Abstract
Background
Given its high surplus and low cost, glycerol has emerged as interesting carbon substrate for the synthesis of value-added chemicals. The soil bacterium Pseudomonas putida KT2440 can use glycerol to synthesize medium-chain-length poly(3-hydroxyalkanoates) (mcl-PHA), a class of biopolymers of industrial interest. Here, glycerol metabolism in P. putida KT2440 was studied on the level of gene expression (transcriptome) and metabolic fluxes (fluxome), using precisely adjusted chemostat cultures, growth kinetics and stoichiometry, to gain a systematic understanding of the underlying metabolic and regulatory network.
Results
Glycerol-grown P. putida KT2440 has a maintenance energy requirement [0.039 (mmolglycerol (gCDW h)−1)] that is about sixteen times lower than that of other bacteria, such as Escherichia coli, which provides a great advantage to use this substrate commercially. The shift from carbon (glycerol) to nitrogen (ammonium) limitation drives the modulation of specific genes involved in glycerol metabolism, transport electron chain, sensors to assess the energy level of the cell, and PHA synthesis, as well as changes in flux distribution to increase the precursor availability for PHA synthesis (Entner–Doudoroff pathway and pyruvate metabolism) and to reduce respiration (glyoxylate shunt). Under PHA-producing conditions (N-limitation), a higher PHA yield was achieved at low dilution rate (29.7 wt% of CDW) as compared to a high rate (12.8 wt% of CDW). By-product formation (succinate, malate) was specifically modulated under these regimes. On top of experimental data, elementary flux mode analysis revealed the metabolic potential of P. putida KT2440 to synthesize PHA and identified metabolic engineering targets towards improved production performance on glycerol.
Conclusion
This study revealed the complex interplay of gene expression levels and metabolic fluxes under PHA- and non-PHA producing conditions using the attractive raw material glycerol as carbon substrate. This knowledge will form the basis for the development of future metabolically engineered hyper-PHA-producing strains derived from the versatile bacterium P. putida KT2440.
Keywords: Pseudomonas putida KT2440, Glycerol metabolism, Transcriptome, Metabolic flux analysis, Flux balance analysis, Elementary flux modes, Polyhydroxyalkanoates, Nitrogen and carbon limitation
Background
Pseudomonas putida is well known for its capacity to use a wide range of carbon sources, including aromatic compounds, sugars, fatty acids and polyols [1]. The broad substrate spectrum elevates the survival rate of bacteria belonging to the genus Pseudomonas in comparison to other microbes, when adverse environmental conditions are present [2, 3]. In addition, P. putida can cope with fluctuations in nutrient availability through the accumulation of polyesters [4, 5], formed as inclusion bodies in the cytoplasm of the cell [6]. During phases of famine, degradation of these polyesters fuels the cellular demand for building blocks, redox power and energy [7]. Most P. putida species can synthesize a wide range of poly(3-hydroxyalkanoates) (PHAs), whereby the monomer composition of the polymer varies with carbon source and other environmental factors [8, 9]. In the past decades, PHAs have attracted considerable attention as sustainable biodegradable materials to replace oil-based polymers, especially because of their mechanical and physical properties, which are similar to conventional plastics. Meanwhile, PHAs are used at industrial scale for bags, containers, and medical devices, among others [10]. PHAs also have potential as drug carriers [11, 12]. Much effort has been poured into the development of new PHAs with tailored monomer composition [13, 14], efficient metabolically engineered strains, and fermentation processes [15, 16]. The latter aims to enhance PHA productivity and to reduce production costs, important pre-requisites for industrial production and further commercialization of PHAs. The use of industrial waste as feedstock for the synthesis of PHAs has opened a new avenue for more sustainable and cheaper microbial fermentation processes [17]. High titers of PHA have been accomplished using animal wastes [18], polyethylene terephthalate (PET) [19], and, particularly, raw glycerol from the biodiesel industry [20, 21]. Recently, we have shown that P. putida KT2440 is most suitable to synthesize PHA from raw glycerol among different P. putida strains due to reduced by-product formation under PHA-producing conditions [21]. Further studies have explored molecular details of glycerol metabolism in P. putida KT2440. This led to the discovery of specific regulatory genes [22, 23]. When grown in batch culture on glycerol, the gene glpR (PP_1074) controls utilization of the substrate. Interestingly, inactivation of this regulator leads to increased synthesis of mcl-PHA in P. putida KT2440 [24]. In addition, transcriptome analyses of glycerol-grown P. putida KT2440 indicate a mixed glycolytic and gluconeogenic pathway use, a rather complex metabolic adjustment, which strongly differs from that of cells, growing on glucose and succinate, respectively [22]. To better understand the mechanisms in P. putida for the production of biopolymers from glycerol, it now appears straightforward to further quantify and integrate metabolic function and regulation as well as physiological parameters in a systematic fashion. Such systems biological approaches have proven valuable to understand cellular physiology [25–27] and enable metabolic engineering approaches, with the purpose of enhancing the synthesis of target chemicals in a rational manner [28–30]. As basis for such analyses in P. putida KT2440, several genome-scale models have been developed from genome-annotation [31–33], unraveling the metabolic capacity of this versatile bacterium. In the present work, we explored the effect of specific growth regimes on global gene expression and carbon flux distribution of pathways of P. putida KT2440 in order to provide an integrated insight into its metabolic and regulatory networks during growth and PHA production on glycerol. A set of fine adjusted chemostat cultures under carbon- and nitrogen-limited conditions, and the integration of experimental and modelling data provided the basis for this systematic analysis.
Results
Growth physiology of P. putida under carbon limitation
The maintenance energy demand is of importance from both a biological and biotechnological point of view [34]. Bacterial population dynamics depend on the capacity of each member of the community to adapt to fluctuations of nutrient availability and the intrinsic metabolic energy demand for cellular functioning [35]. For biotechnological applications, microbes with low maintenance energy requirement can direct more resources for biosynthetic purposes [36], which positively impacts production performance. This, of course, also holds for P. putida. In this regard, the so far unknown maintenance coefficient for P. putida KT2440 on glycerol was quantified. For this purpose, we carefully inspected growth kinetics and stoichiometry of the bacterium, using glycerol-limited chemostat cultures at different dilution rates between 0.044 and 0.21 h−1. At a higher dilution rate of 0.22 h−1, washout of cells from the system occurred, indicating that this value approached the maximum specific growth rate of the strain on glycerol. In addition, accumulation of glycerol in the broth was observed at these elevated dilution rate. From the obtained data, specific rates for glycerol uptake (qglycerol) and biomass yield (YX/glycerol) were calculated (Table 1).
Table 1.
Biomass concentration, glucose uptake rate, observed yield coefficient, and carbon recovery during glycerol-limited continuous cultures of P. putida KT2440 for various dilution rates
| D (h−1) | Biomass (g L−1) | qglycerol [g (gCDW h)−1] | Yieldobserved (g g−1) | Carbon recovery (%) |
|---|---|---|---|---|
| 0.044 | 1.91 | 0.095 | 0.47 | 101.9 |
| 0.066 | 1.93 | 0.137 | 0.48 | 97.6 |
| 0.088 | 2.03 | 0.168 | 0.50 | 101.1 |
| 0.109 | 2.08 | 0.215 | 0.51 | 96.9 |
| 0.120 | 2.08 | 0.237 | 0.51 | 102.4 |
| 0.141 | 2.01 | 0.288 | 0.50 | 101.3 |
| 0.190 | 2.10 | 0.390 | 0.52 | 100.8 |
| 0.210 | 2.11 | 0.410 | 0.53 | 97.4 |
According to the Pirt model (Eq. 1), the maintenance coefficient (mglycerol) can be inferred from the correlation between the specific consumption rate of glycerol (qglycerol) and the corresponding dilution rate, i.e. the specific growth rate (μ) in the chemostat [37]. In addition, the relationship yields the true yield coefficient (Ytruex/glycerol).
| 1 |
As shown in Fig. 1, the specific glycerol consumption rate increased proportional with the dilution rate. Using linear regression analysis, the two coefficients were determined: mglycerol = 0.039 (mmolglycerol) (gCDW)−1 h−1 and Ytruex/glycerol = 0.505 gCDW (gglycerol)−1, respectively. Subsequently, the energy requirement for maintenance (mATP) was derived (see Appendix) as: . The consideration of a phosphate:oxygen (P/O) ratio of 1.75 [38], reflecting the amount of ATP formed per reduced oxygen atom during oxidative phosphorylation yielded 0.175 (mmol ATP) (gCDW)−1 h−1, the ATP amount required for maintenance in glycerol-grown P. putida KT2440. Additionally, the amount of accumulated PHA and of by-products was quantified for each dilution rate. Organic acids were not detected by HPLC. The cellular PHA content was less than 3 % of the cell dry weight (CDW) at all dilution rates, which matches with natural amounts during growth on non-PHA-synthesis-related substrates [23] (Table 2). Even though there is scientific evidence that PHA can be accumulated under carbon limitation in P. putida strains [9, 39], this seems only the case, when fatty acids or related substrates are used as carbon substrates, where PHA precursors are inevitably generated via the β-oxidation pathway. As no by-products were detected under these carbon limiting conditions, consumed glycerol seemed to be purely metabolized into CO2 and biomass. As consistency check, the carbon recovery was determined from the experimental data, involving thorough balancing of carbon consumption and production, respectively. The C-mol content in the biomass was assumed to be constant, corresponding to a molecular weight of 27 g (C-mol biomass)−1 [40]. Within measurement accuracy, the carbon recovery was complete (96.9–102.4 %), which underlines the consistency of the data set (Table 1).
Fig. 1.
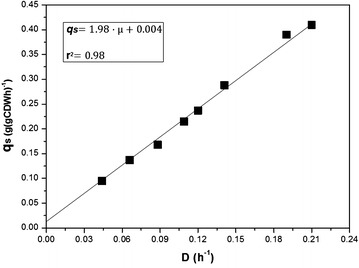
Determination of the maintenance coefficient and the true yield of glycerol-grown P. putida from selected chemostat experiments
Table 2.
Growth kinetics and stoichiometry of P. putida KT2440 under different growth-limiting conditions
| Limitation | D (h−1) | Residual ammonium (mg L−1) | Residual glycerol (g L−1) | YX/S (g g−1) | PHA (wt%) | YPHA/S (g g−1) | Uptake and production rates (mmol gCDW−1 h−1) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Glycerol | Malate | Succinate | CO2 | |||||||
| Glycerol | 0.044 | 68.2 ± 4.8 | ND < 0.1 | 0.47 | 2.5 | – | 1.02 ± 0.10 | ND < 0.1 | ND < 0.1 | 1.5 ± 0.1 |
| Ammonium | 0.044 | ND < 0.1 | 19.9 ± 0.9 | 0.19 | 29.7 | 0.08 | 2.47 ± 0.15 | 0.04 ± 0.01 | 0.24 ± 0.02 | 3.6 ± 0.1 |
| Glycerol | 0.120 | 35.1 ± 2.7 | ND < 0.1 | 0.51 | 2.7 | – | 2.60 ± 0.06 | ND < 0.1 | ND < 0.1 | 3.2 ± 0.1 |
| Ammonium | 0.120 | ND < 0.1 | 25.7 ± 0.7 | 0.34 | 12.8 | 0.05 | 3.71 ± 0.03 | 0.21 ± 0.03 | ND < 0.1 | 4.2 ± 0.1 |
Standard deviation (±) from at least two independent experiments
Yx/s was calculated based on the biomass, excluding PHA
ND not detected in the culture broth
Growth physiology under nitrogen limitation
Next, the PHA production capacity of P. putida on glycerol was evaluated. In P. putida, PHA synthesis is driven by nitrogen limitation [9, 41]. To assess the PHA production capacity on glycerol, chemostat cultures were conducted under nitrogen limitation with glycerol as sole carbon and energy source. In order to cover different growth regimes, a low (0.044 h−1) and a high (0.12 h−1) dilution rate was adjusted, which corresponded to approximately 20 and 50 % of the maximum specific growth rate, respectively. Table 2 shows the physiological parameters, obtained under the two conditions. The biomass yield was higher at high dilution rate in comparison to the one observed at low dilution rate. The opposite result was observed for the PHA content. Here, the PHA yield reached 29.7 %wt/wt of the CDW at low dilution rate and only 12.8 %wt/wt of the CDW at high dilution rate (Table 2). The monomer composition of PHA remained rather constant (Table 3). Two organic acids, i.e. succinate and malate, were produced, when the chemostat was operated at a low dilution rate. This was not the case, when we analyzed the culture at high dilution rate, where malate was the only by-product (Table 2).
Table 3.
Monomer composition of medium chain length PHA produced by P. putida KT2440 under nitrogen limitation
| Nitrogen limitation | Monomer composition (%) | |||||
|---|---|---|---|---|---|---|
| C6 | C8 | C10 | C12:1 | C12 | C14:0 | |
| High dilution rate | <0.1 | 17.2 | 74.3 | 3.1 | 5.3 | <0.1 |
| Low dilution rate | <0.1 | 18.1 | 75.5 | 1.8 | 4.5 | 0.8 |
The data were determined by GC/MS and are given as relative molar fraction (%) of C6: 3-hydroxyexanoate, C8: 3-hydroxyoctanoate, C10:3-hydroxydecanoate, C12: 3-hydroxydodecanoate, C12:1: 3-hydroxy-5-cis-dodecanoate, and C14: 3-hydroxytetradecanoate
Transcriptome analysis of P. putida KT2440 under carbon and nitrogen limiting conditions
To gain a more detailed insight into regulation patterns under different nutrient environments, four scenarios were selected for global gene expression analysis: N-limited growth at high (0.12 h−1) and low (0.044 h−1) dilution rate, and C-limited growth at high (0.12 h−1) and low dilution (0.044 h−1) rate, respectively (Table 2). This setup was designed to allow the extraction of changes at the transcript level originating (i) from the imposed specific growth rate and/or (ii) limitation of a specific nutrient. In Table 4, differentially expressed genes (log2 > 1 classified as up-regulated and log2 < −1 classified as down-regulated, p value <0.05) are listed. Obviously, a larger number of genes was affected by the imposed nutrient limitation than by the growth rate (Table 4). Genes, belonging to pathways involved in energy production and conversion, carbohydrate transport, synthesis of amino acids, nitrogen scavenging, PHA synthesis, cellular processing, and transcriptional regulation were most affected by the type of limitation (N-limiting vs. C-limiting conditions) (Table 4). On the other hand, several genes encoding proteins related to energy metabolism, transporters DNA and RNA repair and synthesis and also putative functions were differentially expressed, when P. putida KT2440 was challenged to different specific growth rates on glycerol. Remarkably, genes encoding signal modulators were highly overexpressed, when shifting from carbon to nitrogen limitation, independently of the set growth rate. A sensor hybrid histidine kinase PAS/PAC (PP_2664) and an integral membrane sensor (PP_2671) showed the highest change in gene expression level, which was more than 30-fold. PAS domains are important elements that sense both, fluctuation of signals in the environment and the overall energy level of the cell [42]. In addition, the LuxR (PP_2672) master gene regulator displayed a high expression level, as also did genes, allocated downstream of PAS/PAC (PP_2065-2069). Up-regulation of PAS and LuxR genes is likely related to the energetic state of the cell, as N-limiting cultures promote not only accumulation of PHAs, but also interactions between PAS domains and electron transport systems [42]. High yields of PHA diminish intracellular level of ATP [5] and to increase the NADH/NAD+ ratio [15, 43]. Probably to meet the ATP requirement under PHA-producing conditions, P. putida overexpressed genes, encoding for proteins of the cytochrome c-type (PP_2675, PP_43233, and PP_4324). These are essential enzymes in the electron transport chain and define the final redox state of the cell.
Table 4.
Genes differentially expressed under various nutrient limitations
| Metabolic function | CH vs. CL* | NH vs. NL | NH vs. CH | NL vs. CL | ||||
|---|---|---|---|---|---|---|---|---|
| Up | Down | Up | Down | Up | Down | Up | Down | |
| Translation, ribosomal structure and biogenesis | 3 | 1 | 2 | 0 | 0 | 1 | 0 | 3 |
| Transcription | 1 | 2 | 0 | 0 | 4 | 1 | 8 | 3 |
| Replication, recombination and repair | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
| Energy production and conversion | 2 | 3 | 2 | 2 | 14 | 6 | 12 | 8 |
| Amino acid transport and metabolism | 12 | 3 | 7 | 1 | 17 | 5 | 7 | 1 |
| Nucleotide transport and metabolism | 1 | 0 | 0 | 0 | 3 | 3 | 5 | 0 |
| Carbohydrate transport and metabolism | 2 | 2 | 1 | 0 | 9 | 6 | 11 | 8 |
| Coenzyme transport and metabolism | 5 | 1 | 3 | 2 | 3 | 2 | 9 | 5 |
| Lipid transport and metabolism | 1 | 1 | 1 | 0 | 2 | 2 | 1 | 2 |
| Inorganic ion transport and metabolism | 4 | 3 | 1 | 2 | 7 | 1 | 11 | 5 |
| Secondary metabolites biosynthesis, transport, and catabolism | 2 | 4 | 2 | 1 | 2 | 0 | 3 | 9 |
| Cellular processing and signaling | 6 | 14 | 3 | 1 | 32 | 9 | 38 | 6 |
| Poorly Characterized | 9 | 13 | 4 | 4 | 27 | 9 | 36 | 20 |
| Total | 48 | 47 | 26 | 13 | 121 | 45 | 142 | 70 |
*Carbon-limitation at high (CH) and low (CL) dilution rate, and nitrogen-limitation at high (NH) and low (NL) dilution rate are compared to extract gene modulation due to limitation (NH vs. CH and NL vs. CL) and growth rate (CH vs. CL and NH vs. NL) changes
Concerning central carbon metabolism (Table 5), increased growth rate specifically caused expression changes of genes encoding for enzymes around the phosphoenolpyruvate and pyruvate node (pgm, eno, acoA and pykA) and in the pentose phosphate (PP) pathway (gnd, pgl and rpiA), whereas the type of nutrient limitation rather impacted the expression of genes of the Embden-Meyerhof–Parnas (EMP) pathway (pgi, fda and gap-1), the Entner–Doudoroff (ED) pathway (eda) and the tricarboxylic acid (TCA) cycle (sucA, sucC, sdhA and mdh), respectively.
Table 5.
Expression profile of genes belonging to PHA biosynthesis and central metabolic pathways in P. putida KT2440 under different conditions
| Gene name | Locus tag | Description | Log2 change | |||
|---|---|---|---|---|---|---|
| CH vs. CL | NH vs. NL | NL vs. CL | NH vs. CH | |||
| PHA synthesis | ||||||
| phaI | PP5008 | PHA granule-associated | −0.69 | −1.10 | 1.41 | 1.00 |
| phaF | PP5007 | PHA granule-associated | −0.49 | −0.89 | 1.11 | 0.72 |
| phaC1 | PP5003 | PHA polymerase | −0.11 | 0 | −0.25 | −0.15 |
| phaC2 | PP5005 | PHA polymerase | −0.27 | −0.29 | 0.48 | 0.46 |
| phaZ | PP5004 | PHA depolymerase | 0.30 | −0.18 | 0.87 | 0.39 |
| phaD | PP5006 | Transcriptional regulator | 0.19 | −0.24 | 0.50 | 0.07 |
| phaG | PP1408 | Acyl-transferase | −1.06 | −1.51 | 2.93 | 2.48 |
| Glycerol metabolism | ||||||
| oprB | PP1019 | Porin B transporter | 0.76 | 0.37 | −0.39 | −0.78 |
| glpF | PP1076 | MIP family channel protein | 0.15 | 0.78 | −1.28 | −0.65 |
| glpK | PP1075 | Glycerol kinase | 0.35 | 0.19 | −0.72 | −0.88 |
| glpR | PP1074 | Transcriptional regulator | 0.81 | 0.40 | −0.97 | −1.39 |
| glpD | PP1073 | Glycerol-3-P dehydrogenase | −0.01 | −0.48 | 0.05 | −0.42 |
| Embden–Meyerhof–Parnas pathway | ||||||
| Glk | PP1011 | Glucokinase | 0.76 | 0.25 | −0.13 | −0.65 |
| gltR | PP1012 | Transcriptional regulator | 0.78 | 0.43 | −0.7 | −1.05 |
| PP1013 | Integral membrane sensor | 0.7 | 0.4 | −1.00 | −1.3 | |
| Pgi | PP1808 | Glucose-6-phosphate isomerase | 0.13 | 0.11 | −0.10 | 0.13 |
| Fbp | PP5040 | Fructose-1,6-bisphosphatase | 0.21 | −0.12 | −0.10 | 0.11 |
| Fda | PP4960 | Fructose-1,6-bisphosphate aldolase | −0.13 | 0.24 | −0.10 | 0.5 |
| tpiA | PP4715 | Triosephosphate isomerase | 0.82 | 0.47 | −0.10 | −0.93 |
| gap1 | PP1009 | GAP dehydrogenase, type I | 0.30 | 0.69 | −1.32 | −0.92 |
| gap2 | PP2149 | GAP dehydrogenase, type II | 0.14 | −0.06 | −0.10 | 0.42 |
| pgk | PP4963 | Phosphoglycerate kinase | 0.08 | 0.37 | −0.10 | 0.46 |
| pgm | PP5056 | Phosphoglyceromutase | 0.25 | 0.24 | −0.1 | 0.04 |
| eno | PP1612 | Phosphopyruvate hydratase | 0.18 | 0.46 | −0.1 | 0.07 |
| pyk | PP1362 | Pyruvate kinase | 0.11 | 0.30 | −0.1 | 0.85 |
| Pentose phosphate pathways | ||||||
| zwf1 | PP1022 | G6P dehydrogenase | 1.33 | 0.39 | 0.85 | −0.09 |
| zwf2 | PP4042 | 0.48 | 0.14 | −0.10 | −0.48 | |
| zwf3 | PP5351 | −0.23 | −0.05 | −0.10 | 0.21 | |
| pgl | PP1023 | 6-P-gluconate dehydrogenase | 0.65 | 0.42 | 1.15 | 0.77 |
| gnd | PP4043 | 6-P-gluconate dehydrogenase | 0.08 | −0.15 | −0.1 | −0.23 |
| gnuK | PP3416 | Carbohydrate kinase | −0.06 | −0.52 | −0.10 | −0.7 |
| kguK | PP3378 | Dehydroglucokinase | −0.02 | −0.63 | −0.10 | −0.21 |
| kguD | PP3376 | 2-Ketogluconate 6-phosphate reductase | −0.15 | −0.17 | −0.10 | −0.2 |
| rpiA | PP5150 | Ribose-5-phosphate isomerase A | −0.09 | 0.06 | −0.10 | 0.06 |
| rpe | PP0415 | Ribulose-phosphate 3-epimerase | −0.04 | 0.20 | −0.74 | −0.5 |
| tktA | PP4965 | Transketolase | 0.28 | 0.29 | −0.10 | 0.46 |
| tal | PP2168 | Transaldolase B | −0.6 | −0.07 | −0.10 | 0.70 |
| Entner-Doudoroff pathway | ||||||
| edd | PP1010 | 6-Phosphogluconate dehydratase | 0.9 | 0.22 | −0.03 | −0.71 |
| eda | PP1024 | KDPG aldolase | 1.24 | 0.29 | 1.65 | 0.98 |
| Pyruvate metabolism | ||||||
| acoA | PP0555 | Pyruvate dehydrogenase | 1.16 | 1.86 | −1.03 | −0.33 |
| acoB | PP0554 | Pyruvate dehydrogenase | 1.5 | 1.88 | −1.04 | −0.66 |
| acoC | PP0553 | Pyruvate dehydrogenase | 1.57 | 1.81 | −0.96 | −0.72 |
| PP0545 | Aldehyde dehydrogenase | 0.2 | 0.11 | −0.91 | −1.01 | |
| acsA | PP4487 | Acetyl-CoA synthetase | −0.13 | 0.23 | −0.15 | 3.56 |
| accC-2 | PP5347 | Pyruvate carboxylase | 0.22 | −0.27 | −0.12 | 0.07 |
| ppsA | PP2082 | Phosphoenolpyruvate synthase | 0.10 | 0.22 | −0.12 | 0.09 |
| ppc | PP1505 | Phosphoenolpyruvate carboxylase | 0.09 | −0.06 | −0.10 | 0.50 |
| PP0154 | Acetyl-CoA hydrolase | 0.21 | 0.23 | 1.42 | 1.41 | |
| TCA cycle | ||||||
| gltA | PP4194 | Citrate synthase | 0.01 | 0.57 | −0.10 | 1.12 |
| acnA | PP2112 | Aconitate hydratase | −0.04 | −0.22 | −0.10 | −0.57 |
| acnB | PP2339 | Aconitate hydratase | −0.33 | 0.2 | −0.10 | 0.74 |
| icd | PP4012 | Isocitrate dehydrogenase | 0.38 | −0.14 | −0.10 | −1.97 |
| sucA | PP4189 | 2-Oxoglutarate dehydrogenase | 0.37 | 0.08 | −0.10 | −0.17 |
| sucD | PP4185 | Succinyl-CoA synthetase sub alpha | 0.6 | 0.35 | −0.10 | −0.13 |
| sucC | PP4186 | Succinyl-CoA synthetase sub beta | 0.47 | 0.35 | −0.10 | 0.14 |
| sdhA | PP4191 | Succinate dehydrogenase | 0.24 | 0.30 | −0.10 | −0.04 |
| fumC | PP0944 | Fumarate hydratase | −2.46 | −0.63 | −1.92 | −0.08 |
| mdh | PP0654 | Malate dehydrogenase | −0.79 | −0.52 | −0.42 | −0.15 |
| Glyoxylate shunt | ||||||
| aceA | PP4116 | Isocitrate lyase | −0.78 | 0.03 | 3.10 | 3.91 |
| glcB | PP0356 | Malate synthase | 0.19 | 0.32 | 0.69 | 0.82 |
Carbon-limitation at high (CH) and low (CL) dilution rate, nitrogen-limitation at high (NH) and low (NL) dilution rate
Boldface represents a differentiated expression pattern
p value <0.05
The metabolic flux profile of glycerol-grown P. putida is affected by the cellular environment
Beyond the demonstrated influence of specific growth rate and nutrient limitation on the regulatory network of P. putida we next performed flux balance analysis to additionally evaluate the performance of the underlying metabolic network for the different scenarios. Metabolic fluxes of central carbon metabolism were inferred by flux balance analysis, based on a detailed metabolic network of P. putida (see Appendix), which was constrained by experimental uptake and production rates to reveal the investigated physiological states (Table 2). Hereby, the relative weight of individual pathways for biomass and PHA synthesis was deduced from the obtained fluxes. Figure 2 shows the distribution of the calculated fluxes for each condition.
Fig. 2.
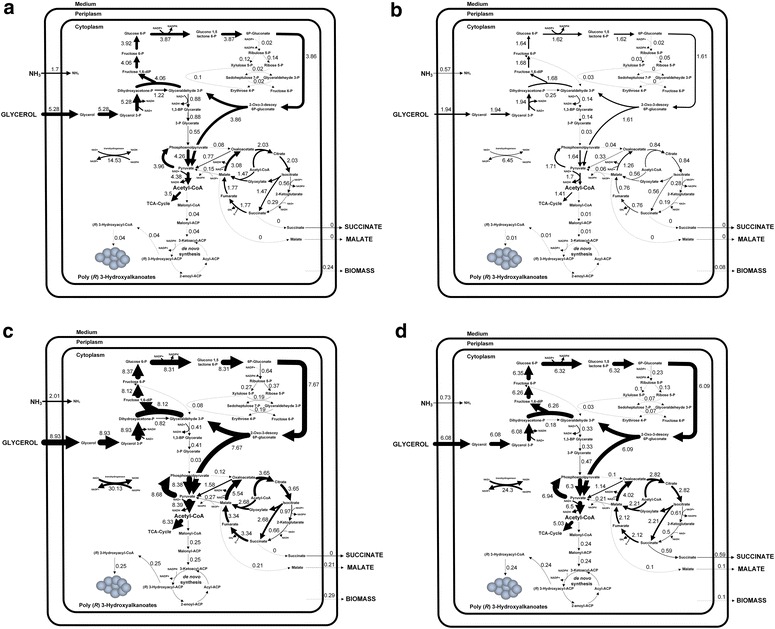
Predicted intracellular fluxes for maximized growth from flux balance analysis under nitrogen and carbon-limiting conditions, both for a high dilution rate (0.12 h−1) and a low dilution rate (0.044 h−1). Intracellular fluxes in P. putida under carbon limitation with a high (a) and a low (b) dilution rate and under nitrogen limitation with a high (c) and a low (d) dilution rate. All molecular fluxes are given next to the respective arrow and are given in mmol L−1 h−1, except for the biomass formation, which is given in mmol g−1CDW h−1. The width of each arrow also represents the size of the corresponding flux, except for the width of the arrow belonging to the transhydrogenase reaction, which, for visualization purposes, correlates to one tenth of the corresponding flux size
As predicted by the modeling approach, P. putida preferentially uses the oxidative PP and the ED pathway under nitrogen limiting conditions. The corresponding flux values were 2.1- and 3.9-fold higher than that under carbon-limitation at high and low dilution rate, respectively. Due to the fact that glycerol uptake only increased by a factor of 1.7 and 3.1, respectively, not only absolutely, but also relatively more carbon was channeled through the ED pathway. Additionally, the TCA cycle was 1.8 and 3.3 times more active at high and low dilution rate, respectively, whereas the carbon partitioning at the isocitrate node, directed relatively more carbon through the glyoxylate shunt (73 and 78 % vs. 72 and 67 %, respectively). No by-product formation was detected under carbon-limitation. In contrast, a minor flux of carbon was funneled to malate and succinate synthesis from the TCA cycle under nitrogen-limiting conditions. These apparent differences in carbon partitioning were associated with a 7- and 20-fold increased PHA biosynthetic flux under nitrogen-limitation at high and low dilution rate, respectively. Concerning redox equivalents, more NADPH was needed to satisfy the increased demand under nitrogen limitation, related to the enhanced PHA synthesis.
When comparing the different growth rates, approximately the same level of PHA synthesis was achieved for both conditions. Apart from a slightly increased ED pathway (2.4- and 1.3-fold under carbon and nitrogen-limitation, respectively) and TCA cycle (2.4- and 1.3-fold under carbon and nitrogen-limitation, respectively) at high dilution rate, no apparent differences were observed in carbon partitioning. The carbon flux through glyoxylate shunt and PHA synthesis remained almost unchanged.
Potential for strain improvement of PHA-producing P. putida grown on glycerol
To evolve into an economically viable candidate for industrial PHA production, P. putida’s productivity needs to be maximized. A first inspection is provided by integration of the achieved performance under the different conditions into the overall flux space (Fig. 3). Obviously, the glycerol-grown wild type accumulated significant amounts of the biopolymer. PHA synthesis was particularly efficient at low dilution rate under nitrogen limitation. Under these conditions, P. putida even approached the upper boundary of the feasible flux space. Ideally, PHA yield improvement would guide P. putida south eastbound, while staying close to the maximal efficiency border. However, even for this best scenario, P. putida spent most of the substrate for growth so that the obtained PHA yield is still far below the theoretical maximum, related to the best PHA-producing elementary flux mode (EFM). Although this extreme value at zero growth seems not reachable in practice and an achievable optimum will be realistically lower, the location of the experimental data in the flux space indicate a significant remaining optimization potential (Table 6).
Fig. 3.

Solution space of the elementary flux mode analysis of glycerol-grown P. putida. Each point represents the biomass and the PHA yield of one unique non-decomposable pathway for P. putida grown on glycerol. All values are given in C-mol per C-mol. Additionally, the red squares display the values found in our chemostat experiments, where C and N are carbon and nitrogen-limited, respectively. The dilution rate is given as the index of C and N
Table 6.
Maintenance coefficient of P. putida and other industrially relevant strains under aerobic conditions
| Strain | Carbon source | Maintenance coefficient [mmolsubstrate·(gCDW·h)−1] | References |
|---|---|---|---|
| P. putida KT2440 | Glycerol | 0.039 | This study |
| A. aerogenes | Glycerol | 0.966 | [46] |
| K. aerogenes | Glycerol | 0.804 | [46] |
| E. coli WT | Glycerol | 0.627 | [47] |
| P. putida KT2440 | Glucose | 0.062 | [78] |
| E. coli MG1655 | Glucose | 0.370 | [79] |
| C. glutamicum | Glucose | 0.08 | [52] |
| B. subtilis | Glucose | 0.45 | [80] |
| B. subtilis | Glucose | 0.39 | [51] |
| K. aerogenes | Glucose | 0.350 | [46] |
Alongside improving culture conditions [16, 21], progress was recently achieved through rational strain engineering by flux design, an elementary flux mode-based correlation analysis for the prediction metabolic engineering targets, involving the full spectrum of amplification, attenuation, deletion and heterologous insertion [44, 45]. Applied here, calculation yielded 1533 unique EFM distributions that define the solution space for P. putida on glycerol (Fig. 3). Based on the cultivation data (Table 2), only the subset of modes that produced PHA, biomass, succinic acid and malic acid was selected for target prediction (Fig. 4). From these remaining modes, (i) a positive effect on PHA synthesis was identified for the ED pathway, (ii) the oxidative PP pathway and (iii) the first three reactions of the EMP pathway, as well as (iv) PHA production from pyruvate. Contrarily, a part of the TCA cycle, the glyoxylate shunt, the reactions of lower glycolysis between dihydroxyacetone phosphate and phosphoenolpyruvate and by-product synthesis had a negative influence on PHA synthesis, suggesting these reactions as targets for elimination and down-regulation, respectively.
Fig. 4.

Genetic engineering targets towards improved PHA production in P. putida as predicted by an elementary flux mode-based correlation strategy (Flux Design). Red arrows represent reactions that are negatively correlated with PHA production and are therefore attenuation targets, whereas green arrows indicate positive correlation and thus overexpression targets. On the right, a priority ranking of all targets is given
Discussion
Low energy requirement of P. putida for maintenance enables versatile metabolic response
Impressively, the maintenance requirement of glycerol-grown P. putida KT440 is 16 times lower in comparison to E. coli and 20–25 times lower than that of other important strains grown on glycerol [46, 47] (Table 6). In addition, the maintenance coefficient was 1.5 times lower than that of P. putida growing on glucose and approximately ten times lower than that of several other glucose-grown industrial strains. As glycerol has a higher degree of reduction than glucose and produces twice as much reducing equivalents, when converted to phosphoenolpyruvate [48], it seems a straightforward substrate for the production of redox-demanding chemicals through fermentation processes. Moreover, glucose is not the preferred carbon source of strains belonging to the Pseudomonas genus [49, 50], making glycerol an attractive raw material for P. putida production processes. Several studies have attempted to reduce the carbon requirement for cell maintenance. Particularly, attention has been poured into industrial strains such as Bacillus subtilis [51], Corynebacterium glutamicum [52], and Escherichia coli [53]. Strains with a low maintenance coefficient can redirect more carbon towards a desired product, which is of utmost importance in white biotechnology as maintenance plays a key role under reduced growth rates [54]. Moreover, the maintenance requirement varies over time [54], thus having direct impact on the economics of the industrial process, particularly when operating the preferred fed-batch cultivation mode [55].
The ED pathway is favored under both carbon and nitrogen limitation
As previously reported, nitrogen limitation promotes synthesis of PHA in P. putida [1]. A similar phenomenon has been observed for metabolically engineered E. coli strains able to accumulate PHB on glucose [56]. This modulation of the PHA flux has been related to several factors including (i) NADPH supply [57], (ii) activity of enzymes of the PHA pathway [56], and (iii) precursor availability [15, 58]. In this study, preferential use of the ED pathway and glyoxylate shunt was associated with increased PHA-production (Fig. 2). Particularly, recycling of resources through the upper EMP pathway, instead of channeling carbon directly through the lower EMP pathway, was associated with increased cofactor and PHA-precursor availability and thus a higher PHA biosynthetic flux. Conversely, this yielded less ATP, indicating that PHA production is constrained to a greater extent by cofactor and precursor availability than by ATP-deficiency. Additionally, the preferential use of the glyoxylate shunt, instead of the reductive TCA cycle between isocitrate and succinate, decreased the loss of carbon to CO2, however less reductive power could be generated. Therefore, it seems that PHA synthesis in P. putida under nitrogen-limitation is mainly constrained by precursor-availability. This also fits nicely with the predicted targets for improved PHA production (Fig. 4). The recycling of resources through the upper EMP pathway and ED pathway promotes PHA synthesis, whereas the reductive TCA-cycle is predicted to influence PHA synthesis adversely. In this regard, it is important to make a clear distinction among metabolic routes fueling PHA synthesis, as each of these pathways yields a specific amount of NADPH and PHA precursors. For instance, C. necator and metabolically engineered E. coli strains both possess a complete EMP and PP pathway. Additionally, the former has an active ED pathway, which works in conjunction with the EMP and PP pathway to yield pyruvate and acetyl-CoA. In this case, improved production of PHB has been achieved by re-directing the carbon flux through the PP pathway instead of the EMP pathway [59, 60]. As more NADPH is produced by the former strain, an increase in cofactor availability led to an improved synthesis of PHB. As P. putida does not have a complete EMP pathway [61], we have previously proven that overexpression of genes of the PP pathway does not improve PHA synthesis in P. putida KT2440, when grown on glucose [15]. On the contrary, our current findings point towards engineering of the ED pathway to improve PHA production capabilities in P. putida. This is also conform with the high natural flux through the ED pathway, when P. putida KT2440 is grown on glucose [62], and as shown here, also on glycerol (Fig. 2). Previous works have also pointed out the ED pathway as the possible main route when synthesizing mcl-PHAs on glycerol. A co-feeding strategy of glycerol and fatty acids was performed to evaluate its activity [24]. Here we fully confirm the use of the ED pathway by P. putida KT2440 under both carbon- and nitrogen-limiting conditions (Fig. 2) and propose its overexpression to improve PHA-productivity.
Evaluation of metabolic responses on different organizational levels is vital to understand an organisms’ survival and success in the environment [63]. Integration of the transcriptome and metabolic fluxes showed that, upon an increased dilution rate under carbon-limiting conditions, P. putida KT2440 exhibits a more active ED pathway and increased flux through the pyruvate node, associated with significant upregulation of pyruvate metabolism (PP0554, acoA) (Fig. 2a, b; Table 5). Also, catabolism and anabolism appear to be tightly coupled as no by-product (citrate, succinate, or malate) formation is found (Table 2). On the other hand, nitrogen-limiting growth drove a major flux of carbon via the hexose-phosphates to the ED pathway when the specific growth rate was increased (Fig. 2).
Interconnection between transcripts and fluxes deciphers regulatory mechanisms of core carbon metabolism
Strong transcriptional regulation of glycerol metabolism
The shift from glycerol limitation to glycerol excess reveals unique flux and gene expression patterns in central carbon metabolism (Fig. 5). At low growth rate, the transcriptional regulator glpR, which represses genes, involved in the uptake and incorporation of glycerol in P. putida [24], did not show transcriptional changes, whereas the transporter glpF was transcriptionally attenuated (Table 5). It has previously been postulated that the presence of glycerol in the medium modulates the expression of glpF [24], which is consistent with our findings. Furthermore, at high growth rates the repression of glpR, mitigating the transcription of glpF and the regulator araC (PP1395) (Table 5), seems responsible for the decreased uptake rate of glycerol. Furthermore, at the transcript and flux level, a more active ED pathway, pyruvate node (anaplerotic reactions), and TCA cycle were found at a low dilution rate. In addition, pyruvate metabolism seems to be a key node, when glycerol is used as carbon source, as this pathway is transcriptionally modulated by the imposed nutrient limitation, a trait that has not been previously described for cells grown on glycerol under PHA-producing conditions (Table 5).
Fig. 5.
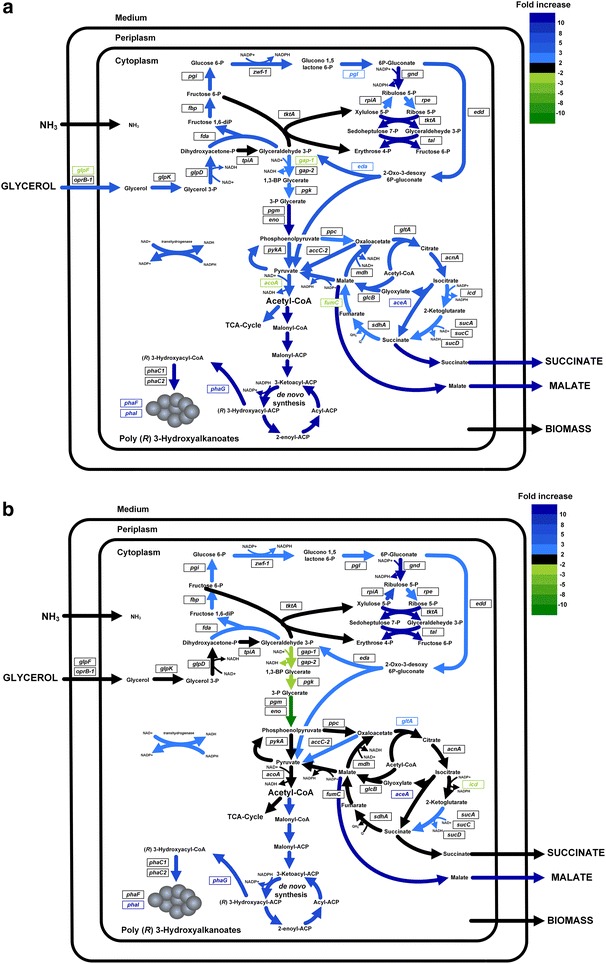
Comparison of fluxes and transcription levels between nitrogen and carbon-limitation under a low dilution rate and b high dilution rate. Significant flux differences are indicated by the color of the arrow, whereas differentially expressed genes are color-coded as gene names next to the respective arrows. Green represents transcription levels or fluxes that are significantly higher under carbon-limiting conditions. Contrastingly, blue indicated values that are increased under nitrogen-limiting conditions. Changes that exceeded a twofold increase or decrease were considered significant when the p value did not exceed 0.05
Complex regulation of isocitrate dehydrogenase mediated flux
Isocitrate dehydrogenase (icd, PP4012) showed no repression at low dilution rate, accompanied by a higher flux through its reaction (Fig. 5a), whereas at high dilution rate a significant decrease in transcription level was linked to a steady flux. Whilst previous studies have shown that at the mRNA level the gene icd is repressed under nitrogen-limiting conditions [9, 15], our findings indicate that regulation of the isocitrate dehydrogenase mediated flux might not be solely transcriptional. In fact, an increased enzymatic activity of Icd under nitrogen-limitation would explain the observed phenomenon.
Compelling correlation between PHA synthesis and expression of several genes from the PHA cluster
With regard to PHA synthesis, the shift from carbon- to nitrogen-limitation promoted accumulation of the biopolymer, whereby the phaG gene was most strongly up-regulated independently of the set dilution rate (Fig. 5). PhaG (transacylase) is the linking enzyme between de novo fatty acid synthesis and PHA biosynthesis in Pseudomonas strains [64, 65] and obviously supported enhanced PHA synthesis. In addition, PHA-granule forming enzymes, encoded by phaI and phaF, were up-regulated among the open reading frames of the PHA cluster, however, to a lesser extent (Table 5). They are known key elements of the PHA synthesis machinery, since they are involved in the segregation and distribution process of PHA [66–68]. Here, we discovered a direct correlation between strong synthesis of PHA and high expression of the phaF gene (Table 5). On the contrary, at a high specific growth rate, mRNA levels of phaF were unaffected, which could explain the observed low PHA production.
PHA biosynthetic enzyme identified as potential bottleneck towards improved PHA production in P. putida
Transcriptome analysis revealed that pyruvate dehydrogenase (encoded by acoA) was overexpressed, when comparing cells growing at a high dilution rate against those at low dilution rate under nitrogen limitation (Table 5). This also correlates well with a high flux through this particular reaction (Fig. 2c, d). Nonetheless, when the carbon flux segregates from acetyl-CoA to various pathways (TCA cycle and PHA synthesis), the PHA flux was the same for both conditions (Fig. 2c, d). This leads to the hypothesis that PHA production in P. putida is restricted at the enzymatic level, probably at some point in the malonyl and/or the synthesis de novo fatty acid pathway. Additionally, the prediction of genetic targets for enhanced PHA synthesis by elementary mode correlation indicated that both the PHA biosynthetic pathways and the recycling of the ED pathway positively influence PHA productivity, as well as the elimination of by-product formation, being malate the target with the highest priority (Fig. 4).
Conclusion
Overall, our results support the view that P. putida KT2440 has evolved to high metabolic versatility by a complex interplay of different molecular layers. This applies not only for the shift from carbon to nitrogen limitation, but also for a given specific growth rate, especially important, when cells are synthesizing PHAs. This study unravels that the Entner–Doudoroff and the glyoxylate pathways, and pyruvate metabolism play a key role when synthesizing mcl-PHA from glycerol as the only carbon and energy source. In addition, P. putida KT2440 modulates the expression of genes responsible for sensing its energetic state of the cell in order to satisfy the ATP requirement under PHA-producing conditions. Predictive metabolic modeling shows that there is still huge potential for improvement of mcl-PHA synthesis, where different metabolic engineering targets are proposed. In this way, genes belonging to the TCA cycle, ED pathway, and the synthesis of de novo fatty acids are identified as promising targets for genetic engineering towards improved PHA synthesis.
Methods
Strain
The wild-type Pseudomonas putida KT2440 (DSMZ, Braunschweig, Germany) was used in this study.
Culture conditions
Cells were kept as frozen stock in 25 % (v/v) glycerol at −80 °C. To obtain single colonies, cells were plated onto Luria–Bertani agar plates. After 1 day incubation at 30 °C, liquid cultures were prepared by picking up a single colony from the plate and inoculating it into a 50 mL shake flask with 10 mL liquid M9 medium. M9 medium contained (per liter): 12.8 g Na2HPO4·7H2O, 3 g KH2PO4, 4.7 g (NH4)2SO4, 0.5 g NaCl, 0.12 g MgSO4·7H2O, 6.0 mg FeSO4·7H2O, 2.7 mg CaCO3, 2.0 mg ZnSO4·H2O, 1.16 mg MnSO4·H2O, 0.37 mg CoSO4·7H2O, 0.33 mg CuSO4·5H2O, 0.08 mg H3BO3, and 3 g glycerol as sole carbon source. Cells were grown under aerobic conditions at 30 °C in an orbital shaker (180 rpm, Innova, New Brunswik, NJ, USA). By taking a calculated volume of the overnight-grown cell suspension, cells were transferred into 500 mL baffled Erlenmeyer flasks with 100 mL M9 medium and cultivated as described above. This second pre-culture was used as an inoculum for the continuous process, operated in a lab scale bioreactor. The feed solution consisted of minimal medium (M9, described above) supplemented with 32.5 and 4.1 g L−1 of glycerol, for the nitrogen- and carbon-limited conditions, respectively.
Continuous cultivation
Continuous cultivations were carried out under aerobic conditions with dilution rates ranging from 0.044 to 0.205 h−1 in a 2.0 L top-bench BIOSTAT B1 bioreactor (Sartorius B Systems GmbH, Melsungen, Germany) with a working volume of 1.0 L at 30 °C. The aeration rate was set to 0.4 L L−1 min−1 (PR4000, MKS instruments, Wilmington, MA, USA). The dissolved oxygen level was kept above 30 % air saturation by control of the agitation speed up to a maximum of 700 rpm. The pH was maintained at 7.0 ± 0.2 by automatic addition of 0.5 M H2SO4 and 1 M KOH, respectively. A gas analyzer, equipped with both a paramagnetic O2 and an infrared bench CO2 analyzer (Servomex Xentra 4100, Sugar Land, TX, USA), which recorded the concentration of carbon dioxide and oxygen, was coupled to the gas outlet of the bioreactor. The working volume of the fermenter was kept constant by removing the fermentation broth through a peristaltic pump and recording the weight with a balance, placed under the bioreactor.
Analytics of substrates and products
Cell growth was recorded as optical density (OD) at 600nm (Ultraspec 2000, Hitachi, Tokio, Japan). Cell dry weight was determined gravimetrically after collection of 10 mL culture broth (10 min, 4 °C, 8.000×g) in pre-weighed tubes, including a washing step with distilled water, and drying of the obtained pellet at 100 °C until constant weight. The ammonium concentration in cultivation supernatant was measured by a photometric test (LCK 303 kit, Hach Lange, Danaher, Washington, DC, USA). The concentration of glycerol and organic acids (succinate, formate, malate, citrate) in cultivation supernatant was analyzed by HPLC (Agilent 1260, Agilent, Krefeld, Germany), equipped with an 8 mm Rezex ROA-organic acid H column (Phenomenex, Torrance, CA, USA), operated with 0.013 N H2SO4 as mobile phase at 0.5 mL min−1 and 65 °C and detection using a refractive index detector (Agilent, Santa Clara, CA, USA).
PHA characterization and quantification
Monomeric composition of PHA, as well as its cellular content, were determined by gas chromatography mass spectrometry (GC/MS) of the methanolyzed polyester. For this purpose, 10 mL culture broth was placed in a falcon tube and centrifuged (10 min, 4 °C, 9000×g), followed by a washing step with distilled water. The supernatant was discarded and the cell pellet was kept at −20 °C for further processing. Methanolysis was then carried out by re-suspending 5–10 mg of lyophilized aliquots in 2 mL chloroform and 2 mL methanol, containing 15 % (v/v) sulfuric acid and 0.5 mg mL−1 3-methylbenzoic acid as internal standard, respectively, followed by incubation at 100 °C for 4 h. After cooling down to room temperature, 1 mL of demineralized water was added and the organic phase, containing the resulting methyl esters of the PHA monomers, was analyzed by GC–MS. Analysis was performed in a Varian 450GC/240MS ion trap mass spectrometer (Varian Inc., Agilent Technologies, Santa Clara, CA, USA) and operated by the software MS Workstation 6.9.3 (Varian Inc., Agilent Technologies). An aliquot (1 mL) of the organic phase was injected into the gas chromatograph at a split ratio of 1:10. Separation of compounds of interest, i.e. the methyl esters of 3-hydroxyexanoate, 3-hydroxyoctanoate, 3-hydroxydecanoate, 3-hydroxydodecanoate, 3-hydroxy-5-cis-dodecanoate, 3-hydroxytetradecanoate, was achieved by a FactorFour VF-5 ms capillary column (30 m × 0.25 mm i.d. × 0.25 mm film thickness, Varian Inc., Agilent Technologies). Helium was used as carrier gas at a flow rate of 0.9 mL min−1. Injector and transfer line temperature were 275 and 300 °C, respectively. The oven temperature program was: initial temperature 40 °C for 2 min, then from 40 °C up to 150 °C at a rate of 5 °C min−1 and finally up to 280 °C at a rate of 10 °C min−1. Positive ions were obtained using electron impact ionization at 70 eV and mass spectra were generated by scanning ions from m/z 50 to m/z 650. The PHA content (wt%) was defined as the percentage of the cell dry weight (CDW), represented by the polyhydroxyalkanoate.
Transcriptome analysis
Aliquots of 10 mL culture broth were placed in RNAprotect buffer (Qiagen, Hilden, Germany) and centrifuged (1 min, 4 °C, 10,000×g). Cell pellets were frozen at −80 °C until further processing. Isolation of total RNA was performed using RNeasy kits (Qiagen, Venlo, The Netherlands), according to instructions by the manufacturer. Extracted RNA (2 μg) was labeled with either Cy3 or Cy5 using the ULS-system (Kreatech, Amsterdam, The Netherlands) according to the manufacturers manual. Equal amounts of Cy3 or Cy5-labelled RNA, one of them corresponding to the control and the other one to the condition to be analyzed, were mixed by pipetting. Labeled RNA (600 ng) was then fragmented and hybridized to the microarray. Agilent 8 × 15 K two-color microarrays (Agilent Technologies, Santa Clara, CA, USA), specifically designed for P. putida KT2440, was used for all transcriptional analyses. The microarrays were scanned using a GenePix Pro 4001 scanner and the GenePix 4.0 software (Axon Instruments, Foster City, CA, USA). Subsequent analysis of the microarrays was conducted with software packages (see below) from the Bioconductor suite. The image analysis results were read using the ‘limma’ packages [69]. The quality of the chips was analyzed with the ‘arrayQualityMetrics’ package [70]. Intensity values were background-corrected using the “‘normexp” method of the ‘limma’ package [71] and were normalized with the variance stabilization method [72]. Differentially expressed genes were identified by fitting the linear model (using the functions ‘lmFit’ and ‘eBayes’ from the ‘limma’ package [69]. Genes for which the adjusted p value (by Benjamini–Hochberg method) was lower than 0.05 and the fold change exceeded 2 in either direction were assumed to be differentially expressed.
Metabolic network analysis
Flux balance analysis (FBA) was performed as described previously [73], using linear programming to maximize a chosen objective function, in our case growth. Shortly, FBA determines the optimal steady-state flux distribution in the metabolic network within a constraint space [74]. The constraints applied here, included experimentally determined uptake and production rates as well as PHA composition (Table 2).
Elementary flux modes (EFM’s) were calculated with efmtool, based on the null space approach and recursive enumeration with bit pattern trees [75]. The matrix, computed by the algorithm, comprises information on all thermodynamically and stoichiometrically possible pathways in the cell, which reduce metabolism into all feasible, unique, non-decomposable biochemical pathways [76]. Normalization of the EFM matrix and subsequent data interpretation was performed as described previously [44]. First, relative fluxes were normalized to the glycerol uptake flux. Subsequent flux correlation analysis investigated the target potential of individual metabolic reactions [44]. If the statistical significance of the correlation was met and the regression coefficient exceeded the cut-off of 0.7, the slope of the linear regression delivered the target potential coefficient to each individual reaction.
The metabolic network model, used for both analyses, was adapted from previous work [58]. Reactions for glycerol utilization, i.e. glycerol transport, glycerol dehydrogenase and dihydroxyacetone kinase, were implemented. Based on experimental observations, formation of gluconate was not considered. However, reactions for the secretion of succinate and malate were included (Table 2). PHA synthesis considered three different PHA types with different monomer composition. Stoichiometry of the reactions was based on the organism specific information provided in the Kyoto Encyclopedia of Genes and Genomes, i.e. the KEGG database [77] and is provided in the Appendix: Table 7. In total, the network consisted of 63 reactions, of which 18 described reversible conversions and 11 were transport reactions. Anabolic pathways for biomass synthesis were merged into a single equation. The precursor demand for growth was taken from previous work [58].
Table 7.
Metabolic network model of P. putida KT2440
| Pathway | Reactions |
|---|---|
| Transport reactions | ‘→ GLY[e]’ ‘→ NH3[c]’ ‘→ SO4[c]’ ‘→ O2[c]’ ‘biomass[c] → ’ ‘FORMATE_ex[e] → ’ ‘MAL_ex[e] → ’ ‘SUCC_ex[e] → ’ ‘PHA[c] → ’ ‘ATPmaintenance[c] →’ ‘CO2[c] → ’ |
| Glycerol uptake and conversion to glycerone-phosphate | ‘GLY[e] → GLY[p]’ ‘GLY[p] → GLY[c]’ ‘GLY[c] + ATP[c → GLY-3P[c] + ADP[c]’ ‘GLY-3P[c] + NAD[c] ↔ DHAP[c] + NADH[c]’ |
| Pentose phosphate pathway | ‘G6P[c] + NADP[c] → 6-P-Gluconate[c] + NADPH[c]’ ‘6-P-Gluconate[c] + NADP[c] → RIB-5P[c] + CO2[c] + NADPH[c]’ ‘RIB-5P[c] ⟺ XYL-5P[c]’ ‘RIB-5P[c] ⟺ RIBO-5P[c]’ ‘S7P[c] + GAP[c] ⟺ RIBO-5P[c] + XYL-5P[c]’ ‘S7P[c] + GAP[c] ⟺ E4P[c] + F6P[c]’ ‘F6P[c] + GAP[c] ⟺ E4P[c] + XYL-5P[c]’ |
| Entner-Doudoroff pathway | ‘6-P-Gluconate[c] → KDPG[c]’ ‘KDPG[c] → GAP[c] + PYR[c]’ |
| Embden-Meyerhof-Parnas pathway | ‘G6P[c] ⟺ F6P[c]’ ‘FBP[c] → F6P[c]’ ‘FBP[c] ⟺ GAP[c] + DHAP[c]’ ‘DHAP[c] ⟺ GAP[c]’ ‘GAP[c] + NAD[c] ⟺ 13-PG[c] + NADH[c]’ ‘ADP[c] + 13-PG[c] ⟺ ATP[c] + 3-PG[c]’ ‘3-PG[c] ⟺ 2-PG[c]’ ‘2-PG[c] ⟺ PEP[c]’ ‘PEP[c] + ADP[c] → PYR[c] + ATP[c]’ ‘PYR[c] + NAD[c] → AcCoA[c] + NADH[c] + CO2[c]’ ‘PYR[c] + 2 ATP[c] → 2 ADP[c] + PEP[c]’ |
| Citric acid cycle | ‘AcCoA[c] + OAA[c] → CIT[c]’ ‘CIT[c] ↔ ICI[c]’ ‘ICI[c] + NADP[c] → AKG[c] + CO2[c] + NADPH[c]’ ‘AKG[c] + NAD[c] → SUCC-CoA[c] + NADH[c] + CO2[c]’ ‘SUCC-CoA[c] + ADP[c] ↔ SUCC[c] + ATP[c]’ ‘SUCC[c] + Q[c] ↔ FUM[c] + QH2[c]’ ‘FUM[c] ↔ MAL[c]’ ‘MAL[c] + NAD[c] ↔ OAA[c] + NADH[c]’ |
| Organic acid production | ‘MAL[c] → MAL_ex[e]’ ‘SUCC[c] → SUCC_ex[e]’ ‘FORMATE[c] → FORMATE_ex[e]’ |
| Glyoxylate metabolism | ‘ICI[c] → Glyoxy[c] + SUCC[c]’ ‘Glyoxy[c] + AcCoA[c] → MAL[c]’ |
| Amphibolic metabolism | ‘OAA[c] → PYR[c] + CO2[c]’ ‘PEP[c] + CO2[c] + ATP[c] → OAA[c] + ADP[c]’ ‘MAL[c] + NADP[c] → PYR[c] + NADPH[c] + CO2[c]’ |
| PHA production | ‘5 AcCoA[c] + 4 ATP[c] + 7 NADPH[c] → C10-PHA[c] + 4 ADP[c] + 7 NADP[c]’ ‘4 AcCoA[c] + 4 ATP[c] + 7 NADPH[c] → C8-PHA[c] + 4 ADP[c] + 7 NADP[c]’ ‘6 AcCoA[c] + 4 ATP[c] + 7 NADPH[c] → C12-PHA[c] + 4 ADP[c] + 7 NADP[c]’ ‘0.75 C10-PHA[c] + 0.17 C8-PHA[c] + 0.08 C12-PHA[c] → PHA[c]’ |
| Energy metabolism | ‘NADPH[c] + NAD[c] → NADP[c] + NADH[c]’ ‘(3) NADH[c] + (3) NADP[c] + ATP[c] → (3) NAD[c] + (3) NADPH[c] + ADP[c]’ ‘(0.5) O2[c] + NADH[c] + (1.33) ADP[c] → NAD[c] + (1.33) ATP[c]’ ‘(0.5) O2[c] + QH2[c] + (0.66) ADP[c] → Q[c] + (0.66) ATP[c]’ ‘ATP[c] → ADP[c] + ATPmaintenance[c]’ ‘SO4[c] + (3) NADPH[c] + (4) ATP[c] → H2S[c] + (3) NADP[c] + (4) ADP[c]’ |
| Biomass production | ‘(1.481) OAA[c] + (1.338) 3-PG[c] + (0.627) RIBO-5P[c] + (17.821) ATP[c] + (16.548) NADPH[c] + (6.965) NH3[c] + (3.548) NAD[c] + (2.930) AcCoA[c] + (2.861) PYR[c] + (1.078) AKG[c] + (0.361) E4P[c] + (0.72) PEP[c] + (0.233) H2S[c] + (0.072) F6P[c] + (0.206) G6P[c] + (0.129) GAP[c] → biomass[c] + (16.548) NADP[c] + (3.548) NADH[c] + (17.821) ADP[c] + (1.678) CO2[c]’ |
Authors’ contributions
IPC conceived and designed the study, performed the experiments including fermentations, analyzed the transcriptome and flux data, interpreted the data and composed the manuscript. JT performed the transcriptome analysis. VB carried out the metabolic flux analysis and composed the manuscript. CW guided the study and wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Ignacio Poblete-Castro acknowledges financial support by the program Fondecyt Inicio N11150174 and “Convenio de Desempeño Apoyo a la Inovación en la Educación Superior” (PMIUAB 1301). Christoph Wittmann acknowledges financial support by the German Ministry of Science and Education (BMBF) within the VIP initiative (FKZ 03V0757).
Competing interests
The authors declare that they have no competing interests.
Abbreviations
- D
dilution rate (h−1)
- µ
specific growth rate (h−1)
- qglycerol
specific glycerol uptake rate [g glycerol (gCDW)−1 h−1]
- Ytruex|glycerol
true biomass yield coefficient on glycerol (–)
- mglycerol
maintenance coefficient on glycerol [mmol glycerol (gCDW)−1 h−1]
- mATP
ATP requirement for maintenance [(mmol ATP) (gCDW)−1 h−1]
- P/O
phosphate:oxygen ratio (–)
Appendix
See Table 7.
Deriving the ATP requirement for maintenance
Overall stoichiometry for synthesis of biomass constituents: (UK = unknown parameter)
The required ATP and NADPH for biomass synthesis are supplied by the catabolic pathways:
- Pentose phosphate pathway, where glycerol is completely oxidized to CO2
- ED pathway and TCA cycle, where glycerol is completely oxidized to CO2
- Oxidative phosphorylation
Consumption of ATP for maintenance
In matrix notation:
With the rate vector of forward reaction rates:
The production and consumption of the three cofactors ATP, NADH and NADPH can now be balanced:
In addition to the three steady state balances above, we can also find equations for the specific glycerol and oxygen uptake and the CO2 production.
By elimation of vPP, vED and vOP we get:
where
or
Footnotes
Veronique Beckers and Ignacio Poblete-Castro contributed equally to this work
Contributor Information
Veronique Beckers, Email: veronique.beckers@basf.com.
Ignacio Poblete-Castro, Email: ignacio.poblete@unab.cl.
Jürgen Tomasch, Email: juergen.tomasch@helmholtz-hzi.de.
Christoph Wittmann, Phone: +49-681-302-71970, Email: christoph.wittmann@uni-saarland.de.
References
- 1.Huijberts GNM, Eggink G. Production of poly(3-hydroxyalkanoates) by Pseudomonas putida KT2442 in continuous cultures. Appl Microbiol Biotechnol. 1996;46:233–239. doi: 10.1007/s002530050810. [DOI] [Google Scholar]
- 2.Timmis KN. Pseudomonas putida: a cosmopolitan opportunist par excellence. Environ Microbiol. 2002;4:779–781. doi: 10.1046/j.1462-2920.2002.00365.x. [DOI] [PubMed] [Google Scholar]
- 3.Martins dos Santos VAP, Heim S, Strätz M, Timmis KN. Insights into the genomic basis of niche specificity of Pseudomonas putida strain KT2440. Environ Microbiol. 2004;6:1264–1286. doi: 10.1111/j.1462-2920.2004.00734.x. [DOI] [PubMed] [Google Scholar]
- 4.Zhao YH, Li HM, Qin LF, Wang HH, Chen GQ. Disruption of the polyhydroxyalkanoate synthase gene in Aeromonas hydrophila reduces its survival ability under stress conditions. FEMS Microbiol Lett. 2007;276:34–41. doi: 10.1111/j.1574-6968.2007.00904.x. [DOI] [PubMed] [Google Scholar]
- 5.Ruiz JA, López NI, Fernández RO, Méndez BS. Polyhydroxyalkanoate degradation is associated with nucleotide accumulation and enhances stress resistance and survival of Pseudomonas oleovorans in natural water microcosms. Appl Environ Microbiol. 2001;67:225–230. doi: 10.1128/AEM.67.1.225-230.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson AJ, Dawes EA. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev. 1990;54:450–472. doi: 10.1128/mr.54.4.450-472.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kadouri D, Jurkevitch E, Okon Y, Castro-Sowinski S. Ecological and agricultural significance of bacterial polyhydroxyalkanoates. Crit Rev Microbiol. 2005;31:55–67. doi: 10.1080/10408410590899228. [DOI] [PubMed] [Google Scholar]
- 8.Hartmann R, Hany R, Geiger T, Egli T, Witholt B, Zinn M. Tailored Biosynthesis of olefinic medium-chain-length Poly[(R)-3-hydroxyalkanoates] in Pseudomonas putida GPo1 with Improved thermal properties. Macromolecules. 2004;37:6780–6785. doi: 10.1021/ma040035+. [DOI] [Google Scholar]
- 9.Poblete-Castro I, Escapa I, Jäger C, Puchalka J, Chi Lam C, Schomburg D, Prieto M, Martins dos Santos VAP. The metabolic response of P. putida KT2442 producing high levels of polyhydroxyalkanoate under single- and multiple-nutrient-limited growth: highlights from a multi-level omics approach. Microb Cell Fact. 2012;11:34. doi: 10.1186/1475-2859-11-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen G-Q. A microbial polyhydroxyalkanoates (PHA) based bio- and materials industry. Chem Soc Rev. 2009;38:2434–2446. doi: 10.1039/b812677c. [DOI] [PubMed] [Google Scholar]
- 11.Yao Y-C, Zhan X-Y, Zhang J, Zou X-H, Wang Z-H, Xiong Y-C, Chen J, Chen G-Q. A specific drug targeting system based on polyhydroxyalkanoate granule binding protein PhaP fused with targeted cell ligands. Biomaterials. 2008;29:4823–4830. doi: 10.1016/j.biomaterials.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 12.Shrivastav A, Kim HY, Kim YR. Advances in the applications of polyhydroxyalkanoate nanoparticles for novel drug delivery system. BioMed Res Int. 2013;2013:581684. doi: 10.1155/2013/581684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Q, Luo G, Zhou XR, Chen GQ. Biosynthesis of poly(3-hydroxydecanoate) and 3-hydroxydodecanoate dominating polyhydroxyalkanoates by β-oxidation pathway inhibited Pseudomonas putida. Metab Eng. 2011;13:11–17. doi: 10.1016/j.ymben.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Escapa IF, Morales V, Martino VP, Pollet E, Avérous L, García JL, Prieto MA. Disruption of β-oxidation pathway in Pseudomonas putida KT2442 to produce new functionalized PHAs with thioester groups. Appl Microbiol Biotechnol. 2011;89:1583–1598. doi: 10.1007/s00253-011-3099-4. [DOI] [PubMed] [Google Scholar]
- 15.Borrero-de Acuña JM, Bielecka A, Häussler S, Schobert M, Jahn M, Wittmann C, Jahn D, Poblete-Castro I. Production of medium chain length polyhydroxyalkanoate in metabolic flux optimized Pseudomonas putida. Microb Cell Fact. 2014;13:88. doi: 10.1186/1475-2859-13-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poblete-Castro I, Rodriguez AL, Lam CMC, Kessler W. Improved production of medium-chain-length polyhydroxyalkanoates in glucose-based fed-batch cultivations of metabolically engineered Pseudomonas putida strains. J Microbiol Biotechnol. 2014;24:59–69. doi: 10.4014/jmb.1308.08052. [DOI] [PubMed] [Google Scholar]
- 17.Castilho LR, Mitchell DA, Freire DMG. Production of polyhydroxyalkanoates (PHAs) from waste materials and by-products by submerged and solid-state fermentation. Bioresour Technol. 2009;100:5996–6009. doi: 10.1016/j.biortech.2009.03.088. [DOI] [PubMed] [Google Scholar]
- 18.Muhr A, Rechberger EM, Salerno A, Reiterer A, Malli K, Strohmeier K, Schober S, Mittelbach M, Koller M. Novel description of mcl-PHA Biosynthesis by Pseudomonas chlororaphis from animal-derived waste. J Biotechnol. 2013;165:45–51. doi: 10.1016/j.jbiotec.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 19.Kenny S, Runic J, Kaminsky W, Woods T, Babu R, O’Connor K. Development of a bioprocess to convert PET derived terephthalic acid and biodiesel derived glycerol to medium chain length polyhydroxyalkanoate. Appl Microbiol Biotechnol. 2012;95:623–633. doi: 10.1007/s00253-012-4058-4. [DOI] [PubMed] [Google Scholar]
- 20.Fu J, Sharma U, Sparling R, Cicek N, Levin DB. Evaluation of medium-chain-length polyhydroxyalkanoate production by Pseudomonas putida LS46 using biodiesel by-product streams. Can J Microbiol. 2014;60:461–468. doi: 10.1139/cjm-2014-0108. [DOI] [PubMed] [Google Scholar]
- 21.Poblete-Castro I, Binger D, Oehlert R, Rohde M. Comparison of mcl-Poly(3-hydroxyalkanoates) synthesis by different Pseudomonas putida strains from crude glycerol: citrate accumulates at high titer under PHA-producing conditions. BMC Biotechnol. 2014;14:962. doi: 10.1186/s12896-014-0110-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nikel PI, Kim J, de Lorenzo V. Metabolic and regulatory rearrangements underlying glycerol metabolism in Pseudomonas putida KT2440. Environ Microbiol. 2014;16:239–254. doi: 10.1111/1462-2920.12224. [DOI] [PubMed] [Google Scholar]
- 23.Wang Q, Nomura CT. Monitoring differences in gene expression levels and polyhydroxyalkanoate (PHA) production in Pseudomonas putida KT2440 grown on different carbon sources. J Biosci Bioeng. 2010;110:653–659. doi: 10.1016/j.jbiosc.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Escapa IF, del Cerro C, García JL, Prieto MA. The role of GlpR repressor in Pseudomonas putida KT2440 growth and PHA production from glycerol. Environ Microbiol. 2013;15:93–110. doi: 10.1111/j.1462-2920.2012.02790.x. [DOI] [PubMed] [Google Scholar]
- 25.Bücker R, Heroven AK, Becker J, Dersch P, Wittmann C. The pyruvate-tricarboxylic acid cycle node: a cofocal point of virulence control in the enteric pathogene yersenia pseudomotuberculosis. J Biol Chem. 2014;289:30114–30132. doi: 10.1074/jbc.M114.581348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohlstedt M, Sappa PK, Meyer H, Maaß S, Zaprasis A, Hoffmann T, Becker J, Steil L, Hecker M, van Dijl JM, et al. Adaptation of Bacillus subtilis carbon core metabolism to simultaneous nutrient limitation and osmotic challenge: a multi-omics perspective. Environ Microbiol. 2014;16:1898–1917. doi: 10.1111/1462-2920.12438. [DOI] [PubMed] [Google Scholar]
- 27.Krömer JO, Sorgenfrei O, Klopprogge K, Heinzle E, Wittmann C. In-Depth profiling of lysine-producing Corynebacterium glutamicum by combined analysis of the transcriptome, metabolome, and fluxome. J Bacteriol. 2004;186:1769–1784. doi: 10.1128/JB.186.6.1769-1784.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Becker J, Wittmann C. Advanced biotechnology: metabolically engineered cells for the bio-based production of chemicals and fuels, materials, and health-care products. Angew Chem Int Ed. 2015;54:3328–3350. doi: 10.1002/anie.201409033. [DOI] [PubMed] [Google Scholar]
- 29.Becker J, Wittmann C. Systems and synthetic metabolic engineering for amino acid production—the heartbeat of industrial strain development. Curr Opin Biotechnol. 2012;23:718–726. doi: 10.1016/j.copbio.2011.12.025. [DOI] [PubMed] [Google Scholar]
- 30.Lee JW, Na D, Park JM, Lee J, Choi S, Lee SY. Systems metabolic engineering of microorganisms for natural and non-natural chemicals. Nat Chem Biol. 2012;8:536–546. doi: 10.1038/nchembio.970. [DOI] [PubMed] [Google Scholar]
- 31.Puchalka J, Oberhardt MA, Godinho M, Bielecka A, Regenhardt D, Timmis KN, Papin JA, Martins dos Santos VAP. Genome-scale reconstruction and analysis of the Pseudomonas putida KT2440 metabolic network facilitates applications in biotechnology. PLoS Comput Biol. 2008;4:e1000210. doi: 10.1371/journal.pcbi.1000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nogales J, Palsson BØ, Thiele I. A genome-scale metabolic reconstruction of Pseudomonas putida KT2440: iJN746 as a cell factory. BMC Syst Biol. 2008;2:79. doi: 10.1186/1752-0509-2-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sohn SB, Kim TY, Park JM, Lee SY. In silico genome-scale metabolic analysis of Pseudomonas putida KT2440 for polyhydroxyalkanoate synthesis, degradation of aromatics and anaerobic survival. Biotechnol J. 2010;5:739–750. doi: 10.1002/biot.201000124. [DOI] [PubMed] [Google Scholar]
- 34.Wang G, Post WM. A theoretical reassessment of microbial maintenance and implications for microbial ecology modeling. FEMS Microbiol Ecol. 2012;81:610–617. doi: 10.1111/j.1574-6941.2012.01389.x. [DOI] [PubMed] [Google Scholar]
- 35.Kaiser C, Franklin O, Dieckmann U, Richter A. Microbial community dynamics alleviate stoichiometric constraints during litter decay. Ecol Lett. 2014;17:680–690. doi: 10.1111/ele.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kayser A, Weber J, Hecht V, Rinas U. Metabolic flux analysis of Escherichia coli in glucose-limited continuous culture. I. Growth-ratedependent metabolic efficiency at steady state. Microbiology. 2005;151:693–706. doi: 10.1099/mic.0.27481-0. [DOI] [PubMed] [Google Scholar]
- 37.Pirt S. Maintenance energy: a general model for energy-limited and energy-sufficient growth. Arch Microbiol. 1982;133:300–302. doi: 10.1007/BF00521294. [DOI] [PubMed] [Google Scholar]
- 38.Zeng A-P, Ross A, Deckwer W-D. A method to estimate the efficiency of oxidative phosphorylation and biomass yield from atp of a facultative anaerobe in continuous culture. Biotechnol Bioeng. 1990;36:965–969. doi: 10.1002/bit.260360912. [DOI] [PubMed] [Google Scholar]
- 39.Sun Z, Ramsay J, Guay M, Ramsay B. Carbon-limited fed-batch production of medium-chain-length polyhydroxyalkanoates from nonanoic acid by Pseudomonas putida KT2440. Appl Microbiol Biotechnol. 2007;74:69–77. doi: 10.1007/s00253-006-0655-4. [DOI] [PubMed] [Google Scholar]
- 40.van Duuren JBJH, Puchałka J, Mars AE, Bücker R, Eggink G, Wittmann C. Martins dos Santos VAP. Reconciling in vivo and in silico key biological parameters of Pseudomonas putida KT2440 during growth on glucose under carbon-limited condition. BMC Biotechnol. 2013;13:93. doi: 10.1186/1472-6750-13-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prieto A, Escapa IF, Martínez V, Dinjaski N, Herencias C, de la Peña F, Tarazona N, Revelles O. A holistic view of polyhydroxyalkanoate metabolism in Pseudomonas putida. Environ Microbiol. 2015;18:341–357. doi: 10.1111/1462-2920.12760. [DOI] [PubMed] [Google Scholar]
- 42.Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63(2):479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ren Q, de Roo G, Ruth K, Witholt B, Zinn M, Thöny-Meyer L. Simultaneous accumulation and degradation of polyhydroxyalkanoates: futile cycle or clever regulation? Biomacromolecules. 2009;10:916–922. doi: 10.1021/bm801431c. [DOI] [PubMed] [Google Scholar]
- 44.Melzer G, Esfandabadi M, Franco-Lara E, Wittmann C. Flux Design: in silico design of cell factories based on correlation of pathway fluxes to desired properties. BMC Syst Biol. 2009;3:120. doi: 10.1186/1752-0509-3-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Becker J, Zelder O, Häfner S, Schröder H, Wittmann C. From zero to hero—Design-based systems metabolic engineering of Corynebacterium glutamicum for l-lysine production. Metab Eng. 2011;13:159–168. doi: 10.1016/j.ymben.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 46.Nielsen J, Villadsen J. Bioreaction Engineering Principles. New York: Plenum Press; 1994. [Google Scholar]
- 47.Farmer IS, Jones CW. The energetics of Escherichia coli during aerobic growth in continuous culture. Eur J Biochem. 1976;67:115–122. doi: 10.1111/j.1432-1033.1976.tb10639.x. [DOI] [PubMed] [Google Scholar]
- 48.Durnin G, Clomburg J, Yeates Z, Alvarez PJ, Zygourakis K, Campbell P, Gonzalez R. Understanding and harnessing the microaerobic metabolism of glycerol in Escherichia coli. Biotechnol Bioeng. 2009;103(1):148–161. doi: 10.1002/bit.22246. [DOI] [PubMed] [Google Scholar]
- 49.Görke B, Stülke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Micro. 2008;6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 50.Rojo F. Carbon catabolite repression in Pseudomonas: optimizing metabolic versatility and interactions with the environment. FEMS Microbiol Rev. 2010;34:658–684. doi: 10.1111/j.1574-6976.2010.00218.x. [DOI] [PubMed] [Google Scholar]
- 51.Tännler S, Decasper S, Sauer U. Maintenance metabolism and carbon fluxes in Bacillus species. Microb Cell Fact. 2008;7:19. doi: 10.1186/1475-2859-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koch-Koerfges A, Pfelzer N, Platzen L, Oldiges M, Bott M. Conversion of Corynebacterium glutamicum from an aerobic respiring to an aerobic fermenting bacterium by inactivation of the respiratory chain. Biochim Biophys Acta. 2013;1827(6):699–708. doi: 10.1016/j.bbabio.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 53.Aristidou AA, San KY, Bennett GN. Metabolic flux analysis of Escherichia coli expressing the Bacillus subtilis acetolactate synthase in batch and continuous cultures. Biotechnol Bioeng. 1999;63(6):737–749. doi: 10.1002/(SICI)1097-0290(19990620)63:6<737::AID-BIT12>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 54.Zhang W, Bevins MA, Plantz BA, Smith LA, Meagher MM. Modeling Pichia pastoris growth on methanol and optimizing the production of a recombinant protein, the heavy-chain fragment C of botulinum neurotoxin, serotype A. Biotechnol Bioeng. 2000;70(1):1–8. doi: 10.1002/1097-0290(20001005)70:1<1::AID-BIT1>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 55.Lee J, Lee SY, Park S, Middelberg AP. Control of fed-batch fermentations. Biotechnol Adv. 1999;17(1):29–48. doi: 10.1016/S0734-9750(98)00015-9. [DOI] [PubMed] [Google Scholar]
- 56.Tyo KEJ, Fischer CR, Simeon F, Stephanopoulos G. Analysis of polyhydroxybutyrate flux limitations by systematic genetic and metabolic perturbations. Metab Eng. 2010;12:187–195. doi: 10.1016/j.ymben.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 57.Sánchez AM, Andrews J, Hussein I, Bennett GN, San K-Y. Effect of overexpression of a soluble pyridine nucleotide transhydrogenase (udha) on the production of Poly(3-hydroxybutyrate) in Escherichia coli. Biotechnol Prog. 2006;22:420–425. doi: 10.1021/bp050375u. [DOI] [PubMed] [Google Scholar]
- 58.Poblete-Castro I, Binger D, Rodrigues A, Becker J, Martins dos Santos VAP, Wittmann C. In-silico-driven metabolic engineering of Pseudomonas putida for enhanced production of poly-hydroxyalkanoates. Metab Eng. 2013;15:113–123. doi: 10.1016/j.ymben.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 59.Lee J-N, Shin H-D, Lee Y-H. Metabolic engineering of pentose phosphate pathway in Ralstoniaeutropha for enhanced biosynthesis of Poly-β-hydroxybutyrate. Biotechnol Prog. 2003;19:1444–1449. doi: 10.1021/bp034060v. [DOI] [PubMed] [Google Scholar]
- 60.Lim S-J, Jung Y-M, Shin H-D, Lee Y-H. Amplification of the NADPH-related genes zwf and gnd for the oddball biosynthesis of PHB in an E. coli transformant harboring a cloned phbCAB operon. J Biosci Bioeng. 2002;93:543–549. doi: 10.1016/S1389-1723(02)80235-3. [DOI] [PubMed] [Google Scholar]
- 61.Nelson KE, Weinel C, Paulsen IT, Dodson RJ, Hilbert H, Martins dos Santos VAP, Fouts DE, Gill SR, Pop M, Holmes M, et al. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol. 2002;4(12):799–808. doi: 10.1046/j.1462-2920.2002.00366.x. [DOI] [PubMed] [Google Scholar]
- 62.del Castillo T, Ramos JL, Rodríguez-Herva JJ, Fuhrer T, Sauer U, Duque E. Convergent peripheral pathways catalyze initial glucose catabolism in Pseudomonas putida: genomic and flux analysis. J Bacteriol. 2007;189:5142–5152. doi: 10.1128/JB.00203-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trautwein K, Lahme S, Wöhlbrand L, Feenders C, Mangelsdorf K, Harder J, Steinbüchel A, Blasius B, Reinhardt R, Rabus R. Physiological and proteomic adaptation of “Aromatoleum aromaticum” EbN1 to low growth rates in benzoate-limited, anoxic chemostats. J Bacteriol. 2012;194:2165–2180. doi: 10.1128/JB.06519-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rehm BHA, Krüger N, Steinbüchel A. A new metabolic link between fatty acid de novo synthesis and polyhydroxyalkanoic acid synthesis. The phaG gene from Pseudomonas putida KT2440 encodes a 3-hydroxyacyl-acyl carrier protein-coenzyme a transferase. J Biol Chem. 1998;273:24044–24051. doi: 10.1074/jbc.273.37.24044. [DOI] [PubMed] [Google Scholar]
- 65.Rehm BHA, Mitsky TA, Steinbüchel A. Role of Fatty acid de novo biosynthesis in Polyhydroxyalkanoic Acid (PHA) and rhamnolipid synthesis by pseudomonads: establishment of the transacylase (phag)-mediated pathway for pha biosynthesis in Escherichia coli. Appl Environ Microbiol. 2001;67:3102–3109. doi: 10.1128/AEM.67.7.3102-3109.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Galán B, Dinjaski N, Maestro B, De Eugenio LI, Escapa IF, Sanz JM, García JL, Prieto MA. Nucleoid-associated PhaF phasin drives intracellular location and segregation of polyhydroxyalkanoate granules in Pseudomonas putida KT2442. Mol Microbiol. 2011;79:402–418. doi: 10.1111/j.1365-2958.2010.07450.x. [DOI] [PubMed] [Google Scholar]
- 67.Dinjaski N, Prieto MA. Swapping of phasin modules to optimize the in vivo immobilization of proteins to medium-chain-length polyhydroxyalkanoate granules in Pseudomonas putida. Biomacromolecules. 2013;14:3285–3293. doi: 10.1021/bm4008937. [DOI] [PubMed] [Google Scholar]
- 68.Prieto MA, Bühler B, Jung K, Witholt B, Kessler B. PhaF, a polyhydroxyalkanoate-granule-associated protein of Pseudomonas oleovorans GPo1 involved in the regulatory expression system for pha genes. J Bacteriol. 1999;181:858–868. doi: 10.1128/jb.181.3.858-868.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smyth GK. limma: Linear Models for Microarray Data. In: Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S, editors. Bioinformatics and computational biology solutions using R and bioconductor SE-23. Springer: New York; 2005. pp. 397–420. [Google Scholar]
- 70.Kauffmann A, Gentleman R, Huber W. arrayQualityMetrics—A bioconductor package for quality assessment of microarray data. Bioinformatics. 2009;25:415–416. doi: 10.1093/bioinformatics/btn647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, Smyth GK. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23:2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- 72.Huber W, Von Heydebreck A, Sültmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18:S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.S96. [DOI] [PubMed] [Google Scholar]
- 73.Lotz K, Hartmann A, Grafahrend-Belau E, Schreiber F, Junker BH. Elementary flux modes, flux balance analysis, and their application to plant metabolism. In: Sriram G, editor. Plant metabolism SE-14. New York: Humana Press; 2014. pp. 231–252. [DOI] [PubMed] [Google Scholar]
- 74.Chen X, Alonso AP, Allen DK, Reed JL, Shachar-Hill Y. Synergy between 13C-metabolic flux analysis and flux balance analysis for understanding metabolic adaption to anaerobiosis in E. coli. Metab Eng. 2011;13:38–48. doi: 10.1016/j.ymben.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 75.Terzer M, Stelling J. Large-scale computation of elementary flux modes with bit pattern trees. Bioinformatics. 2008;24:2229–2235. doi: 10.1093/bioinformatics/btn401. [DOI] [PubMed] [Google Scholar]
- 76.Schuster S, Dandekar T, Fell DA. Detection of elementary flux modes in biochemical networks: a promising tool for pathway analysis and metabolic engineering. Trends Biotechnol. 1999;17:53–60. doi: 10.1016/S0167-7799(98)01290-6. [DOI] [PubMed] [Google Scholar]
- 77.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36(Database issue):D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ebert BE, Kurth F, Grund M, Blank LM, Schmid A. Response of Pseudomonas putida KT2440 to increased NADH and ATP demand. Appl Environ Microbiol. 2011;77:6597–6605. doi: 10.1128/AEM.05588-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nanchen A, Schicker A, Sauer U. Nonlinear dependency of intracellular fluxes on growth rate in miniaturized continuous cultures of Escherichia coli. Appl Environ Microbiol. 2006;72:1164–1172. doi: 10.1128/AEM.72.2.1164-1172.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sauer U, Hatzimanikatis V, Hohmann HP, Manneberg M, van Loon AP, Bailey JE. Physiology and metabolic fluxes of wild-type and riboflavin-producing Bacillus subtilis. Appl Environ Microbiol. 1996;62(10):3687–3696. doi: 10.1128/aem.62.10.3687-3696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]


