Many medical and biological genetics and functional genomics studies include genome-wide analysis. Due to the coordination of cellular functions, the behavior of groups of genes rather than of a single gene can be more informative in these studies. Experimental and technical developments now allow genome-wide measurement of many molecular components of the cell, including mRNA transcripts (1), DNA sequence (2–5) and structure (6–10), DNA binding by transcriptional regulators (11), microRNAs, proteins, and metabolites (12). For each type of data, analysis software has been developed, much of it available within the R/Bioconductor framework (13).
A major issue remains for data mining or statistical inference on high-throughput data due to the “curse of dimensionality” arising from the tens of thousands of molecular components generally being measured in only tens or hundreds of conditions. A logical approach to this problem is the use of Bayesian statistics (14), where prior information developed from many years of targeted biological studies can be used to reduce the search space during model fitting.
For many analyses, there are several steps required for data processing, from image acquisition and processing through normalization to data mining or statistical inference. Often, it is necessary to create a pipeline for the analysis. The ideal pipeline would allow the integration of both prior knowledge and potentially the use of measurements in one molecular domain to guide inference in another. For instance, genes known a priori to function in parallel redundant pathways may be more likely to show genetic interactions in a genome-wide association study (GWAS). Alternatively genes that share transcription factor binding determined by ChIP-seq measurements may be more likely to show correlated expression. The Bayesian framework is quite natural for data exchange in this case, especially for programs that handle different forms of gene-related information and different representations of the data.
XML (eXtendible Markup Language) was invented in the late 1990’s (15) as a way to represent documents in a machine-readable hypertext form. The represented information is organized as a tree, and a pre-given description of the tree allows verifying the data. The tree nodes are XML elements. Elements can contain each other. If a node A is a child of node B, the element corresponding to B contains that corresponding to A. Each of the elements belongs to a type, and the list of the types and their possible relations is the essence of the description (XML schema) mentioned above. Each schema corresponds to a definite data type, e.g. a book, an image, a worksheet, etc. Over the past decade, XML became the most common way of Internet data exchange.
Current bioinformatics practice uses a large variety of XML-based languages that describe different data types (e.g., for a review see (15)). We mention a few of them that are most applicable to this domain. XEMBL (16) is an XML format for EMBL data. CisML (17) and SmallBisMark (18) are for sequence motif information such as transcription factor binding sites, while MAGE-ML (19, 20) is intended for microarray metadata representation. SBML (21) and CellML (22) capture biological network models, and MFAML (23) describes metabolic fluxes.
In addition, there are XML formats (24, 25) that represent Bayesian information in a very general form. However, we require a format for Bayesian information that is suited to biological systems, but which is not too specialized, unlike the biological XMLs noted above. Our goal is an XML to encode relationships as probabilities of interactions for the purposes of genetics and bioinformatics, with the interpretation of the message in the XML depending on the context of the parser. This will permit the interchange of probabilistic information between bioinformatics frameworks that refer to different aspects of genomics knowledge.
Materials and Methods
The OnionTree XML language is an XML-based markup language that is intended for the interchange of biomolecule-related information in a Bayesian probabilistic paradigm. Here we assume these biomolecules are related to genes, for purposes of illustration, however the framework is general. The most basic information is a probabilistic (joint and/or conditional) statement between the specific predicate relating one, two or more genes (loci and/or variants), like “gene G is differentially expressed given phenotype A”, “gene G is associated with disease X with probability 0.5” or “gene G is expressed if gene H is expressed with probability 0.9”. Each portion of the information is characterized with its reliability which states the a priori evaluation of the probability that information form the source is correct, so that the reader can combine different, even contradictory, statements in a Bayesian paradigm.
The main motivation for the format is the view that the interpretation of the information is mainly the XML receiver’s role, as different receivers may have different approaches to using the information. The XML file mentions a gene, and, for example, the receiver interprets this as indicating that the gene is differentially expressed in a specific context. The biological objects that are mentioned in the message, e.g., loci, genetic variants, etc., are called atoms. All the mentioned atoms are listed in the dictionary in the beginning of the file. Statements comprise linking of these atoms to probabilities that a predicate (e.g. a disease association) holds for these atoms. Technically, the precise meaning of a predicate is out of the scope of the format, as the format assumes that the reader of the message knows the purpose. Actually, the essence of the predicate could be noted within the encoded data by the attributes, but it is used only to validate the data.
Results
The format is defined by an XML Schema that is available at http://onion-xml.sourceforge.net/oniontree.xsd. If future developments require extensions to the OnionTree standard, they will be added in a “backwards-compatible” way, i.e., each XML data file that is valid for a version of the OnionTree format will be valid for all the subsequent versions.
A set of examples of the XML application are at http://onion-xml.sourceforge.net/. Two examples of linkage disequilibrium (LD; Figure 2, A and B) describe the linkage between rs2112979 and rs6870870 SNP’s in Utah residents with Northern and Western European ancestry from the CEPH collection (CEU). The data is extracted from the HapMap (27) database (http://hapmap.ncbi.nlm.nih.gov/), the exact URL is shown in the XML files. The atoms are rare alleles of the SNP’s and the predicate is the occurrence of the alleles. The difference between the two examples is stylistic: the first one provides atoms in unconditional statements defining the allelic frequencies and then provides a conditional statement that shows that that allele rs6870870:A occurs with probability 1 given the allele rs2112979:G occurs; the second example defines a Boolean conjunction (tuple) on the two atoms and then mention the atoms and the tuple in three unconditional statements. The information content of the two messages is identical.
Figure 2.
An OnionTree XML data representing linkage disequilibrium (LD) of two SNP’s (rs2112979 and rs6870870) in Utah residents with Northern and Western European ancestry from the CEPH collection (CEU).
The example is also available at: http://onion-xml.sourceforge.net/onion_LD_example_1.xml. A and B differ in style.
The KEGG-related example (http://onion-xml.sourceforge.net/onion_KEGG_example.xml) shows the presence of the NADH and NADHA genes in H. Sapiens pathways according to KEGG. The predicate here is ‘a gene is a member of the pathway’. The XML is automatically generated by the ‘db/KeggOnionTree’ plan (scenario) by the Automated Sequence Annotation Pipeline (ASAP) (28) that is publicly available at http://hammurabi.onc.jhmi.edu/cgi-bin/ASAP/login.pl. This plan implements the following steps:
Specify the type of identifier in use (e.g. NCBI, UNIPROT, gene name, etc.). If this is left blank, the plan will check for matches in all possible fields.
List the genes of interest (tab or newline delimited). Alternatively, a file with the gene ids can be uploaded (see 3).
Upload a file here with the gene identifiers if necessary. This is just a text file and it is treated the same as if you copied all of the content into the field in 2.
Optionally filter the results to pathways in the given KEGG pathway category.
Choose the organism that is the source of the genes. The default is human.
You can query all of the genes by leaving the gene list blank. Note that this will not query all of the genes in KEGG (this would take too long), but rather all of the genes in KEGG from the selected organism.
The value of this example is not obvious until we realize that it is common for two databases to provide different versions of a pathway, leading to different analytic results. In this case, the possibility to ascribe individual probabilities to the statements like ‘gene G is mentioned in the pathway P’ provides a powerful way to encode a prior belief.
Short Format Description
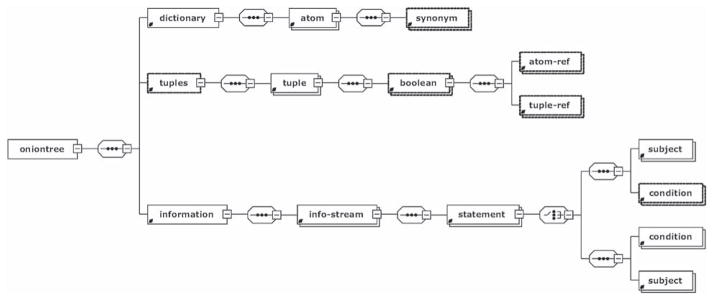
The root XML element <oniontree> (Figure 1) contains three parts: a <dictionary> that describes the biological objects (atoms) of interest (e.g. loci and their alleles), <tuples> that define a set of boolean combinations referring to the atoms, and the <information> section that describes the probabilities, possibly conditional, of the validities of the specific predicate on the atoms and of the boolean combinations of them. Each element in the XML that is an indivisible component of the message is described by an <atom> XML element.
Figure 1.
OnionTree XML Schema diagram created by XSD Diagram xml schema definition diagram viewer (http://regis.cosnier.free.fr).
An example of a <dictionary> is a list of loci (i.e., genes) of interest and alleles for which information is presented. Both the genes and alleles can be invoked in expressions. This full list of possible words is included, as it could be useful for event-driven XML readers to know the full list before receiving the probability data.
Each <tuple> that is stored in <tuples> contains a Boolean function referring to atoms or other tuples. Its main element, <boolean>, is a two-place or one-place Boolean function, that refers to atoms by an <atom-ref> element, or to another tuple by <tuple-ref>. The <tuple> container represents a Boolean function on the specific predicate about its atoms. The function itself (AND, OR, etc.) and its arguments are defined in the container <boolean>. Primary key integrity in references to atoms and tuples and primary key uniqueness are checked by the XML schema.
The main part of an OnionTree file is the <information> element. It is a sequence of <info-stream> containers that represent structured portions of Bayesian information. An <info-stream> is a representation of a data stream, i.e. an output of a single run of a program of a database query. Also, the <information > element carries attributes. Each <info-stream> is a set of statements (<statement> elements) that all represent information from a common source. The source is referred to by the attributes ‘source’ and ‘date’, which provides a unique resource identifier and the date, when the information was gathered. The ‘reliability’ shows how much we trust the source of information, and this can be specified by the user. For instance, if the source is a predictive algorithm, such as used for miRNA target prediction (26), the reliability will be lower than for a curated database of pathways, such as KEGG (27). The reliability value is effectively treated as a Bayesian probability on the issue of the validity of the information from the source.
Every <statement> represents a Bayesian probabilistic equation, e.g. P(predicate(gene1) | predicate(expr2)) = P. The probability P is given by the ‘probability’ attribute. The subject of the statement (gene1 in our example) is referred to by <subject> element and the condition (gene2), if any, is referred to by <condition>. Each element contains one <tuple-ref> or one <atom-ref> element. Of course, a <statement> without a <condition > is valid and it represents an unconditional probability. All the internal connections inside the file (<atom-ref>’s to <atom>’s, etc.) are organized by addressing unique identifiers that all elements carry as attributes.
Potential Immediate Applications
There have been a number of recent uses of prior and integrated knowledge within the biological community that could make use of the XML format described here.
Bayesian networks have been used to encode prior information from protein-protein interactions and the literature to refine gene expression analysis (28). Similarly, from cancer studies it is now known that miRNAs play an important role, and since miRNAs often target multiple genes, the expected expression of miRNAs in cancer subtypes can be used as prior information on gene expression changes.
Additional areas, including high-throughput sequencing and genotyping, linkage disequilibrium, differential arrays, data on definite disease predisposition, data on definite SNP harmfulness like that provided by PolyPhen (29), all provide examples where prior knowledge can guide analysis and could be encoded in Onion- Tree format. For GWAS studies it is highly useful to identify potential interactions between SNPs, as the combinatorics overwhelm any imaginable sample size. One approach is to identify missense mutations that affect protein structure, perhaps even focusing on SNPs predicted to have a direct impact on disease development (30). The probability framework described here allows us to assign high probability to SNPs likely to be drivers of disease, medium probability to SNPs that cause missense mutations, and low probability to SNPs that do not affect protein structure. Likewise, we can use identification of parallel pathways to identify sets of genes whose interaction would be more likely to lead to disease due to loss of natural redundancy in the biological system.
Discussion
The XML language we present is developed to transfer probabilistic biomolecule-related information between applications, which can handle very different aspects of genomic data and implement different approaches for the use of prior information. Our original purpose was to provide information about differential expression or protein structure to APSampler (31) for refining our Markov chain Monte Carlo transition probabilities based on probabilities that SNPs in a GWAS study were related to disease. We realized, however, that the task was applicable to genomics studies in general, and we believe this OnionTree XML format should find wide use in knowledge-based data analysis (32).
In order to maintain the generality of our approach, we do not apply restrictive rules for the representation of the context of information. Context can be provided within the XML file by its creator using the ‘event’ attributes, or it can be determined by the algorithm parsing the XML if appropriate. Also, we do not place any constraints on the probabilistic framework employed by the algorithm parsing the XML nor on its method of handling the information. This makes the format quite general for different biological applications, but it limits our ability to produce a standard library for its interpretation. Our goal is to have an exchange format of sufficient flexibility and adequate brevity that exchange of prior probabilities is enabled. We have found it relatively simple to implement code to generate the XML within our own ASAP systems, and we are presently implementing a parser within our APSampler tool.
Conclusion
The XML framework provides a convenient format for the transfer of probabilistic information between diverse systems. We have developed an XML language for encoding both joint and conditional probabilities for biological relationships based on biomolecules. These relationships can be quite general, involving results of coexpression experiments, links through protein-protein interactions, sequencing results, and other experimental and validated data.
Acknowledgments
The authors would like to thank NIH/NLM (LM008932), UEPHA*MS FP7 Marie Curie Initial Training Network (FP7/2007-2013, grant agreement 212877), Russian Foundation for Basic Research (11-04-02016-a) and the Johns Hopkins University Framework for the Future for support of this work.
References
- 1.Allison DB, Cui X, Page GP, Sabripour M. Nat Rev Genet. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 2.Shendure J, Ji H. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 3.Anbazhagan P, Purushottam M, Kumar HBK, Mukherjee O, Jain S, Sowdhamini R. J Biomol Struct Dyn. 2010;27:581–598. doi: 10.1080/07391102.2010.10508573. [DOI] [PubMed] [Google Scholar]
- 4.Putta P, Mitra CK. J Biomol Struct Dyn. 2010;27:599–610. doi: 10.1080/07391102.2010.10508574. [DOI] [PubMed] [Google Scholar]
- 5.Cao J, Shi F, Liu X, Jia J, Zeng J, Huang G. J Biomol Struct Dyn. 2011;28:535–544. doi: 10.1080/07391102.2011.10508593. [DOI] [PubMed] [Google Scholar]
- 6.Sabbia V, Romero H, Musto H, Naya H. J Biomol Struct Dyn. 2009;27:361–369. doi: 10.1080/07391102.2009.10507322. [DOI] [PubMed] [Google Scholar]
- 7.Mukhopadhyay P, Ghosh TC. J Biomol Struct Dyn. 2010;27:477–488. doi: 10.1080/07391102.2010.10507332. [DOI] [PubMed] [Google Scholar]
- 8.De Santis P, Morosetti S, Scipioni A. J Biomol Struct Dyn. 2010;27:747–764. doi: 10.1080/07391102.2010.10508583. [DOI] [PubMed] [Google Scholar]
- 9.Johnson SM. J Biomol Struct Dyn. 2010;27:795–802. doi: 10.1080/073911010010524946. [DOI] [PubMed] [Google Scholar]
- 10.Rapoport AE, Frenkel ZM, Trifonov EN. J Biomol Struct Dyn. 2011;28:567–574. doi: 10.1080/07391102.2011.10531243. [DOI] [PubMed] [Google Scholar]
- 11.Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Nat Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu ZR, Seo E, Yan L, Yin SJ, Si YX, Qian GY, Park YD, Yang JM. J Biomol Struct Dyn. 2010;28:259–276. doi: 10.1080/07391102.2010.10507358. [DOI] [PubMed] [Google Scholar]
- 13.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JYH, Zhang J. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rannala B. Am J Pharmacogenomics. 2001;1:203–221. doi: 10.2165/00129785-200101030-00005. [DOI] [PubMed] [Google Scholar]
- 15.Strömbäck L, Hall D, Lambrix P. Proteomics. 2007;7:857–867. doi: 10.1002/pmic.200600438. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, Riethoven JJ, Robinson A. Bioinformatics. 2002;18:1147–1148. doi: 10.1093/bioinformatics/18.8.1147. [DOI] [PubMed] [Google Scholar]
- 17.Haverty PM, Weng Z. Bioinformatics. 2004;20:1815–1817. doi: 10.1093/bioinformatics/bth162. [DOI] [PubMed] [Google Scholar]
- 18.Kulakovskiy IV, Favorov AV, Makeev VJ. Bioinformatics. 2009 doi: 10.1093/bioinformatics/btp434. [DOI] [PubMed] [Google Scholar]
- 19.Spellman PT, Miller M, Stewart J, Troup C, Sarkans U, Chervitz S, Bernhart D, Sherlock G, Ball C, Lepage M, Swiatek M, Marks W, Goncalves J, Markel S, Iordan D, Shojatalab M, Pizarro A, White J, Hubley R, Deutsch E, Senger M, Aronow BJ, Robinson A, Bassett D, Stoeckert CJ, Brazma A. Genome Biol. 2002;3:research0046.1–research0046.9. doi: 10.1186/gb-2002-3-9-research0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durinck S, Allemeersch J, Carey VJ, Moreau Y, De Moor B. Bioinformatics. 2004;20:3641–3642. doi: 10.1093/bioinformatics/bth396. [DOI] [PubMed] [Google Scholar]
- 21.Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, Cuellar AA, Dronov S, Gilles ED, Ginkel M, Gor V, Goryanin II, Hedley WJ, Hodgman TC, Hofmeyr JH, Hunter PJ, Juty NS, Kasberger JL, Kremling A, Kummer U, Le Novère N, Loew LM, Lucio D, Mendes P, Minch E, Mjolsness ED, Nakayama Y, Nelson MR, Nielsen PF, Sakurada T, Schaff JC, Shapiro BE, Shimizu TS, Spence HD, Stelling J, Takahashi K, Tomita M, Wagner J, Wang J. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 22.Cuellar AA, Lloyd CM, Nielsen PF, Bullivant DP, Nickerson DP, Hunter PJ. Simulation. 2003;79:740–747. [Google Scholar]
- 23.Yun H, Lee DY, Jeong J, Lee S, Lee SY. Bioinformatics. 2005;21:3329–3330. doi: 10.1093/bioinformatics/bti502. [DOI] [PubMed] [Google Scholar]
- 24.Kimelfeld B, Sagiv Y. SIGMOD Rec. 2008;37:69–77. [Google Scholar]
- 25.Zhao W, Dekhtyar A, Goldsmith J. Department of Computer Science, University of Kentucky 2004. 2003:219. [Google Scholar]
- 26.Friedman RC, Farh KKH, Burge CB, Bartel DP. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M, Goto S, Kawashima S, Nakaya A. Nucleic Acids Res. 2002;30:42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Djebbari A, Quackenbush J. BMC Syst Biol. 2008;2:57. doi: 10.1186/1752-0509-2-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramensky V, Bork P, Sunyaev S. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carter H, Chen S, Isik L, Tyekucheva S, Velculescu VE, Kinzler KW, Vogelstein B, Karchin R. Cancer Res. 2009;69:6660–6667. doi: 10.1158/0008-5472.CAN-09-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Favorov AV, Andreewski TV, Sudomoina MA, Favorova OO, Parmigiani G, Ochs MF. Genetics. 2005;171:2113–2121. doi: 10.1534/genetics.105.048090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ochs MF. Briefings in Bioinformatics. 2010;11:30–39. doi: 10.1093/bib/bbp044. [DOI] [PMC free article] [PubMed] [Google Scholar]