Abstract
Clathrin coated vesicles mediate endocytosis and transport between the trans Golgi network (TGN) and endosomes in eukaryotic cells. Clathrin adaptors play central roles in coat assembly, interacting with clathrin, cargo, and membranes. Two major types of clathrin adaptors act in TGN-endosome traffic, Gga proteins and the AP-1 complex. Here we characterize the relationship between Gga proteins, AP-1, and other TGN clathrin adaptors using live cell and superresolution microscopy in yeast. We present evidence that Gga proteins and AP-1 are recruited sequentially in two waves of coat assembly at the TGN. Mutations that decrease phosphatidylinositol 4-phosphate (PI4P) levels at the TGN slow or uncouple AP-1 coat assembly from Gga coat assembly. Conversely, enhanced PI4P synthesis shortens the time between adaptor waves. Gga2p binds directly to the TGN PI4-kinase Pik1p and contributes to Pik1p recruitment. These results identify a PI4P-based mechanism for regulating progressive assembly of adaptor-specific clathrin coats at the TGN.
Introduction
The last subcompartment of the Golgi complex, the trans Golgi network (TGN), sorts proteins into distinct transport carriers that are targeted to different destinations including the plasma membrane and the endosome/lysosome system. A major class of TGN-derived transport carriers are clathrin-coated vesicles (ccv), which select cargo for delivery to endosomes. Ccv form through the concerted action of three types of highly conserved proteins: clathrin, which forms the outer coat scaffold; adaptors, which link clathrin to membranes by binding to clathrin, phosphoinositides, and/or cargo proteins; and accessory proteins which contribute to coat assembly, membrane invagination, scission, and uncoating1.
The principal adaptors that participate in TGN ccv formation are GGA proteins, Golgi-localized epsin-related proteins, and the heterotetrameric AP-1 complex (β1, γ1, μ1, and σ1)1,2. Yeast cells express AP-1, two GGA proteins, Gga1p and Gga2p, and two epsin-related proteins, Ent3p and Ent5p2. Physical and genetic interaction studies indicate that these adaptors are part of an extended clathrin-based network in which AP-1 and Gga proteins appear to constitute distinct network hubs3-8.
The relationship between AP-1 and Gga proteins during TGN ccv formation is uncertain. Both proteins appear to rely on similar sets of low affinity (≈low μM Kd) multivalent interactions for recruitment to the TGN, including binding to the activated form of the ADP-ribosylation factor 1 (ARF1) GTPase, phosphatidylinositol 4-phosphate (PI4P), and cargo1,9. However, whether the adaptors act sequentially, in parallel, or in distinct pathways remains unresolved, and the extent of colocalization has varied in different studies1. Here we have taken advantage of the dispersed nature of Golgi cisternae in the yeast Saccahromyces cerevisiae to assess the distribution and relative dynamics of fluorescently-tagged AP-1 and Gga proteins expressed at endogenous levels. Our results provide evidence for sequential waves of adaptor-specific coat assembly, coupled by synthesis of PI4P.
Results
Gga2p and AP-1 assemble sequentially
Movies of cells expressing clathrin coat proteins fused to GFP or mRFP were acquired by spinning disk confocal microscopy. Clathrin coat protein fusions, expressed from the normal chromosomal loci, localized as heterogeneous puncta throughout the cell, similar to patterns observed by immunofluorescence of fixed cells6,13. Fluorescently tagged clathrin heavy chain (Chc1p-mRFP) localized as transient puncta at the plasma membrane and at internal sites (Supplemental Information Movie S1) as observed by others14, normally persisting for 2 minutes or less. Internal clathrin foci grew to relatively large sizes (0.3-1.5μm), assumed irregular and often changing shapes, and moved in random directions. TGN clathrin adaptors formed puncta with characteristics of the internal clathrin structures (Fig 1).
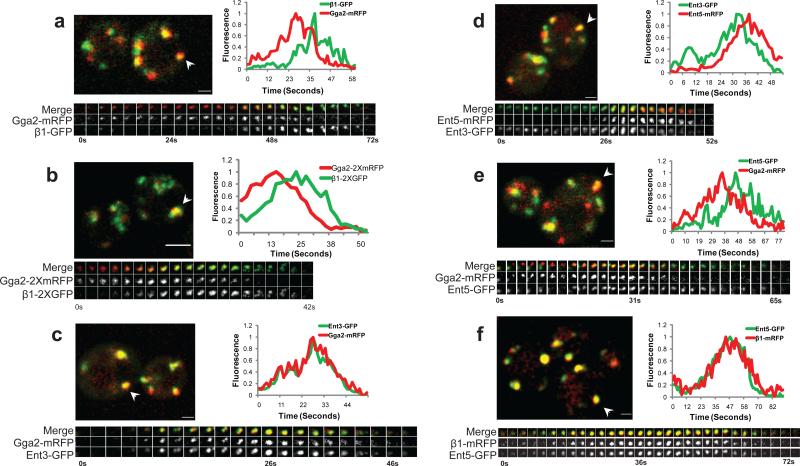
Figure 1. Sequential assembly of clathrin adaptors.
Pairs of clathrin coat proteins were assessed in two-color movies of single optical sections (a, c-f) or in 3 dimensions (b) by tracking puncta that remained separate from other puncta throughout their lifetimes and were present for at least 7 frames (~8.4 s). (a, c-f) Upper panel: merged image of live cells co-expressing GFP- and mRFP-tagged versions of the indicated clathrin adaptors; arrowhead indicates a puncta selected for kymograph in the bottom panel. Scale bar = 1 μm. (b) Upper panel: Five optical sections along the Z axis were imaged at 0.3 μm intervals. Each z-stack was collected every 2.1s. Scale bar = 2μm. (a-f) Bottom panel: three channel kymograph (merged, mRFP and GFP) of the selected puncta; time to acquire one image pair was 1.2s. Every other image pair is shown in the kymograph. (a-f) Graph: normalized level of GFP and mRFP fluorescence intensity in the puncta as a function of time. (a) GPY3109 (b) GPY4974 (c) GPY3954 (d) GPY3912 (e) GPY3962 (f) GPY3900.
The relative dynamics of specific protein pairs were assessed in two-color movies of single optical sections by tracking individual puncta. In cells expressing Gga2p-mRFP and β1-GFP (ß1 subunit of AP-1), Gga2p foci appeared first, increased in intensity, and then became AP-1 positive as the Gga2p intensity declined (Fig 1a, Supplemental Information Movie S2). 98% of Gga2p puncta transitioned to AP-1 (n=101; 38 cells). Conversely, 95.8% of AP-1 puncta were preceded by the appearance of Gga2p (n=142, 51 cells). In contrast, imaging of cells expressing clathrin light chain (GFP-Clc1p) and heavy chain (Chc1-RFP) revealed complete colocalization and coincident profiles, as expected for subunits of the same protein complex (Supplementary Information Fig. S1a). We presume that declines in coat protein fluorescence reflect budding of ccv. Infrequently we observed release of smaller puncta, but were unable to effectively track budding events.
The Gga2p to AP-1 sequence was also observed by 3D live cell imaging where optical sections were collected along the z-axis over time (Fig. 1b), suggesting that the appearance of puncta in single optical sections represents coat assembly rather than movement of pre-existing coats into the focal plane. Because of better temporal and spatial resolution and reduced photobleaching, subsequent analyses were carried out with single optical sections.
We determined the times between peak intensities of Gga2p-mRFP and ß1-GFP in puncta (peak-to-peak time). Gga2p-mRFP fluorescence peaked 10.4s±0.55s before AP-1 reached peak fluorescence (Table 1 row#1; Supplementary Information Fig. S1b). Similar results were obtained when the σ1 subunit was tagged with GFP and compared to Gga2p-mRFP (L.D., unpublished).
Table 1. Dynamics of clathrin adaptors, phosphoinositide binding reporters, and Golgi markers at the TGN.
Haploid cells expressing the indicated fluorescent proteins were imaged using live-cell microscopy. Multiple puncta from each strain were analyzed for changes in fluorescent intensity of the indicated proteins over time. Time between the points of peak fluorescence intensity for each protein was determined and the mean peak-to-peak time calculated for all the analyzed puncta in a given strain. The data are presented as the mean ±S.E.M. for each strain. The order of tagged proteins in each row represents the order of assembly. Some rows are repeated to facilitate comparison (1,15,24; 4,26; 8,13; 17,22).
| # | Strain | Number of puncta (n) in total cells (c) | Peak to Peak time (seconds) | ||
|---|---|---|---|---|---|
| mutations | labeled proteins | n | c | ||
| 1 | wt | Gga2-mRFP β1-GFP | 102 | 59 | 10.4 ± 0.55 |
| 2 | wt | Gga2-mRFP Ent3-GFP | 91 | 38 | 0.01 ± 0.16 |
| 3 | wt | Ent3-GFP β1-mRFP | 30 | 21 | 10.2 ± 1.40 |
| 4 | wt | Ent3-GFP Ent5-mRFP | 99 | 52 | 8.4 ± 0.70 |
| 5 | wt | Gga2-mRFP Ent5-GFP | 91 | 46 | 8.0 ± 0.14 |
| 6 | wt | Ent5-GFP β1-mRFP | 58 | 30 | 1.3 ± 0.62 |
| 7 | wt | Gga2-GFP Sec7-mRFP | 97 | 25 | 0.29 ±0.39 |
| 8 | wt | Sec7-mRFP β1-GFP | 76 | 48 | 10.4 ± 1.18 |
| 9 | wt | Gga2-GFP Chc1-mRFP | 95 | 49 | 1.3 ± 0.4 |
| 10 | wt | Chc1-mRFP β1-GFP | 59 | 34 | 8.0 ± 0.7 |
| 11 | gga1Δ gga2Δ | Chc1-mRFP β1-GFP | 71 | 36 | 0.79 ± 0.73 |
| 12 | β1Δ (apl2Δ) | Chc1-mRFP Gga2-GFP | 22 | 40 | 0.9 ±0.64 |
| 13 | wt | Sec7-mRFP β1-GFP | 76 | 48 | 10.4 ± 1.18 |
| 14 | gga1Δ gga2Δ | Sec7-mRFP β1-GFP | 49 | 25 | 26.1 ± 1.9 |
| 15 | wt | Gga2-mRFP β1-GFP | 102 | 59 | 10.4 ± 0.55 |
| 16 | arf1Δ | Gga2-mRFP β1-GFP | 47 | 41 | 20.5 ± 2.7 |
| 17 | wt | Gga2-mRFP GFP-PHOSH1 | 104 | 52 | 5.3 ± 0.44 |
| 18 | wt | GFP-PHOSH1 β1-mRFP | 87 | 52 | 4.1 ± 0.4 |
| 19 | wt | Sec7-mRFP GFP-PHOSH1 | 103 | 61 | 3.8 ± 0.54 |
| 20 | gga1Δ gga2Δ | Sec7-mRFP GFP-PHOSH1 | 117 | 68 | 11.3 ± 0.86 |
| 21 | arf1Δ | Sec7-mRFP GFP-PHOSH1 | 45 | 33 | 11.0 ± 2.1 |
| 22 | wt | Gga2-mRFP GFP-PHOSH1 | 104 | 52 | 5.3 ± 0.6 |
| 23 | GPD-PIK1 GPD-FRQ1 | Gga2-mRFP GFP-PHOSH1 | 48 | 27 | 0.0 ±0.81 |
| 24 | wt | Gga2-mRFP β1-GFP | 102 | 59 | 10.4 ± 0.55 |
| 25 | GPD-PIK1 GPD-FRQ1 | Gga2-mRFP β1-GFP | 67 | 35 | 5.7 ±0.77 |
| 26 | wt | Ent3-GFP Ent5-mRFP | 99 | 52 | 8.4 ± 0.70 |
| 27 | GPD-PIK1 GPD-FRQ1 | Ent3-GFP Ent5-mRFP | 30 | 17 | 3.9 ±1.5 |
| 28 | wt | Sec7-mRFP GFP-Pik1 | 75 | 47 | 4.1 ± 0.6 |
| 29 | gga1Δ gga2Δ | Sec7-mRFP GFP-Pik1 | 61 | 36 | 12.6 ± 1.3 |
| 30 | wt | Frq1-GFP Sec7-mRFP | 55 | 38 | 1.5 ± 0.8 |
| 31 | gga1Δ gga2Δ | Frq1-GFP Sec7-mRFP | 73 | 48 | 0.4 ± 0.9 |
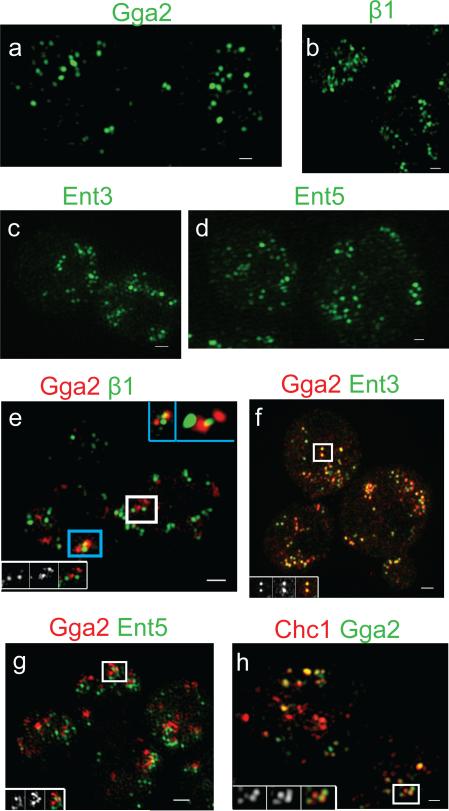
Chemically fixed cells were imaged by structured illumination microscopy (SIM), a super-resolution technique15. Gga2p-GFP or β1-GFP were localized as clusters of ≈100-200nm structures, consistent with the size expected for individual or closely-spaced coated pits (Fig. 2a,b). This suggests that puncta observed by confocal microscopy can be composed of multiple individual coats below the resolution limit of the microscope. In SIM images of cells expressing both Gga2p-mRFP and β1-GFP, most of the Gga2p structures were adjacent to, but distinct from, the β1 structures (Fig 2e). Our data provide evidence that the consecutive fluorescence peaks of Gga2p and AP-1 represent a wave of semi-synchronous Gga2p coat formation followed by a wave of AP-1 coats. Multiple coated structures in each fluorescent puncta likely accounts for heterogeneity in peak-to-peak values.
Figure 2. Spatial relationships of clathrin adaptors by structured illumination microscopy.
Cells expressing the GFP- and mRFP-tagged adaptors were imaged by structured illumination microscopy. (a-d) Maximum image projection of the GFP channel is shown for the following strains: (a) GGA2-GFP CHC1-mRFP (GPY4931), (b) β1-GFP CHC1-mRFP (GPY4932), (c) ENT3-GFP ENT5-mRFP (GPY3912), (d) ENT5-GFP β1-mRFP (GPY3900). (e-h) Maximum image projection of the merged channel is shown for (e) GGA2-mRFP β1-GFP (GPY3109). The blue box inset (top right) denotes the 3D volume view of the blue boxed region with no rotation (left) and with rotation around the x- and z-axis (right). (f) GGA2-mRFP ENT3-GFP (GPY3954), (g) GGA2-2XmRFP ENT5-2XGFP (GPY4962), (h) CHC1-mRFP GGA2-GFP (GPY4931). For e-h, insets contain (left to right) the GFP, mRFP and merged maximum image projection for the puncta in the white box. Scale bar = 400 nm.
Two waves of clathrin adaptor assembly
Ent3p binds to Gga2p and depends on Gga2p for localization4,6. Ent3p and Gga2p fluorescence peaked simultaneously (Figure 1c; Figure S1c; Table 1, row#2; Supplemental Information Movie S3). Moreover, Ent3p localized as 100-200nm puncta that substantially colocalized with Gga2p by SIM (Fig. 2c, f). In comparison to AP-1 [either ß1 (Table 1, row#3), or σ1 (L.D., unpublished)], Ent3p peaked on average 10.2s earlier, like Gga2p. Little colocalization between Ent3p and AP-1 was evident by SIM (Supplementary Information Fig. S2a).
Ent5p binds clathrin, Gga2p, and AP-14,6. Genetic interactions indicate that Ent5p function is more important for AP-1-mediated transport4. Ent5p appeared as puncta after Ent3p (Fig. 1d); 98.2% of Ent3p puncta became positive for Ent5p (n=114 puncta, 38 cells) and almost all Ent5p puncta derived from Ent3p puncta (98.5% , n=67 puncta, 23 cells). The peak-to-peak time between Ent3p and Ent5p was 8.4s (Table 1, row#4), slightly shorter than observed between Gga2p/Ent3p and AP-1. This difference can be attributed to a small population (≈20%) of puncta in which Ent5p intensity peaked close to that of Ent3p (Supplementary Information Fig. S1d), and a major population that peaked 9.8s after Ent3p. Similar results were obtained with Gga2p-mRFP and Ent5p-GFP (Fig 1e; Table 1, row#5; Supplementary Information Movie S4). In accord with these findings, most but not all Ent5p peaked coincident with AP-1 (Fig. 1f; Table 1, row#6; Supplementary Information Fig. S1e). By SIM, 100-200nm Ent5p puncta infrequently colocalized with Gga2p and more commonly overlapped with ß1 (Fig.2g; Supplementary Information Fig. S2d).
Together, our results reveal two waves of adaptor assembly: Gga2p and Ent3p assemble first, peaking along with a minor fraction of Ent5p, followed 10 seconds later by a spatially distinct peak of AP-1 assembly and most of Ent5p. These relationships correspond well with known physical and genetic interactions4.
Sequential adaptor assembly occurs at the TGN
Sec7p, an ARF GTPase nucleotide exchange factor, is a TGN peripheral membrane protein16-18. Sec7p-mRFP and Gga2p-GFP fluorescence profiles were coincident (Fig. 3a; Table 1, row#7), providing evidence that the major site of Gga2p assembly is the TGN. Sec7p-mRFP peaked 10.4s before β1-GFP, the same time separation observed between Gga2p and AP-1 (Fig. 3b; Table 1, rows#1,8). 95.4% of Sec7p-mRFP puncta became positive for AP-1 (n=154 puncta, 68 cells) and 94.4% of AP-1 puncta arose from Sec7p puncta (n=108 puncta, 53 cells). These data indicate that the process of sequential clathrin adaptor assembly originates at the TGN.
Figure 3. Adaptor and clathrin dynamics at the TGN.
(a-f) Panels presenting the indicated proteins as in Figure 1. Scale bar = 2 μm. Time to acquire one image pair was 1.1-1.3 s. (a) GPY4933, (b) GPY4934, (c) GPY4935, (d) GPY4931, (e) GPY4932, (f) GPY4936.
Clathrin and AP-1 dynamics depend on Gga proteins
Clathrin assembly, monitored with Chc1p-mRFP, peaked at about the same time as Gga2p (Fig. 3d; Table 1, row#9). In static images of cells, 56% of Gga2p colocalized with Chc1p and significant overlap was observed by SIM, as expected for assembly of individual Gga2p-containing clathrin coats (Fig. 2h). In cells expressing Chc1p-mRFP and β1-GFP, the peak of Chc1p preceded the AP-1 peak by 8s (Table 1, row#10), consistent with the relative timing of Gga2p and AP-1 assembly. However, in contrast to Gga2p, substantial clathrin fluorescence persisted through the AP-1 peak (Fig. 3e). AP-1 colocalization with Chc1p in static images was lower than observed for Gga2p; 44% (n=75 cells) vs. 56% (n=99 cells), p=.002. Similarly, the overlap between AP-1 and clathrin was less apparent by SIM, suggesting that at steady state clathrin is preferentially associated with Gga2p-enriched coats (Supplementary Fig. S2b).
In contrast to wild-type cells, cells lacking Gga proteins exhibited Chc1p peak intensity at virtually the same time as AP-1 (0.79s; Fig. 3f; Table 1, row#11) and colocalization between AP-1 and clathrin increased from 44% (n=75 cells) to 58% (n=78 cells), p=.001. Similar results were obtained in cells lacking Gga proteins and Ent3p (L.D., unpublished). As expected from these findings, SIM analysis of gga1Δ gga2Δ ent3Δ cells revealed greater colocalization between clathrin and AP-1 (Supplementary Information Fig. S2c). Thus, Gga proteins appear to establish the initial timing and localization of clathrin assembly at the TGN. Consistent with this interpretation, inactivation of AP-1 (β1Δ) did not alter the relative timing of Gga2p and Chc1p assembly, except to eliminate the shoulder of clathrin fluorescence that corresponds to the peak of AP-1 in wild-type cells (Table 1, row#12; Supplementary Fig. S3).
In gga1Δ gga2Δ cells, AP-1 assembly relative to Sec7p-mRFP was delayed by ≈2.5-fold compared to wild-type (Fig 3b,c; Table 1, rows#13-14). Thus, timing of AP-1 assembly depends on Gga proteins, suggesting that AP-1 recruitment is coupled to prior assembly of Gga/clathrin coats.
Arf1p influences adaptor dynamics
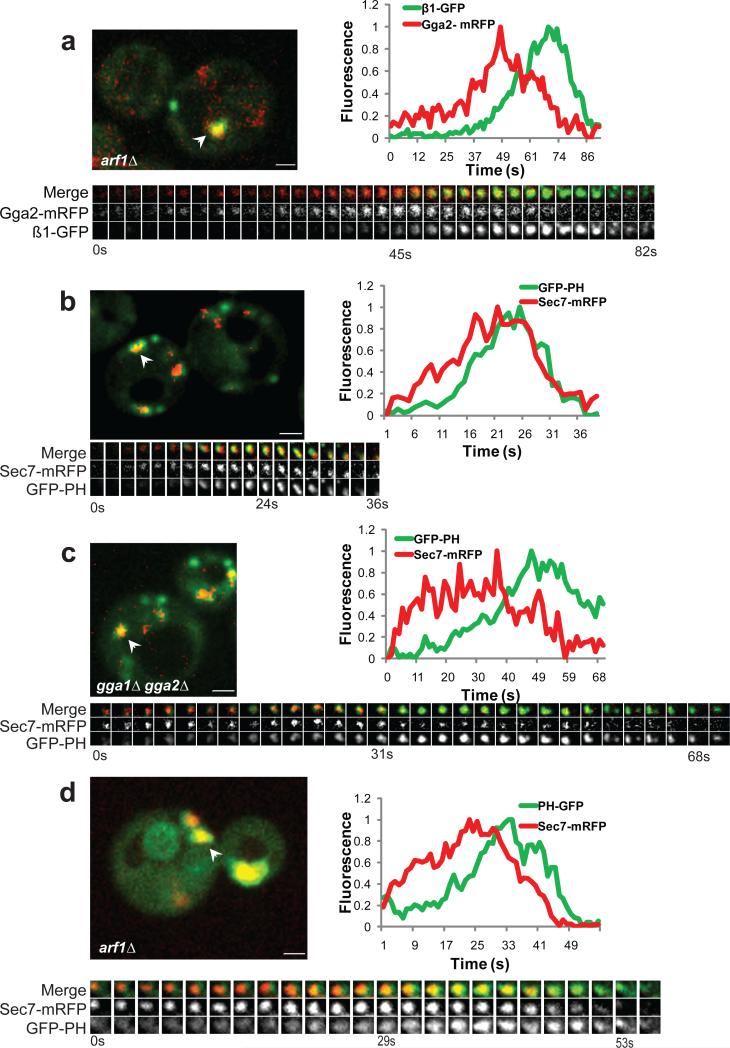
Arf GTPases are associated with Gga- and AP-1-mediated protein transport from the TGN1. There are two Golgi-localized Arf proteins in yeast, Arf1p and Arf2p. Both Yeast Gga proteins bind to Arf-GTP but this interaction is not absolutely required for Gga protein localization and function7,13. In contrast, AP-1 localization is more dependent on Arf-GTP7. Adaptor dynamics were investigated in cells carrying a deletion of ARF1, which is expressed at ten times the level of ARF2 (arf1Δ arf2Δ cells are inviable19). Although Golgi elements coalesce into a limited number of large structures in arf1Δ cells20, there was a clear progression from Gga2p to AP-1 (Fig. 4a, Supplementary Information Movie S5). However, AP-1 peak fluorescence was delayed by two-fold compared to wild-type (Supplementary Information Fig. S1f; Table 1, rows#15-16).
Figure 4. TGN PI4P dynamics depend on Arf1p and Gga proteins.
Panels presenting the indicated strains as described for Figure 1. Scale bar = 1 μm. Time to acquire one image pair was between 1.0-1.2s. GFP-PH (GFP-PHOSH1). (a) GPY4937, (b) GPY4938, (c) GPY4939, (d) GPY4955.
Temporal modulation of PI4P levels by Gga and Arf1 proteins
PI4P binds to, and promotes localization of, GGA proteins and AP-1 in mammalian cells21,22. A similar low affinity interaction between PI4P and Gga protein occurs in yeast23. To monitor PI4P levels, we used GFP fused to the PI4P-binding PH domain of Osh1p (GFP-PHOSH1; ref. 24). GFP-PHOSH1 reached maximum intensity 5.3s after the peak of Gga2p and 4.1s before the peak of AP-1 (Table 1, rows#17-18). Consistent with this sequence, GFP-PHOSH1 also peaked after Sec7p-mRFP (Fig. 4b; Table 1, row#19). In gga1Δ gga2Δ cells, GFP-PHOSH1 exhibited a ≈3-fold delay in maximum recruitment compared to Sec7p-mRFP (Figure 4c; Table 1, rows#19-20). Similarly, GFP-PHOSH1 recruitment was slowed in arf1Δ cells (Fig. 4d; Table 1, row#21), known to have reduced PI4P levels25. These results suggest that PI4P increases at the TGN after recruitment of Sec7p and Gga2p and peaks prior to the maximum levels of AP-1. Like AP-1, the normal kinetics of PI4P accumulation depends on Gga and Arf1 proteins.
Depletion of PI4P inhibits sequential assembly of clathrin adaptors
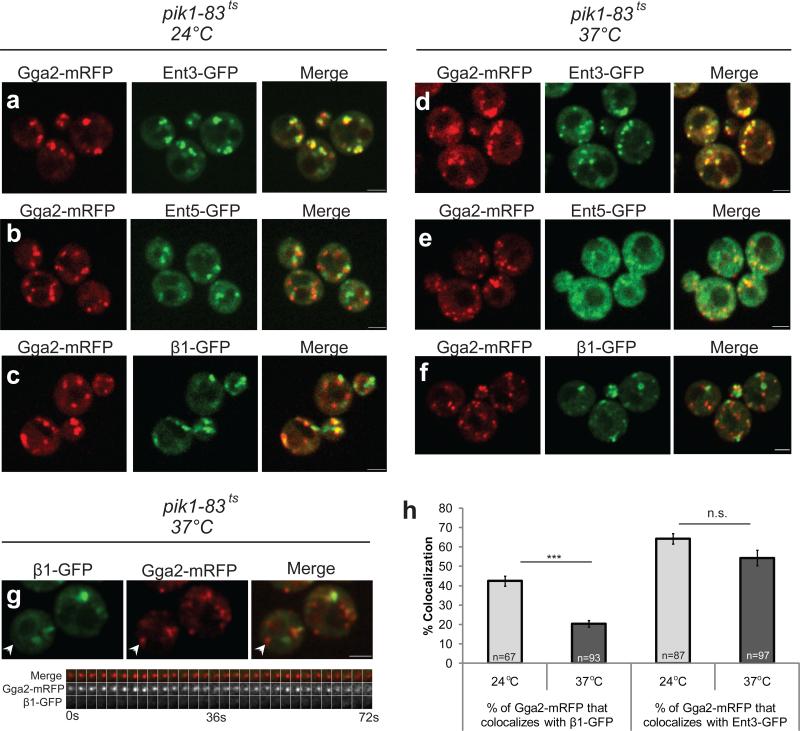
PI4P levels at the Golgi were lowered using a temperature-sensitive allele of PIK1 (pik1-83ts), encoding the PI4-kinase associated with Golgi function in yeast25,26. At the permissive temperature, GFP-PHOSH1 displayed a wild-type localization pattern. After 30 minutes at 37°C, the reporter was primarily cytoplasmic (Supplementary Information Fig. S4a-d), indicative of reduced PI4P levels. There was no significant effect of Pik1p inactivation on localization of a PI3P reporter, GFP-FYVE (Supplementary Information Fig. S4e-h).
In pik1-83ts cells shifted to 37°C, Gga2p and Ent3p maintained punctate colocalization (Fig. 5a,d). By comparison, a significant fraction of Ent5p was redistributed to the cytoplasm (Fig. 5b,e). Under the same conditions, AP-1 assembled, although little AP-1 was recruited to Gga2p puncta (Fig. 5c,f,g). This uncoupling resulted in a significant decrease in Gga2p/AP-1 colocalization in static images whereas there was little change in Gga2p/Ent3p colocalization (Fig. 5h). Gga2p/AP-1 colocalization was not restored by introducing an enzymatically inactive Pik1p mutant27 (Supplementary Information Fig. S5a). The relative colocalization of Gga2p and AP-1 was equivalent to wild-type in ent5Δ cells (G.C., unpublished) making it unlikely that the effect of Pik1p inactivation on the relationship between Gga2p and AP-1 was due to Ent5p mislocalization.
Figure 5. Depletion of PI4P alters localization of AP-1 and Ent5p.
Representative still images from live cells of (a, d) GGA2-mRFP ENT3-GFP pik1-83ts (GPY4940), (b, e) GGA2-mRFP ENT5-GFP pik1-ts83 (GPY4941) and (c, f) GGA2-mRFP β1-GFP pik1-ts83 (GPY4942) incubated at 24°C (a-c) or shifted for 30 min. to 37°C (d-f). (g) Top panel: still image from live cells of GPY4942 shifted to 37°C for 30 min. White arrowhead highlights puncta in kymograph in the bottom panel. Scale bar = 2 μm. Bottom panel: three channel kymograph of the selected puncta; time to acquire one image pair was 1.2s. Every other image pair is shown in the kymograph. (h) Gga2p-mRFP colocalization with β1-GFP (GPY4942) or Ent3-GFP (GPY4940) was quantified in pik1-83ts cells at 24°C (grey bars) or after shift to 37°C for 30 min. (black bars). Error bars are standard error (S.E.M.); n is the number of events; *** denotes p < 0.001, n.s. denotes non-significant (two-tailed t-test).
PI3P synthesis was eliminated by deleting VPS34, which encodes the only PI3-kinase in yeast28. The PI3P reporter GFP-FYVE, but not GFP-PHOSH1, was primarily cytoplasmic in vps34Δ cells, demonstrating a specific loss of PI3P (Supplementary Information Fig. S5b,c). In vps34Δ cells, we detected punctate localization and sequential assembly of Gga2p and AP-1 as well as Ent3p and Ent5p, similar to wild-type cells (Supplementary Information Fig. S5d,e). Together our findings support a specific role for PI4P in recruitment of Ent5p and AP-1 to Gga2p/Ent3p-positive Golgi membranes.
Pik1p overexpression increases the rate of adaptor progression
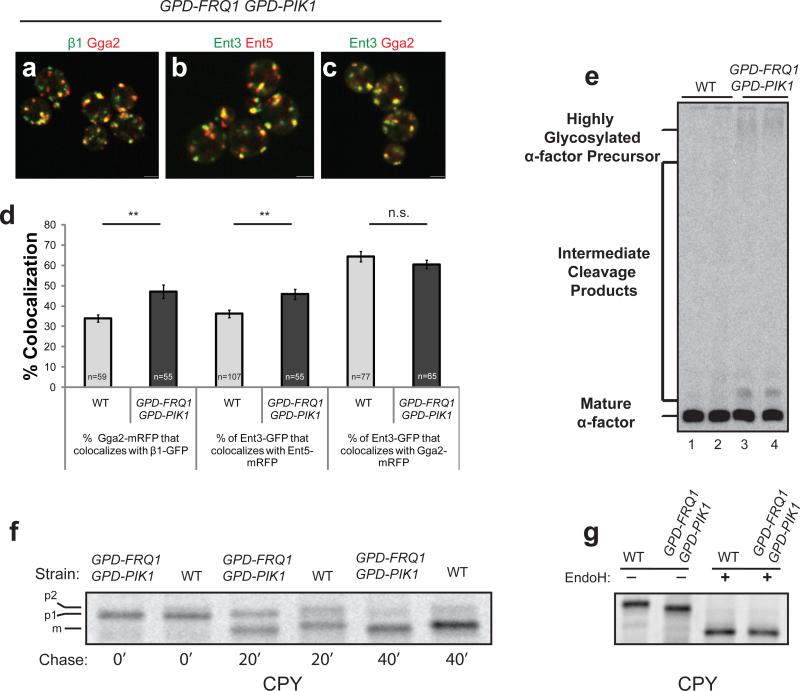
To elevate PI4P levels, we used strong constitutive promoters to drive expression of PIK1 and FRQ1, an adaptor necessary for Pik1p localization29. In these cells, GFP-PHOSH1 peaked simultaneously with Gga2p, about 5s earlier than in wild-type cells, providing evidence for increased rates of PI4P synthesis at the TGN (Table 1, rows#22-23). Under these conditions, we observed significantly higher Gga2p/AP-1 colocalization and a two-fold reduction in the peak to peak separation (Fig. 6a,d; Supplementary Information Fig. S1g; Table 1, rows#24-25). Similar results were obtained with Ent3p and Ent5p (Fig. 6b,d; Table 1, rows#26-27). Levels of colocalization between Ent3p and Gga2p were not altered (Fig. 6c-d). The diametric effects of elevating or reducing PI4P levels identify this phosphoinositide as a critical factor controlling TGN clathrin adaptor progression.
Figure 6. Pik1p/Frq1p overexpression enhances co-localization of sequentially-recruited clathrin adaptors with functional consequences.
(a-c) Representative still images from live cells of the following strains: (a) GPD-PIK1 GPD-FRQ1 β1-GFP GGA2-mRFP (GPY4943), (b) GPD-PIK1 GPD-FRQ1 ENT3-GFP ENT5-mRFP (GPY4944), (c) GPD-PIK1 GPD-FRQ1 ENT3-GFP GGA2-mRFP (GPY4945). Scale bar = 2 μm. (d) Colocalization of the indicated adaptors was quantified in wild-type (wt) and Pik1p/Frq1p overexpressing strains. Left two bars: GPY3109 (grey bar) vs GPY4943 (black bar); Middle 2 bars: GPY3912 (grey bar) vs GPY4944 (black bar); Right two bars: GPY3954 (grey bar) vs GPY4945 (black bar). Error bars are standard error (S.E.M.); n is the number of events; ** denotes p < 0.005, n.s. denotes non-significant (two tailed t-test). (e) Secreted α-factor immunoprecipitated from the media of radiolabelled cells. Lane 1: wild-type (WT, SEY6210), Lane 2: GGA2-mRFP β1-GFP (WT, GPY3109), Lane 3: GPD-PIK1 GPD-FRQ1 (GPY 4961), Lane 4: GPD- PIK1 GPD-FRQ1 GGA2-mRFP β1-GFP (GPY4943). (f) Pulse-chase immunoprecipitation of CPY. Cells from WT (SEY 6210) and GPD-PIK1 GPD-FRQ1 (GPY4961) were metabolically labeled with [35S]methionine- cysteine for 10 min at 24°C then labeling was quenched with nonradioactive amino acids. Samples of cells were removed at the indicated time points and CPY was immunoprecipitated and analyzed by SDS-PAGE and autoradiography. (g) Endoglycosidase H treatment of CPY. CPY was immunoprecipitated from a 40 min. time point of a pulse-chase as in (f) and analyzed directly (EndoH −) or treated with endoglycosidase H (EndoH +).
To assess the fidelity of clathrin-mediated transport between the TGN and endosomes in Pik1p/Frq1p-overexpressing cells, maturation of the mating pheromone α-factor was evaluated. Pheromone maturation is initiated in the TGN by the Kex2p protease, which relies on clathrin, AP-1 and Gga-dependent cycling between the TGN and endosomes for localization5,30-32. Compared to wild-type cells, Pik1p/Frq1p-overexpressing cells displayed partial α-factor maturation defects [Fig. 6e; 8.6±1.3% (n=4) precursor forms in Pik1p/Frq1p-overexpressing cells vs. 3.2±1.2% (n=4) in wild-type cells (p<0.01)]. This result provides evidence that clathrin-mediated TGN localization of Kex2p is perturbed in Pik1p/Frq1p-overexpressing cells.
We also observed a defect in glycosylation of carboxypeptidase Y (CPY). The core oligosaccharides added to CPY in the endoplasmic reticulum (p1 CPY) are extended in the Golgi apparatus (p2 CPY). p2 CPY is proteolytically matured in the vacuole (mCPY). The Golgi α1-3 mannosyltransferase Mnn1p adds the final sugars to generate p2 CPY33. Localization of Mnn1p to the TGN, like Kex2p, depends on clathrin34. In pulse-chase experiments, p2 and mCPY in Pik1p/Frq1p-overexpressing cells were slightly smaller than in wild-type cells (Fig. 6f,g). The p1 forms in the two strains were identical (Fig. 6f) and endoglycosidase H treatment eliminated the difference between mCPY species (Fig. 6g), indicating that the smaller sizes of p2 and mCPY in Pik1p/Frq1p-overexpressing cells is due to incomplete glycosylation in the Golgi. The defects in α-factor maturation and CPY glycosylation suggest that increased synthesis of PI4P and shortened adaptor progression times are associated with compromised clathrin-mediated TGN-endosome traffic, likely because precocious AP-1 assembly sorts Kex2p and Mnn1p away from their substrates in the TGN.
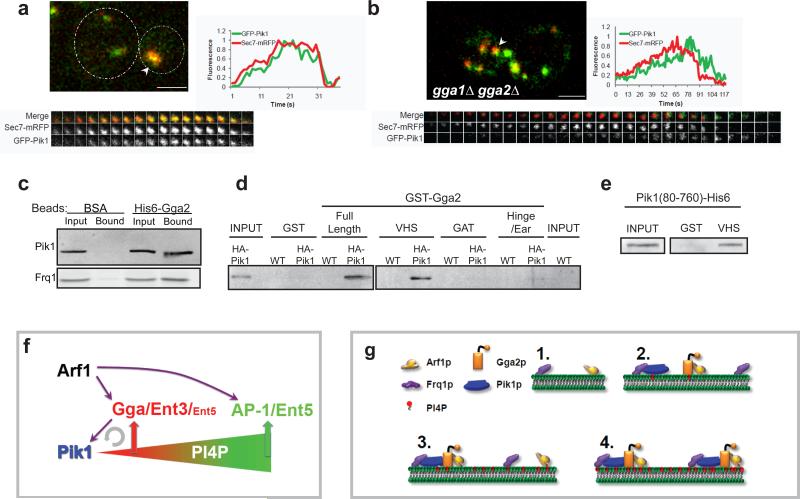
Gga2p acts in Pik1p recruitment and directly binds Pik1p
In wild-type cells, Frq1p-GFP peaked at nearly the same time as Sec7p-mRFP whereas GFP-Pik1p reached maximum levels 4.1s later, similar to the peak of PI4P (Fig. 7a; Table 1 rows#19,28,30). In gga1Δ gga2Δ cells, Pik1p recruitment was delayed, like PI4P levels, by 3-fold compared to Sec7p (12.6s, Fig. 7b, Table 1 rows#28-29). The profile of Frq1p was not altered (Table 1, rows#30-31). Pik1p and Frq1p in cell extracts bound to full length Gga2p (Fig. 7c, Supplementary Information S6a,b). Moreover, Pik1p from cell extracts interacted specifically with the Gga2p VHS domain (Fig. 7d, Supplementary Information S6c) and the Gga2p VHS domain bound directly to a recombinant Pik1p fragment (Fig. 7e, Supplementary Information S6d). Together these results provide evidence that direct physical interactions between Gga proteins and Pik1p contribute to Pik1p recruitment to the TGN.
Figure 7. Gga2p acts in Pik1p recruitment and binds Pik1p.
(a,b) Panels presenting the indicated proteins and strains as for Figure 1. Time to acquire one image pair was 1.3 s. (a) GPY 4968. Dotted lines in upper left panel indicate cell boundaries; (b) GPY 4969. Scale bar =2 μm. (c) Extracts from cells expressing HA-Pik1p (GPY 4966, top panel) or Frq1p-GFP (GPY4967, bottom panel) were incubated with BSA-conjugated or His6-Gga2p-conjugated Sepharose. Bound proteins were eluted and analyzed by SDS-PAGE and immunoblotting. (d) Extracts of cells expressing HA-Pik1p (GPY 4966) were applied to glutathione beads coupled to GST, the indicated GST-Gga2p domains or to GST-Gga2p full length. Bound proteins were analyzed as in c. (e) Recombinant Pik1(aa80-720)-His6 was incubated with glutathione-Sepharose coupled to GST or to GST-Gga2pVHS (VHS). Bound proteins were analyzed as in c. (f) Model for regulation of PI4P and adaptor progression at the TGN. Arrows represent known regulatory interactions. Semicircular arrow indicates positive feedback loop. See text for details. (g) Model for positive feedback between Pik1p and Gga2p. 1. Arf1p and Frq1p are recruited to TGN membranes. 2. Frq1p recruits Pik1p, initiating low levels of PI4P production. Arf1p and PI4P (and cargo, not shown) recruit Gga2p. 3. The VHS domain of Gga2p interacts with Pik1p, helping to recruit or stabilize Pik1p at TGN membranes, thereby promoting PI4P production. 4. Increased PI4P stimulates recruitment of additional Gga2p which in turn further enhances Pik1p association with the TGN.
Discussion
Our results demonstrate two sequential waves of clathrin coat assembly that originate at the TGN and are distinguished by adaptor type. Gga2p, Ent3p and a minor population of Ent5p assemble in the first wave. AP-1 and most Ent5p are recruited in the second wave. The relative timing of AP-1-enriched coat assembly is dependent on Gga proteins and Arf1p, and the progression between coat types is controlled by PI4P. This coupled progression of adaptor-specific clathrin coat formation reveals a hitherto unrecognized process of TGN maturation.
The relationship between Gga and AP-1 adaptor function has not been clearly defined. Although the two adaptors share a number of interaction partners, including Arf, PI4P, and clathrin, they recognize different cargo sorting signals and certain accessory proteins, and only partially colocalize in static images1,4,9. We observed that the major populations of Gga2p and AP-1 were separated in time and space. These results suggest that most clathrin coats forming at the TGN consist primarily of one or the other type of adaptor, and so would be enriched with the corresponding cargo selectivity. This organization provides a simple mechanism to generate ccv at the TGN targeted to different compartments based on adaptor-directed incorporation of targeting/fusion proteins. In accord with this view, Gga proteins recruit Ent3p, which in turn binds SNAREs involved in targeting/fusion to late endosomes, thereby specifying the destination of Gga-enriched ccv35-37. Thus, temporally-regulated genesis of adaptor-specific coats represents a means to diversify the trafficking repertoire of coated vesicles budding from a particular compartment.
Our results indicate that PI4P is a key regulator of the TGN adaptor assembly sequence. Changes in PI4P levels that alter adaptor progression are accompanied by defects associated with adaptor function. For example, in combination with pik1-ts alleles that lower PI4P levels, deletions of Gga proteins or AP-1 subunits result in synthetic growth and/or partial α-factor maturation defects (ref. 23; L.D. and G.C., unpublished). Importantly, increased PI4P synthesis due to Pik1p/Frq1p overexpression also leads to incomplete α-factor maturation and CPY glycosylation. These findings support the view that adaptor progression contributes to optimal function of clathrin-mediated traffic from the TGN.
The effects of PI4P on adaptor assembly are likely to be, at least in part, direct. Mammalian AP-1 binds PI4P and the residues in the mammalian AP-1 γ subunit necessary for phosphoinositide binding are conserved in the yeast protein22. Ent5p contains an N-terminal domain homologous to phosphoinositide-binding ANTH domains. Binding of Ent5p (and Ent3p) to PI3P and PI(3,5)P2 has been reported39-41, however the specificity of this interaction has been questioned42. Our findings are most consistent with a primary role for PI4P in Ent5p localization in vivo.
Like AP-1, Gga2p binds to PI4P and Arf1p through low affinity interactions that cooperate to enhance Gga2p membrane association23. However, Gga2p does not absolutely require Arf interaction for localization13 and our findings indicate that acute PI4P reduction does not drastically alter Gga2p recruitment. Thus, our in vivo analyses reveal differential dependencies of Gga2p and AP-1 on shared interaction partners, providing a basis for the observed temporal separation of adaptor assembly. Adaptor-specific interactions may also contribute to the spatial and temporal patterns of assembly7,43. We favor the view that spatial separation of adaptor-specific coats reflects temporally-distinct assembly events (Supplementary Information Fig. S7).
The Gga2p VHS domain directly binds Pik1p and deletion of the GGA genes delayed recruitment of Pik1p to the TGN, providing evidence that binding of Pik1p to Gga proteins contributes to Pik1p recruitment. A defect in Gga-mediated Pik1p recruitment accounts for our finding that cells lacking Gga proteins or Arf1p accumulated PI4P more slowly and displayed a corresponding delay in AP-1 assembly. These results support a model in which assembly of Gga-enriched coats with Arf1p at the TGN stimulates Pik1p recruitment and the attendant increase in PI4P synthesis in turn promotes assembly of AP-1/Ent5p coats (Fig. 7f). Notably, recruitment of Pik1p by the VHS domain of Gga proteins, and the combined contribution of Pik1p-generated PI4P and Arf1p to Gga2p localization, constitute a positive feedback pathway to drive PI4P accumulation at the TGN (Fig. 7g). Thus, our model posits a regulatory network converging on Pik1p to generate a temporal gradient of PI4P that controls adaptor progression.
PI4P-coupled progression of Gga2p- to AP-1-enriched coats represents a previously unrecognized maturation process at the TGN (Fig. 7f, Supplementary Fig. S7). Although there are some differences in localization mechanisms of yeast and mammalian Gga proteins and AP-1, the strong conservation of binding partners, including ARF and PI4P, favors the view that a similar progression occurs in mammalian cells. Supporting this possibility, mammalian GGA proteins often localize with a more compact perinuclear distribution than AP-1, and by immuno-electron microscopy are more prevalent than AP-1 on uncoated TGN membranes, consistent with assembly of GGA proteins at an earlier TGN stage than AP-138,44,45.
A PI3P-controlled maturation process occurs in the endocytic pathway at early endosomes46 that, although mechanistically distinct, is analogous to what we describe for the TGN. Taken together, these findings reveal phosphoinositide-based maturation as a mechanism that allows temporal subspecialization within major organelles in both the secretory and endocytic pathways.
Methods
Media and Strains
Strains used in this study are listed in Supplemental Table 14,7,25,47,48. Yeast strains were grown in standard rich medium (YPD) or synthetic dextrose medium (SD) with the appropriate supplements. Fluorescent tags and deletions were introduced at endogenous loci using standard PCR-based homologous recombination. All tagged genes were fully functional as assessed by growth of cells harboring a tagged gene in a genetic background where deletion of the gene causes severe growth defects.
Except where noted here, all strains were generated from diploid cells by mating, sporulation, and isolation of haploid spores. Strains expressing GFP-PHOSH1 under control of the PHO5 promoter were generated by integrating pGFP-PHOSH1 at the URA3 locus of haploid cells24. FYVE-GFP expressing strains were obtained by transformation of haploid cells with pRS316-FYVE-GFP. Strains overexpressing Pik1p and Frq1p were generated by integrating the glucose phosphate dehydrogenase promoter at the 5’ end of both PIK1 and FRQ49 and then carrying out the appropriate crosses. The VPS34 gene disruption was generated by replacing the first 2190bp of the coding region with TRP1 in diploid cells, which were then sporulated for isolation of haploids. All integrations were confirmed by PCR. All primers used for integrations and deletions are available upon request.
Plasmids
pFA6a-2xGFP(S65T)-HIS3MX6
Primers 5’gctgcaggtcgacggatccccgggctaattaacagtaaaggagaagaacttttc3’ and 5’gtattcgttaattaaagatctgagtccggatttgtatagttcatccatgccatgtgtaatcccagcagctgt3’ were used with pFA6a-GFP(S65T)-His3MX650 to amplify GFP(S65T), introducing restriction sites for SalI and PacI. The PCR product was then treated with SalI and PacI and introduced into the same sites in front of GFP(S65T) on pFA6a-GFP(S65T)-His3MX6 to create pFA6a-2xGFP(S65T)-His3MX6. The construct was confirmed by sequencing.
pFA6a-2XmRFP-TRP1
Primers 5’aattcacaccggcgcctccggactcagatctatggcctcctccgaggacgtcatcaaggagttcatgcgc3’ and 5’agaagtggcgcgccttaggcgccggttgagtggcggccctcggcgcgctcg3’ were used with pFA6a-mRFP-TRP151 to amplify mRFP, introducing restriction sites for SgrAI and AscI. The PCR product was then treated with SgrAI and AscI and introduced into the same sites at the end of mRFP on pFA6a-mRFP-TRP1 to create pFA6a-2xmRFP-TRP1. The construct was confirmed by sequencing.
pRS316-GFP-FYVE
A BamH1 fragment encompassing GFP-FYVE from pGFP-FYVEEEA1-URA352 was inserted into the BamHI sites of pRS31653.
pPik1-GFP(A8)-PIK1
A BamHI/EcoRI fragment from pFA6a-natNT2 (Euroscarf, accession #p30346) was ligated into pBluescript KS(+) to generate pB-natNT2. Then, PCR fragments containing nucleotides −550 to −351 from the promoter region of PIK1 and −350 to +96 were inserted into SacII/NotI and EcoRI sites of pB-natN2 to generate pB-natN2-PIK1(−550+97). The EcoRI fragment was checked by sequencing. Finally, a PCR fragment containing GFP from pGFP-FYVE and nucleotides coding for an 8 Ala linker insertion at the 3’ was cloned into an existing NsiI site at the ATG start site of PIK1 to generate pB-NatN2-GFP(A8)-PIK1. To integrate into the PIK1 genomic locus, pB-NatN2-GFP(A8)-PIK1 was cut with SacII/EcoRV prior to transformation.
pGex4T1-Gga2(1-169), pGex4T1-Gga2(170-336), pGex4T1-Gga2(337-585)
Relevant regions of GGA2 [aa1-169 (VHS), aa170-336 (GAT), and aa337-585 (hinge/γ-ear)] were amplified by PCR and inserted into BamHI/SalI sites of pGex-4T-1 (GE Healthcare, UK). The plasmids were checked by restriction enzyme analysis and DNA sequencing.
pRS316-pik1(D918A)
An NheI/SacI fragment from pRS314-GAL1-mycPIK1(D918A) (PTS12 27) was inserted into pRS316-PIK1 29. The plasmid was checked by restriction analysis and DNA sequencing.
pET28a-Pik1(80-760)
A fragment encoding aa 250-760 of Pik1 was amplified by PCR and inserted into NcoI/SalI sites of pET28(a) (Novagen, EMD Biosciences) in frame with the carboxy-terminal His6 tag, creating pET28a-Pik1(250-760). The plasmid was checked by restriction analysis and DNA sequencing . An NcoI/NheI fragment from pRS316-PIK1 was then inserted into pET28a-Pik1(80-760) to generate pet28a-Pik1(80-760), checked by restriction analysis.
Cell Fixation and Structured Illumination Microscopy (SIM)
Cells were grown overnight in YPD media to 0.4-0.8 × 107 cells/ml. Cells were sedimented at 750g for 3 min and resuspended in 4% paraformaldehyde, 42mM NaOH, 100mM KH2PO4, 100mM sucrose, 1mM MgCl2 pH5.5 at room temperature for 15 minutes. Fixed cells were sedimented, washed once in 1.2M Sorbitol, 0.1M KPO4, pH7.4 and resuspended in 1.2M Sorbitol, 0.1M KPO4 pH7.4.
Structured Illumination Microscopy achieves resolution of fluorescent objects below the diffraction limit by passing structured light through a diffraction grating to generate multiple interfering beams of light, collecting images, then repeating the process twice with the diffraction grating rotated 60° from the previous position15. The resulting super resolution image is computationally reconstructed based on changes in the fluorescent patterns between images15. Images were captured using a 60X, 1.42 NA objective on an OMX microscope (Applied Precision, Issaquah, Wa) equipped with two Photometrics Cascade II 512 cameras for the GFP and mRFP channels. Each channel was acquired simultaneously by excitation at 488 and 561.5 nm, respectively. A diffraction grating illuminated samples at the indicated wavelengths at 125 nm intervals, at three different 60° angles15. 3D images were reconstructed using SoftWoRx (Applied Precision).
Live Cell Microscopy
Cells were grown in SD complete or SD-uracil at room temperature to a density of 0.1–0.3 × 107 cells/ml. Cells were sedimented by centrifugation at 750g for 2 minutes, resuspended in 100μl of the appropriate media and then imaged at room temperature. GFP and mRFP channels were exposed for between 100-500ms per frame, and a mRFP/GFP frame pair was collected every 1-2 seconds. Typically 100-200 GFP/mRFP frame pairs were collected per time lapse movie.
Image Acquisition and Analysis
Images were captured using a 100X/1.45NA objective on a Marianas spinning disc confocal microscope equipped with a Zeiss AxioObserver Z1, Yokogawa CSU-22 Confocal Head, and a Hamamatsu EMCCD C9100-13 camera, all controlled by Slidebook 4.2 software. GFP and mRFP images were acquired by excitation at 488 nm and 561nm from a high speed AOTF laser launch line. The microscope and all of its components were controlled by Slidebook 4.2.
Time-lapse movies were analyzed using Slidebook 4.2. Movies were first photobleach-corrected and then the average fluorescent intensity of a tracked puncta was measured for each frame over its lifetime. The time point with the highest average value was taken as the time point of peak fluorescence and the time point with the lowest average value was taken as the minumum. Normalized fluorescent intensity at time point X = (intensity at time point X – minimum intensity) / (maximum intensity - minimum intensity). Two criteria were applied to select puncta for data collection: 1) the structure had to be present for more than 7 frames (~8.4s); 2) each structure had to remain distinct from other puncta.
Colocalization was quantified by creating masks from the GFP and mRFP channels after sequential Gaussian and Laplacian filtering. The accuracy of each mask was checked by visual comparison to the original image. Colocalization between proteins was expressed as the percentage of pixels from the corresponding channel that overlap with the other channel from the same image7.
Statistical Analysis
Statistical significance of peak-to-peak and colocalization data was tested using the unpaired, students t-test. Significance of the α-factor data was tested using the Chi-Squared Test. Results are expressed as the mean ± S.E.M.
Metabolic Labeling, Immunoprecipitation, and Affinity Binding
Metabolic labeling and immunoprecipitation of α-factor from the media was carried out at 24°C as described5. Pulse-chase immunoprecipitation of CPY was performed as described for carboxypeptidase S4 except that antibody to CPY was used. EndoH treatment was as described in ref. 54.
For binding experiments, N-terminal GST fusions of Gga2p aa1-169 (VHS domain), aa170-336 (GAT domain), aa337-585 (Hinge/Ear domain), full length GST-Gga2, full-length His6-Gga2 and Pik1(aa80-760)-His6 were expressed in BL21-DE3 (Codon +) E. coli. Bacteria were lysed in PBS pH7.4 (and 30 mM imidazole for Pik1 fragments) with a protease inhibitor mixture (Roche Diagnostics GmbH, Mannheim, Germany) by sonication. The resulting lysate was cleared by centrifugation for 20 min. at 3000 × g. Glutathione-Sepharose (Gga2 fragments) or Ni-NTA beads (Pik1 fragment or His6-Gga2) were added to the supernatant and allowed to bind for 1 hour at 4°C. GST fusions bound to glutathione were used directly. The bound Pik1 fragment or His6-Gga2p were washed 4 times with PBS, then 3 times with 50mM Hepes, 50mM NaCl, 30mM imidazole, and then eluted in 50mM Hepes, 50 mM NaCl and 300mM imidazole. His6-Gga2p and BSA were coupled to CNBr-activated Sepharose following manufacturer instructions (GE Healthcare). Proteins were analyzed by SDS-polyacrylamide gel electrophoresis and immunoblotting with anti-HA antibody (Covance) at a 1:1000 dilution.
For affinity binding, yeast cells were grown to mid-logarithmic phase in YPD medium. A total of 15 × 108 cells per sample were converted to spheroplasts and lysed by resuspension in a final volume of 1 ml of lysis buffer : 100mM MES-NaOH pH6.5, 1.5mM MgCl2, 2mM CaCl2, 0.2mM DTT, 2mM NaN3 containing 1% Triton X-100 and a protease inhibitor mixture5. The extract was clarified by centrifugation for 30 min at 16,000 × g at 4°C. The supernatant was incubated for 30 min in the presence of Protein A Sepharose (Amersham Pharmacia Biotech) followed by centrifugation for 20s at 16,000 × g. The resulting supernatant was brought to 1 ml in lysis buffer and BSA-conjugated or His6-Gga2-conjugated Sepharose was added; the samples were incubated for 1.5 hours at 4°C, washed twice in lysis buffer, then twice in lysis buffer without Triton X-100, and eluted with Sample Buffer at 100°C for 3 min. (2% SDS, 10% Glycerol, 62.4 mM Tris-HCl pH 6.8, 0.1 mg/ml Bromophenol Blue, 0.4% β-Mercaptoethanol). Proteins were analyzed by SDS-polyacrylamide gel electrophoresis and immunoblotting with anti-GFP (Scantibodies, 1:1000) or anti-HA (Covance, 1:1000) antibody and goat anti-rabbit IgG conjugated with alkaline phosphatase (Bio-Rad Richmond, Ca). Yeast lysates for affinity binding experiments with N-terminal GST fusions of Gga2 were carried out as described above except that yeast were lysed in 50mM Hepes, 50mM NaCl, 1% Triton X-100. Beads containing GST only or GST-Gga2 fragments were incubated with the yeast lysate for 2 hours at 4°C. Washing and elution of the beads were carried out as described above.
Direct binding experiments were carried out by incubating purified Pik1(80-760)-His6 with GST or GST-VHS (Gga2p) bound glutathione-Sepharose in 50mM Hepes, 50mM NaCl and 300mM imidazole for 1 hour at 4°C. Washing and elution were carried out as described above. To detect Pik1(80-760)-His6, penta His monoclonal antibody (Qiagen) was used for immunoblot (1:1000 dilution).
Supplementary Material
Acknowledgements
We thank Jeremy Thorner, Scott Emr and Timothy Levine for plasmids and strains, Jennifer Atkins for help with SIM, and members of the lab for helpful discussions. We are especially grateful to Karl Kilborn for advice and assistance with confocal microscopy and Kelsey Martin, Gerry Weinmaster, and Alex van der Bliek for comments on the manuscript. This work was supported by NIH NRSA T32 GM-007104 and a UCLA Dissertation Year Fellowship to L.D. and NIH GM39040 to G.P.
Footnotes
Author Contributions L.D. and G.P. conceived the experiments. L.D. carried out all microscopy, protein interaction, and CPY experiments. G.C. carried out the α-factor experiments, and generated recombinant expression and integration constructs. L.D. and G.P. wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Traub LM. Common principles in clathrin-mediated sorting at the Golgi and the plasma membrane. Biochim Biophys Acta. 2005;1744:415–437. doi: 10.1016/j.bbamcr.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Duncan MC, Payne GS. ENTH/ANTH domains expand to the Golgi. Trends Cell Biol. 2003;13:211–215. doi: 10.1016/s0962-8924(03)00076-x. [DOI] [PubMed] [Google Scholar]
- 3.Black MW, Pelham HRB. A selective transport route from Golgi to late endosomes that requires the yeast GGA proteins. J. Cell Biol. 2000;151:587–600. doi: 10.1083/jcb.151.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costaguta G, Duncan MC, Fernandez GE, Huang GH, Payne GS. Distinct roles for TGN/endosome epsin-like adaptors Ent3p and Ent5p. Mol. Biol. Cell. 2006;17:3907–3920. doi: 10.1091/mbc.E06-05-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costaguta G, Stefan CJ, Bensen ES, Emr SD, Payne GS. Yeast Gga coat proteins function with clathrin in Golgi to endosome transport. Mol. Biol. Cell. 2001;12:1885–1896. doi: 10.1091/mbc.12.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duncan MC, Costaguta G, Payne GS. Yeast epsin-related proteins required for Golgi-endosome traffic define a gamma-adaptin ear-binding motif. Nat. Cell Biol. 2003;5:77–81. doi: 10.1038/ncb901. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez GE, Payne GS. Laa1p, a conserved AP-1 accessory protein important for AP-1 localization in yeast. Mol. Biol. Cell. 2006;17:3304–3317. doi: 10.1091/mbc.E06-02-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ha SA, et al. The synaptojanin-like protein Inp53/Sjl3 functions with clathrin in a yeast TGN-to-endosome pathway distinct from the GGA protein-dependent pathway. Mol. Biol. Cell. 2003;14:1319–1333. doi: 10.1091/mbc.E02-10-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham TR, Burd CG. Coordination of Golgi functions by phosphatidylinositol 4-kinases. Trends Cell Biol. 2011;21:113–121. doi: 10.1016/j.tcb.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polishchuk RS, San Pietro E, Di Pentima A, Tete S, Bonifacino JS. Ultrastructure of long-range transport carriers moving from the trans Golgi network to peripheral endosomes. Traffic. 2006;7:1092–1103. doi: 10.1111/j.1600-0854.2006.00453.x. [DOI] [PubMed] [Google Scholar]
- 11.Puertollano R, et al. Morphology and dynamics of clathrin/GGA1-coated carriers budding from the trans-Golgi network. Mol. Biol. Cell. 2003;14:1545–1557. doi: 10.1091/mbc.02-07-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waguri S, et al. Visualization of TGN to endosome trafficking through fluorescently labeled MPR and AP-1 in living cells. Mol. Biol. Cell. 2003;14:142–155. doi: 10.1091/mbc.E02-06-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boman AL, et al. ADP-ribosylation factor (ARF) interaction is not sufficient for yeast GGA protein function or localization. Mol. Biol. Cell. 2002;13:3078–3095. doi: 10.1091/mbc.E02-02-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newpher TM, Smith RP, Lemmon V, Lemmon SK. In vivo dynamics of clathrin and its adaptor-dependent recruitment to the actin-based endocytic machinery in yeast. Dev. Cell. 2005;9:87–98. doi: 10.1016/j.devcel.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 15.Schermelleh L, et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–1336. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franzusoff A, Redding K, Crosby J, Fuller RS, Schekman R. Localization of components involved in protein transport and processing through the yeast Golgi apparatus. J. Cell Biol. 1991;112:27–37. doi: 10.1083/jcb.112.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rossanese OW, et al. A role for actin, Cdc1p, and Myo2p in the inheritance of late Golgi elements in Saccharomyces cerevisiae. J. Cell Biol. 2001;153:47–62. doi: 10.1083/jcb.153.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Losev E, et al. Golgi maturation visualized in living yeast. Nature. 2006;441:1002–1006. doi: 10.1038/nature04717. [DOI] [PubMed] [Google Scholar]
- 19.Stearns T, Kahn RA, Botstein D, Hoyt MA. ADP ribosylation factor is an essential protein in Saccharomyces cerevisiae and is encoded by two genes. Mol. Cell Biol. 1990;10:6690–6699. doi: 10.1128/mcb.10.12.6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaynor EC, Chen CY, Emr SD, Graham TR. ARF is required for maintenance of yeast Golgi and endosome structure and function. Mol. Biol. Cell. 1998;9:653–670. doi: 10.1091/mbc.9.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, et al. PI4P promotes the recruitment of the GGA adaptor proteins to the trans-Golgi network and regulates their recognition of the ubiquitin sorting signal. Mol. Biol. Cell. 2007;18:2646–2655. doi: 10.1091/mbc.E06-10-0897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang YJ, et al. Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell. 2003;114:299–310. doi: 10.1016/s0092-8674(03)00603-2. [DOI] [PubMed] [Google Scholar]
- 23.Demmel L, et al. The clathrin adaptor Gga2p is a phosphatidylinositol 4-phosphate effector at the Golgi exit. Mol. Biol. Cell. 2008;19:1991–2002. doi: 10.1091/mbc.E06-10-0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loewen CJ, Roy A, Levine TP. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 2003;22:2025–2035. doi: 10.1093/emboj/cdg201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Audhya A, Foti M, Emr SD. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol. Biol. Cell. 2000;11:2673–2689. doi: 10.1091/mbc.11.8.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walch-Solimena C, Novick P. The yeast phosphatidylinositol-4-OH kinase pik1 regulates secretion at the Golgi. Nat. Cell Biol. 1999;1:523–525. doi: 10.1038/70319. [DOI] [PubMed] [Google Scholar]
- 27.Strahl T, Hama H, DeWald DB, Thorner J. Yeast phosphatidylinositol 4-kinase, Pik1, has essential roles at the Golgi and in the nucleus. J. Cell Biol. 2005;171:967–979. doi: 10.1083/jcb.200504104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schu PV, et al. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- 29.Hendricks KB, Wang BQ, Schnieders EA, Thorner J. Yeast homologue of neuronal frequenin is a regulator of phosphatidylinositol-4-OH kinase. Nat. Cell Biol. 1999;1:234–241. doi: 10.1038/12058. [DOI] [PubMed] [Google Scholar]
- 30.Fuller RS, Sterne RE, Thorner J. Enzymes required for yeast prohormone processing. Annual Review of Physiology. 1988;50:345–362. doi: 10.1146/annurev.ph.50.030188.002021. [DOI] [PubMed] [Google Scholar]
- 31.Payne GS, Schekman R. Clathrin - a Role in the Intracellular Retention of a Golgi Membrane Protein. Science. 1989;245:1358–1365. doi: 10.1126/science.2675311. [DOI] [PubMed] [Google Scholar]
- 32.Phan HL, et al. The Saccharomyces cerevisiae APS1 gene encodes a homolog of the small subunit of the mammalian clathrin AP-1 complex: evidence for functional interaction with clathrin at the Golgi complex. EMBO J. 1994;13:1706–1717. doi: 10.1002/j.1460-2075.1994.tb06435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dean N. Asparagine-linked glycosylation in the yeast Golgi. Biochim Biophys Acta. 1999;1426:309–322. doi: 10.1016/s0304-4165(98)00132-9. [DOI] [PubMed] [Google Scholar]
- 34.Graham TR, Seeger M, Payne GS, MacKay VL, Emr SD. Clathrin-dependent localization of a1,3 mannosyltransferase to the Golgi complex of Saccharomyces cerevisiae. J. Cell Biol. 1994;127:667–678. doi: 10.1083/jcb.127.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Black MW, Pelham HR. A selective transport route from Golgi to late endosomes that requires the yeast GGA proteins. J. Cell Biol. 2000;151:587–600. doi: 10.1083/jcb.151.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chidambaram S, Zimmermann J, von Mollard GF. ENTH domain proteins are cargo adaptors for multiple SNARE proteins at the TGN endosome. J. Cell Sci. 2008;121:329–338. doi: 10.1242/jcs.012708. [DOI] [PubMed] [Google Scholar]
- 37.Wang J, et al. Epsin N-terminal homology domains bind on opposite sides of two SNAREs. Proc Natl Acad Sci U S A. 2011;108:12277–12282. doi: 10.1073/pnas.1013101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doray B, Ghosh P, Griffith J, Geuze HJ, Kornfeld S. Cooperation of GGAs and AP-1 in packaging MPRs at the trans-Golgi network. Science. 2002;297:1700–1703. doi: 10.1126/science.1075327. [DOI] [PubMed] [Google Scholar]
- 39.Chidambaram S, Mullers N, Wiederhold K, Haucke V, von Mollard GF. Specific interaction between SNAREs and epsin N-terminal homology (ENTH) domains of epsin-related proteins in trans-Golgi network to endosome transport. J. Biol. Chem. 2004;279:4175–4179. doi: 10.1074/jbc.M308667200. [DOI] [PubMed] [Google Scholar]
- 40.Eugster A, et al. Ent5p is required with Ent3p and Vps27p for ubiquitin-dependent protein sorting into the multivesicular body. Mol. Biol. Cell. 2004;15:3031–3041. doi: 10.1091/mbc.E03-11-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Friant S, et al. Ent3p Is a PtdIns(3,5)P2 effector required for protein sorting to the multivesicular body. Dev. Cell. 2003;5:499–511. doi: 10.1016/s1534-5807(03)00238-7. [DOI] [PubMed] [Google Scholar]
- 42.Narayan K, Lemmon MA. Determining selectivity of phosphoinositide-binding domains. Methods. 2006;39:122–133. doi: 10.1016/j.ymeth.2006.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singer-Kruger B, et al. Yeast and human Ysl2p/hMon2 interact with Gga adaptors and mediate their subcellular distribution. EMBO J. 2008;27:1423–1435. doi: 10.1038/emboj.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hirst J, et al. A family of proteins with gamma-adaptin and VHS domains that facilitate trafficking between the trans-Golgi network and the vacuole/lysosome. J. Cell Biol. 2000;149:67–80. doi: 10.1083/jcb.149.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poussu A, Lohi O, Lehto VP. Vear, a novel Golgi-associated protein with VHS and gamma-adaptin “ear” domains. J. Biol. Chem. 2000;275:7176–7183. doi: 10.1074/jbc.275.10.7176. [DOI] [PubMed] [Google Scholar]
- 46.Zoncu R, et al. A phosphoinositide switch controls the maturation and signaling properties of APPL endosomes. Cell. 2009;136:1110–1121. doi: 10.1016/j.cell.2009.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeung BG, Phan HL, Payne GS. Adaptor complex-independent clathrin function in yeast. Mol. Biol. Cell. 1999;10:3643–3659. doi: 10.1091/mbc.10.11.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Janke C, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 50.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 51.Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 52.Burd CG, Emr SD. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol. Cell. 1998;2:157–162. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- 53.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vowels JJ, Payne GS. A role for the lumenal domain in Golgi localization of the Saccharomyces cerevisiae guanosine diphosphatase. Mol. Biol. Cell. 1998;9:1351–1365. doi: 10.1091/mbc.9.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.