Summary
Mitochondrial ribosomes of Trypanosoma brucei are composed of 9S and 12S rRNAs, eubacterial-type ribosomal proteins, polypeptides lacking discernible motifs and approximately 20 pentatricopeptide repeat (PPR) RNA binding proteins. Several PPRs also populate the polyadenylation complex; among these, KPAF1 and KPAF2 function as general mRNA 3′ adenylation/uridylation factors. The A/U-tail enables mRNA binding to the small ribosomal subunit and is essential for translation. The presence of A/U-tail also correlates with requirement for translation of certain mRNAs in mammalian and insect parasite stages. Here, we inquired whether additional PPRs activate translation of individual mRNAs. Proteomic analysis identified KRIPP1 and KRIPP8 as components of the small ribosomal subunit in mammalian and insect forms, but also revealed their association with the polyadenylation complex in the latter. RNAi knockdowns demonstrated essential functions of KRIPP1 and KRIPP8 in the actively respiring insect stage, but not in the mammalian stage. In the KRIPP1 knockdown, A/U-tailed mRNA encoding cytochrome c oxidase subunit 1 declined concomitantly with the de novo synthesis of this subunit whereas polyadenylation and translation of cyb mRNA were unaffected. In contrast, the KRIPP8 knockdown inhibited A/U-tailing and translation of both CO1 and cyb mRNAs. Our findings indicate that ribosome-associated PPRs may selectively activate mRNAs for translation.
Introduction
Trypanosomes belong to a kinetoplastid group of protists characterized by the presence of the kinetoplast – a dense nucleoprotein structure containing the mitochondrial genome. The kinetoplast DNA is composed of catenated maxicircles and minicircles; the former encode ribosomal RNAs (mt-rRNAs), a single ribosomal protein PRS12, and subunits of respiratory complexes while the latter carry guide RNA genes. Polycistronic precursors are transcribed from maxicircles and then processed into rRNA and individual pre-mRNAs by an unknown mechanism. In Trypanosoma brucei, 12 of the 18 annotated pre-mRNAs are referred to as ‘pre-edited’ as they must undergo U-insertion/deletion editing to acquire open reading frames; for the remaining six ‘unedited’ transcripts the functional protein coding sequence is encoded in the maxicircle. Mitochondrial mRNAs possess short 5′ untranslated regions (UTRs) lacking apparent ribosome binding sites and are extensively 3′ modified by addition of adenosine and uridine residues (Bhat et al., 1990). The mRNA 3′ extension processes can be defined as A-tailing, in which KPAP1 poly(A) polymerase adds 20–30 adenylate residues (Etheridge et al., 2008), and A/U-tailing in which KPAP1 and RET1 TUTase aided by KPAF1/KPAF2 pentatricopeptide repeat (PPR) proteins further extend the A-tail into 200–300 nucleotide long A/U heteropolymer (Aphasizheva et al., 2011). Existing evidence indicates that the short A-tail is required for mRNA stabilization while the A/U-tail is critical for mRNA binding to the small ribosomal subunit. It appears that only A/U-tailed mRNAs are populated by ribosomes (Aphasizheva et al., 2011; Aphasizheva et al., 2013; Ridlon et al., 2013).
In the actively respiring developmental form of T. brucei (insect or procyclic form, PF), the length of the 3′ extension correlates with the mRNA's editing status: pre-edited and partially edited mRNAs possess short A-tails while fully edited mRNAs exist as short A-tailed and long A/U-tailed populations (Etheridge et al., 2008). Similarly to fully edited mRNAs, most unedited mRNAs bear short A-tails or extended A/U-tails. Remarkably, the mRNA's A/U-tailing state correlates with anticipated production of respective proteins in procyclic and bloodstream (BF) developmental forms. For example, in the oxidative phosphorylation-deficient mitochondrion of BF parasites, ATP is produced via substrate level phosphorylation and is hydrolyzed by ATP synthase to generate mitochondrial trans-membrane potential (Schnaufer et al., 2005; Brown et al., 2006). Therefore, translation of the mitochondrial mRNA encoding subunit 6 (A6) of the F1FO-ATPase complex is expected to occur (Schnaufer et al., 2001). Likewise, functional ribosome in PF and BF parasites likely requires mitochondrially encoded small subunit ribosomal protein S12 (RPS12), which is highly conserved among eubacteria (Aphasizheva et al., 2013; Lin et al., 2015). Coincidentally, pan-edited A6 and RPS12 mRNAs are A/U-tailed in the BF parasites whereas mRNAs encoding apparently dispensable subunits of NADH dehydrogenase, cytochrome b and cytochrome c oxidase are not. Accordingly, most fully edited and unedited mRNAs possess long A/U-tails in actively respiring PF parasites (Aphasizheva et al., 2011). Proteomic studies of RNA editing, polyadenylation and translation processes revealed extensive physical interactions between macromolecular complexes involved in mRNA processing and protein synthesis (Etheridge et al., 2008; Weng et al., 2008; Zikova et al., 2008; Aphasizheva et al., 2014; Aphasizheva et al., 2011). In particular, polyadenylation complex has been shown to interact with RNA editing substrate binding complex and with the small ribosomal subunit (SSU), and share several PPR RNA binging proteins with the latter (Aphasizheva et al., 2014). Among PPR proteins identified in the polyadenylation complex, only KPAF1 and KPAF2 have been characterized as a stable heterodimer that stimulates mRNA polyadenylation/uridylation reaction (Aphasizheva et al., 2011).
Phylogenetic analyses suggest that kinetoplastids and euglenoids may have acquired plastids by endosymbiosis before their divergence (Hannaert et al., 2003). The secondary loss of photosynthetic organelle in kinetoplastids is indicated by retention of numerous plant-like genes, such as PPR proteins (Delannoy et al., 2007). These polypeptides contain arrays of 35 amino acids repeats and are often involved in a sequence-specific RNA binding serving as adaptors for RNA editing enzymes in plant organelles (Delannoy et al., 2007; Yin et al., 2013). In T. brucei, genome-wide searches identified 39 PPR proteins, with approximately half of these having been detected in affinity-purified ribosomal particles and termed kinetoplast ribosomal PPR proteins, KRIPPs (Aphasizheva et al., 2011). Indeed, a reported decline of 9S or 12S rRNA levels upon repression of a several PPR genes (Mingler et al., 2006; Pusnik et al., 2007) correlates with association of respective proteins with either small or large ribosomal subunit (Zikova et al., 2008; Aphasizheva et al., 2011). The related tetratricopeptide repeat containing proteins (TPR) have also been detected in the mitochondrial ribosome and termed kinetoplast ribosomal TPR proteins, KRITs (Aphasizheva et al., 2011).
In this work, we investigated functions of two PPR proteins previously annotated as KRIPP1 and KRIPP8 (Aphasizheva et al., 2011). These proteins have been identified in the affinity purified polyadenylation complex (Etheridge et al., 2008) and in the small ribosomal subunit (Zikova et al., 2008; Aphasizheva et al., 2011). We show that protein composition of KRIPP1- and KRIPP8-associated complexes isolated from procyclic parasites resembles that of the small ribosomal subunit purified via TAP-tagged protein S17. Furthermore, we provide the first proteomic analysis of the mitochondrial ribosomal subunits from the bloodstream form and demonstrate that KRIPP1 and KRIPP8 are present at similar levels in the small ribosomal subunit from PF and BF, and in the assembled ribosome from PF parasites. However, KRIPP1 and KRIPP8 appear to be absent in the assembled ribosome isolated from BF cells. Inducible RNAi knockdowns of KRIPP1 triggered selective downregulation of 9S small ribosomal RNA although the integrity of the assembled ribosome remained uncompromised. In KRIPP1 knockdown, steady-state levels of translationally competent A/U-tailed mRNAs encoding cytochrome c oxidase subunit 1 declined concomitantly with CO1 protein synthesis while most other mRNAs, such as cytochrome b mRNA (cyb), remained largely unaffected. KRIPP8 RNAi, conversely, caused downregulation of 9S and 12S mitochondrial rRNAs, A/U-tailed CO1 and cytochrome b mRNAs, and their translation. Nonetheless, we found that two mRNAs whose translation is expected to be essential for mitochondrial function in the bloodstream form, A6 and RPS12, were properly edited and 3′ A/U-tailed in KRIPP1 and KRIPP8 RNAi cell lines. In agreement with these observations, expression of KRIPP1 and KRIPP8 was found to be essential for the actively respiring procyclic form of the parasite, but not for the bloodstream developmental form. Collectively, our findings provide further evidence for the existence of the SSU-like particle, previously termed 45S* SSU (Ridlon et al., 2013), which is responsible for translational activation of specific transcripts, for example, CO1 mRNA. The 45S* SSU-like particle most likely participates in ribosome assembly, but its abundance is developmentally downregulated in the bloodstream form. It follows that the mRNAs required for cell viability in both developmental stages, such as A6 and RPS12, are not subject to 45S*-dependent regulation and are translated constitutively. We also find that PPR proteins at the interface of SSU-like particle and the polyadenylation complex, such as KRIPP1, are essential for translational mRNA activation in a developmental stage-specific manner.
Results
KRIPP1- and KRPP8-associated complexes resemble the small ribosomal subunit
KRIPP1 and KRIPP8 were identified in the affinity purified small ribosomal subunit (Zikova et al., 2008; Aphasizheva et al., 2011) and in the polyadenylation complex (Etheridge et al., 2008) from procyclic form of T. brucei. To unambiguously assign these PPR proteins to either SSU or to the KPAP1 complex, complete coding sequences of KRIPP1, KRIPP8, S17 and L3 ribosomal protein, and KPAP1 poly(A) polymerase were overexpressed under control of tet-operator as C-terminally TAP-tagged fusion proteins (Jensen et al., 2007). Tandem affinity purification from enriched mitochondrial fractions was carried out under uniform conditions and resultant samples were analyzed by SDS PAGE (Fig. 1A) and by LC-MS/MS (Fig. 1B and Supporting Information Table S1). A label-free quantitative MS strategy was applied to calculate the relative abundance of a given protein in each complex and to quantify interactions based on distributed normalized spectral abundance factor (dNSAF) (Zhang et al., 2010; Fang et al., 2012). Quantitative clustering of protein interaction network revealed a significant proteomic overlap between all five complexes that could be separated into ‘weak’, ‘moderate’ and ‘strong’ interactions based on relative abundance of shared peptides (Fig. 1B). Within pentavalent contacts, proteins with highest representation in KRIPP1, KRIPP8 and S17 have been grouped together. Such clustering is consistent with previously reported interactions of ribosomal subunits with each other and between polyadenylation complex and the ribosome (Maslov et al., 2006; Maslov et al., 2007; Etheridge et al., 2008; Zikova et al., 2008; Aphasizheva et al., 2011; Aphasizheva et al., 2014). However, within these networks equally strong tripartite interactions between KRIPP1-, KRIPP8- and S17-associated complexes were also apparent as well as nearly identical binary contacts of KRIPP1, KRIPP8 and S17 with the large ribosomal subunit. The important difference was observed in representation of KRIPP1 and KRIPP8 versus S17 in the KPAP1 complex: whereas both PPR proteins were enriched in the polyadenylation complex, S17 protein was markedly absent in the latter. Collectively, these results typify KRIPP1 and KRIPP8 polypeptides as components of the small ribosomal subunit-like particle that are positioned at the interface with the polyadenylation complex.
Fig. 1.
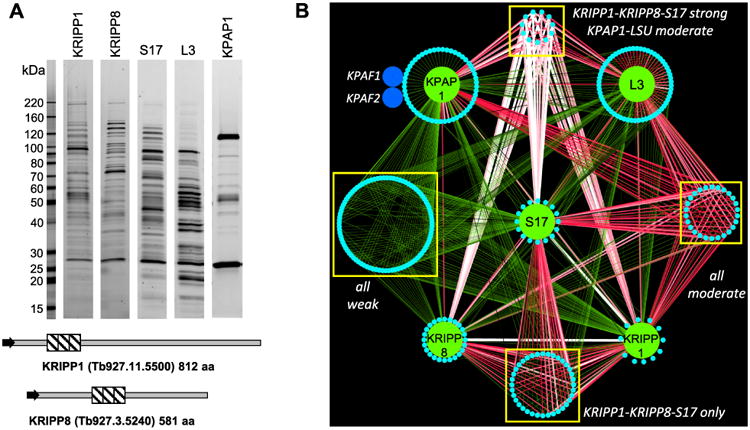
Interaction network of mitochondrial ribosomal, KRIPP-associated and polyadenylation complexes.
A. Tandem affinity purifications were carried out in parallel under uniform conditions (100 mM of KCl and 10 mM of MgCl2). Final fractions were separated in 8–16% SDS gel and stained with Sypro Ruby.
B. Visual representation of LC-MS/MS analyses of affinity purified complexes. Interaction network was generated in Cytoscape software from bait-prey pairs in which the prey protein was identified with at least 4 unique peptides (Supporting Information Table S1). The edge thickness and color intensity correlate with dNSAF values for bait-prey interactions; reciprocal contacts (i.e. bait-bait interactions captured in both purifications) are depicted by a single edge as the sum of two dNSAF values. Proteins shared by all five complexes are clustered as ‘weak’ and ‘moderate’ based on dNSAF values of each bait-prey pair; polypeptides overrepresented in KRIPP1, KRIPP8 and S17 are grouped as ‘KRIPP1-KRIPP8-S17 strong’. Proteins unique to each bait are positioned on a periphery of each named node.
This figure is available in colour online at wileyonlinelibrary.com.
A growing evidence suggests that elimination of the same protein may differentially affect mitochondrial RNA processing complexes in PF and BF proliferative forms (McDermott et al., 2015a,b). However, protein composition of the mitochondrial ribosome in bloodstream parasites has not been established to date. To examine association of KRIPP1 and KRIPP8 with mitochondrial ribosomes in BF, we expressed TAP-tagged ribosomal proteins L3 and S5 in Lister 427 ‘single marker’ cell line under control of inducible T7 RNA polymerase promoter. We then used a rapid pulldown technique to isolate large and small subunit ribosomal proteins under uniform conditions from PF and BF parasites (Supporting Information Table S2). While the tandem affinity purification predominantly yields individual ribosomal subunits, the rapid pulldown method enables isolation of relatively intact assembled ribosomes with bound mRNAs (Aphasizheva et al., 2011). LC-MS/MS analysis of pulldown fractions revealed that KRIPP1 and KRIPP8 co-purify with small ribosomal subunit proteins S17 and S5 from both PF and BF parasites, and with L3 protein from procyclic form (Table 1, Supporting Information Table S2). Provided that the KRIPP1- and KRIPP8-associated particles closely resemble SSU, these data indicate that in procyclic parasites KRIPP1 and KRIPP8 are also found in the assembled ribosome, as judged by their representation in the L3 PF fraction. However, KRIPP1 and KRIPP8 were markedly absent from assembled ribosome in bloodstream parasites while a core SSU protein S17 was readily detectable. Together, these findings suggest that the KRIPP1 and KRIPP8 are stably associated with two types of macromolecular complexes: small ribosomal subunit and SSU-like particle (45S SSU*). Importantly, the 45S SSU* along with associated KRIPP1 and KRIPP8 is downregulated in bloodstream parasites while the SSU remains largely unaffected.
Table 1.
Pentatricopeptide (PPR) and tetratricopeptide (TPR) repeat containing proteins identified in affinity-purified large ribosomal subunit L3 and small ribosomal subunit S5 and S17 protein samples.
| Gene ID | Name | Motif | L3-PF | L3-BF | S17-PF | S5-BF |
|---|---|---|---|---|---|---|
| Tb927.7.3460 | KRIT2 | TPR | 1235 | 637 | 418 | |
| Tb927.10.380 | KRIPP5 | PPR | 1159 | 504 | 227 | |
| Tb927.8.3170 | KRIPP9 | PPR | 1040 | 564 | 232 | |
| Tb927.11.5500 | KRIPP1 | PPR | 980 | 1254 | 485 | |
| Tb927.6.3930 | KRIT1 | TPR | 935 | 549 | 224 | |
| Tb927.11.11470 | KRIPP14 | PPR | 625 | 844 | 375 | |
| Tb927.3.4550 | KRIPP6 | PPR | 561 | 254 | ||
| Tb927.3.5240 | KRIPP8 | PPR | 556 | 961 | 541 | |
| Tb927.1.1160 | KRIPP3 | PPR | 496 | 188 | 171 | |
| Tb927.4.2790 | KRIPP12 | PPR | 494 | 318 | ||
| Tb927.9.5280 | KRIPP16 | PPR | 470 | 815 | 376 | |
| Tb927.9.12750 | KRIT3 | TPR | 389 | |||
| Tb927.2.4460 | KRIPP21 | PPR | 325 | 222 | ||
| Tb927.4.4720 | KRIPP7 | PPR | 305 | 322 | ||
| Tb927.11.4650 | KRIPP13 | PPR | 280 | 101 | ||
| Tb927.1.2990 | KRIPP2 | PPR | 267 | 638 | 293 | |
| Tb927.11.7680 | KRIPP22 | PPR | 241 | 301 | ||
| Tb927.8.4860 | KRIPP10 | PPR | 144 | 562 | 248 | |
| Tb927.10.11820 | KRIPP17 | PPR | 103 | 641 | 205 | |
| Tb927.7.2490 | KRIPP15 | PPR | 263 | |||
| Tb927.6.610 | KRIPP18 | PPR | 385 | |||
| Tb927.10.13200 | KRIPP4 | PPR | 274 | |||
| Tb927.8.6040 | KRIPP11 | PPR | 967 | 149 | ||
| Tb927.3.5610 | L3 | L3 ribo prot | 1855 | 600 | 182 | |
| Tb927.9.11280 | S17 | S17 ribo prot | 562 | 197 | 1685 | 337 |
| Tb927.10.6300 | S5 | S5 ribo prot | 640 | 1080 | 1300 |
Respective TAP-tagged proteins (in bold) were overexpressed in procyclic (PF) and bloodstream (BF) parasites. SAF values for each polypeptide are provided for each preparation. Previously, identified KRIPP19 and KRIPP20 (Aphasizheva et al., 2011) were excluded under cutoff implemented in this study (SAF < 100). Conversely, association of additional PPR (KRIPP22) and TPR (KRIT3) proteins with the large ribosomal subunit has been detected. KRIPP, kinetoplast ribosome-associated PPR protein; KRIT, kinetoplast associated TPR protein. Motifs are from Pfam.
KRIPP1 and KRIPP8 are essential for viability of procyclic but not bloodstream parasites
The roles of KRIPP1 and KRIPP8 were examined by inducible RNAi knockdowns in procyclic and bloodstream developmental forms of T. brucei. Repression of both genes caused growth inhibition phenotypes in PF, but not in BF parasites indicating essential function(s) of these proteins in the actively respiring procyclic form and a lack thereof in the oxidative phosphorylation-deficient bloodstream form (Fig. 2A–C). Quantitative RT-PCR was used to evaluate changes in relative abundance of mitochondrial RNAs (Fig. 2D). At the steady-state transcript level in PF parasites, the most uniform effect was the downregulation of 9S small ribosomal RNA in both RNAi cell lines. KRIPP1 ablation also caused moderate upregulation of several pan-edited mRNAs (CO3, ND3, ND7, ND8, ND9, A6 and RPS12) and decline of the cis-edited CO2 mRNA. Transcript-specific adverse effects of KRIPP8 knockdown were most apparent among 9S and 12S rRNAs, unedited mRNAs (CO1, ND1, ND4, ND5 and Murf1) and cis-edited CO2 mRNA whereas pan-edited mRNAs remained largely unaffected. Although the relative abundance of KRIPP1 and KRIPP8 mRNAs and RNAi efficiency were similar in PF and BF cell lines (Fig. 2B,C), no significant changes have been observed in relative abundance of mitochondrial RNAs (data not shown) or cell growth of bloodstream parasites (Fig. 2A). To conclude, partial loss of 9S rRNA in KRIPP1 and KRIPP8 RNAi is consistent with association of these proteins with SSU-like subunit in procyclic form of T. brucei. Our results are also in agreement with previous report of ribosomal RNA downregulation in knockdowns of several PPR proteins (Pusnik et al., 2007) that subsequently have been detected in mitochondrial ribosomes (Zikova et al., 2008; Aphasizheva et al., 2011).
Fig. 2.
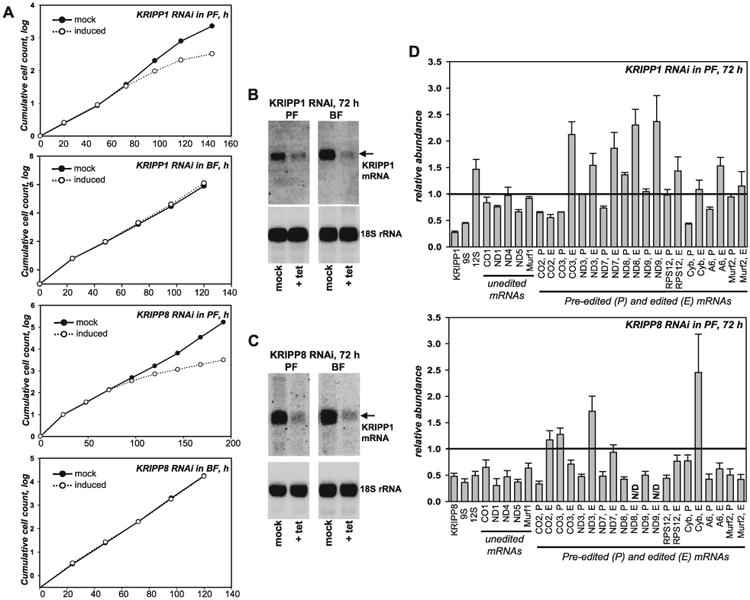
Effects of KRIPP1 and KRIPP8 repression on cell viability and steady-state levels of mitochondrial rRNAs and mRNAs.
A. Growth kinetics of procyclic (PF) and bloodstream (BF) parasite cultures after mock (closed circles) or RNAi induction (open circles) for indicated periods of time.
B. Northern blotting analysis of KRIPP1 nuclear encoded mRNA repression. RNAi was induced with tetracycline for 72 h, which corresponds to approximately six and 12 cell divisions for PF and BF parasites respectively. Loading control: 18S cytosolic rRNA.
C. Northern blotting analysis of KRIPP8 mRNA repression.
D. Quantitative real-time RT-PCR analysis of RNAi-targeted nuclear mRNAs, and mitochondrial rRNAs, edited and unedited mRNAs in respective RNAi knockdowns. RNA levels were normalized to β-tubulin mRNA. RNAi was induced for 72 h, a time point at which growth rate has not yet been affected. Error bars represent the standard deviation from at least three replicates. The thick line at ‘1’ reflects no change in relative abundance upon RNAi induction; bars above or below represent an increase or decrease respectively. N/D: not determined.
KRIPP1 and KRIPP8 RNAi knockdowns trigger loss of specific translatable mRNAs
A partial loss of 9S rRNA in procyclic KRIPP1 and KRIPP8 knockdowns, and, by inference, the functional small ribosomal subunit, and a lack of such effects in BF would be a simplest explanation for the observed growth phenotypes (Fig. 2A). However, notable presence of KRIPP1 and KRIPP8 in the polyadenylation complex (Supporting Information Table S1) warranted a more detailed analysis of mRNA's 3′ modifications in both RNAi cell lines. Select mRNAs that are expected to be essential either in PF only (CO1, CO2 and CO3), or in both PF and BF (A6 and RPS12 (Schnaufer et al., 2005; Brown et al., 2006; Aphasizheva et al., 2013), have been analyzed along with mt-rRNAs. Because rRNAs and mRNAs are also subjects to 3′ uridylation by RET1 TUTase (Aphasizheva and Aphasizhev, 2010), gRNAs that bear 3′ U-tails have been investigated likewise (Fig. 3). In agreement with qRT-PCR data (Fig. 2D), high resolution Northern blotting showed moderate upregulation of short A-tailed A6 and CO3 pan-edited mRNAs and steady levels of RPS12 and cyb mRNAs in KRIPP1 RNAi cells (Fig. 3A). It is noteworthy that 9S mt-rRNA declined by ∼50% within 48 h of RNAi induction, but then remained stable at later time points (Fig. 3B). Remarkably, the translation-competent, that is, A/U-tailed, unedited CO1 and cis-edited CO2 mRNAs underwent a reduction by 50% and 90% respectively while the corresponding short A-tailed form remained unaffected. The lack of detectable changes in uridylated RNAs, such as 12S rRNA and gRNAs, and the absence of a similar effect on A/U-tailed RPS12 and cyb mRNA indicates that KRIPP1 repression effects are restricted to a subset of mitochondrial mRNAs. In line with unimpeded growth of bloodstream cells (Fig. 2A), no significant changes in mitochondrial RNAs have been detected upon KRIPP1 knockdown in those cells except for decline of 9S rRNA (Fig. 3D,E).
Fig. 3.
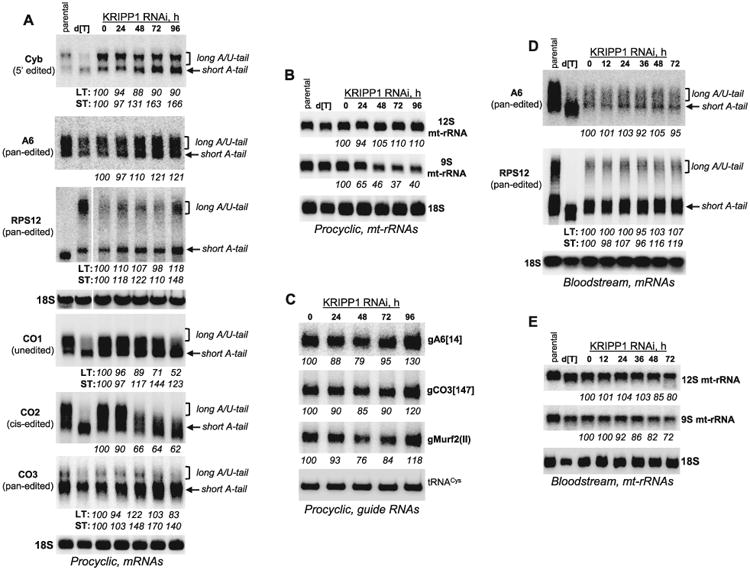
Effects of KRIPP1 repression on mRNA, rRNAs and gRNA 3′ end modifications and abundance.
A. Northern blotting of mitochondrial mRNAs.
B. Ribosomal RNAs. RNAi was induced for indicated periods of time in procyclic clonal cell line. For RPS12 mRNA detection, total RNA was separated in a 5% polyacrylamide/8M urea gel, transferred onto membrane and probed for fully-edited sequence. All other transcripts were visualized by separation in 1.8% agarose-formaldehyde gel. Cytosolic 18S rRNA served as loading control. [dT], RNA was treated with RNase H in the presence of 18-mer [dT] to remove poly(A) tails. Relative change in abundance was calculated separately wherever possible for long and short mRNA tails in reference to mock induction as 100%. LT, long tail; ST, short tail.
C. Guide RNAs were separated on 10% polyacrylamide/8M urea gel and detected by hybridization with oligonucleotide probes. Mitochondrially-targeted tRNACys was used as loading control.
D and E. Similar approaches were applied to analyze of KRIPP1 RNAi outcomes in BF parasites as described for panels A and B.
As expected, KRIPP8 RNAi induced downregulation of 9S mt-rRNA by approximately 50% although the effects became apparent at a later data point (72 h vs. 48 h for KRIPP1); a minor loss of 12S rRNA was also confirmed (Figs. 2D and 4B). Similarly to KRIPP1 knockdown, RPS12 and A6 mRNAs largely persisted while the A/U-tailed form of cyb and CO2 mRNAs decreased by ∼50% and the A/U-tailed CO1 mRNA was effectively eliminated (Fig. 4). We conclude that mRNAs affected by KRIPP1 and KRIPP8 knockdowns include overlapping (CO1, CO2) and distinct (cyb) sets of mitochondrial mRNAs.
Fig. 4.
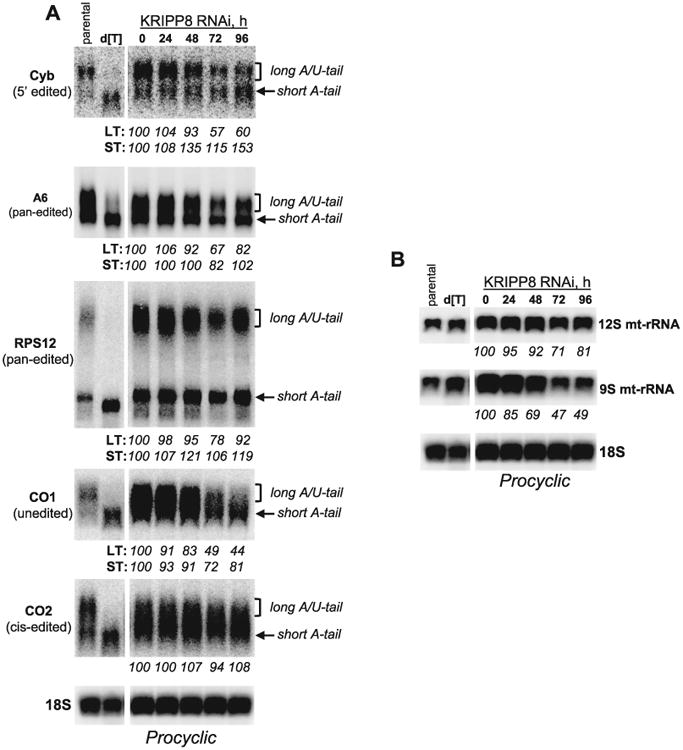
Effects of KRIPP8 repression on 3′ end modifications in mRNAs and rRNAs.
A. High-resolution Northern blotting of mitochondrial mRNAs and (B) ribosomal RNAs. RNAi was induced for indicated periods of time in procyclic cell line. For RPS12 mRNA detection, total RNA was separated in a 5% polyacrylamide/8M urea gel, transferred onto membrane and probed for fully edited sequence. All other transcripts were visualized by separation in 1.8% agarose-formaldehyde gel. Relative change in abundance was calculated separately wherever possible for long and short mRNA tails in reference to mock induction as 100%. LT, long tail; ST, short tail. Cytosolic 18S rRNA served as loading control. [dT], RNA was treated with RNase H in the presence of 18-mer [dT] to remove poly(A) tails.
Ribosome integrity is not affected by KRIPP1 and KRIPP8 knockdowns
Partial loss of 9S rRNAs in KRIPP1/KRIPP8 knockdowns and overlapping mRNA-specific effects raised the question of whether the general translational apparatus was inactivated due to compromised small ribosomal subunit, or the translation of specific mRNAs was blocked. We selected pan-edited RPS12 mRNA to test the formation of functional translating complexes because: (i) this transcript exists in two forms, A-tailed and A/U-tailed, which are not substantially affected by KRIPP1 or KRIPP8 knockdowns (Figs. 2 and 3), (ii) in glycerol gradients, the A/U-tailed form is found in translating ribosomes in >70S region, (iii) the A/U-tailed form is specifically enriched in affinity-purified ribosomes (Aphasizheva et al., 2011) and (iv) translation of RPS12 mRNA is expected to be essential in both PF and BF parasites (Aphasizheva et al., 2013). In agreement with previous reports (Aphasizheva et al., 2011; Ridlon et al., 2013), fully edited mRNA was clearly segregated in the parental cell line: a size-heterogeneous fraction sedimented in 25S-50S region while a predominantly long-tailed form was enriched in the >70S region (Fig. 5A, upper panel). The ratio between long- and short-tailed mRNAs in gradient fraction 15 was approximately threefold higher as compared with input mitochondrial RNA.
Fig. 5.
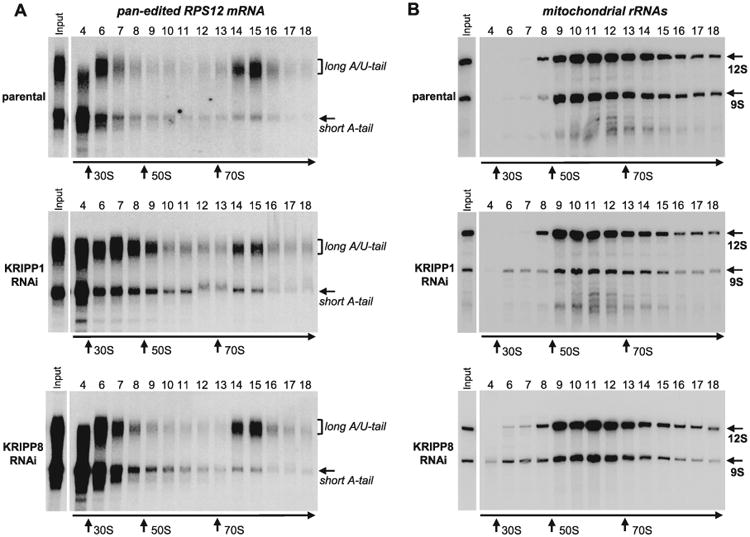
Sedimentation analysis of RPS12 mRNA and ribosomal RNAs in KRIPP1 and KRIPP8 knockdown cell lines. Detergent-extracted cell lysate from approximately 2 × 108 parasites was separated on 10–30% glycerol gradient. RNA was extracted from each fraction and separated in 5% polyacrylamide/8M urea gels for Northern blotting of mRNAs and rRNAs. All three membranes were hybridized simultaneously with probes specific for fully-edited RPS12 mRNA (A) and mitochondrial rRNAs (B). Purified bacterial ribosomal subunits and ribosomes were separated in parallel experiments as sedimentation standards. Arrows indicate peaks of optical density for apparent S value markers.
The peak of long tailed mRNA co-sedimented with a heavier fraction of ribosomal RNA-containing particles while the bulk of 9S and 12S mt-rRNA was detected in the 60S–70S region that corresponds to an assembled monosome (Fig. 5B, upper panel, [Sharma et al., 2009; Aphasizheva et al., 2013]). Remarkably, partial loss of 9S ribosomal RNA in KRIPP1 or KRIPP8 knockdowns exerted minor effect on sedimentation properties of either 12S rRNA or A/U-tailed RPS12 mRNA (Fig. 5A, middle panel). Assuming stoichiometric presence of small and large subunits in the assembled monosome, these data indicate that the loss of KRIPP1- and KRIPP8-associated SSU-like particles did not affect the monosome assembly. Rather, the affected complexes that resemble the 45S* SSU-like particle previously described in L. tarentolae (Maslov et al., 2007) and T. brucei (Ridlon et al., 2013).
KRIPP1 complex is associated with translating ribosome and contains predominantly 9S rRNA
Similar protein compositions of KRIPP1/KRIPP8-associated complexes and selective downregulation of 9S mt-rRNA in both knockdowns suggested a high degree of similarity between these particles and the small ribosomal subunit (Fig. 1). However, the correlations were less perfect considering the overlapping and distinct sets of mitochondrial mRNAs affected by KRIPP1 and KRIPP8 RNAi (Figs. 2–4). To investigate the relationship between small ribosomal subunit and KRIPP1/KRIPP8-associtated complexes further, lysates from cell lines expressing TAP-tagged canonical ribosomal proteins, such as small subunit protein S17 and large subunit protein L3, were separated on glycerol gradients along with parental, KRIPP1- and KRIPP8-expressing cells. Gradient fractions were separated on native gel and subjected to Northern blotting to visualize ribosomal RNAs and to Western blotting with peroxidase-antiperoxidase conjugates to detect TAP-tagged proteins (Fig. 6A). By monitoring 9S and 12S mt-rRNAs, a peak of free monosomes (fractions 10–13) was distinguished from the population of translating ribosomes (fractions 14–18). The latter are defined by co-migration with edited mRNA (Fig. 5 and Refs. [Aphasizheva et al., 2013; Ridlon et al., 2013]) and by co-immunoprecipitation with tRNA (Aphasizheva et al., 2011). Under these conditions, the bulk of TAP-tagged KRIPP8 was localized to the small ribosomal subunit as delineated by sedimentation patterns of S17 protein and 9S mt-rRNA. In contrast, KRIPP1 was detected not only in the monosome (fractions 10–13, 50–70S), but also in 30S–50S region (fractions 4–8), and in the >70S gradient region that contains active translation complexes (fractions 13–18).
Fig. 6.
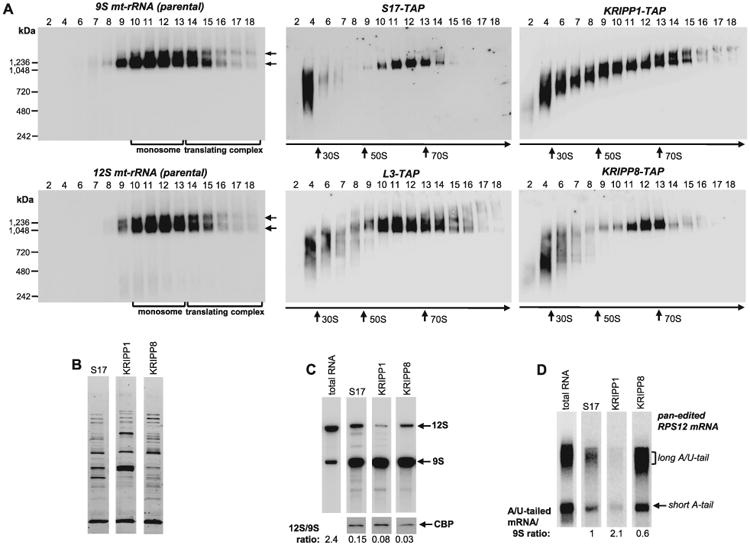
Distribution of KRIPP1 and KRIPP8 between ribosomal subunits, monosomes and mRNA-bound ribosomes.
A. Total cell lysates obtained from procyclic cell lines expressing indicated TAP-tagged fusions proteins were fractioned on glycerol gradients and each fraction was further separated on 3–12% Bis-Tris NuPAGE native gel. Native gels were transferred to nylon membrane, subjected to UV-crosslinking and hybridized with oligonucleotide probes specific for 9S and 12S mt-rRNAs (left panels). In parallel experiment, TAP-tagged proteins were detected by Western blotting with PAP reagent (central and right panels). NativeMARK unstained protein standards (Invitrogen) were separated on native gel alongside gradient fractions and stained separately. Sedimentation patterns of monosomes and translating complexes are indicated by arrows.
B. S17, KRIPP1 and KRIPP8 complexes were purified by rapid pulldowns, separated by SDS PAGE and stained with Sypro Ruby.
C. RNA was extracted from the parental cell line and from affinity purified complexes depicted in panel (B). RNA was separated on 5% polyacrylamide/8M urea gels for Northern blotting of mt-rRNAs (C) and edited RPS12 mRNA.
D. RNA amounts were normalized by protein bait in respective purified fractions, as quantitated by Western blotting with antibody against calmodulin binding peptide affinity tag. The 12S/9S ratio was calculated in relative intensity units by hybridizing the membrane with both probes simultaneously. The A/U-tailed mRNA ratio to 9S mt-rRNA was calculated from relative intensities of A/U-forms assuming such value in S17 preparation as 1.
Allowing for the significant proteomic overlap between KRIPP1- and KRIPP8-associated complexes (Fig. 1B, Supporting Information Table S1) and the differential distribution of KRIPP1 and KRIPP8 among ribosomal particles (Fig. 6A), we next inquired whether respective complexes contain similar amounts of ribosomal RNAs and translation-competent mRNA. The rapid pulldown strategy (Aphasizheva et al., 2011) was applied to isolate S17 as marker for the SSU, and KRIPP1 and KRIPP8 complexes (Fig. 6B). RNA was extracted from affinity purified complexes and analyzed for mt-rRNAs (Fig. 6C) and edited RPS12 mRNA (Fig. 6D) in comparison to total cellular RNA. As expected, purification of SSU via S17 pulldown led to ∼16-fold enrichment of 9S mt-rRNA indicating that a mixture of SSU and assembled ribosome had been purified. Surprisingly KRIPP1 pulldown contained ∼twofold lower relative amount of 12S mt-rRNA and ∼threefold less of A/U-tailed RPS12 mRNA. Although the KRIPP8 pulldown contained similar relative amount of the 12S mt-rRNA as the S17 pulldown, it contained an increased (about threefold) relative amount of A/U-tailed RPS12 mRNA. In sum, we find that in procyclic parasites KRIPP1 belongs to an SSU-like ∼45S particle that can enter the >70S actively translating ribosomal fraction, but does not interact with RPS12 mRNA. Properties of KRIPP8, conversely, are more consistent with this protein being an authentic component of the small ribosomal subunit in terms of association with the12S mt-rRNA and edited RPS12 mRNA.
Transcript-specific inhibition of translation in KRIPP1 RNAi cell line
The concurrent loss of select A/U-tailed mRNAs and 9S mt-rRNA upon KRIPP1 knockdown led us to inquire whether these changes affect mRNA-ribosome interactions and cause transcript-specific or general inhibition of translation. Among 18 predicted mitochondrial proteins, the de novo synthesis of only two polypeptides can be traced by metabolic labelling of live cells and two dimensional (2D) SDS-PAGE (Horváth et al., 2000). Encoded by unedited CO1 and 5′-edited cyb mRNAs, these proteins migrate abnormally in the second dimension and appear as off-diagonal spots. Incidentally, the unedited CO1 mRNA was strongly downregulated in both KRIPP1 and KRIPP8 knockdowns while the A/U-tailed edited cyb mRNA was unaffected in the former and noticeably downregulated in the latter genetic background (Figs. 3 and 4). Therefore, we next compared sedimentation patterns of CO1 (Fig. 7A) and cyb (Fig. 7B) mRNAs after 96 h of RNAi induction with the synthesis of respective polypeptides after 72, 96 and 120 h of KRIPP1 RNAi induction (Fig. 7C). Similarly to RPS12 mRNA (Fig. 5), in the parental cell line the A/U-tailed forms of both CO1 and cyb mRNAs were enriched in fractions 14 and 15 that correspond to sedimentation region of translating ribosomes. As expected, KRIPP1 RNAi caused significant downregulation of the A/U-tailed CO1 mRNA (Figs. 3A and 7A) and gradual loss of the corresponding protein (Fig. 7C). Accordingly, neither cyb mRNA sedimentation nor syntheses of the encoded polypeptide were comparably affected by KRIPP1 depletion. A minor relative reduction of the long A/U-tailed cyb mRNA in fractions 14–15 at 96 h and its redistribution to heavier fractions (Fig. 7B) sharply contrast with a near complete elimination of the long tailed CO1 mRNA from the fractions corresponding to translating ribosomes (Fig. 7A). In the case of KRIPP8 RNAi, we observed an unambiguous correlation between the loss of A/U-tailed forms in CO1 and cyb mRNA (Fig. 4A) and the translation inhibition of both polypeptides (Fig. 8). Although technical limitations prevent a comprehensive analysis of all mitochondrially encoded proteins, our results demonstrate that KRIPP1 is required for translation of specific mitochondrial transcripts, typified by CO1 mRNA. Remarkably, the ensued decreased level of 9S mt-rRNA exerted only insignificant effects on cyb mRNA translation further suggesting the existence of two KRIPP1-containig particles: the ‘canonical’ SSU, which remained unaffected by KRIPP1 repression and participated in translation of cyb and likely some other mRNAs, and SSU-like particle (45S SSU*). The latter complex appears to be compromised in KRIPP1 RNAi background causing downregulation of select translation-competent mRNAs, such as A/U-tailed CO1 and CO2 transcripts.
Fig. 7.
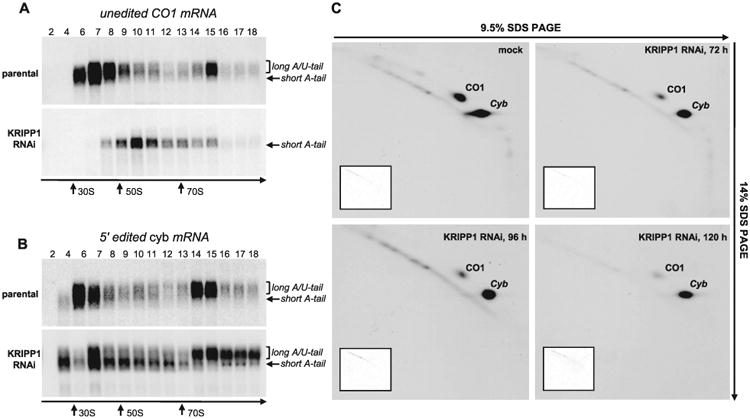
Transcript-specific inhibition of translation by KRIPP1 RNAi. Sedimentation analysis of unedited CO1 mRNA (A) and 5′ edited cyb mRNA (B) in KRIPP1 knockdown cell line. Detergent-extracted lysate from approximately 2 × 108 parasites was separated in 10–30% glycerol gradient. RNA was extracted from each fraction and separated on 1.8% agarose-formaldehyde gels for Northern blotting.
C. Analysis of translation products in KRIPP1-depleted parasites. RNAi was induced for indicated periods of time and cells were labelled with EasyTag labelling mix (PerkinElmer Life Sciences) in isotonic buffer supplemented with 0.1 mg/ml of cycloheximide. Cells were collected by centrifugation, dissolved in SDS gel loading buffer and fractionated by 2D electrophoresis. Gels were stained with Coomassie Brilliant Blue R250 (inset panels) and exposed to X-ray film (large panels). Based on previous protein identifications (Nebohacova et al., 2004), major spots indicated by arrows represent CO1 and Cyb subunits.
Fig. 8.
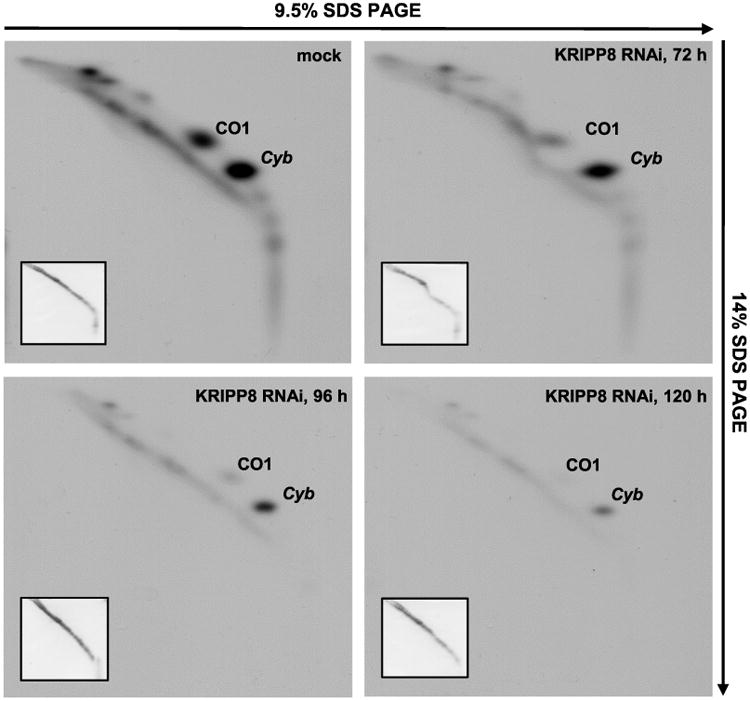
Indiscriminate inhibition of CO1 and cyb mRNA translation by KRIPP8 RNAi. Analysis of translation products in KRIPP8-depleted parasites was performed as described in Fig. 7C.
Discussion
Mitochondrial transcription and mRNA processing in trypanosomes generate relatively few functional transcripts that nonetheless represent a logistical challenge for translation initiation. It is assumed that the nucleolytic cleavage of multicistronic precursors generates monophosphorylated pre-mRNAs with short 5′ UTR lacking any discernible features that may be used for recognition by the ribosome. Furthermore, the U-insertion/deletion editing introduces additional level of complexity. It seems plausible that a mechanism may exist to prevent productive ribosome binding to pre-edited mRNA that sometimes differs from the edited form by as little as four missing uridines in the middle of a transcript (as in cis-edited CO2 mRNA). Further, unbiased recognition of the initiation codon is unlikely to be a uniform pathway since AUG may be created by U-insertion, such as in cyb mRNA, or may be encoded. In addition, it has been estimated that for a pan-edited mRNA only few percent of a given transcript actually possess a perfect open reading frame (Suematsu and Aphasizhev, unpublished). Lastly, the mRNA polyadenylation and editing processes are interconnected temporally and functionally: the pre-editing addition of a short A-tail is dispensable for mRNA stability until few initial editing events take place in the 3′ region. After few changes in the coding region, the same short A-tail becomes an essential stabilizing element ensuring that only adenylated molecules proceed through the editing pathway. The postediting extension of the short A-tail into long A/U-tail at the 3′ end appears to be tightly correlated with completion of editing at the mRNA's 5′ region. The first insights into functional significance of the A/U-tail were gained by identification of two PPR factors, KPAF1 and KPAF2, which are universally required for A/U-tail addition to all mRNAs tested, and demonstration of preferential binding of such modified mRNAs to the small ribosomal subunit (Aphasizheva et al., 2011). In addition, the A/U-tailing process, rather than editing, shows remarkable correlation with expected requirements for mitochondrially encoded proteins in insect and bloodstream developmental stages of the parasite.
Discovery of general polyadenylation/uridylation PPR factors and enzymes involved in the A/U-tails addition, however, posed the question of the mechanism that recruits the 3′ modification machinery to specific transcripts depending on their editing state or in developmentally regulated manner. Presence of multiple PPR proteins in the mitochondrial ribosome and polyadenylation complex (Aphasizhev and Aphasizheva, 2013), suggests an existence of a regulatory network based on protein adaptors that channel mRNAs containing specific sequences, whether encoded or created de novo by editing, for 3′ A/U-tailing and translation. To test this hypothesis, we focused on KRIPP1 and KRIPP8 PPR proteins that have been identified in the polyadenylation complex and in the small ribosomal subunit. Here, we show that these proteins are essential for viability of procyclic, but not bloodstream form of T. brucei and present evidence that KRIPP1 may be considered as translational activator for a subset of mitochondrial mRNAs. Cross-tagging and mass spectrometry analysis of affinity purified KRIPP1- and KRIPP8-associated complexes along with polyadenylation complex and ribosomal subunits from PF and BF parasites established similar proteomic profiles of both PPRs and conserved small ribosomal subunit protein S17. However, our results suggest that KRIPP1-associated particle resembles the 45S SSU-like complex which contains the 9S mt-rRNA, a set of SSU ribosomal proteins and several PPR proteins (Maslov et al., 2006; Maslov et al., 2007; Ridlon et al., 2013). Conversely, KRIPP8 appears to be a genuine small ribosome subunit protein. Importantly, our proteomics data indicate that the 45S SSU* particle is downregulated in bloodstream parasites; incidentally mRNAs affected by KRIPP1 knockdown are required only in procyclic trypanosomes. Selective downregulation of A/U-tailed mRNAs that encode cytochrome oxidase subunits 1, 2 and 3 was observed in KRIPP1 RNAi cell line and corroborated by experiments that monitor association of the CO1 mRNA with the ribosomes and the de novo synthesis of CO1 polypeptide. Conversely, KRIPP8 RNAi negatively affected a broader spectrum of mitochondrial RNAs and presumably their translation, as has been directly shown for cyb and CO1 protein products. Remarkably, two pan-edited mRNAs that encode subunit 6 of ATP-synthase and RSP12 ribosomal protein apparently are not the subject to KRIPP1-or KRIPP8-dependent regulation. This observation is consistent with a lack of growth phenotype on depletion of these PPR proteins in the bloodstream form of T. brucei.
Although not as numerous as in plants and trypanosomes, PPR proteins from other organisms participate in translational control of mitochondrial mRNAs as RNA recognition components of divergent molecular mechanisms. In yeast, PPR proteins facilitate initiation of translation by binding to the 5′ UTRs of specific mRNAs, thus, mediating the feedback control loop that adjusts the translation rate to the assembly of nascent polypeptides into respective respiratory complexes (reviewed in [Herrmann et al., 2013]). In humans, the well-studied Leu-rich PPR motif-containing factor (LRPPRC) protects virtually all protein-encoding transcripts from the mitochondrial degradosome activity by stimulating poly(A) polymerase and mRNA polyadenylation (reviewed in [Richter-Dennerlein et al., 2015]). However, an example of the human mitochondrial ribosome small subunit protein mS39, which is bound to the solvent side of the subunit's head and extends the helix-turn-helix array towards the entrance into the mRNA channel (Amunts et al., 2015), illustrates that ribosome-bound PPRs may directly recognize specific mRNAs and facilitate their translation. In this context, our work together with earlier studies (Aphasizheva et al., 2011; Aphasizheva et al., 2013; Ridlon et al., 2013) puts forward two alternative, albeit not mutually exclusive, hypotheses regarding the potential roles of PPR proteins in selective translational activation of mitochondrial mRNAs. Their prominent presence in the polyadenylation complex, much less so than that of canonical SSU proteins, argues for direct involvement in the A/U-tailing process; hence translational activation may occur by virtue of creating a structural feature required for efficient ribosome binding. This scenario is supported by identification of KPAF1, a general A/U-tailing factor, in KRIPP1 and KPAP1, but not in KRIPP8, S17 or L3-associated complexes (Supporting Information Table S1). Alternatively, selective translational activation may occur by other means, such as direct mRNA binding by 45S SSU* or SSU-embedded PPR proteins. In this case, active translation would be expected to stabilize mRNAs and inhibition thereof would lead to selective degradation of the A/U-tailed mRNA. The outcomes of this study emphasize the transcript-specific stimulation of translation by PPR proteins, but further work is required to distinguish their roles in mRNA 3′ modification or direct binding to the ribosome.
Experimental procedures
Trypanosome culture, RNAi and protein expression
RNAi plasmids were generated by cloning ∼500-bp gene fragments into p2T7-177 vector for tetracycline-inducible expression (Wickstead et al., 2002). For tetracycline-inducible protein expression in PF parasites, full-length genes were cloned into pLEW-MHTAP vector (Jensen et al., 2007). For expression in BF, TAP-tagged L3 and S5 ribosomal protein were cloned into pLew100v5 vector. Genetic constructs were transfected into procyclic 29-13 and bloodstream ‘single marker’ Lister 427 T. brucei strains (Wirtz et al., 1999) by electroporation and clonal cell lines were obtained by limiting dilution. The RNAi knockdowns were performed as described (Weng et al., 2008) and verified by qRT-PCR.
RNA analysis and mitochondrial extract fractionations
Total RNA isolation, qRT-PCR and Northern blotting were performed as described (Aphasizheva et al., 2011). The change in relative abundance was calculated assuming the ratio between analyzed transcripts and loading control in mock-induced cells as 100%. Membranes and gels were exposed to phosphor storage screens and volume quantitation was performed with Quantity One software (BioRad). The oligonucleotides A304 (TGAACAATCAATCATGGTAA-TAAGTAGACGATG) and A504 (ACGGCTGGCATCCA TTTC) served as hybridization probes specific for the 12S rRNA and the 9S rRNA respectively. The probes were 5′ labelled with [γ-32P]ATP (6000 Ci/mmol) and T4 polynucleotide kinase. Pre-hybridizations and hybridizations were performed at 42°C in 6 × SSPE, 5 × Denhardt's solution, 0.5% SDS and 0.2 mg/ml sheared denatured salmon sperm DNA. For KRIPP1 and KRIPP8 mRNA detection by Northern blotting, gene fragments 756–973 and 908-1021 respectively, were amplified with oligonucleotides containing T7 RNA polymerase promoter. RNA hybridization probes were synthesized from purified PCR products in the presence of [α-32P]CTP and used for hybridization in ULTRAhyb solution (Thermo-Fisher) at 65°C according to manufacturer's protocol. For glycerol gradient fractionation, 2 × 108 parasites were collected, washed once with PBS and resuspended in buffer containing 50 mM of HEPES (pH 7.6), 125 mM of KCl, 12 mM of MgCl2 and 1.2% Nonidet P-40. After 20 min incubation on ice, the extract was clarified at 21,000 g for 10 min and fractionated on 10–30% glycerol gradient in SW41 rotor for 4 h at 178,000 g. Subsequently, 580 μl fractions were collected from the top and separated on 4–12% gradient precast nNuPAGE Bis-Tris gels (Life technologies), transferred onto nitrocellulose membrane and probed with peroxidase-anti-peroxidase (PAP, Sigma) conjugate to detect tagged proteins. Quantitative chemiluminescent images were acquired with LAS-4000 digital analyzer (GE Healthcare).
Tandem affinity purification
Mitochondrial fraction was enriched by hypotonic lysis of ∼2 × 109 parasites followed by addition of sucrose to 250 mM, DNase digestion and differential centrifugation. Two-step affinity purification was performed in the presence of 100 mM of KCl and 10 mM of MgCl2 as described (Aphasizhev and Aphasizheva, 2007).
Rapid affinity pulldowns
Cell pellets were subjected to cryogenic homogenization using Retsch Cryomill and TAP-tagged ribosomal proteins were isolated by rapid pulldown with rabbit IgG-coated Dynabeads (Thermo-Fisher) as described in (Aphasizheva et al., 2011).
LC MS/MS analysis, protein identification and network construction
Proteomic analysis and interactions network building were performed as described previously (Kaake et al., 2010; Aphasizheva et al., 2011; Fang et al., 2012) with following modifications. Tandem affinity-purified complexes were precipitated by addition of trichloroacetic acid and deoxycholate to 20% and 0.1% respectively, washed three times with ice-cold acetone and digested with 2% (w/w) LysC peptidase in 8 M urea for 4 h at 37°C. Reaction was diluted fivefold with 50 mM of ammonium bicarbonate and further digested with 1% trypsin (w:w) for 16 h. Peptides were purified on Vivapure spin columns (Sartorius). LC MS/MS was carried out by nanoflow reverse phase liquid chromatography (Eksigent, CA) coupled on-line to a Linear Ion Trap (LTQ)-Orbitrap XL mass spectrometer (Thermo Scientific) as described (Fang et al., 2012). The LC analysis was performed using a capillary column (100 μm ID × 150 mm long) packed with Inertsil ODS-3 resin (GL Sciences) and the peptides were eluted using a linear gradient of 2–35% B in 85 min at a flow of 400 nl/min (solvent A: 100% H2O/0.1% formic acid; solvent B: 100% acetonitrile/0.1% formic acid). A cycle of one full FT scan mass spectrum (350– 1800 m/z, resolution of 60,000 at m/z 400) followed by 10 data-dependent MS/MS scans was acquired in the linear ion trap with normalized collision energy (setting of 35%). Target ions already selected for MS/MS were dynamically excluded for 30 s. Monoisotopic masses of parent ions and corresponding fragment ions, parent ion charge states and ion intensities from the tandem mass spectra (MS/MS) were obtained using in-house software with Raw_Extract script from Xcalibur v2.4. Following automated data extraction, resultant peak lists for each LC MS/MS experiment were submitted to the development version of Protein Prospector (UCSF) for database searching as described (Kaake et al., 2010). Each project was searched against a normal form concatenated with the random form of the T. brucei database (www.genedb.org, v5, accessed 12/31/2015). Trypsin was set as the enzyme with a maximum of two missed cleavage sites. The mass tolerance for parent ion was set as ±20 ppm, whereas ±0.6 Da tolerance was chosen for the fragment ions. Chemical modifications such as protein N-terminal acetylation, methionine oxidation, N-terminal pyroglutamine, and deamidation of asparagine were selected as variable modifications during database search. The search compare program in protein prospector was used for summarization, validation and comparison of results. Protein identification was based on at least three unique peptides with expectation value ≤0.05. Protein's spectrum abundance factor (SAF) is calculated as described (Zhang et al., 2010) with modification. It is defined as a protein's total peptide spectral counts divided by its molecular weight and then multiplied by 1000. SAF = (SUM #PSM/MW [kDa]) × 1000. The calculation of dNSAF, uses label-free strategy based on the quantitative information stored in MS/MS data as spectral counts, and converts peptide counts into a normalized value, for subsequent quantitation (Fang et al., 2012). Using an in-house generated analysis platform, we calculated the dNSAF for each putative interactor in each bait purification. Total unique and shared peptide counts were calculated from the peptide report generated from Search Compare in Protein Prospector (v5.8.0) and analyzed as described (Fang et al., 2012).
Mitochondrial translation
Cell labelling and gel separations were performed as described (Aphasizheva et al., 2011). In brief, procyclic trypanosomes (107 cells) were washed with SoTE (20 mM Tris-HCl, pH 7.5, 600 mM of sorbitol, 2 mM of EDTA) and resuspended in the same buffer. Cytosolic translation was inhibited with 100 μg/ml of cycloheximide and the products of mitochondrial translation were labelled with [35S]-Easy-Tag Express Protein Labelling Mix (Perkin Elmer) by incubation at 26°C for 1 h with mild agitation. Labelled products were analyzed on 2D (9 vs. 14% polyacrylamide) Tris-Glycine-SDS gels.
Supplementary Material
Acknowledgments
This work was supported in part by NIH grants AI091914 to RA, AI088292 to DM, AI113157 to IA, GM074830 to LH and P41 GM104603 to CEC. We thank members of our laboratories for discussions and George Cross for a kind gift of pLew100v5 vector.
Footnotes
Authors declare no conflict of interest.
Supporting information: Additional supporting information may be found in the online version of this article at the publisher's web-site.
References
- Amunts A, Brown A, Toots J, Scheres SH, Ramakrishnan V. Ribosome. The structure of the human mitochondrial ribosome. Science. 2015;348:95–98. doi: 10.1126/science.aaa1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I. RNA Editing Uridylyltransferases of Trypanosomatids. Methods Enzymol. 2007;424:51–67. doi: 10.1016/S0076-6879(07)24003-0. [DOI] [PubMed] [Google Scholar]
- Aphasizheva I, Aphasizhev R. RET1-catalyzed Uridylylation Shapes the Mitochondrial Transcriptome in Trypanosoma brucei. Mol Cell Biol. 2010;30:1555–1567. doi: 10.1128/MCB.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I. Emerging roles of PPR proteins in trypanosomes: switches, blocks, and triggers. RNA Biol. 2013;10:1679–1688. doi: 10.4161/rna.26215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizheva I, Maslov D, Wang X, Huang L, Aphasizhev R. Pentatricopeptide Repeat Proteins Stimulate mRNA Adenylation/Uridylation to Activate Mitochondrial Translation in Trypanosomes. Mol Cell. 2011;42:106–117. doi: 10.1016/j.molcel.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizheva I, Maslov DA, Aphasizhev R. Kinetoplast DNA-encoded ribosomal protein S12: a possible functional link between mitochondrial RNA editing and translation in Trypanosoma brucei. RNA Biol. 2013;10:1679–1688. doi: 10.4161/rna.26733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizheva I, Zhang L, Wang X, Kaake RM, Huang L, Monti S, Aphasizhev R. RNA binding and core complexes constitute the u-insertion/deletion editosome. Mol Cell Biol. 2014;34:4329–4342. doi: 10.1128/MCB.01075-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat GJ, Koslowsky DJ, Feagin JE, Smiley BL, Stuart K. An extensively edited mitochondrial transcript in kinetoplastids encodes a protein homologous to ATPase subunit 6. Cell. 1990;61:885–894. doi: 10.1016/0092-8674(90)90199-o. [DOI] [PubMed] [Google Scholar]
- Brown SV, Hosking P, Li J, Williams N. ATP synthase is responsible for maintaining mitochondrial membrane potential in bloodstream form Trypanosoma brucei. Eukaryot Cell. 2006;5:45–53. doi: 10.1128/EC.5.1.45-53.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delannoy E, Stanley WA, Bond CS, Small ID. Pentatricopeptide repeat (PPR) proteins as sequence-specificity factors in post-transcriptional processes in organelles. Biochem Soc Trans. 2007;35:1643–1647. doi: 10.1042/BST0351643. [DOI] [PubMed] [Google Scholar]
- Etheridge RD, Aphasizheva I, Gershon PD, Aphasizhev R. 3′ adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria. EMBO J. 2008;27:1596–1608. doi: 10.1038/emboj.2008.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Kaake RM, Patel VR, Yang Y, Baldi P, Huang L. Mapping the protein interaction network of the human COP9 signalosome complex using a label-free QTAX strategy. Mol Cell Proteomics. 2012;11:138–147. doi: 10.1074/mcp.M111.016352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannaert V, Saavedra E, Duffieux F, Szikora JP, Rigden DJ, Michels PA, Opperdoes FR. Plant-like traits associated with metabolism of Trypanosoma parasites. Proc Natl Acad Sci USA. 2003;100:1067–1071. doi: 10.1073/pnas.0335769100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann JM, Woellhaf MW, Bonnefoy N. Control of protein synthesis in yeast mitochondria: the concept of translational activators. Biochim Biophys Acta. 2013;1833:286–294. doi: 10.1016/j.bbamcr.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Horváth A, Berry EA, Maslov DA. Translation of the edited mRNA for cytochrome b in trypanosome mitochondria. Science. 2000;287:1639–1640. doi: 10.1126/science.287.5458.1639. [DOI] [PubMed] [Google Scholar]
- Jensen BC, Kifer CT, Brekken DL, Randall AC, Wang Q, Drees BL, Parsons M. Characterization of protein kinase CK2 from Trypanosoma brucei. Mol Biochem Parasitol. 2007;151:28–40. doi: 10.1016/j.molbiopara.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaake RM, Milenkovic T, Przulj N, Kaiser P, Huang L. Characterization of cell cycle specific protein interaction networks of the yeast 26S proteasome complex by the QTAX strategy. J Proteome Res. 2010;9:2016–2029. doi: 10.1021/pr1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Gagnon MG, Bulkley D, Steitz TA. Conformational changes of elongation factor G on the ribosome during tRNA translocation. Cell. 2015;160:219–227. doi: 10.1016/j.cell.2014.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslov DA, Sharma MR, Butler E, Falick AM, Gingery M, Agrawal RK, et al. Isolation and characterization of mitochondrial ribosomes and ribosomal subunits from Leishmania tarentolae. Mol Biochem Parasitol. 2006;148:69–78. doi: 10.1016/j.molbiopara.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Maslov DA, Spremulli LL, Sharma MR, Bhargava K, Grasso D, Falick AM, et al. Proteomics and electron microscopic characterization of the unusual mitochondrial ribosome-related 45S complex in Leishmania tarentolae. Mol Biochem Parasitol. 2007;152:203–212. doi: 10.1016/j.molbiopara.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott SM, Carnes J, Stuart K. Identification by random mutagenesis of functional domains in KREPB5 that differentially affect RNA editing between life cycle stages of Trypanosoma brucei. Mol Cell Biol. 2015a;35:3945–3961. doi: 10.1128/MCB.00790-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott SM, Guo X, Carnes J, Stuart K. Differential editosome protein function between life cycle stages of trypanosoma brucei. J Biol Chem. 2015b;290:24914–24931. doi: 10.1074/jbc.M115.669432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingler MK, Hingst AM, Clement SL, Yu LE, Reifur L, Koslowsky DJ. Identification of pentatricopeptide repeat proteins in Trypanosoma brucei. Mol Biochem Parasitol. 2006;150:37–45. doi: 10.1016/j.molbiopara.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Nebohacova M, Maslov DA, Falick AM, Simpson L. The effect of RNA interference Down-regulation of RNA editing 3′-terminal uridylyl transferase (TUTase) 1 on mitochondrial de novo protein synthesis and stability of respiratory complexes in Trypanosoma brucei. J Biol Chem. 2004;279:7819–7825. doi: 10.1074/jbc.M311360200. [DOI] [PubMed] [Google Scholar]
- Pusnik M, Small I, Read LK, Fabbro T, Schneider A. Pentatricopeptide repeat proteins in Trypanosoma brucei function in mitochondrial ribosomes. Mol Cell Biol. 2007;27:6876–6888. doi: 10.1128/MCB.00708-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter-Dennerlein R, Dennerlein S, Rehling P. Integrating mitochondrial translation into the cellular context. Nat Rev Mol Cell Biol. 2015;16:586–592. doi: 10.1038/nrm4051. [DOI] [PubMed] [Google Scholar]
- Ridlon L, Skodova I, Pan S, Lukes J, Maslov DA. The Importance of the 45S Ribosomal Small Subunit-related Complex for Mitochondrial Translation in Trypanosoma brucei. J Biol Chem. 2013;288:32963–32978. doi: 10.1074/jbc.M113.501874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaufer A, Panigrahi AK, Panicucci B, Igo RP, Salavati R, Stuart K. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science. 2001;291:2159–2161. doi: 10.1126/science.1058955. [DOI] [PubMed] [Google Scholar]
- Schnaufer A, Clark-Walker GD, Steinberg AG, Stuart K. The F1-ATP synthase complex in bloodstream stage trypanosomes has an unusual and essential function. EMBO J. 2005;24:4029–4040. doi: 10.1038/sj.emboj.7600862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma MR, Booth TM, Simpson L, Maslov DA, Agrawal RK. Structure of a mitochondrial ribosome with minimal RNA. Proc Natl Acad Sci USA. 2009;106:9637–9642. doi: 10.1073/pnas.0901631106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng J, Aphasizheva I, Etheridge RD, Huang L, Wang X, Falick AM, Aphasizhev R. Guide RNA-binding complex from mitochondria of trypanosomatids. Mol Cell. 2008;32:198–209. doi: 10.1016/j.molcel.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B, Ersfeld K, Gull K. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol Biochem Parasitol. 2002;125:211–216. doi: 10.1016/s0166-6851(02)00238-4. [DOI] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- Yin P, Li Q, Yan C, Liu Y, Liu J, Yu F, et al. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature. 2013;504:168–171. doi: 10.1038/nature12651. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wen Z, Washburn MP, Florens L. Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal Chem. 2010;82:2272–2281. doi: 10.1021/ac9023999. [DOI] [PubMed] [Google Scholar]
- Zikova A, Panigrahi AK, Dalley RA, Acestor N, Anupama A, Ogata Y, et al. Trypanosoma brucei mitochondrial ribosomes: affinity purification and component identification by mass spectrometry. Mol Cell Proteomics. 2008;7:1286–1296. doi: 10.1074/mcp.M700490-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


