Abstract
A classic metabolic concept posits that insulin promotes energy storage and adipose expansion, while catecholamines stimulate release of adipose energy stores by hydrolysis of triglycerides through β-adrenergic receptor (βARs) and protein kinase A (PKA) signaling. Here, we have shown that a key hub in the insulin signaling pathway, activation of p70 ribosomal S6 kinase (S6K1) through mTORC1, is also triggered by PKA activation in both mouse and human adipocytes. Mice with mTORC1 impairment, either through adipocyte-specific deletion of Raptor or pharmacologic rapamycin treatment, were refractory to the well-known βAR-dependent increase of uncoupling protein UCP1 expression and expansion of beige/brite adipocytes (so-called browning) in white adipose tissue (WAT). Mechanistically, PKA directly phosphorylated mTOR and RAPTOR on unique serine residues, an effect that was independent of insulin/AKT signaling. Abrogation of the PKA site within RAPTOR disrupted βAR/mTORC1 activation of S6K1 without affecting mTORC1 activation by insulin. Conversely, a phosphomimetic RAPTOR augmented S6K1 activity. Together, these studies reveal a signaling pathway from βARs and PKA through mTORC1 that is required for adipose browning by catecholamines and provides potential therapeutic strategies to enhance energy expenditure and combat metabolic disease.
Introduction
Adipose tissue depots in both humans and rodents perform a remarkable spectrum of contrasting functions, including sequestration of excess caloric energy in the form of triglyceride in white adipose tissue (WAT) and rapid conversion of oxidative energy to heat for survival in the cold in brown adipose tissue (BAT). Various fat depots also secrete specific signature profiles of proteins and other factors that play key roles in overall systemic energy metabolism and glucose homeostasis (1). A general paradigm in the field is that 2 major opposing hormone systems, insulin (Ins) and catecholamines, are prominent regulators of these adipocyte functions. The catecholamines norepinephrine and epinephrine activate β-adrenergic receptors (βARs) to increase cAMP levels and cAMP-dependent protein kinase A (PKA) activity. PKA phosphorylates and regulates several important targets in adipocytes, including hormone-sensitive lipase and the lipid droplet–associated perilipins, which collectively promote triglyceride hydrolysis and liberation of free fatty acids (2–4). The opposing metabolic regulator is Ins, which antagonizes the action of the catecholamines to stimulate lipolysis by activating phosphodiesterases that degrade cAMP (5) and by activating lipid synthesis pathways through actions of the protein kinase AKT (6).
Signaling by catecholamines to stimulate lipolysis in WAT provides fatty acid substrates to fuel peripheral tissues, while, in BAT, lipolysis provides fatty acid substrates for generating heat. Brown adipocytes are highly enriched in mitochondria and express the unique protein uncoupling protein-1 (UCP1), which “uncouples” the mitochondrial proton gradient from adenosine triphosphate (ATP) production during fatty acid oxidation to produce thermal energy (7). The result is net energy expenditure. UCP1-containing “brown-like” adipocytes can also be recruited within WAT depots through prolonged βAR stimulation of lipolysis (8, 9) and are called “brite” or “beige” adipocytes (10, 11). Studies using mouse models show that the increase in these beige adipocytes closely correlates with resistance to obesity (12, 13), and in humans, the amount of detectable brown/beige adipocytes is significantly correlated with reduced percent body fat and circulating triglycerides, as well as greater Ins sensitivity (14–21). As with lipolysis, where Ins opposes the actions of βARs, Ins also appears to antagonize catecholamine stimulation of adipose browning (22) secondary to its suppression of lipolysis.
A major signaling node for the anabolic actions of Ins that strongly promote lipogenesis and protein synthesis downstream of AKT are the mTOR complexes. mTOR is a 250 kDa conserved Ser/Thr kinase that regulates cell growth and metabolism in response to environmental cues such as growth factors and nutrients, in addition to Ins (23). There are 2 structurally and functionally distinct mTOR-containing protein complexes, mTORC1 and mTORC2, required for activation of AKT (24–26). A defining characteristic of mTORC1 is its inhibition by the macrolide antibiotic rapamycin (26, 27), and it contains the partner protein RAPTOR (regulatory-associated protein of mTOR). By contrast, at least acutely, mTORC2 is not directly affected by rapamycin, and it contains the partner protein RICTOR (rapamycin-insensitive companion of mTOR). A well-characterized downstream target of mTORC1, but not mTORC2, is p70 ribosomal S6 kinase 1 (S6K1), which is thus highly activated by Ins.
Ins activation of mTOR would suggest that catecholamines may oppose this action and inhibit this complex. Indeed, data showing that the cAMP pathway can inhibit mTOR exists not only in adipocytes (28, 29), but also in other cells such as lymphoblasts (30) and smooth muscle cells (31, 32). However, catecholamine action to increase the abundance of beige adipocytes within WAT entails increased biosynthetic capacity, including enhanced mitochondrial and cell protein mass, for which mTOR1 is a central player. We therefore further investigated this connection and show here that β-adrenergic stimulation activates the mTORC1/S6K1 pathway. Remarkably, we found that βAR-mediated browning of white adipose depots in mice requires the activation of mTORC1 and S6K1 through a mechanism that is distinct from that of Ins and growth factors. Specifically, both mTOR and the mTORC1 component RAPTOR can be phosphorylated by PKA, and interference with phosphorylation of RAPTOR by PKA prevents activation of S6K1 by β-agonists, without affecting its stimulation by Ins. These findings illustrate the existence of a new signaling pathway from PKA to mTORC1 and its requirement for the sympathetic nervous system–driven (SNS-driven) recruitment and expansion of the UCP1-containing beige cells in WAT.
Results
βAR agonists activate mTORC1 in adipocytes
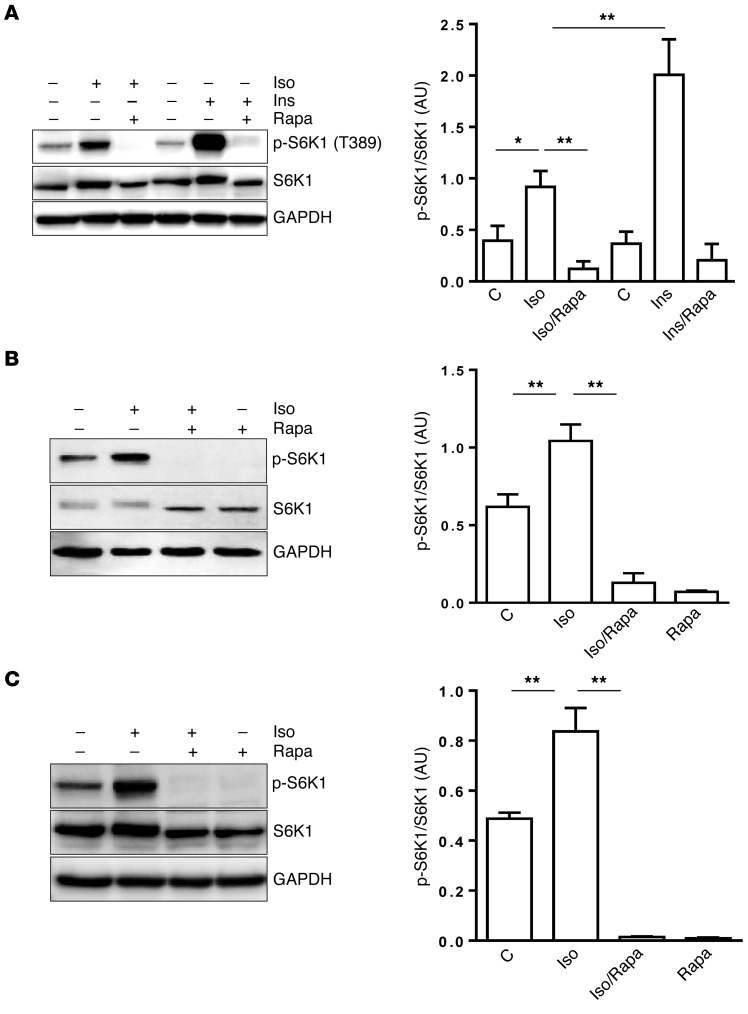
During preliminary studies comparing the effects of Ins and βAR agonists on signal transduction and metabolism in adipocytes, we observed that when 3T3-L1 adipocytes were treated with the nonselective βAR agonist isoproterenol (Iso), S6K1 became phosphorylated, which is best known to be activated by Ins. We then compared the ability of Ins and Iso to activate S6K1 in adipocytes. As shown in Figure 1A, Ins was more potent than Iso in mediating this effect, while rapamycin, an inhibitor of mTORC1, blocked the actions of both agents. Phosphorylation of S6K1 in response to Iso was both potent (≤10 nM) and brisk (≤10 minutes) (Supplemental Figure 1, A and B; supplemental material available online with this article; doi:10.1172/JCI83532DS1) and was mediated by all 3 βARs in adipocytes (Supplemental Figure 1, C–F). The ability of βARs to activate S6K1 is also observed in mouse brown adipocytes (Figure 1B), as well as in human primary s.c. adipocytes (Figure 1C). In addition, treatment of cells with the direct adenylyl cyclase activator forskolin (Fsk) increased S6K1 phosphorylation (Supplemental Figure 2A). Therefore, although Ins and β-adrenergic agents classically serve opposing roles in adipocytes for managing metabolic fuel usage, including an antagonistic role of cAMP to blunt Ins-stimulated mTORC1 (Supplemental Figure 3), they are both capable of activating mTORC1.
Figure 1. βAR activation of mTORC1 in adipocytes.
(A–C) 3T3-L1 adipocytes were pretreated or not with rapamycin (Rapa, 100 nM) for 30 minutes prior to the addition of Iso (1 μM) or Ins (10 nM) for 1 hour (A); HIB-1B brown adipocytes were pretreated or not with rapamycin. Iso was added for 1 hour (B); Human primary s.c. adipocytes were treated or not with Rapa for 30 minutes, followed by Iso for 1 hour (C), and levels of phosphorylated, total S6K1, and internal control GAPDH were measured by Western blotting. Bar graphs show image quantification of p-S6K1 normalized to total S6K1 (n = 3). One-way ANOVA followed by post-hoc comparisons. C, control. *P < 0.05, **P < 0.01.
mTORC1 activation by βARs depends on PKA
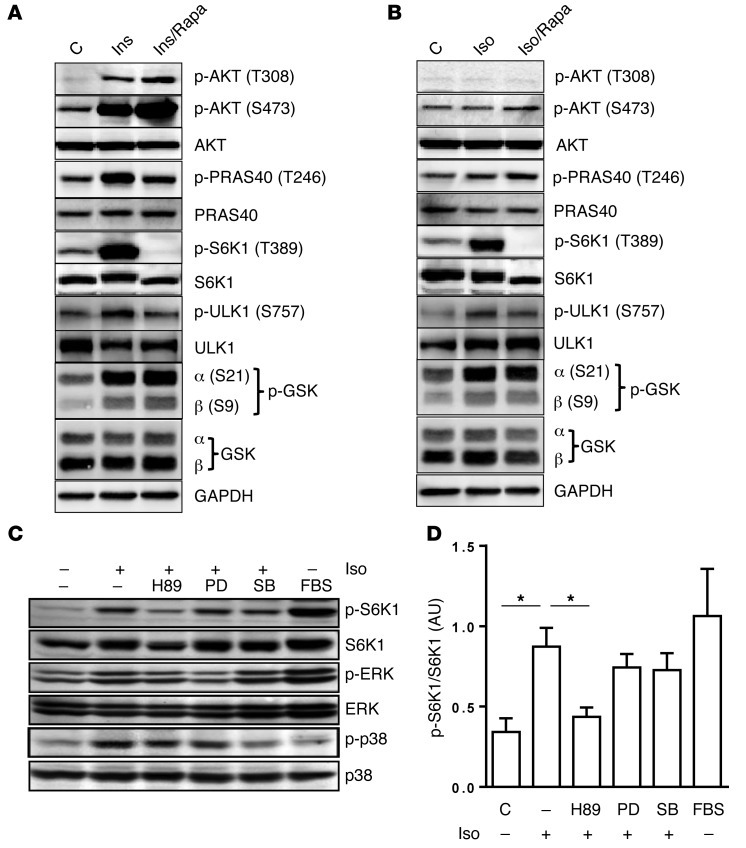
Next, we used a set of in vitro approaches to gain insight into the molecular signaling events connecting βAR activation and mTORC1. It is well known that Ins and growth factors activate mTORC1 through an AKT-dependent pathway (33). We compared signaling events mediating S6K1 activation in response to Iso versus Ins in 3T3-L1 adipocytes. As shown in Figure 2A, Ins (10 nM) increased phosphorylation of AKT (Thr308, Ser473) and S6K1 (Thr389), as established by others (6, 34). Phosphorylation of S6K1 by Ins was fully suppressed by rapamycin. In contrast to Ins, Iso activated S6K1 but did not trigger phosphorylation of AKTT308 or AKTS473 (Figure 2B). In addition, proline-rich AKT substrate 40 (PRAS40) was phosphorylated by Ins treatment but not by Iso. We also observed that GSK3α and -β were phosphorylated by either Ins or Iso, which was unaffected by rapamycin. ULK1, another established mTORC1 target (35), was also activated by both Ins and Iso. To further confirm the activation of mTORC1 by Iso, the pan-mTOR inhibitor torin1 (36) blocked both Iso- and Ins-stimulated S6K1 activation (Supplemental Figure 4). We also found that rapamycin treatment in L1 adipocytes did not affect Iso-mediated lipolysis (Supplemental Figure 5). Considering that G protein–coupled receptors including βARs can also activate MAPKs such as ERK and p38 MAPK (37–41), we examined these pathways. As shown in Figure 2, C and D, the MEK inhibitor PD98059 (PD) and the p38 MAPK inhibitor SB202190 (SB) fully blocked activation of their respective kinases following Iso stimulation, but these inhibitors had no effect on Iso-mediated S6K1 phosphorylation. Only the PKA inhibitors H89 (Figure 2C) and KT5720 (Supplemental Figure 2B) blocked Iso-induced S6K1 activation, suggesting that PKA itself, and not these other downstream kinases, is responsible for βAR-stimulated mTORC1-S6K1 activation.
Figure 2. βAR activation of S6K1 in adipocytes is PKA dependent, and independent of AKT.
(A and B) 3T3-L1 adipocytes were pretreated or not with Rapa for 30 minutes followed by either Ins (A) or Iso (B) for 1 hour, and levels of total and phosphorylated AKT, PRAS40, S6K1, ULK1, and GSK3 in lysates were measured by Western blotting. (C) 3T3-L1 adipocytes were treated with the following kinase inhibitors for 30 min: PKA inhibitor H89 (20 μM), MEK inhibitor PD (40 μM), and p38α/β inhibitor SB (20 μM), followed by Iso (1 μM). The addition of 10% FBS to medium for 1 hr served as a positive control to confirm activation of S6K1 and ERK by growth factors. Cell lysates were prepared, and total and phospho-S6K1, ERK, and p38 MAPKs were measured by immunoblot. (D) Bar graphs show image quantification of p-S6K1 normalized to total S6K1 of n = 3 experiments from C. One-way ANOVA followed by post-hoc group comparisons; *P < 0.05.
mTORC1 activity contributes to white adipose browning
Cold-temperature challenge.
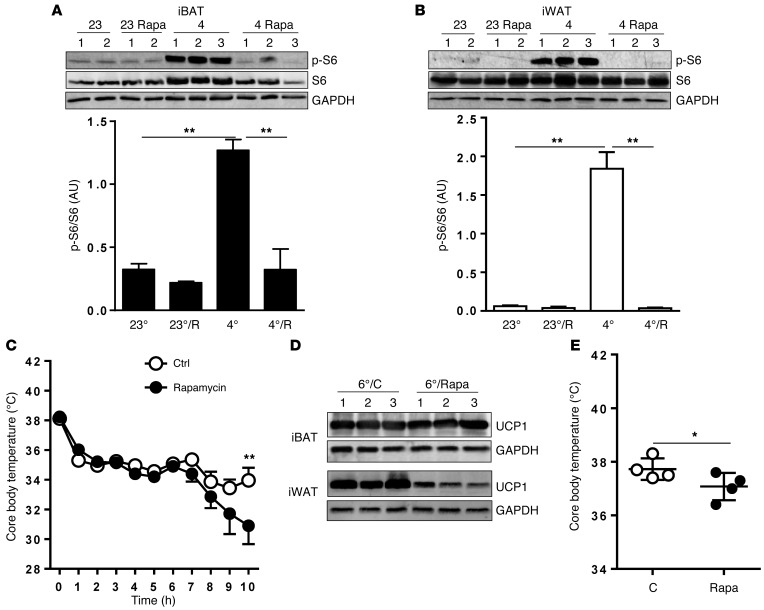
A cold-temperature challenge is a classic maneuver that triggers an increase in SNS outflow to tissues including white and brown fat and generates heat from stored energy (42–45). When placed in a cold environment, mice exhibit a moderate decrease in core body temperature within the first few hours, followed by a temperature plateau. In this early phase, the animal generates heat largely through muscle shivering, which is followed by nonshivering thermogenesis (NST) in BAT (7). Mice were treated with rapamycin or vehicle for 48 hours prior to the cold (4°C) exposure. There was a dramatic increase in S6 phosphorylation in interscapular BAT (iBAT) (Figure 3A) and in s.c. inguinal WAT (iWAT) (Figure 3B). In mice that were pretreated with rapamycin, this phosphorylation was fully blocked. Similar results were observed in gonadal WAT (gWAT; not shown). Figure 3C shows that core body temperature dropped slightly in both groups as expected, but after 7 hours, the rapamycin-treated group exhibited a further decline in temperature, which became significant by 10 hours. At this time point, UCP1 protein level in iBAT tended to be lower with rapamycin treatment, with no effect on Ins signaling as measured by AKT phosphorylation (Supplemental Figure 6). However, given the whole-body exposure to rapamycin, this temperature change cannot be ascribed specifically to adipose tissue. In general, during this acute exposure period, detectable levels of beige adipocytes are not observed in WAT depots. However, following longer exposure times, the presence of these cells becomes quite evident (46, 47). Figure 3D shows that, after 7 days of cold exposure, the strong UCP1 expression in iBAT was unaffected by continuous rapamycin treatment, while the robust levels of UCP1 in iWAT were greatly diminished by rapamycin. Although there is no apparent difference in iBAT UCP1 by Western blot between the vehicle and rapamycin groups, there nevertheless could be a net increase in total tissue mass and protein, including more overall UCP1 (42). However, the weights of the iBAT were not different between the groups (4°C saline: 0.085 ± 0.021 mg vs. 4°C rapamycin: 0.077 ± 0.014 mg) (n = 4; t test; P = 0.605). Additionally, the rapamycin-treated cold-adapted mice displayed lower core body temperature, shown in Figure 3E.
Figure 3. Elevating SNS activity via cold exposure activates mTORC1 signaling in BATs and WATs.
Male mice (n = 5/group) received either vehicle or rapamycin (Rapa; 4 mg/kg BW) by i.p. injection for 2 days followed by exposure to 4°C or 23°C for 6 hrs. (A and B) iBAT (A) and iWAT (B) were collected, homogenized, and assayed by immunoblot for p-S6 and total S6. GAPDH served as an internal control. Bar graphs show image quantification of p-S6 normalized to total S6. One-way ANOVA followed by post-hoc group comparisons; **P < 0.01. (C) Core body temperature was monitored and recorded for vehicle- or Rapa-treated mice during cold exposure. One-way ANOVA followed by post-hoc Bonferroni analysis. **P < 0.01. (D) iBAT and iWAT were dissected from cold-acclimated mice receiving either vehicle or Rapa for immunoblotting detection of UCP1 and internal-control GAPDH. (E) Core body temperature was recorded for vehicle- or Rapa-treated mice during cold adaptation (n = 4). C, control. One-way ANOVA followed by post-hoc Bonferroni analysis. *P < 0.05.
Selective stimulation of adipose tissue by β3AR agonist.
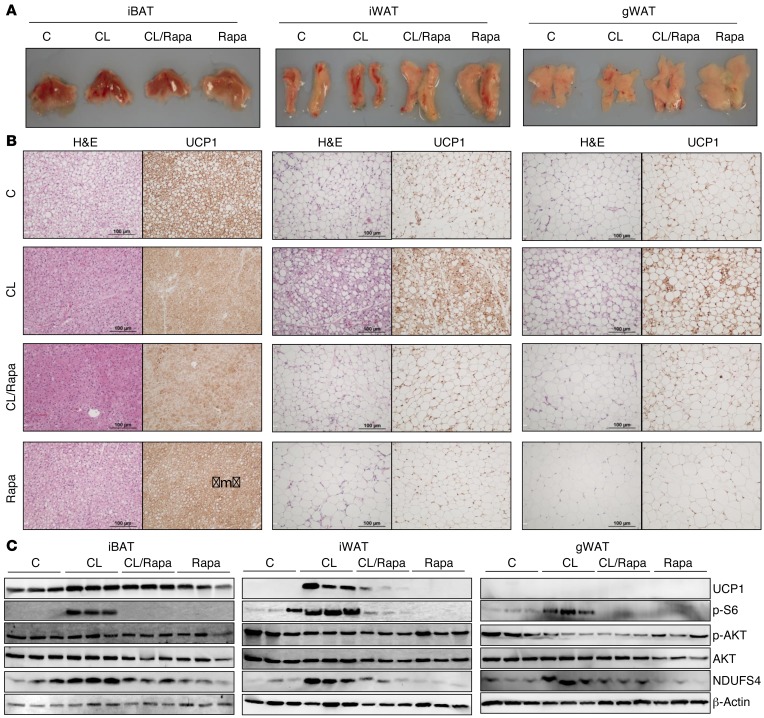
To more specifically activate βAR signaling only in adipose tissue, we used the well-established model of β3AR activation with the highly selective agonist CL316,243 (CL), in which chronic β3AR agonist treatment leads to the robust browning of WAT that is often accompanied by increased energy expenditure and resistance to obesity and its metabolic complications (48–51). Mice received CL either with or without rapamycin for 1 week as detailed in Methods. Upon dissection, gross examination of the adipose tissues revealed noticeable differences among the treatment groups. Adipose tissues from the CL-treated mice appeared to have a slightly darker complexion than those of the vehicle or rapamycin groups (Figure 4A). Histologically, CL treatment decreased lipid droplet size in iBAT and caused a substantial increase in multilocular adipocytes within the iWAT, along with a robust increase in UCP1+ staining (Figure 4B). Rapamycin dramatically suppressed the numbers of UCP1-staining cells in iWAT, and rapamycin alone was observed to have no effect. In gWAT, a similar suppression of UCP1 by rapamycin was observed as in iWAT but to a lesser extent. Figure 4C presents Western blots showing that, similar to cold exposure, CL treatment led to rapamycin-sensitive S6 phosphorylation in all 3 adipose depots. iWAT exhibited the most robust increase in UCP1 expression, which was prevented by rapamycin. Levels of NDUFS4, a component of respiratory complex I, measured here as a surrogate indicator of mitochondrial mass, was also increased in a rapamycin-sensitive manner. This is consistent with the need for significant mitochondrial biogenesis to support the increased respiration and uncoupling that occurs in brown adipocytes. In addition to Ucp1, peroxisome proliferation–activated nuclear receptor-α (PPARα), which is abundant in brown adipocytes, was increased in iWAT and gWAT by CL treatment, as were a number of established PPARα gene targets involved in fatty acid oxidation. The same pattern was observed in iWAT and gWAT: increased by CL and essentially blocked by rapamycin (Supplemental Figure 7). Taken together, these data from cold exposure and CL treatment suggest that mTORC1 signaling is necessary for the browning of WAT.
Figure 4. mTORC1 signaling participates in adipose browning induced by β3AR activation.
Male mice (n = 5/group) were pretreated or not with rapamycin (Rapa; 2.5 mg/kg/BW) for 1 day, followed by daily treatment of either vehicle control, CL (1 mg/kg BW/day), CL with Rapa, or Rapa alone for 7 days. (A) Representative images of the iBAT, iWAT, and gWAT tissues. (B) Histochemical staining by H&E (left panels) and UCP1 antisera (right panel) from iBAT, iWAT, and gWAT sections. (C) iBAT (20 μg protein; left panels), iWAT (30 μg protein; middle panels), and gWAT (50 μg protein; right panels) were analyzed by Western blotting for p-S6, p-AKT (S473) and total AKT, mitochondrial proteins UCP1, mitochondrial complex I component NDUFS4, and internal loading control β-actin.
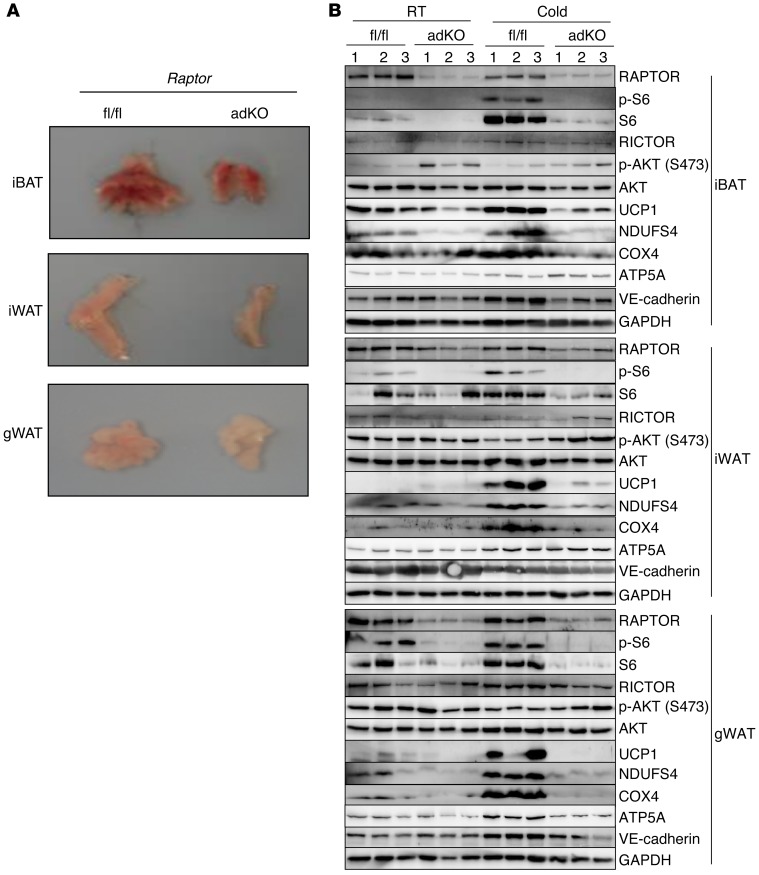
Adipose-specific deletion of Raptor blocks acquisition of brown/beige adipocytes.
We generated mice with adipose-specific deletion of Raptor by crossing Raptorfl/fl mice with adiponectin-Cre–expressing mice. The Raptorfl/fl and Raptorfl/fl adiponectin-Cre (herein referred to as adKO) mice were subsequently subjected to a cold challenge. As reported earlier by Hall and colleagues (52), the adipose depots lacking RAPTOR were smaller in size (Figure 5A). They also contained fully mature adipocytes and expressed adipogenic markers such as PPARγ2 and fatty acid binding protein 4 (aP2) (Supplemental Figure 8, A and B). Further analysis by Western blotting confirmed that Raptor was deleted in adipose tissues (Figure 5B), while it remained intact in other tissues (not shown). As expected with a lack of RAPTOR, levels of p-S6 were almost undetectable. For reasons that are unclear, total S6 was also reduced in the Raptor-null adipose tissues, while total amounts of AKT and VE-cadherin were unchanged between control and adKO adipose tissues. Importantly, the response to the cold challenge was lost in the adKO adipose tissues. The levels of UCP1 and indicators of mitochondrial content such as NDUSF4 and COX4 were much lower in all adipose depots of the Raptor-null mice after chronic cold exposure, while another mitochondrial marker, ATP5A, was unchanged in these adipose tissues between the adKO and Raptorfl/fl control mice. We also noted that, unlike treating adult mice with rapamycin, there was a blunted response observed in iBAT of adKO mice. This is most likely because the deletion of Raptor occurs prenatally at the time the cells are becoming adipocytes during late gestation (53) and adiponectin-Cre begins to be expressed. This genetic evidence confirms a role for mTORC1 in the process of adipose browning and also suggests that there is a role for mTORC1 in the original development of the interscapular depot, which may or may not be dependent on PKA signaling.
Figure 5. Adipose-specific deletion of Raptor impairs brown/beige adipocytes during cold challenge.
(A) Representative images of the iBAT, iWAT, and gWAT tissues from female floxed control (fl/fl) and adipose-specific Raptor KO (adKO). (B) Raptor, p-S6 and total S6, p-AKT (S473), total AKT, mitochondrial proteins UCP1, NDUFS4, COX4, endothelial marker VE-cadherin, and internal loading control GAPDH were detected by Western blotting in fat tissues of female Raptorfl/fl control and the adipose tissue–specific KO (adKO) at room temperature (RT) or cold exposure.
mTOR and its component RAPTOR are phosphorylated by PKA
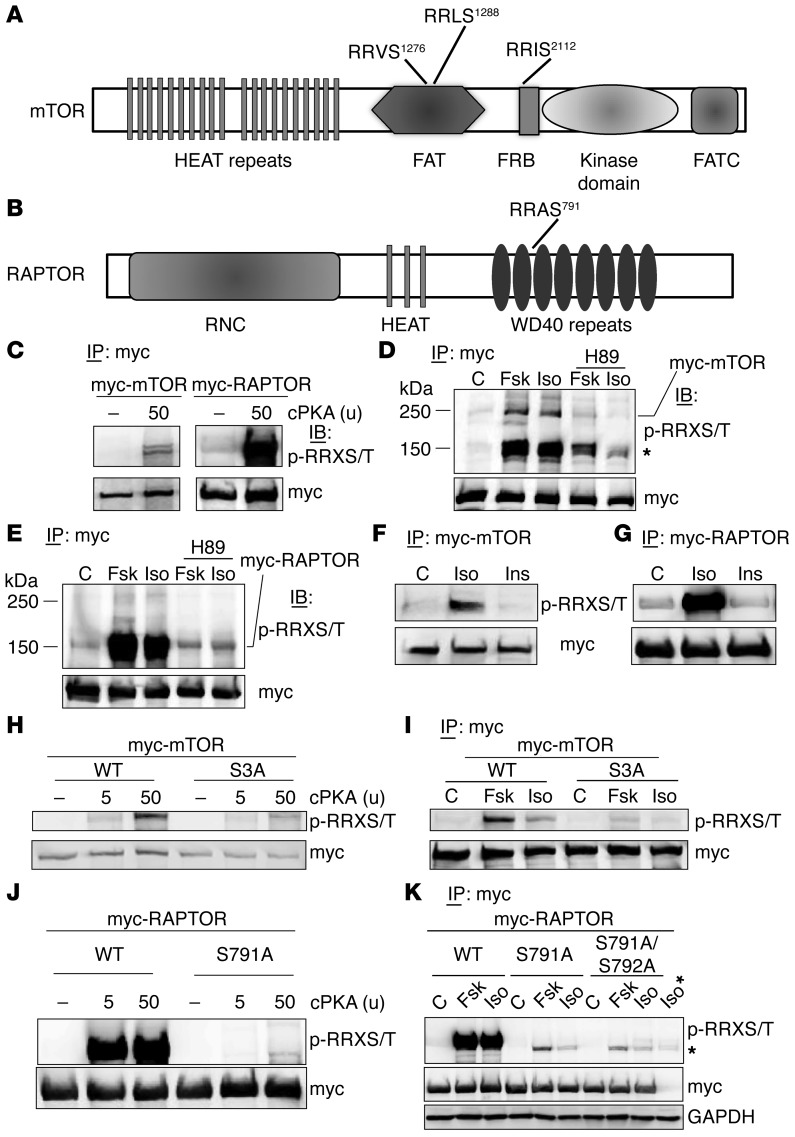
We next tested the possibility that mTORC1 might itself be a target of PKA. By inspecting the amino acid sequences of mTOR and RAPTOR across species, we indeed found that all mammalian mTOR proteins contain 3 strong and completely conserved RRXS motifs (Figure 6A) and that there is 1 motif in RAPTOR, as well (Figure 6B). Figure 6C shows that purified catalytic subunit of PKA (cPKA) can phosphorylate mTOR and RAPTOR, detected with an antibody that recognizes phosphorylated Ser or Thr in the context of the RRXS/T motif. Next, we expressed a myc-tagged mTOR in HEK293 cells and treated these cells either with Fsk or Iso for 30 minutes in the presence or absence of H89. As shown in Figure 6D, both Fsk and Iso increased PKA-substrate motif phosphorylation of myc-mTOR, which was blocked by H89. Figure 6D also shows a heavily phosphorylated band of approximately 150 kDa, which was also phosphorylated following PKA activation and suppressed by H89. Since cPKA can phosphorylate RAPTOR (Figure 6C), which is 150 kDa in size (54), we expressed myc-tagged RAPTOR in HEK293 cells and treated the cells with Fsk or Iso, with or without H89. Following IP of myc-RAPTOR and Western blotting with the PKA substrate antibody, Fsk and Iso treatment induced robust phosphorylation of myc-RAPTOR (Figure 6E), which was blocked by H89 treatment. We compared Iso with Ins treatment and examined this phosphorylation. While Iso clearly increased the PKA-motif phosphorylation of mTOR (Figure 6F) and Raptor (Figure 6G), there was no such effect of Ins treatment. These results indicate that the mTORC1 components mTOR and RAPTOR can be directly phosphorylated by PKA.
Figure 6. PKA phosphorylates mTOR and Raptor.
(A and B) Schematic of mTOR (A) and RAPTOR (B) illustrate the domains and locations of candidate PKA sites. (C) myc-mTOR (left panel) and myc-RAPTOR (right panel) were transfected into HEK293 cells for 24 hours; myc-tagged proteins were IP from cell lysates with anti–c-myc–conjugated beads, washed twice with the lysis buffer, and incubated with PKA catalytic (cPKA) subunit, followed by Western blotting for PKA substrate and myc detection. (D) HEK293 cells transfected with myc-mTOR plasmid and treated 24 hrs later with the PKA inhibitor H89 (15 μM) for 30 minutes, followed by Fsk (10 μM) or Iso (10 μM) for 30 minutes treatment. Myc-mTOR was IP from cell lysates with anti–c-myc–conjugated beads, and Western blots were probed with PKA substrate antisera or anti-myc antisera as indicated. (E) HEK293 cells expressing myc-RAPTOR were treated and analyzed as in panel D. (F and G) HEK293 cells were transfected with either myc-mTOR plasmid (F) or myc-RAPTOR plasmid (G) and treated 24 hours later with Iso (1 μM) or Ins (100 nM) for 30 min, followed by process as in D. (H) HEK293 cells were transfected with myc-mTOR or triple Ser mutant (S3A: S1276A/S1288A/S2112A), followed by the procedure as in C. (I) WT myc-mTOR or the S3A vector was expressed in HEK293 cells, treated as in D, and processed to measure PKA motif phosphorylation or myc as indicated. (J) HEK293 cells were transfected with myc-RAPTOR or S791A plasmid for 24 hrs and proceeded as in C. (K) WT myc-RAPTOR and the Ser mutants S791A and S791A/S792A were expressed in HEK293 cells, treated as in D, and processed to measure PKA motif phosphorylation or myc as indicated. The sample indicated as Iso* are nontransfected cells treated with Iso, revealing a nonspecific band (*). GAPDH served as loading input control.
We next mutated the 3 Ser in mTOR (S1276, S1288, and S2112) and the Ser in RAPTOR (S791) to Ala. First, shown in Figure 6H, the triple mutant of mTOR, designated as S3A, could no longer be phosphorylated in vitro by cPKA. Next, WT mTOR and the S3A mutant were introduced into HEK293 cells. Following treatment of the cells with Iso or Fsk for 30 minutes, mTOR was IP and analyzed for phosphorylation. Figure 6I shows that the S3A mutant of mTOR is refractory to the cAMP-elevating agents in cells. Second, in sharp contrast with WT RAPTOR, the Ser791A mutant could no longer be phosphorylated in vitro by cPKA (Figure 6J). We then compared the effects of WT RAPTOR and S791A RAPTOR in cells. In addition, although Ser792 is known to be an AMPK phosphorylation site (55), some protein kinase substrate sequence prediction algorithms (e.g., Kinexus PhosphoNet) include Ser792 as a candidate PKA site. Therefore, we also generated the double-mutant S791A/S792A. All 3 of these myc-RAPTOR constructs (WT, S791A, and S791A/S792A) were introduced into HEK293 cells and were subsequently treated with Iso or Fsk. Compared with the robustly phosphorylated WT myc-RAPTOR, phosphorylation was eliminated in the mutants (Figure 6K) to an equal degree. The residual phosphorylation that is seen is not RAPTOR, but rather is a protein that is slightly smaller than RAPTOR and is captured nonspecifically by myc antibody–conjugated beads alone, since cells lacking a myc-RAPTOR construct still contain this faint phosphorylated band. Since mutation of Ser791 to Ala already disrupted the PKA-dependent phosphorylation of RAPTOR, AMPK and Ser792 are unlikely to be involved in β-agonist–dependent phosphorylation of RAPTOR. From this series of experiments, we conclude that mTOR and RAPTOR are direct substrates of PKA.
Mutations of RAPTOR PKA site Ser791 have functional consequences for βAR signaling
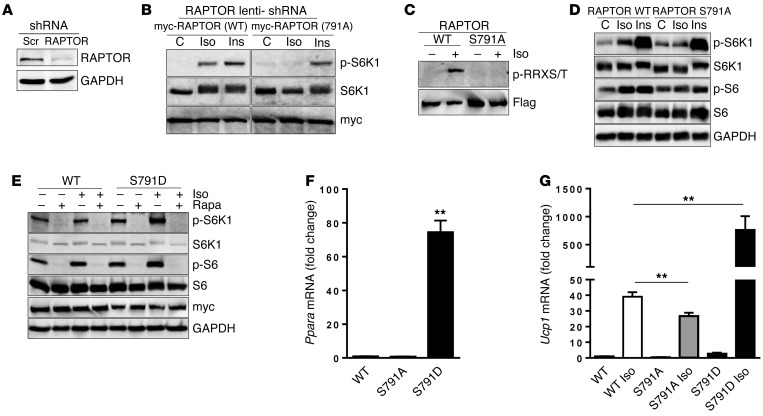
To determine if phosphorylation of RAPTOR S791 is necessary for S6K1 activation mediated by βAR signaling, a lentivirus harboring RAPTOR shRNA was used to infect NIH3T3 cells in order to knock down endogenous RAPTOR, as shown in Figure 7A. Using this maneuver, WT myc-RAPTOR or the S791A mutant myc-RAPTOR was introduced into these RAPTOR knockdown cells, followed by treatment with either Iso or Ins to evaluate S6K1 activation. Figure 7B shows that S6K1 phosphorylation was abolished in response to Iso, while the Ins response remained comparable in both groups of cells. To next test whether phosphorylation of RAPTOR Ser791 conveys the signal originating from βAR action in adipocytes, we employed HIB-1B cells stably expressing Flag-tagged RAPTOR (WT) or S791A. As shown in Figure 7C, while Iso treatment led to WT RAPTOR phosphorylation, this phosphorylation was completely lost in the S791A mutant. Figure 7D shows that, similar to the NIH3T3 cell model in Figure 7B, Iso increased p-S6K1 as well as p-S6 in WT Raptor cells but not in RAPTOR S791A adipocytes, while the response to Ins remained unaffected. Together, these results support the conclusion that mTOR and RAPTOR are targets of PKA and that their phosphorylation is necessary to relay the signal from βARs to S6K1 activation.
Figure 7. Mutations of RAPTOR PKA site Ser791 have functional consequences for βAR signaling but not for Ins.
(A) NIH3T3 cells were infected with lentivirus harboring mouse RAPTOR shRNA or scramble (Scr) control. Four days later, cells were lysed and assayed for endogenous RAPTOR and internal control GAPDH by immunoblotting. (B) NIH3T3 cells infected with lentivirus harboring RAPTOR shRNA were transfected with WT or S791A mutant myc-RAPTOR plasmid followed by either Iso (10 μM) or Ins (100 nM) treatment for 30 minutes. Phospho- and total S6K1 were measured by immunoblotting. (C) HIB-1B cells stably expressing Flag-RAPTOR (WT or S791A) were differentiated and treated with Iso for 30 minutes, followed by anti-Flag IP. They were detected by PKA substrate and reprobed with anti-Flag. (D) Cells as in C were treated with Iso (1 μM) or Ins (10 nM) for 30 minutes. The lysates were detected by Western blotting for p-S6K1 and p-S6 and their respective total antibodies. GAPDH serves as loading control. (E) HIB-1B cells stably expressing myc-RAPTOR (WT or S791D) were pretreated with rapamycin (Rapa; 100 nM), followed by Iso (1 μM), for 1 hour and analyzed as in B. (F and G) Transcripts of Ppara in stably expressing RAPTOR (WT), S791A, and S791D were detected by qPCR (F), while Ucp1 was detected after 6 hours treatment by Iso (G) (100 nM). One-way ANOVA followed by post-hoc group comparison tests; **P < 0.01. (In HIB-1B cells, endogenous RAPTOR still present.)
To further test the functional consequences of RAPTOR phosphorylation at Ser791, a phosphomimetic RAPTOR was generated by changing Ser at 791 to Asp, and once again HIB-1B cells stably expressing myc-RAPTOR or the myc-RAPTOR S791D mutant were created. As shown in Figure 7E, even under basal conditions without the addition of Iso, levels of p-S6K1 and p-S6 tended to be higher in the cells expressing the S791D mutant. Provision of Iso increased their levels further. However, the more striking effect of the presence of RAPTOR S791D in these cells is shown in Figure 7, F and G. Under basal conditions, the level of PPARα expression is massively increased (Figure 7F), and basal levels of Ucp1 transcripts are 3-fold higher in S791D cells compared with WT cells (Figure 7G). Also shown in Figure 7G is that, when treated with Iso, Ucp1 transcripts were increased approximately 40-fold in WT cells, but in S791D cells, the increase over its basal level was an impressive 250-fold. In the S791A cells, Ppara was expressed at a comparable level to WT cells, but the Iso-stimulated Ucp1 transcripts were significantly lower. Together, these results argue for an important role of PKA phosphorylation of RAPTOR in the acquisition of the Ucp1-expressing cell phenotype.
Discussion
The SNS is well known to activate PKA through βARs not only for lipolysis and uncoupled respiration in brown adipocytes through UCP1, but also for increasing the net amount of browned adipocytes present within white fat depots (8, 56–58). It has been repeatedly noted in laboratory animal models that the ability to generate these beige adipocytes is closely correlated with resistance to weight gain and preserved Ins sensitivity when challenged with a high-fat diet (48–50, 59–61). With the current intense interest in brown/beige adipose tissue in adult humans as a therapeutic target for increasing energy expenditure in metabolic disease, understanding the molecular events that drive their appearance and maintenance are critical. While the underlying cell lineage distinctions or molecular cues need further clarification (reviewed by ref. 62), increasing this UCP1-expressing adipocyte population is clearly a process that requires considerable synthetic and remodeling capacity within the tissue. This remodeling includes producing additional mitochondria and respiratory chain components, fatty acid oxidizing capacity, and, of course, UCP1. Here, we have shown that a direct signaling pathway from PKA to mTORC1 is involved in the β-adrenergic recruitment of these beige adipocytes in response to cold adaptation or to treatment with the β3AR agonist CL. In addition, our studies in mice lacking RAPTOR in adipocytes show impaired expression of UCP1 and other mitochondrial components, suggesting that there may be a role for mTORC1 even in the early development of the iBAT depot.
We previously showed that, downstream of βARs and PKA, other kinases are activated in adipocytes, which together coordinately maximize fuel metabolism. These kinases include ERK (39) to contribute to overall lipolysis (41), as well as p38 MAPK (40, 63). While these MAPKs, particularly p38, have an important functional role for driving the transcription of the Ucp1 and Pgc1a genes (60, 63, 64), blockade of these MAPKs did not interfere with βAR activation of mTORC1. Instead, key components of mTORC1, specifically mTOR and RAPTOR, are directly phosphorylated by PKA. In mTOR, mutation of the 3 candidate PKA sites prevented its phosphorylation. Similarly, mutation of Ser791 in the RAPTOR PKA site eliminated its phosphorylation and functionally prevented activation of S6K1 by Iso but not by Ins.
In addition to UCP1 and mitochondrial genes, key components of the machinery necessary for the metabolic needs of brown and beige cells are also under the control of mTORC1. For example, PPARα is a master nuclear receptor for fatty acid β oxidation, and PPARα has been shown to also participate in UCP1 expression either directly or indirectly through ERRα (65–70). In addition, several downstream target genes of PPARα and ERRα are similarly under the control of mTORC1. These include Pdk4, Cpt1b, Fabp3, and Elovl3. Clearly, a number of molecular events are set in motion to achieve the complex cellular process of adipose browning; some are dependent upon mTORC1, while others are not. Given that translational control is one of the hallmarks of biological regulation by mTOR (71), it would not be surprising if, along with direct transcriptional events, a layer of translational regulation turns out to be part of this mTORC1-driven effect on adipose tissue. This will be an important direction to explore in expanding these studies in the future.
The finding that PKA directly activates mTORC1 to drive adipose browning is both unexpected and rather extraordinary for several reasons. First, Ins and catecholamines are best known for their ability to antagonize one another’s metabolic effects, yet Ins is a well-known activator of mTORC1 for promoting growth and energy storage. From much work conducted in this field, inhibiting mTORC1 with rapamycin seems to be beneficial in terms of metabolism under some circumstances, but there are also complications. For example, while mTORC1 inhibition with rapamycin can increase longevity in mice (72, 73), chronic rapamycin treatment causes glucose intolerance and hyperlipidemia in both mice (74) and humans (75, 76); this appears to be caused by disturbances in mTORC2 function (77). Mice with aP2-driven deletion of Raptor in adipose tissue were reported to be resistant to diet-induced obesity, with smaller adipose tissue, increases in UCP1, and apparent improved Ins sensitivity (52, 78, 79). However, unlike our results described here, these earlier studies did not assess the effects of RAPTOR on the recruitment of brown adipocytes in response to cold or β-agonist stimulation. Importantly, the use of the aP2-Cre driver has subsequently been largely abandoned because it has been discovered that, in this animal, Cre recombinase can be expressed in other nonadipose tissues, including the brain, and other tissues during early development (64, 80, 81). Therefore, one cannot rule out the possibility that the loss of RAPTOR in some other tissue or cell type might contribute to the reported lean phenotype (52).
While it remains to be tested, our results here may contribute to understanding some other observations in the literature that suggest cAMP can activate mTORC1. For instance, thyroid-stimulating hormone (TSH) has been reported to activate S6K1 in thyroid epithelia in a cAMP-dependent manner (82), and in pancreatic islets, GLP-1–dependent increases in cAMP — in the context of elevated glucose — were observed to activate S6K1 (83), which the authors concluded was due to cAMP-evoked intracellular Ca+2 release. More recently, in a disease model of primary pigmented nodular adrenocortical disease (PPNAD), which inactivates the regulatory subunit of PKA, it was reported that mTORC1 activity was increased, resulting in prevention of apoptosis (84). In 2 of these studies, the authors pointed to PRAS40 phosphorylation as a mediator, but whether this is the sole factor involved was not tested. In our studies here in adipocytes, phosphorylation of PRAS40 does not occur in response to β-agonists, thus eliminating it as a regulatory factor.
We believe that our study makes 2 important biological observations. First, that mTORC1 is a necessary component in the process of the browning of white adipose depots. The second point is that there exists a signaling pathway from PKA to mTORC1 activation via direct phosphorylation of mTOR and RAPTOR. Given these findings and the ubiquitous expression of mTOR and PKA in many cell types, we speculate that this PKA-mTORC1 pathway may have broader significance by regulating a variety of cell signaling events in other tissues. Therefore, our studies illustrate that an inherently catabolic process of energy expenditure in adipose tissue utilizes what is most commonly considered to be the molecular driver of growth and anabolic metabolism. When considered together, our findings suggest that the control of mTORC1 activity, and its contributions to metabolic regulation, deserves fresh appraisal.
Methods
Reagents and antibodies.
Iso, Fsk, Ins, dexamethasone (Dex), isobutylmethylxanthine (IBMX), and the small molecule kinase inhibitors H89, SB, and PD were obtained from Sigma-Aldrich. The β3AR-selective agonist CL was a gift from Elliott Danforth Jr. (American Cyanamid Co., Pearl River, New York, USA). Rapamycin and torrin1 were from LC Laboratory. KT5720 was from Enzo Life Sciences. The protease inhibitor cocktail (complete Mini) and a phosphatase inhibitor cocktail (PhosSTOP) were from Roche Diagnostics. The following antisera were obtained from Cell Signaling Technology: p-S6 (Ser240/244, catalog 2215), total S6 (catalog 2217), p-S6K1(Thr389, catalog 9205), total S6K1 (catalog 9202), p-AKT (T308, catalog 9275), p-AKT (Ser473, catalog 4060), total AKT (catalog 9272), p-PRAS40 (Thr246, catalog 2640), total PRAS40 (catalog 2610), p-ULK1 (S757, catalog 6888), total ULK1 (catalog 8054), p-ERK (Thr202/Tyr204, catalog 4370), total ERK(catalog 9102), p38 MAPK(catalog 9202), p-GSK3 (S9 and S21, catalog 8566), total GSK3 (catalog 5676), phospho-RRXS/T motif antibody (PKA substrate, catalog 9624), RAPTOR (catalog 2280), RICTOR (catalog 9476), “myc tag” (catalog 2272), COX4 (catalog 4844), and β-actin (catalog 4967). Other antisera used include UCP1 (catalog ab23841) and ATP5A (catalog ab14748) (both from Abcam). p-p38 MAPK (catalog ref-V121-A) and purified PKA catalytic subunit were ordered from Promega, plus NDUFS4 (Pierce Biotechnology, catalog PA5-21677), VE-cadherin (catalog sc-28644) and GAPDH (catalog sc-25788) (both from Santa Cruz Biotechnology). The EZview Red Anti–c-myc Affinity Gel (catalog E6654) and the secondary antibodies conjugated with alkaline phosphatase (anti-rabbit, catalog A3687; anti-mouse, catalog A3562) were from Sigma-Aldrich.
Animal experiments.
Male C57BL/6J mice were obtained from the Jackson Laboratory. Experiments were conducted in mice that were 12–14 weeks of age. For the cold-challenge experiments, mice were administered rapamycin (5 mg/kg body weight [BW]/day) delivered in 100 μl/20 g BW or vehicle (75% saline, 10% ethanol, 10% PEG300, 5% Tween 80) (85) once a day for 2 consecutive days. On the third day, the mice were housed individually and placed at 4°C for up to 10 hours, with fresh bedding and free access to water. Control mice were maintained at room temperature (23°C). For chronic cold challenge, mice were placed at 4°C for 7 days with daily rapamycin (2.5 mg/kg BW/day) administration. For selective activation of β3AR, mice were injected i.p. with CL (1 mg/kg BW/day) (86, 87) with or without rapamycin (2.5 mg/kg BW) for 1 week, after which they were euthanized, and iBAT, iWAT, and gWAT were dissected and immediately frozen in liquid nitrogen. Mice with a Raptorfl/fl allele (JAX-013188), as well as adiponectin-Cre (JAX-010803), were obtained from the Jackson Laboratory and bred to generate mice with a Raptor adipose–specific deletion. Tail DNA genotyping and Western blotting of lysate from adipose tissues, liver, and heart were used to confirm the specific deletion of Raptor in adipose.
Adipocyte cell culture.
3T3-L1 preadipocytes were obtained from ATCC. They were maintained and differentiated into adipocytes as described (88), with modifications (89). HEK293 cells were purchased from ATCC and cultured in DMEM with 10% FBS. Human primary s.c. adipocytes were provided by Zen-Bio. NIH3T3 cells were purchased from ATCC and cultured in DMEM supplemented with 10% calf bovine serum. Prior to treatment, cells were washed twice with serum-free DMEM and maintained overnight in DMEM with 0.1% fatty-acid–free BSA cells. HIB-1B preadipocytes were cultured and differentiated as described (87, 90). HIB-1B cells stably expressing RAPTOR WT, S791A (alanine) or S791D (aspartic acid) were generated as previously described (91, 92).
Plasmids and mutagenesis.
The WT myc-mTOR and myc-RAPTOR mammalian expression vectors were from Addgene. To mutate candidate PKA sites in mTOR, the following primers were used to change the Ser residues to Ala: S2112A forward: 5′-TTC AGA CGG ATC GCC AAG CAG CTA-3′ and S2112A reverse: 5′-TAG CTG CTT GGC GAT CCG TCT GAA-3′; for S1276A/S1288A/S2112A triple mutation, the pair of primers forward: 5′-CTG CGA CGC TTG GCT CTG GAG CTG CTG AAG-3′ and reverse: 5′-CTT CAG CAG CTC CAG AGC CAA GCG TCG CAG-3′ was used and combined with 5′-GCC AGA AGG GTC GCC AAG GAC GAC TGG-3′. To mutate RAPTOR Ser791 to Ala, the primers were RAPTOR S791A forward: 5′-GAC AAG ATG CGC CGC GCC GCC TCC TAC TCC TCC CTC AAC TCC-3′ and RAPTOR S791A reverse: 5′-GGA GTT GAG GGA GTA GGA GGC GGC GCG GCG CAT CTT GTC. For the RAPTOR S791A/S792A mutation, forward: 5′-GAC AAG ATG CGC CGC GCC GCC GCC TAC TCC TCC CTC AAC TCC-3′ and reverse: 5′-GGA GTT GAG GGA GGA GTA GGC GGC GGC GCGGCG CAT CTT GTC-3′. To mutate RAPTOR S791 to aspartic acid (S791D), we changed S791A forward primer from GCC to GAC, and the reverse primer GGC to GTC. PCR was performed according to the instructions of the QuickChange Multi Site-Directed Mutagenesis kit (Agilent Technologies) using WT myc-mTOR or RAPTOR as template. Mutations were all confirmed by DNA sequencing.
Transfection, IP, and Western blotting.
Cells or tissues were lysed and sonicated in buffer containing 25 mM HEPES (pH 7.4), 150 mM NaCl, 5 mM EDTA, 5 mM EGTA, 5 mM glycerophosphate, 0.9% Triton X-100 (Sigma-Aldrich), 0.1% IGEPAL CA630, 5 mM sodium pyrophosphate, and 10% glycerol, plus 1 tablet each of complete protease inhibitor cocktail and PhoSTOP phosphatase inhibitors per 10 ml. For IP, HEK293 cells were transfected with myc-tagged mTOR or RAPTOR expression plasmid using lipofectamine 2000 (Invitrogen) according to the instructions provided. Cells were collected and lysed in the above buffer, and 2 mg total protein was incubated with anti–c-myc affinity gel overnight (Sigma-Aldrich). Beads were washed thoroughly in lysis buffer. For the in vitro PKA phosphorylation assay, 2 mg total protein was incubated with anti–c-myc affinity gel for 4 hours. Beads were washed twice with the lysis buffer and once with the PKA reaction buffer (40 mM Tris-HCL [pH 7.4], 20 mM magnesium acetate, 0.2 mM ATP), and PKA catalytic subunit was added to the beads. Protein phosphorylation by PKA was measured with PKA substrate antibody (phospho-RRXS/T motif antibody; Cell Signaling Technology). Flag-tagged WT RAPTOR and the S791A mutant RAPTOR, or myc-tagged WT RAPTOR and S791D mutant, were introduced into HIB-1B cells, and single clones were selected with 5 μg/ml blastcidin for FLAG-RAPTOR cells or 100 μg/ml hygromycin for myc-RAPTOR cells to generate stable cell lines. Unless otherwise indicated, 50 μg total protein was resolved in 4%–20% Tris-glycine gels (Invitrogen) and transferred to nitrocellulose membranes (Bio-Rad) and incubated overnight at 4°C with specific primary antibodies. Secondary antisera conjugated with alkaline phosphatase were used for specific protein detection. To reprobe blots, membranes were stripped in buffer (62.5 mM Tris, 2% SDS, 100 mM β-mercaptoethanol) at 37°C for 60 minutes. Image acquisition was performed on a Typhoon FLA 9000 imager (GE Healthcare), and data were analyzed using ImageQuant TL software.
RNA isolation and quantitative PCR.
RNA from adipose tissues were first extracted by Trizol and purified by RNA mini-columns from QIAGEN. Reverse transcription and SYBR green quantitative PCR (qPCR; Invitrogen) were performed according to the manufacturer protocols. Target primer sequences are shown in Supplemental Table 1.
Tissue histology and microscopy.
Adipose tissues were fixed with 4% paraformaldehyde in PBS, dehydrated, embedded in paraffin, and sectioned (5-μm thickness). Sections were stained with H&E and examined under bright-field microscopy with a Nikon 80i. UCP1 staining was performed as described previously (60, 93).
Statistics.
All data are presented as mean ±SEM. Comparisons between groups were assessed by 1-way ANOVA with Tukey’s post-hoc test or unpaired Student’s t test (2-tailed) as indicated in figure legends.
Study approval.
All animal experiments were approved by the Sanford Burnham Prebys Medical Discovery Institute’s Institutional Animal Care and Use Committee in accordance with the 8th edition of the NIH Guide for the Care and Use of Laboratory Animals.
Note added in proof.
While this manuscript was in press, an independent report confirmed our finding of a positive role for mTORC1 signaling in the browning of white fat (94).
Author contributions
DL and SC conceived, designed, and performed experiments; analyzed the data; prepared figures; and wrote the manuscript. MB and WW performed cell culture and mouse studies. CZ and HF managed the mouse colony and performed experiments with DL and MB. JLL conducted the data analyses. AG and KG performed experiments. MPC contributed to experimental design and data analysis.
Supplementary Material
Acknowledgments
We thank the following individuals at the Sanford Burnham Prebys Medical Discovery Institute: Timothy Osborne’s laboratory for providing the mouse Raptor shRNA lentivirus, Lea Dib and Crystal Woodard for their critical reading of the manuscript, and Rebecca Kaercher and Sarah Lardizabal for administrative assistance. This work was supported in part by NIH grant R21DK85412 (to S. Collins).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(5):1704–1716. doi:10.1172/JCI83532.
Contributor Information
Dianxin Liu, Email: dliu@sanfordburnham.org.
Marica Bordicchia, Email: marica.bordicchia@gmail.com.
Chaoying Zhang, Email: zhangchaoying@yahoo.com.
Huafeng Fang, Email: hfang@sbpdiscovery.org.
Wan Wei, Email: wwei@sbpdiscovery.org.
Jian-Liang Li, Email: jlli@sbpdiscovery.org.
Adilson Guilherme, Email: adilson.Guilherme@umassmed.edu.
Kalyani Guntur, Email: kguntur@mattek.com.
Sheila Collins, Email: scollins@sbpdiscovery.org.
References
- 1.Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nat Med. 2013;19(10):1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- 2.Lafontan M, Berlan M. Fat cell adrenergic receptors and the control of white and brown fat cell function. J Lipid Res. 1993;34(7):1057–1091. [PubMed] [Google Scholar]
- 3.Collins S, Cao W, Robidoux J. Learning new tricks from old dogs: beta-adrenergic receptors teach new lessons on firing up adipose tissue metabolism. Mol Endocrinol. 2004;18(9):2123–2131. doi: 10.1210/me.2004-0193. [DOI] [PubMed] [Google Scholar]
- 4.Granneman JG, Moore HP. Location, location: protein trafficking and lipolysis in adipocytes. Trends Endocrinol Metab. 2008;19(1):3–9. doi: 10.1016/j.tem.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 5.Manganiello VC, Smith CJ, Degerman E, Vasta V, Tornqvist H, Belfrage P. Molecular mechanisms involved in the antilipolytic action of insulin: phosphorylation and activation of a particulate adipocyte cAMP phosphodiesterase. Adv Exp Med Biol. 1991;293:239–248. doi: 10.1007/978-1-4684-5949-4_22. [DOI] [PubMed] [Google Scholar]
- 6.Summers SA, Whiteman EL, Birnbaum MJ. Insulin signaling in the adipocyte. Int J Obes Relat Metab Disord. 2000;24(suppl 4):S67–S70. doi: 10.1038/sj.ijo.0801509. [DOI] [PubMed] [Google Scholar]
- 7.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84(1):277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 8.Cousin B, et al. Occurrence of brown adipocytes in rat white adipose tissue: molecular and morphological characterization. J Cell Sci. 1992;103(pt 4):931–942. doi: 10.1242/jcs.103.4.931. [DOI] [PubMed] [Google Scholar]
- 9.Loncar D. Convertible adipose tissue in mice. Cell Tissue Res. 1991;266(1):149–161. doi: 10.1007/BF00678721. [DOI] [PubMed] [Google Scholar]
- 10.Giralt M, Villarroya F. White, brown, beige/brite: different adipose cells for different functions? Endocrinology. 2013;154(9):2992–3000. doi: 10.1210/en.2013-1403. [DOI] [PubMed] [Google Scholar]
- 11.Sidossis L, Kajimura S. Brown and beige fat in humans: thermogenic adipocytes that control energy and glucose homeostasis. J Clin Invest. 2015;125(2):478–486. doi: 10.1172/JCI78362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xue B, Rim JS, Hogan JC, Coulter AA, Koza RA, Kozak LP. Genetic variability affects the development of brown adipocytes in white fat, but not in interscapular brown fat. J Lipid Res. 2006;48(1):41–51. doi: 10.1194/jlr.M600287-JLR200. [DOI] [PubMed] [Google Scholar]
- 13.Schulz TJ, et al. Brown-fat paucity due to impaired BMP signalling induces compensatory browning of white fat. Nature. 2013;495(7441):379–383. doi: 10.1038/nature11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nedergaard J, Bengtsson T, Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. Am J Physiol Endocrinol Metab. 2007;293(2):E444–E452. doi: 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]
- 15.Cypess AM, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360(15):1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Virtanen KA, et al. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360(15):1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 17.Saito M, et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes. 2009;58(7):1526–1531. doi: 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ouellet V, et al. Brown adipose tissue oxidative metabolism contributes to energy expenditure during acute cold exposure in humans. J Clin Invest. 2012;122(2):545–552. doi: 10.1172/JCI60433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orava J, et al. Blunted metabolic responses to cold and insulin stimulation in brown adipose tissue of obese humans. Obesity (Silver Spring) 2013;21(11):2279–2287. doi: 10.1002/oby.20456. [DOI] [PubMed] [Google Scholar]
- 20.Kajimura S, Saito M. A new era in brown adipose tissue biology: molecular control of brown fat development and energy homeostasis. Annu Rev Physiol. 2014;76:225–249. doi: 10.1146/annurev-physiol-021113-170252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blondin DP, et al. Increased brown adipose tissue oxidative capacity in cold-acclimated humans. J Clin Endocrinol Metab. 2014;99(3):E438–EE46. doi: 10.1210/jc.2013-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehran AE, et al. Hyperinsulinemia drives diet-induced obesity independently of brain insulin production. Cell Metab. 2012;16(6):723–737. doi: 10.1016/j.cmet.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 23.Dann SG, Selvaraj A, Thomas G. mTOR Complex1-S6K1 signaling: at the crossroads of obesity, diabetes and cancer. Trends Mol Med. 2007;13(6):252–259. doi: 10.1016/j.molmed.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17(6):596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 25.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124(3):471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 26.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12(1):9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 27.Fingar DC, Richardson CJ, Tee AR, Cheatham L, Tsou C, Blenis J. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol Cell Biol. 2004;24(1):200–216. doi: 10.1128/MCB.24.1.200-216.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott PH, Lawrence JC., Jr Attenuation of mammalian target of rapamycin activity by increased cAMP in 3T3-L1 adipocytes. J Biol Chem. 1998;273(51):34496–34501. doi: 10.1074/jbc.273.51.34496. [DOI] [PubMed] [Google Scholar]
- 29.Mullins GR, et al. Catecholamine-induced lipolysis causes mTOR complex dissociation and inhibits glucose uptake in adipocytes. Proc Natl Acad Sci U S A. 2014;111(49):17450–17455. doi: 10.1073/pnas.1410530111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monfar M, et al. Activation of pp70/85 S6 kinases in interleukin-2-responsive lymphoid cells is mediated by phosphatidylinositol 3-kinase and inhibited by cyclic AMP. Mol Cell Biol. 1995;15(1):326–337. doi: 10.1128/MCB.15.1.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graves LM, et al. cAMP- and rapamycin-sensitive regulation of the association of eukaryotic initiation factor 4E and the translational regulator PHAS-I in aortic smooth muscle cells. Proc Natl Acad Sci U S A. 1995;92(16):7222–7226. doi: 10.1073/pnas.92.16.7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott PH, et al. A regulatory role for cAMP in phosphatidylinositol 3-kinase/p70 ribosomal S6 kinase-mediated DNA synthesis in platelet-derived-growth-factor-stimulated bovine airway smooth-muscle cells. Biochem J. 1996;318(pt 3):965–971. doi: 10.1042/bj3180965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12(1):21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alessi DR, et al. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15(23):6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 35.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thoreen CC, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284(12):8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pierce KL, Luttrell LM, Lefkowitz RJ. New mechanisms in heptahelical receptor signaling to mitogen activated protein kinase cascades. Oncogene. 2001;20(13):1532–1539. doi: 10.1038/sj.onc.1204184. [DOI] [PubMed] [Google Scholar]
- 38.Lefkowitz RJ, Shenoy SK. Transduction of receptor signals by β-arrestins. Science. 2005;308(5721):512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- 39.Soeder KS, et al. The β3-adrenergic receptor activates mitogen-activated protein kinase in adipocytes through a Gi-dependent mechanism. J Biol Chem. 1999;274(17):12017–12022. doi: 10.1074/jbc.274.17.12017. [DOI] [PubMed] [Google Scholar]
- 40.Cao W, Medvedev AV, Daniel KW, Collins S. β-Adrenergic activation of p38 MAP kinase in adipocytes: cAMP induction of the uncoupling protein-1 (UCP1) gene requires p38 MAP kinase. J Biol Chem. 2001;276(29):27077–27082. doi: 10.1074/jbc.M101049200. [DOI] [PubMed] [Google Scholar]
- 41.Robidoux J, et al. Maximal β3-adrenergic regulation of lipolysis involves Src and epidermal growth factor receptor-dependent ERK1/2 activation. J Biol Chem. 2006;281(49):37794–37802. doi: 10.1074/jbc.M605572200. [DOI] [PubMed] [Google Scholar]
- 42.Bukowiecki L, Collet AJ, Follea N, Guay G, Jahjah L. Brown adipose tissue hyperplasia: a fundamental mechanism of adaptation to cold and hyperphagia. Am J Physiol. 1982;242(6):E353–E359. doi: 10.1152/ajpendo.1982.242.6.E353. [DOI] [PubMed] [Google Scholar]
- 43.Géloën A, Collet AJ, Guay G, Bukowiecki LJ. β-Adrenergic stimulation of brown adipocyte proliferation. Am J Physiol. 1988;254(1 pt 1):C175–C182. doi: 10.1152/ajpcell.1988.254.1.C175. [DOI] [PubMed] [Google Scholar]
- 44.Park IR, Himms-Hagen J. Neural influences on trophic changes in brown adipose tissue during cold acclimation. Am J Physiol. 1988;255(6 pt 2):R874–R881. doi: 10.1152/ajpregu.1988.255.6.R874. [DOI] [PubMed] [Google Scholar]
- 45.Géloën A, Collet AJ, Bukowiecki LJ. Role of sympathetic innervation in brown adipocyte proliferation. Am J Physiol. 1992;263(6 pt 2):R1176–R1181. doi: 10.1152/ajpregu.1992.263.6.R1176. [DOI] [PubMed] [Google Scholar]
- 46.Rosenwald M, Perdikari A, Rulicke T, Wolfrum C. Bi-directional interconversion of brite and white adipocytes. Nat Cell Biol. 2013;15(6):659–657. doi: 10.1038/ncb2740. [DOI] [PubMed] [Google Scholar]
- 47.Barbatelli G, et al. The emergence of cold-induced brown adipocytes in mouse white fat depots is determined predominantly by white to brown adipocyte transdifferentiation. Am J Physiol Endocrinol Metab. 2010;298(6):E1244–E1253. doi: 10.1152/ajpendo.00600.2009. [DOI] [PubMed] [Google Scholar]
- 48.Arch JRS, et al. Atypical β-adrenoceptor on brown adipocytes as target for anti-obesity drugs. Nature. 1984;309(5964):163–165. doi: 10.1038/309163a0. [DOI] [PubMed] [Google Scholar]
- 49.Himms-Hagen J, et al. Effect of CL-316,243, a thermogenic β3-agonist, on energy balance and brown and white adipose tissues in rats. Am J Physiol. 1994;266(4 pt 2):R1371–R1382. doi: 10.1152/ajpregu.1994.266.4.R1371. [DOI] [PubMed] [Google Scholar]
- 50.Collins S, Petro AE, Surwit RS. Strain-specific response to β3-adrenergic receptor agonist treatment of diet-induced obesity in mice. Endocrinology. 1997;138(1):405–413. doi: 10.1210/endo.138.1.4829. [DOI] [PubMed] [Google Scholar]
- 51.Inokuma K, et al. Indispensable role of mitochondrial UCP1 for antiobesity effect of beta3-adrenergic stimulation. Am J Physiol Endocrinol Metab. 2006;290(5):E1014–E1021. doi: 10.1152/ajpendo.00105.2005. [DOI] [PubMed] [Google Scholar]
- 52.Polak P, Cybulski N, Feige JN, Auwerx J, Ruegg MA, Hall MN. Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab. 2008;8(5):399–410. doi: 10.1016/j.cmet.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 53.Hahn P, Novak M. Development of brown and white adipose tissue. J Lipid Res. 1975;16(2):79–91. [PubMed] [Google Scholar]
- 54.Hara K, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110(2):177–189. doi: 10.1016/S0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 55.Gwinn DM, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30(2):214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petrovic N, Walden TB, Shabalina IG, Timmons JA, Cannon B, Nedergaard J. Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J Biol Chem. 2010;285(10):7153–7164. doi: 10.1074/jbc.M109.053942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walden TB, Hansen IR, Timmons JA, Cannon B, Nedergaard J. Recruited vs. nonrecruited molecular signatures of brown, “brite,” and white adipose tissues. Am J Physiol Endocrinol Metab. 2012;302(1):E19–E31. doi: 10.1152/ajpendo.00249.2011. [DOI] [PubMed] [Google Scholar]
- 58.Wu J, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150(2):366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Almind K, Kahn CR. Genetic determinants of energy expenditure and insulin resistance in diet-induced obesity in mice. Diabetes. 2004;53(12):3274–3285. doi: 10.2337/diabetes.53.12.3274. [DOI] [PubMed] [Google Scholar]
- 60.Bordicchia M, et al. Cardiac natriuretic peptides act via p38 MAPK to induce the brown fat thermogenic program in mouse and human adipocytes. J Clin Invest. 2012;122(3):1022–1036. doi: 10.1172/JCI59701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stanford KI, et al. Brown adipose tissue regulates glucose homeostasis and insulin sensitivity. J Clin Invest. 2013;123(1):215–223. doi: 10.1172/JCI62308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sanchez-Gurmaches J, Guertin DA. Adipocyte lineages: tracing back the origins of fat. Biochim Biophys Acta. 2014;1842(3):340–351. doi: 10.1016/j.bbadis.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cao W, et al. p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Mol Cell Biol. 2004;24(7):3057–3067. doi: 10.1128/MCB.24.7.3057-3067.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jeffery E, et al. Characterization of Cre recombinase models for the study of adipose tissue. Adipocyte. 2014;3(3):206–211. doi: 10.4161/adip.29674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Debevec D, Christian M, Morganstein D, Seth A, Parker M, White R. RIP140 regulates expression of Uncoupling Protein 1 in adipocytes through specific PPAR isoforms and ERRa. Mol Endocrinol. 2007;21(7):1581–1592. doi: 10.1210/me.2007-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morganstein DL, Wu P, Mane MR, Fisk NM, White R, Parker MG. Human fetal mesenchymal stem cells differentiate into brown and white adipocytes: a role for ERRα in human UCP1 expression. Cell Res. 2010;20(4):434–444. doi: 10.1038/cr.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ahmadian M, et al. Desnutrin/ATGL is regulated by AMPK and is required for a brown adipose phenotype. Cell Metab. 2011;13(6):739–748. doi: 10.1016/j.cmet.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bostrom P, et al. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 2012;481(7382):463–468. doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huss JM, Torra IP, Staels B, Giguere V, Kelly DP. Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor alpha signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol Cell Biol. 2004;24(20):9079–9091. doi: 10.1128/MCB.24.20.9079-9091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cresci S, et al. A PPARalpha promoter variant impairs ERR-dependent transactivation and decreases mortality after acute coronary ischemia in patients with diabetes. PLoS One. 2010;5(9): doi: 10.1371/journal.pone.0012584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485(7396):109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harrison DE, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460(7253):392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Neff F, et al. Rapamycin extends murine lifespan but has limited effects on aging. J Clin Invest. 2013;123(8):3272–3291. doi: 10.1172/JCI67674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Houde VP, et al. Chronic rapamycin treatment causes glucose intolerance and hyperlipidemia by upregulating hepatic gluconeogenesis and impairing lipid deposition in adipose tissue. Diabetes. 2010;59(6):1338–1348. doi: 10.2337/db09-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brattstrom C, Wilczek H, Tyden G, Bottiger Y, Sawe J, Groth CG. Hyperlipidemia in renal transplant recipients treated with sirolimus (rapamycin) Transplantation. 1998;65(9):1272–1274. doi: 10.1097/00007890-199805150-00023. [DOI] [PubMed] [Google Scholar]
- 76.Morrisett JD, et al. Effects of sirolimus on plasma lipids, lipoprotein levels, and fatty acid metabolism in renal transplant patients. J Lipid Res. 2002;43(8):1170–1180. [PubMed] [Google Scholar]
- 77.Lamming DW, et al. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science. 2012;335(6076):1638–1643. doi: 10.1126/science.1215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Um SH, et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431(7005):200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- 79.Ruvinsky I, et al. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 2005;19(18):2199–2211. doi: 10.1101/gad.351605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee KY, et al. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes. 2013;62(3):864–874. doi: 10.2337/db12-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mullican SE, Tomaru T, Gaddis CA, Peed LC, Sundaram A, Lazar MA. A novel adipose-specific gene deletion model demonstrates potential pitfalls of existing methods. Mol Endocrinol. 2013;27(1):127–134. doi: 10.1210/me.2012-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Blancquaert S, et al. cAMP-dependent activation of mammalian target of rapamycin (mTOR) in thyroid cells. Implication in mitogenesis and activation of CDK4. Mol Endocrinol. 2010;24(7):1453–1468. doi: 10.1210/me.2010-0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kwon G, Marshall CA, Pappan KL, Remedi MS, McDaniel ML. Signaling elements involved in the metabolic regulation of mTOR by nutrients, incretins, and growth factors in islets. Diabetes. 2004;53(suppl 3):S225–S232. doi: 10.2337/diabetes.53.suppl_3.s225. [DOI] [PubMed] [Google Scholar]
- 84.de Joussineau C, et al. mTOR pathway is activated by PKA in adrenocortical cells and participates in vivo to apoptosis resistance in primary pigmented nodular adrenocortical disease (PPNAD) Hum Mol Genet. 2014;23(20):5418–5428. doi: 10.1093/hmg/ddu265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chang GR, et al. Rapamycin protects against high fat diet-induced obesity in C57BL/6J mice. J Pharmacol Sci. 2009;109(4):496–503. doi: 10.1254/jphs.08215FP. [DOI] [PubMed] [Google Scholar]
- 86.Guerra C, Koza RA, Yamashita H, Walsh K, Kozak LP. Emergence of brown adipocytes in white fat in mice is under genetic control. Effects on body weight and adiposity. J Clin Invest. 1998;102(2):412–420. doi: 10.1172/JCI3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Robidoux J, et al. Selective activation of mitogen-activated protein (MAP) kinase kinase 3 and p38α MAP kinase is essential for cyclic AMP-dependent UCP1 expression in adipocytes. Mol Cell Biol. 2005;25(13):5466–5479. doi: 10.1128/MCB.25.13.5466-5479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Green H, Meuth M. An established pre-adipose cell line and its differentiation in culture. Cell. 1974;3(2):127–133. doi: 10.1016/0092-8674(74)90116-0. [DOI] [PubMed] [Google Scholar]
- 89.Kumar N, Liu D, Wang H, Robidoux J, Collins S. Orphan nuclear receptor NOR-1 enhances 3′,5′-cyclic adenosine 5′-monophosphate-dependent uncoupling protein-1 gene transcription. Mol Endocrinol. 2008;22(5):1057–1064. doi: 10.1210/me.2007-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ross SR, et al. Hibernoma formation in transgenic mice and isolation of a brown adipocyte cell line expressing the uncoupling protein gene. Proc Natl Acad Sci U S A. 1992;89(16):7561–7565. doi: 10.1073/pnas.89.16.7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell. 1994;79(7):1147–1156. doi: 10.1016/0092-8674(94)90006-X. [DOI] [PubMed] [Google Scholar]
- 92.El-Jack AK, Hamm JK, Pilch PF, Farmer SR. Reconstitution of insulin-sensitive glucose transport in fibroblasts requires expression of both PPARγ and C/EBPα. J Biol Chem. 1999;274(12):7946–7951. doi: 10.1074/jbc.274.12.7946. [DOI] [PubMed] [Google Scholar]
- 93.Dib L, Bugge A, Collins S. LXRα fuels fatty acid-stimulated oxygen consumption in white adipocytes. J Lipid Res. 2014;55(2):247–257. doi: 10.1194/jlr.M043422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tran CM, et al. Rapamycin blocks induction of the thermogenic program in white adipose tissue [published online ahead of print February 8, 2016. Diabetes. doi: 10.2337/db15-0502. doi: 10.2337/db15-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.