Abstract
The NFAT (nuclear factor of activated T cells) family of transcription factors consists of four Ca2+-regulated members (NFAT1–NFAT4), which were first described in T lymphocytes. In addition to their well-documented role in T lymphocytes, where they control gene expression during cell activation and differentiation, NFAT proteins are also expressed in a wide range of cells and tissue types and regulate genes involved in cell cycle, apoptosis, angiogenesis and metastasis. The NFAT proteins share a highly conserved DNA-binding domain (DBD), which allows all NFAT members to bind to the same DNA sequence in enhancers or promoter regions. The same DNA-binding specificity suggests redundant roles for the NFAT proteins, which is true during the regulation of some genes such as IL-2 and p21. However, it has become increasingly clear that different NFAT proteins and even isoforms can have unique functions. In this review, we address the possible reasons for these distinct roles, particularly regarding N- and C-terminal transactivation regions (TADs) and the partner proteins that interact with these TADs. We also discuss the genes regulated by NFAT during cell cycle regulation and apoptosis and the role of NFAT during tumorigenesis.
Facts
NFAT proteins regulate cell cycle-, apoptosis-, angiogenesis- and metastasis-related genes;
Although they bind to the same DNA sequence, different NFAT members and even isoforms have both redundant and opposite functions;
NFAT proteins have a weak DNA-binding capacity and frequently cooperate with other transcriptional partners;
NFAT signaling and expression are deregulated in cancer.
Open Questions
The binding of partner proteins to the less conserved transactivation region (TAD) domains of different NFAT members and isoforms might be responsible for the non-redundant functions of NFAT. What are these proteins?
T-cell activation is an example where the presence/absence of a specific partner, AP-1, dictates the program of gene expression driven by NFAT, determining the mode of immune response. It remains to be shown whether other partners are also pivotal for the functions mediated by NFAT proteins in specific contexts or cell types.
NFAT proteins are relevant factors in tumorigenesis and tumor progression. What are the subsets of genes regulated by NFAT in the different stages of tumor progression? There are specific NFAT isoforms expressed in tumor samples? Is it possible to modulate NFAT activity in order to get therapeutic response in cancer?
The NFAT (nuclear factor of activated T cells) family of transcription factors consists of five members (NFAT1–NFAT5) encoded by different genes, first described in T lymphocytes as an inducible nuclear factor that could bind and transactivate the IL-2 promoter.1 Currently, it is clear that these proteins are also expressed in other immune cells2, 3 and non-immune cells, such as cartilage cells,4 adipocytes,5 pancreatic,6 breast7 and cardiac cells.8 In addition to their well-documented role in T lymphocytes, where they control gene expression during cell activation and differentiation,9, 10 NFAT also regulates genes involved in the cell cycle, apoptosis, angiogenesis and metastasis.7, 10, 11, 12
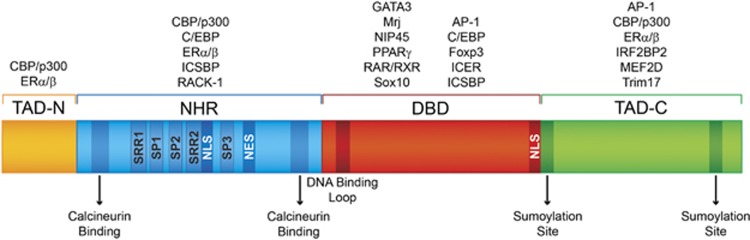
Besides NFAT5, which is activated in response to osmotic stress,13 all the other NFAT proteins (NFAT1–4) are regulated by the calcium and calcineurin signaling pathways.14, 15 The conserved regions of calcium-regulated NFAT proteins consist of two tandem domains: (1) the regulatory domain, which is also known as the NFAT-homology region (NHR) and (2) the DNA-binding domain (DBD), also known as the Rel-homology region. The NHR is moderately conserved, sharing 22–36% sequence identity among the different NFAT members (Figure 1). The DBD comprises ~270 amino acids and shares 64–72% sequence identity among the different NFAT members (Figure 1). This highly conserved domain confers the specificity to bind the DNA core sequence (A/T)GGAAA14 to all the NFAT proteins. Flanking the NHR and the DBD domains are two transcriptional activation domains (TAD) at the N- and C termini (Figure 1), which are highly variable among the different NFAT members and isoforms.14, 15
Figure 1.
Schematic primary structure of NFAT. The calcium-regulated NFAT proteins are composed of the NHR, the DBD and the N- and C-terminus transactivation domains (TAD-N and TAD-C, respectively). The NHR displays two calcineurin-binding sites, a nuclear localization sequence (NLS) and a nuclear export signal (NES). Another NLS is located in the DBD region. The NFAT serine residues that are dephosphorylated upon calcineurin activation are located in the SRR1, SRR2, SP1, SP2 and SP3 regions. The NFAT DBD region that directly contacts DNA is indicated as the DNA-binding loop. The TAD-C region of NFAT1 and NFAT2 has two sumoylation sites. Different NFAT partner proteins interact with specific domains of NFAT, and these partners are listed above each NFAT domain
The NHR is highly phosphorylated in resting cells, keeping NFAT in an inactive state and restricted to the cytoplasm (Figure 2). An increase in intracellular calcium levels activates calcineurin, a calcium/calmodulin-dependent serine/threonine phosphatase, which dephosphorylates the NHR, exposing the NFAT1–4 nuclear localization signal and inducing NFAT translocation to the nucleus14, 16, 17 (Figure 2). Once in the nucleus, NFAT proteins can bind to their target promoter elements and regulate the transcription of specific responsive genes, either alone or in combination with other nuclear partner proteins9 (Figure 2).
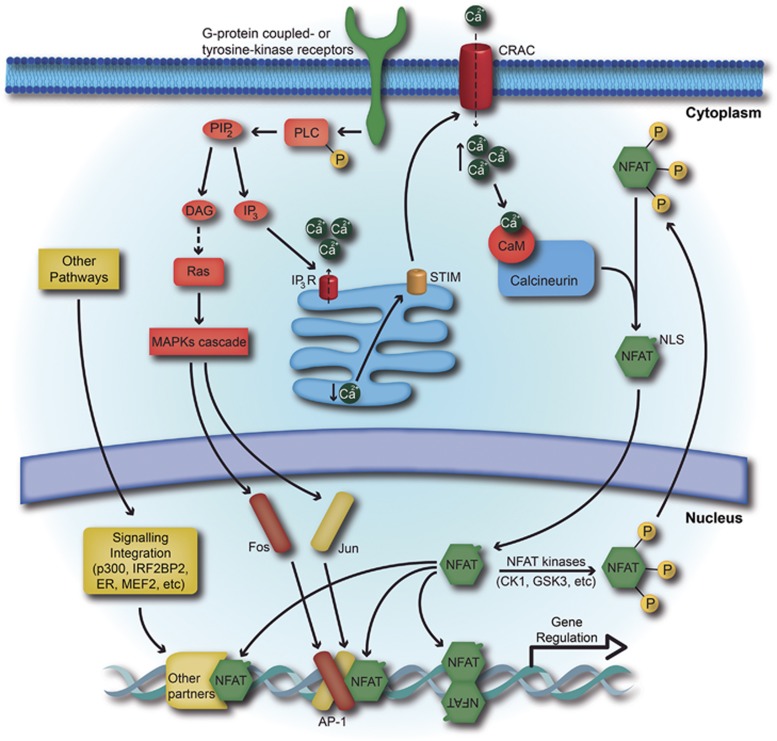
Figure 2.
The NFAT signaling pathway. Representation of NFAT activation and translocation to the nucleus. The triggering of a G-protein-coupled or tyrosine-kinase receptor leads to the activation of phospholipase C, which promotes the hydrolysis of phosphatidylinositol-4,5-biphosphate (PIP2), producing diacylglycerol and inositol-1,4,5-triphosphate (IP3). Then, IP3 binds to IP3R and induces the release of Ca2+ ions from the endoplasmic reticulum to the cytoplasm, depleting the intracellular stores. The depletion of calcium stores induces the activation of the stromal interaction molecule, which promotes the opening of calcium release-activated channel in the plasma, leading to the sustained increase in the cytoplasmic calcium levels. Next, the Ca2+ ions bind to calmodulin and activate the phosphatase calcineurin. Calcineurin dephosphorylates specific serine residues in NFAT. Dephosphorylated NFAT undergoes a conformational change exposing its NLS and translocates to the nucleus. In the nucleus, NFAT binds to DNA alone or in cooperation with other nuclear partner proteins, activated by other signaling pathways, to regulate gene expression. To reverse its activation, NFAT kinases rephosphorylate NFAT and promote the concealment of its NLS and exposure of the NES, redirecting NFAT to the cytoplasm
Although NFAT proteins bind to the same DNA sequence, these proteins have both redundant and non-redundant roles during the regulation of different processes. In this review, we discuss the non-conserved TAD regions that could explain at least in part this dual role, together with the NFAT partners that interact with the variable TADs. In addition, we highlight the regulation of cell cycle and apoptosis genes by the different NFAT proteins and the function of NFAT in malignant transformation and tumorigenesis.
TAD-conserved Regions
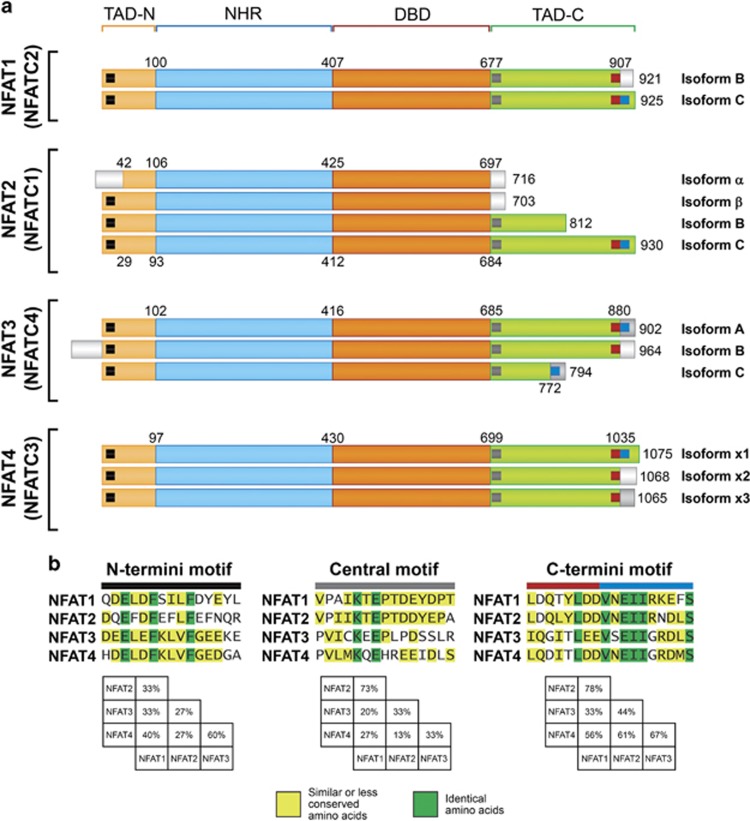
The TADs are putative sites where distinct NFAT proteins interact with specific partners and control important biological functions such as proliferation and cell death. They are the least conserved domains of NFAT. However, even these regions have some degree of conservation (Figure 3). Three main small conserved regions can be identified among the TADs of NFAT variants and will be referred to as the N-terminus motif (black square, Figure 3), central motif (gray square) and C-terminus motif (sub-motif 1, red square; sub-motif 2, blue square).
Figure 3.
The NFAT family of transcription factors. (a) Schematic primary structure of human calcium-regulated NFAT proteins and their isoforms. The alternative nomenclature for the NFAT genes is shown in parenthesis. As in Figure 1, NHR, DBD and TADs are represented. In addition, the TAD-conserved regions are indicated by a black square (N-terminus motif); a gray square (central motif); and by red and blue squares (C-terminus motif). The NCBI reference numbers for the NFAT isoforms are: NP_036472.2 (NFAT1-B); NP_775114.1 (NFAT1-C); NP_765978.1 (NFAT2-α); NP_001265604.1 (NFAT2-β); NP_765977.1 (NFAT2-B); NP_765975.1 (NFAT2-C); NP_004545.2 (NFAT3-A); NP_001129494.1 (NFAT3-B); NP_001185894.1 (NFAT3-C); NP_775188.1 (NFAT4-x1); NP_004546.1 (NFAT4-x2); NP_775186.1 (NFAT4-x3). (b) Alignment of the NFAT TAD-conserved regions, designated as the N-terminus motif, central motif and C-terminus motif. Amino acids marked with yellow indicate that residues are similar or occur more than 50% of the time at that position; green indicates identical residues. The similarity percentage for each region between the NFAT proteins is also shown
The N-terminus motif is present in NFAT variants depending on the initial exon used for transcription. The NFAT2-α autoregulatory loop is one mechanism where the N-terminus motif is excluded. In a resting state, the transcription initiation of NFAT2 is in exon 2, which encodes the N-terminus motif. After activation, NFAT binds to the NFAT2 promoter, leading to initiation of transcription from exon 1 and skipping exon 2.18 The N-terminus motif is rich in acidic and hydrophobic residues (Figure 3b), and it can act as an acidic activation domain (AAD).14, 19 The AADs are among the most potent transcriptional activators described.20 In fact, it has been shown that the NFAT2-β amino-acid region from 1 to 30 (region containing the N-terminus motif, Figure 3a) is necessary and sufficient to elicit high in vitro transcription.19 Furthermore, the NFAT1-C amino-acid region 1–144 also contains a potent transactivation domain.21 Moreover, the importance of this motif is not restricted to transcriptional activation. It was shown that casein kinase 1 specifically binds to the N-terminus motif and induces NHR phosphorylation and nuclear export of NFAT,22 with consequent NFAT inactivation. Because several NFAT isoforms lack the N-terminus motif (Figure 3a), this splicing variation might facilitate the nuclear localization and function of NFAT.
The NFAT small central motif is located immediately after the DBD and is composed of a highly conserved region of 15 amino acids present in most of NFAT family members besides NFAT2-α and β (gray square; Figure 3). Apparently, the central motif is not involved in transcriptional activation.23 However, it was shown that the sumoylation of the well-conserved lysine residue (K; Figure 3b) in the central motif of both NFAT1 and NFAT2 is tightly linked to NFAT nuclear localization and transcriptional function.24, 25 Therefore, the presence of this small central motif might be important for the function mediated by different NFATs.
The C-terminus motif is located in the last 50 amino acids of C-terminus transactivation domain (TAD-C; Figure 3b) and appears to be important for transactivation.21, 23 This motif has a sequence identity of ~50% among different NFATs and can be divided into two sub-motifs that are present in different exons and are subjected to splicing variation: sub-motif 1 and 2 (red and blue squares, respectively; Figure 3). NFAT1-C contains both sub-motifs and induces CAT expression at least two times greater than NFAT1-B, which only has the first sub-motif.21 The same phenotype was observed for NFAT4 isoforms that lack sub-motif 2.26 Although several NFAT splicing variants have been established, as reviewed by Vihma et al.,27 little is known about the functional meaning of these different proteins; however, it is reasonable to expect that they exert specific functions depending on the conserved regions present in each isoform.
NFAT Partners
NFAT proteins can interact with several transcription factors and regulators in the nucleus, being important integrators of calcium signals with other signaling pathways.15, 28 The interaction of NFAT with other proteins through their conserved DBD and NHR regions can result in synergistic activation29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46 (such as with GATA-3;38 ICSBP;42 C/EBP;43 AP-141, 44) or repression47, 48, 49, 50, 51, 52, 53 (PPARγ47, 48 Mrj;49 Foxp3;50, 51 ICER52) of NFAT-mediated transcription (Table 1). Although the interactions with DBD and NHR regions are important for NFAT function, in this section, we will focus on the NFAT partners that interact with the less conserved NFAT TAD regions.
Table 1. NFAT partners in transcription.
| Partner | NFAT member | Techniquea | Region of interaction on NFAT | Effect on transcription | Gene regulated | Reference |
|---|---|---|---|---|---|---|
| AP-1 (c-Fos/c-Jun) | NFAT1; 2; 3; 4 | A; B; E | DBD | Activation | Several cytokines | 41 |
| TAD-C | IL-2 | 44 | ||||
| CBP/p300 | NFAT1-C | A; B | TAD-N | Activation | IL-2; TNF-α | 54, 55, 56 |
| NFAT2-β | NHR | |||||
| NFAT3-A | TAD-C | |||||
| C/EBP | NFAT1-C | A | NHR-DBD | Activation | PPAR-γ IGF-2 | 43 |
| NFAT3-A | ||||||
| c-Maf | NFAT1 | C | ND | Activation | IL-4 | 29 |
| c-rel (NFκB) | NFAT2-α | A | ND | Activation | CD40L | 35 |
| EGR1; EGR4 | NFAT1; 2 | A; B | ND | Activation | IL-2; TNF-α | 31 |
| ERα/ERβ | NFAT3-A | B; C | TAD-N | Inhibition | IL-2 | 57, 58 |
| NHR | ||||||
| TAD-C | ||||||
| Foxp3 | NFAT1-C | A; E | DBD | Inhibition | IL-2; IL-4; IFN-γ | 50, 51 |
| GATA-2 | NFAT2 | A | ND | ND | ND | 36 |
| GATA-4 | NFAT3 | A; C | ND | Activation | BNP | 37 |
| GATA-3 | NFAT1-C | A | DBD | Activation | IL-5 | 38 |
| NFAT2-α | ||||||
| ICER | NFAT1-C | B | DBD | Inhibition | IL-2; TNF-α | 52 |
| ICSBP | NFAT2-α | A; B | NHR-DBD | Activation | IL-12 | 42 |
| IRF2BP2 | NFAT1-C | B; C | TAD-C | Inhibition | IL-2; IL-4; TNF-α | 62 |
| IRF4 | NFAT1 | A; B | ND | Activation | IL-4 | 30 |
| MEF2D | NFAT1-C | A | TAD-C | Activation | Nur77 | 59 |
| Mrj | NFAT4-x1 | A; C | DBD | Inhibition | TNF-α | 49 |
| NIP45 | NFAT1-C | A; C | DBD | Activation | IL-4 | 39 |
| PPAR-γ | NFAT2-α | A; B | DBD | Inhibition | IL-4; IL-2 | 47, 48 |
| PU.1 | NFAT2 | A | ND | Activation | Cathepsin K | 33, 34 |
| Rack-1 | NFAT2 | A; C | DBD | Inhibition | 3x NFAT | 53 |
| Raf-1 | NFAT4 | A | ND | Activation | CXCR5 | 45 |
| RAR/RXR | NFAT1 | B | DBD | Activation | ||
| NFAT2 | ND | Inhibition of NFAT1-dependent activation | CCR9 | 134 | ||
| Sp1/Sp3 | NFAT1; 2 | A | ND | Activation | p21 | 32 |
| Sox10 | NFAT3-A | A; D | DBD | Activation | Krox20 | 40 |
| STAT3 | NFAT1 | A | ND | Activation | CDK6 | 46 |
| Trim17 | NFAT3 | C | TAD-C | Inhibition | BDNF | 63 |
| NFAT4 |
Abbreviation: ND, non determined.
Co-immune, A; GST pull-down, B; two hybrid, C; mass spectrometry, D; X-ray analysis, E.
Besides the conserved motifs in the TAD regions discussed in the previous section, both the amino- and carboxyl-terminal ends of the NFAT family proteins show low sequence conservation. It is possible that different NFAT TADs could account for the non-redundant functions of these transcription factors by distinct control of transcription or through cooperation with the different protein partners, such as described below:
CBP/p300
CBP and p300 directly interact with amino acids 1–145 of NFAT1 and 1–108 of NFAT2.54, 55 NFAT2 further interacts with CBP/p300 at the NHR amino acids 113–205.55 CBP also interacts with the NHR (amino acids 130–237) and with the TAD-C (amino acids 771–902) regions of the NFAT3 protein.56 Both NFAT3 regions seem to be fundamental for CBP binding.56 Because NFAT TAD is a highly variable region of NFAT proteins (Figure 3a), it would be of interest to analyze CBP/p300 interactions with different NFAT splice variants. Once CBP/p300 interacts with multiple NFAT regions, a differential transcriptional activity may be achieved by various NFAT splicing isoforms.
ERα/ERβ
Both ERα and ERβ interact with NFAT3-A through residues 1–261 (TAD-N (N-terminus transactivation domain)/NHR), 261–450 (NHR) and 613–902 (TAD-C).57, 58 The NFAT3–ER complex dramatically reduced the transactivation of the IL-2 promoter, suggesting that ER can function as an NFAT co-repressor.58 Interestingly, NFAT3 can act in synergy with ER to transactivate elements containing ER- but not NFAT-binding sites, suggesting cross-talk between NFAT and ER: when the NFAT3–ER complex is bound to a NFAT element, it functions as a repressor of transcription, and, once bound to the ER element, as an activator.57
MEF2
The transcription factor MEF2 interacts with NFAT1-C TAD-C (residues 679–927)59 but not with NFAT2-α or NFAT3-A.60 Because NFAT2-C and NFAT1-C TAD-C regions exhibit some conservation (Figure 3), it would be interesting to verify whether MEF2 can interact with the NFAT2-C protein as well. The MEF2–NFAT1 interaction leads to synergistic activation of pathways involved in cell death, muscle development,60, 61 thymocyte-negative selection and apoptosis.59, 60
IRF2BP2
IRF2BP2 interacts with the C-terminal region of NFAT1 and strongly inhibits its transcriptional activity during the regulation of cytokine genes.62 Interestingly, no interaction was detected between IRF2BP2 and the other NFAT family members, indicating that IRF2BP2 is a NFAT1-specific partner and could be responsible for some repression functions mediated by NFAT1 but not by the other NFAT members.
Trim17
It was shown that Trim17 interacts with the C-terminal end of both NFAT3 and NFAT4 in neuronal cells.63 In addition, sumoylated sites of NFAT4 but not NFAT3 are necessary for the interaction. Trim17 inhibits the activity of NFAT3–4 by favoring their cytoplasmic localization. Thus, this interaction shows a new mechanism of NFAT3–4 regulation. The effect of Trim17 on the NFAT1–2 cellular localization was not investigated.
The interaction between all of these proteins and the TAD regions of distinct NFAT members can strongly contribute to our knowledge of the non-redundant phenotypes observed both in mice lacking individual NFAT proteins and in many in vitro studies as described below.
Cell Cycle and Apoptosis Regulation by NFAT
The specific role of each NFAT member in the control of gene transcription during cell cycle and apoptosis is not completely clear, mainly because most of the studies did not evaluate different NFAT members in the same model. We clearly demonstrated that ectopic expression of constitutively active forms of NFAT proteins (CA-NFAT1-C and CA-NFAT2-α) produces opposite phenotypes in the regulation of the cell cycle and apoptosis in NIH3T3 cells. Whereas NFAT2 acts as a positive regulator of cell proliferation and a repressor of cell death, NFAT1 induces a slight cell cycle arrest and a significant increase in cell death.64 A truncated CA-NFAT1 protein that lacks the TAD-C domain was unable to induce apoptosis, showing that the C-terminal domain of NFAT1 is responsible for its pro-apoptotic characteristic.64 More recently, it became clear that not just the NFAT isoform, but also the cell type and the signaling pathways activated contribute for the distinct functions exerted by the NFAT proteins. Below, we discuss the role of NFAT proteins during the regulation of genes involved in the control of the cell cycle and apoptosis.
NFAT and Cell Cycle Regulation
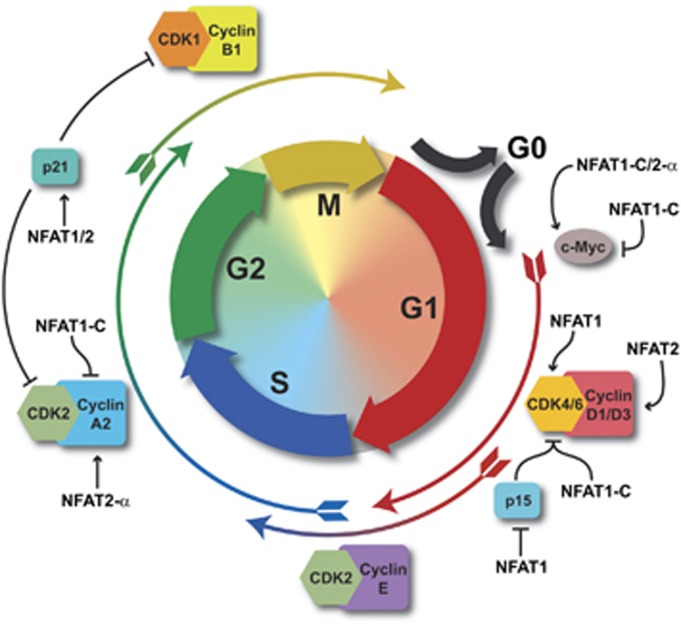
Evidence that NFAT proteins are involved in cell cycle control emerged from observations that DNA synthesis due to stimulation with growth factors is Ca2+-dependent and is markedly inhibited by CsA and FK506,65 drugs that inhibit calcineurin activity and, consequently, NFAT activation.14 Calcineurin has a major role in the regulation of cell cycle progression during the early stages of the G1 phase.11, 66 Thereafter, several studies have determined the specific molecules regulated by different NFAT members (Figure 4 and Table 2).
Figure 4.
Cell cycle regulation by NFAT proteins. Schematic representation of the cell cycle regulation. The cell cycle phases G1, S, G2 and M are indicated. Non-dividing cells are in G0. The specific complexes of Cyclin/Cyclin-dependent kinases (CDK) and the CDK inhibitors p15 and p21 are indicated in specific cell cycle phases. The proto-oncogene c-Myc is also shown. NFAT proteins control cell cycle progression by regulating the expression of some cell cycle genes. The negative or/and positive regulations driven by NFAT proteins on these cell cycle-related genes are indicated
Table 2. Genes regulated by NFAT.
| Function | Gene | NFAT member or isoform | Effect on transcription regulation | Reference |
|---|---|---|---|---|
| Cell cycle | CDK4 | NFAT1-C | Downregulation | 11 |
| NFAT3 and 4 | No effect | |||
| CDK6 | NFAT1 | Upregulation | 46 | |
| c-Myc | NFAT2-α | Upregulation | 6 | |
| NFAT1-C | Down/upregulation | 80 | ||
| Cyclin A2 | NFAT2-α | Upregulation | 73 | |
| NFAT1-C | Downregulation | 72 | ||
| Cyclin D1 | NFAT2-α | Upregulation | 74 | |
| NFAT1 and 3 | No effect | |||
| Cyclin D3 | NFAT2 | Upregulation | 76 | |
| p15 | NFAT1 | Downregulation | 77 | |
| p21 | NFAT1 and 2 | Upregulation | 32 | |
| NFAT3 and 4 | No effect | |||
| Anti-apoptotic | A1 | NFAT1-C | Upregulation | 93 |
| NFAT2-β | No effect | |||
| Bcl-2 | NFAT2 | Upregulation | 135, 136 | |
| BDNF | NFAT3 | Upregulation | 86 | |
| NFAT4 | No effect | |||
| DDIAS | NFAT2 | Upregulation | 94 | |
| Pro-apoptotic | c-FLIP | NFAT1 and 2 | Upregulation | 91 |
| NFAT3 | No effect | |||
| FasL | NFAT1, 2 and 4 | Upregulation | 96, 97 | |
| Nur77 | NFAT1-C | Upregulation | 59, 101 | |
| TNF-α | NFAT1, 2-β and 2-C | Upregulation | 103, 107, 108, 111 | |
| NFAT2-α | No effect | |||
| TRAIL | NFAT2 | Upregulation | 137 | |
| Trim17 | NFAT3 | No effect | 112 | |
| NFAT4 | Upregulation |
The phenotypes of NFAT knockout mice provided strong evidence for the important role of NFAT in cell cycle regulation. Cells from the lymph nodes and spleen of NFAT1−/− mice hyperproliferate in response to different antigen stimulations.67, 68 NFAT1−/− lymphocytes have a shorter cell division period upon activation, which is related to an overexpression of cyclins A2, B1, E and F. Moreover, upon activation, NFAT1−/− lymphocytes seem to exit the resting state faster than NFAT1+/+ cells. These data suggest that NFAT1 acts as an inhibitor of cell proliferation, regulating very early inducible genes that control the commitment of resting cells to become activated upon stimulation.69 NFAT2 deficiency in mice is lethal as a consequence of defects in cardiac valve formation;8 nevertheless, in the RAG-deficient complementation system, NFAT2−/− T and B cells showed reduced proliferation when compared with wild-type (WT) mice,70, 71 suggesting that, in contrast to NFAT1, NFAT2 might activate genes involved in cell cycle progression.
NFAT1 was shown to directly downregulate cyclin A2 expression by binding to and suppressing the cyclin A2 promoter.72 On the other hand, NFAT2 upregulates cyclin A2 expression.73 Furthermore, NFAT2 is also involved in upregulation of cyclin D174, 75 and cyclin D3 expression.76
The ectopic expression of NFAT1 also inhibits the activity of the human CDK4 promoter. This repression seems to be specific for NFAT1, as no repression was observed with NFAT3 or NFAT4. Apparently, the mechanism by which NFAT1 represses the CDK4 promoter is by competing with E2F family members because the overexpression of NFAT1 inhibited the ability of E2F1–3 to transactivate the CDK4 promoter.11 Notably, the NFAT-binding site found in the cyclin A2 promoter lies in close proximity to a putative E2F-binding site.72 It would be interesting to evaluate whether the function of NFAT1 in repressing cyclin A2 also occurs by displacing E2F.
Another target for NFAT regulation is the CDK inhibitor p21. p21 is induced by both NFAT1 and NFAT2 in primary mouse keratinocytes, and CsA inhibits the p21 promoter activity associated with Ca2+-induced differentiation.32 Interestingly, the direct binding of NFAT to the p21 promoter is dispensable and can be replaced by the interaction of NFAT with Sp1/3.32
Although the majority of the studies describes NFAT1 as a negative regulator of cell cycle, a few reports show NFAT1 as a positive regulator. Ectopic expression of NFAT1 promotes heterochromatin formation on CDK inhibitor p15 promoter and silencing of p15 expression, resulting in an increased expression of D-type cyclins and their partner kinases CDK4/6, consequently contributing to tumor growth.77 The same group has also demonstrated NFAT1 cooperating with STAT3 to promote CDK6 upregulation and pancreatic cancer cell proliferation.46 Remarkably, loss of STAT3 expression reversed the NFAT1-dependent stimulation of proliferation, emphasizing how the signaling pathways present when NFAT is activated might convert NFAT from activators to repressors or vice versa.46
Finally, NFAT also regulates the c-Myc gene. It has been shown that NFAT2 induces c-Myc transcriptional activity by binding to a specific element within the c-Myc proximal promoter.6, 78, 79 We demonstrated that NFAT1 also regulates the c-Myc promoter.80 In addition to this proximal site previously described, NFAT1 directly binds to distal sites of the c-Myc promoter with different affinities. NFAT1 positively regulates the proximal site but acts as a negative regulator once bound to distal elements of the c-Myc promoter. The extent of NFAT1 regulation depends on the cooperation with other partners, such as p300,80 again highlighting the importance of protein partners to modulate NFAT function.
It has not been demonstrated whether NFAT3 and NFAT4 directly regulate genes involved in cell cycle progression; however, they do have a role in cell death and survival, as discussed in the following section.
NFAT and Apoptosis Regulation
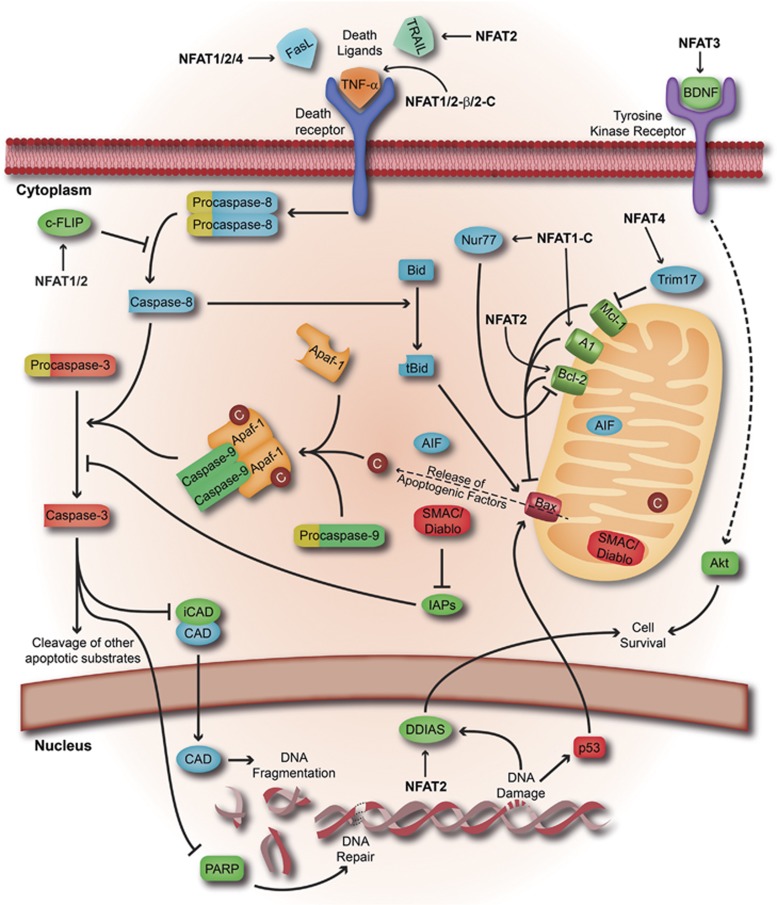
The hyperproliferative disorder observed in NFAT1−/− lymphocytes,68, 69 as well as the lymphadenopathy and splenomegaly exhibited by double-knockout NFAT1/NFAT4 mice,81 has been linked to resistance to cell death. In contrast, whereas peripheral NFAT2−/− T cells show impaired proliferation but no apparent defects in apoptosis,82 NFAT2−/− B cells exhibit increased activation-induced cell death compared with WT cells.83 Altered proliferation or apoptosis patterns have not been reported in NFAT3−/−,84 but it was shown that NFAT3 does promote survival in neuronal cells.85, 86 The NFAT4 single knockout mouse displays increased apoptosis of both double-positive thymocytes and peripheral T cells.28, 87 Likewise, NFAT4 protects pulmonary cells from apoptosis under hypoxia.88 A more recent study, however, reports the involvement of NFAT4 with the promotion of cardiomyocyte apoptosis.89 Whether specific NFAT4 isoforms or protein partners expressed in different cell types explain the distinct phenotypes induced by NFAT4 remains to be demonstrated. All of these data indicate that each NFAT gene exhibits individual properties, suggesting that the cellular threshold level for each protein and the cell type might determine which set of target genes will be expressed. Next, we discuss the pro- and anti-apoptotic genes regulated by individual NFAT proteins and their roles in cell physiology (Figure 5 and Table 2).
Figure 5.
Regulation of the apoptosis signaling pathway by NFAT proteins. Schematic representation of the apoptosis regulation. The extrinsic and intrinsic apoptotic pathways are represented. NFAT proteins control cell survival and cell death by regulating the expression of some apoptotic genes. The negative or/and positive regulations driven by NFAT proteins on these apoptotic-related genes are indicated
Anti-apoptotic Genes
c-FLIP
This protein acts as an apoptosis inhibitor through modulation of caspase-8/10 activity.90 Recently activated T cells are resistant to FasL-mediated cell death, presumably because of increased expression of anti-apoptotic molecules.91 In contrast to naive or long-term activated T cells, short-term activated T cells strongly upregulate the short splice variant of c-FLIP, c-FLIPs. The induction of c-FLIPs in T cells is primarily mediated by the NFAT pathway, and blockage of NFAT-mediated c-FLIPs expression rendered T cells sensitive toward FasL-induced cell death.91 Interestingly, whereas NFAT1 and NFAT2 proteins bind and transactivate the c-FLIP promoter, NFAT3 is unable to promote c-FLIP expression.91
A1
The FcɛRI activation-induced survival of mast cells is dependent on the prosurvival protein A1, a member of the anti-apoptotic Bcl-2 family.92 A NFAT-binding site was characterized in the A1 promoter, and inhibition of NFAT activation by CsA treatment abrogated the expression of A1 in mast cells.93 Furthermore, CA-NFAT1-C but not CA-NFAT2-β overexpression increases A1 mRNA levels.93 This difference might be explained by the presence of the long TAD-C present in NFAT1-C that is absent in NFAT2-β. As previously discussed, the TAD-C domain works as a transactivation domain and can bind NFAT partner proteins important for A1 transactivation.
DDIAS
NFAT2 directly induces the transcription of DDIAS,94 an anti-apoptotic protein highly expressed in lung cancers.95 A positive correlation between DDIAS and NFAT2 expression was found in human lung tumors, and inhibition of NFAT2 or DDIAS increased cell death in the presence of cisplatin, providing insights into DDIAS-associated chemoresistance via NFAT2 activation.94
Pro-apoptotic Genes
FasL
The resistance to apoptosis observed in T lymphocytes from double-knockout NFAT1/NFAT4 mice could be explained by the remarkably impaired expression of the FasL gene in these cells.81 Treatment of lymphocytes with CsA inhibits TCR-mediated FasL expression.96, 97, 98 Both NFAT1 and NFAT2 proteins bind to two sites within the FasL promoter,96 and ectopically expressed NFAT1-C transactivates the FasL promoter in T cells.97, 99 The effect of NFAT in the transcription of FasL is at least in part indirect, via Egr3 (early growth response-3). NFAT1 or NFAT4 directly transactivates the Egr3 promoter, which, in turn, transactivates the FasL promoter.98 However, the magnitude of Egr3 induction in activated T cells isolated from single NFAT1−/− or NFAT4−/− mice is normal, suggesting that NFAT1 and NFAT4 could have a compensatory role in the transactivation of the Egr3 gene and, consequently, on FasL expression.98
Nur77
Nur77 mediates negative selection in thymocytes after strong TCR engagement in the thymus.100 The Ca2+-responsive element in the Nur77 promoter contains two binding sites for MEF2, which cooperates with coactivators to recruit histone acetylases and activates the Nur77 expression.101 One of these coactivators might be NFAT1, because NFAT1–MEF2 can cooperatively bind to the Nur77 promoter and induce its expression. Interestingly, this NFAT1–MEF2 cooperation does not depend on NFAT binding to DNA. Moreover, the complex MEF2D–NFAT1 still recruits p300 for the full transcriptional activation of Nur77.59 Because MEF2 and p300 interact with both NFAT1-C TAD-N and TAD-C, it becomes evident that splicing of NFAT members is an important step in the regulation of Nur77 and, consequently, in determining the cell fate.
TNF-α
Four putative NFAT-binding sites have been demonstrated in the TNF-α promoter.102, 103, 104, 105 Although all NFAT members bind to the TNF-α promoter,106 NFAT1 binds to all the elements, whereas NFAT2 apparently only binds to the distal site.103, 104, 107, 108, 109 It was shown that NFAT1-C can induce TNF-α transcription more efficiently than NFAT2-α.109 This difference was mapped to the long TAD-C domain present in the NFAT1-C protein because a hybrid of NFAT2-α fused to NFAT1-C TAD-C increases the TNF-α promoter transactivation. Furthermore, the NFAT2-C long isoform also transactivates the TNF-α promoter less efficiently than the NFAT1-C protein.109 These results are in accordance with a previous work, which demonstrated that NFAT2-α does not promote cell death of primary T lymphocytes, whereas the NFAT1 and NFAT2 long isoforms do induce apoptosis.110 Recently, our group showed that NFAT2-β also induces apoptosis through the upregulation of TNF-α expression. Similarly to NFAT2-α, NFAT2-β also lacks the long TAD-C but it has a highly acidic domain in the TAD-N, which confers high capacity to transactivate the TNF-α promoter (black square, Figure 3).111 Thus, the inability of NFAT2-α to induce apoptosis lies in the lack of both the long TAD-C and the acidic domain present in TAD-N.
Trim17
Trim17 is an essential protein for neuronal apoptosis,112 which interacts with NFAT3–4 and inhibits their function by decreasing NFAT nuclear localization. NFAT4 binds to a NFAT site in the Trim17 promoter, and overexpression of NFAT4 increases Trim17 expression and aggravates cell death. Accordingly, shRNA against NFAT4 protects neurons from apoptosis and reduces Trim17 mRNA. Conversely, NFAT3 overexpression protects neurons from apoptosis, with no effect in Trim17 expression.63 Thus, this study suggests a mechanism that explains the opposite role for NFAT3 and NFAT4 on neuronal apoptosis.
The effect of individual NFAT proteins in apoptosis regulation might rely on their ability to control the expression of both pro- and anti-apoptotic genes at the appropriate stage of activation and differentiation. Different NFAT proteins are specific and prominent regulators of apoptosis and cell cycle genes. It is important to have more studies comparing the function of different members and isoforms in the same model to elucidate the role of each one.
NFAT and Tumorigenesis
As described in the previous sections, the NFAT family has an important role in the regulation of cell cycle progression, gene expression and apoptosis. All these studies indicate that the Ca2+/NFAT signaling pathway is essential to maintain normal cell physiology, and, therefore, the deregulation of this pathway can be associated with malignant cell transformation. In fact, deregulation of calcineurin/NFAT signaling and/or abnormal expression of its components have been reported in solid tumors of mesenchymal and epithelial origin, lymphoma and leukemia.113 In this section, we focus on the different roles described for the NFAT members in tumor development.
To investigate the effects of sustained NFAT2 signaling in preadipocyte cells, Neal and Clipstone114 reported that the expression of the CA-NFAT2-α leads these cells to adopt the well-established hallmarks of cellular transformation, providing direct evidence for the oncogenic potential of NFAT2.114 Later, NFAT2 was found to be ectopically expressed and highly activated in pancreatic cancer cells.6 Importantly, the proliferation and the anchorage-independent growth of cultured pancreatic cancer cells were significantly attenuated by CsA or siRNA against NFAT2.6 Active nuclear NFAT2 was also found in cases of Burkitt lymphoma, diffuse large B-cell lymphoma, aggressive T-cell lymphoma and bladder cancer.35, 115, 116, 117 More recently, NFAT2 was also implicated in colon cancer cell invasion and metastasis118 and in prostate cancer progression.119, 120 Of note, besides all those studies reporting NFAT2 as an oncogene, it was shown that one of the antitumor effects of arachidonic acid treatment in mammary and pancreatic cancer cell lines was through the induction of NFAT2, which downregulates GLI-1 transcription, resulting in decreased expression of anti-apoptotic genes such as Bcl-2 and 4-1BB.121 However, it remains to be clarified whether a specific NFAT2 isoform is induced in this context or whether the arachidonic acid modifies the expression of putative NFAT2 partners.
Those phenotypes of oncogenicity induced by CA-NFAT2-α were also demonstrated by our group in NIH3T3 fibroblast cells.64 Remarkably, we showed that, in contrast to CA-NFAT2-α, CA-NFAT1-C was neither able to transform NIH3T3 cells nor to induce tumors in athymic mice. Furthermore, CA-NFAT1-C expression reduced cell growth, the number of foci in culture and induced apoptosis of cells transformed with either CA-NFAT2-α or H-rasV12 oncogene. Strikingly, when challenged with a chemical carcinogen, the NFAT1−/− mice were more susceptible to tumor development than the WT mice, suggesting an important role for NFAT1 in tumor suppression.64 The tumor-suppressor role for the NFAT1-C protein was further mapped to the TAD-C domain.64, 122 The antitumor activity of NFAT1 was first proposed in a chondrocyte lineage, based on observations of uncontrolled proliferation of abnormal extra-articular cartilage cells in NFAT1−/− mice.4 Recently, it was shown that NFAT1−/− mice also develop spontaneous B-cell lymphomas.123 Taken together, these results support an opposite role for NFAT1 and NFAT2 in the regulation of tumorigenesis.
Despite the fact that a cell-intrinsic tumor-suppressor role for NFAT1 has been established during the last decade,4, 64 a second role for NFAT1 in tumor progression has been demonstrated. NFAT proteins also promote migration and invasion through α6β4 integrin signaling.7 High levels of NFAT1 and NFAT5 were found in tissue sections from patients with invasive ductal breast carcinoma, where the expression of the β4 integrin was restricted to tumor tissue and was absent in normal breast tissue. The signaling through α6β4 integrin activates NFAT1, NFAT4 and NFAT5.7 Consistent with these data, the Akt-dependent NFAT1 ubiquitination and its subsequent degradation proteasome resulted in impaired migratory and invasive potential of the breast cancer cell lines.124, 125 NFAT1 also regulates the invasion of glioblastoma multiforme cells126 and metastasis of mammary cancer cells.127 NFAT1 was also found ectopically expressed in the nucleus of pancreatic tumors. It was shown that an inflammatory microenvironment induces NFAT1 activity, which in turn contributes to tumor growth.46, 77 Furthermore, a crucial role for NFAT1 in initiation and promotion of inflammation-associated colorectal tumors was reported.128 In addition, some recent studies have implicated NFAT factors with the dedifferentiation of cancer cells. NFAT1 promotes the dedifferentiation of melanoma cells,129 whereas NFAT3 induces acinar to ductal dedifferentiation and pancreatic cancer initiation.130 Moreover, in pancreatic cancer, NFAT2 drives epithelial–mesenchymal transition and seems to be responsible for the appearance of stem-cell features.131 These studies demonstrate that NFAT factors control the plasticity of some cancer cells and might be involved in the maintenance of cancer stem cells. However, the mechanisms linking cancer cell dedifferentiation driven by NFAT and stemness remain to be demonstrated.
These findings also show that changes in the transducing pathways are the key elements to pathogenesis of cancer. NFAT proteins may be overexpressed or inactivated, becoming dangerous to the cell. During tumor development, alterations in NFAT regulation may occur, leading to the expression of different protein levels and/or isoforms as well as alterations in the expression of partner proteins. Consequently, NFAT proteins can contribute to distinct aspects of the tumorigenic phenotype, including the initial deregulation of cellular growth control, the recruitment of an adequate blood supply and the regulation of tumor metastasis. Indeed, the importance of NFAT as a cancer therapeutic target has been raised in the last few years (reviewed by Qin et al.132 and Metzelder et al.133). However, the subsets of genes that are regulated by NFAT proteins during the different stages of tumor progression remain to be elucidated.
Concluding Remarks
NFAT proteins have been described in many cell types and regulate genes involved in cell cycle progression, cell differentiation and apoptosis. The role of NFAT proteins in these pathways reveals a fundamental function for this family in normal cell physiology. Although NFAT members share conserved domains, the TADs are highly variable regions, which can have a critical role in NFAT function because they can contribute to non-redundant functions. In fact, it has been shown that the TADs are sites of interaction between distinct NFAT family members and several partners. The differential levels and splicing variants of NFAT proteins and the cell type in which they are being expressed can determine whether the cell will live, proliferate or die. Beyond that, the deregulation of NFAT signaling is clearly involved in tumorigenesis, where NFAT2 acts as an oncogene, whereas NFAT1 acts as an oncogene or a tumor-suppressor gene depending on the cells and tissue types that were evaluated. The detailed identification of the isoforms expressed in different tumors will elucidate whether different NFAT isoforms have distinct roles during the tumorigenesis process.
Acknowledgments
We thank Dr Miriam BF Werneck for critical reading of this manuscript. This work was supported by grants to JPBV from CNPq (307296/2011-3 and 476314/2012-7), FAPERJ (112.056/2012, 102.308/2013, 110.794/2013 and 101.147/2013) and INCT-Cancer (573806/2008-0 and 170.026/2008). G.P.M. was supported by a CNPq fellowship, Brazil and D.V.F. was supported by a FAPERJ fellowship, Brazil.
Glossary
- NFAT
nuclear factor of activated T cells
- CA-NFAT
constitutively active NFAT
- NHR
NFAT-homology region
- DBD
DNA-binding domain
- TAD
transactivation domain
- AAD
acidic activation domain
- AP-1
activator protein-1
- IL
interleukin
- ER
estrogen receptor
- MEF
myocyte enhancer factor
- IRF2BP2
interferon regulatory factor 2-binding protein 2
- CBP
CREB-binding protein
- TNF
tumor necrosis factor
- c-FLIP
cellular FLICE-like inhibitory protein
- STAT
signal transducer and activator of transcription
- DDIAS
DNA damage-induced apoptosis suppressor
- CsA
Cyclosporin A
- CDK
cyclin-dependent kinase
- CKI
CDK inhibitor
- SRR
serine-rich region
- SP
serine-proline motif
- NLS
nuclear localization signal
- NES
nuclear export sequence
- WT
wild type
The authors declare no conflict of interest.
Footnotes
Edited by G Melino
References
- Shaw JP, Utz PJ, Durand DB, Toole JJ, Emmel EA, Crabtree GR. Identification of a putative regulator of early T cell activation genes. Science 1988; 241: 202–205. [DOI] [PubMed] [Google Scholar]
- Shukla U, Hatani T, Nakashima K, Ogi K, Sada K. Tyrosine phosphorylation of 3BP2 regulates B cell receptor-mediated activation of NFAT. J Biol Chem 2009; 284: 33719–33728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanoni I, Ostuni R, Capuano G, Collini M, Caccia M, Ronchi AE et al. CD14 regulates the dendritic cell life cycle after LPS exposure through NFAT activation. Nature 2009; 460: 264–268. [DOI] [PubMed] [Google Scholar]
- Ranger AM, Gerstenfeld LC, Wang J, Kon T, Bae H, Gravallese EM et al. The nuclear factor of activated T cells (NFAT) transcription factor NFATp (NFATc2) is a repressor of chondrogenesis. J Exp Med 2000; 191: 9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho IC, Kim JH, Rooney JW, Spiegelman BM, Glimcher LH. A potential role for the nuclear factor of activated T cells family of transcriptional regulatory proteins in adipogenesis. Proc Natl Acad Sci USA 1998; 95: 15537–15541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz M, Schatz A, Wagner M, Michl P, Linhart T, Adler G et al. Overexpression of c-myc in pancreatic cancer caused by ectopic activation of NFATc1 and the Ca2+/calcineurin signaling pathway. EMBO J 2006; 25: 3714–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauliac S, Lopez-Rodriguez C, Shaw LM, Brown LF, Rao A, Toker A. The role of NFAT transcription factors in integrin-mediated carcinoma invasion. Nat Cell Biol 2002; 4: 540–544. [DOI] [PubMed] [Google Scholar]
- de la Pompa JL, Timmerman LA, Takimoto H, Yoshida H, Elia AJ, Samper E et al. Role of the NF-ATc transcription factor in morphogenesis of cardiac valves and septum. Nature 1998; 392: 182–186. [DOI] [PubMed] [Google Scholar]
- Macian F. NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol 2005; 5: 472–484. [DOI] [PubMed] [Google Scholar]
- Viola JP, Carvalho LD, Fonseca BP, Teixeira LK. NFAT transcription factors: from cell cycle to tumor development. Braz J Med Biol Res 2005; 38: 335–344. [DOI] [PubMed] [Google Scholar]
- Baksh S, DeCaprio JA, Burakoff SJ. Calcineurin regulation of the mammalian G0/G1 checkpoint element, cyclin dependent kinase 4. Oncogene 2000; 19: 2820–2827. [DOI] [PubMed] [Google Scholar]
- Hernandez GL, Volpert OV, Iniguez MA, Lorenzo E, Martinez-Martinez S, Grau R et al. Selective inhibition of vascular endothelial growth factor-mediated angiogenesis by cyclosporin A: roles of the nuclear factor of activated T cells and cyclooxygenase 2. J Exp Med 2001; 193: 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rodriguez C, Aramburu J, Rakeman AS, Rao A. NFAT5, a constitutively nuclear NFAT protein that does not cooperate with Fos and Jun. Proc Natl Acad Sci USA 1999; 96: 7214–7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol 1997; 15: 707–747. [DOI] [PubMed] [Google Scholar]
- Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev 2003; 17: 2205–2232. [DOI] [PubMed] [Google Scholar]
- Clipstone NA, Crabtree GR. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature 1992; 357: 695–697. [DOI] [PubMed] [Google Scholar]
- Shibasaki F, Price ER, Milan D, McKeon F. Role of kinases and the phosphatase calcineurin in the nuclear shuttling of transcription factor NF-AT4. Nature 1996; 382: 370–373. [DOI] [PubMed] [Google Scholar]
- Zhou B, Cron RQ, Wu B, Genin A, Wang Z, Liu S et al. Regulation of the murine Nfatc1 gene by NFATc2. J Biol Chem 2002; 277: 10704–10711. [DOI] [PubMed] [Google Scholar]
- Hock MB, Brown MA. Nuclear factor of activated T cells 2 transactivation in mast cells: a novel isoform-specific transactivation domain confers unique FcepsilonRI responsiveness. J Biol Chem 2003; 278: 26695–26703. [DOI] [PubMed] [Google Scholar]
- Triezenberg SJ. Structure and function of transcriptional activation domains. Curr Opin Genet Dev 1995; 5: 190–196. [DOI] [PubMed] [Google Scholar]
- Luo C, Burgeon E, Carew JA, McCaffrey PG, Badalian TM, Lane WS et al. Recombinant NFAT1 (NFATp) is regulated by calcineurin in T cells and mediates transcription of several cytokine genes. Mol Cell Biol 1996; 16: 3955–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura H, Garcia-Rodriguez C, Martinson H, Qin J, Virshup DM, Rao A. A conserved docking motif for CK1 binding controls the nuclear localization of NFAT1. Mol Cell Biol 2004; 24: 4184–4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuvpilo S, Avots A, Berberich-Siebelt F, Glockner J, Fischer C, Kerstan A et al. Multiple NF-ATc isoforms with individual transcriptional properties are synthesized in T lymphocytes. J Immunol 1999; 162: 7294–7301. [PubMed] [Google Scholar]
- Terui Y, Saad N, Jia S, McKeon F, Yuan J. Dual role of sumoylation in the nuclear localization and transcriptional activation of NFAT1. J Biol Chem 2004; 279: 28257–28265. [DOI] [PubMed] [Google Scholar]
- Nayak A, Glockner-Pagel J, Vaeth M, Schumann JE, Buttmann M, Bopp T et al. Sumoylation of the transcription factor NFATc1 leads to its subnuclear relocalization and interleukin-2 repression by histone deacetylase. J Biol Chem 2009; 284: 10935–10946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura R, Masuda ES, Naito Y, Imai S, Fujino T, Takano T et al. Carboxyl-terminal 15-amino acid sequence of NFATx1 is possibly created by tissue-specific splicing and is essential for transactivation activity in T cells. J Immunol 1998; 161: 3455–3463. [PubMed] [Google Scholar]
- Vihma H, Pruunsild P, Timmusk T. Alternative splicing and expression of human and mouse NFAT genes. Genomics 2008; 92: 279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells. Cell 2002; 109: S67–S79. [DOI] [PubMed] [Google Scholar]
- Ho IC, Hodge MR, Rooney JW, Glimcher LH. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell 1996; 85: 973–983. [DOI] [PubMed] [Google Scholar]
- Rengarajan J, Mowen KA, McBride KD, Smith ED, Singh H, Glimcher LH. Interferon regulatory factor 4 (IRF4) interacts with NFATc2 to modulate interleukin 4 gene expression. J Exp Med 2002; 195: 1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker EL, Nehmann N, Kampen E, Eibel H, Zipfel PF, Skerka C. Early growth response proteins (EGR) and nuclear factors of activated T cells (NFAT) form heterodimers and regulate proinflammatory cytokine gene expression. Nucleic Acids Res 2003; 31: 911–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini MP, Talora C, Seki T, Bolgan L, Dotto GP. Cross talk among calcineurin, Sp1/Sp3, and NFAT in control of p21(WAF1/CIP1) expression in keratinocyte differentiation. Proc Natl Acad Sci USA 2001; 98: 9575–9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Kogawa M, Wada S, Takayanagi H, Tsujimoto M, Katayama S et al. Essential role of p38 mitogen-activated protein kinase in cathepsin K gene expression during osteoclastogenesis through association of NFATc1 and PU.1. J Biol Chem 2004; 279: 45969–45979. [DOI] [PubMed] [Google Scholar]
- Sharma SM, Bronisz A, Hu R, Patel K, Mansky KC, Sif S et al. MITF and PU.1 recruit p38 MAPK and NFATc1 to target genes during osteoclast differentiation. J Biol Chem 2007; 282: 15921–15929. [DOI] [PubMed] [Google Scholar]
- Pham LV, Tamayo AT, Yoshimura LC, Lin-Lee YC, Ford RJ. Constitutive NF-kappaB and NFAT activation in aggressive B-cell lymphomas synergistically activates the CD154 gene and maintains lymphoma cell survival. Blood 2005; 106: 3940–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musaro A, McCullagh KJ, Naya FJ, Olson EN, Rosenthal N. IGF-1 induces skeletal myocyte hypertrophy through calcineurin in association with GATA-2 and NF-ATc1. Nature 1999; 400: 581–585. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Lu JR, Antos CL, Markham B, Richardson J, Robbins J et al. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 1998; 93: 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein-Hessling S, Bopp T, Jha MK, Schmidt A, Miyatake S, Schmitt E et al. Cyclic AMP-induced chromatin changes support the NFATc-mediated recruitment of GATA-3 to the interleukin 5 promoter. J Biol Chem 2008; 283: 31030–31037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge MR, Chun HJ, Rengarajan J, Alt A, Lieberson R, Glimcher LH. NF-AT-Driven interleukin-4 transcription potentiated by NIP45. Science 1996; 274: 1903–1905. [DOI] [PubMed] [Google Scholar]
- Kao SC, Wu H, Xie J, Chang CP, Ranish JA, Graef IA et al. Calcineurin/NFAT signaling is required for neuregulin-regulated Schwann cell differentiation. Science 2009; 323: 651–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macian F, Lopez-Rodriguez C, Rao A. Partners in transcription: NFAT and AP-1. Oncogene 2001; 20: 2476–2489. [DOI] [PubMed] [Google Scholar]
- Zhu C, Rao K, Xiong H, Gagnidze K, Li F, Horvath C et al. Activation of the murine interleukin-12 p40 promoter by functional interactions between NFAT and ICSBP. J Biol Chem 2003; 278: 39372–39382. [DOI] [PubMed] [Google Scholar]
- Yang TT, Chow CW. Transcription cooperation by NFAT.C/EBP composite enhancer complex. J Biol Chem 2003; 278: 15874–15885. [DOI] [PubMed] [Google Scholar]
- Nguyen TN, Kim LJ, Walters RD, Drullinger LF, Lively TN, Kugel JF et al. The C-terminal region of human NFATc2 binds cJun to synergistically activate interleukin-2 transcription. Mol Immunol 2010; 47: 2314–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geil WM, Yen A. Nuclear Raf-1 kinase regulates the CXCR5 promoter by associating with NFATc3 to drive retinoic acid-induced leukemic cell differentiation. FEBS J 2014; 281: 1170–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart S, Chen NM, Zhang JS, Billadeau DD, Gaisina I, Kozikowski AP et al. GSK-3beta governs inflammation-induced NFATc2 signaling hubs to promote pancreatic cancer progression. Mol Cancer Ther 2016; 15: 491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SW, Kang BY, Kim TS. Inhibition of interleukin-4 production in CD4+ T cells by peroxisome proliferator-activated receptor-gamma (PPAR-gamma) ligands: involvement of physical association between PPAR-gamma and the nuclear factor of activated T cells transcription factor. Mol Pharmacol 2003; 64: 1169–1179. [DOI] [PubMed] [Google Scholar]
- Yang XY, Wang LH, Chen T, Hodge DR, Resau JH, DaSilva L et al. Activation of human T lymphocytes is inhibited by peroxisome proliferator-activated receptor gamma (PPARgamma) agonists. PPARgamma co-association with transcription factor NFAT. J Biol Chem 2000; 275: 4541–4544. [DOI] [PubMed] [Google Scholar]
- Dai YS, Xu J, Molkentin JD. The DnaJ-related factor Mrj interacts with nuclear factor of activated T cells c3 and mediates transcriptional repression through class II histone deacetylase recruitment. Mol Cell Biol 2005; 25: 9936–9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelli E, Dastrange M, Oukka M. Foxp3 interacts with nuclear factor of activated T cells and NF-kappa B to repress cytokine gene expression and effector functions of T helper cells. Proc Natl Acad Sci USA 2005; 102: 5138–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Borde M, Heissmeyer V, Feuerer M, Lapan AD, Stroud JC et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell 2006; 126: 375–387. [DOI] [PubMed] [Google Scholar]
- Bodor J, Habener JF. Role of transcriptional repressor ICER in cyclic AMP-mediated attenuation of cytokine gene expression in human thymocytes. J Biol Chem 1998; 273: 9544–9551. [DOI] [PubMed] [Google Scholar]
- Han J, Kim M, Oum JH, Myung H, Lee SW, Jeong S et al. RACK-1, a receptor for activated C kinase, interacts with the transcription factor NFAT and represses its transactivation. Mol Cells 2002; 14: 420–424. [PubMed] [Google Scholar]
- Garcia-Rodriguez C, Rao A. Nuclear factor of activated T cells (NFAT)-dependent transactivation regulated by the coactivators p300/CREB-binding protein (CBP). J Exp Med 1998; 187: 2031–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avots A, Buttmann M, Chuvpilo S, Escher C, Smola U, Bannister AJ et al. CBP/p300 integrates Raf/Rac-signaling pathways in the transcriptional induction of NF-ATc during T cell activation. Immunity 1999; 10: 515–524. [DOI] [PubMed] [Google Scholar]
- Yang T, Davis RJ, Chow CW. Requirement of two NFATc4 transactivation domains for CBP potentiation. J Biol Chem 2001; 276: 39569–39576. [DOI] [PubMed] [Google Scholar]
- Zhang H, Xie X, Zhu X, Zhu J, Hao C, Lu Q et al. Stimulatory cross-talk between NFAT3 and estrogen receptor in breast cancer cells. J Biol Chem 2005; 280: 43188–43197. [DOI] [PubMed] [Google Scholar]
- Qin X, Wang XH, Yang ZH, Ding LH, Xu XJ, Cheng L et al. Repression of NFAT3 transcriptional activity by estrogen receptors. Cell Mol Life Sci 2008; 65: 2752–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn HD, Chatila TA, Liu JO. Integration of calcineurin and MEF2 signals by the coactivator p300 during T-cell apoptosis. EMBO J 2000; 19: 4323–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaeser F, Ho N, Prywes R, Chatila TA. Ca(2+)-dependent gene expression mediated by MEF2 transcription factors. J Biol Chem 2000; 275: 197–209. [DOI] [PubMed] [Google Scholar]
- Wu H, Naya FJ, McKinsey TA, Mercer B, Shelton JM, Chin ER et al. MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fiber type. EMBO J 2000; 19: 1963–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro FR, Ramalho-Oliveira R, Mognol GP, Viola JP. Interferon regulatory factor 2 binding protein 2 is a new NFAT1 partner and represses its transcriptional activity. Mol Cell Biol 2011; 31: 2889–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojsa B, Mora S, Bossowski JP, Lassot I, Desagher S. Control of neuronal apoptosis by reciprocal regulation of NFATc3 and Trim17. Cell Death Differ 2015; 22: 274–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbs BK, Cruz AL, Werneck MB, Mognol GP, Viola JP. Dual roles for NFAT transcription factor genes as oncogenes and tumor suppressors. Mol Cell Biol 2008; 28: 7168–7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomono M, Toyoshima K, Ito M, Amano H. Calcineurin is essential for DNA synthesis in Swiss 3T3 fibroblasts. Biochem J 1996; 317: 675–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fric J, Lim CX, Mertes A, Lee BT, Vigano E, Chen J et al. Calcium and calcineurin-NFAT signaling regulate granulocyte-monocyte progenitor cell cycle via Flt3-L. Stem Cells 2014; 32: 3232–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xanthoudakis S, Viola JP, Shaw KT, Luo C, Wallace JD, Bozza PT et al. An enhanced immune response in mice lacking the transcription factor NFAT1. Science 1996; 272: 892–895. [DOI] [PubMed] [Google Scholar]
- Hodge MR, Ranger AM, Charles de la Brousse F, Hoey T, Grusby MJ, Glimcher LH. Hyperproliferation and dysregulation of IL-4 expression in NF-ATp-deficient mice. Immunity 1996; 4: 397–405. [DOI] [PubMed] [Google Scholar]
- Caetano MS, Vieira-de-Abreu A, Teixeira LK, Werneck MB, Barcinski MA, Viola JP. NFATC2 transcription factor regulates cell cycle progression during lymphocyte activation: evidence of its involvement in the control of cyclin gene expression. FASEB J 2002; 16: 1940–1942. [DOI] [PubMed] [Google Scholar]
- Ranger AM, Grusby MJ, Hodge MR, Gravallese EM, de la Brousse FC, Hoey T et al. The transcription factor NF-ATc is essential for cardiac valve formation. Nature 1998; 392: 186–190. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Nishina H, Takimoto H, Marengere LE, Wakeham AC, Bouchard D et al. The transcription factor NF-ATc1 regulates lymphocyte proliferation and Th2 cytokine production. Immunity 1998; 8: 115–124. [DOI] [PubMed] [Google Scholar]
- Carvalho LD, Teixeira LK, Carrossini N, Caldeira AT, Ansel KM, Rao A et al. The NFAT1 transcription factor is a repressor of cyclin A2 gene expression. Cell Cycle 2007; 6: 1789–1795. [DOI] [PubMed] [Google Scholar]
- Karpurapu M, Wang D, Singh NK, Li Q, Rao GN. NFATc1 targets cyclin A in the regulation of vascular smooth muscle cell multiplication during restenosis. J Biol Chem 2008; 283: 26577–26590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpurapu M, Wang D, Van Quyen D, Kim TK, Kundumani-Sridharan V, Pulusani S et al. Cyclin D1 is a bona fide target gene of NFATc1 and is sufficient in the mediation of injury-induced vascular wall remodeling. J Biol Chem 2010; 285: 3510–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundumani-Sridharan V, Singh NK, Kumar S, Gadepalli R, Rao GN. Nuclear factor of activated T cells c1 mediates p21-activated kinase 1 activation in the modulation of chemokine-induced human aortic smooth muscle cell F-actin stress fiber formation, migration, and proliferation and injury-induced vascular wall remodeling. J Biol Chem 2013; 288: 22150–22162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart S, Chen NM, Siveke JT, Konig A, Zhang JS, Singh SK et al. Inflammation-induced NFATc1-STAT3 transcription complex promotes pancreatic cancer initiation by KrasG12D. Cancer Discov 2014; 4: 688–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart S, Glesel E, Singh G, Chen NM, Reutlinger K, Zhang J et al. Restricted heterochromatin formation links NFATc2 repressor activity with growth promotion in pancreatic cancer. Gastroenterology 2012; 142: 388–398.e1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig A, Linhart T, Schlengemann K, Reutlinger K, Wegele J, Adler G et al. NFAT-induced histone acetylation relay switch promotes c-Myc-dependent growth in pancreatic cancer cells. Gastroenterology 2010; 138: 1189–1199.e1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Singh SK, Konig A, Reutlinger K, Nye MD, Adhikary T et al. Sequential activation of NFAT and c-Myc transcription factors mediates the TGF-beta switch from a suppressor to a promoter of cancer cell proliferation. J Biol Chem 2010; 285: 27241–27250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mognol GP, de Araujo-Souza PS, Robbs BK, Teixeira LK, Viola JP. Transcriptional regulation of the c-Myc promoter by NFAT1 involves negative and positive NFAT-responsive elements. Cell Cycle 2012; 11: 1014–1028. [DOI] [PubMed] [Google Scholar]
- Ranger AM, Oukka M, Rengarajan J, Glimcher LH. Inhibitory function of two NFAT family members in lymphoid homeostasis and Th2 development. Immunity 1998; 9: 627–635. [DOI] [PubMed] [Google Scholar]
- Ranger AM, Hodge MR, Gravallese EM, Oukka M, Davidson L, Alt FW et al. Delayed lymphoid repopulation with defects in IL-4-driven responses produced by inactivation of NF-ATc. Immunity 1998; 8: 125–134. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S, Deb J, Patra AK, Thuy Pham DA, Chen W, Vaeth M et al. NFATc1 affects mouse splenic B cell function by controlling the calcineurin—NFAT signaling network. J Exp Med 2011; 208: 823–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef IA, Chen F, Chen L, Kuo A, Crabtree GR. Signals transduced by Ca(2+)/calcineurin and NFATc3/c4 pattern the developing vasculature. Cell 2001; 105: 863–875. [DOI] [PubMed] [Google Scholar]
- Benedito AB, Lehtinen M, Massol R, Lopes UG, Kirchhausen T, Rao A et al. The transcription factor NFAT3 mediates neuronal survival. J Biol Chem 2005; 280: 2818–2825. [DOI] [PubMed] [Google Scholar]
- Quadrato G, Benevento M, Alber S, Jacob C, Floriddia EM, Nguyen T et al. Nuclear factor of activated T cells (NFATc4) is required for BDNF-dependent survival of adult-born neurons and spatial memory formation in the hippocampus. Proc Natl Acad Sci USA 2012; 109: E1499–E1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oukka M, Ho IC, de la Brousse FC, Hoey T, Grusby MJ, Glimcher LH. The transcription factor NFAT4 is involved in the generation and survival of T cells. Immunity 1998; 9: 295–304. [DOI] [PubMed] [Google Scholar]
- Ran Y, Wu H, Wei L, Yu X, Chen J, Li S et al. NFATc3 pathway participates in the process that 15-LO/15-HETE protects pulmonary artery smooth muscle cells against apoptosis during hypoxia. J Recept Signal Transduct Res 2014; 34: 270–282. [DOI] [PubMed] [Google Scholar]
- Wang K, Zhang DL, Long B, An T, Zhang J, Zhou LY et al. NFAT4-dependent miR-324-5p regulates mitochondrial morphology and cardiomyocyte cell death by targeting Mtfr1. Cell Death Dis 2015; 6: e2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmler M, Thome M, Hahne M, Schneider P, Hofmann K, Steiner V et al. Inhibition of death receptor signals by cellular FLIP. Nature 1997; 388: 190–195. [DOI] [PubMed] [Google Scholar]
- Ueffing N, Schuster M, Keil E, Schulze-Osthoff K, Schmitz I. Up-regulation of c-FLIP short by NFAT contributes to apoptosis resistance of short-term activated T cells. Blood 2008; 112: 690–698. [DOI] [PubMed] [Google Scholar]
- Xiang Z, Ahmed AA, Moller C, Nakayama K, Hatakeyama S, Nilsson G. Essential role of the prosurvival bcl-2 homologue A1 in mast cell survival after allergic activation. J Exp Med 2001; 194: 1561–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulleras E, Karlberg M, Moller Westerberg C, Alfredsson J, Gerondakis S, Strasser A et al. NFAT but not NF-kappaB is critical for transcriptional induction of the prosurvival gene A1 after IgE receptor activation in mast cells. Blood 2008; 111: 3081–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im JY, Lee KW, Won KJ, Kim BK, Ban HS, Yoon SH et al. DNA damage-induced apoptosis suppressor (DDIAS), a novel target of NFATc1, is associated with cisplatin resistance in lung cancer. Biochim Biophys Acta 2016; 1863: 40–49. [DOI] [PubMed] [Google Scholar]
- Won KJ, Im JY, Yun CO, Chung KS, Kim YJ, Lee JS et al. Human Noxin is an anti-apoptotic protein in response to DNA damage of A549 non-small cell lung carcinoma. Int J Cancer 2014; 134: 2595–2604. [DOI] [PubMed] [Google Scholar]
- Latinis KM, Carr LL, Peterson EJ, Norian LA, Eliason SL, Koretzky GA. Regulation of CD95 (Fas) ligand expression by TCR-mediated signaling events. J Immunol 1997; 158: 4602–4611. [PubMed] [Google Scholar]
- Holtz-Heppelmann CJ, Algeciras A, Badley AD, Paya CV. Transcriptional regulation of the human FasL promoter-enhancer region. J Biol Chem 1998; 273: 4416–4423. [DOI] [PubMed] [Google Scholar]
- Rengarajan J, Mittelstadt PR, Mages HW, Gerth AJ, Kroczek RA, Ashwell JD et al. Sequential involvement of NFAT and Egr transcription factors in FasL regulation. Immunity 2000; 12: 293–300. [DOI] [PubMed] [Google Scholar]
- Pedrosa AM, Weinlich R, Mognol GP, Robbs BK, Viola JP, Campa A et al. Melatonin protects CD4+ T cells from activation-induced cell death by blocking NFAT-mediated CD95 ligand upregulation. J immunol 2010; 184: 3487–3494. [DOI] [PubMed] [Google Scholar]
- Liu ZG, Smith SW, McLaughlin KA, Schwartz LM, Osborne BA. Apoptotic signals delivered through the T-cell receptor of a T-cell hybrid require the immediate-early gene nur77. Nature 1994; 367: 281–284. [DOI] [PubMed] [Google Scholar]
- Youn HD, Liu JO. Cabin1 represses MEF2-dependent Nur77 expression and T cell apoptosis by controlling association of histone deacetylases and acetylases with MEF2. Immunity 2000; 13: 85–94. [DOI] [PubMed] [Google Scholar]
- Goldfeld AE, McCaffrey PG, Strominger JL, Rao A. Identification of a novel cyclosporin-sensitive element in the human tumor necrosis factor alpha gene promoter. J Exp Med 1993; 178: 1365–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey PG, Goldfeld AE, Rao A. The role of NFATp in cyclosporin A-sensitive tumor necrosis factor-alpha gene transcription. J Biol Chem 1994; 269: 30445–30450. [PubMed] [Google Scholar]
- Tsai EY, Jain J, Pesavento PA, Rao A, Goldfeld AE. Tumor necrosis factor alpha gene regulation in activated T cells involves ATF-2/Jun and NFATp. Mol Cell Biol 1996; 16: 459–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai EY, Yie J, Thanos D, Goldfeld AE. Cell-type-specific regulation of the human tumor necrosis factor alpha gene in B cells and T cells by NFATp and ATF-2/JUN. Mol Cell Biol 1996; 16: 5232–5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esensten JH, Tsytsykova AV, Lopez-Rodriguez C, Ligeiro FA, Rao A, Goldfeld AE. NFAT5 binds to the TNF promoter distinctly from NFATp, c, 3 and 4, and activates TNF transcription during hypertonic stress alone. Nucleic Acids Res 2005; 33: 3845–3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Takeuchi A, Sharma S. Characterization of a new isoform of the NFAT (nuclear factor of activated T cells) gene family member NFATc. J Biol Chem 1996; 271: 20914–20921. [DOI] [PubMed] [Google Scholar]
- Oum JH, Han J, Myung H, Hleb M, Sharma S, Park J. Molecular mechanism of NFAT family proteins for differential regulation of the IL-2 and TNF-alpha promoters. Mol Cells 2002; 13: 77–84. [PubMed] [Google Scholar]
- Kaminuma O, Kitamura F, Kitamura N, Hiroi T, Miyoshi H, Miyawaki A et al. Differential contribution of NFATc2 and NFATc1 to TNF-alpha gene expression in T cells. J Immunol 2008; 180: 319–326. [DOI] [PubMed] [Google Scholar]
- Chuvpilo S, Jankevics E, Tyrsin D, Akimzhanov A, Moroz D, Jha MK et al. Autoregulation of NFATc1/A expression facilitates effector T cells to escape from rapid apoptosis. Immunity 2002; 16: 881–895. [DOI] [PubMed] [Google Scholar]
- Lucena PI, Faget DV, Pachulec E, Robaina MC, Klumb CE, Robbs BK et al. NFAT2 isoforms differentially regulate gene expression, cell death, and transformation through alternative N-terminal domains. Mol Cell Biol 2015; 36: 119–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassot I, Robbins I, Kristiansen M, Rahmeh R, Jaudon F, Magiera MM et al. Trim17, a novel E3 ubiquitin-ligase, initiates neuronal apoptosis. Cell Death Differ 2010; 17: 1928–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medyouf H, Ghysdael J. The calcineurin/NFAT signaling pathway: a novel therapeutic target in leukemia and solid tumors. Cell Cycle 2008; 7: 297–303. [DOI] [PubMed] [Google Scholar]
- Neal JW, Clipstone NA. A constitutively active NFATc1 mutant induces a transformed phenotype in 3T3-L1 fibroblasts. J Biol Chem 2003; 278: 17246–17254. [DOI] [PubMed] [Google Scholar]
- Marafioti T, Pozzobon M, Hansmann ML, Ventura R, Pileri SA, Roberton H et al. The NFATc1 transcription factor is widely expressed in white cells and translocates from the cytoplasm to the nucleus in a subset of human lymphomas. Br J Haematol 2005; 128: 333–342. [DOI] [PubMed] [Google Scholar]
- Medyouf H, Alcalde H, Berthier C, Guillemin MC, dos Santos NR, Janin A et al. Targeting calcineurin activation as a therapeutic strategy for T-cell acute lymphoblastic leukemia. Nat Med 2007; 13: 736–741. [DOI] [PubMed] [Google Scholar]
- Kawahara T, Kashiwagi E, Ide H, Li Y, Zheng Y, Miyamoto Y et al. Cyclosporine A and tacrolimus inhibit bladder cancer growth through down-regulation of NFATc1. Oncotarget 2015; 6: 1582–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi MK, Deane NG, Zhu J, An H, Mima S, Wang X et al. Nuclear factor of activated T-cell activity is associated with metastatic capacity in colon cancer. Cancer Res 2014; 74: 6947–6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manda KR, Tripathi P, Hsi AC, Ning J, Ruzinova MB, Liapis H et al. NFATc1 promotes prostate tumorigenesis and overcomes PTEN loss-induced senescence. Oncogene 2015; (doi:10.1038/onc.2015.389; e-pub ahead of print). [DOI] [PMC free article] [PubMed]
- Kawahara T, Kashiwagi E, Ide H, Li Y, Zheng Y, Ishiguro H et al. The role of NFATc1 in prostate cancer progression: cyclosporine A and tacrolimus inhibit cell proliferation, migration, and invasion. Prostate 2015; 75: 573–584. [DOI] [PubMed] [Google Scholar]
- Comba A, Almada LL, Tolosa EJ, Iguchi E, Marks DL, Vara Messler M et al. Nuclear factor of activated T cells-dependent down-regulation of the transcription factor glioma-associated protein 1 (GLI1) underlies the growth inhibitory properties of arachidonic acid. J Biol Chem 2016; 291: 1933–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faget DV, Lucena PI, Robbs BK, Viola JP. NFAT1 C-terminal domains are necessary but not sufficient for inducing cell death. PLoS One 2012; 7: e47868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May SL, Zhou Q, Lewellen M, Carter CM, Coffey D, Highfill SL et al. Nfatc2 and Tob1 have non-overlapping function in T cell negative regulation and tumorigenesis. PLoS One 2014; 9: e100629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoeli-Lerner M, Yiu GK, Rabinovitz I, Erhardt P, Jauliac S, Toker A. Akt blocks breast cancer cell motility and invasion through the transcription factor NFAT. Mol Cell 2005; 20: 539–550. [DOI] [PubMed] [Google Scholar]
- Yoeli-Lerner M, Chin YR, Hansen CK, Toker A. Akt/protein kinase b and glycogen synthase kinase-3beta signaling pathway regulates cell migration through the NFAT1 transcription factor. Mol Cancer Res 2009; 7: 425–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie X, Han S, Meng L, Wang Y, Wu A. NFAT1 is highly expressed in, and regulates the invasion of, glioblastoma multiforme cells. PLoS One 2013; 8: e66008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quang CT, Leboucher S, Passaro D, Fuhrmann L, Nourieh M, Vincent-Salomon A et al. The calcineurin/NFAT pathway is activated in diagnostic breast cancer cases and is essential to survival and metastasis of mammary cancer cells. Cell Death Dis 2015; 6: e1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach K, Daniel C, Lehr HA, Nikolaev A, Gerlach T, Atreya R et al. Transcription factor NFATc2 controls the emergence of colon cancer associated with IL-6-dependent colitis. Cancer Res 2012; 72: 4340–4350. [DOI] [PubMed] [Google Scholar]
- Perotti V, Baldassari P, Molla A, Vegetti C, Bersani I, Maurichi A et al. NFATc2 is an intrinsic regulator of melanoma dedifferentiation. Oncogene 2015; (doi:10.1038/onc.2015.355; e-pub ahead of print). [DOI] [PubMed]
- Hessmann E, Zhang JS, Chen NM, Hasselluhn M, Liou GY, Storz P et al. NFATc4 regulates Sox9 gene expression in acinar cell plasticity and pancreatic cancer initiation. Stem Cells Int 2016; 2016: 5272498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Chen NM, Hessmann E, Siveke J, Lahmann M, Singh G et al. Antithetical NFATc1-Sox2 and p53-miR200 signaling networks govern pancreatic cancer cell plasticity. EMBO J 2015; 34: 517–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin JJ, Nag S, Wang W, Zhou J, Zhang WD, Wang H et al. NFAT as cancer target: mission possible? Biochim Biophys Acta 2014; 1846: 297–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzelder SK, Michel C, von Bonin M, Rehberger M, Hessmann E, Inselmann S et al. NFATc1 as a therapeutic target in FLT3-ITD-positive AML. Leukemia 2015; 29: 1470–1477. [DOI] [PubMed] [Google Scholar]
- Ohoka Y, Yokota A, Takeuchi H, Maeda N, Iwata M. Retinoic acid-induced CCR9 expression requires transient TCR stimulation and cooperativity between NFATc2 and the retinoic acid receptor/retinoid X receptor complex. J Immunol 2011; 186: 733–744. [DOI] [PubMed] [Google Scholar]
- Patra AK, Avots A, Zahedi RP, Schuler T, Sickmann A, Bommhardt U et al. An alternative NFAT-activation pathway mediated by IL-7 is critical for early thymocyte development. Nat Immunol 2013; 14: 127–135. [DOI] [PubMed] [Google Scholar]
- Kawamura T, Ono K, Morimoto T, Akao M, Iwai-Kanai E, Wada H et al. Endothelin-1-dependent nuclear factor of activated T lymphocyte signaling associates with transcriptional coactivator p300 in the activation of the B cell leukemia-2 promoter in cardiac myocytes. Circ Res 2004; 94: 1492–1499. [DOI] [PubMed] [Google Scholar]
- Wang Q, Zhou Y, Weiss HL, Chow CW, Evers BM. NFATc1 regulation of TRAIL expression in human intestinal cells. PLoS One 2011; 6: e19882. [DOI] [PMC free article] [PubMed] [Google Scholar]