Abstract
Hemodynamic shear forces are intimately linked with cardiac development, during which trabeculae form a network of branching outgrowths from the myocardium. Mutations that alter Notch signaling also result in trabeculation defects. Here, we assessed whether shear stress modulates trabeculation to influence contractile function. Specifically, we acquired 4D (3D + time) images with light sheets by selective plane illumination microscopy (SPIM) for rapid scanning and deep axial penetration during zebrafish morphogenesis. Reduction of blood viscosity via gata1a morpholino oligonucleotides (MO) reduced shear stress, resulting in downregulation of Notch signaling and attenuation of trabeculation. Arrest of cardiomyocyte contraction either by troponin T type 2a (tnnt2a) MO or in weak atriumm58 (wea) mutants resulted in reduced shear stress and downregulation of Notch signaling and trabeculation. Integrating 4D SPIM imaging with synchronization algorithm demonstrated that coinjection of neuregulin1 mRNA with gata1 MO rescued trabeculation to restore contractile function in association with upregulation of Notch-related genes. Crossbreeding of Tg(flk:mCherry) fish, which allows visualization of the vascular system with the Tg(tp1:gfp) Notch reporter line, revealed that shear stress–mediated Notch activation localizes to the endocardium. Deleting endocardium via the clochesk4 mutants downregulated Notch signaling, resulting in nontrabeculated ventricle. Subjecting endothelial cells to pulsatile flow in the presence of the ADAM10 inhibitor corroborated shear stress–activated Notch signaling to modulate trabeculation.
Introduction
Hemodynamic forces such as shear stress are intimately linked with cardiac morphogenesis and function (1, 2). In addition to cardiogenic differentiation and transcription factors, intracardiac fluid forces are essential for embryonic cardiogenesis (3–9). Occlusion of flow at either the cardiac inflow or outflow tract resulted in hearts with an abnormal third chamber, diminished looping, and impaired valve formation (4). The myocardium differentiates into 2 layers, an outer compact zone and an inner trabeculated zone. The trabeculae form a network of branching outgrowths from the myocardial wall (10), and both trabeculation and compaction are essential for normal cardiac contractile function. A significant reduction in trabeculation is usually associated with ventricular compact zone deficiencies, whereas hypertrabeculation (noncompaction) is closely associated with left ventricular noncompaction (LVNC) (11). LVNC is the third most common cardiomyopathy after dilated and hypertrophic cardiomyopathy in the pediatric population (12). Its prevalence was estimated from at 4.5 to 26 per 10,000 adult patients referred for echocardiographic diagnosis (12, 13). However, the mechanotransduction mechanisms underlying trabeculation during cardiac development remain elusive (11).
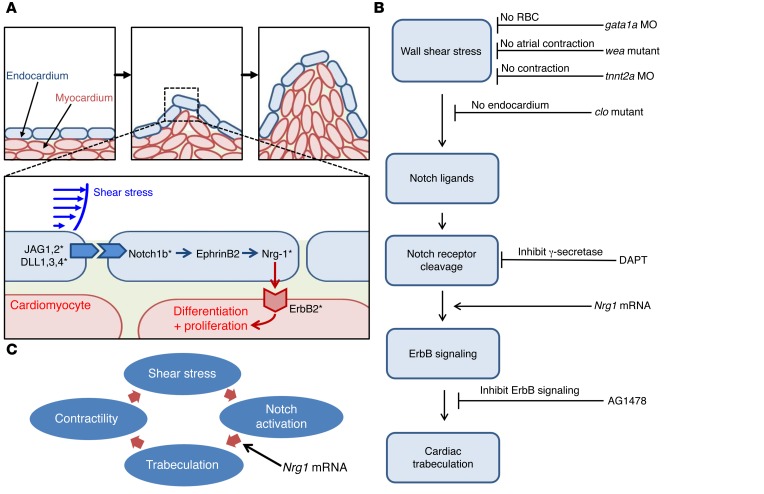
We have demonstrated that peristaltic contractions of the embryonic heart tube produce time-varying shear stress (∂τ/∂t) and pressure gradients (∇P) across the atrioventricular (AV) canal in a zebrafish model of cardiac development (14). The advent of genetic manipulation in zebrafish has enabled the application of the fli1 promoter to drive expression of EGFP throughout the vasculature during embryogenesis (Tg[fli1a:EGFP]y1) (15), thereby allowing for 3D visualization of the moving boundary conditions (2D + time) for computational fluid dynamics (CFD) simulation. However, 4D (3D + time) imaging throughout the cardiac cycle requires fast tissue scanning and deep axial penetration. For these reasons, a laser light-sheet approach known as selective plane illumination microscopy (SPIM) coupled with nongated 4D synchronization algorithm enabled us to capture live 3D cardiac morphogenesis. By illuminating with a thin plane of light and detecting with a high-speed sCMOS camera, we were able to acquire a stack of cardiac sections with both high axial and temporal resolution (Figure 1 and Supplemental Figures 1 and 2; supplemental material available online with this article; doi:10.1172/JCI83496DS1). The 4D SPIM imaging technique revealed dynamic architecture in response to changes in hemodynamic forces, cardiomyocyte contraction, and Notch signaling.
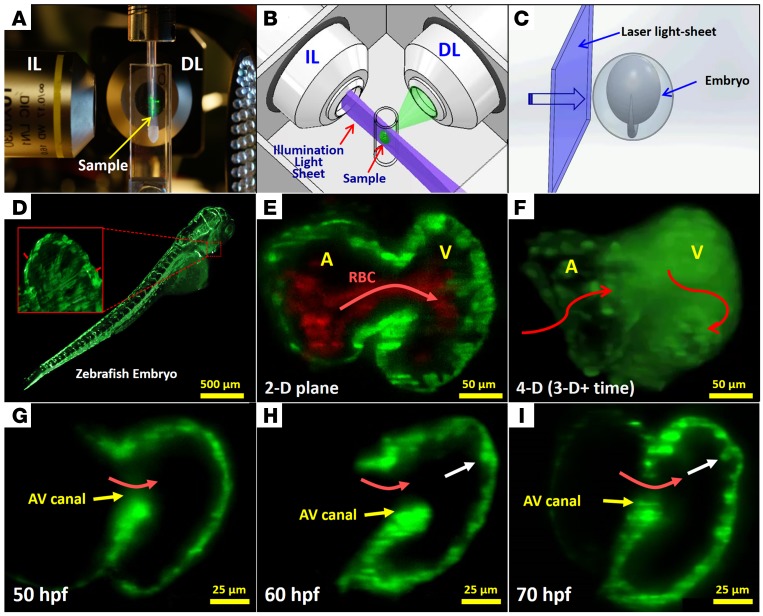
Figure 1. Fluorescent light sheets to image cardiac morphogenesis.
(A) The sample is placed at the orthogonal intersection between the illumination lens (IL) and the detection lens (DL) in the SPIM system. (B) A cylindrical lens generates a thin laser light sheet, and the illumination lens excites a thin slice of the sample in a 2D plane. The fluorescence from the illuminated planes is orthogonally detected by the detection lens. (C) A schematic diagram illustrates the laser light-sheet sectioning across a zebrafish embryo. (D) The entire embryo can be imaged within 30 seconds at a single cellular resolution. Inset reveals the trabecular endocardium. (E) Magnification of the heart reveals the contracting cmlc2-gfp–labeled myocardium and flowing DsRed-labeled red blood cells across the atriventricular valve. (F) The integration of SPIM image with 4D synchronization algorithm reveals a beating 4D heart. A, atrium; V, ventricle. (G) Trabecular ridges were absent in the myocardium at approximately 50 hpf. (H) Ridges protruding into the ventricular cavity occurred at approximately 60 hpf near the myocardial lining experienced high ventricular inflow. (I) Distinct trabecular ridges occurred at approximately 70 hpf.
To elucidate hemodynamic forces underlying the initiation of trabeculation, we lowered hemodynamic shear forces via (a) microinjection of gata1a morpholino oligonucleotides (MO) at the 1- to 4-cell stage to reduce hematopoiesis and viscosity by 90% (16, 17), (b) microinjection of troponin T type 2a (tnnt2a) MO to arrest cardiomyocyte contraction in embryos (18, 19), and (c) genetic mutation of the weak atriumm58 (wea) to inhibit atrial contraction (10, 20). A myocardium-specific GFP transgene in zebrafish allowed for visualization of the ventricular wall. SPIM imaging characterized the attenuated trabecular myocardial network in response to both gata1a MO and tnnt2a MO injection. Both the wea mutants with noncontractile atrium and clochesk4 (clo) mutants with deletion of endocardium expressed substantially lower levels of Notch-related genes as compared with those of WT, and both developed nontrabeculated myocardium. Integrating computation with quantitative analyses, we demonstrated that attenuation of trabeculation in response to gata1a MO, erb-b2 receptor tyrosine kinase 2 (ErbB2) inhibitor (AG1478), and wea mutation was associated with decreased ventricular strain and ejection fraction (EF). The aforementioned loss-of-function was rescued with neuregulin1 (Nrg1) mRNA to restore trabeculation and contraction in association with upregulation of Notch-related genes. We further crossbred Tg(flk:mCherry) with the Tg(tp1:gfp) Notch reporter line to localize shear stress–mediated endocardial Notch activation. In addition, we demonstrated using a dynamic flow system that pulsatile shear stress (PSS) upregulated Notch ligands and target genes in human aortic endothelial cells (HAEC). Overall, interfacing 4D light-sheet imaging with the zebrafish system opens a fundamental direction for demonstrating shear stress modulation of trabeculation to influence contractile function via Notch signaling.
Results
Trabeculation formation.
Trabeculation was visualized with the transgenic zebrafish line, Tg(cmlc2:gfp), in which a cardiac-specific promoter drives the expression of GFP in cardiomyocytes. Trabeculation starts to appear after cardiac looping (21). At approximately 50 hours post fertilization (hpf), 3D SPIM imaging revealed a nontrabecular myocardium (Figure 1G). At approximately 60 hpf, trabecular ridges started to form in the region exposed to ventricular inflow across the AV canal (Figure 1H). At approximately 70 hpf, these ridges developed into a network of trabecular myocardium (Figure 1I and ref. 22).
Reduced hemodynamic shear stress attenuated trabecular formation.
The gata1a MO microinjection reduced hematopoiesis and viscosity by 90% (16, 17), leading to a reduction in hemodynamic shear stress and a delayed initiation of trabecular network at 75 and 100 hpf when compared with control Tg(cmlc2:gfp) zebrafish (Figure 2, C–F, and Supplemental Videos 1 and 2). Furthermore, gata1a MO injection significantly downregulated Notch ligands (delta-like 4 [Dll4], jagged 1 [Jag1], and Jag2), receptor (Notch1b), and downstream signaling components (Nrg1 and ErbB2) (P < 0.05, n = 5) (Figure 3A and refs. 10, 23, 24), whereas coinjection with Nrg1 mRNA (5 pg/nl) rescued trabecular formation at 75 and 100 hpf in association with upregulation of Notch signaling–related genes (Figure 2, G and H; Figure 3A; and Supplemental Video 3). Tg(tp1:gfp) zebrafish allow assessment of Notch activation in response to shear stress (25). Crossbreeding Tg(flk:mCherry) with Tg(tp1:gfp) lines further localized altered shear stress–activated Notch signaling in endocardium in response to gata1a MO injection (Figure 4), whereas tp1-gfp signal was present outside of the endocardium in response to coinjection of Nrg1 mRNA with gata1a MO. Thus, gata1a MO injection reduced hemodynamic shear forces, leading to downregulation of Notch signaling, whereas coinjection with Nrg1 mRNA restored trabecular formation and resulted in the upregulation of Notch-related genes.
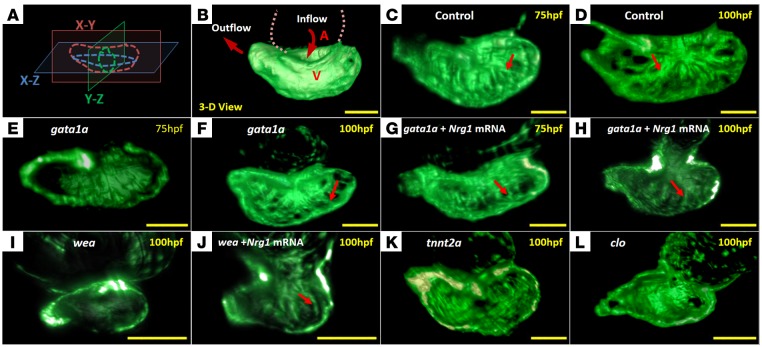
Figure 2. The dynamics of 3D cardiac architecture in response to genetic manipulations of the Tg(cmlc:gfp) zebrafish system.
(A) A 3D Cartesian coordinate system provides the orientation of the ventricle. (B) The ventricular inflow tract from the atrium relates to the outflow tract from the 3D ventricle. (C) Trabecular network developed in the ventricle at 75 hpf in the WT (control). (D) Trabeculae formed a prominent sponge-like structure at 100 hpf in the WT. (E) gata1a MO microinjection at the 1- to 4-cell stage attenuated trabeculation at 75 hpf. (F) gata1a MO–treated fish delayed trabecular formation at 100 hpf. (G) Coinjection of Nrg1 mRNA promoted trabeculation at 75 hpf. (H) Coinjection of Nrg1 mRNA nearly restored trabecular network at 100 hpf. (I) Trabeculation was absent in the wea mutants with a small ventricle. (J) Injection of Nrg1 mRNA into the wea mutants promoted trabecular formation. (K) The tnnt2a MO microinjection inhibited trabeculation at both 75 hpf (not shown) and 100 hpf. (L) The clo mutants developed nontrabeculated endocardium at 100 hpf. Red arrows indicate trabecular ridges. Scale bars: 50 μm. Original magnification, ×10.
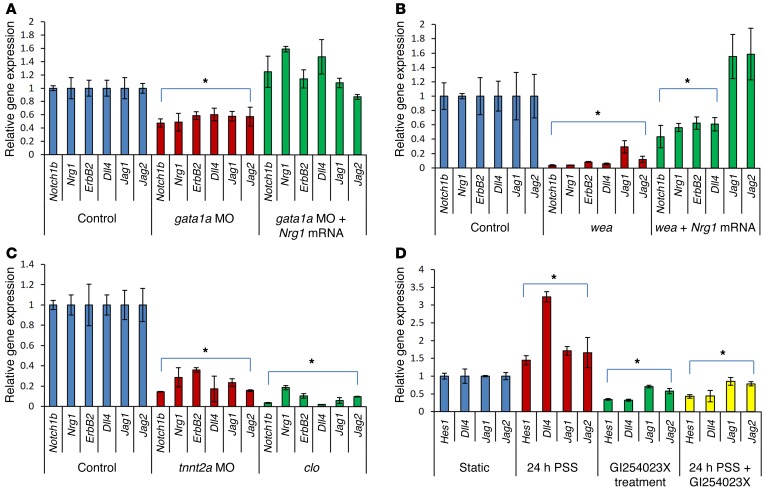
Figure 3. Hemodynamic shear stress modulation of Notch ligand, receptor, and target gene expression.
(A) At 100 hpf, gata1a treatment significantly downregulated the mRNA expression of Notch1, Nrg1, and Jag2 (t test, *P < 0.05, n = 5 vs. WT). Coinjection of Nrg1 mRNA with gata1a MO significantly increased the expression of Notch1b, Nrg1, ErbB, and Jag1 genes compared with the WT. (B) The wea mutations resulted in downregulation of Notch-related gene expression (t test, *P < 0.05, n = 5 vs. WT). Nrg1 mRNA injection into the wea mutants increased Notch-related gene expression; in particular, Jag1 and Jag2 were significantly upregulated compared with the WT. (C) tnnt2a MO injection and clo mutation significantly downregulated Notch-related gene expression (t test, *P < 0.05, n = 5 vs. WT). (D) PSS (τaverage = 30 dyn × cm–2 × s–1 at 1 Hz) upregulated Notch target genes and ligands in HAEC, which were downregulated in the presence of ADAM10 inhibitor, which blocks the extracellular cleavage of NICD. (t test, *P < 0.05, n = 5).
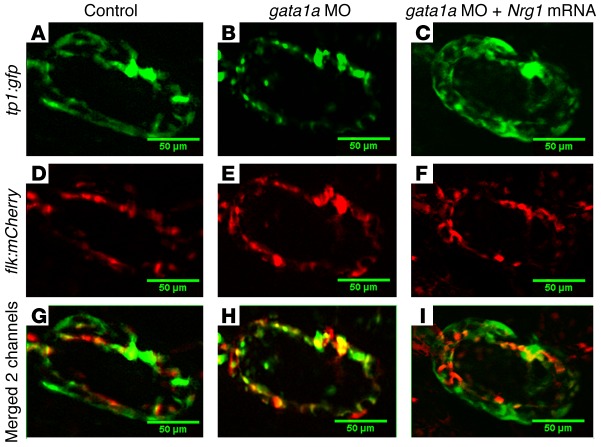
Figure 4. Endocardial Notch activation in Tg(cmlc2:mCherry;tp1:gfp) zebrafish.
(A) Tp1-gfp signal was present throughout the ventricle. (B) Following gata1a MO injection, Notch signaling was diminished in the endocardium, as demonstrated by reduced tp1 signals. (C) Nrg1 mRNA coinjection restored Notch signaling signals. (D–F) flk:mCherry transgene was used to delineate the endocardium. (G–I) The merged gfp and mCherry channels revealed endocardial Notch signaling. Scale bars: 50 μm. Original magnification, ×20.
To further elucidate hemodynamic modulation of trabeculation via Notch signaling, we introduced the wea mutants to reduce atrial contraction, leading to reduced ventricular inflow. The wea mutants developed downregulation of cardiac mRNA levels of Notch signaling–related genes as compared with those of WT and developed small, nontrabeculated ventricles (Figure 2I and Figure 3B). However, microinjection of Nrg1 mRNA (10 pg/nl) to wea mutants at the 1- to 4-cell stage partially rescued trabecular ridges to enhance contractile ventricular function, accompanied with upregulation of Notch signaling genes (Figure 2J and ref. 10). In this context, the wea mutants demonstrated that a noncontractile atrium engendered a nontrabeculated ventricle, underscoring the need for synchronized alternating atrial and ventricular contraction to generate hemodynamic forces to upregulate Notch signaling for trabeculation.
The tnnt2a MO inhibited trabecular formation.
The tnnt2a MO microinjection inhibited cardiac tnnt2a, leading to the arrest of myocardial contraction and the absence of hemodynamic forces (18, 19). Microinjection of tnnt2a MO significantly downregulated Notch signaling (P < 0.05 for all comparisons, n = 5) (Figure 3C). The tnnt2a MO–injected zebrafish persistently harbored a nontrabeculated and thin ventricular wall at 100 hpf (Figure 2K and Supplemental Video 4). These observations further support hemodynamic forces underlying the initiation of trabeculation.
The clo mutants attenuated notch signaling and trabeculation.
To demonstrate hemodynamic force–mediated Notch signaling in the endocardium, we abolished endothelial lining with the clo mutants (26, 27). At 100 hpf, clo mutants displayed a small and thin ventricle as previously described (refs. 14, 27, Figure 2L, and Supplemental Video 5), accompanied by a reduction in cardiac mRNA for Notch ligands, receptor, and target genes as compared with those of WT (Figure 3C). Using the well-calibrated in vitro flow system, HAEC were exposed to PSS with the time-averaged shear stress (τavg) at 23 dyn × cm–2 and 1 Hz cycle (28–31). We were able to recapitulate PSS-mediated upregulation in endothelial Notch signaling–related genes, which were inhibited in the presence of the ADAM10 inhibitor, GI254023X, which blocks proteolytic cleavage of Notch and formation of Notch intracellular domain (NICD) (Figure 3D). In addition, we demonstrated Notch signaling–mediated trabecular formation by inhibiting the translocation of NICD to the nuclei with the γ-secretase inhibitor (DAPT) (32, 33), where we observed downregulation of cardiac Notch-related gene expression and absence of trabecular formation (Supplemental Figure 3). Taken together, hemodynamic forces were implicated in the initiation of trabeculation via endocardial-dependent Notch signaling.
EPO-augmented viscosity induced no additional trabecular network.
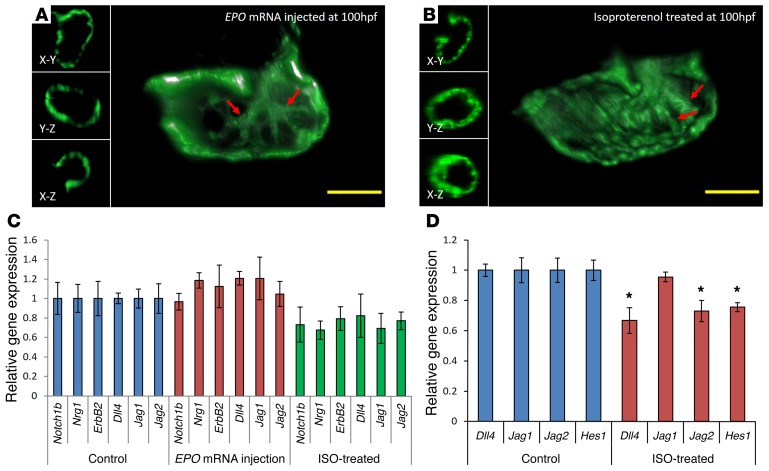
We further cloned erythropoietin (EPO) mRNA, which was injected at the 1- to 2-cell stage at 20 pg/nl to increase hematopoiesis and, thus, blood viscosity (Supplemental Videos 6 and 7, and ref. 34). Since heart rates remained regular, assuming that pressure gradients are not affected by injection of EPO mRNA, shear stress increased by keeping the tangential velocity gradient normal. Despite an EPO-augmented time-averaged shear stress, the Notch-related genes were not upregulated, and the trabecular network appeared to be the same as that of WT (Figure 5, A and C). We further used isoproterenol (ISO), a β1- and β2- adrenoreceptor agonist, at 50 μM to increase myocardial contractility and heart rate. However, neither Notch-related genes nor trabecular network was significantly affected (Figure 5, B and C, and ref. 35). Moreover, ISO treatment moderately reduced Notch activation. We further corroborated the effect of ISO in HAEC, where treatment at 24 hours resulted in reduced Notch signaling (Figure 5D).
Figure 5. EPO-augmented shear stress and Notch signaling and trabeculation at 100 hpf.
(A) EPO mRNA injection and (B) ISO treatment were ineffective in increasing trabecular network. (C) Augmented shear stress failed to increase Notch signaling (t test, P not significant for all gene transcripts, n = 5). (D) Treatment with 50 μM of ISO for 24 hours moderately downregulated Notch target genes and ligands in HAEC (t test, *P < 0.05, n = 5). Scale bars: 50 μm. Original magnification, ×10.
Overexpression of Notch signaling induced abnormal ventricular morphogenesis.
We showed that coinjection of Nrg1 mRNA (10 pg/nl) with gata1a MO and Nrg1 mRNA alone engendered thickened ventricular wall and disrupted trabeculation (Figure 6A). Furthermore, both treatments increased Notch signaling and Notch-related genes (Figure 6B). Zhao et al. reported that, while Notch signaling in both endo- and epicardium is considered important in cardiac regeneration in response to ventricular amputation, hyperactivation of Notch signaling is found to suppress cardiomyocyte proliferation and heart regeneration in zebrafish (36). Therefore, our data also suggested that a well-defined level of Notch signaling is indicated as inducing trabeculation during cardiac morphogenesis.
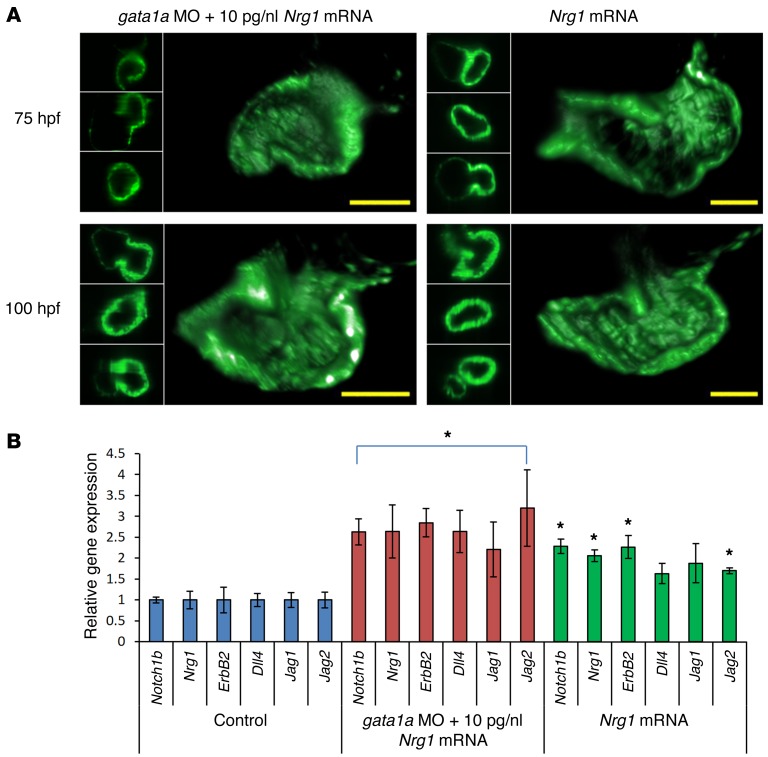
Figure 6. High-dose Nrg1 mRNA (10 μM) did not restore trabeculation in the gata1a MO–injected zebrafish.
(A) While gata1a MO injection alone attenuated trabeculation, coinjection of gata1a MO with 5 μM of Nrg1 mRNA partially restored trabeculation (Figure 3). However, coinjection with 10 μM of Nrg1 mRNA promoted the development of a thick and small ventricular wall rather than restoring trabeculation at 75 hpf and 100 hpf. (B) Injection of 5 μM Nrg1 mRNA alone into WT upregulated Notch signaling–related mRNA expression and failed to develop trabecular formation (t test, *P < 0.05, n = 5).
Trabeculation contributed to contractile function.
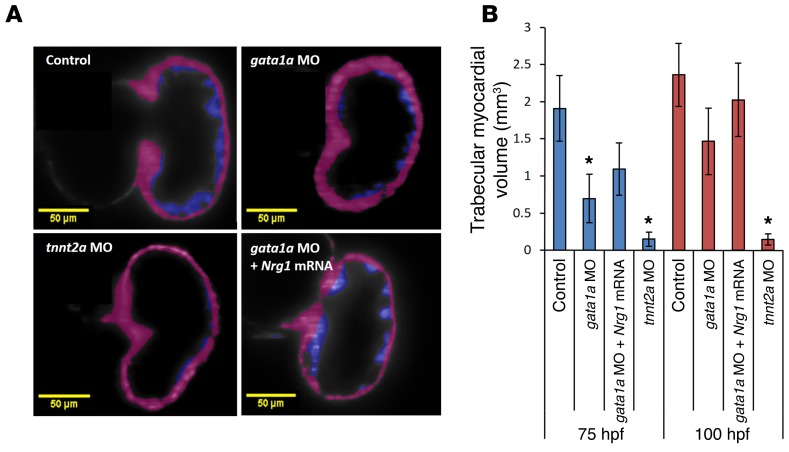
To demonstrate the role of trabeculation on ventricular function, we quantified the volume of trabecular myocardial ridges in response to gata1a MO and tnnt2a MO injection and assessed the effects of Nrg1 mRNA rescue. At 75 hpf, the volume of trabecular myocardial ridges was reduced by approximately 2.7 ± 0.4-fold in response to gata1a MO (0.7 × 10–4 ± 0.3 × 10–4 μm3), by approximately 19.0 ± 0.3-fold in response to tnnt2a MO (0.1 × 10–4 ± 0.1 × 10–4 μm3), and by approximately 1.7 ± 0.4-fold in response to coadministration of Nrg1 mRNA with gata1a MO (1.1 × 10–4 ± 0.4 × 10–4 μm3) as compared with the WT (1.9 × 10–4 ± 0.4 × 10–4 μm3) (Figure 7). At 100 hpf, the volume of trabecular myocardial ridges was reduced by approximately 1.5 ± 0.4-fold in response to gata1a MO (1.5 × 10–4 ± 0.3 × 10–4 μm3), by approximately 23.0 ± 0.3-fold in response to tnnt2a MO (0.1 × 10–4 ± 0.1 × 10–4 μm3), and by approximately 1.0 ± 0.5-fold in response to coadministration of Nrg1 mRNA with gata1a MO (2.0 × 10–4 ± 0.5 × 10–4 μm3) as compared with the WT (2.3 × 10–4 ± 0.4 × 10–4 μm3) (Figure 7). This reduction in volume remained persistent in the tnnt2a MO–injected group at 100 hpf. However, Nrg1 mRNA coinjection partially rescued trabecular formation in the gata1a MO group.
Figure 7. 3D quantification of myocardium from the trabecular ridges.
(A) Transgenic cardiac myosin light chain–labeled GFP Tg(cmlc:gfp) lines were visualized by SPIM imaging. Myocardium from trabecular ridges was highlighted in violet and the outside layer in magenta. (B) gata1a MO and tnnt2a MO significantly reduced myocardium volume from trabecular ridges (μm3) at 75 hpf and 100 hpf, respectively. The tnnt2a MO had a stronger effect than the gata1a MO (t test, *P < 0.05, n = 5). Coinjection of Nrg1 mRNA with gata1a MO showed a trend toward restoring the trabecular myocardial volume as compared with the WT. Scale bars: 50 μm. Original magnification, ×10.
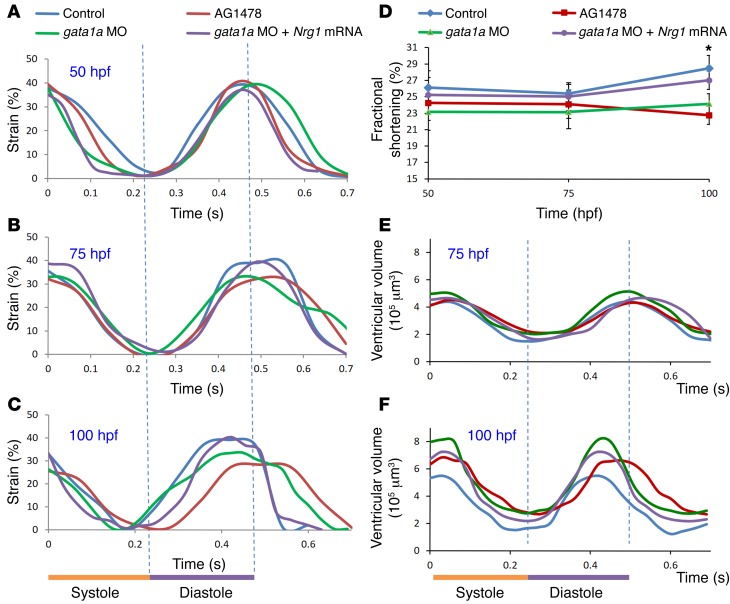
To assess contractile function, we analyze time-dependent changes in ventricular strain in terms of circumferential displacement (D) during cardiac cycles in response to gata1a MO and ErbB signaling inhibitor (AG1478) at 50 hpf, 75 hpf, and 100 hpf (Figure 8, A–C). Treatment with AG1478 significantly reduced strain during ventricular diastole at 100 hpf (Figure 8C), which was accompanied by a decreased fractional shortening (Figure 8D). Both gata1a MO and AG1478 treatment reduced ventricular EF, as assessed by the changes in 4D SPIM-acquired ventricular volume during the cardiac cycle (Figure 8, E and F). Both groups developed an increased end systolic volume (ESV) and end diastolic volume (EDV) (Table 1). Nrg1 mRNA rescue to gata1a MO injection normalized the ventricular strain, fractional shortening, and EF. Interestingly, trabeculated myocardium in the control zebrafish demonstrated higher local strain and local deformation than the nontrabeculated myocardium (Supplemental Figure 4, A, C, and D). In the AG1478 group, trabeculation was absent, and the local strain was similar to that of the nontrabeculated regions of control zebrafish (Supplemental Figure 4, B–D).
Figure 8. Effects of trabeculation on strain and fractional shortening.
Strains were depicted at (A) 50 hpf, (B) 75 hpf, and (C) 100 hpf in response to AG1478 treatment and gata1a MO injection. (C) Significant decrease and delay in strain were observed in response to AG1478 (red) and gata1a MO (green) as compared with the WT (control, black) at 100 hpf. Coinjection of Nrg1 mRNA (blue) with gata1a MO improved strain at 100 hpf. (D) AG1478 and gata1a MO significantly decreased fractional shortening at 100 hpf. Coinjection of Nrg1 mRNA with gata1a MO improved FS. (E and F) Integrating 4D synchronization algorithm with SPIM revealed that AG1478 and gata1a MO groups developed increased ESV and EDV.
Table 1. Ventricular volume and EF in response to gata1a MO– and AG1478-treated zebrafish.
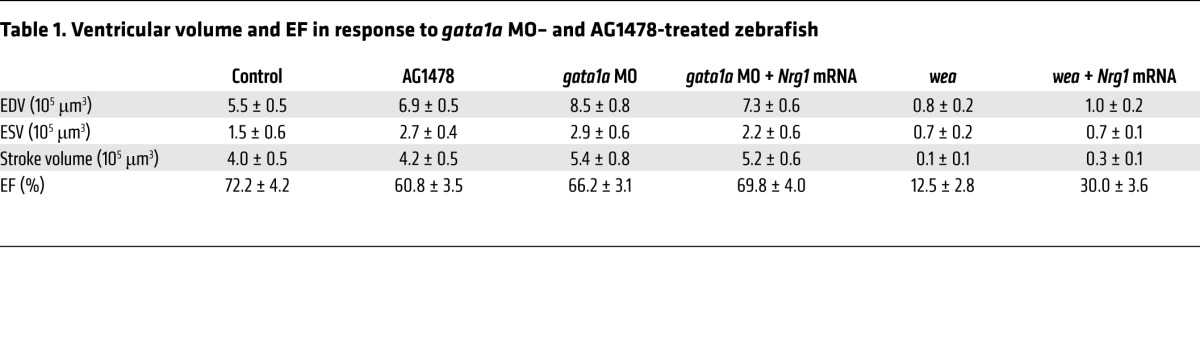
The combination of reduced atrial contractility and absent active ventricular filling in the wea mutants resulted in a significant reduction in ventricular volume (Table 1 and Supplemental Figure 5). Injection of Nrg1 mRNA to the wea mutants at the 1- to 4-cell stage partially restored trabecular formation (Figure 2J) and partially improved the strain, stroke volume, and ventricular contraction (Supplemental Figure 5B). Thus, the reduced-contractile atrium in the wea mutants is associated with (a) an absence of hemodynamic force to generate active ventricular filling, (b) downregulation of Notch-related genes, and (c) absence of trabeculation to contribute to contractile function. Taken together, hemodynamic shear force–mediated trabeculation contributed to cardiac contractile function (Figure 9).
Figure 9. Shear stress activation of Notch signaling to promote trabeculation.
(A) Shear stress modulates trabecular formation in the myocardium. The inset reveals the activated Notch signaling in the endocardial cells (blue). Asterisks indicate tested genes. (B) A host of genetic manipulations elucidates the mechanotransduction of Notch signaling pathway underlying shear stress and initiation of trabeculation. (C) Nrg1 mRNA injection rescues trabeculation, which, in turn, restores cardiac contractile function. The gain of function in contractility generates hemodynamic shear forces to activate Notch signaling.
Discussion
The main contribution of integrating 4D SPIM technique with hemodynamic forces resides in the elucidation of the interplay between shear stress and Notch activation in initiating trabeculation during cardiac morphogenesis. By implementing the nongated 4D synchronization algorithm with SPIM, we have unraveled the trabecular network in association with ventricular contractile function. While the development of trabeculation is conserved between the zebrafish and chick, mouse, and human embryos (3, 23, 37–39), there are numerous layers of cardiomyocytes in mice (23), and there is a thin layer of cardiomyocytes for trabeculation in zebrafish embryos. For this reason, 4D SPIM imaging enables uniform illumination to uncover shear stress–induced endocardial Notch signaling to form a trabecular network for contractile function with high axial, spatial, and temporal resolution. We demonstrated that (a) gata1a MO decreased hematopoiesis to reduce shear stress, (b) tnnt2a MO inhibited ventricular contractile function to reduce hemodynamic shear forces, and (c) wea mutants lacked atrial contraction, resulting in a reduction of hemodynamic forces to the ventricle. All 3 conditions resulted in downregulation of Notch-related genes and attenuation of trabeculation. As a corollary, clo mutants lacked endocardium, resulting in the absence of trabeculation. Subjecting endothelial cells to PSS in the presence of the ADAM10 inhibitor corroborated that shear stress activated Notch signaling.
The use of the Tg(flk:mCherry;tp1:gfp) line further localized and corroborated Notch signaling in endocardium (Figure 4). In addition, Notch activation was present beyond endocardium in both the WT and Nrg1 mRNA–rescued groups (Figure 4, A, C, G, and I). Notch1a is recognized as present in the endothelium during angiogenesis (40) and vascular development (41). Endothelial cells lining the coronary arteries communicate from endocardium to myocardium to epicardium (42). As development proceeds, the trabecular myocardium collapses toward the myocardial wall, forming a thick and compact ventricular wall (43). Recently, Tian et al. reported that trabecular compaction traps endocardial cells to mature into the inner-wall vasculature (44). Furthermore, Notch signaling in both endo- and epicardium is considered important in cardiac regeneration in response to ventricular amputation (36). In this context, we speculate that Notch signaling is present from endocardium to myocardium and epicardium via the coronary vasculature. Nevertheless, there remains a complicated role of Notch signaling beyond endocardium to activate Nrg1, and the precise mechanism remains to be investigated.
Current imaging techniques, including confocal microscopy, are limited by their intrinsic depth penetration, axial resolution, and long scanning time (45). Confocal microscopy is oftentimes limited from capturing the entire live zebrafish embryo and the beating hearts due to its small working distance for the objective lens and the long acquisition time. Digital particle image velocimetry (DPIV) is inherently limited in analyzing 3D cardiac mechanics due to its time-dependent 2D image (2D + time domain) and assumption needed for volumetric analysis. Images acquired with the conventional microscopy techniques incur (a) serious background noise due to out-of-focus illumination and (b) low axial resolution due to a large depth of field, whereas our SPIM-based imaging applies 2 separate sets of lenses for illumination and detection through selective plane excitation via laser light sheet, which greatly reduces background noise with the use of long working distance objectives (Supplemental Figure 2 and refs. 46, 47). Furthermore, the SPIM-based system is able to visualize the neonatal mouse hearts, following euthanasia and optical clearing (48). SPIM is superior to ultra–high frequency ultrasound (U-HFU) for its capability in tracking fluorescently labeled proteins, cellular structures, and cells of interest with superior spatial resolution (0.6 μm for SPIM vs. ~30 μm for U-HFU) as well as deep penetration into tissues cleared by ClARITY (49, 50). Thus, we integrated 4D SPIM (3D + time domain) with a postimaging synchronization algorithm to address the irregular periodicity of zebrafish heart rhythms, thereby providing a basis to overcome time-dependent 3D CFD simulation (14). In this context, our SPIM system has advanced optical imaging for (a) deep axial resolution, (b) large dynamic ranges, (c) fast acquisition, and thus, (d) reduced photo-bleaching/toxicity (Supplemental Figure 1).
During heart development, the myocardium differentiates into 2 layers: an outer compact zone and an inner trabeculated zone. In the developing nonzebrafish embryonic hearts, trabeculation facilitates oxygenation and nutrition to the myocardium and enhances cardiac contractile function (37). As development proceeds, the trabecular myocardium collapses toward the myocardial wall, forming a thick and compact ventricular wall (43). The formation of a multilayered spiral myocardium during the late fetal and neonatal stage is essential for cardiac contractile function (51). A recent study of the highly trabeculated zebrafish heart demonstrates that Nrg1 and ErbB2, in addition to their role in promoting cell proliferation, have another important function in regulating cardiomyocyte delamination to initiate ventricular trabeculation (23).
Biomechanical forces upregulate Notch ligands and receptors, and Notch upregulation establishes force-induced proteolysis as a mechanism of cellular mechanotransduction (52, 53). We demonstrated that PSS (23 dyne × cm–2 at 1 Hz for 24 hours) upregulated Notch activation in HAEC in vitro (Figure 3D). We further recapitulated hemodynamic forces–mediated endocardial Notch activation by genetic manipulation of hematopoiesis (gata1a MO), cardiac contraction (tnnt2a Mo), atrial contraction (wea), endocardial endothelial deletion (clo mutants), and localization of Notch activation in endocardium (crossbreeding Tg[flk:mCherry] with Tg[tp1:gfp] Notch reporter line). We further demonstrated that Nrg1 mRNA rescue restored trabeculation in association with an improved ventricular strain and EF. Concomitant upregulation of Notch-related genes, including Dll4, Jag1, and Jag2, suggested that Nrg1 mRNA–restored trabeculation and contractile function is implicated in activation of Notch ligands (Jag1, Jag2, and Dll4) and receptor (Notch 1b) to promote proteolytic cleavage of Notch (52, 53). Masumura et al. have demonstrated that shear stress further induced time-dependent Notch signaling in murine embryonic stem cell–derived vascular endothelial cells (54). de la Pompa et al. reported a positive feedback loop between Notch ligands and target genes (55), and inactivation of Notch in embryonic endocardium results in a decrease in Dll4 expression (24, 56).
Notch activation by Dll1 or Dll4 in the endocardium results in the transcription of EphrinB2, which in turn regulates Nrg1 (57). As a secreted factor, Nrg1 signals to the adjacent cells to promote their differentiation into trabecular myocytes. In a parallel pathway, Notch activity in the endocardium activates BMP10 expression in the adjacent myocytes to promote proliferation (57). Unlike mouse and chick development, ErbB2 contributes to both proliferation and differentiation in zebrafish cardiomyocytes to form a relatively thin layer of myocardium (23).
Nrg1 contributes to cardiac contractility (57, 58). Disruption of Nrg1 expression after ischemic insult impairs cardiac contractility (59), whereas Nrg1 preconditioning confers cardiac protection from ischemic injury (60). Nrg1 mRNA injection rescued trabeculation to restore cardiac strain (Figure 8). This gain of function is suggested by the increase in contractile function–mediated hemodynamic forces to activate Notch signaling (Figure 3). We further demonstrated that shear stress–activated Notch signaling is endocardium dependent. Coinjection of Nrg1 mRNA with gata1a MO restored both trabecular formation (Figure 2) and contractile function (Figure 8). Although increasing Nrg1 mRNA injection from 5 to 10 pg/nl increased Notch-related gene expression by approximately 2-fold, it led to abnormal ventricular morphogenesis, suggesting that a well-defined level of Notch signaling is required for trabeculation (Figure 6). In response to gata1a MO, tnnt2a MO, wea mutation, or AG1478 treatment, Notch-related genes were significantly downregulated, and trabeculation was attenuated or inhibited (Figures 2 and 3). However, in response to EPO mRNA–augmented shear stress, neither Notch-related genes nor trabeculation were altered (Figure 5).
gata1a MO and AG1478 treatment reduced strain and fractional shortening, leading to decreased EF. Interestingly, both gata1a MO– and AG1478-treated embryos developed large ventricular volumes. However, the wea mutants developed small ventricles and low ventricular volumes (Table 1).
Overall, we provide new mechanotransduction insights into shear stress modulation of cardiac trabeculation via endocardial Notch signaling (Figure 9). Interfacing 4D light-sheet imaging with the genetically engineered zebrafish system introduces a dynamic model to establish the significance of blood flow underlying cardiac architecture and function with clinical relevance to noncompaction cardiomyopathy.
Methods
4D cardiac SPIM imaging with synchronization algorithm.
We have integrated our in-house 4D SPIM imaging system with postprocessing synchronization (Supplemental Methods) to visualize the dynamic cardiac architecture with high axial resolution (Supplemental Figure 1). Using the SPIM technique, we scanned 300 sections from the rostral to the caudal end of the zebrafish heart. Each section was captured with 300 x-y planes (frames) at 10-ms exposure time per frame via a sCMOS camera (Hamamatsu Photonics). The thickness of the light sheet was tuned to approximately 5 μm to provide a high axial (z axis) resolution for adequate reconstruction of the 3D cardiac morphology, and the Z scanning was set to 1 μm for lossless digital sampling according to the Nyquist sampling principle. To synchronize with the cardiac cycle, we determined the cardiac periodicity on a frame-to-frame basis by comparing the pixel intensity from the smallest during peak systole to the largest ventricular volume during end diastole (61, 62). The reconstructed 4D images were processed by Amira software (Supplemental Video 8).
Zebrafish embryos.
Zebrafish were bred and maintained at the UCLA Core Facility, and experiments were performed in compliance with UCLA IACUC protocols (63). The transgenic Tg(cmlc2:gfp), Tg(flk:mCherry;tp1:gfp), and Tg(cmlc2:mCherry;tp1:gfp) zebrafish lines were used under the following conditions: (a) control (WT), (b) gata1a MO, and (c) tnnt2a MO microinjection (64). The gata1a MO reduced hematopoiesis and blood viscosity by 90%, as previously reported (16, 17). Fluid shear stress (τ) is characterized as the frictional force that acts tangentially on the surface of endothelial cells (65). For Newtonian fluids, shear stress is defined as follows: τ = μ × du/dy, where μ represents viscosity, and du/dy is the velocity gradient along the y axis perpendicular to the wall (66). In Newtonian fluid mechanics, reduction in viscosity (μ) by 90% resulted in a proportional reduction in wall shear stress (τ) (22, 67). The tnnt2a MO inhibited cardiac tnnt2a to prevent cardiomyocyte contraction, leading to noncontractile atrium and ventricle (18, 19). The clo mutants with specific deletion of endocardium were used to verify endocardium-dependent Notch signaling in response to shear stress. The noncontractile wea mutants (a gift from Deborah Yelon at UCSD) were used to inhibit the development of AV gradients. Tricaine mesylate (0.5%) was used to humanely sacrifice the embryos.
Chemical treatment to modulate trabeculation.
ErbB signaling inhibitor AG1478 at 5 μM (Sigma-Aldrich) in 1% DMSO was diluted in E3 medium at 30 hpf. Similarly, DAPT (Sigma-Aldrich) at 100 μM in 1% DMSO was diluted in E3 medium to inhibit Notch signaling at 40 hpf. GI254023X, an ADAM10 inhibitor at 5 μM, was added in HAEC culture medium and incubated for 30 minutes. 5 μM of premixed GI254023X was added to the culture medium for HAEC subject to PSS in the ensuing experiments. To increase shear stress in the ventricular cavity, 50 μM of isoprenaline hydrochloride (DL-ISO) (I5627, Sigma-Aldrich) was applied at 50 hpf, thereby increasing heart rate and contractility (35).
Blood shear stress modulation.
Shear stress (τ) is characterized as dynamic viscosity (μ) of fluid multiplied by shear rate (r), defined as a gradient of velocity between 2 adjacent fluid layers (68).
(Equation 1)
where (∂ux/∂y) is the tangential velocity gradient between 2 adjacent fluid layers. Since shear stress is a function of velocity (∂ux), injection of tnnt2a MO and use of wea mutants reduced ventricular contractility, which in turn decreased shear stress to the endocardium. In addition, we applied ISO treatment to increase heart rate and contractility, thereby augmenting shear stress. Furthermore, shear stress is also interpreted as time rate of momentum change (ṗ) as the rate of fluid mass (ṁ) acting on the surface per unit area (A) as follows:
(Equation 2)
(Equation 3)
(Equation 4)
where ρ is the density of blood, ū is the average molecular speed, ⟨ux⟩ is the mean velocity along the x axis of fluid molecule hitting the unit area, and λ is the mean free path defined as the average traveling distance of moving particles between collisions. Injecting gata1a MO inhibited hematopoiesis to decrease the hematocrit, leading to a decrease in viscosity, whereas injecting EPO mRNA increased hematopoiesis, leading to an increase in viscosity and shear stress to the endocardium (Supplemental Videos 7 and 8).
Quantification of strain and fractional shortening.
To measure 2D ventricular diameter change, we used SPIM imaging to follow the ventricular developmental stages at 50, 75, and 100 hpf via a sCMOS camera. The captured images were segmented to create the 2D moving boundary conditions with 600 nodes (14). The nodes were guided to replicate cardiac wall motion captured by SPIM segmentation as described previously (14). Matlab (Mathworks) was employed to calculate the strain as defined by the time-dependent changes in displacement (D) between the individual time steps:
(Equation 5)
The changes in displacement between end diastole and systole were used to calculate the fractional shortening (69):
(Equation 6)
where Do denotes the initial displacement at time = 0, and D at time = Δt. Accuracy of repetition error was tested by calculating the strain of coefficient of variation at end diastole (Supplemental Table 1).
3D quantification of the volume of trabecular ridges.
To quantify changes in the volume of trabecular myocardial ridges (μm3), we employed Amira 3D software to reconstruct the cardiac volume. The ventricular volume without trabeculation was simulated by removing trabecular ridges. The volume of trabecular myocardial ridges was derived by subtracting the volume of smooth curve, ignoring trabecular ridges from the total myocardial volume.
Stroke volume and EF.
Based on SPIM images with nongated 4D synchronization computational algorithm (61, 62), the time-dependent changes in ventricular chamber volume throughout the cardiac cycle were measured by Amira. ESV and EDV were obtained by determining the ventricular volume during systole and diastole, respectively. EF was calculated using ESV and EDV (69).
Preparation of Nrg1 and EPO mRNA for rescue.
Human Nrg1 cDNA (a gift from William Talbot, Stanford University, Stanford, California, USA) was amplified from a donor plasmid (purchased from GE Health) and cloned into the plasmid pCS2+ at the BamHI and EcoRI sites (Supplemental Figure 6A). Clones with the human Nrg1 cDNA insert were selected by PCR screening. Two clones with human Nrg1 cDNA insert were verified by transfecting the plasmids into HEK-293 cells, and Nrg1 protein expression was detected by Western blot with anti-Nrg1 antibody (Supplemental Figure 6B). mRNA was made from the clone 1 plasmid using the mMessage SP6 Kit (Invitrogen) following the manufacturer’s instructions. In vitro–transcribed Nrg1 mRNA was purified with Bio-Rad’s Total RNA Isolation Kit for in vivo rescue experiments. Zebrafish EPO cDNA was amplified from a donor plasmid and cloned into pCS2+ at the EcoRI and XhoI sites (Creative Biogene). Zebrafish EPO mRNA was prepared and purified with the same procedure as described above.
Genes knocked down by morpholinos and rescued by Nrg1 mRNA.
MO (GeneTools) were designed against the ATG of gata1a (5′-CTGCAAGTGTAGTATTGAAGATGTC-3′) and tnnt2a (5′-CGCGTGGACAGATTCAAGAGCCCTC-3′). Morpholinos were resuspended in nuclease-free water and injected at 8 ng/nl and 4 ng/nl at 1- to 4-cell stages, respectively. Nrg1 mRNA at 5 or 10 pg/nl was coinjected at 1- to 4-cell stages with gata1a MO to overexpress Notch target genes and restore trabeculation. 20 pg/nl of EPO mRNA was also injected at the 1- to 4-cell stage to increase hematopoiesis (34).
Dynamic shear stress model.
Confluent HAEC were exposed to PSS at 23 dyn × cm–2 at 1 Hz at a slew rate (∂τ/∂t) of 71 dyn × cm–2s–1 for 24 hours (29) in a well-defined dynamic flow system (29). The pulsatile flow simulates hemodynamic shear stress in the arterial system (70, 71). Notch ligands (Dll4, Jag1, and Jag2) and target genes (hairy and enhancer of split [Hes]) were quantified by quantitative real-time PCR (qRT-PCR) as previously described (31).
Zebrafish heart RNA isolation for Notch ligands, receptor, and target gene expression.
Zebrafish embryos were humanely sacrificed by overdosing with tricaine methylate. The embryonic hearts were isolated under a dissecting microscope as previously described (72). Total RNA was isolated from the extracted hearts using Aurum Total RNA Mini Kit (Bio-Rad), and cDNA was synthesized using iScript cDNA Synthesis Kit (Bio-Rad). PCR primers for Notch ligands (Jag1 and Jag2, Dll4), receptor (Notch1b), and signaling related genes (Nrg1 and ErbB2) were designed (Supplemental Tables 2–4). The mRNA expression levels were determined by qRT-PCR and normalized to zebrafish α-actin.
Statistics.
All the values were expressed as mean ± SD. For statistical comparisons between 2 experimental conditions, unpaired 2-tailed t test was used. P < 0.05 was considered significant. Comparisons of multiple mean values were performed by 1-way ANOVA, and statistical significance among multiple groups was determined using Tukey’s method.
Study approval.
Zebrafish were maintained in accordance with UCLA Institutional Animal Care and Use Committee (IACUC) protocols under a project license also approved by the UCLA IACUC (ARC no. 2015-055).
Author contributions
JL and PF set up the SPIM system and imaging. JL, PF, RRSP, and TKH wrote the manuscript. JL and PF performed postimaging processing. JL, PF, HX, and RRSP performed 4D beating zebrafish heart imaging and analysis. HK and RL prepared DNA clones and in vitro–transcribed RNAs. JL, HK, and KIB performed gene-expression studies. JL, KIB, and NJ performed in vitro experiments. JL, KIB, JC, and HY performed microinjections. CCJK, NCC, CMH, RL, and TKH designed, supervised, revised, and supported the study.
Supplementary Material
Acknowledgments
The authors would like to express gratitude to William Talbot from Stanford University for providing the human Nrg1 plasmid and to Deborah Yelon from UCSD for providing the wea mutants. In addition, the authors also thank David Traver at UCSD and Nathan Lawson at the University of Massachusetts Medical School (Worcester, Massachusetts, USA) for generously providing the Tg(tp1:gfp) line. This study was supported by grants NIH HL118650 (to T.K. Hsiai), HL083015 (to T.K. Hsiai), HD069305 (to N.C. Chi and T.K. Hsiai.), HL111437 (to T.K. Hsiai and N.C. Chi), HL129727 (to T.K. Hsiai), T32HL007895 (to R.R. Sevag Packard), and American Heart Association Pre-Doctoral Fellowship 15PRE21400019 (to J. Lee).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(5):1679–1690. doi:10.1172/JCI83496.
Contributor Information
Juhyun Lee, Email: juhyunlee@ucla.edu.
Peng Fei, Email: feipeng82@gmail.com.
Hanul Kang, Email: hanullkang@gmail.com.
Hao Xu, Email: hxu1@usc.edu.
Kyung In Baek, Email: Qorruddls122@gmail.com.
Nelson Jen, Email: njen@ucla.edu.
Junjie Chen, Email: roy.chen0404@gmail.com.
Hilary Yen, Email: hyen@ucla.edu.
C.-C. Jay Kuo, Email: cckuo@ee.usc.edu.
Chih-Ming Ho, Email: chihming@g.ucla.edu.
Rongsong Li, Email: RongsongLi@mednet.ucla.edu.
References
- 1.Auman HJ, Coleman H, Riley HE, Olale F, Tsai HJ, Yelon D. Functional modulation of cardiac form through regionally confined cell shape changes. PLoS Biol. 2007;5(3): doi: 10.1371/journal.pbio.0050053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hove JR, Koster RW, Forouhar AS, Acevedo-Bolton G, Fraser SE, Gharib M. Intracardiac fluid forces are an essential epigenetic factor for embryonic cardiogenesis. Nature. 2003;421(6919):172–177. doi: 10.1038/nature01282. [DOI] [PubMed] [Google Scholar]
- 3.Chen H, et al. BMP10 is essential for maintaining cardiac growth during murine cardiogenesis. Development. 2004;131(9):2219–2231. doi: 10.1242/dev.01094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hove JR, Köster RW, Forouhar AS, Acevedo-Bolton G, Fraser SE, Gharib M. Intracardiac fluid forces are an essential epigenetic factor for embryonic cardiogenesis. Nature. 2003;421(6919):172–177. doi: 10.1038/nature01282. [DOI] [PubMed] [Google Scholar]
- 5.Li J, et al. Piezo1 integration of vascular architecture with physiological force. Nature. 2014;515(7526):279–282. doi: 10.1038/nature13701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banjo T, et al. Haemodynamically dependent valvulogenesis of zebrafish heart is mediated by flow-dependent expression of miR-21. Nat Commun. 2013;4: doi: 10.1038/ncomms2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ten Dijke P, Egorova AD, Goumans MJ, Poelmann RE, Hierck BP. TGF-β signaling in endothelial-to-mesenchymal transition: the role of shear stress and primary cilia. Sci Signal. 2012;5(212): doi: 10.1126/scisignal.2002722. [DOI] [PubMed] [Google Scholar]
- 8.Santhanakrishnan A, Miller LA. Fluid dynamics of heart development. Cell Biochem Biophys. 2011;61(1):1–22. doi: 10.1007/s12013-011-9158-8. [DOI] [PubMed] [Google Scholar]
- 9.Lucitti JL, Jones EA, Huang C, Chen J, Fraser SE, Dickinson ME. Vascular remodeling of the mouse yolk sac requires hemodynamic force. Development. 2007;134(18):3317–3326. doi: 10.1242/dev.02883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peshkovsky C, Totong R, Yelon D. Dependence of cardiac trabeculation on neuregulin signaling and blood flow in zebrafish. Dev Dyn. 2011;240(2):446–456. doi: 10.1002/dvdy.22526. [DOI] [PubMed] [Google Scholar]
- 11.Zhang W, Chen H, Qu X, Chang CP, Shou W. Molecular mechanism of ventricular trabeculation/compaction and the pathogenesis of the left ventricular noncompaction cardiomyopathy (LVNC) Am J Med Genet C Semin Med Genet. 2013;163C(3):144–156. doi: 10.1002/ajmg.c.31369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nugent AW, et al. The epidemiology of childhood cardiomyopathy in Australia. N Engl J Med. 2003;348(17):1639–1646. doi: 10.1056/NEJMoa021737. [DOI] [PubMed] [Google Scholar]
- 13.Hoedemaekers YM, et al. The importance of genetic counseling, DNA diagnostics, and cardiologic family screening in left ventricular noncompaction cardiomyopathy. Circ Cardiovasc Genet. 2010;3(3):232–239. doi: 10.1161/CIRCGENETICS.109.903898. [DOI] [PubMed] [Google Scholar]
- 14.Lee J, et al. Moving domain computational fluid dynamics to interface with an embryonic model of cardiac morphogenesis. PLoS One. 2013;8(8): doi: 10.1371/journal.pone.0072924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic Zebrafish. Dev Biol. 2002;248(2):307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- 16.Vermot J, et al. Reversing blood flows act through klf2a to ensure normal valvulogenesis in the developing heart. PLoS Biol. 2009;7(11): doi: 10.1371/journal.pbio.1000246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galloway JL, Wingert RA, Thisse C, Thisse B, Zon LI. Loss of gata1 but not gata2 converts erythropoiesis to myelopoiesis in zebrafish embryos. Dev Cell. 2005;8(1):109–116. doi: 10.1016/j.devcel.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Arnaout R, et al. Zebrafish model for human long QT syndrome. Proc Natl Acad Sci U S A. 2007;104(27):11316–11321. doi: 10.1073/pnas.0702724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chi NC, et al. Genetic and physiologic dissection of the vertebrate cardiac conduction system. PLoS Biol. 2008;6(5): doi: 10.1371/journal.pbio.0060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berdougo E, Coleman H, Lee DH, Stainier DY, Yelon D. Mutation of weak atrium/atrial myosin heavy chain disrupts atrial function and influences ventricular morphogenesis in zebrafish. Development. 2003;130(24):6121–6129. doi: 10.1242/dev.00838. [DOI] [PubMed] [Google Scholar]
- 21.Hu N, Sedmera D, Yost HJ, Clark EB. Structure and function of the developing zebrafish heart. Anat Rec. 2000;260(2):148–157. doi: 10.1002/1097-0185(20001001)260:2<148::AID-AR50>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 22.Liu E, et al. [Hyperconjugation, characteristic infrared absorption of methylsulfones and crystal structures of selected aromaticsulfones] Guang Pu Xue Yu Guang Pu Fen Xi. 2000;20(1):31–39. [PubMed] [Google Scholar]
- 23.Liu J, et al. A dual role for ErbB2 signaling in cardiac trabeculation. Development. 2010;137(22):3867–3875. doi: 10.1242/dev.053736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grego-Bessa J, et al. Notch signaling is essential for ventricular chamber development. Dev Cell. 2007;12(3):415–429. doi: 10.1016/j.devcel.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parsons MJ, et al. Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mech Dev. 2009;126(10):898–912. doi: 10.1016/j.mod.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao W, et al. The zebrafish gene cloche acts upstream of a flk-1 homologue to regulate endothelial cell differentiation. Development. 1997;124(2):381–389. doi: 10.1242/dev.124.2.381. [DOI] [PubMed] [Google Scholar]
- 27.Stainier DY, Weinstein BM, Detrich HW, 3rd, Zon LI, Fishman MC. Cloche, an early acting zebrafish gene, is required by both the endothelial and hematopoietic lineages. Development. 1995;121(10):3141–3150. doi: 10.1242/dev.121.10.3141. [DOI] [PubMed] [Google Scholar]
- 28.Li R, et al. Disturbed flow induces autophagy, but impairs autophagic flux to perturb mitochondrial homeostasis. Antioxid Redox Signal. 2015;23(15):1207–1219. doi: 10.1089/ars.2014.5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jen N, et al. Atrial fibrillation pacing decreases intravascular shear stress in a New Zealand white rabbit model: implications in endothelial function. Biomech Model Mechanobiol. 2013;12(4):735–745. doi: 10.1007/s10237-012-0437-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takabe W, et al. Oscillatory shear stress induces mitochondrial superoxide production: implication of NADPH oxidase and c-Jun NH2-terminal kinase signaling. Antioxid Redox Signal. 2011;15(5):1379–1388. doi: 10.1089/ars.2010.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li R, et al. Shear stress-activated Wnt-angiopoietin-2 signaling recapitulates vascular repair in zebrafish embryos. Arterioscler Thromb Vasc Biol. 2014;34(10):2268–2275. doi: 10.1161/ATVBAHA.114.303345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geling A, Steiner H, Willem M, Bally-Cuif L, Haass C. A gamma-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO Rep. 2002;3(7):688–694. doi: 10.1093/embo-reports/kvf124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mumm JS, Kopan R. Notch signaling: from the outside in. Dev Biol. 2000;228(2):151–165. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- 34.Paffett-Lugassy N, et al. Functional conservation of erythropoietin signaling in zebrafish. Blood. 2007;110(7):2718–2726. doi: 10.1182/blood-2006-04-016535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Luca E, et al. ZebraBeat: a flexible platform for the analysis of the cardiac rate in zebrafish embryos. Sci Rep. 2014;4: [Google Scholar]
- 36.Zhao L, et al. Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration. Proc Natl Acad Sci U S A. 2014;111(4):1403–1408. doi: 10.1073/pnas.1311705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sedmera D, Pexieder T, Vuillemin M, Thompson RP, Anderson RH. Developmental patterning of the myocardium. Anat Rec. 2000;258(4):319–337. doi: 10.1002/(SICI)1097-0185(20000401)258:4<319::AID-AR1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 38.Chen H, Zhang W, Li D, Cordes TM, Mark Payne R, Shou W. Analysis of ventricular hypertrabeculation and noncompaction using genetically engineered mouse models. Pediatr Cardiol. 2009;30(5):626–634. doi: 10.1007/s00246-009-9406-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ben-Shachar G, Arcilla RA, Lucas RV, Manasek FJ. Ventricular trabeculations in the chick embryo heart and their contribution to ventricular and muscular septal development. Circ Res. 1985;57(5):759–766. doi: 10.1161/01.RES.57.5.759. [DOI] [PubMed] [Google Scholar]
- 40.Ramasamy SK, Kusumbe AP, Wang L, Adams RH. Endothelial Notch activity promotes angiogenesis and osteogenesis in bone. Nature. 2014;507(7492):376–380. doi: 10.1038/nature13146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hofmann JJ, Iruela-Arispe ML. Notch signaling in blood vessels: who is talking to whom about what? Circ Res. 2007;100(11):1556–1568. doi: 10.1161/01.RES.0000266408.42939.e4. [DOI] [PubMed] [Google Scholar]
- 42.Brutsaert DL. Cardiac endothelial-myocardial signaling: its role in cardiac growth, contractile performance, and rhythmicity. Physiol Rev. 2003;83(1):59–115. doi: 10.1152/physrev.00017.2002. [DOI] [PubMed] [Google Scholar]
- 43.Risebro CA, Riley PR. Formation of the ventricles. ScientificWorldJournal. 2006;6:1862–1880. doi: 10.1100/tsw.2006.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tian X, et al. Vessel formation. Science. 2014;345(6192):90–94. doi: 10.1126/science.1251487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EH. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305(5686):1007–1009. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- 46.Engelbrecht CJ, Stelzer EH. Resolution enhancement in a light-sheet-based microscope (SPIM) Opt Lett. 2006;31(10):1477–1479. doi: 10.1364/OL.31.001477. [DOI] [PubMed] [Google Scholar]
- 47.Verveer PJ, Swoger J, Pampaloni F, Greger K, Marcello M, Stelzer EH. High-resolution three-dimensional imaging of large specimens with light sheet-based microscopy. Nat Methods. 2007;4(4):311–313. doi: 10.1038/nmeth1017. [DOI] [PubMed] [Google Scholar]
- 48.Dodt HU, et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat Methods. 2007;4(4):331–336. doi: 10.1038/nmeth1036. [DOI] [PubMed] [Google Scholar]
- 49.Chung K, Deisseroth K. CLARITY for mapping the nervous system. Nat Methods. 2013;10(6):508–513. doi: 10.1038/nmeth.2481. [DOI] [PubMed] [Google Scholar]
- 50.Yang B, et al. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell. 2014;158(4):945–958. doi: 10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rumyantsev PP, Krylova MI. Ultrastructure of myofibers and cells synthesizing DNA in the developing and regenerating lymph-heart muscles. Int Rev Cytol. 1990;120:1–52. doi: 10.1016/s0074-7696(08)61598-3. [DOI] [PubMed] [Google Scholar]
- 52.Shergill B, Meloty-Kapella L, Musse AA, Weinmaster G, Botvinick E. Optical tweezers studies on Notch: single-molecule interaction strength is independent of ligand endocytosis. Dev Cell. 2012;22(6):1313–1320. doi: 10.1016/j.devcel.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gordon WR, et al. Mechanical allostery: evidence for a force requirement in the proteolytic activation of Notch. Dev Cell. 2015;33(6):729–736. doi: 10.1016/j.devcel.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Masumura T, Yamamoto K, Shimizu N, Obi S, Ando J. Shear stress increases expression of the arterial endothelial marker ephrinB2 in murine ES cells via the VEGF-Notch signaling pathways. Arterioscler Thromb Vasc Biol. 2009;29(12):2125–2131. doi: 10.1161/ATVBAHA.109.193185. [DOI] [PubMed] [Google Scholar]
- 55.de la Pompa JL, Epstein JA. Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev Cell. 2012;22(2):244–254. doi: 10.1016/j.devcel.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Timmerman LA, et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004;18(1):99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.High FA, Epstein JA. The multifaceted role of Notch in cardiac development and disease. Nat Rev Genet. 2008;9(1):49–61. doi: 10.1038/nrg2279. [DOI] [PubMed] [Google Scholar]
- 58.Ge W, Ren J. mTOR-STAT3-notch signalling contributes to ALDH2-induced protection against cardiac contractile dysfunction and autophagy under alcoholism. J Cell Mol Med. 2012;16(3):616–626. doi: 10.1111/j.1582-4934.2011.01347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hedhli N, et al. Endothelium-derived neuregulin protects the heart against ischemic injury. Circulation. 2011;123(20):2254–2262. doi: 10.1161/CIRCULATIONAHA.110.991125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fang SJ, et al. Neuregulin-1 preconditioning protects the heart against ischemia/reperfusion injury through a PI3K/Akt-dependent mechanism. Chin Med J (Engl) 2010;123(24):3597–3604. [PubMed] [Google Scholar]
- 61.Liebling M, Forouhar AS, Gharib M, Fraser SE, Dickinson ME. Four-dimensional cardiac imaging in living embryos via postacquisition synchronization of nongated slice sequences. J Biomed Opt. 2005;10(5): doi: 10.1117/1.2061567. [DOI] [PubMed] [Google Scholar]
- 62.Mickoleit M, et al. High-resolution reconstruction of the beating zebrafish heart. Nat Methods. 2014;11(9):919–922. doi: 10.1038/nmeth.3037. [DOI] [PubMed] [Google Scholar]
- 63.Westerfield M. The Zebrafish Book: A Guide For The Laboratory Use Of Zebrafish (Danio Rerio) Corvallis, Oregon, USA: University of Oregon Press; 2000. [Google Scholar]
- 64.Choi WY, et al. In vivo monitoring of cardiomyocyte proliferation to identify chemical modifiers of heart regeneration. Development. 2013;140(3):660–666. doi: 10.1242/dev.088526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fung Y, Liu S. Elementary mechanics of the endothelium of blood vessels. J Biomech Eng. 1993;115(1):1–12. doi: 10.1115/1.2895465. [DOI] [PubMed] [Google Scholar]
- 66.Fung YC. Biomechanics: Motion, Flow, Stress, and Growth. New York, New York, USA: Springer; 1990. [Google Scholar]
- 67.Kay JM, Nedderman RM. Fluid Mechanics And Transfer Processes. Cambridge, United Kingdom: Cambridge University Press; 1985. [Google Scholar]
- 68.Lee J, Packard RR, Hsiai TK. Blood flow modulation of vascular dynamics. Curr Opin Lipidol. 2015;26(5):376–383. doi: 10.1097/MOL.0000000000000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mayet J, et al. Improvement in midwall myocardial shortening with regression of left ventricular hypertrophy. Hypertension. 2000;36(5):755–759. doi: 10.1161/01.HYP.36.5.755. [DOI] [PubMed] [Google Scholar]
- 70.Hwang J, et al. Pulsatile versus oscillatory shear stress regulates NADPH oxidase subunit expression: implication for native LDL oxidation. Circ Res. 2003;93(12):1225–1232. doi: 10.1161/01.RES.0000104087.29395.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hsiai TK, et al. Monocyte recruitment to endothelial cells in response to oscillatory shear stress. FASEB J. 2003;17(12):1648–1657. doi: 10.1096/fj.02-1064com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Singleman C, Holtzman NG. Heart dissection in larval, juvenile and adult zebrafish, Danio rerio. J Vis Exp. 2011;(55): doi: 10.3791/3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.