Significance
Alzheimer’s disease, the most common cause of dementia, has been associated with a complex transcriptional response. To define the nature of this response, we carried out a comprehensive analysis that reveals a set of differentially expressed genes encoding proteins at risk for aggregation. These results identify a transcriptional signature of Alzheimer’s disease consisting of down-regulated genes corresponding to a highly expressed “metastable subproteome” prone to aggregation. Our analysis of this metastable subproteome singles out a small number of biochemical pathways enriched in proteins that are simultaneously supersaturated in control brains and encoded by genes down-regulated in Alzheimer’s disease.
Keywords: neurodegenerative diseases, amyloid formation, protein misfolding, protein aggregation, protein supersaturation
Abstract
It is well-established that widespread transcriptional changes accompany the onset and progression of Alzheimer’s disease. Because of the multifactorial nature of this neurodegenerative disorder and its complex relationship with aging, however, it remains unclear whether such changes are the result of nonspecific dysregulation and multisystem failure or instead are part of a coordinated response to cellular dysfunction. To address this problem in a systematic manner, we performed a meta-analysis of about 1,600 microarrays from human central nervous system tissues to identify transcriptional changes upon aging and as a result of Alzheimer’s disease. Our strategy to discover a transcriptional signature of Alzheimer’s disease revealed a set of down-regulated genes that encode proteins metastable to aggregation. Using this approach, we identified a small number of biochemical pathways, notably oxidative phosphorylation, enriched in proteins vulnerable to aggregation in control brains and encoded by genes down-regulated in Alzheimer’s disease. These results suggest that the down-regulation of a metastable subproteome may help mitigate aberrant protein aggregation when protein homeostasis becomes compromised in Alzheimer’s disease.
Alzheimer’s disease (AD) is a neurodegenerative condition responsible for the majority of reported cases of dementia, affecting over 44 million people worldwide (1–6). Although the exact nature of this disease has not been defined fully, its onset and progression have been associated with a multitude of factors, including mitochondrial dysfunction, disruption of the endoplasmic reticulum and membrane trafficking, disturbances in protein folding and clearance, and the activation of the inflammatory response (1–6). More generally, however, it is clear that AD belongs to a class of protein conformational disorders whose characteristic feature is that specific peptides and proteins misfold and aggregate to form amyloid assemblies (1, 3, 6). The presence of such aberrant aggregate species generates a cascade of pathological events, leading to the loss of the ability of protein homeostasis mechanisms to preserve normal biological function and to avoid the formation of toxic species (1, 3, 6).
The appearance of protein aggregates in living systems is increasingly recognized as being common, as growing evidence indicates that proteins are only marginally stable against aggregation in their native states (1, 7) and that the molecular processes that prevent protein aggregation decline with aging (8–12). Thus, protein aggregation is emerging as a widespread biological phenomenon, in which hundreds of different proteins can aggregate in aging, stress, or disease (9, 13–23). To understand why some proteins aggregate whereas others remain soluble, we recently observed that many proteins in the proteome are insufficiently soluble relative to their expression levels (24). Such proteins are metastable to aggregation as their concentrations exceed their solubilities, that is, they are supersaturated (24–27). Upon formation of aggregate seeds by nucleation events, a supersaturated protein will form insoluble deposits until the concentration of its soluble fraction is reduced to match its solubility (24–28). We found that the proteins that coaggregate with inclusion bodies, those that aggregate in aging, and those in the major biochemical pathways associated with neurodegenerative diseases tend to be supersaturated (24). The observation that these metastable proteins appear to be a common feature in aging, stress, and disease prompts the question of whether or not their supersaturation levels are altered in AD. These levels are particularly crucial, as supersaturation represents a major driving force for aggregation (25). It is thus interesting to ask whether the down-regulation of supersaturated proteins may limit their aggregation in response to compromised protein homeostasis.
In the present study, we examined the experimental information acquired in the last decade about transcriptional changes in AD (29–43). We aimed specifically to determine the relationship between protein supersaturation and the transcriptional changes that occur during normal aging and in AD. We found that distinct but partially overlapping transcriptional changes take place in aging and AD. Moreover, down-regulated genes generally correspond to metastable proteins at risk for aggregation, as they are supersaturated and encoded by highly expressed genes. Accordingly, the biochemical pathways down-regulated in AD are nearly identical to those previously identified as highly enriched in supersaturated proteins (24). These changes are also accompanied by a transcriptional down-regulation of certain components of the protein homeostasis network. The down-regulation of genes corresponding to supersaturated proteins may thus represent a specific mechanism to limit widespread aggregation by regulating cellular concentrations in a compromised protein-folding environment.
Results
Analysis of the Transcriptional Changes in Aging and AD.
A long-standing question is whether AD represents an acceleration of the normal aging process or a qualitatively distinct phenomenon. Determining changes in gene expression can offer important insights into this problem. The complications associated with obtaining human tissue samples, however, constrain the extent to which confounding variables such as age, gender, and tissue type can be controlled in a transcriptional analysis of AD. In the present work, the control samples (mean 70.8 ± 16.4 y) are younger than the disease samples (mean 81.1 ± 9.5 y), necessitating the use of techniques to account for these disparities (SI Materials and Methods and Table S1).
Table S1.
List of the studies used for the microarray meta-analyses carried out in this work for Alzheimer's disease and clinical depression
| Series | Platform | Refs. | Control samples | Disease samples | Disease set |
| GSE1297 | GPL96 | (30) | 74 | 87 | AD |
| GSE5281 | GPL96 | (32, 33) | 9 | 22 | AD |
| GSE15222 | GPL2700 | (34) | 187 | 174 | AD |
| GSE26927 | GPL6255 | (38) | 7 | 11 | AD |
| GSE29378 | GPL6947 | (41) | 32 | 31 | AD |
| GSE29652 | GPL570 | (36) | 6 | 12 | AD |
| GSE36980 | GPL6244 | (40) | 47 | 32 | AD |
| GSE37263 | GPL5175 | (35) | 8 | 8 | AD |
| GSE44772 | GPL4272 | (42) | 299 | 388 | AD |
| GSE12654 | GPL8300 | (63) | 15 | 11 | CD |
| GSE53987 | GPL570 | (61) | 55 | 50 | CD |
| GSE54562, GSE54563, GSE54564 | GPL6947 | (60) | 56 | 56 | CD |
| GSE54565, GSE54566 | GPL570 | (60) | 29 | 30 | CD |
| GSE54567, GSE54568, GSE54571, GSE54572 | GPL570 | (60) | 54 | 54 | CD |
| GSE24095 | GPL10907 | (64) | 30 | 30 | CD |
In rows that include multiple series, we pooled different series and treated them as one for analysis.
For the human genes examined in our analysis, we constructed a linear model of expression differences across a range of factors (SI Materials and Methods). We thus obtained the overall median magnitude and statistical significance of expression changes by combining these individual values across different studies. In this analysis, microarray probes were mapped onto UniProt IDs to determine the corresponding protein (SI Materials and Methods). Using this procedure, we determined the transcriptional changes associated with 19,254 genes. An important aspect of this approach is that the effects on gene expression of different factors are considered as additive. Because the occurrence of AD increases with age, Alzheimer’s subjects exhibit specific disease-related transcriptional changes in addition to those associated with natural aging. We considered a gene to be differentially expressed if it undergoes a change in expression of at least 10% with a Benjamini–Hochberg–corrected P value ≤0.01. We then tested over 18,000 other combinations of thresholds and found our results to be robust to changes in these thresholds (Figs. S1 and S2). In the model used here the aging component is a linear variable, and therefore estimating the magnitude of change requires specifying a range of ages. Because the assumption of linearity is expected to hold best near the average age, we used the change in expression for an age range of approximately two SDs, namely 25 y.
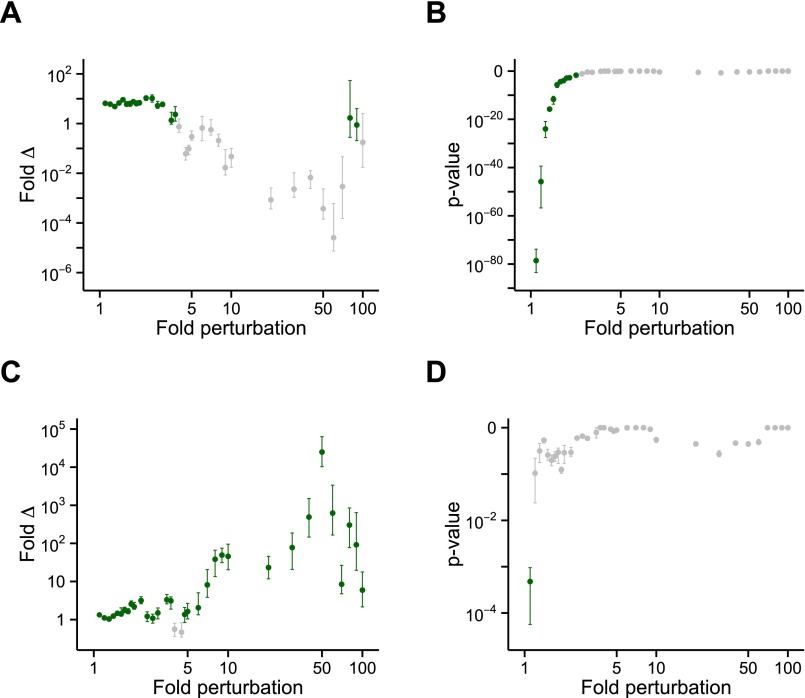
Fig. S1.
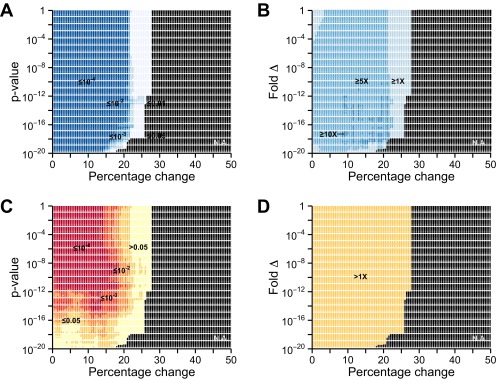
Differences in metastability between transcriptionally regulated proteins in aging are robust against changes in differential expression thresholds. A range of values for thresholds of minimum percentage change (0.5–50%) and P value (10−20 to 1) was used to determine which genes are increased, decreased, or unchanged in expression upon aging. A total of 18,100 combinations were considered. Supersaturation scores were then calculated for the proteins corresponding to differentially expressed genes. The corresponding protein supersaturation was assessed in terms of (A and C) P value and (B and D) median fold difference. This analysis was performed for down-regulated (A and B) and up-regulated (C and D) genes.
Fig. S2.
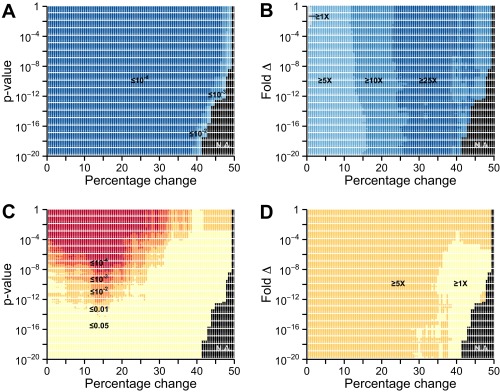
Differences in metastability between transcriptionally regulated proteins in AD are robust against changes in differential expression thresholds. A range of values for thresholds of minimum percentage change (0.5–50%) and P value (10−20 to 1) was used to determine which genes are increased, decreased, or unchanged in expression in AD. A total of 18,100 combinations were considered. Supersaturation scores were then calculated for the proteins corresponding to differentially expressed genes. The corresponding protein supersaturation was assessed in terms of (A and C) P value and (B and D) median fold difference. This analysis was performed for down-regulated (A and B) and up-regulated (C and D) genes.
Proteins That Aggregate in AD Correspond to Transcriptionally Down-Regulated Genes.
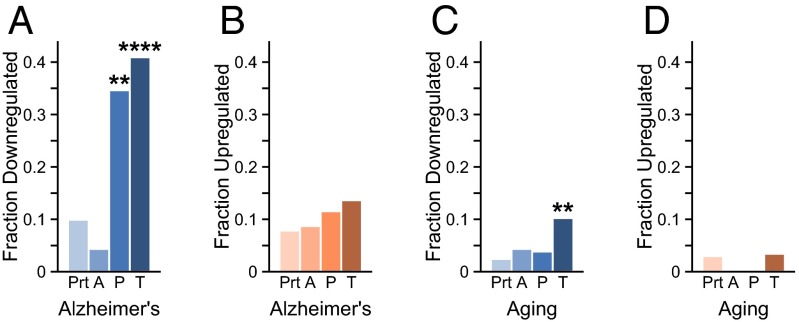
We next asked how the transcriptional changes identified in aging and AD might be associated with protein aggregation. First, we considered the set of disease-related amyloid proteins, that is, those annotated as “amyloid” in UniProt, which include those associated with neurodegenerative diseases (24). On average, we could not detect an overall connection between amyloid proteins and proteins corresponding to differentially expressed genes (Fig. 1 A and B). We also note, however, that this analysis does not imply that individual genes in the amyloid class may not have important roles in AD. As an example, the down-regulation of the APP gene (in our analysis by 9.5%, with P =0.011) has been reported in neurons containing neurofibrillary tangles (44).
Fig. 1.
Proteins that aggregate in AD correspond to transcriptionally down-regulated genes. (A and B) Fraction of proteins corresponding to transcriptionally down-regulated (A) or up-regulated (B) genes in AD in the whole proteome (Prt; down-regulated fraction 1,907/19,254; up-regulated fraction 1,509/19,254) and for amyloid deposits (A; 1/23; 2/23), plaques (P; 9/26; 3/26), and tangles (T; 36/88; 9/88). (C and D) Fraction of proteins corresponding to transcriptionally down-regulated (C) or up-regulated (D) genes in aging in the whole proteome (432/17,833; 534/17,833), and for amyloid deposits (1/23; 0/23), plaques (1/26; 0/26), and tangles (9/88; 3/88). The statistical significance of the difference with the proteome (first column) was assessed with a Fisher’s exact test with Holm–Bonferroni corrections (**P < 0.001, ****P < 0.0001).
We identified, however, a clear signal for another set of proteins associated with AD, namely those that coaggregate with amyloid plaques (13) and neurofibrillary tangles (14) in human autopsy samples as identified by mass spectrometry. Among the proteins that coaggregate with plaques (35%, P = 4.7⋅10−3) and tangles (41%, P = 1.7⋅10−13), a disproportionate number correspond to down-regulated genes in AD (Fig. 1A) in addition to those that are down-regulated during natural aging (Fig. 1C). Proteins corresponding to genes down-regulated in aging are overrepresented among tangle coaggregators (10%, P = 2.5⋅10−3) but not plaque coaggregators (4%, P = 1.0) (Fig. 1C). By contrast, only an insignificant number of genes encoding proteins aggregating in plaques and tangles were observed to be up-regulated in AD (Fig. 1B) or aging (Fig. 1D).
Metastable Proteins Correspond to Transcriptionally Down-Regulated Genes in Aging and AD.
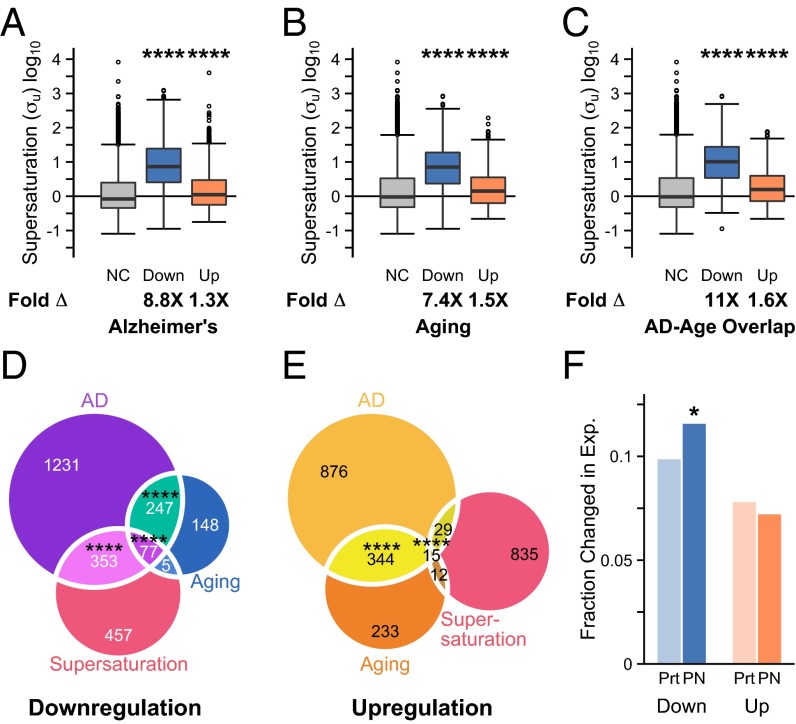
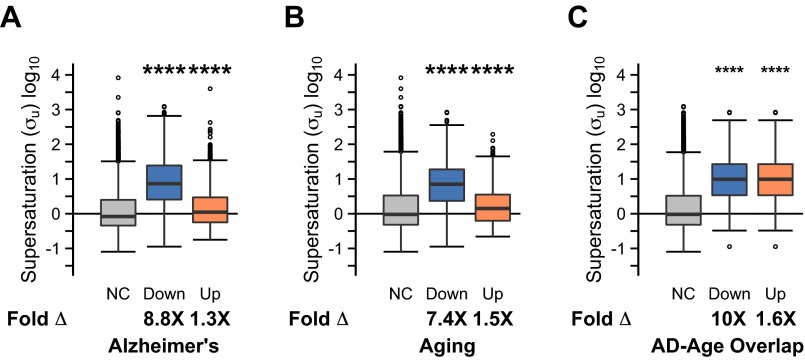
We next investigated whether the fact that so many proteins that coaggregate with plaques and tangles correspond to genes down-regulated in AD could be a consequence of their metastability to aggregation. We previously observed that these metastable proteins tend to be supersaturated, having concentrations exceeding their solubility limits (24). Here we calculated the metastability of proteins to aggregation in terms of supersaturation scores (σu), which represent the risk of proteins aggregating from their unfolded states (24). We assessed proteins corresponding to genes down-regulated in AD to be about 8.8-fold (8.8×, P < 2.2⋅10−16) more metastable than those for which the expression levels of the corresponding genes do not change significantly in disease (Fig. 2A). Similarly, we found proteins encoded by genes down-regulated in aging to be more metastable (7.4×, P < 2.2⋅10−16) than those whose expression does not change (Fig. 2B).
Fig. 2.
Transcriptionally regulated genes in aging and AD correspond to proteins metastable against aggregation. (A–C) Assessment of the metastability to aggregation of the proteins associated with differentially expressed genes in (A) AD, (B) aging, and (C) the overlap between the two groups. The median fold difference in supersaturation (which is a measure of metastability to aggregation) is indicated by Fold Δ. NC, Down, and Up indicate, respectively, no change in expression, down-regulation, and up-regulation. Whiskers range from the lowest to highest value data points within 150% of the interquartile ranges. (D and E) Overlap between the 5% most supersaturated proteins and the corresponding genes either (D) down-regulated or (E) up-regulated in aging and AD. The number of proteins in each subset is indicated. (F) Fraction of genes down-regulated (blue) and up-regulated (orange) in the whole proteome (down-regulated fraction 1,907/19,254; up-regulated fraction 1,509/19,254) and the protein homeostasis network (PN; 1,509/19,254; 148/2,041). For A–C, ****P ≤ 0.0001, one-sided Wilcoxon/Mann–Whitney test with Holm–Bonferroni correction. For D and E, *P ≤ 0.05, ****P ≤ 0.0001, one-sided Fisher’s exact test with Holm–Bonferroni correction.
We also found that proteins corresponding to genes up-regulated in AD (1.3×, P = 9.7⋅10−13) (Fig. 2A) and in aging (1.5×, P < 8.8⋅10−7) (Fig. 2B) are modestly, but significantly, more metastable than those with unchanged expression in AD. These up-regulated genes are almost exclusively associated with an inflammatory response (Dataset S1). For example, of those genes that encode metastable proteins, the most highly up-regulated gene (123% increase in expression) in AD is alpha-1 antichymotrypsin, which inhibits serine proteases, particularly those active in inflammation (45).
Despite the fact that only 16% of down-regulated genes are common to aging and AD (Fig. 2D), in both cases the transcriptional response appears to be associated with metastability to aggregation (Fig. 2 A–C). Indeed, we observed a significant overlap (P < 2.2⋅10−16) between the most metastable proteins (≥95th percentile), proteins corresponding to genes down-regulated in AD, and proteins corresponding to genes down-regulated in aging, as well as between any two of these categories (Fig. 2D). The proteins that are supersaturated proteins and encoded by genes down-regulated in AD make up a metastable subproteome specific to AD (Dataset S1), which is here referred to as the “metastable subproteome.” By contrast, the most transcriptionally up-regulated genes in AD and in aging overlap significantly with each other, but neither group is significantly enriched in genes encoding metastable proteins (Fig. 2E). As a control, we divided the down-regulated and up-regulated genes into low, medium, and high levels and calculated the supersaturation scores at each of these levels (Fig. 3). Our results indicated a trend toward increasing levels of supersaturation with increasing levels of down-regulation in AD (Fig. 3A). This correlation is weaker in aging (Fig. 3C), and weaker still among up-regulated genes (Fig. 3 B and D). The negative correlation between protein supersaturation and gene down-regulation also persists at the individual level for AD, but much less so for aging (Fig. S3).
Fig. 3.
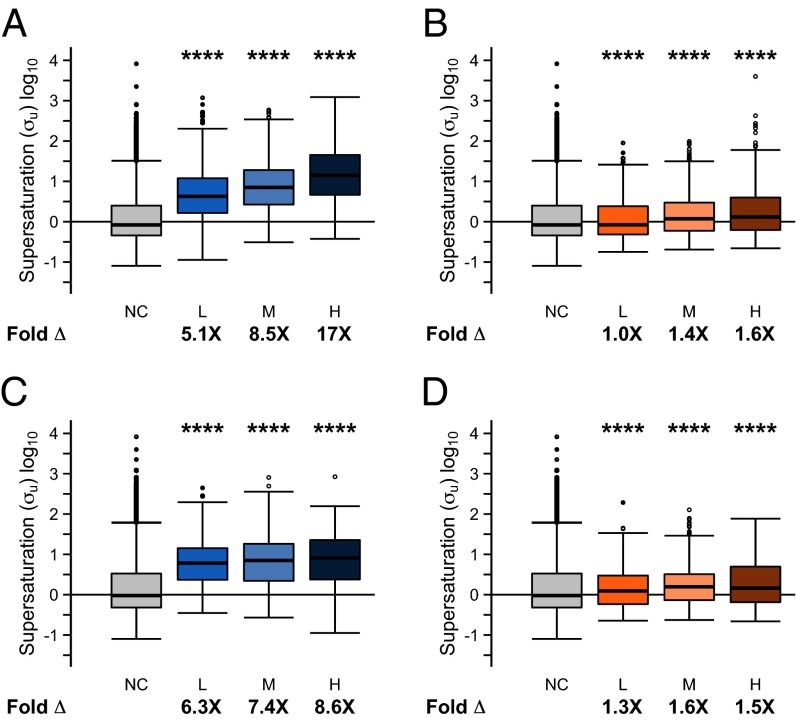
Metastability of proteins to aggregation is correlated with the down-regulation of the corresponding genes in AD. Metastability levels, assessed by supersaturation scores, for proteins associated with differentially expressed genes: (A) down-regulated in AD, (B) up-regulated in AD, (C) down-regulated in aging, and (D) up-regulated in aging. Differentially expressed genes are divided into thirds (low, L; medium, M; high, H) based on the fold change of expression. The median fold difference in supersaturation is indicated by Fold Δ. NC indicates no change in expression. ****P ≤ 0.0001, one-sided Wilcoxon/Mann–Whitney test with Holm–Bonferroni correction. Whiskers range from the lowest to highest value data points within 150% of the interquartile ranges.
Fig. S3.
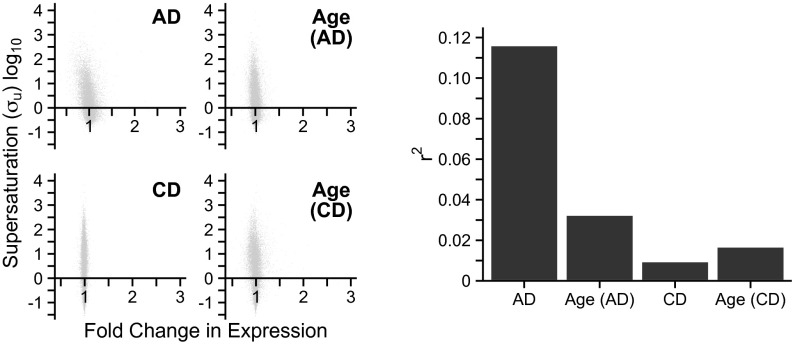
Metastability levels are correlated with average expression levels for genes down-regulated in AD. (Left) Plot of protein supersaturation scores against the fold change in expression for the corresponding genes in AD (AD, Upper Left), aging based on the AD studies [Age (AD), Upper Right], clinical depression (CD, Lower Left), and aging based on the clinical depression studies [Age (CD), Lower Right]. (Right) Pearson’s correlation coefficient (r2) for the categories plotted (Left).
Elevated supersaturation scores of differentially expressed genes may result from an easier detection of the differences in highly expressed genes than in genes of low expression. To control for this possibility, however, we excluded low-expression genes from our analysis, finding the median supersaturation of proteins corresponding to differentially expressed genes to be elevated even after this procedure (Fig. S4). We also tested the robustness of our results against changes in the details of our analysis. We found that our results on the metastability of the proteins corresponding to differentially expressed genes are stable across a wide range of thresholds for defining the groups of up-regulated and down-regulated genes (Figs. S1 and S2), and also against the introduction of Gaussian noise into the supersaturation score (Figs. S5 and S6).
Fig. S4.
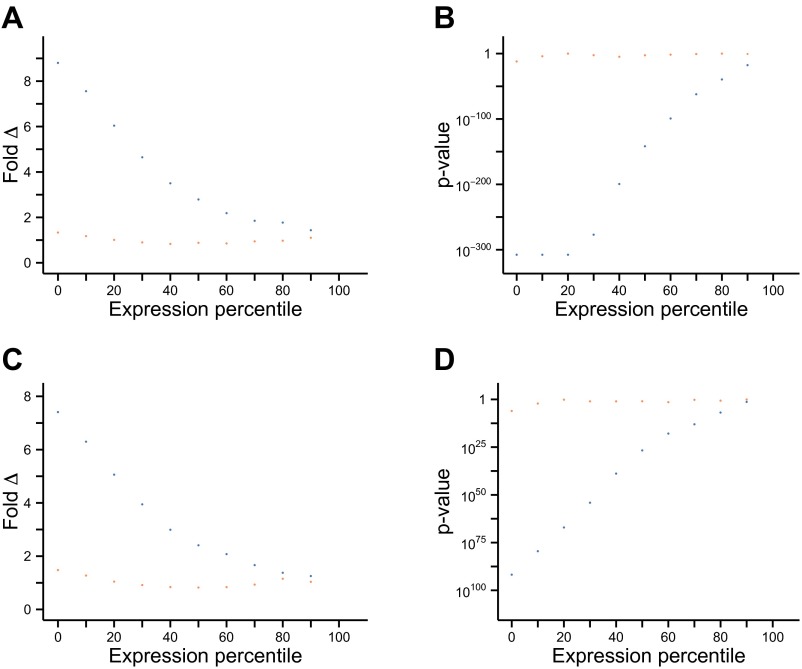
Metastability of proteins encoded by differentially expressed genes is elevated in AD and aging for a range of expression values. Supersaturation of proteins associated with downregulated (A and B) and upregulated (C and D) genes in AD (circles) and aging (triangles) was determined after restricting the genes of interest to those above a range of expression levels plotted by expression percentile rank. (A and B) Fold Δ and (C and D) P value are plotted. Orange points represent values for down-regulated genes; blue points represent values for up-regulated genes. The median fold difference in supersaturation is indicated by Fold Δ. P values are calculated using the one-sided Wilcoxon/Mann–Whitney test with Holm–Bonferroni correction.
Fig. S5.
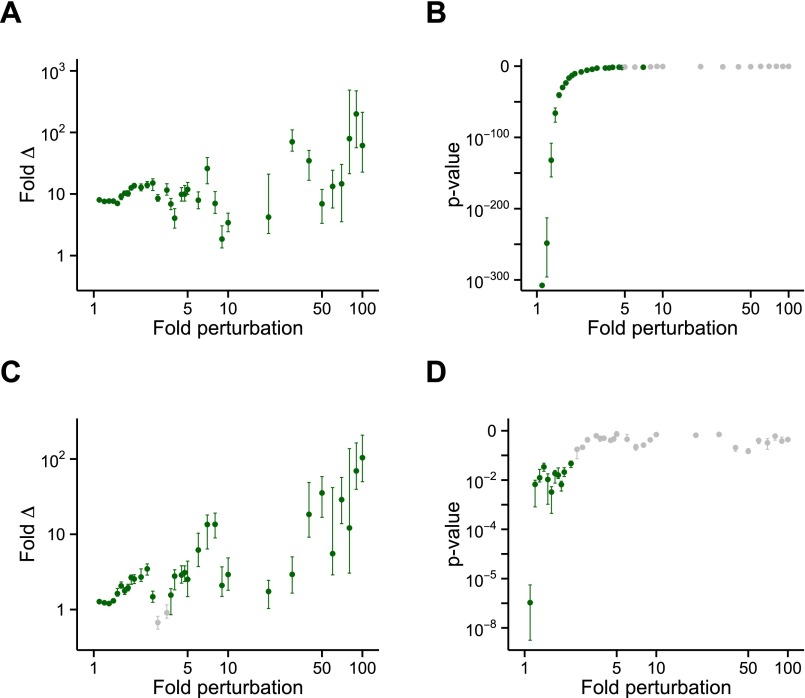
Differences in metastability between transcriptionally regulated proteins in AD are robust against Gaussian noise in the supersaturation score. Test of the robustness of the significance of the (A and C) median fold difference and (B and D) P value of supersaturation for proteins transcriptionally (A and B) down-regulated or (C and D) up-regulated in AD. Gaussian noise was introduced 100 independent times into the proteome scores at levels ranging from 1.1× to 100× (where 1× signifies no noise). Tests were performed at each noise level to determine whether the 100 median fold differences obtained were significantly greater than 1 and the 100 P values obtained were significantly below 0.05. For down-regulated genes, supersaturation (A) median fold difference is robust up to 100× and (B) P value is robust up to 7×. For up-regulated genes, supersaturation (C) median fold difference is robust up to 100× and (D) P value is robust up to 2.25×. Error bars indicate interquartile ranges; green points indicate P ≤ 0.05 by the one-sided Wilcoxon/Mann–Whitney test.
Fig. S6.
Differences in metastability between transcriptionally regulated proteins in aging are robust against Gaussian noise in the supersaturation score. Test of the robustness of the significance of the (A and C) median fold difference and (B and D) P value of supersaturation for proteins transcriptionally (A and B) down-regulated or (C and D) up-regulated in aging (AD dataset). Gaussian noise was introduced 100 independent times into the proteome scores at levels ranging from 1.1× to 100× (where 1× signifies no noise). Tests were performed at each noise level to determine whether the 100 median fold differences obtained were significantly greater than 1 and the 100 P values obtained were significantly below 0.05. For down-regulated genes, supersaturation (A) median fold difference is robust up to 3.75× and (B) P value is robust up to 2.25×. For up-regulated genes, supersaturation (C) median fold difference is robust up to 100× and (D) P value is robust up to 1.1×. Error bars indicate interquartile ranges; green points indicate P ≤ 0.05 by the one-sided Wilcoxon/Mann–Whitney test.
Specific Protein Homeostasis Components Correspond to Genes Down-Regulated in AD.
As we have discussed above, widespread down-regulation of genes corresponding to metastable proteins may represent a general mechanism to maintain protein homeostasis upon aging and AD. An additional transcriptional response, however, may also involve specific components of the protein homeostasis network (8). Following a recent study that showed an enrichment in genes down-regulated in aging in this network (8), we examined whether or not particular subnetworks in the overall protein homeostasis network correspond to genes particularly down-regulated in aging and AD (Fig. 2F). We found a significant number of protein homeostasis network genes in the “trafficking” subnetwork to be down-regulated in AD (14%, P = 1.1⋅10−2).
We then investigated whether or not the cell is endowed with transcriptional mechanisms to regulate the solubility burden in register with the protein homeostasis capacity. If so, there may be transcriptional regulators that coordinate such a response by modulating protein homeostasis. To determine in particular whether specific transcription factors and histone modifiers are up-regulated or down-regulated in AD and aging, we generated a map of transcriptional regulators and their targets using Encyclopedia of DNA Elements (ENCODE) regulator binding site data (46). Here we considered a gene to be regulated by a particular transcription factor or histone modifier if the regulator has a binding site at least half of which is within 1,000 bp of the start codon of the gene itself. We identified 23 transcription factors and histone modifiers associated with a significant number of genes down-regulated in AD (Dataset S2), including EGR1 (47), NRF1 (48), and REST (49). By contrast, we found only one regulator associated with a significant number of genes down-regulated in aging, the histone modifier EZH2 (Dataset S3). In addition, four regulators were found to be associated with a significant number of genes up-regulated in AD, and none was found to be associated with a significant number of genes up-regulated in aging (Datasets S2 and S3).
Biochemical Pathways Enriched in Metastable Proteins Are Also Enriched in Proteins Corresponding to Genes Down-Regulated in AD.
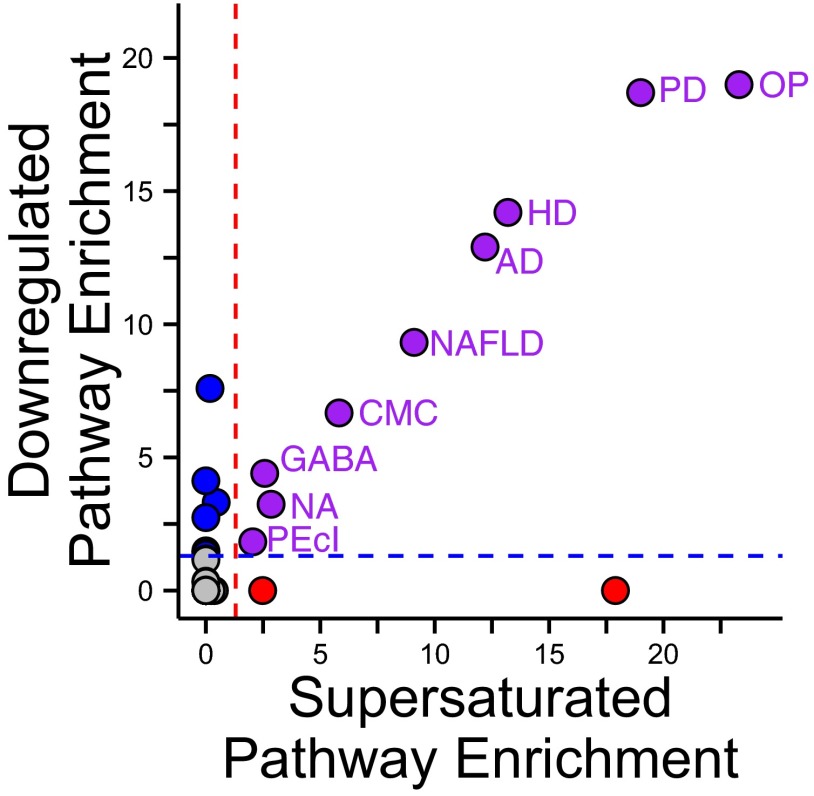
To determine the functional implications of the transcriptional regulation of metastable proteins in AD, we conducted an unbiased search of the entire set of 284 pathways in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (50), a repository of biochemical pathways and protein networks. We found a close correspondence between the pathways down-regulated in AD and pathways that we previously found to be supersaturated based on independent data (24, 25) (Fig. 4 and Table S2). Remarkably, most of these KEGG pathways fall along a band in which increasing metastability levels correspond to increasing down-regulation (Fig. 4, purple circles). The overlap between metastable and down-regulated pathways is highly significant (P = 8.7⋅10−11). Among the simultaneously metastable and down-regulated KEGG pathways, we found oxidative phosphorylation (OP), Parkinson’s disease (PD), Huntington’s disease (HD), Alzheimer’s disease, nonalcoholic fatty liver disease (NAFLD), cardiac muscle contraction (CMC), nicotine addiction (NA), GABAergic synapse (GABA), and pathogenic Escherichia coli infection (PEcI). These results reveals pathological (AD, PD, HD, and NAFLD) and functional (OP, CMC, and PEcI) networks and pathways enriched in physiological complexes, as well as pathways involved in neuronal signaling (NA and GABA). In particular, our analysis identified certain proteins in the oxidative phosphorylation pathway as being particularly metastable, including all of the components of the mitochondrial ATP synthase complex for which we have data, consistent with the reported involvement of this complex in AD (51). In addition, 43% of the genes in our analysis that encode for mitochondrial ATP synthase complex are transcriptionally repressed. The most repressed is the alpha subunit of the F1 catalytic core (whose expression is reduced by 26% in AD), which has been observed to accumulate in degenerating neurons in AD and to be associated with neurofibrillary tangles (52). We also verified that, although oxidative phosphorylation is central to the pathways down-regulated in AD, the signal for metastability in AD and aging is robust against the exclusion of proteins in this pathway from our analysis (Fig. S7).
Fig. 4.
Comparison between down-regulated and metastable biochemical pathways and networks. We found that the biochemical pathways and networks down-regulated in AD correspond closely to those enriched in supersaturated proteins (purple circles). Using the KEGG classification, these biochemical pathways and networks are oxidative phosphorylation, Parkinson’s disease, Huntington’s disease, Alzheimer’s disease, nonalcoholic fatty liver disease, cardiac muscle contraction, nicotine addiction, GABAergic synapse, and pathogenic E. coli infection.
Table S2.
KEGG pathways in which transcriptionally regulated genes are overrepresented in Alzheimer’s disease, clinical depression, or aging
| KEGG pathway | AD | Age[AD] | CD (P ≤ 0.01) | Age[CD] (P ≤ 0.01) | CD (P ≤ 0.05) | Age[CD] (P ≤ 0.05) |
| Down-regulated | ||||||
| Staphylococcus aureus infection | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Complement and coagulation cascades | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Toxoplasmosis | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Mineral absorption | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Legionellosis | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Proteoglycans in cancer | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Malaria | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Pertussis | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Allograft rejection | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Alzheimer’s disease | 1.39E-13 | N.S. | N.S. | N.S. | N.S. | 9.07E-06 |
| Antigen processing and presentation | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Asthma | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Axon guidance | N.S. | 1.14E-02 | N.S. | N.S. | N.S. | N.S. |
| Cardiac muscle contraction | 2.16E-07 | N.S. | N.S. | N.S. | N.S. | N.S. |
| Citrate cycle (TCA cycle) | 1.84E-03 | N.S. | N.S. | N.S. | N.S. | N.S. |
| Cysteine and methionine metabolism | 3.59E-02 | N.S. | N.S. | N.S. | N.S. | N.S. |
| GABAergic synapse | 3.99E-05 | N.S. | N.S. | N.S. | N.S. | N.S. |
| Huntington’s disease | 6.00E-15 | N.S. | N.S. | N.S. | N.S. | 1.65E-03 |
| Leishmaniasis | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Morphine addiction | 3.26E-02 | N.S. | N.S. | N.S. | N.S. | N.S. |
| NOD-like receptor signaling pathway | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Nicotine addiction | 5.71E-04 | N.S. | N.S. | N.S. | N.S. | N.S. |
| Nonalcoholic fatty liver disease | 4.77E-10 | N.S. | N.S. | N.S. | N.S. | 1.59E-02 |
| Olfactory transduction | N.S. | N.S. | N.S. | N.S. | 4.52E-03 | N.S. |
| Oxidative phosphorylation | 9.67E-20 | N.S. | N.S. | N.S. | N.S. | 3.35E-07 |
| Parkinson’s disease | 2.12E-19 | N.S. | N.S. | N.S. | N.S. | 1.36E-06 |
| Pathogenic Escherichia coli infection | 1.50E-02 | N.S. | N.S. | N.S. | N.S. | N.S. |
| Phagosome | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Proteasome | 4.86E-04 | N.S. | N.S. | N.S. | N.S. | 1.57E-02 |
| Pyrimidine metabolism | N.S. | N.S. | N.S. | N.S. | N.S. | 1.19E-01 |
| Retrograde endocannabinoid signaling | 7.59E-05 | N.S. | N.S. | N.S. | N.S. | N.S. |
| Ribosome | N.S. | N.S. | N.S. | N.S. | N.S. | 2.10E-01 |
| Synaptic vesicle cycle | 2.59E-08 | N.S. | N.S. | N.S. | N.S. | N.S. |
| Systemic lupus erythematosus | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Vibrio cholerae infection | 4.33E-02 | N.S. | N.S. | N.S. | N.S. | N.S. |
| Up-regulated | ||||||
| S. aureus infection | 2.12E-06 | 4.69E-11 | N.S. | N.S. | N.S. | N.S. |
| Complement and coagulation cascades | 1.91E-04 | 3.75E-07 | N.S. | N.S. | N.S. | N.S. |
| Toxoplasmosis | 7.92E-04 | 6.00E-03 | N.S. | N.S. | N.S. | N.S. |
| Mineral absorption | 2.67E-03 | N.S. | N.S. | 2.40E-03 | N.S. | 4.94E-03 |
| Legionellosis | 7.73E-03 | 1.79E-02 | N.S. | N.S. | N.S. | N.S. |
| Proteoglycans in cancer | 2.74E-02 | N.S. | N.S. | N.S. | N.S. | N.S. |
| Malaria | 3.04E-02 | N.S. | N.S. | N.S. | N.S. | N.S. |
| Pertussis | 4.07E-02 | 4.39E-04 | N.S. | N.S. | N.S. | N.S. |
| Allograft rejection | N.S. | 2.74E-02 | N.S. | N.S. | N.S. | N.S. |
| Alzheimer’s disease | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Antigen processing and presentation | N.S. | 2.86E-03 | N.S. | N.S. | N.S. | N.S. |
| Asthma | N.S. | 4.54E-02 | N.S. | N.S. | N.S. | N.S. |
| Axon guidance | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Cardiac muscle contraction | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Citrate cycle (TCA cycle) | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Cysteine and methionine metabolism | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| GABAergic synapse | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Huntington’s disease | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Leishmaniasis | N.S. | 2.61E-04 | N.S. | N.S. | N.S. | N.S. |
| NOD-like receptor signaling pathway | N.S. | 2.46E-02 | N.S. | N.S. | N.S. | N.S. |
| Nicotine addiction | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Nonalcoholic fatty liver disease | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Olfactory transduction | N.S. | N.S. | N.S. | N.S. | 4.91E-02 | N.S. |
| Oxidative phosphorylation | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Parkinson’s disease | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Pathogenic E. coli infection | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Phagosome | N.S. | 1.31E-02 | N.S. | N.S. | N.S. | N.S. |
| Proteasome | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Pyrimidine metabolism | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Retrograde endocannabinoid signaling | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Ribosome | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Synaptic vesicle cycle | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Systemic lupus erythematosus | N.S. | 2.47E-05 | N.S. | N.S. | N.S. | N.S. |
| V. cholerae infection | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
In the case of aging, the pathways identified based on analyses of the Alzheimer’s disease (Age[AD]) dataset and the depression (Age[CD]) dataset are listed separately. For the depression and aging–depression sets, pathways enriched with expression significance cutoffs of P ≤ 0.01 and P ≤ 0.05 are both shown. Values shown are Holm–Bonferroni–corrected one-sided Fisher’s exact test P values, where the Holm–Bonferroni correction was applied to each dataset separately. Only P values ≤0.05 are listed. N.S., not significant; NOD, nucleotide oligomerization domain.
Fig. S7.
Elevated metastability of proteins encoded by differentially expressed genes in AD and aging is not dependent on oxidative phosphorylation proteins. Supersaturation of proteins associated with differentially expressed genes in (A) AD, (B) aging, and (C) the overlap between the two, but with those proteins found in the KEGG pathway for oxidative phosphorylation excluded. The median fold difference in supersaturation is indicated by Fold Δ. NC indicates genes that do not change significantly in expression. ****P ≤ 0.0001, one-sided Wilcoxon/Mann–Whitney test with Holm–Bonferroni correction. Whiskers range from the lowest to highest value data points within 150% of the interquartile ranges.
In this comprehensive analysis of KEGG pathways, we also found other pathways that are significantly enriched in either metastable proteins (Fig. 4, red circles) or in proteins corresponding to down-regulated genes (Fig. 4, blue circles), but not both (Table S2). However, the large majority of these pathways have significance values that are lower than the average jointly metastable and down-regulated pathway (Fig. 4, purple circles), the exceptions being the “ribosome,” which is highly metastable but not down-regulated, and the “synaptic vesicle cycle,” “proteasome,” and “retrograde endocannabinoid signaling,” which are down-regulated but not metastable. A similar analysis for up-regulated pathways in AD did not provide particularly significant results, although one may expect genes associated with the immune response to be up-regulated, as, for example, complement C1q subcomponent subunit C and plasma protease C1 inhibitor in the “complement and coagulation cascade” pathway.
Thus, the observation that in AD there is a highly specific down-regulation of metastable biochemical pathways and networks suggests the presence of a robust transcriptional response to protein aggregation in AD.
Widespread Down-Regulation of the Metastable Subproteome Is Not a General Feature of Disease.
Because the genes corresponding to the metastable subproteome are, on average, highly expressed, we considered the possibility that their widespread down-regulation could be a general feature of cellular dysfunction in disease. If this were the case, any process that disrupts normal cellular function could impair transcription, preferentially affecting those genes that are highly expressed. To investigate this possibility, we performed a meta-analysis of expression changes in another cognitive disorder, clinical depression. We considered 470 microarrays, including 239 from control patients and 231 from those with clinical depression (Table S1). As with our analysis of AD, we restricted our analysis to brain samples from cases in which the gender and age (for which we controlled) were known and Gene Expression Omnibus (GEO) database series that included at least 10 total cases. Among the 19,190 genes for which we evaluated changes in expression, we found 7 genes down-regulated and 11 genes up-regulated in clinical depression at the thresholds of 10% change in expression and P ≤ 0.01 (Dataset S4). Overall, we did not observe the same widespread transcriptional repression of the metastable subproteome found in AD, and we found no KEGG pathways significantly enriched in proteins corresponding to those genes differentially expressed in AD.
We then considered the possibility that we had only identified a small number of genes as being differentially expressed in clinical depression because of low statistical power. Although our meta-analysis of clinical depression included only 22% as many arrays as that of AD, this is unlikely to explain the fact that only 0.6% as many genes are differentially regulated in clinical depression. In addition, our separate analysis for aging provided a control to assess the statistical power of the clinical depression dataset relative to that for AD. At the thresholds of 10% change in expression and P ≤ 0.01, we found 196 genes down-regulated and 122 genes up-regulated in aging in the clinical depression dataset. This is 23% as many genes as we found differentially regulated in aging based on the AD dataset, consistent with the smaller number of microarrays in the clinical depression analysis. As a further control, we reanalyzed these data after relaxing the significance threshold for differential expression to P ≤ 0.05. At this threshold, we found 24 genes down-regulated and 17 genes up-regulated in clinical depression and 569 genes down-regulated and 291 genes up-regulated in aging (Dataset S4). At the relaxed threshold, the KEGG pathway for “olfactory transduction” was enriched in proteins corresponding both to genes down-regulated (P = 4.5⋅10−3) and genes up-regulated (P = 4.9⋅10−2) in clinical depression (Dataset S4). Only “mineral absorption” was enriched in proteins corresponding to genes up-regulated in aging in the clinical depression dataset (Table S2). We also assessed the overall relationship between metastability and transcriptional regulation, and found little correlation between the two.
SI Materials and Methods
Array Normalization.
We performed array normalization using the limma, affy, gcrma, and text itxps packages for the statistical programming language R (https://www.r-project.org). Affymetrix gene arrays read using the ReadAffy function in affy were normalized using the GC Robust Multiarray Average (GCRMA) using the gcrma function, which uses estimates of cross-hybridization based on the GC content of mismatch (MM) probes. It is a modification of the robust multiarray average (RMA) algorithm using the rma function in xps, which was used here to normalize Affymetrix exon arrays. GCRMA cannot be used on these arrays because they do not have the MM probes needed to estimate cross-hybridization. We read Illumina arrays using the read.ilmn function in LIMMA and background-corrected with the neqc function in LIMMA. Preprocessed arrays, if they have accompanying significance values, were filtered to remove those with a significance score <0.95 (where significance scores were available), and expression values were scaled to log10. We treated each two-color array that we encountered in the clinical depression dataset as two separate arrays, analyzing them using the backgroundCorrect, normalizeWithinArrays, and normalizeBetweenArrays (by the Aquantile method) functions. One other significant difference between our analysis of two-color arrays and one-color arrays is that when performing backgroundCorrect, we used the “minimum” method for two-color arrays whereas we used the normexp method for one-color arrays. The reason for the use of the minimum method here is to eliminate negative values, which are not compatible with the normalization we perform to correct for the fact that each channel in a given two-color array is on a single chip exposed to the same sample. For the clinical depression meta-analysis, we grouped certain series together, as indicated in Dataset S1.
Construction of the Linear Model.
We fitted a linear model to the expression of each gene that included the cofactors of tissue type, gender, age, and disease status in all cases, and subject ID and technical replication when these were relevant. If there was technical replication, we used the function duplicateCorrelation in limma to account for this replication. From LIMMA, we then used the function lmFit to generate the fit, and the function eBayes to generate statistical significance values.
Determination of Significance and Magnitude Values.
We obtained adjusted P values (i.e., q values) using the Benjamini–Hochberg method (56). In addition, we obtained the coefficients for the disease status and age cofactors to estimate the magnitude of the contribution of these parameters to gene expression. We converted Probe IDs to human-reviewed UniProt IDs, based on a mapping of those probes that unambiguously mapped to a single UniProt ID. If multiple probes mapped to a single UniProt ID, we used the median parameter P value and coefficient. Because aging is a continuous variable, the magnitude of the expression change attributable to aging is the product of the aging cofactor coefficient and some age range. In this study, we used an age range of 25 y for two reasons. First, this equals approximately two SDs of the age distribution, which is a reasonable range over which to assume linearity. Second, this value reflects an age range from about 63 y to about 88 y, a period over which the prevalence of AD increases dramatically.
For the clinical depression meta-analysis, the ages of the control samples were mean 50.3 ± 12.8 y and those of the disease samples were mean 49.4 ± 15.6 y. We used an age range of 25 y for these samples, as well, for consistency, and because this was also approximately two SDs of the age distribution in the clinical depression meta-analysis. As described below, varying the magnitude threshold significance of aging has the same effect as changing the age range used for our analysis, and the results are robust against such changes.
Combination of Significance and Magnitude.
There are several methods to combine significances across a series of studies. Here the question was whether or not the change in expression of a gene corresponding to a given protein is statistically significant. In general, we had 10 P values because we analyzed 10 microarray studies separately, although in some cases there were fewer than 10 P values because some genes are represented in some arrays but not others. The goal was to estimate the probability of obtaining a set of cofactor coefficients in each of the studies that are at least as extreme as those observed, assuming the null hypothesis that there is no change in gene expression is valid. One way to address this issue is to combine P values from various studies. Perhaps the most popular method to combine P values is Fisher’s method, which in essence yields a significant result if at least one of the studies can reject the null hypothesis (57). By contrast, Pearson’s method can be interpreted as assessing a result as insignificant if at least one of the studies fails to reject the null hypothesis (58). Stouffer et al.’s method is attractive because it is somewhat less sensitive to extreme values (59). In this method, P values are first converted to Z scores, which are standard normal variables. These Z scores can be combined to give a composite Z score, based on the property that the sum of k standard normal variables has mean 0 and variance p(k). This sum can then be converted unambiguously back into a P value. A common variant of this method was proposed by Liptak and involves weighting each individual Z score by the sample size of the study, an approach that has been shown to be superior by simulation. In the current analysis, we used Liptak’s method (50). This method requires that the P values be one-tailed and, although the P values obtained from the LIMMA functions lmFit and eBayes are two-tailed, they can be converted into one-tailed P values. To obtain a combined magnitude, we used the median of magnitudes per cofactor per gene.
Calculation of Basal Expression Levels for Supersaturation Scores.
We estimated metastability using our previously defined supersaturation scores, and estimated basal mRNA expression in control subjects. Because of normalization differences, it is challenging to obtain values for basal expressions by combining data from different studies, and so we selected a single study to obtain these levels. For AD, the study GSE44772 (Table S1) included the most control samples (299) but used the Rosetta/Merck Human 44k 1.1 microarray (42), in which expression values reported for each array are relative to the expression of a pooled background array, thus making between-gene comparisons impossible. The study GSE1297 (Table S1) used the Affymetrix Human Genome U133 array (30), which reports raw array expression values that we are then able to renormalize. This Affymetrix array is also the most commonly used array in the GEO database among those arrays represented in this analysis. The study GSE1297 also has a relatively large number of control samples (74), although this number is smaller than that available in the study GSE44772. However, given that the Affymetrix platform is more common, better-characterized, and amenable to renormalization within this analysis, we estimated basal expression levels from the control expression values in GSE1297. These values are the log2 average of all of the samples, as obtained from the LIMMA function lmFit. For clinical depression, the GSE54562/GSE54563/GSE54564 set of series included the most control samples (56; ref. 60), but GSE53987 had the advantage of deriving all 55 of its samples from the same series. It also used a similar platform to that for basal expression AD, Affymetrix Human Genome U133 Plus 2.0 Array (61).
We also note that the use of proteome-level mass spectrometry (9), when applied to brain tissues, could provide a quantitative way to measure supersaturation levels directly as the ratio between the actual soluble and total amounts of individual proteins observed in vivo.
Multiple Hypothesis Correction.
In addition to the multiple hypothesis correction described above, the following families of tests were corrected using the Holm–Bonferroni method (62): (i) overlap of aging and disease down-regulation with most supersaturated proteins; (ii) overlap of aging and disease up-regulation with most supersaturated proteins; (iii) enrichment of disease-related up-regulated and down-regulated proteins in disease-related amyloid proteins, plaque coaggregators, tangle coaggregators, the most supersaturated proteins [included in this family and family (i) above], and the proteostasis network; (iv) enrichment of age-related up-regulated and down-regulated proteins in disease-related amyloid proteins, plaque coaggregators, tangle coaggregators, the most supersaturated proteins, and the proteostasis network; (v) supersaturation scores of proteins up-regulated and down-regulated in disease; (vi) supersaturation scores of proteins up-regulated and down-regulated in aging; (vii) supersaturation scores of proteins up-regulated and down-regulated in disease divided into low, medium, and high categories; (viii) supersaturation scores of proteins up-regulated and down-regulated in aging divided into low, medium, and high categories; (ix) disease-related down-regulation of subcategories of the protein homeostasis network; and (x) overlap of KEGG pathways for up-regulation and down-regulation in aging and disease. KEGG pathway enrichment was corrected using the Holm–Bonferroni method (62), considering aging up-regulation, aging down-regulation, disease up-regulation, and disease down-regulation each as a separate family. Transcription factor target enrichment was corrected using the Benjamini–Hochberg method, considering aging up-regulation, aging down-regulation, disease up-regulation, and disease down-regulation each as a separate family. Analyses that excluded oxidative phosphorylation genes were considered as separate families. Analyses of clinical depression and AD were included in separate families.
KEGG Analysis.
KEGG analysis was performed by first assembling a database of the components of each KEGG pathway (50) from publicly available data. The KEGG gene identifiers were then converted to UniProt IDs to make it possible to compare them with the rest of our data. Enrichment was calculated using a one-sided Fisher’s exact test (57) and corrected using the Holm–Bonferroni method (62). Results for the most metastable proteins made use of previously published supersaturation scores, but two aspects of the current analysis of that data differed. First, the revised method of deriving KEGG pathways resulted in the analysis of 85 KEGG pathways not analyzed in the previous study. Second, the revised method used a one-sided Fisher’s exact test instead of the modified Expression Analysis Systematic Explorer (EASE, https://david.ncifcrf.gov/ease/ease.jsp) score to calculate significance. This resulted in some differences in the pathways identified as being enriched in metastable proteins.
Overlap Analysis.
The significance of the overlaps between aging, AD, and metastability were calculated using a one-sided Fisher’s exact test, corrected using the Holm–Bonferroni method. For the significance of the triple intersection, the P value was estimated as being less than or equal to the minimum P value of any double overlap.
Transcription Factor Analysis.
We used transcription factor binding site data from the ENCODE database (46) to identify transcription factors whose targets are enriched in the genes that we identified as being differentially expressed in aging and AD. ENCODE provides the genome address for binding sites for each transcription factor (46). We defined a gene as being regulated by a transcription factor if its binding site was less than 1,000 nt upstream of its start codon. Using this method, we generated a map of transcription factors and their targets. We converted the identifiers for the target genes to human-reviewed UniProt accession numbers and did the same for the UniProt IDs in our expression analysis. We then used a one-sided Fisher’s exact test to determine the significance of enrichment, correcting this P value using the Benjamini–Hochberg method (56).
Threshold Sensitivity Analysis.
To test the sensitivity of our results for aging and AD to variations in the threshold for differential expression, we varied the expression change threshold between 0.5% and 50% and the significance threshold between P = 10−20 and P = 1, for a total of 18,100 combinations of thresholds. At these thresholds, we determined which genes were up-regulated, down-regulated, or unchanged in expression. For those threshold levels at which there were at least five genes in each category, we then recalculated the median fold difference in supersaturation between the proteins encoded by up-regulated/down-regulated genes and those encoded by genes unchanged in expression, as well as the corresponding statistical significance. Because the aging results scale linearly with the age range selected, changing the magnitude threshold for aging has the same effect as varying the width of the age range used to calculate expression changes in aging.
Sensitivity to Gaussian Noise in the Supersaturation Score.
In a method similar to that which we previously described (24), we introduced random error into the supersaturation scores we calculated drawn from 34 increasingly wide Gaussian distributions with SD ranging from 1.1× error to 100× error. At each level, we performed 100 independent trials. At each level, we calculated the median fold difference in supersaturation and the corresponding significance, and then performed a one-sided Wilcoxon/Mann–Whitney test on these sets of median fold differences and P values to assess whether they were significantly greater than 1 or less than 0.05, respectively.
Discussion
A major area of investigation into the molecular origins of AD concerns the chemical and physical instability of the proteins associated with the disease, and the mechanisms by which the cell responds to such a situation. A number of studies have reported biophysical features, environmental conditions, and molecular partners that promote or repress the initial aggregation of specific proteins (1, 3, 7, 13–15). More recently, it has been recognized that the regulation of many other proteins is disrupted as a consequence of these initial aggregation events (8, 16–25). In a complementary approach, the origins of AD have been studied by analyzing the transcriptional response associated with its onset and progression (29–43). These studies have revealed that this transcriptional response involves genes corresponding to proteins that can cause the disease and those associated with the cellular processes engaged in combating it (29–43).
In the present study, we have brought together these two approaches, finding that the transcriptional changes that occur in AD can be rationalized, at least in part, on the basis of the presence of an AD-specific metastable subproteome at risk for aggregation (Fig. 2). This metastable subproteome is defined as the overlap between the proteins that are most supersaturated and that correspond to highly expressed genes, and those encoded by genes most transcriptionally down-regulated in AD (Fig. 2D). These proteins are intrinsically at risk for aggregation and, as we found here, tend to be the target of the transcriptional response in aging and AD. These results are consistent with previous observations that the expression of oxidative phosphorylation genes is suppressed in AD (53, 54), but suggest in addition that such suppression may be part of a broader response to the disease.
Having previously shown that the proteins associated with AD tend to be metastable to aggregation because they are supersaturated (24, 25), we have now reported a response to this intrinsic metastability of the proteome in the face of disruptions to protein homeostasis through the transcriptional down-regulation of their respective genes. The close correspondence of the biochemical pathways associated with metastability and those down-regulated in AD (Fig. 4) supports this conclusion, as do the tendency for proteins that coaggregate in plaques and tangles to correspond to down-regulated genes (Fig. 1) and the high overall metastability level of proteins encoded by down-regulated genes (Fig. 2). We found these results to be stable against a range of potentially confounding factors, including the choice of thresholds for differential expression (Figs. S1 and S2), noise in the supersaturation score (Figs. S5 and S6), and the large contribution of oxidative phosphorylation (Fig. S7).
Analysis of the transcriptional response to the collapse of protein homeostasis in terms of a metastable subproteome at risk for aggregation has also enabled us to address another central question about the progression of AD, namely the way in which changes occurring in this disease are related to the natural process of aging. These results indicate that aging and AD are very different at the transcriptional level, as over three-quarters of the transcriptional changes that occur in AD do not occur in aging (Fig. 2 D and E). In addition, many cellular processes down-regulated in AD are not significantly down-regulated in aging (Fig. S2). Although the differences between regulation in aging and AD are profound, there are important commonalities, as shown by the significant overlap in the specific transcriptional changes that occur in AD and in aging (Fig. 3). AD therefore appears to involve an acceleration in the decline of protein homeostasis associated with aging, and also an extension of its scope and significance. Overall, such an acceleration makes the metastable subproteome that we have identified in this work more susceptible to aggregation. This conclusion offers an explanation of why a transcriptional down-regulation of genes corresponding to metastable proteins is observed in both aging and AD.
We also observe that these phenomena are unlikely to be a general feature of cellular dysfunction. Our results indicate that a different transcriptional response is present in the case of clinical depression (Table S2 and Dataset S4), consistent also with results for epilepsy derived considering the differentially expressed genes in hippocampal samples from five patients with mesial temporal lobe epilepsy with hippocampal sclerosis (55). In that study, 518 genes were found to be differentially expressed between the subjects. Functional enrichment using the Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/) showed enrichment for KEGG pathways associated with neuroactive ligand receptor interaction, drug metabolism, and cytokine interaction, among others. The KEGG pathways of oxidative phosphorylation and of Alzheimer’s, Parkinson’s, and Huntington’s diseases were not, however, seen in epilepsy.
The findings that we have reported here, therefore, suggest that the widespread down-regulation of genes corresponding to metastable proteins at risk for aggregation may represent an important aspect of the strategy for cellular regulation in the face of disruptions in protein homeostasis. More generally, understanding the physicochemical implications of transcriptional regulation in aging, AD, and other protein-misfolding disorders has important implications both for a fundamental biological understanding of the origins of the disease and for clinical practice. Because the maintenance of protein homeostasis is an essential function in the cell, determining how the overall proteome composition is managed and modulated is a central question in biology. At the same time, understanding endogenous strategies for handling supersaturated, metastable, and potentially misfolding proteins may provide an avenue for improved therapies. If widespread aggregation is associated with AD, then determining how to regulate this phenomenon is of great value and practical importance.
Conclusions
We have shown that AD is associated with the transcriptional regulation of a metastable subproteome at risk for aggregation. The presence of these poorly soluble proteins in the cellular environment is inherently dangerous, in particular because these proteins tend to cluster into specific biochemical pathways, and only limited molecular chaperones and other protective resources are available at any given time to prevent their misfolding and aggregation. In conjunction with emerging insights into the molecular chaperone functions and the regulation of protein translation and degradation, our results indicate that the study of protein metastability may clarify how failures in maintaining proteins in their normal functional states could result in protein aggregation and in multifactorial disorders such as AD.
Despite the great complexity of aging processes and neurodegenerative disorders, protein solubility may underlie many aspects of their resultant cellular dysfunction. In this work, we have adopted this idea to investigate how the levels of poorly soluble proteins are regulated, finding that the overall transcriptional response to AD is associated with a global down-regulation of the expression of the genes encoding proteins that are metastable to aggregation. We anticipate that interventions that target the metastable subproteome at risk for aggregation that we have identified in this work may provide novel opportunities for the early diagnosis and treatment of AD.
Materials and Methods
The method of array normalization, construction of the linear model, and determination of significance and magnitude values are described in SI Materials and Methods. The calculation of basal expression levels for supersaturation scores and the sensitivity analysis are also described in SI Materials and Methods. The multiple hypothesis correction, KEGG analysis, and transcription factor analysis are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
P.C. was supported by grants from the US-UK Fulbright Commission, St. John’s College, University of Cambridge, and the National Institutes of Health (Northwestern University Medical Scientist Training Program Grant T32 GM8152-28). R.I.M. was supported by grants from the National Institutes of Health (National Institute of General Medical Sciences, National Institute on Aging, National Institute of Neurological Disorders and Stroke), the Ellison Medical Foundation, the Glenn Foundation, and the Daniel F. and Ada L. Rice Foundation. C.M.D. and M.V. were supported by the Wellcome Trust.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1516604113/-/DCSupplemental.
References
- 1.Knowles TP, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 2014;15(6):384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- 2.Querfurth HW, LaFerla FM. Alzheimer’s disease. N Engl J Med. 2010;362(4):329–344. doi: 10.1056/NEJMra0909142. [DOI] [PubMed] [Google Scholar]
- 3.Selkoe D, Mandelkow E, Holtzman D. Deciphering Alzheimer disease. Cold Spring Harb Perspect Med. 2012;2(1):a011460. doi: 10.1101/cshperspect.a011460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443(7113):780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 5.Glass CK, Saijo K, Winner B, Marchetto MC, Gage FH. Mechanisms underlying inflammation in neurodegeneration. Cell. 2010;140(6):918–934. doi: 10.1016/j.cell.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science. 2002;297(5580):353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 7.Tartaglia GG, Pechmann S, Dobson CM, Vendruscolo M. Life on the edge: A link between gene expression levels and aggregation rates of human proteins. Trends Biochem Sci. 2007;32(5):204–206. doi: 10.1016/j.tibs.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 8.Brehme M, et al. A chaperome subnetwork safeguards proteostasis in aging and neurodegenerative disease. Cell Reports. 2014;9(3):1135–1150. doi: 10.1016/j.celrep.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walther DM, et al. Widespread proteome remodeling and aggregation in aging C. elegans. Cell. 2015;161(4):919–932. doi: 10.1016/j.cell.2015.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci USA. 2009;106(35):14914–14919. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Labbadia J, Morimoto RI. The biology of proteostasis in aging and disease. Annu Rev Biochem. 2015;84:435–464. doi: 10.1146/annurev-biochem-060614-033955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Labbadia J, Morimoto RI. Repression of the heat shock response is a programmed event at the onset of reproduction. Mol Cell. 2015;59(4):639–650. doi: 10.1016/j.molcel.2015.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao L, et al. Proteomic characterization of postmortem amyloid plaques isolated by laser capture microdissection. J Biol Chem. 2004;279(35):37061–37068. doi: 10.1074/jbc.M403672200. [DOI] [PubMed] [Google Scholar]
- 14.Wang Q, et al. Proteomic analysis of neurofibrillary tangles in Alzheimer disease identifies GAPDH as a detergent-insoluble paired helical filament tau binding protein. FASEB J. 2005;19(7):869–871. doi: 10.1096/fj.04-3210fje. [DOI] [PubMed] [Google Scholar]
- 15.Xia Q, et al. Proteomic identification of novel proteins associated with Lewy bodies. Front Biosci. 2008;13:3850–3856. doi: 10.2741/2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olzscha H, et al. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell. 2011;144(1):67–78. doi: 10.1016/j.cell.2010.11.050. [DOI] [PubMed] [Google Scholar]
- 17.Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311(5766):1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- 18.Chapman E, et al. Global aggregation of newly translated proteins in an Escherichia coli strain deficient of the chaperonin GroEL. Proc Natl Acad Sci USA. 2006;103(43):15800–15805. doi: 10.1073/pnas.0607534103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.David DC, et al. Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol. 2010;8(8):e1000450. doi: 10.1371/journal.pbio.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reis-Rodrigues P, et al. Proteomic analysis of age-dependent changes in protein solubility identifies genes that modulate lifespan. Aging Cell. 2012;11(1):120–127. doi: 10.1111/j.1474-9726.2011.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koga H, Kaushik S, Cuervo AM. Protein homeostasis and aging: The importance of exquisite quality control. Ageing Res Rev. 2011;10(2):205–215. doi: 10.1016/j.arr.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koplin A, et al. A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. J Cell Biol. 2010;189(1):57–68. doi: 10.1083/jcb.200910074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narayanaswamy R, et al. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc Natl Acad Sci USA. 2009;106(25):10147–10152. doi: 10.1073/pnas.0812771106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciryam P, Tartaglia GG, Morimoto RI, Dobson CM, Vendruscolo M. Widespread aggregation and neurodegenerative diseases are associated with supersaturated proteins. Cell Reports. 2013;5(3):781–790. doi: 10.1016/j.celrep.2013.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ciryam P, Kundra R, Morimoto RI, Dobson CM, Vendruscolo M. Supersaturation is a major driving force for protein aggregation in neurodegenerative diseases. Trends Pharmacol Sci. 2015;36(2):72–77. doi: 10.1016/j.tips.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hofrichter J, Ross PD, Eaton WA. Supersaturation in sickle cell hemoglobin solutions. Proc Natl Acad Sci USA. 1976;73(9):3035–3039. doi: 10.1073/pnas.73.9.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikenoue T, et al. Heat of supersaturation-limited amyloid burst directly monitored by isothermal titration calorimetry. Proc Natl Acad Sci USA. 2014;111(18):6654–6659. doi: 10.1073/pnas.1322602111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muta H, et al. Supersaturation-limited amyloid fibrillation of insulin revealed by ultrasonication. J Biol Chem. 2014;289(26):18228–18238. doi: 10.1074/jbc.M114.566950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho L, et al. Altered expression of a-type but not b-type synapsin isoform in the brain of patients at high risk for Alzheimer’s disease assessed by DNA microarray technique. Neurosci Lett. 2001;298(3):191–194. doi: 10.1016/s0304-3940(00)01753-5. [DOI] [PubMed] [Google Scholar]
- 30.Blalock EM, et al. Incipient Alzheimer’s disease: Microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci USA. 2004;101(7):2173–2178. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Umemura K, et al. Autotaxin expression is enhanced in frontal cortex of Alzheimer-type dementia patients. Neurosci Lett. 2006;400(1-2):97–100. doi: 10.1016/j.neulet.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 32.Liang WS, et al. Gene expression profiles in anatomically and functionally distinct regions of the normal aged human brain. Physiol Genomics. 2007;28(3):311–322. doi: 10.1152/physiolgenomics.00208.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang WS, et al. Alzheimer’s disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc Natl Acad Sci USA. 2008;105(11):4441–4446. doi: 10.1073/pnas.0709259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Webster JA, et al. NACC-Neuropathology Group Genetic control of human brain transcript expression in Alzheimer disease. Am J Hum Genet. 2009;84(4):445–458. doi: 10.1016/j.ajhg.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan MG, et al. Genome wide profiling of altered gene expression in the neocortex of Alzheimer’s disease. J Neurosci Res. 2010;88(6):1157–1169. doi: 10.1002/jnr.22290. [DOI] [PubMed] [Google Scholar]
- 36.Simpson JE, et al. MRC Cognitive Function and Ageing Neuropathology Study Group Microarray analysis of the astrocyte transcriptome in the aging brain: Relationship to Alzheimer’s pathology and APOE genotype. Neurobiol Aging. 2011;32(10):1795–1807. doi: 10.1016/j.neurobiolaging.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 37.Cooper-Knock J, et al. Gene expression profiling in human neurodegenerative disease. Nat Rev Neurol. 2012;8(9):518–530. doi: 10.1038/nrneurol.2012.156. [DOI] [PubMed] [Google Scholar]
- 38.Durrenberger PF, et al. Selection of novel reference genes for use in the human central nervous system: A BrainNet Europe study. Acta Neuropathol. 2012;124(6):893–903. doi: 10.1007/s00401-012-1027-z. [DOI] [PubMed] [Google Scholar]
- 39.Antonell A, et al. A preliminary study of the whole-genome expression profile of sporadic and monogenic early-onset Alzheimer’s disease. Neurobiol Aging. 2013;34(7):1772–1778. doi: 10.1016/j.neurobiolaging.2012.12.026. [DOI] [PubMed] [Google Scholar]
- 40.Hokama M, et al. Altered expression of diabetes-related genes in Alzheimer’s disease brains: The Hisayama study. Cereb Cortex. 2014;24(9):2476–2488. doi: 10.1093/cercor/bht101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller JA, Woltjer RL, Goodenbour JM, Horvath S, Geschwind DH. Genes and pathways underlying regional and cell type changes in Alzheimer’s disease. Genome Med. 2013;5(5):48. doi: 10.1186/gm452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang B, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153(3):707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding B, et al. Gene expression profiles of entorhinal cortex in Alzheimer’s disease. Am J Alzheimers Dis Other Demen. 2014;29(6):526–532. doi: 10.1177/1533317514523487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ginsberg SD, Hemby SE, Lee VM, Eberwine JH, Trojanowski JQ. Expression profile of transcripts in Alzheimer’s disease tangle-bearing CA1 neurons. Ann Neurol. 2000;48(1):77–87. [PubMed] [Google Scholar]
- 45.Baker C, Belbin O, Kalsheker N, Morgan K. SERPINA3 (aka alpha-1-antichymotrypsin) Front Biosci. 2007;12:2821–2835. doi: 10.2741/2275. [DOI] [PubMed] [Google Scholar]
- 46.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koldamova R, et al. Genome-wide approaches reveal EGR1-controlled regulatory networks associated with neurodegeneration. Neurobiol Dis. 2014;63:107–114. doi: 10.1016/j.nbd.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheng B, et al. Impaired mitochondrial biogenesis contributes to mitochondrial dysfunction in Alzheimer’s disease. J Neurochem. 2012;120(3):419–429. doi: 10.1111/j.1471-4159.2011.07581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu T, et al. REST and stress resistance in ageing and Alzheimer’s disease. Nature. 2014;507(7493):448–454. doi: 10.1038/nature13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whitlock MC. Combining probability from independent tests: The weighted Z-method is superior to Fisher’s approach. J Evol Biol. 2005;18(5):1368–1373. doi: 10.1111/j.1420-9101.2005.00917.x. [DOI] [PubMed] [Google Scholar]
- 51.Terni B, Boada J, Portero-Otin M, Pamplona R, Ferrer I. Mitochondrial ATP-synthase in the entorhinal cortex is a target of oxidative stress at stages I/II of Alzheimer’s disease pathology. Brain Pathol. 2010;20(1):222–233. doi: 10.1111/j.1750-3639.2009.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sergeant N, et al. Association of ATP synthase α-chain with neurofibrillary degeneration in Alzheimer’s disease. Neuroscience. 2003;117(2):293–303. doi: 10.1016/s0306-4522(02)00747-9. [DOI] [PubMed] [Google Scholar]
- 53.Chandrasekaran K, Hatanpää K, Rapoport SI, Brady DR. Decreased expression of nuclear and mitochondrial DNA-encoded genes of oxidative phosphorylation in association neocortex in Alzheimer disease. Brain Res Mol Brain Res. 1997;44(1):99–104. doi: 10.1016/s0169-328x(96)00191-x. [DOI] [PubMed] [Google Scholar]
- 54.Manczak M, Park BS, Jung Y, Reddy PH. Differential expression of oxidative phosphorylation genes in patients with Alzheimer’s disease: Implications for early mitochondrial dysfunction and oxidative damage. Neuromolecular Med. 2004;5(2):147–162. doi: 10.1385/NMM:5:2:147. [DOI] [PubMed] [Google Scholar]
- 55.Johnson MR, et al. Systems genetics identifies Sestrin 3 as a regulator of a proconvulsant gene network in human epileptic hippocampus. Nat Commun. 2015;6:6031. doi: 10.1038/ncomms7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289–300. [Google Scholar]
- 57.Fisher RA. Statistical Methods for Research Workers. Oliver and Boyd; London: 1925. [Google Scholar]
- 58.Pearson ES. The probability integral transformation for testing goodness of fit and combining independent tests of significance. Biometrika. 1938;30(1-2):134–148. [Google Scholar]
- 59.Stouffer SA, Suchman EA, DeVinney LC, Star SA, Williams RM., Jr . The American Soldier: Adjustment During Army Life. Princeton Univ Press; Princeton, NJ: 1949. [Google Scholar]
- 60.Lanz TA, et al. STEP levels are unchanged in pre-frontal cortex and associative striatum in post-mortem human brain samples from subjects with schizophrenia, bipolar disorder and major depressive disorder. PLoS One. 2015;10(3):e0121744. doi: 10.1371/journal.pone.0121744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6(2):65–70. [Google Scholar]
- 62.Iwamoto K, Kakiuchi C, Bundo M, Ikeda K, Kato T. Molecular characterization of bipolar disorder by comparing gene expression profiles of postmortem brains of major mental disorders. Mol Psychiatry. 2004;9(4):406–416. doi: 10.1038/sj.mp.4001437. [DOI] [PubMed] [Google Scholar]
- 63.Chang L-C, et al. A conserved BDNF, glutamate- and GABA-enriched gene module related to human depression identified by coexpression meta-analysis and DNA variant genome-wide association studies. PLoS One. 2014;9(3):e90980. doi: 10.1371/journal.pone.0090980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Duric V, et al. A negative regulator of MAP kinase causes depressive behavior. Nat Med. 2010;16(11):1328–1332. doi: 10.1038/nm.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.