IpaC to mediate increased phagosomal escape might be a shared trait among Inv/Mxi-Spa translocator orthologs from cytosolic pathogens, we generated sipC::cipC Salmonella, a strain whereby Salmonella SipC is replaced with Chromobacterium CipC. CipC has greater amino acid sequence identity to SipC (41%) than does IpaC (30%), which likely explains its enhanced ability to substitute for ΔsipC::FRT Salmonella in both effector translocation (Fig. S6B) and invasion of epithelial cells (Fig. S6C). As observed for IpaC, the introduction of CipC into Salmonella resulted in a strain that escaped into the cytosol of both epithelial cells and macrophages at substantially elevated levels compared with WT Salmonella, with a 2.5-, 2.8-, and 6.4-fold increase in vacuole lysis in HeLa, HCT-8, and J774A.1 cells, respectively (Fig. 5). These observations demonstrate that the IpaC/SipC/CipC class of translocators plays a major role in determining the degree to which intracellular pathogens establish phagosomal versus cytoplasmic replicative niches within infected epithelial cells and macrophages.
Fig. S7.
C. violaceum resides within the cytosol of epithelial cells. Shown are representative electron micrographs of cytosolic and vacuolar Chromobacterium. HeLa cells were infected with a spontaneous NalR mutant of C. violaceum ATCC 12472 (CVN WT) or a CVN vioA::KAN mutant. At 1 h p.i., cells were fixed and processed for TEM. Approximately half of the WT (14 of 30 bacteria) and vioA::KAN mutant (19 of 36) bacteria are found free in the cytosol, with a clear absence of any vacuolar membrane remnants. The remaining bacteria are membrane-bound, in tight-fitting vacuoles. Asterisks denote bacteria. N, nucleus. Insets show 2.5× enlargement of the boxed regions. Arrowheads indicate vacuole membranes around bacteria. (Scale bars, 500 nm.)
SI Materials and Methods
Plasmid Construction.
pmT3SS∆eff.
The multistep recombineering approach outlined in Fig. S2 was used to generate pmT3SSΔeff. (i) The FRT KAN cassette present with VP_∆ipaJ::FRT-KANR-FRT (VP) (9) was removed via the FLP recombinase (34) to generate VP_∆ipaJ::FRT. (ii) The following two-step “landing pad” technology described by Kuhlman and Cox (35) was then used to replace the region of the VP between virB and ipgD with one that contains ipaD through ipgC. In step 1, the LP/SceI TETR cassette of pTKS/CS was PCR amplified with oligonucleotides virB-LP2 and icsB-LP1 to add flanking homology to sequences that flank virB and icsB. The λ-Red recombinase system was then used to replace the DNA fragment between virB and the region upstream of icsB on the VP_∆ipaJ::KANR with a landing pad integration site, generating a version of VP (pVP_LP_ΔipaA-icsB) that no longer expresses the translocon apparatus as well as IpaA, IpgB1, and IcsB. In step 2, a region of DNA that encodes ipgCipaBCD was PCR amplified from the Shigella VP with oligonucleotides ipaD-XhoI and ipgC-KpnI to introduce this DNA fragment into the MCS (multicloning site) of pTKIP-HYG and generate a construct that carries an ipgCipaBCD-FRT-HYGR-FRT cassette flanked by LP and SceI restriction sites (pTKIP-ipgCipaBCD). After sequence verification of the insert, this plasmid was introduced into E. coli that carry VP_LP_ΔipaA-icsB and pTKRED, a plasmid that encodes both pBAD SceI and plac lambda-Red (35). Landing pad technology was then used to replace the LP1-TETR-LP2 cassette of VP_LP_ΔipaA-icsB with the LP1-ipgCipaBCD-FRT-HYGR-FRT-LP2 cassette to generate VP_LP_ipgCipaBCD-HYGR. (iii) The FLP recombinase was used to resolve the FRT-HYGR-FRT cassette. However, given the close proximity of the remnant FRT of the resolved FRT-KANR-FRT cassette and the FRT sites flanking the HYGR cassette, this manipulation resulted in virB loss as well as the HYGR cassette, thus generating pVP_ΔicsB/ipgB1/ipaA/virB. (iv) The λ-Red recombinase system was used to replace ipgD with an FRT-KANR-FRT cassette to generate pVP_ΔicsB/ipgB1/ipaA/VirB/ipgD::KANR (oligos ipgD-F and R). (v) The previously described capture vector strategy (9, 36) was next used to transfer the region between IpaD and Spa40 of pVP_ΔicsB/ipgB1/ipaA/VirB/ipgD::KANR onto pLLX13 (36) to generate pmT3SSΔeff.
Capture vector strategy.
Initially, a capture vector was assembled via homologous recombination between (i) pLLX13 vector backbone, linearized with NheI [pLLX13 is a yeast/E. coli shuttle vector that carries a yeast-selectable ura3 marker and a TETR marker for bacteria (9, 36)]; (ii) ipaD PCR product amplified from purified Shigella VP DNA using oligos LP1-ipaD-F and ipaD-R; (iii) spa40 PCR product using oligos spa40-F and LP2-spa40-R; and (iv) a PCR fragment amplified from the vector pLLX8, which encodes the ampicillin resistance gene cassette bla (pLLX8 amp-F and pLLX8 amp-R). ipaD and spa40 were amplified by PCR with primers that add homology to both the pLLX8-derived bla PCR fragment and the Landing Pad sequence. These initial PCRs were followed by a second round of nested PCR to add a region of homology to the pLLX13 vector backbone to ipaD and spa40 using oligos pLLX13-LP1-F or pLLX13-LP2-R, respectively. The bla-carrying fragment from pLLX8 was amplified with primers that provide homology to ipaD and spa40. The flanking homology on these DNA sequences enables their assembly by homologous recombination when cotransformed into competent Saccharomyces cerevisiae. The assembled capture vector pLLX13-IpaD-Spa40 was linearized and introduced into DH10B E. coli containing the pVP_ΔicsB/ipgB1/ipaA/virB/ipgD::KANR plasmid. Induction of λ-Red recombinase led to recombination and transfer of genes between ipaD and spa40 from the modified VP onto pLLX13 (Fig. S2).
pmT3SS∆eff_IpaA.
pmT3SS∆eff_IpaA was generated by sequentially deleting fragments of DNA from pmT3SS (9) via homologous recombination using the λ-Red recombinase system (34) (Fig. S4). First, the FRT-KANR-FRT cassette in pmT3SS was removed via the FLP recombinase. The region of DNA encoding icsB, ipgA, and ipgB1 was then replaced via homologous recombination with a second FRT-KANR-FRT cassette PCR amplified with oligonucleotides icsB-F and ipgB1-R. After confirming the correct location of this cassette via PCR, the FRT-KANR-FRT cassette was removed via the FLP recombinase. Subsequently, using homologous recombination, a FRT-KANR-FRT was PCR amplified with oligonucleotides ipgD-F and ipgD-R to generate pmT3SS∆eff_IpaA.
Each of the intermediate and final plasmids generated above were extensively analyzed via PCR to ensure not only that the correct recombination events took place but also that regions of interest outside of that manipulated were not affected. Furthermore, the vector/inset junctions of pmT3SSΔeff and pmT3SS∆eff_ipaA were confirmed by DNA sequencing.
CyaA plasmids.
Two adenylate cyclase (CyaA) fusions were used to measure type III effector translocation by Salmonella. pBAD18-SopA-FLAG-CyaA has been described previously (37). pBAD18-SipA-CyaA was constructed by excising the fragment encoding SipA-CyaA from pB500 (38) with SacI and HindIII and ligating into the corresponding sites of pBAD18-Cm.
Strain Construction.
∆sipB::FRT, ∆sipC::FRT, and ∆sipD::FRT strains were constructed in Salmonella typhimurium SL1344 (Salmonella) using λ-Red recombinase technology (34) with the oligonucleotides sipB-KO-F and sipB-KO-R, sipC-KO-F and sipC-KO-R, and sipD-KO-F and sipD-KO-R, respectively. Replacement of each gene with the FRT-KANR-FRT cassette was verified by PCR with flanking primers, and the allele was moved into a clean Salmonella background via P22 phage transduction. The FLP recombinase (34) was then used to eliminate the FRT-flanked KANR gene.
sipB::ipaB Salmonella was generated by first using homologous recombination to replace Salmonella sipB with Shigella ipaB. Approximately 800 bp of region upstream and downstream of sipB ORF were amplified from Salmonella genomic DNA with the oligonucleotides sipBipaB-SacIF1 and sipBipaB-R1, and sipBipaB-F3 and sipBipaB-KpnR3. The ipaB ORF was amplified from S. flexneri 2457T (Shigella) genomic DNA with the oligonucleotides sipBipaB-F2 and sipBipaB-R2. The three amplicons were mixed together and subject to a second round of amplification with sipBipaB-SacIF1 and sipBipaB-KpnR3. The resulting product was cloned into SacI/KpnI-digested pRE112 (39), followed by electroporation into E. coli SY327λpir. After sequence verification, the pRE112-sipB::ipaB plasmid was transferred into E. coli SM10λpir to generate the conjugative donor strain. The sipB::ipaB strain was then constructed by allelic exchange into WT Salmonella as described previously (40) and confirmed by PCR analysis.
sipC::ipaC Salmonella was generated by first using homologous recombination to replace Salmonella sipC with Shigella ipaC. Approximately 1.5 kb and 1 kb of region upstream and downstream, respectively, of the sipC ORF were amplified from Salmonella genomic DNA with the oligonucleotides sipCipaC-Xma and sipCipaC-R1, and sipCipaC-F3 and sipCipaC-Xba. The ipaC ORF was amplified from S. flexneri 2457T genomic DNA with the oligonucleotides sipCipaC-F2 and sipCipaC-R2. The three amplicons were mixed together and subject to a second round of amplification with sipCipaC-Xma and sipCipaC-Xba. The resulting product was cloned into XmaI/XbaI-digested pRE112 (39) to create pRE112-sipC::ipaC, and allelic exchange proceeded as described above to construct the sipC::ipaC strain.
sipD::ipaD Salmonella was generated by first using homologous recombination to replace Salmonella sipD with Shigella ipaD. Approximately 700 bp and 750 bp of region upstream and downstream, respectively, of the sipD ORF were amplified from Salmonella DNA with the oligonucleotides sipDipaD-XmaF and sipDipaD-OLR1, and sipDipaD-OLF2 and sipDipaD-XbaR. The ipaD ORF was amplified from Shigella genomic DNA with the oligonucleotides sipDipaD-OLF1 and sipDipaD-OLR2. The three amplicons were mixed together and subject to a second round of amplification with sipDipaD-XmaF and sipDipaD-XbaR. The resulting product was cloned into XmaI/XbaI-digested pRE112 to create pRE112-sipD::ipaD, and allelic exchange proceeded as described above to construct the sipD::ipaD strain.
sipC::cipC Salmonella was generated by first using homologous recombination to replace Salmonella sipC with C. violaceum cipC. Approximately 1.5 kb and 1 kb of region upstream and downstream, respectively, of the sipC ORF were amplified from Salmonella genomic DNA with the oligonucleotides sipCipaC-Xma and sipCcipC-R1, and sipCcipC-F3 and sipCipaC-Xba. The cipC ORF was amplified from C. violaceum Bergonzini (ATCC 12472) genomic DNA with the oligonucleotides sipCcipC-F2 and sipCcipC-R2. The three amplicons were mixed together and subject to a second round of amplification with sipCipaC-Xma and sipCipaC-Xba. The resulting product was cloned into XmaI/XbaI-digested pRE112 to create pRE112-sipC::cipC, and allelic exchange proceeded as described above to construct the sipC::cipC strain.
Secretion Assays.
CR type III secretion assay.
CR secretion assays were performed as previously described (41) with the following modifications. All incubations were conducted at 37 °C with aeration. Overnight cultures of Shigella grown in TCS (trypticase soy) broth or E. coli grown in LB (Luria broth) were back diluted between 1:50 and 1:200 to account for the slower growth observed with E. coli strains associated with introduction of the larger plasmid (i.e., VP DNA). The diluted cultures were grown for 90 min, at which point 1 mM IPTG was added to the cultures to induce expression of VirB, hence T3SS expression, as well as any heterologously expressed IPTG-driven FLAG-tagged effectors. After an additional 45 min, the bacteria were resuspended for 30 min in PBS containing 10 μM CR (Sigma), the presence of which triggers the secretion of translocator proteins (IpaB, IpaC, IpaD) and type III effectors into the media. To account for differences in bacterial titers at this point, the number of bacteria harvested for each culture were normalized based on OD600 readings. After the incubation in CR, the bacteria were centrifuged and the pellets were saved as whole-cell lysates. After an additional centrifugation step to ensure that intact bacteria were removed from the supernatant fractions, the proteins present were precipitated by the addition of TCA [10% (vol/vol)]. Proteins present in the whole-cell lysate and supernatant fractions were separated via SDS/PAGE and analyzed by immunoblotting with antibodies against IpaB, IpaC, and IpaD (generous gifts of Wendy Picking, University of Kansas, Lawrence, KS), secreted components of the Shigella TS3A. Levels of DnaK, a conserved cytoplasmic protein found in Shigella and E. coli, were assessed as controls for bacterial lysis using a commercial anti-E. coli DnaK antibody (Abcam ab69617).
Yersinia type III secretion assay.
Yersinia T3SS assays were conducted as previously described (42). Briefly, WT and YPIIIΔ6 Y. pseudotuberculosis (Yersinia) were grown overnight with aeration at 26 °C; diluted 1:50 in fresh 2XYT media with or without 20 mM NaOxalate and 20 mM MgCl2, the presence of which induces Yersinia T3SS; and incubated with shaking at 37 °C for 5 h. To account for differences in bacterial titers at this point, the number of cells harvested for each culture were normalized based on OD600 readings. Proteins in the secreted supernatant fractions were TCA precipitated, separated by SDS/PAGE, and visualized by staining with GelCode Blue (Life Technologies).
Salmonella type III secretion assay.
Bacteria were grown overnight with aeration at 37 °C in LB–Miller broth and then subcultured for 3.5 h to induce maximal SPI1 expression (43). An aliquot was taken and centrifuged at 16,000 × g for 5 min, and the pellet was resuspended in SDS/PAGE sample buffer (whole-cell lysate). The remaining subculture was centrifuged at 16,000 × g for 10 min, the supernatant collected and passed through 0.22-µm low protein-binding filters (Whatman) to remove any residual bacteria, and proteins precipitated with 10% (vol/vol) TCA (secreted). Proteins were separated by SDS/PAGE and analyzed by probing with polyclonal antibodies against SipB and SipC (kindly provided by Daoguo Zhou, Purdue University, West Lafayette, IN) or monoclonal antibodies against SipD (kindly provided by Francisco Martinez-Becerra, University of Kansas, Lawrence, KS), IpaB (2F1), IpaC (2G2), and IpaD (16F8) (kindly provided by Robert Kaminski, Walter Reed Army Institute of Research, Silver Springs, MD) and DnaK.
Cell culture conditions.
HeLa cervical epithelial cells (ATCC CCL-2), HCT-8 colonic epithelial cells (ATCC CCL-244), and J774A.1 mouse macrophage-like cells (ATCC TIB-67) were purchased from ATCC and grown as recommended by ATCC at 37 °C in a 5% CO2 incubator.
Internalization Assays.
Inside/outside microscopy assay.
One day before infection, six-well plates containing acid-washed glass coverslips (Corning) were seeded with 3 × 105 HeLa cells per well. Bacteria were grown overnight in LB (E. coli) or TCS (Shigella), then subcultured and grown for 90 min, after which 1 mM IPTG was added to the media for 45 min to induce VirB expression. HeLa cells were infected with bacteria at an MOI of 100 or 10 (the lower MOI was used for strains that express an adhesin, either AfaI or Inv). The plates were centrifuged at 800 × g for 10 min at room temperature (RT) and then incubated for 1 h at 37 °C. Postfixation with 3.7% (wt/vol) paraformaldehyde (PFA) (20 min), the extracellular bacteria were labeled with rabbit anti-E. coli (Abcam ab137967) or rabbit anti-Shigella (Abcam ab65282) polyclonal antibodies at 1:100 in TBS (Tris buffered saline) buffer plus 2% (wt/vol) BSA (Fisher Scientific) for 30 min followed by goat–anti-rabbit Alexa Fluor 488 (green) secondary antibodies (Life Technologies) diluted 1:100 for 30 min. The infected cells were then permeabilized with 0.5% Triton X-100 for 10 min and stained with DAPI (100 μM, 5 min) to label the DNA of intra- and extracellular bacteria and host cell nuclei. All fixation and immunostaining steps were carried out at RT. Coverslips mounted on glass slides were then examined with a Nikon TE300 microscope with Chroma Technology filters. Images were captured digitally using a black and white Sensys charge-coupled device camera and IP LAB software (Scanalytics). Color figures were assembled by separately capturing signals with each of the appropriate filter sets and digitally pseudocoloring the images using IP LAB and Adobe Photoshop. The percentage of infected cells was defined as the number of HeLa cells that contain intracellular bacteria.
Gent protection assay.
HeLa cells were seeded in 24-well plates (Nunc) 18–24 h before infection at 5 × 104 cells per well. Salmonella were grown overnight with aeration at 37 °C in LB–Miller broth and then subcultured for 3.5 h to induce maximal SPI1 expression (43), and infection conditions were as described previously (15). Gent (100 µg/mL) was added at 30 min p.i., and monolayers were washed once in PBS and lysed at 1 h p.i. in 0.2% (wt/vol) sodium deoxycholate. Serial dilutions were plated and the number of internalized bacteria enumerated. Bacterial internalization (% invasion) reflects the ratio of gentR internalized bacteria/initial inoculum.
Phagosomal Escape Assays.
CHQ resistance assay.
HeLa cells seeded in 96-well plates (3 × 104 cells per well) were infected with the Shigella, E. coli, and Yersinia strains grown at MOIs experimentally determined such that on the order of at least 2,000 bacteria per well were isolated after treatment with gent: MOI 10 for Yersinia and Inv_E. coli and MOI 100 for Shigella and E. coli strains that lack an adhesin. Before infection, each of the strains was grown under conditions that ensure activation of its respective T3SS as described in Secretion Assays. After a 1-h infection, gent (100 μg/mL) ± CHQ (200 μg/mL, 400 µM) was added to the media for 1 h to kill extracellular versus phagosomal bacteria, respectively. The infected cells were then washed three times with PBS and then lysed with PBS containing 1% (vol/vol) Triton X-100. For Salmonella strains, CHQR assay was performed in 24-well plates (5 × 104 cells per well for HeLa cells, 1.2 × 105 cells per well for HCT-8 epithelial cells and J774A.1 macrophages), and antibiotics were added at 30 min p.i. for 1 h before cell monolayers were washed once in PBS and lysed with sodium deoxycholate. Serial dilutions of the lysates were plated and the number of bacteria present enumerated. Percent cytosolic bacteria were determined by calculating the ratio of (CHQR + gentR)/gentR bacteria. Percentages > 100% were capped at 100%.
Digitonin permeablization assay.
Based on published protocols (15, 16), the following modified digitonin permeabilization assay was performed. HeLa cells were infected with WT Shigella or genetically modified strains of E. coli, each of which carries the pBR322 mCherry expression plasmid. One hour p.i., extracellular bacteria were labeled with Alexa Fluor 647 (far-red) goat–anti-rabbit secondary antibodies (Life Technologies) conjugated to either rabbit anti-E. coli or rabbit anti-Shigella primary antibodies at 1:100 for 30 min. The cells were then washed three times with KHM buffer (110 mM potassium acetate, 20 mM Hepes, 2 mM MgCl2, pH 7.3) and then incubated for 1 min in KHM buffer with freshly prepared 35 μg/mL digitonin (Sigma) to specifically permeabilize the plasma membrane. Cells were then blocked with KHM buffer containing 2% BSA for 15 min, followed by staining with Alexa Fluor 555 (orange)-conjugated GM130 antibody (BD Biosciences) at 1:10 for 1 h. (Only coverslips that exhibit <85% GM130-positive cells were subsequently analyzed to ensure that the cells were not overpermeabilized by digitonin.) Post-digitonin permeabilization, the infected cells were again labeled with the anti-E. coli and anti-Shigella antibodies at 1:100 for 30 min to label both extracellular and cytoplasmic bacteria, followed by PFA fixation for 20 min and Alexa Fluor 488 (green) goat–anti-rabbit secondary antibodies (Life Technologies) at 1:100 for 30 min. All fixation and immunostaining steps were carried out at RT. The stained coverslips were then visualized as described above. Quantification of the percentage of cytoplasmic bacteria as defined by the ratio of intracytoplasmic to total intracellular (green/red)/(green/red + red) bacteria was associated with digitonin-permeabilized (GM130-positive) cells. For each infection, 100 intracellular bacteria were counted, and extracellular bacteria (far-red channel) were excluded from calculations.
Translocation assays.
J774A.1 cells were seeded in 24-well plates at 1.4 × 105 cells per well 24 h before infection. Salmonella strains bearing the pBAD18-SipA-CyaA and pBAD18-SopA-CyaA plasmids were subcultured from overnight cultures as described above. l-arabinose [0.2% (wt/vol) final concentration] was added to the subcultures after 2.5 h of growth to induce the production of SipA–CyaA and SopA–CyaA for 1 h. Macrophages were infected at an MOI of ∼20 for 10 min, washed thrice in Hanks’ balanced salt solution (HBSS), and incubated in growth medium without antibiotics for 20 min. At 30 min p.i., growth medium containing 100 µg/mL gent was added to kill any noninternalized bacteria. At 1 h p.i., monolayers were lysed in 0.1 M HCl for 10 min, heated to 95 °C for 5 min, and neutralized with NaOH. cAMP levels were determined using the Amersham cAMP Biotrak enzymeimmunoassay system (GE Healthcare Life Sciences) with the nonacetylation enzymeimmunoassay procedure.
TEM.
HeLa cells were seeded in T-25 tissue culture flasks at 7 × 105 cells per flask the day before infection. A spontaneous NalR mutant of C. violaceum ATCC 12472 (CVN) and CVN vioA::KAN mutant were grown overnight in LB–Miller at 37 °C with shaking and then subcultured 1:33 the next day in 10 mL LB–Miller broth, and growth continued at 37 °C for 3 h with shaking (OD600, ∼2.0). Bacteria were pelleted (6,000 × g for 1.5 min), resuspended in an equal volume of HBSS, and incubated with HeLa cells for 30 min at 37 °C in a 5% CO2 incubator. Monolayers were washed thrice in HBSS and growth medium containing 50 µg/mL gent added for 30 min. At 1 h p.i., infected monolayers were washed with PBS and trypsinized to collect. Cells were washed once in PBS, then fixed in 2% (vol/vol) glutaraldehyde/2% (wt/vol) PFA in 0.1 M cacodylate buffer, pH 7.2, overnight at 4 °C, pelleted at 900 × g for 5 min, and rinsed three times with 0.1 M cacodylate buffer with 3.5% (wt/vol) sucrose for 10 min. Osmium tetraoxide [2% (wt/vol)] was added in 0.1 M cacodylate buffer and incubated at RT for 2 h. Cells were then rinsed thrice with double-distilled water (ddH2O), 5 min each, and 1% aqueous (vol/vol) tannic acid added and incubated for 1 h at RT. The cell pellet was rinsed with ddH2O and dehydrated in an ascending ethanol series for 10 min each. Finally, samples were treated with 100% propylene oxide for 10 min followed by a 1:1 mixture of propylene oxide and Spurr’s embedding media overnight. Next, samples were treated with Spurr’s embedding media and placed in a 70 °C oven overnight for polymerization. Ultrathin sections (∼70 nm) were prepared with glass knives and a Leica Reichert Ultracut R Microtome and placed on grids, which were stained for 6 min with 4% (vol/vol) aqueous uranyl acetate and 6 min with Reynold’s lead reagent. Samples were imaged on FEI Technai G2 20 Twin at 200 kv.
Discussion
Despite its recognition as a model professional intracytoplasmic pathogen, our understanding of how Shigella rapidly escape from phagosomes into the host cell cytosol is currently limited. Here, to separate the established roles of the Shigella T3SS in effector delivery from phagosomal escape, we established a platform to study the behavior of the Shigella T3SA in the absence of all effectors. This technical feat was accomplished using recombineering and synthetic biology-based approaches to capture and transfer all of the genes needed to form a Shigella T3SA, but none of its effectors, into DH10B, a laboratory strain of E. coli. Our observation that these type 3 secretion-competent E. coli efficiently escape from phagosomes into the cytosol of infected cells demonstrates for the first time, to our knowledge, that the Shigella T3SA alone is sufficient to mediate phagosomal escape, a property we did not observe to be a conserved trait of the Yersinia T3SA.
To complement these reductionist-based studies, as well as to identify which component of the Shigella T3SA mediates escape, we examined the behavior of Shigella IpaB and IpaC, the two components of the T3SA that directly contact host cells and form a translocon pore in host cell membranes, when expressed at physiologic levels in place of their Salmonella orthologs. Shigella IpaB and IpaC each restored the ability of their corresponding Salmonella deletion strains to translocate effectors and invade nonphagocytic cells, but only expression of Shigella IpaC in Salmonella substantially altered the intracellular fate of this bacterium in epithelial cells and macrophages. This indicates that Shigella IpaC and Salmonella SipC confer different inherent vacuole lysis abilities to these two bacteria (6). Furthermore, enhanced phagosomal escape is not unique to Shigella IpaC, as complementation with the translocator protein CipC from another cytosolic pathogen, Chromobacterium, similarly enhanced vacuole lysis by Salmonella. Taken together, the results of our reductionist- and functional interchangeability-based experiments provide strong evidence that a single component of the T3SA translocon plays a major role in controlling the degree to which bacteria enter the cytosol of host cells.
The exact mechanism by which these proteins regulate phagosomal escape remains to be discovered. It is possible that translocon pore formation plays a role in this process, akin to the means by which Listeria pore-forming toxins mediate vacuole lysis. However, this seems unlikely, as the T3SSs of Shigella, Salmonella, and Yersinia are all well-known to form pores in host cell membranes (8, 24, 28), yet only Shigella efficiently lyses its phagosome. Our Salmonella complementation experiments indicate that it is not likely to be related to T3SS functionality, as there is no correlation between effector translocation and phagosomal escape; the sipC::ipaC Salmonella strain is much less efficient at translocating effectors and invading host cells than sipC::cipC Salmonella, yet both strains lyse their vacuole to a comparable extent. While it is possible that differences in the physical properties of the translocon pores formed by the bacteria result in altered stability of the phagosomal membrane, pore size does not seem to account for this, as the diameter of Shigella and Salmonella translocon pores is very similar (8, 24, 28).
Rather, we favor the possibility that interactions between IpaC, SipC, and CipC with host cell proteins play major roles in mediating phagosome escape. Besides being inserted into host cell membranes, each of these proteins is delivered directly into the cytosol of host cells. Interestingly, IpaC and SipC interact with both actin and intermediate filaments (29–31). It is possible that differences in the recruitment of cytoskeletal proteins to the internalization vacuole by IpaC and SipC play a role in determining phagosome stability. In this regard, it is intriguing that chemicals that disrupt the cytoskeletal cages that normally envelop Chlamydia-containing vacuoles, known as inclusions, result in the release of these pathogens into the host cell cytosol (32). Furthermore, SipC has been demonstrated to interact with Exo70, a component of the exocyst (33). Interactions with Exo70 promote localized membrane expansion at sites of Salmonella entry, perhaps resulting in differences in the lipid contents of Salmonella and Shigella-containing vacuoles, which could differentially affect the efficiency of phagosomal escape.
In summary, here we present complementary experimental approaches that together implicate a single component of the translocon, the outermost portion the T3SA, in mediating the efficiency of bacterial escape from phagosomes into the host cell cytosol. This work has broad scientific and technical implications, as it not only suggests a paradigm regarding how the intracellular niche of Gram-negative pathogens, many of which adopt an intracellular lifestyle as part of their colonization strategy, use their T3SA to mediate vacole lysis but also describes how an innovative “bottom–up” approach can be used to circumvent potential issues with effectors (and translocase) functional redundancy.
Materials and Methods
Bacterial Plasmids, Strains, and Cell Lines.
Plasmids, strains, and oligonucleotides are summarized in Tables S1–S3. Cloning strategy and strain construction details are described in SI Materials and Methods. HeLa and HCT-8 epithelial cells and J774A.1 mouse macrophage-like cells were grown as recommended by American Type Culture Collection (ATCC).
Table S1.
Plasmid summary
| Name | Description | Source |
| pVP ∆ipaJ::FRT-KANR-FRT | S. flexneri 2457T VP carrying deletion of ipaJ | (9, 44) |
| pmT3SS | pLLX13 ipaJ thru spa40, TETR, KANR | (9) |
| pmT3SSΔeff | pLLX13 ipaJ thru spa40∆ipaA,∆ ipgB1,∆icsB,∆ipgD, TETR, KANR | This study |
| pmT3SSΔeff_IpaA | pLLX13 ipaJ thru spa40∆ipgB1,∆icsB,∆ipgD, TETR, KANR | This study |
| pRI203 | Invasin expression plasmid, CMR, AMPR | (14) |
| pIL22 | pBR322 encoding E. coli afimbrial adhesin AFA-I, AMPR | (18) |
| pNG162-VirB | IPTG-inducible VirB allele, SPECR | (9) |
| pDSW206-OspD1-FLAG | IPTG-inducible OspD1-FLAG, AMPR | (41) |
| pDSW206-OspF-FLAG | IPTG-inducible OspF-FLAG, AMPR | (41) |
| pDSW206-OspB-FLAG | IPTG-inducible OspB-FLAG, AMPR | (41) |
| pRE112 | sacB1 suicide vector, CMR | (39) |
| pBAD18-SipA-CyaA | Arabinose-inducible SipA(1–158)-CyaA, CMR | This study |
| pBAD18-SopA-FLAG-CyaA | Arabinose-inducible SopA(1–95)-FLAG-CyaA, KANR, AMPR | (37) |
Table S3.
Oligonucleotide summary
| Oligo | Sequences, 5′–3′ |
| virB-LP2 | tgatgtgagagctccatagatgaaaaaatgcttatcaattttggcttcagggatgaggcg |
| icsB-LP1 | gttaataaagtatgatcctcaaaattagcaatttcattgatacggccccaaggtccaaac |
| ipaD-XhoI | ccggctcgagtcagaaatggagaaaaagtttatct |
| ipgC-KpnI | ggcggtaccatcttttatattaaatcttatactt |
| ipgD-F | caatgccccatttatgactgaggataattaaatgcacatagtgtaggctggagctgcttc |
| ipgD-R | taacatctgctaaatcttccatatattcctcttatacaaacatatgaatatcctccttag |
| LP1-ipaD-F | cgatctcgagtacggccccaaggtccaaacggtgaggcgcacctcagaaatggag |
| ipaD-R | catatatactttagattttaattaaacgcgttctagaaaactctgactaatagtatttcc |
| spa40-F | cattttcaccgttttttgtttaaacgttaactctagagggatggcaaataaaacagaaaagccg |
| LP2-spa40-R | gcaggagctcttggcttcagggatgaggcgccatcttaactcccatataacacatctcc |
| pLLX8 amp-F | ttttctagaacgcgtttaattaaaatctaaagtatatatgagtaaac |
| pLLX8 amp-R | ccctctagagttaacgtttaaacaaaaaacggtgaaaatgggtgatag |
| pLLX13-LP1-F | atattaccctgttatccctagcgtaactatcgatctcgagtacggccccaaggtc |
| pLLX13-LP2-R | taacagggtaatatagagatctggtaccctgcaggagctcttggcttcagggatg |
| icsB-F | tggagagttaataaagtatgatcctcaaaattagcaatttgtgtaggctggagctgcttc |
| ipgB1-R | ctttactttaataagtataagatttaatataaaagatttaacatatgaatatcctccttag |
| sipB-KO-F | agcacagtgaacaagaaaaggaataattatggtaaatgacgctgtaggctggagctgcttcg |
| sipB-KO-R | atttaaataagcggcgggatttattcccacattactaattaacatatgaatatcctccttag |
| sipC-KO-F | aaaactgccaaaataaagggagaaaaatatgttaattagtaatgtaggctggagctgcttcg |
| sipC-KO-R | gattaaatcacacccatgatggcgtatagatgacctttcagacatatgaatatcctccttag |
| sipD-KO-F | ctcctgatggcgaactggggatattatgcttaatattcaaaatgtaggctggagctgcttcg |
| sipD-KO-R | acctggcattatgacggggggctgagtccttacacttgtaaccatatgaatatcctccttag |
| sipBipaB-SacIF1 | cgagctcaaactggcgcgcagtctt |
| sipBipaB-R1 | tgtggtgcttacattatgcataattattccttttcttgttca |
| Sipbipab-F3 | ttgcaacaaactactgcttgaaaactgccaaaataaagggag |
| sipBipaB-KpnR3 | ggggtaccggttttcttcatatt |
| sipBipaB-F2 | tgaacaagaaaaggaataattatgcataatgtaagcaccaca |
| sipBipaB-R2 | ctccctttattttggcagttttcaagcagtagtttgttgcaa |
| sipCipaC-Xma | tccccccgggtcactatgctcatggcc |
| sipCipaC-R1 | gtctgggttggttttgtgttttgaatttccatatttttctccctttattttggcag |
| sipCipaC-F3 | attgctggtaacattcgagcttaatctgaaaggtcatctatacgc |
| sipCipaC-Xba | gctctagagttatccttgcaggaagct |
| sipCipaC-F2 | ctgccaaaataaagggagaaaaatatggaaattcaaaacacaaaaccaacccagac |
| sipCipaC-R2 | gcgtatagatgacctttcagattaagctcgaatgttaccagcaat |
| sipDipaD-XmaF | ccccccgggcttaaacataatgccgccaag |
| sipDipaD-OLR1 | ctattagtcagagttgttatattcataatatccccagttcgcca |
| sipDipaD-OLF2 | gataaactttttctccatttctgacagaagaggatattaataatgg |
| sipDipaD-XbaR | gctctagatgccgggcttcccgtcctacg |
| sipDipaD-OLF1 | tggcgaactggggatattatgaatataaacaactctgactaatag |
| sipDipaD-OLR2 | ccattattaatatcctcttctgtcagaaatggagaaaaagtttatc |
| sipCcipC-R1 | cggggaagaggtaatggcagtcatatttttctccctttattttggcag |
| sipCcipC-F3 | gcgatcgccggcaacatccgcgtctgatctgaaaggtcatctatacgc |
| sipCcipC-F2 | ctgccaaaataaagggagaaaaatatgactgccattacctcttccccg |
| sipCcipC-R2 | gcgtatagatgacctttcagatcagacgcggatgttgccggcgatcgc |
Shigella/Salmonella/Chromobacterium-specific sequences are italicized, and engineered restriction sites are underlined.
Table S2.
Bacterial strain summary
| Name | Description/plasmids | Source |
| WT Shigella | S. flexneri 2457T | (45) |
| WT E. coli | E. coli DH10B | Life Technologies |
| WT Salmonella | S. typhimurium SL1344 | (46) |
| WT Yersinia | Y. pseudotuberculosis YPIII | (21) |
| C. violaceum | ATCC 12472 | ATCC |
| mT3_E. coli (mT3_VirBIPTG DH10B E. coli) | E. coli: pmT3SS + pNG162-VirB | (9) |
| VP_E. coli | E. coli: pVP ∆ipaJ::FRT-KANR-FRT | (9) |
| Inv_E. coli | E. coli: pRI203 | This study |
| mT3Δeff_E. coli (mT3 Δeff_VirBIPTG DH10B E. coli) | E. coli: pmT3SSΔeff + pNG162-VirB | This study |
| mT3Δeff_AfaI_E. coli | E. coli: pmT3SSΔeff + pNG162-VirB + pIL22 | This study |
| mT3Δeff_Inv_E. coli | E. coli: pmT3SSΔeff + pNG126-VirB + pRI203 | This study |
| VP_Inv_E. coli | E. coli: pVP ∆ipaJ::FRT-KANR-FRT + pRI203 | This study |
| mT3Δeff_IpaA_E. coli (mT3Δeff_IpaA_VirBIPTG DH10B E. coli) | DH10B E. coli + pmT3Δeff_IpaA + pNG162-VirB | This study |
| YersiniaΔ6 | Yersinia YPIII ΔyopE, ΔyopH, ΔyopM, ΔyopO, ΔyopT, ΔyopJ | (21) |
| ∆sipB::FRT Salmonella | S. typhimurium deleted for sipB | This study |
| ΔsipC::FRT Salmonella | S. typhimurium SL1344 deleted for sipC | This study |
| ΔsipD::FRT Salmonella | S. typhimurium SL1344 deleted for sipD | This study |
| sipB::ipaB Salmonella | S. typhimurium SL1344 with sipB replaced by S. flexneri ipaB | This study |
| sipC::ipaC Salmonella | S. typhimurium SL1344 with sipC replaced by S. flexneri ipaC | This study |
| sipC::cipC Salmonella | S. typhimurium SL1344 with sipC replaced by C. violaceum cipC | This study |
| sipD::ipaD Salmonella | S. typhimurium SL1344 with sipD replaced by S. flexneri ipaD | This study |
| C. violaceum (CVN) | Spontaneous NalR-strain of ATCC 12472 | (47) |
| CVN vioA::KAN | C. violaceum insertion mutant of vioA, lacking violacein pigment | (47) |
Inside/Outside Microscopy Assay.
As previously described (9), HeLa cells were infected with the designated bacteria grown under conditions that induce T3SSs. At 1 h p.i., infected cells were fixed and stained with rabbit anti-E. coli or anti-Shigella antibodies followed by goat–anti-rabbit Alexa Fluor 488 (green) secondary antibody to label extracellular bacteria. The cells were subsequently permeabilized and treated with DAPI (4',6-diamidino-2-phenylindole) to label intra- and extracellular bacteria as well as host cell nuclei. Images were captured digitally using a Nikon TE3000 microscope with Chroma Technology filters and a 60× objective. See SI Materials and Methods for additional details.
CHQR Assay.
At 0.5–1 h p.i., infected cells were exposed to gentamicin (gent) (100 μg/mL) ± CHQ (200 μg/mL) for 1 h. Cell monolayers were then lysed and bacteria enumerated. See SI Materials and Methods for additional details.
Digitonin Permeablization Assay.
HeLa cells were infected with bacterial strains that constitutively express mCherry. At 1 h p.i., the extracellular bacteria were labeled with Alexa Fluor 647 (far-red)-conjugated anti-E. coli or anti-Shigella antibodies, and the monolayers were then treated with digitonin (35 μg/mL) for 1 min to selectively permeabilize the plasma membrane. Cells were then labeled with Alexa Fluor 555 (orange)-conjugated anti-GM130 and Alexa Flour 488 (green)-labeled anti-E. coli or anti-Shigella antibodies to identify Golgi in permeabilized cells as well as label intracytoplasmic and extracellular bacteria. The percentage of cytoplasmic bacteria was determined by quantifying the ratio of intracytoplasmic to total intracellular (green/red)/(green/red + red) bacteria associated with digitonin-permeabilized cells. Far-red bacteria were not counted. See SI Materials and Methods for additional details.
Acknowledgments
We thank W. Spears for help with plasmid and strain construction; N. Ernst and U. Powale for critically reading the manuscript; and J. Mecsas, W. L. Picking, F. Martinez-Becerra, D. Zhou, R. W. Kaminski, J. Wilson, E. A. Miao, T. Miki, and M. B. Goldberg for sharing reagents/strains. We thank the Franceschi Microscopy and Imaging Center at Washington State University for TEM and C. Davitt, V. Lynch-Holm, D. Mullendore, and J. Benavides-Montaño for their assistance with sample preparation. This work was supported by NIH Grants AI064285 and AI103882 (to C.F.L.) and startup funds from the Paul G. Allen School for Global Animal Health and the Stanley L. Adler Research Fund (to L.A.K.). J.D. was supported by a Svenska Sällskapet för Medicinsk Forskning Fellowship and J.A.K. by a Poncin Scholarship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1520699113/-/DCSupplemental.
References
- 1.Schnupf P, Portnoy DA. Listeriolysin O: A phagosome-specific lysin. Annu Microbes Infect. 2007;9(10):1176–1187. doi: 10.1016/j.micinf.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Galán JE, Lara-Tejero M, Marlovits TC, Wagner S. Bacterial type III secretion systems: Specialized nanomachines for protein delivery into target cells. Annu Rev Microbiol. 2014;68:415–438. doi: 10.1146/annurev-micro-092412-155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchrieser C, et al. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol Microbiol. 2000;38(4):760–771. doi: 10.1046/j.1365-2958.2000.02179.x. [DOI] [PubMed] [Google Scholar]
- 4.Mellouk N, et al. Shigella subverts the host recycling compartment to rupture its vacuole. Cell Host Microbe. 2014;16(4):517–530. doi: 10.1016/j.chom.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 5.De Geyter C, et al. Characterization of the interaction of IpaB and IpaD, proteins required for entry of Shigella flexneri into epithelial cells, with a lipid membrane. Eur J Biochem. 2000;267(18):5769–5776. doi: 10.1046/j.1432-1327.2000.01649.x. [DOI] [PubMed] [Google Scholar]
- 6.Osiecki JC, et al. IpaC from Shigella and SipC from Salmonella possess similar biochemical properties but are functionally distinct. Mol Microbiol. 2001;42(2):469–481. doi: 10.1046/j.1365-2958.2001.02654.x. [DOI] [PubMed] [Google Scholar]
- 7.Senerovic L, et al. Spontaneous formation of IpaB ion channels in host cell membranes reveals how Shigella induces pyroptosis in macrophages. Cell Death Dis. 2012;3:e384. doi: 10.1038/cddis.2012.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blocker A, et al. The tripartite type III secreton of Shigella flexneri inserts IpaB and IpaC into host membranes. J Cell Biol. 1999;147(3):683–693. doi: 10.1083/jcb.147.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reeves AZ, et al. Engineering Escherichia coli into a protein delivery system for mammalian cells. ACS Synth Biol. 2015;4(5):644–654. doi: 10.1021/acssynbio.5b00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tobe T, Yoshikawa M, Mizuno T, Sasakawa C. Transcriptional control of the invasion regulatory gene virB of Shigella flexneri: Activation by virF and repression by H-NS. J Bacteriol. 1993;175(19):6142–6149. doi: 10.1128/jb.175.19.6142-6149.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valencia-Gallardo CM, Carayol N, Tran Van Nhieu G. Cytoskeletal mechanics during Shigella invasion and dissemination in epithelial cells. Cell Microbiol. 2015;17(2):174–182. doi: 10.1111/cmi.12400. [DOI] [PubMed] [Google Scholar]
- 12.Finlay BB, Falkow S. Comparison of the invasion strategies used by Salmonella cholerae-suis, Shigella flexneri and Yersinia enterocolitica to enter cultured animal cells: Endosome acidification is not required for bacterial invasion or intracellular replication. Biochimie. 1988;70(8):1089–1099. doi: 10.1016/0300-9084(88)90271-4. [DOI] [PubMed] [Google Scholar]
- 13.Ray K, et al. Tracking the dynamic interplay between bacterial and host factors during pathogen-induced vacuole rupture in real time. Cell Microbiol. 2010;12(4):545–556. doi: 10.1111/j.1462-5822.2010.01428.x. [DOI] [PubMed] [Google Scholar]
- 14.Isberg RR, Falkow S. A single genetic locus encoded by Yersinia pseudotuberculosis permits invasion of cultured animal cells by Escherichia coli K-12. Nature. 1985;317(6034):262–264. doi: 10.1038/317262a0. [DOI] [PubMed] [Google Scholar]
- 15.Knodler LA, Nair V, Steele-Mortimer O. Quantitative assessment of cytosolic Salmonella in epithelial cells. PLoS One. 2014;9(1):e84681. doi: 10.1371/journal.pone.0084681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meunier E, Broz P. Quantification of cytosolic vs. vacuolar salmonella in primary macrophages by differential permeabilization. J Vis Exp. 2015;(101):e52960. doi: 10.3791/52960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parsot C, Ménard R, Gounon P, Sansonetti PJ. Enhanced secretion through the Shigella flexneri Mxi-Spa translocon leads to assembly of extracellular proteins into macromolecular structures. Mol Microbiol. 1995;16(2):291–300. doi: 10.1111/j.1365-2958.1995.tb02301.x. [DOI] [PubMed] [Google Scholar]
- 18.Clerc P, Sansonetti PJ. Entry of Shigella flexneri into HeLa cells: Evidence for directed phagocytosis involving actin polymerization and myosin accumulation. Infect Immun. 1987;55(11):2681–2688. doi: 10.1128/iai.55.11.2681-2688.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cossart P, Sansonetti PJ. Bacterial invasion: The paradigms of enteroinvasive pathogens. Science. 2004;304(5668):242–248. doi: 10.1126/science.1090124. [DOI] [PubMed] [Google Scholar]
- 20.Viboud GI, Bliska JB. A bacterial type III secretion system inhibits actin polymerization to prevent pore formation in host cell membranes. EMBO J. 2001;20(19):5373–5382. doi: 10.1093/emboj/20.19.5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Logsdon LK, Mecsas J. Requirement of the Yersinia pseudotuberculosis effectors YopH and YopE in colonization and persistence in intestinal and lymph tissues. Infect Immun. 2003;71(8):4595–4607. doi: 10.1128/IAI.71.8.4595-4607.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwuan L, Adams W, Auerbuch V. Impact of host membrane pore formation by the Yersinia pseudotuberculosis type III secretion system on the macrophage innate immune response. Infect Immun. 2013;81(3):905–914. doi: 10.1128/IAI.01014-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pujol C, Bliska JB. The ability to replicate in macrophages is conserved between Yersinia pestis and Yersinia pseudotuberculosis. Infect Immun. 2003;71(10):5892–5899. doi: 10.1128/IAI.71.10.5892-5899.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Håkansson S, et al. The YopB protein of Yersinia pseudotuberculosis is essential for the translocation of Yop effector proteins across the target cell plasma membrane and displays a contact-dependent membrane disrupting activity. EMBO J. 1996;15(21):5812–5823. [PMC free article] [PubMed] [Google Scholar]
- 25.Diepold A, Armitage JP. Type III secretion systems: The bacterial flagellum and the injectisome. Philos Trans R Soc Lond B Biol Sci. 2015;370(1679):pii:20150020. doi: 10.1098/rstb.2015.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem. 2006;281(16):11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- 27.Miki T, Akiba K, Iguchi M, Danbara H, Okada N. The Chromobacterium violaceum type III effector CopE, a guanine nucleotide exchange factor for Rac1 and Cdc42, is involved in bacterial invasion of epithelial cells and pathogenesis. Mol Microbiol. 2011;80(5):1186–1203. doi: 10.1111/j.1365-2958.2011.07637.x. [DOI] [PubMed] [Google Scholar]
- 28.Miki T, Okada N, Shimada Y, Danbara H. Characterization of Salmonella pathogenicity island 1 type III secretion-dependent hemolytic activity in Salmonella enterica serovar Typhimurium. Microb Pathog. 2004;37(2):65–72. doi: 10.1016/j.micpath.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 29.Scherer CA, Cooper E, Miller SI. The Salmonella type III secretion translocon protein SspC is inserted into the epithelial cell plasma membrane upon infection. Mol Microbiol. 2000;37(5):1133–1145. doi: 10.1046/j.1365-2958.2000.02066.x. [DOI] [PubMed] [Google Scholar]
- 30.Hayward RD, Koronakis V. Direct nucleation and bundling of actin by the SipC protein of invasive Salmonella. EMBO J. 1999;18(18):4926–4934. doi: 10.1093/emboj/18.18.4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tran Van Nhieu G, Caron E, Hall A, Sansonetti PJ. IpaC induces actin polymerization and filopodia formation during Shigella entry into epithelial cells. EMBO J. 1999;18(12):3249–3262. doi: 10.1093/emboj/18.12.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar Y, Valdivia RH. Actin and intermediate filaments stabilize the Chlamydia trachomatis vacuole by forming dynamic structural scaffolds. Cell Host Microbe. 2008;4(2):159–169. doi: 10.1016/j.chom.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nichols CD, Casanova JE. Salmonella-directed recruitment of new membrane to invasion foci via the host exocyst complex. Curr Biol. 2010;20(14):1316–1320. doi: 10.1016/j.cub.2010.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuhlman TE, Cox EC. Site-specific chromosomal integration of large synthetic constructs. Nucleic Acids Res. 2010;38(6):e92. doi: 10.1093/nar/gkp1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolfgang MC, et al. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2003;100(14):8484–8489. doi: 10.1073/pnas.0832438100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cangelosi C, Hannagan S, Santiago CP, Wilson JW. Transfer of the cloned Salmonella SPI-1 type III secretion system and characterization of its expression mechanisms in Gram negative bacteria in comparison with cloned SPI-2. Microbiol Res. 2015;180:57–64. doi: 10.1016/j.micres.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 38.Bronstein PA, Miao EA, Miller SI. InvB is a type III secretion chaperone specific for SspA. J Bacteriol. 2000;182(23):6638–6644. doi: 10.1128/jb.182.23.6638-6644.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edwards RA, Keller LH, Schifferli DM. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene. 1998;207(2):149–157. doi: 10.1016/s0378-1119(97)00619-7. [DOI] [PubMed] [Google Scholar]
- 40.Coombes BK, Brown NF, Kujat-Choy S, Vallance BA, Finlay BB. SseA is required for translocation of Salmonella pathogenicity island-2 effectors into host cells. Microbes Infect. 2003;5(7):561–570. doi: 10.1016/s1286-4579(03)00094-7. [DOI] [PubMed] [Google Scholar]
- 41.Schmitz AM, Morrison MF, Agunwamba AO, Nibert ML, Lesser CF. Protein interaction platforms: Visualization of interacting proteins in yeast. Nat Methods. 2009;6(7):500–502. doi: 10.1038/nmeth.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davis AJ, Mecsas J. Mutations in the Yersinia pseudotuberculosis type III secretion system needle protein, YscF, that specifically abrogate effector translocation into host cells. J Bacteriol. 2007;189(1):83–97. doi: 10.1128/JB.01396-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ibarra JA, et al. Induction of Salmonella pathogenicity island 1 under different growth conditions can affect Salmonella-host cell interactions in vitro. Microbiology. 2010;156(Pt 4):1120–1133. doi: 10.1099/mic.0.032896-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slagowski NL, Kramer RW, Morrison MF, LaBaer J, Lesser CF. A functional genomic yeast screen to identify pathogenic bacterial proteins. PLoS Pathog. 2008;4(1):e9. doi: 10.1371/journal.ppat.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Labrec EH, Schneider H, Magnani TJ, Formal SB. Epithelial cell penetration as an essential step in the pathogenesis of bacillary dysentery. J Bacteriol. 1964;88(5):1503–1518. doi: 10.1128/jb.88.5.1503-1518.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoiseth SK, Stocker BA. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291(5812):238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 47.Miki T, et al. Chromobacterium pathogenicity island 1 type III secretion system is a major virulence determinant for Chromobacterium violaceum-induced cell death in hepatocytes. Mol Microbiol. 2010;77(4):855–872. doi: 10.1111/j.1365-2958.2010.07248.x. [DOI] [PubMed] [Google Scholar]