Abstract
The ubiquitin-proteasome protein degradation system is involved in many essential cellular processes including cell cycle regulation, cell differentiation, and the unfolded protein response.The anaphase-promoting complex/cyclosome (APC/C), an evolutionary conserved E3 ubiquitin ligase, was discovered 15 years ago because of its pivotal role in cyclin degradation and mitotic progression. Since then, we have learned that the APC/C is a very large, complex E3 ligase composed of 13 subunits, yielding a molecular machine of approximately 1 MDa. The intricate regulation of the APC/C is mediated by the Cdc20 family of activators, pseudosubstrate inhibitors, protein kinases and phosphatases and the spindle assembly checkpoint. The large size, complexity, and dynamic nature of the APC/C represent significant obstacles toward high-resolution structural techniques; however, over the last decade, there have been a number of lower resolution APC/C structures determined using single particle electron microscopy. These structures, when combined with data generated from numerous genetic and biochemical studies, have begun to shed light on how APC/C activity is regulated. Here, we discuss the most recent developments in the APC/C field concerning structure, substrate recognition, and catalysis.
Keywords: Ubiquitination, anaphase-promoting complex, spindle assembly checkpoint, mitosis, cyclosome, phosphorylation, electron microscopy
Introduction
The ubiquitin-proteasome system is a highly regulated signaling network that targets specific proteins for degradation. Ubiquitination, the covalent addition of a ubiquitin (Ub) or Ub chain to a substrate protein, is carried out through a cascade of enzymes. Ub-activating enzymes (E1) covalently bind ubiquitin through a thioester linkage and transfer it to a Ub-conjugating enzyme (E2), which ultimately cooperates with a Ub ligase (E3) to mark a lysine on a substrate protein (Hershko, 1997). After assembly of a ubiquitin chain on a substrate, the poly-Ub protein is shuttled to the 26S proteasome and degraded. The ubiquitination process is counteracted by deubiquitinating enzymes that remove Ub and/or disassemble free Ub chains (Reyes-Turcu et al., 2009). Thorough reviews on the topics of proteasome structure, assembly, and mechanism of proteolysis are available (Gallastegui & Groll, 2010; Bedford et al., 2010; Tanaka, 2009; Kim et al., 2010b) and will not be discussed further here. Targets of the Ub-proteasome system include key cell cycle machinery, as well as improperly folded and/or damaged proteins (housekeeping protein turnover).
Proper cell cycle progression requires oscillation of regulatory protein levels in all eukaryotes. Two highly conserved E3 ubiquitin ligases, the anaphase-promoting complex/cyclosome (APC/C) and Skp1-Cul1-F-box protein (SCF), are differentially regulated over the course of the cell cycle. The APC/C is activated during M phase and remains active during G1 phase (reviewed in Peters, 2006; Thornton & Toczyski, 2006; van Leuken et al., 2008; Matyskiela et al., 2009), while the SCF is active throughout the cell cycle. Excellent review articles describing the details of SCF structure, regulation, cellular targets, and cross-regulation with the APC/C are available (Peters, 1998; Cardozo & Pagano, 2004; Vodermaier, 2004; Skaar & Pagano, 2009; Ang & Harper, 2005).
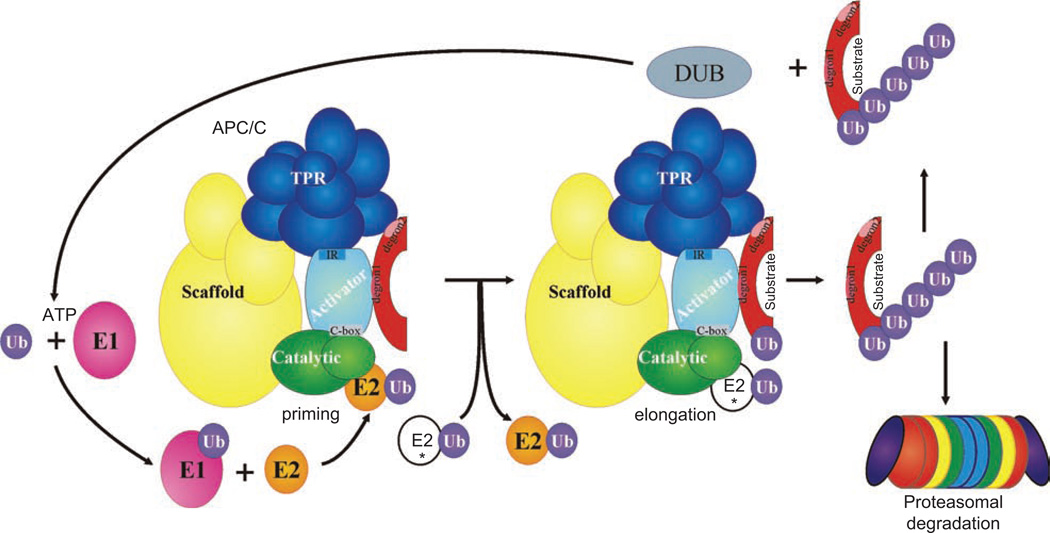
The APC/C is perhaps the most complex ubiquitin ligase known. It is composed of more than a dozen subunits (Table 1), yielding a molecular machine approximately 1 MDa in size. The APC/C can be divided into four modules: (1) catalytic (2) tetrico-peptide repeats (TPRs) (3) scaffolding, and (4) activator (Figure 1). The catalytic subunits of the APC/C include Apc11, a protein that contains a Zn+2-binding RING (really interesting new gene) domain, and Apc2, a cullin-like protein; thus, the APC/C is classified as a RING-cullin E3 ligase (Gmachl et al., 2000; Tang et al., 2001b). The TPR repeat proteins modulate activator interactions (Vodermaier et al., 2003; Kraft et al., 2005; Thornton et al., 2006; Matyskiela & Morgan, 2009). The scaffolding proteins bridge the catalytic and TPR modules, perhaps to optimize spacing between these regions for efficient catalysis. Finally, APC/C activators are members of the Cdc20 family of tryptophan-aspartate (WD) repeat proteins and are necessary for efficient substrate binding and ubiquitination (reviewed in Visintin et al., 1997; Kramer et al., 1998; Yu, 2007). Multiple recent reviews on the APC/C are available, for example (Peters, 2006; Thornton & Toczyski, 2006; van Leuken et al., 2008; Matyskiela et al., 2009). Here, we have emphasized the newest developments in the APC/C literature, particularly APC/C structure and modes of activator substrate recognition. (Note that we will use mammalian protein names except where species-specific names are required; please refer to Table 1 for clarification of nomenclature.)
Table 1.
Nomenclature of APC/C components and other proteins across species.
| S. pombe | kDa | S. cerevisiae | kDa | D. melanogaster | kDa | H. sapiens | kDa | Domain |
|---|---|---|---|---|---|---|---|---|
| APC/C subunits | ||||||||
| Cut4 | 165 | Apc1 | 196 | Shattered A/B | 227/56 | Apc1 | 217 | PC repeats |
| Apc2 | 81 | Apc2 | 100 | Morula | 92 | Apc2 | 94 | Cullin-like |
| Nuc2* | 53 | Cdc27 | 79 | Makos | 101 | Apc3 | 92 | TPR repeats |
| Lid1 | 76 | Apc4 | 85 | Apc4 | 87 | Apc4 | 92 | |
| Apc5 | 83 | Apc5 | 75 | Ida A/B | 89/81 | Apc5 | 85 | TPR repeats |
| Cut9* | 76 | Cdc16 | 95 | Cdc16 | 82 | Apc6 | 72 | TPR repeats |
| — | — | Apc7A/B | 60/70 | Apc7 | 63 | TPR repeats | ||
| Cut23* | 60 | Cdc23 | 79 | Cdc23 | 78 | Apc8 | 69 | TPR repeats |
| — | Apc9 | 31 | — | — | ||||
| Apc10 | 22 | Doc1 | 33 | Apc10 | 22 | Apc10 | 21 | |
| Apc11 | 11 | Apc11 | 19 | Lemming | 10 | Apc11 | 10 | RING domain |
| Hcn1 | 9 | Cdc26 | 14 | — | Cdc26 | 10 | ||
| Apc13 | 16 | Swm1 | 19 | — | Apcl3 | 8 | ||
| Apc14 | 12 | — | — | — | ||||
| Apc15 | 16 | Mnd2 | 43 | — | — | |||
| — | — | — | Apc16 | 12 | ||||
| Ubiquitin conjugating enzymes | ||||||||
| Ubc11 | 20 | Ubc11 | 18 | vihar/UbcH10 | 20 | UbcH10/Ube2C | 20 | E2 |
| — | — | Ube2S | 23 | Ube2S | 24 | E2 | ||
| Ubc4 | 16 | Ubc4/Ubc5 | 16 | effete/UbcD1 | 17 | UbcH5 | 17 | E2 |
| Ubc1 | 24 | Ubc1 | 24 | Ube2K | 22 | Ube2K/E2-25K | 22 | E2 |
| Critical yeast substrates | ||||||||
| Cdc13 | 56 | Clb2 | 56 | cyclin B | 60 | cyclin B | 48 | cyclin box |
| Cut2 | 33 | Pds1 | 42 | Pirn | 58 | securin | 22 | securin |
Denotes two copies of subunit present in each Schizosaccharomyces pombe APC/C complex (Ohi et al. 2007). — Denotes that subunit has not been identified in this species. Proteins listed in the same row indicate homologs.
Figure 1.
Theoretical model of APC/C ubiquitination pathway. The four APC/C modules (Catalytic=Apc2, Apc11; Scaffold = Apc1, Apc4, Apc5; TPR=Apc3, Apc6, Apc8, Cdc26, Apc10, and species-specific subunits; and Activator=Cdc20 family member) are labeled. El = Ub-activating enzyme, E2 = Ub-conjugating enzyme, IR = IR motif in Cdc20 family members, C-box= conserved motif in Cdc20 family members, degrons=APC/C degradation motifs (e.g., D box, KEN box), DUB = deubiquitinating enzyme, * indicates elongation E2.
Functions of the APC/C
Mitosis and meiosis
The yeast APC/C has two essential mitotic targets—cyclin and securin (see Table 1 for species-specific names) (Thornton & Toczyski, 2003). The APC/C ubiquitinates A and B type cyclins to downregulate mitotic cyclin-dependent kinase (CDK) activity and is essential for mitotic exit. In fact, the APC/C was discovered in yeast, frog, and clam oocyte extracts as one factor required for cyclin ubiquitination (Irniger et al., 1995; King et al., 1995; Sudakin et al., 1995). Thus, initially, the APC/C was termed the ‘cyclosome’ because of its pivotal role in cell cycle regulation (Sudakin et al., 1995). During mitosis, the APC/C is sequentially activated by two Cdc20 family members—first Cdc20 and then Cdh1 (see Table 1 for species-specific names and Regulation of the APC/C section for more detail) (reviewed in Pesin & Orr-Weaver, 2008). APC/CCdc20 promotes the metaphase to anaphase transition by ubiquitinating securin, an inhibitor of separase, the enzyme responsible for cleavage of the chromosome cohesin complex, initiating separation of sister chromatids (Cohen-Fix et al., 1996; Funabiki et al., 1996) as well as cyclin B (Holloway et al., 1993; Gorr et al., 2005). After anaphase onset, Cdc20 is ubiquitinated by APC/CCdh1 and degraded (Visintin et al., 1997; Fang et al., 1998b; Prinz et al., 1998; Shirayama et al., 1998). APC/CCdh1 then targets cyclin B and other substrates for degradation promoting mitotic exit (Fang et al., 1998b; Kramer et al., 1998; Zachariae et al., 1998; Fang et al., 1999; Blanco et al., 2000; Listovsky et al., 2000). APC/CCdh1 remains active through G1 phase to continue degradation of cyclins and perhaps limit expression of Cdc20 until mitosis (Huang et al., 2001). During mitosis, the cellular localization of the human APC/C is dynamic; it localizes to centrosomes and the mitotic spindle, but active (phosphorylated) APC/C is enriched on centrosomes (Tugendreich et al., 1995; Kraft et al., 2003). Unfortunately, APC/C localization remains difficult to study in yeasts.
Although there is less information available on the role of the APC/C during meiosis, it is clear that APC/C activity is carefully modulated to achieve the special chromosome cohesion requirements of meiotic progression. That is sister chromatid cohesion must be maintained through meiosis I, but dissolved during meiosis II. The APC/C regulates cyclin and securin levels as in mitosis, but its activity is modified during meiosis by specific activators and inhibitors (see Regulation of the APC/C section) (Table 2) to prevent complete cyclin degradation between meiosis I and meiosis II, premature chromosome segregation and meiotic exit (Cooper et al., 2000; Asakawa et al., 2001; Blanco et al., 2001; Diamond et al., 2009) (reviewed in Irniger, 2006). The APC/C is required for homologous chromosome separation (meiosis I) in Saccharomyces cerevisiae, and securin (separase inhibitor) is targeted for destruction by the APC/C during both meioses in yeasts and mammals (Jin et al., 2010; Salah & Nasmyth, 2000); however, shugoshin protects sister chromatid cohesin from separase cleavage during meiosis I (Katis et al., 2004; Kitajima et al., 2004; Marston et al., 2004; Rabitsch et al., 2004) and is then targeted by the APC/C for degradation (Salic et al., 2004; Penkner et al., 2005; Karamysheva et al., 2009) to promote efficient and timely chromosome segregation in meiosis II. In mammals, where female meiosis is especially protracted (oocytes are blocked in prophase I before birth and resume meiosis in sexually mature females, often years later), loss of cohesin and shugoshin in older females correlates with increased homolog separation errors during meiosis I, resulting in more trisomy births (Lister et al., 2010; Revenkova et al., 2010).
Table 2.
APC/C activators and inhibitors.
| S. pombe | S. cerevisiae | D. melanogaster | X. laevis | H. sapiens | Phase | |||
|---|---|---|---|---|---|---|---|---|
| Activators | ||||||||
| * | Slp1 | Cdc20 | Fizzy | Cdc20/Fizzy | Cdc20/Fzy/p55CDC | M/Me | ||
| Ste9/Srw1 | Cdh1/Hct1 | Fizzy-related | Fizzy-related | Cdh1/Fzr1 | M/Me/G1 | |||
| Mfr1 | — | — | — | — | Me | |||
| — | Ama1 | — | — | — | Me | |||
| — | — | Cortex | — | — | Me | |||
| Inhibitors | Form inhibited | |||||||
| Rca1 | Emi1 | Emi1 | M/G1 | APC/CCdh1 | ||||
| — | — | — | Erp1 | Emi2 | Me | |||
| Mad2 | Mad2 | Mad2 | Mad2 | Mad2 | M | Apc/cCdc20 | ||
| Mad3 | Mad3 | BubR1 | BubR1 | BubR1 | M | |||
| * | Mes1 | — | — | — | — | Me | ||
| * | — | Acm1 | — | — | — | M/G1 | Apc/CCdh1 | |
Also an APC/C substrate.
M = mitosis; Me = meiosis; G1 = G1 phase.
Neuronal differentiation
In postmitotic neurons, the APC/C regulates axon and dendrite morphogenesis depending on its localization and bound activator (Konishi et al., 2004). While APC/CCdh1 localizes to the nucleus and inhibits axon growth, APC/CCdc20 localizes to dendrites and promotes dendrite growth and arborization (Yang et al., 2010). For a detailed review on the role of the APC/C in neuronal development and patterning, we refer the reader to a recent article by Yang and co-workers (2010).
Cancer, viruses, and the APC/C
Given the crucial roles played by the APC/C in maintenance of genetic fidelity through cell division, it is not surprising that many cancers and viruses have deregulated or usurped the APC/C, its activators, or conjugate E2 enzymes to escape cell death and/or promote viral reproduction. Mutations in APC/C subunits have been found in colon cancer cell lines and primary human colon cancer cells (Wang et al., 2003), and Cdc20 is overexpressed in some cancers, perhaps to promote cell division even in the presence of spindle defects and genetic aberrations (Chen et al., 2006; Yuan et al., 2006; Thirthagiri et al., 2007). The human APC/C’s cognate E2s (see Catalysis section), UbcH10, and Ube2S, are overexpressed in primary tumors (Okamoto et al., 2003; Tedesco et al., 2007; van Ree et al., 2010) and ectopic expression of either UbcH10 or Ube2S promotes cell transformation and tumor invasion (Okamoto et al., 2003; Jung et al., 2006). Ube2S expression inversely correlates with the stability of the tumor suppressor von Hippel-Lindau E3 ligase (Jung et al., 2006), providing one potential mechanism of cell transformation for this E2. However, both of these E2 enzymes likely affect cell transformation and proliferation via the APC/C.
The APC/C is reported to regulate a number of genes involved in tumor suppression. For example, the APC/CCdc20 directly regulates p21, a CDK inhibitor, in prometaphase (Amador et al., 2007). APC/CCdh1 interacts with the tumor suppressor Rb (retinoblastoma) protein, presumably to direct the APC/C to specific substrates (Binne et al., 2007). Both Cdc20- and Cdh1-bound APC/C may be inhibited by RASSF1A, a centrosomal tumor suppressor protein (Mathe, 2004; Song & Lim, 2004; Song et al., 2004; Liu et al., 2007; Whitehurst et al., 2008). Furthermore, APC/CCdh1 is required for stable G1 phase and governs the switch between cell proliferation and differentiation (Garcia-Higuera et al., 2008; Skaar & Pagano, 2008) and misregulation of APC/CCdh1 in differentiated cells can lead to cell cycle reentry, dedifferentiation, and potentially carcinogenesis (Wirth et al., 2004; Wasch et al., 2010).
Viruses also appear to highjack the APC/C or its regulators to promote viral replication and/or cell transformation and carcinogenesis. The Human T lymphoma virus type-1 (HTLV-1) protein Tax is reported to impact APC/C function by interacting with Mad1, Cdc20, and securin, effectively mitigating the spindle assembly checkpoint (SAC, an inhibitor of anaphase and the APC/C, see Regulation of the APC/C section) while activating the APC/C to allow mitosis to proceed unchecked (Grassmann et al., 2005). Tax binding to the APC/C causes premature ubiquitination and degradation of cyclin B and securin, promoting aneuploidy and cancer initiation (Grassmann et al., 2005; Liu et al., 2005). An adenoviral protein, E4orf4, also interacts with Cdc20 and may also promote carcinogenesis in a similar way (Mui et al., 2010). Apoptin, another viral protein, is reported to inhibit the APC/C to induce G2/M arrest and p53-independent apoptosis, facilitating viral egress and infection of new cells (Teodoro et al., 2004). Human cytomegalovirus infection triggers a G1/S arrest (and E2F-dependent transcription), and appears to induce degradation of core APC/C subunits Apc4 and Apc5 (Wiebusch et al., 2005; Tran et al., 2008; Tran et al., 2010), promoting viral infection and replication. Finally, a poxvirus protein, with homology to Apc11 (APC/cyclosome regulator or PACR) causes cell cycle deregulation and accumulation of APC/C substrates (Mo et al., 2009).
Substrates and their recognition
Destruction signals
The recognition and destruction of many APC/C subunits depends on the presence of short conserved sequence motifs referred to as degrons or destruction motifs. A combination of genetic and biochemical studies has led to the characterization of a number of these specific sequences. At least eight different APC/C targeting motifs in substrates have now been identified (see Table 3). Few of the destruction sequences contain ubiquitination sites; however, in the case of cyclin B, the number of lysines (site of ubiquitin attachment) is highly enriched in the region C-terminal to the destruction box (D box) (Glotzer et al., 1991). Mutations in degron sequences often stabilize APC/C substrates, while inserting a degron into a protein that is not an APC/C substrate promotes its ubiquitination (Glotzer et al., 1991). Interestingly, degrons are most often found in disordered regions of proteins, as was found for both securin and cyclin B (Cox et al., 2002). APC/C degrons are very short sequences that are relatively common in the proteome, making it clear that the mere presence of a degron consensus sequence does not signify a bona fide APC/C substrate. Structural studies examining APC/C-substrate interactions will be required to understand the specific criteria of APC/C degron recognition.
Table 3.
APC/C destruction motifs.
| Destruction motif | Sequence | References |
|---|---|---|
| D box | RxxLxxxN | Glotzer et al., 1991 |
| KEN box | KENxxxN | Pfleger Kirschner, 2000 |
| A box or DAD (D box activating domain) |
RxLxPSN | Castro et al., 2002; Littlepage Ruderman, 2002 |
| CRY box | CRYxPS | Reis et al., 2006 |
| GxEN | GxEN | Castro et al., 2003 |
| O box | LxEKN | Araki et al., 2005 |
| Spo13 | LxExxxN | Sullivan Morgan, 2007 |
| TEK box | R/KxxTxKT | Jin et al., 2008 |
“x” indicates any amino acid.
The most common degradation motifs found in APC/C substrates are the D box and the KEN box. The D box was first characterized in cyclin B (Glotzer et al., 1991) (RxxLxxxN) and contains at a minimum an arginine and a leucine separated by two residues. This destruction signal is found in both APC/CCdc20 and APC/CCdh1 substrates. The KEN box (KENxxxE/D/N) is recognized by APC/CCdh1 and is often found in APC/C substrates targeted for destruction after Cdc20 dependent substrates (Visintin et al., 1997; Pfleger & Kirschner, 2000; Bashir & Pagano, 2004).
In addition to the D box and KEN box, six other APC/C destruction motifs have been characterized including the TEK box (Jin et al., 2008), the A box (Littlepage & Ruderman, 2002), the GXEN box (Castro et al., 2002), the CRY box (Reis et al., 2006), the O box (Araki et al., 2005) and a sequence specific to Spo13 (Sullivan & Morgan, 2007) (Table 3). The TEK box consensus (R/KxxTxKT) forms a charged patch around K11 in Ub and human APC/C target lysines, directing K11 Ub chain formation (Jin et al., 2008); however, this motif is not found in yeast APC/C substrates. The other motifs listed above are not as common in APC/C substrates and, in general, they are often found in substrates that contain additional degrons making it unclear how they contribute to APC/C-substrate recognition. For example, the mitotic kinase Aurora A contains an A box, a D box and a KEN box. Although it is not clear whether the A box directly interacts with APC/CCdh1, this sequence is required for activation of the D box in Aurora A (Crane et al., 2004). It has been postulated that dephosphorylation of a conserved serine in the A box during mitotic exit allows APC/CCdh1 to recognize Aurora A’s previously silent D box (and possibly the A box too) and precisely control the timing of Aurora A destruction (Littlepage & Ruderman, 2002; Crane et al., 2004; Kitajima et al., 2007). Thus, it is likely that important auxiliary sequences help the APC/C recognize its degrons.
Modification of APC/C substrates by phosphorylation near or in degron sequences has also been shown to modify APC/C-substrate interaction. For example, in certain cases phosphorylation in or near a degron protects a substrate from APC/C-dependent ubiquitination, as is the case for Aurora A (Littlepage et al., 2002; Crane et al., 2004; Kitajima et al., 2007) and securin (Holt et al., 2008). However, there are other examples where phosphorylation in or near degrons may be necessary for APC/C-mediated ubiquitination, such as for Mcl1-1 (Harley et al., 2010) and S. cerevisiae Cdc5 (Simpson-Lavy et al., 2009). Because degrons are often found in disordered regions of proteins (Cox et al., 2002), it is possible that changes in phosphorylation affect the overall structure of the degron region making the motif more or less accessible to the APC/C.
Activators mediate substrate recruitment
The activation of the APC/C is mediated in part by the binding of transiently available subunits typified by the founding member of the family, Cdc20 (reviewed in Harper et al., 2002; Peters, 2006; Thornton & Toczyski, 2006; Yu, 2007). Cdc20 binds the core APC/C in mitosis and meiosis and is critical for anaphase initiation. A second family member, Cdh1, plays distinct roles in mitotic exit and G1 phase. Meiotic, species-specific APC/C activators (Mfr1 and Ama1) have been characterized in yeasts (Cooper et al., 2000; Blanco et al., 2001) and Drosophila (Cortex) (Pesin & Orr-Weaver, 2007; Swan & Schupbach, 2007) and likely exist in higher eukaryotes.
APC/C activators could facilitate substrate ubiquitination through a number of mechanisms including simple tethering of substrates or E2 enzymes to the APC/C to modulation of APC/C E3 ligase activity through conformational changes. Indeed, the Cdc20 family of activators has been shown to bind APC/C substrates (Burton & Solomon, 2001; Hilioti et al., 2001; Pfleger et al., 2001; Schwab et al., 2001; Kraft et al., 2005; Kimata et al., 2008b; da Fonseca et al., 2010; Buschhorn et al., 2010), activate ubiquitination (Kimata et al., 2008a), and recruit one of its conjugate E2 enzymes, Ube2S (Williamson et al., 2009). Cdc20 proteins contain three domains important for their role in modulation of APC/C activity. The largest and most prominent are the C-terminal WD40 repeats that can be directly cross-linked to substrates (Kraft et al., 2005; Kimata et al., 2008b) making this region a substrate-binding platform. The C-terminus also contains a short IR-motif that interacts directly with the S. cerevisiae TPR-containing proteins Cdc23 (Apc8) and Cdc27 (Apc3) (Vodermaier et al., 2003; Matyskiela & Morgan, 2009), suggesting that this domain tethers Cdc20 and Cdh1 to the APC/C. However, the IR domain cannot represent the only and/or most important APC/C-activator interaction because this Cdc20 domain is nonessential (Yamada et al., 2000; Thornton et al., 2006). The N-terminus contains an eight amino acid motif called the C-box (Schwab et al., 2001) that has been shown to both bind the APC/C and modulate its E3 ligase activity (Thornton et al., 2006; Kimata et al., 2008a), making this conserved motif of the Cdc20 family the only region currently shown to actually increase APC/C activity toward substrates. Interestingly, two structural studies have shown that adding recombinant Cdh1 or Cdc20 to the apo-APC/C (APC/C core subunits) causes the APC/C to adopt a more ‘open’ conformation (Dube et al., 2005; Herzog et al., 2009). How this more ‘open’ structure affects APC/C activity is still not clear, but these results confirm that both Cdh1 and Cdc20 function as more than simple substrate tethers.
APC/C-substrate interaction
Although Cdc20 and Cdh1 are clearly important for facilitating APC/C-substrate interactions, there is accumulating evidence that substrates can also interact directly with core components, often in a degron-dependent manner (Yamano et al., 1998; Meyn et al., 2002; Passmore et al., 2003; Carroll et al., 2005; Eytan et al., 2006; Hayes et al., 2006; da Fonseca et al., 2010; Buschhorn et al., 2010). These findings suggest that the APC/C core contains degron binding and/or substrate recognition domains, in addition to those found on the activators. One prime candidate for mediating the interaction between the APC/C core and substrates is Doc1/Apc10, which is essential for APC/C interaction with substrates and processivity of the ubiquitination reaction (Carroll and Morgan, 2002; Passmore et al., 2003). Indeed, two recent cryo-EM structural studies of the S. cerevisiae APC/C have shown that the D box directly contacts both Cdh1 and Doc1, creating a bridge between these two components (da Fonseca et al., 2010; Buschhorn et al., 2010). In addition, results from a recent analysis of the S. cerevisiae APC/C showed that the TPR subunits facilitate activator and perhaps substrate binding, with the TPR proteins serving as a platform for substrates to bivalently interact with the activator and core APC/C subunits (Matyskiela & Morgan, 2009).
Thus, although the activators are important for initially recruiting substrates to the APC/C, binding affinity for the substrate and processivity of the ubiquitination reaction are likely dictated by interaction of substrates with APC/C core components and activators. Furthermore, APC/C core-substrate interactions may be strengthened or modified by structural changes to the APC/C core induced by activator binding (Kimata et al., 2008a; Herzog et al., 2009). The ability of some substrates to bind more tightly to the APC/C, perhaps by engaging in multivalent interactions with core subunits, could also affect the order of substrate recognition by the APC/C and processivity of ubiquitination as proposed by Rape and co-workers (Rape et al., 2006). Finally, it has been known for decades that the small phosphobinding protein of Cdc2-cyclin, termed Suc1/Cks1, is required for the onset of anaphase (Hayles et al., 1986; Kaiser et al., 1999) and that active APC/C can be purified using Suc1 (Sudakin et al., 1997). Several studies have shown that Cks1 is necessary for efficient cyclin recruitment and ubiquitination by the APC/C (Patra & Dunphy, 1998; Wolthuis et al., 2008; Di Fiore & Pines, 2010; van Zon et al., 2010). A recent study by Di Fiore and Pines showed that Cdc20-bound cyclin A is targeted to the APC/C by Csk1 in a CDK-independent manner when the SAC is activated (Di Fiore & Pines, 2010). Another recent study showed that cyclin B-CDK complexes can be targeted by Cks1 to the SAC-inhibited APC/C in prometaphase, such that the complex is poised to ubiquitinate cyclin B in metaphase once the SAC has been satisfied (van Zon et al., 2010). The exact mechanism by which Cks1 selectively facilitates cyclin destruction is not known but might involve the need to extract cyclin from its Cdk1 partner at some point in the ubiquitin-mediated degradation process. In summary, the complicated web of interactions required for efficient ubiquitination of a large number of structurally diverse substrates at variable cell cycle times may explain why the APC/C is so complex compared to other E3 ligases.
Regulation of the APC/C
Activator role
As discussed in previous sections, the APC/C is activated by the Cdc20 family of proteins (Table 2). During mitosis, the APC/C is temporally regulated by binding to the activators Cdc20 and Cdh1 (reviewed in Pesin & Orr-Weaver, 2008). During metaphase/anaphase, APC/CCdc20 promotes cyclin B and securin degradation to trigger anaphase (Holloway et al., 1993; Cohen-Fix et al., 1996; Funabiki et al., 1996; Gorr et al., 2005). Degradation of cyclin B downregulates CDK activity, leading to the removal of inhibitory CDK phosphorylation sites on Cdh1 by Cdc14 phosphatases (Jaspersen et al., 1999; Kotani et al., 1999; Visintin et al., 1998), promoting association of Cdh1 with the APC/C. APC/CCdh1 marks Cdc20, cyclin B, and other key substrates to facilitate mitotic exit and then remains active during G1 phase to maintain low mitotic cyclin levels (Fang et al., 1998b; Kramer et al., 1998; Zachariae et al., 1998; Fang et al., 1999; Blanco et al., 2000; Listovsky et al., 2000). Cdh1 is nonessential from yeast to man for mitotic exit, but loss of Cdh1 in human RNAi experiments and mouse knockout models resulted in increased incidence of tumors and genome instability (Engelbert et al., 2008; Garcia-Higuera et al., 2008; Li et al., 2008). Cdc20 is essential in yeasts (Hartwell et al., 1973; Kim et al., 2010a) and required for mitosis during mouse embryogenesis, confirming that Cdc20 is also essential in mammals (Li et al., 2007).
During meiosis, the APC/C is activated by Cdc20 and other Cdc20 family members to tailor ubiquitin-mediated degradation events to meiotic progression (see Functions of the APC/C and Table 2). In the fission yeast, Schizosaccharomyces pombe, a Cdc20 family member, Mfr1 (meiotic fizzy-related 1), is required for cyclin degradation at the end of meiosis II and coordinates nuclear division with spore formation (Blanco et al., 2001). In budding yeast, Ama1 (activator of meiotic APC 1), another Cdc20 family activator only expressed during meiosis, is required for cyclin degradation (Cooper et al., 2000), securin degradation (Oelschlaegel et al., 2005), and spore wall formation (Coluccio et al., 2004). Although it is clear that Ama1 plays a role in meiotic progression, there are conflicting reports of the ama1Δ phenotype; Cooper et al. report that ama1Δ cells are stalled in meiosis I, whereas Coluccio et al. find that ama1Δ cells complete meiosis but not spore formation (Cooper et al., 2000; Coluccio et al., 2004). A recent report suggests that the essential role of Ama1 is actually to coordinate meiotic exit and cytokinesis during spore formation (Diamond et al., 2009). Drosophila melanogaster utilizes a meiosis-specific Cdc20 family activator called Cortex (see Table 2) (Swan & Schupbach, 2007) and it is likely that higher eukaryotes also utilize meiosis-specific APC/C activators that have not yet been identified.
Phosphorylation
Another layer of APC/C regulation is provided by post-translational modification of the core subunits, especially by protein kinases and phosphatases. Multiple subunits of the APC/C are phosphorylated in mitosis when the APC/C is active (Kraft et al., 2003; Steen et al., 2008; Wilson-Grady et al., 2008; Beltrao et al., 2009; Holt et al., 2009) and phosphatase treatment of the APC/C inactivates it (King et al., 1995; Lahav-Baratz et al., 1995; Kramer et al., 2000). Phosphorylation of the core APC/C has been correlated with an increased affinity for Cdc20 (Fang et al., 1998b; Shteinberg et al., 1999; Kramer et al., 2000; Yamada et al., 2000; Kraft et al., 2003), a change in APC/C localization (Huang et al., 2007; Torres et al., 2010), and binding to Cks1-Cdk-cyclin complexes (Di Fiore & Pines, 2010; van Zon et al., 2010).
Cdk1 is required for APC/C activation but is not the only protein kinase involved in APC/C regulation (Lahav-Baratz et al., 1995; Kotani et al., 1998). In vitro, purified Cdk1 phosphorylates and activates the conserved mammalian kinase, Plk1 (see Table 1), which in turn can lead to APC/C activation (Kotani et al., 1998). Both protein kinases can phosphorylate multiple subunits of the complex in vitro (Harper et al., 2002). Protein kinase A (PKA) can also phosphorylate several members of the complex but PKA phosphorylation events are inhibitory (Kotani et al., 1998), consistent with the observed genetic interactions between S. pombe mutants affecting PKA activity and APC/C function (Yamashita et al., 1996; Yamada et al., 1997). While several components of the purified complex are phosphoproteins (Peters et al., 1996; Yamada et al., 1997; Kotani et al., 1998; Rudner & Murray, 2000; Steen et al., 2008; Holt et al., 2009), it is not yet clear which phosphorylation events on which components are important for altering activity and/or protein-protein interactions.
One notable investigation of APC/C phosphorylation was that of Rudner and Murray (2000). In this study, all consensus Cdk1 phosphorylation sites in S. cerevisiae Cdc27 (Apc3), Cdc16 (Apc6), and Cdc23 (Apc8) were altered to alanines. There were some defects in mitotic exit, Cdc20 binding, and increased sensitivity to the spindle checkpoint when cells produced the APC/C complex missing these serine and threonine residues. Similarly, loss of the sole Cdk1 site on S. pombe Hcn1/Cdc26 led to a mild defect in APC/C function (Yoon et al., 2006). However, the lack of significant defects in the context of previous data indicating an essential role of APC/C phosphorylation argued that critical phosphorylation sites had not been eliminated in these studies.
One comprehensive investigation of human APC/C phosphorylation indicated that there are at least 43 sites of phosphorylation within the human APC/C, 34 of which are specific to mitosis (Kraft et al., 2003). The approach taken to identify sites was mass spectrometry; however, the entire APC/C was not represented in the data (Kraft et al., 2003) indicating that the phosphorylation landscape of the APC/C might be even more complex. Indeed, other large-scale mass spectrometry studies revealed more APC/C phosphorylation sites (Steen et al., 2008; Wilson-Grady et al., 2008; Beltrao et al., 2009; Holt et al., 2009; Mazanek et al., 2010). However, many valuable lessons were learned by the analysis of phosphosites by Kraft and colleagues (2003). Cdk1 was able to phosphorylate many of the identified sites, Plk1 was able to phosphorylate others and still others were not phosphorylated by either of these protein kinases. While Kraft and co-workers proposed that Cdk1 phosphorylation alone can activate the APC/C to some extent in vitro, this and previous studies indicated that Cdk1 and Plk1 most likely cooperate to activate the APC/C (Kraft et al., 2003). The fact that Cdk1 phosphorylation can provide a docking site for Plk1 (Elia et al., 2003) raises the possibility that APC/C phosphorylation events might be ordered (Cdk1 followed by Plk1) and thus, have different consequences. It is also evident from the work of Kraft et al. (2003) and other studies that additional protein kinases are involved in APC/C phosphorylation. It is of course possible that some phosphorylation events inhibit aspects of APC/C function. Indeed mitotic phosphorylation of the APC/C core component and meiotic inhibitor, Mnd2, during mitosis has been implicated as a regulatory switch for its role in APC/C inhibition during meiosis (Torres & Borchers, 2007). Clearly, it will require a comprehensive analysis of the phosphorylation sites of several subunits in order to understand an apparently complex phospho-regulatory scheme.
Inhibition of the APC/C by the SAC
Prior to anaphase, the APC/C is kept inactive by the spindle assembly checkpoint (SAC) (reviewed in Malmanche et al., 2006; Chen, 2007; Ciliberto & Shah, 2009; Musacchio & Salmon, 2007). The SAC ensures that a proper mitotic spindle is formed and all chromosomes are attached and aligned at the metaphase plate prior to chromosome segregation, a key to maintenance of genetic fidelity and prevention of aneuploidy and carcinogenesis. The spindle checkpoint mediates APC/C activity by inhibiting Cdc20; however, the mechanism of inhibition remains an area of intense research. SAC proteins, Mad2 and BubR1 (Mad3), bind Cdc20 either independently or as a single inhibitory complex called the mitotic checkpoint complex (MCC) (Fang et al., 1998a; Sudakin et al., 2001; Tang et al., 2001a; Millband & Hardwick, 2002; Tang et al., 2004) and a checkpoint kinase, Bub1, phosphorylates and inhibits Cdc20 activity (activation of the APC/C) (Tang et al., 2004). The ability of Mad2 and BubR1 to form complexes with Cdc20 suggests a model wherein the checkpoint proteins bind and sequester Cdc20, preventing it from activating the APC/C. However, there is also evidence that the MCC interacts directly with the APC/C core (Morrow et al., 2005; Sczaniecka et al., 2008; Herzog et al., 2009), converting the APC/C’s overall structure to a more ‘closed’ conformation and inhibiting catalysis (Herzog et al., 2009). In addition, Mad3 (BubR1) contains a KEN box and is proposed to act as a pseudosubstrate, blocking interaction of the APC/C with bona fide substrates (Burton & Solomon, 2007; King et al., 2007; Sczaniecka et al., 2008). In fact, recent structural studies showing that the location of D box binding to the APC/C overlaps with the binding position of the MCC provide even further evidence supporting this model (Buschhorn et al., 2010; da Fonseca et al., 2010). Yet, another proposed model suggests that MCC interaction with the APC/C leads to Cdc20 degradation, preventing APC/C activity toward substrates such as cyclin B and securin (Pan & Chen, 2004; Nilsson et al., 2008). Finally, SAC activation and maintenance is affected by a class of CDK activators (RINGO/Speedy) distinct from cyclins that act in recruitment of SAC components and Aurora kinase to unattached kinetochores (Mouron et al., 2010).
From the above discussion, it is clear that there are substrates, such as Cdc20, cyclin A, Nek2A, and HOXC10 that are ubiquitinated and degraded even when the SAC is active (den Elzen & Pines, 2001; Geley et al., 2001; Hames et al., 2001; Fry, 2002; Gabellini et al., 2003; Nilsson et al., 2008; Wolthuis et al., 2008). How these substrates are distinguished from substrates such as cyclin B is still actively being investigated; however, there are a number of hypotheses that could explain this phenomenon. For example, it is possible that checkpoint independent substrates have distinct features that allow them to interact and be ubiquitinated by SAC-inhibited APC/C. This explanation is not completely satisfactory, because SAC-independent substrates, such as Nek2A and Cdc20, have degron sequences that are required for ubiquitination and degradation (Fry & Yamano, 2006; Hayes et al., 2006). Another theory is that checkpoint-independent APC/C substrates may be more efficiently targeted to APC/CCdc20 particles that have yet to be inhibited by the SAC. Further analysis of APC/C structure and cellular localization patterns is required to improve our understanding of how SAC signals selectively inhibit APC/C activity toward some but not all substrates.
Release of the APC/C from SAC inhibition is also not completely understood. One model of APC/C inhibition by the SAC describes an anaphase switch in which the human APC/C ubiquitinates Cdc20 to dissociate the inhibitory MCC and allow activation of the APC/C, while the deubiquitinating enzyme USP44 reverses this modification to strengthen APC/C inhibition by stabilizing the Cdc20-MCC interaction (Reddy et al., 2007; Stegmeier et al., 2007). It is not yet clear whether this is a general mechanism because deubiquitinating enzymes have not been implicated in SAC release in other organisms. A potentially parallel mechanism of SAC inactivation is catalyzed by competitive binding of p31comet to Mad1- or Cdc20- associated Mad2, which results in dissociation of the MCC and activation of the APC/C (Xia et al., 2004; Mapelli et al., 2006; Yang et al., 2007). Other groups have confirmed that polyUb by the APC/C is required for checkpoint release in human cells and in particular that the E2, Ube2S, is required for release from prolonged SAC inhibition (Garnett et al., 2009; Williamson et al., 2009; Miniowitz-Shemtov et al., 2010). Miniowitz-Shemto and co-workers (2010) also showed that an unknown ATP-dependent process involving a β-γ cleavage of ATP is necessary for SAC release. Finally, the protein phosphatase, PP1 (Dis2), has been shown to effectively release the SAC in two divergent yeast by antagonizing Aurora kinase, an essential SAC component (Pinsky et al., 2009; Vanoosthuyse and Hardwick, 2009). A cohesive model of SAC release from yeast to humans has yet to be elucidated and will require better understanding of APC/C localization, binding partners and modification status before, during, and after checkpoint release.
During meiosis, APC/C activity must be temporally restricted by inhibitory proteins to prevent (1) complete degradation of cyclin B between meiosis I and meiosis II and (2) premature chromosome segregation (Shonn et al., 2000; Bernard et al., 2001; Izawa et al., 2005; Oelschlaegel et al., 2005; Penkner et al., 2005; Kimata et al., 2008b) (reviewed in Irniger, 2006). Meiotic inhibition of the APC/C is mediated by the SAC (Shonn et al., 2000; Bernard et al., 2001) and other meiosis specific factors (see section below). SAC components are essential for timing meiotic progression from yeast to mammals; loss of SAC function during meiosis accelerates meiotic progression and/or chromosome missegregation (Shonn et al., 2000; Tsurumi et al., 2004; Homer et al., 2005a; Homer et al., 2005b; Wei et al., 2010) and overexpression induces a meiosis I arrest (He et al., 1997; Sironi et al., 2001; Wassmann et al., 2003a; Wassmann et al., 2003b; Niault et al., 2007; Li et al., 2009; Wei et al., 2010).
Other APC/C inhibitors
As mentioned above, APC/C activity is restrained during meiosis by inhibitory proteins to permit efficient and faithful meiotic progression. In the fission yeast S. pombe, APC/CCdc20 is partially inhibited by Mes1 (meiosis II protein), preventing the complete degradation of cyclin during meiosis I and making the cell competent for meiosis II (Izawa et al., 2005; Kimata et al., 2008b). Mes1 restrains APC/CCdc20 by competing with other substrates (e.g., cyclin) for Cdc20 binding and this inhibition is released by ubiquitin-mediated degradation of Mes1 (Kimata et al., 2008b). In budding yeast, the activity of APC/CAma1 is inhibited during meiosis by an APC/C subunit called Mnd2 (meiotic nuclear divisions 2) to prevent premature chromosome segregation prior to meiosis II (Oelschlaegel et al., 2005; Penkner et al., 2005). The APC/C is inhibited in similar ways in higher eukaryotes. For instance, Emi2 (XErp1), an inhibitor of the APC/C, is an essential regulator of meiosis (Reimann and Jackson, 2002; Ohsumi et al., 2004; Tung and Jackson, 2005) and, as an essential component of cytostatic factor, blocks unfertilized eggs in metaphase II (Rauh et al., 2005; Schmidt et al., 2005; Tung et al., 2005; Schmidt et al., 2006).
Yeast and higher eukaryotes utilize mitotic inhibitors to restrain APC/C activity until the appropriate time(s) in mitosis (Reimann et al., 2001; Martinez et al., 2006; Dial et al., 2007; Enquist-Newman et al., 2008). Acm1 inhibits APCCdh1 in interphase and is then degraded in late mitosis (Martinez et al., 2006; Dial et al., 2007; Enquist-Newman et al., 2008; Hall et al., 2008). In higher eukaryotes, Emi1 (Rca1), initially discovered in Drosophila is essential for regulation of G1 phase during eye development and asymmetric cell division in the central nervous system (Dong et al., 1997; Lear et al., 1999; Wai et al., 1999). Emi1 couples mitosis and DNA replication by inhibiting APC/CCdh1 to stabilize cyclin and geminin and promote mitosis (Grosskortenhaus & Sprenger, 2002; Hsu et al., 2002; Di Fiore & Pines, 2007) and prevents re-replication (Machida and Dutta, 2007; Zielke et al., 2008). The Emi proteins (Emi1 and Emi2) may both mediate APC/C inhibition via competitive pseudosubstrate binding (Miller et al., 2006) and/or interaction of their C-termini with the APC/C core (Ohe et al., 2010; Tang et al., 2010) preventing substrate binding or APC/C activation, respectively.
Catalysis
E2 interaction and role of priming and elongation
E2-E3 interactions are tailored to provide substrate and ubiquitin chain specificity (see Ub chain topology section below) (Kirkpatrick et al., 2006; Jin et al., 2008; Matsumoto et al., 2010). The APC/C subunit Apc11 binds E2 ubiquitin-conjugating enzymes via its RING domain and mediates specific ubiquitination events (Leverson et al., 2000; Ye and Rape, 2009). In yeasts and vertebrates, the APC/C interacts with more than one E2 to accomplish substrate and site-specific processive ubiquitination (Osaka et al., 1997; Seino et al., 2003; Rodrigo-Brenni & Morgan, 2007; Garnett et al., 2009; Williamson et al., 2009). Evidence in yeast and human systems suggests that the roles of the E2s are complementary—one for initial substrate modification (priming) and another for ubiquitin chain elongation (Seino et al., 2003; Rodrigo-Brenni & Morgan, 2007; Garnett et al., 2009; Williamson et al., 2009) (Figure 1).
The first evidence for the priming and elongation model was reported by Seino and co-workers (2003) in S. pombe. Ubc4 and Ubc11, the S. pombe homologs of human UbcH5 (84% identity) and UbcH10 (61% identity), respectively, are required for mitotic progression in S. pombe and depletion of either E2 results in mitotic arrest and cyclin accumulation (Seino et al., 2003). Furthermore, in the absence of Ubc4, Ubc11 forms short ubiquitin chains on APC/C substrates. Conversely, Ubc4 cannot ubiquitinate APC/C substrates in the absence of Ubc11, suggesting that Ubc11 is required for initial substrate ubiquitination. Similar models have been reported in budding yeast (Ubc1 and Ubc4) (Rodrigo-Brenni & Morgan, 2007) and human (UbcH10 and Ube2S) systems (Garnett et al., 2009; Williamson et al., 2009). It has long been thought that human UbcH5, a promiscuous E2 that can ubiquitinate APC/C substrates in vitro (Yu et al., 1996; Summers et al., 2008), is the chain elongation APC/C cognate, but recent evidence suggests that the true human APC/C cognate E2s are UbcH10 (priming) and Ube2S (elongation) (Garnett et al., 2009; Williamson et al., 2009).
UbcH10 was the first E2 enzyme identified as an important player in cyclin degradation (Townsley et al., 1997) and knockdown of UbcH10 phenocopies loss of APC/C function (Townsley et al., 1997; Bastians et al., 1999). UbcH10 is degraded in an APC/-dependent manner at the end of mitosis, effectively de-activating the APC/C as other APC/C substrates become limiting (Rape & Kirschner, 2004; Walker et al., 2008; Williamson et al., 2009). However, this mechanism of APC/C regulation does not appear to be conserved in yeasts because Ubc11 (UbcH10 homolog) protein expression is not cell cycle regulated (data not shown (Osaka et al., 1997)).
Interestingly, UbcH10 has a distinctive N-terminal extension (ca. 30 amino acids), which affects its ability to be charged with ubiquitin and in turn alters the regulation and substrate specificity of the APC/C (Huang et al., 2008; Summers et al., 2008). The UbcH10 N-terminal extension is conserved from S. pombe to humans, but is not present in S. cerevisiae, and it might contact APC/C subunits other than Apc11 (RING finger protein) (Tang et al., 2001b; Summers et al., 2008).
Ub chain topology
Ub chains are assembled by a condensation reaction catalyzed by an E2-E3 pair that forms an isopeptide bond between the C-terminal glycine of one Ub molecule to lysine residues of subsequent Ub molecules. Ub contains seven lysine residues (K6, K11, K27, K29, K33, K48, and K63) and chains formed via specific lysine linkages elicit unique cellular responses (Pickart & Fushman, 2004). It has long been known that K48 chains of at least four Ub molecules target substrates to the 26S proteasome for degradation (Thrower et al., 2000), but recent evidence strongly suggests that both K11 and K48 are capable of targeting proteins for degradation (Rodrigo-Brenni & Morgan, 2007; Jin et al., 2008; Xu et al., 2009). In fact, all polyUb chain structures can be targeted by the proteasome, albeit with different efficiencies (Xu et al., 2009). The topology of APC/C-catalyzed Ub chains differs between budding yeast (K48) and human (K11) (Kirkpatrick et al., 2006; Rodrigo-Brenni & Morgan, 2007; Jin et al., 2008; Garnett et al., 2009; Williamson et al., 2009; Bremm et al., 2010; Wu et al., 2010). This switch in topology is likely due to the absence of K11 specificity factors (TEK box and Ube2S) in budding yeast (Jin et al., 2008).
Structure
As discussed in previous sections, many questions remain regarding the mechanism of APC/C action and regulation, including how the core APC/C interacts with substrates, how processive transfer of Ub to substrates is accomplished, and how APC/C regulators stimulate or inhibit E3 ligase activity. Just as structural studies of other large macromolecular machines, such as the proteasome, RNA polymerase and the ribosome, have proven instrumental in unraveling the mechanisms of protein degradation, transcription, and protein synthesis, respectively, generating a detailed three-dimensional (3D) map of the APC/C will be crucial in developing a mechanistic model for its function.
Efforts to structurally characterize the holo-APC/C using X-ray crystallography have been difficult due to cell cycle specific changes in composition and posttranslational modifications of the APC/C, as well as its low cellular abundance. In addition, the large size of the complex has made reconstituting the APC/C using purified components a daunting task. For these reasons, atomic-resolution structural studies have been limited to only a few individual components. These include the winged-helix motif found in the Apc2 cullin domain (Zheng et al., 2002), Apc10/Doc1 (Wendt et al., 2001; Au et al., 2002), the N-terminal domain of human Apc7 (Han et al., 2009), a subcomplex between the TPR repeats in Apc6 and a short N-terminal region of Cdc26 (Wang et al., 2009), the N-terminal TPR domains of Cdc27 (Apc3) (Zhang et al., 2010b), and a subcomplex of S. pombe Cut9 (Apc6) and Hcn1 (Cdc26) (Zhang et al., 2010a; Zhang et al., 2010b). These structures have provided some information about how distinct subcomplexes within the APC/C, especially the TPR repeats, may assemble and perhaps interact with substrates and/or activators. However, these small snapshots of individual domains have not provided direct answers to the fundamental questions about how the APC/C mechanistically functions.
Single particle cryo-electron microscopy (EM) has proven to be a viable approach for gaining insights into the overall shape and organization of the holo-complex. Cryo-EM studies of S. pombe, S. cerevisiae, Xenopus, and human APC/C have now been reported (Gieffers et al., 2001; Dube et al., 2005; Passmore et al., 2005; Ohi et al., 2007; Herzog et al., 2009; da Fonseca et al., 2010; Buschhorn et al., 2010). In addition, there are also 3D structures of the APC/C bound to the activators Cdh1 and Cdc20 (Dube et al., 2005; Ohi et al., 2007; Herzog et al., 2009), to the MCC inhibitory complex (Herzog et al., 2009), as well as substrates (da Fonseca et al., 2010; Buschhorn et al., 2010). These cryo-EM 3D maps have provided an essential starting point for understanding how the structural organization of the APC/C translates into function. In the following paragraphs, we describe each 3D model, taking note of how each complex was purified and visualized by EM, as well as discuss some of the similarities and differences found between the different APC/C 3D models.
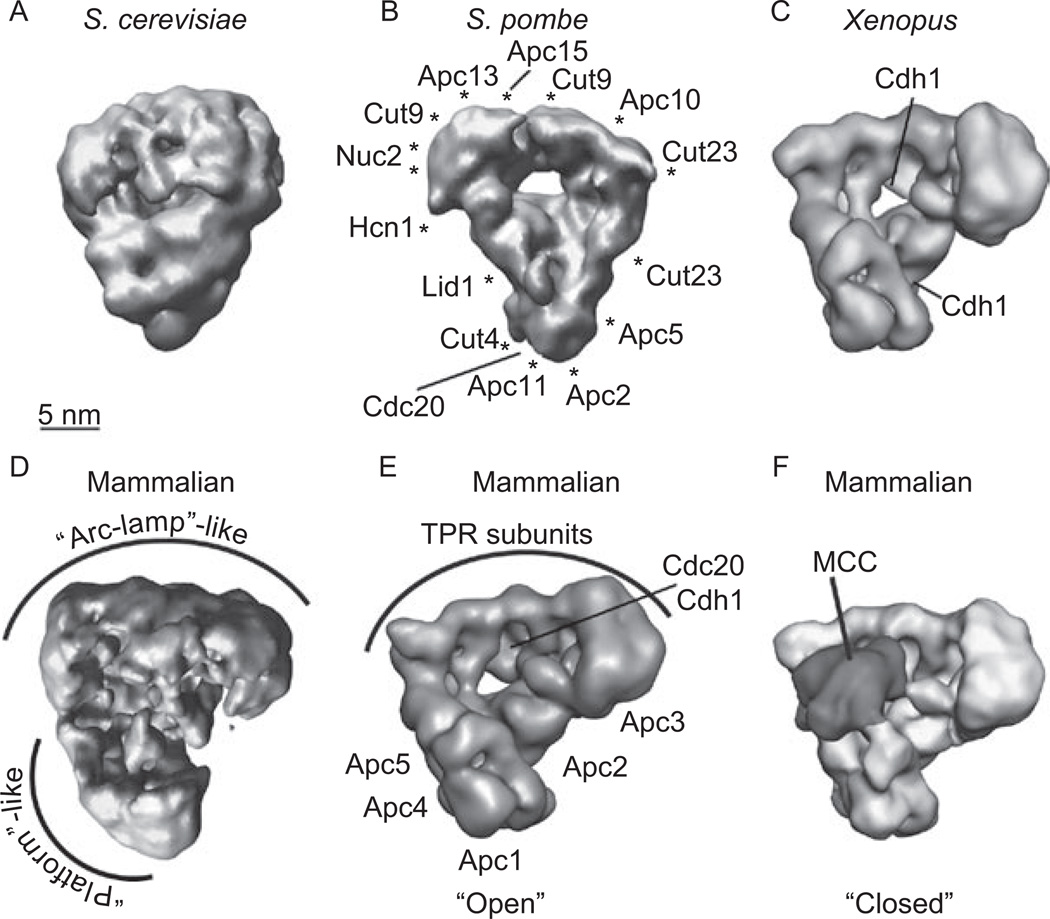
There are currently three published 3D structures of the S. cerevisiae APC/C. The first ~20 Å structure was determined using particles purified from asynchronously growing cells and imaged in unstained vitrified ice (Passmore et al., 2005) (Figure 2A). Very recently, the same group published a second structure of the S. cerevisiae APC/C in vitrified ice bound to Cdh1 and a D box peptide (da Fonseca et al., 2010). This map reached the much higher resolution of ~10 Å allowing the visualization of a triangular shaped structure that contains a central cavity lined with a lattice-like scaffold. Importantly, the authors localized the position of a D box peptide bound in the central cavity and determined that the D box peptide contacts both Cdh1 and Doc1, a core APC/C subunit previously shown to be required for Ub processivity (Carroll and Morgan, 2002; Passmore et al., 2003). This work provides the first structural model for understanding how the APC/C interacts with substrates and further strengthens the hypothesis that APC/C-substrate binding results from multivalent interactions with both activators and core APC/C components. This work was closely followed by a third ~25 Å cryo-negative stain structure determined from particles bound to Cdh1 and a D box peptide (Buschhorn et al., 2010). As with the structure from da Fonseca et al., Buschhorn et al. showed that the D box binds in the central cavity of the APC/C and contacts both the activator and Doc1. Unfortunately, the APC/C density maps from both of the above studies were not publically released at the time of this review and, thus, are not included in Figure 1. There are a number of conclusions that can be drawn from these two new S. cerevisiae APC/C structures. First, both Passmore et al., (2005) and Buschhorn et al., (2010) found evidence that the S. cerevisiae APC/C forms dimers; however, the presence of dimerized APC/C has thus far only been observed in budding yeast purifications making it unclear whether this is truly a physiologically relevant phenomenon. Second, the conformational changes seen in both the Xenopus and human APC/C upon activator binding (Herzog et al., 2009) were not observed in the 10 Å S. cerevisiae structure (da Fonseca et al., 2010) making it unclear whether this is a conserved mechanism of APC/C regulation. Third, the new S. cerevisiae APC/C structures (da Fonseca et al., 2010; Buschhorn, et al., 2010) look very similar in shape and size to structures of the APC/C determined from S. pombe and vertebrates (Ohi et al., 2007; Herzog et al., 2009), clearly showing that this complex has been conserved during evolution. Fourth, both S. cerevisiae structures clearly show that the D box peptide interacts with the APC/C by binding to both the activator and to Doc1, a core APC/C component (da Fonseca et al., 2010; Buschhorn et al., 2010), providing the first structural models of APC/C substrate recognition and binding modes.
Figure 2.
Structural analysis of the APC/C spanning from yeasts to vertebrates. (A) ~20Å structure of the Saccharomyces cerevisiae APC/C purified from asynchronous cells (Passmore et al., 2005) (EMDB1174). (B) Single-particle cryo-EM analysis of the mitotic Schizosaccharomyces pombe APC/CCdc20 (Ohi et al., 2007). Position of APC/C subunits and the activator Cdc20, found by antibody labeling and difference mapping, respectively, are labeled. Cut4 corresponds to human Apc1, Nuc2 corresponds to human Apc3, and Lid1 corresponds to human Apc4. (C) ~24Å structure of the Xenopus APC/C. Approximate locations of recombinant Cdh1 are labeled (Herzog et al., 2009). (D) Human APC/C (~26Å) purified from asynchronous cells (EMDB 1139) (Dube et al., 2005). (E) Human apo-APC/C (~20 Å) purified from checkpoint active lysates (EMDB 1592) (Herzog et al., 2009). Position of APC/C subunits as well as the activators Cdc20 and Cdh1, localized using antibody labeling and recombinant proteins, respectively, are noted. (F) ~20Å structure of the human APC/C bound to the MCC inhibitory complex (APC/CMCC, EMDB 1591) (Herzog et al., 2009). Position of MCC labeled in dark gray. Images for (D) and (F) were kindly provided by Franz Herzog (IMP-Vienna), Jan-Michael Peters (IMP-Vienna), and Holger Stark (Max Planck Institute). Scale bar for all panels, 5nm.
The 27Å structure of the S. pombe APC/CCdc20 was determined using particles purified from mitotically blocked cells and visualized in unstained vitrified ice (Ohi et al., 2007). The S. pombe APC/CCdc20 adopts an asymmetric, tricorn-shaped structure, ~ 19 × 17 × 15 nm in size, with a deep internal cavity and a prominent horn-like protrusion emanating from the bottom of the cavity lip (Figure 2B). The size of the central cavity is ~11.5 × 9.5 × 6.5 nm and is large enough to hold both an E2-Ub conjugated complex and an APC/C substrate, such as the CDK-cyclin complex. Using antibody labeling and mutant analysis, this study localized the C-terminus of 12 of the 13 core APC/C components, as well the position of the activator Cdc20, generating the most comprehensive map of APC/C organization to date. From the position of the RING component Apc11 and the activator Cdc20, the authors proposed a model where Cdc20 could initially recruit the substrate to the complex for ubiquitination, but the processivity of the reaction would be determined by interactions between the substrate and core components lining the central cavity (Ohi et al., 2007).
There have been a number of cryo-negative stain structures reported of vertebrate APC/C (Figure 2C–F). These include Xenopus APC/C bound and unbound to recombinant Cdh1 (APC/CCdh1 and apo-APC/C) (Dube et al., 2005; Herzog et al., 2009), mammalian apo-APC/C purified either from asynchronous lysates (Gieffers et al., 2001; Dube et al., 2005) or from spindle checkpoint active lysates (Herzog et al., 2009), mammalian APC/C bound to either recombinant Cdh1 or Cdc20 (hAPC/CCdh1 and hAPC/CCdc20) (Herzog et al., 2009), and mammalian APC/C bound to the MCC purified from human cells arrested with an active SAC (hAPC/CMCC) (Herzog et al., 2009). As the same group has determined all of these structures, following the progression of their APC/C density maps, starting with the first reported structure in 2001 (Gieffers et al., 2001) highlights the steady improvements made in single particle EM methodologies over the last decade.
As with the S. cerevisiae and S. pombe APC/C (Passmore et al., 2005; Ohi et al., 2007; da Fonseca et al., 2010; Buschhorn et al., 2010) (Figure 2A and B), the vertebrate structures (~24–20Å resolution, Figure 2C–E) reveal a triangular-shaped complex. Unlike the original S. cerevisiae structure (Passmore et al., 2005), but similar to the S. pombe APC/C (Ohi et al., 2007) and new S. cerevisiae structures (da Fonesca et al., 2010; Buschhorn et al., 2010), the vertebrate APC/C contains a central cavity surrounded by what the authors label the ‘arc-lamp’- and platform-like domains (Dube et al., 2005; Herzog et al., 2009) (Figure 2C–E). Antibody labeling studies were used to localize the position of Apc1, Apc2, Apc3 (Cdc27), Apc4, Apc5, Apc6, Apc7 (Dube et al., 2005; Herzog et al., 2009) and found that the ‘arc-lamp’-like domain is composed of the TPR subunits Apc6 and Apc7, while Apc1, Apc2, Apc4, and Apc5 are located in or near the ‘platform’-like domain. This subunit organization closely resembles that of S. pombe APC/C (Ohi et al., 2007) (Figure 2B and E). In fact, other than the mirrored handedness of the S. pombe with the S. cerevisiae and vertebrate structures, the overall structural organization of the APC/C appears very similar (Figure 2B, C, E, and data not shown), as would be predicted by the functional conservation of APC/C subunits across species (Table 1) (Thornton & Toczyski, 2006).
One apparent difference between the S. pombe and other structures is where Cdc20 and Cdh1 bind to the core complex (Figure 2B, C, and E). Structural analysis of the S. pombe APC/C purified from a mutant strain that precludes Cdc20 binding at the restrictive temperature, as well as antibody labeling studies of an endogenously tagged Cdc20 bound to the APC/C, localized the position of Cdc20 at the lip of the APC/C cavity close to the C-terminus of Apc2 (Figure 2B) (Ohi et al.). This position was very similar to the original localization of recombinant Cdh1 in Xenopus APC/C (Dube et al., 2005). In contrast, the densities corresponding to recombinant Cdc20 or Cdh1 bound to mammalian apo-APC/C and S. cerevisiae APC/C were found in the central cavity close to the TPR subunit (or ‘arc-lamp’-like) domain and Cdc27 (Apc3) (Herzog et al., 2009), a subunit involved in activator binding (Figure 2E) (Vodermaier et al., 2003; Matyskiela & Morgan, 2009; da Fonseca et al., 2010; Buschhorn et al., 2010). Since this localization was different from what was previously published for Xenopus APC/CCdh1 (Dube et al., 2005), the authors re-examined Xenopus APC/CCdh1 using improved sample preparation and EM imaging techniques. This new analysis found that addition of recombinant Cdh1 to the Xenopus structure generated two extra densities, one on the side of the structure and one overlapping with the activator binding position of mammalian APC/C (Figure 2C) (Herzog et al., 2009). This suggests that the APC/C may contain multiple activator-binding sites. Why two sites are only detected in the Xenopus structure and not the mammalian or S. pombe APC/C remains to be examined, although it should be noted that variance mapping of S. pombe APC/C showed one high difference peak in a central location similar to that found for mammalian activator(Ohi et al., 2007). In retrospect, this variance peak could represent a second binding event for Cdc20 the S. pombe structure.
Interestingly, three major conformations of mammalian apo-APC/C were observed that differ mainly in the position of the ‘arc-lamp’ and ‘platform’-like domains relative to each other (Herzog et al., 2009). Thus, apo-APC/C appears to adopt a continuum of flexible states ranging from open to closed states. Although this range of structural flexibility was not apparent in the yeasts, Xenopus, or earlier mammalian structures (Gieffers et al., 2001; Dube et al., 2005; Passmore et al., 2005; Ohi et al., 2007; da Fonseca et al., 2010), it is still tempting to speculate that different conformations may directly correlate with APC/C E3 ligase activity and that one role of APC/C activators and inhibitors is to shift the structural equilibrium between these open and closed states. At least for mammalian APC/C, this model is supported by the comparison of the 3D density maps of apo-APC/C, APC/CCdc20, and APC/CMCC (Herzog et al., 2009). In these structures, the binding of the inhibitory MCC ‘locks’ the APC/C into a closed or more compact conformation (Figure 2F), while the presence of Cdc20 alone shifts the equilibrium to a more open state (Herzog et al., 2009). Interestingly, the position of Cdc20 in the APC/CMCC density map is shifted by ~2 nm as compared to its position in APC/CCdc20 (Herzog et al., 2009). This suggests that MCC binding may also affect APC/C activity by modulating how Cdc20 interacts with the complex, perhaps adding another layer of structural regulation to APC/C activity. It remains to be determined what, if any, structural effects MCC binding has on APC/C-substrate interactions.
Although the above reported structures have provided an important first glimpse into overall APC/C structure and organization, it is clear that more in-depth structural analyses will be required before we reach a comprehensive understanding of how the APC/C works and is regulated. In particular, it is still unclear how the APC/C transfers Ub to substrates and regulates processivity. Structures of the APC/C bound to substrates and activators and higher resolution EM maps to allow precise docking of atomic resolution structures of individual APC/C components and/or subcomplexes, are required to begin to understand the mechanisms of APC/C catalysis and regulation.
Acknowledgments
This work was supported by the Howard Hughes Medical Institute (http://www.hhmi.org/), of which K.L.G. is an investigator. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of this article. Funding for J.R.M. was provided by the NIH training grant T32CA009385-25.
Footnotes
Declaration of interest
The authors report no declaration of interest.
References
- Amador V, Ge S, Santamaría PG, Guardavaccaro D, Pagano M. APC/C(Cdc20) controls the ubiquitin-mediated degradation of p21 in prometaphase. Mol Cell. 2007;27:462–473. doi: 10.1016/j.molcel.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang XL, Harper JW. SCF-mediated protein degradation and cell cycle control. Oncogene. 2005;24:2860–2870. doi: 10.1038/sj.onc.1208614. [DOI] [PubMed] [Google Scholar]
- Araki M, Yu H, Asano M. A novel motif governs APC-dependent degradation of Drosophila ORC1 in vivo. Genes Dev. 2005;19:2458–2465. doi: 10.1101/gad.1361905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa H, Kitamura K, Shimoda C. A novel Cdc20-related WD-repeat protein, Fzr1, is required for spore formation in Schizosaccharomyces pombe. Mol Genet Genomics. 2001;265:424–435. doi: 10.1007/s004380000429. [DOI] [PubMed] [Google Scholar]
- Au SW, Leng X, Harper JW, Barford D. Implications for the ubiquitination reaction of the anaphase-promoting complex from the crystal structure of the Doc1/Apc10 subunit. J Mol Biol. 2002;316:955–968. doi: 10.1006/jmbi.2002.5399. [DOI] [PubMed] [Google Scholar]
- Bashir T, Pagano M. Don’t skip the G1 phase: how APC/CCdh1 keeps SCFSKP2 in check. Cell Cycle. 2004;3:850–852. [PubMed] [Google Scholar]
- Bastians H, Topper LM, Gorbsky GL, Ruderman JV. Cell cycle-regulated proteolysis of mitotic target proteins. Mol Biol Cell. 1999;10:3927–3941. doi: 10.1091/mbc.10.11.3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford L, Paine S, Sheppard PW, Mayer RJ, Roelofs J. Assembly, structure, and function of the 26S proteasome. Trends Cell Biol. 2010;20:391–401. doi: 10.1016/j.tcb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrao P, Trinidad JC, Fiedler D, Roguev A, Lim WA, Shokat KM, Burlingame AL, Krogan NJ. Evolution of phosphoregulation: comparison of phosphorylation patterns across yeast species. PLoS Biol. 2009;7:e1000134. doi: 10.1371/journal.pbio.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Javerzat JP. Fission yeast Bub1 is essential in setting up the meiotic pattern of chromosome segregation. Nat Cell Biol. 2001;3:522–526. doi: 10.1038/35074598. [DOI] [PubMed] [Google Scholar]
- Binné UK, Classon MK, Dick FA, Wei W, Rape M, Kaelin WG, Jr, Näär AM, Dyson NJ. Retinoblastoma protein and anaphase-promoting complex physically interact and functionally cooperate during cell-cycle exit. Nat Cell Biol. 2007;9:225–232. doi: 10.1038/ncb1532. [DOI] [PubMed] [Google Scholar]
- Blanco MA, Sánchez-Díaz A, de Prada JM, Moreno S. APC(ste9/srw1) promotes degradation of mitotic cyclins in G(1) and is inhibited by cdc2 phosphorylation. EMBO J. 2000;19:3945–3955. doi: 10.1093/emboj/19.15.3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco MA, Pelloquin L, Moreno S. Fission yeast mfr1 activates APC and coordinates meiotic nuclear division with sporulation. J Cell Sci. 2001;114:2135–2143. doi: 10.1242/jcs.114.11.2135. [DOI] [PubMed] [Google Scholar]
- Bremm A, Freund SM, Komander D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat Struct Mol Biol. 2010;17:939–947. doi: 10.1038/nsmb.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JL, Solomon MJ. D box and KEN box motifs in budding yeast Hsl1p are required for APC-mediated degradation and direct binding to Cdc20p and Cdh1p. Genes Dev. 2001;15:2381–2395. doi: 10.1101/gad.917901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JL, Solomon MJ. Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes Dev. 2007;21:655–667. doi: 10.1101/gad.1511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschhorn BA, Petzold G, Galova M, Dube P, Kraft C, Herzog F, Stark H, Peters JM. Substrate binding on the APC/C occurs between the coactivator Cdh1 and the processivity factor Doc1. Nat Struct Mol Biol. 2010 doi: 10.1038/nsmb.1979. 2010 Dec 26. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Carroll CW, Morgan DO. The Doc1 subunit is a processivity factor for the anaphase-promoting complex. Nat Cell Biol. 2002;4:880–887. doi: 10.1038/ncb871. [DOI] [PubMed] [Google Scholar]
- Carroll CW, Enquist-Newman M, Morgan DO. The APC subunit Doc1 promotes recognition of the substrate destruction box. Curr Biol. 2005;15:11–18. doi: 10.1016/j.cub.2004.12.066. [DOI] [PubMed] [Google Scholar]
- Castro A, Arlot-Bonnemains Y, Vigneron S, Labbé JC, Prigent C, Lorca T. APC/Fizzy-Related targets Aurora-A kinase for proteolysis. EMBO Rep. 2002;3:457–462. doi: 10.1093/embo-reports/kvf095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E, Lim MS, Rosic-Kablar S, Liu J, Jolicoeur P, Dubé ID, Hough MR. Dysregulated expression of mitotic regulators is associated with B-cell lymphomagenesis in HOX11-transgenic mice. Oncogene. 2006;25:2575–2587. doi: 10.1038/sj.onc.1209285. [DOI] [PubMed] [Google Scholar]
- Chen RH. Dual inhibition of Cdc20 by the spindle checkpoint. J Biomed Sci. 2007;14:475–479. doi: 10.1007/s11373-007-9157-3. [DOI] [PubMed] [Google Scholar]
- Ciliberto A, Shah JV. A quantitative systems view of the spindle assembly checkpoint. EMBO J. 2009;28:2162–2173. doi: 10.1038/emboj.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Coluccio A, Bogengruber E, Conrad MN, Dresser ME, Briza P, Neiman AM. Morphogenetic pathway of spore wall assembly in Saccharomyces cerevisiae. Eukaryotic Cell. 2004;3:1464–1475. doi: 10.1128/EC.3.6.1464-1475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KF, Mallory MJ, Egeland DB, Jarnik M, Strich R. Ama1p is a meiosis-specific regulator of the anaphase promoting complex/cyclosome in yeast. Proc Natl Acad Sci USA. 2000;97:14548–14553. doi: 10.1073/pnas.250351297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox CJ, Dutta K, Petri ET, Hwang WC, Lin Y, Pascal SM, Basavappa R. The regions of securin and cyclin B proteins recognized by the ubiquitination machinery are natively unfolded. FEBS Lett. 2002;527:303–308. doi: 10.1016/s0014-5793(02)03246-5. [DOI] [PubMed] [Google Scholar]
- Crane R, Kloepfer A, Ruderman JV. Requirements for the destruction of human Aurora-A. J Cell Sci. 2004;117:5975–5983. doi: 10.1242/jcs.01418. [DOI] [PubMed] [Google Scholar]
- da Fonseca PC, Kong EH, Zhang Z, Schreiber A, Williams MA, Morris EP, Barford D. Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature. 2010 doi: 10.1038/nature09625. 2010 Nov 24. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Elzen N, Pines J. Cyclin A is destroyed in prometaphase and can delay chromosome alignment and anaphase. J Cell Biol. 2001;153:121–136. doi: 10.1083/jcb.153.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore B, Pines J. Emi1 is needed to couple DNA replication with mitosis but does not regulate activation of the mitotic APC/C. J Cell Biol. 2007;177:425–437. doi: 10.1083/jcb.200611166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore B, Pines J. How cyclin A destruction escapes the spindle assembly checkpoint. J Cell Biol. 2010;190:501–509. doi: 10.1083/jcb.201001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dial JM, Petrotchenko EV, Borchers CH. Inhibition of APCCdh1 activity by Cdh1/Acm1/Bmh1 ternary complex formation. J Biol Chem. 2007;282:5237–5248. doi: 10.1074/jbc.M606589200. [DOI] [PubMed] [Google Scholar]
- Diamond AE, Park JS, Inoue I, Tachikawa H, Neiman AM. The anaphase promoting complex targeting subunit Ama1 links meiotic exit to cytokinesis during sporulation in Saccharomyces cerevisiae. Mol Biol Cell. 2009;20:134–145. doi: 10.1091/mbc.E08-06-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Zavitz KH, Thomas BJ, Lin M, Campbell S, Zipursky SL. Control of G1 in the developing Drosophila eye: rca1 regulates Cyclin A. Genes Dev. 1997;11:94–105. doi: 10.1101/gad.11.1.94. [DOI] [PubMed] [Google Scholar]
- Dube P, Herzog F, Gieffers C, Sander B, Riedel D, Müller SA, Engel A, Peters JM, Stark H. Localization of the coactivator Cdh1 and the cullin subunit Apc2 in a cryo-electron microscopy model of vertebrate APC/C. Mol Cell. 2005;20:867–879. doi: 10.1016/j.molcel.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Elia AE, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, Mohammad D, Cantley LC, Smerdon SJ, Yaffe MB. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 2003;115:83–95. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- Engelbert D, Schnerch D, Baumgarten A, Wäsch R. The ubiquitin ligase APC(Cdh1) is required to maintain genome integrity in primary human cells. Oncogene. 2008;27:907–917. doi: 10.1038/sj.onc.1210703. [DOI] [PubMed] [Google Scholar]
- Enquist-Newman M, Sullivan M, Morgan DO. Modulation of the mitotic regulatory network by APC-dependent destruction of the Cdh1 inhibitor Acm1. Mol Cell. 2008;30:437–446. doi: 10.1016/j.molcel.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eytan E, Moshe Y, Braunstein I, Hershko A. Roles of the ana-phase-promoting complex/cyclosome and of its activator Cdc20 in functional substrate binding. Proc Natl Acad Sci USA. 2006;103:2081–2086. doi: 10.1073/pnas.0510695103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 1998a;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol Cell. 1998b;2:163–171. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. Control of mitotic transitions by the anaphase-promoting complex. Philos Trans R Soc Lond, B, Biol Sci. 1999;354:1583–1590. doi: 10.1098/rstb.1999.0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry AM. The Nek2 protein kinase: a novel regulator of centrosome structure. Oncogene. 2002;21:6184–6194. doi: 10.1038/sj.onc.1205711. [DOI] [PubMed] [Google Scholar]
- Fry AM, Yamano H. APC/C-mediated degradation in early mitosis: how to avoid spindle assembly checkpoint inhibition. Cell Cycle. 2006;5:1487–1491. doi: 10.4161/cc.5.14.3003. [DOI] [PubMed] [Google Scholar]
- Funabiki H, Yamano H, Kumada K, Nagao K, Hunt T, Yanagida M. Cut2 proteolysis required for sister-chromatid seperation in fission yeast. Nature. 1996;381:438–441. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- Gabellini D, Colaluca IN, Vodermaier HC, Biamonti G, Giacca M, Falaschi A, Riva S, Peverali FA. Early mitotic degradation of the homeoprotein HOXC10 is potentially linked to cell cycle progression. EMBO J. 2003;22:3715–3724. doi: 10.1093/emboj/cdg340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallastegui N, Groll M. The 26S proteasome: assemble and function of a destructive machine. Trends Biochem Sci. 2010;35:634–642. doi: 10.1016/j.tibs.2010.05.005. [DOI] [PubMed] [Google Scholar]
- García-Higuera I, Manchado E, Dubus P, Cañamero M, Méndez J, Moreno S, Malumbres M. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat Cell Biol. 2008;10:802–811. doi: 10.1038/ncb1742. [DOI] [PubMed] [Google Scholar]
- Garnett MJ, Mansfeld J, Godwin C, Matsusaka T, Wu J, Russell P, Pines J, Venkitaraman AR. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat Cell Biol. 2009;11:1363–1369. doi: 10.1038/ncb1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geley S, Kramer E, Gieffers C, Gannon J, Peters JM, Hunt T. Anaphase-promoting complex/cyclosome-dependent prote-olysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J Cell Biol. 2001;153:137–148. doi: 10.1083/jcb.153.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieffers C, Dube P, Harris JR, Stark H, Peters JM. Three-dimensional structure of the anaphase-promoting complex. Mol Cell. 2001;7:907–913. doi: 10.1016/s1097-2765(01)00234-9. [DOI] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Gmachl M, Gieffers C, Podtelejnikov AV, Mann M, Peters JM. The RING-H2 finger protein APC11 and the E2 enzyme UBC4 are sufficient to ubiquitinate substrates of the anaphase-promoting complex. Proc Natl Acad Sci USA. 2000;97:8973–8978. doi: 10.1073/pnas.97.16.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorr IH, Boos D, Stemmann O. Mutual inhibition of separase and Cdk1 by two-step complex formation. Mol Cell. 2005;19:135–141. doi: 10.1016/j.molcel.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Grassmann R, Aboud M, Jeang KT. Molecular mechanisms of cellular transformation by HTLV-1 Tax. Oncogene. 2005;24:5976–5985. doi: 10.1038/sj.onc.1208978. [DOI] [PubMed] [Google Scholar]
- Grosskortenhaus R, Sprenger F. Rca1 inhibits APC-Cdh1(Fzr) and is required to prevent cyclin degradation in G2. Dev Cell. 2002;2:29–40. doi: 10.1016/s1534-5807(01)00104-6. [DOI] [PubMed] [Google Scholar]
- Hall MC, Jeong DE, Henderson JT, Choi E, Bremmer SC, Iliuk AB, Charbonneau H. Cdc28 and Cdc14 control stability of the anaphase-promoting complex inhibitor Acm1. J Biol Chem. 2008;283:10396–10407. doi: 10.1074/jbc.M710011200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hames RS, Wattam SL, Yamano H, Bacchieri R, Fry AM. APC/C-mediated destruction of the centrosomal kinase Nek2A occurs in early mitosis and depends upon a cyclin A-type D-box. EMBO J. 2001;20:7117–7127. doi: 10.1093/emboj/20.24.7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Kim K, Kim Y, Kang Y, Lee JY, Kim Y. Crystal structure of the N-terminal domain of anaphase-promoting complex subunit 7. J Biol Chem. 2009;284:15137–15146. doi: 10.1074/jbc.M804887200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley ME, Allan LA, Sanderson HS, Clarke PR. Phosphorylation of Mcl-1 by CDK1-cyclin B1 initiates its Cdc20-dependent destruction during mitotic arrest. EMBO J. 2010;29:2407–2420. doi: 10.1038/emboj.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Burton JL, Solomon MJ. The anaphase-promoting complex: it’s not just for mitosis any more. Genes Dev. 2002;16:2179–2206. doi: 10.1101/gad.1013102. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Mortimer RK, Culotti J, Culotti M. Genetic Control of the Cell Division Cycle in Yeast: V. Genetic Analysis of cdc Mutants. Genetics. 1973;74:267–286. doi: 10.1093/genetics/74.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes MJ, Kimata Y, Wattam SL, Lindon C, Mao G, Yamano H, Fry AM. Early mitotic degradation of Nek2A depends on Cdc20-independent interaction with the APC/C. Nat Cell Biol. 2006;8:607–614. doi: 10.1038/ncb1410. [DOI] [PubMed] [Google Scholar]
- Hayles J, Aves S, Nurse P. suc1 is an essential gene involved in both the cell cycle and growth in fission yeast. EMBO J. 1986;5:3373–3379. doi: 10.1002/j.1460-2075.1986.tb04653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Patterson TE, Sazer S. The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc Natl Acad Sci USA. 1997;94:7965–7970. doi: 10.1073/pnas.94.15.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A. Roles of ubiquitin-mediated proteolysis in cell cycle control. Curr Opin Cell Biol. 1997;9:788–799. doi: 10.1016/s0955-0674(97)80079-8. [DOI] [PubMed] [Google Scholar]
- Herzog F, Primorac I, Dube P, Lenart P, Sander B, Mechtler K, Stark H, Peters JM. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science. 2009;323:1477–1481. doi: 10.1126/science.1163300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilioti Z, Chung YS, Mochizuki Y, Hardy CF, Cohen-Fix O. The anaphase inhibitor Pds1 binds to the APC/C-associated protein Cdc20 in a destruction box-dependent manner. Curr Biol. 2001;11:1347–1352. doi: 10.1016/s0960-9822(01)00399-2. [DOI] [PubMed] [Google Scholar]
- Holloway SL, Glotzer M, King RW, Murray AW. Anaphase is initiated by proteolysis rather than by the inactivation of maturation-promoting factor. Cell. 1993;73:1393–1402. doi: 10.1016/0092-8674(93)90364-v. [DOI] [PubMed] [Google Scholar]
- Holt LJ, Krutchinsky AN, Morgan DO. Positive feedback sharpens the anaphase switch. Nature. 2008;454:353–357. doi: 10.1038/nature07050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt LJ, Tuch BB, Villén J, Johnson AD, Gygi SP, Morgan DO. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homer HA, McDougall A, Levasseur M, Murdoch AP, Herbert M. Mad2 is required for inhibiting securin and cyclin B degradation following spindle depolymerisation in meiosis I mouse oocytes. Reproduction. 2005a;130:829–843. doi: 10.1530/rep.1.00856. [DOI] [PubMed] [Google Scholar]
- Homer HA, McDougall A, Levasseur M, Yallop K, Murdoch AP, Herbert M. Mad2 prevents aneuploidy and premature proteolysis of cyclin B and securin during meiosis I in mouse oocytes. Genes Dev. 2005b;19:202–207. doi: 10.1101/gad.328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JY, Reimann JD, Sørensen CS, Lukas J, Jackson PK. E2F-dependent accumulation of hEmi1 regulates S phase entry by inhibiting APC(Cdh1) Nat Cell Biol. 2002;4:358–366. doi: 10.1038/ncb785. [DOI] [PubMed] [Google Scholar]
- Huang DT, Zhuang M, Ayrault O, Schulman BA. Identification of conjugation specificity determinants unmasks vestigial preference for ubiquitin within the NEDD8 E2. Nat Struct Mol Biol. 2008;15:280–287. doi: 10.1038/nsmb.1387. [DOI] [PubMed] [Google Scholar]
- Huang JN, Park I, Ellingson E, Littlepage LE, Pellman D. Activity of the APC(Cdh1) form of the anaphase-promoting complex persists until S phase and prevents the premature expression of Cdc20p. J Cell Biol. 2001;154:85–94. doi: 10.1083/jcb.200102007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JY, Morley G, Li D, Whitaker M. Cdk1 phosphorylation sites on Cdc27 are required for correct chromosomal localisation and APC/C function in syncytial Drosophila embryos. J Cell Sci. 2007;120:1990–1997. doi: 10.1242/jcs.006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irniger S, Piatti S, Michaelis C, Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin prote-olysis in budding yeast. Cell. 1995;81:269–278. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- Irniger S. Preventing fatal destruction: inhibitors of the anaphase-promoting complex in meiosis. Cell Cycle. 2006;5:405–415. doi: 10.4161/cc.5.4.2476. [DOI] [PubMed] [Google Scholar]
- Izawa D, Goto M, Yamashita A, Yamano H, Yamamoto M. Fission yeast Mes1p ensures the onset of meiosis II by blocking degradation of cyclin Cdc13p. Nature. 2005;434:529–533. doi: 10.1038/nature03406. [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Charles JF, Morgan DO. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr Biol. 1999;9:227–236. doi: 10.1016/s0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- Jin F, Hamada M, Malureanu L, Jeganathan KB, Zhou W, Morbeck DE, van Deursen JM. Cdc20 is critical for meiosis I and fertility of female mice. PLoS Genet. 2010;6:e1001147. doi: 10.1371/journal.pgen.1001147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Williamson A, Banerjee S, Philipp I, Rape M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133:653–665. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CR, Hwang KS, Yoo J, Cho WK, Kim JM, Kim WH, Im DS. E2-EPF UCP targets pVHL for degradation and associates with tumor growth and metastasis. Nat Med. 2006;12:809–816. doi: 10.1038/nm1440. [DOI] [PubMed] [Google Scholar]
- Kaiser P, Moncollin V, Clarke DJ, Watson MH, Bertolaet BL, Reed SI, Bailly E. Cyclin-dependent kinase and Cks/Suc1 interact with the proteasome in yeast to control proteolysis of M-phase targets. Genes Dev. 1999;13:1190–1202. doi: 10.1101/gad.13.9.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamysheva Z, Diaz-Martinez LA, Crow SE, Li B, Yu H. Multiple anaphase-promoting complex/cyclosome degrons mediate the degradation of human Sgo1. J Biol Chem. 2009;284:1772–1780. doi: 10.1074/jbc.M807083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katis VL, Galova M, Rabitsch KP, Gregan J, Nasmyth K. Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr Biol. 2004;14:560–572. doi: 10.1016/j.cub.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, Han S, Jeffery L, Baek ST, Lee H, Shim YS, Lee M, Kim L, Heo KS, Noh EJ, Lee AR, Jang YJ, Chung KS, Choi SJ, Park JY, Park Y, Kim HM, Park SK, Park HJ, Kang EJ, Kim HB, Kang HS, Park HM, Kim K, Song K, Song KB, Nurse P, Hoe KL. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2010a;28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HM, Yu Y, Cheng Y. Structure characterization of the 26s proteasome. Biochim Biophys Acta. 2010b doi: 10.1016/j.bbagrm.2010.08.008. 26 August 2010 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y, Baxter JE, Fry AM, Yamano H. A role for the Fizzy/Cdc20 family of proteins in activation of the APC/C distinct from substrate recruitment. Mol Cell. 2008a;32:576–583. doi: 10.1016/j.molcel.2008.09.023. [DOI] [PubMed] [Google Scholar]
- Kimata Y, Trickey M, Izawa D, Gannon J, Yamamoto M, Yamano H. A mutual inhibition between APC/C and its substrate Mes1 required for meiotic progression in fission yeast. Dev Cell. 2008b;14:446–454. doi: 10.1016/j.devcel.2007.12.010. [DOI] [PubMed] [Google Scholar]
- King EM, van der Sar SJ, Hardwick KG. Mad3 KEN boxes mediate both Cdc20 and Mad3 turnover, and are critical for the spindle checkpoint. PLoS ONE. 2007;2:e342. doi: 10.1371/journal.pone.0000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- Kitajima S, Kudo Y, Ogawa I, Tatsuka M, Kawai H, Pagano M, Takata T. Constitutive phosphorylation of aurora-a on ser51 induces its stabilization and consequent overexpression in cancer. PLoS ONE. 2007;2:e944. doi: 10.1371/journal.pone.0000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- Konishi Y, Stegmüller J, Matsuda T, Bonni S, Bonni A. Cdh1-APC controls axonal growth and patterning in the mammalian brain. Science. 2004;303:1026–1030. doi: 10.1126/science.1093712. [DOI] [PubMed] [Google Scholar]
- Kotani S, Tugendreich S, Fujii M, Jorgensen PM, Watanabe N, Hoog C, Hieter P, Todokoro K. PKA and MPF-activated polo-like kinase regulate anaphase-promoting complex activity and mitosis progression. Mol Cell. 1998;1:371–380. doi: 10.1016/s1097-2765(00)80037-4. [DOI] [PubMed] [Google Scholar]
- Kotani S, Tanaka H, Yasuda H, Todokoro K. Regulation of APC activity by phosphorylation and regulatory factors. J Cell Biol. 1999;146:791–800. doi: 10.1083/jcb.146.4.791. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kraft C, Herzog F, Gieffers C, Mechtler K, Hagting A, Pines J, Peters JM. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J. 2003;22:6598–6609. doi: 10.1093/emboj/cdg627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C, Vodermaier HC, Maurer-Stroh S, Eisenhaber F, Peters JM. The WD40 propeller domain of Cdh1 functions as a destruction box receptor for APC/C substrates. Mol Cell. 2005;18:543–553. doi: 10.1016/j.molcel.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Kramer ER, Gieffers C, Hölzl G, Hengstschläger M, Peters JM. Activation of the human anaphase-promoting complex by proteins of the CDC20/Fizzy family. Curr Biol. 1998;8:1207–1210. doi: 10.1016/s0960-9822(07)00510-6. [DOI] [PubMed] [Google Scholar]
- Kramer ER, Scheuringer N, Podtelejnikov AV, Mann M, Peters JM. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol Biol Cell. 2000;11:1555–1569. doi: 10.1091/mbc.11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahav-Baratz S, Sudakin V, Ruderman JV, Hershko A. Reversible phosphorylation controls the activity of cyclosome-associated cyclin-ubiquitin ligase. Proc Natl Acad Sci USA. 1995;92:9303–9307. doi: 10.1073/pnas.92.20.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear BC, Skeath JB, Patel NH. Neural cell fate in rca1 and cycA mutants: the roles of intrinsic and extrinsic factors in asymmetric division in the Drosophila central nervous system. Mech Dev. 1999;88:207–219. doi: 10.1016/s0925-4773(99)00190-2. [DOI] [PubMed] [Google Scholar]
- Leverson JD, Joazeiro CA, Page AM, Huang H, Hieter P, Hunter T. The APC11 RING-H2 finger mediates E2-dependent ubiquitination. Mol Biol Cell. 2000;11:2315–2325. doi: 10.1091/mbc.11.7.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, York JP, Zhang P. Loss of Cdc20 causes a securin-dependent metaphase arrest in two-cell mouse embryos. Mol Cell Biol. 2007;27:3481–3488. doi: 10.1128/MCB.02088-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Shin YH, Hou L, Huang X, Wei Z, Klann E, Zhang P. The adaptor protein of the anaphase promoting complex Cdh1 is essential in maintaining replicative lifespan and in learning and memory. Nat Cell Biol. 2008;10:1083–1089. doi: 10.1038/ncb1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Li S, Yuan J, Wang ZB, Sun SC, Schatten H, Sun QY. Bub3 is a spindle assembly checkpoint protein regulating chromosome segregation during mouse oocyte meiosis. PLoS ONE. 2009;4:e7701. doi: 10.1371/journal.pone.0007701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister LM, Kouznetsova A, Hyslop LA, Kalleas D, Pace SL, Barel JC, Nathan A, Floros V, Adelfalk C, Watanabe Y, Jessberger R, Kirkwood TB, Höög C, Herbert M. Age-related meiotic segregation errors in Mammalian oocytes are preceded by depletion of cohesin and Sgo2. Curr Biol. 2010;20:1511–1521. doi: 10.1016/j.cub.2010.08.023. [DOI] [PubMed] [Google Scholar]
- Listovsky T, Zor A, Laronne A, Brandeis M. Cdk1 is essential for mammalian cyclosome/APC regulation. Exp Cell Res. 2000;255:184–191. doi: 10.1006/excr.1999.4788. [DOI] [PubMed] [Google Scholar]
- Littlepage LE, Ruderman JV. Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Genes Dev. 2002;16:2274–2285. doi: 10.1101/gad.1007302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlepage LE, Wu H, Andresson T, Deanehan JK, Amundadottir LT, Ruderman JV. Identification of phosphorylated residues that affect the activity of the mitotic kinase Aurora-A. Proc Natl Acad Sci USA. 2002;99:15440–15445. doi: 10.1073/pnas.202606599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Hong S, Tang Z, Yu H, Giam CZ. HTLV-I Tax directly binds the Cdc20-associated anaphase-promoting complex and activates it ahead of schedule. Proc Natl Acad Sci USA. 2005;102:63–68. doi: 10.1073/pnas.0406424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Baier K, Dammann R, Pfeifer GP. The tumor suppressor RASSF1A does not interact with Cdc20, an activator of the anaphase-promoting complex. Cell Cycle. 2007;6:1663–1665. doi: 10.4161/cc.6.13.4435. [DOI] [PubMed] [Google Scholar]
- Machida YJ, Dutta A. The APC/C inhibitor, Emi1, is essential for prevention of rereplication. Genes Dev. 2007;21:184–194. doi: 10.1101/gad.1495007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmanche N, Maia A, Sunkel CE. The spindle assembly checkpoint: preventing chromosome mis-segregation during mitosis and meiosis. FEBS Lett. 2006;580:2888–2895. doi: 10.1016/j.febslet.2006.03.081. [DOI] [PubMed] [Google Scholar]
- Mapelli M, Filipp FV, Rancati G, Massimiliano L, Nezi L, Stier G, Hagan RS, Confalonieri S, Piatti S, Sattler M, Musacchio A. Determinants of conformational dimerization of Mad2 and its inhibition by p31 comet. EMBO J. 2006;25:1273–1284. doi: 10.1038/sj.emboj.7601033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston AL, Tham WH, Shah H, Amon A. A genome-wide screen identifies genes required for centromeric cohesion. Science. 2004;303:1367–1370. doi: 10.1126/science.1094220. [DOI] [PubMed] [Google Scholar]
- Martinez JS, Jeong DE, Choi E, Billings BM, Hall MC. Acm1 is a negative regulator of the CDH1-dependent anaphase-promoting complex/cyclosome in budding yeast. Mol Cell Biol. 2006;26:9162–9176. doi: 10.1128/MCB.00603-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Máthé E. RASSF1A, the new guardian of mitosis. Nat Genet. 2004;36:117–118. doi: 10.1038/ng0204-117. [DOI] [PubMed] [Google Scholar]
- Matsumoto ML, Wickliffe KE, Dong KC, Yu C, Bosanac I, Bustos D, Phu L, Kirkpatrick DS, Hymowitz SG, Rape M, Kelley RF, Dixit VM. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol Cell. 2010;39:477–484. doi: 10.1016/j.molcel.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Matyskiela ME, Morgan DO. Analysis of activator-binding sites on the APC/C supports a cooperative substrate-binding mechanism. Mol Cell. 2009;34:68–80. doi: 10.1016/j.molcel.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyskiela ME, Rodrigo-Brenni MC, Morgan DO. Mechanisms of ubiquitin transfer by the anaphase-promoting complex. J Biol. 2009;8:92. doi: 10.1186/jbiol184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazanek M, Roitinger E, Hudecz O, Hutchins JR, Hegemann B, Mitulovic G, Taus T, Stingl C, Peters JM, Mechtler K. A new acid mix enhances phosphopeptide enrichment on titanium-and zirconium dioxide for mapping of phosphorylation sites on protein complexes. J Chromatogr B Analyt Technol Biomed Life Sci. 2010;878:515–524. doi: 10.1016/j.jchromb.2009.12.017. [DOI] [PubMed] [Google Scholar]
- Meyn MA, 3rd, Melloy PG, Li J, Holloway SL. The destruction box of the cyclin Clb2 binds the anaphase-promoting complex/cyclosome subunit Cdc23. Arch Biochem Biophys. 2002;407:189–195. doi: 10.1016/s0003-9861(02)00467-8. [DOI] [PubMed] [Google Scholar]
- Millband DN, Hardwick KG. Fission yeast Mad3p is required for Mad2p to inhibit the anaphase-promoting complex and localizes to kinetochores in a Bub1p-, Bub3p-, and Mph1p–dependent manner. Mol Cell Biol. 2002;22:2728–2742. doi: 10.1128/MCB.22.8.2728-2742.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JJ, Summers MK, Hansen DV, Nachury MV, Lehman NL, Loktev A, Jackson PK. Emi1 stably binds and inhibits the anaphase-promoting complex/cyclosome as a pseudosubstrate inhibitor. Genes Dev. 2006;20:2410–2420. doi: 10.1101/gad.1454006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miniowitz-Shemtov S, Teichner A, Sitry-Shevah D, Hershko A. ATP is required for the release of the anaphase-promoting complex/cyclosome from inhibition by the mitotic checkpoint. Proc Natl Acad Sci USA. 2010;107:5351–5356. doi: 10.1073/pnas.1001875107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo M, Fleming SB, Mercer AA. Cell cycle deregulation by a pox-virus partial mimic of anaphase-promoting complex subunit 11. Proc Natl Acad Sci USA. 2009;106:19527–19532. doi: 10.1073/pnas.0905893106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow CJ, Tighe A, Johnson VL, Scott MI, Ditchfield C, Taylor SS. Bub1 and aurora B cooperate to maintain BubR1-mediated inhibition of APC/CCdc20. J Cell Sci. 2005;118:3639–3652. doi: 10.1242/jcs.02487. [DOI] [PubMed] [Google Scholar]
- Mourón S, de Cárcer G, Seco E, Fernández-Miranda G, Malumbres M, Nebreda AR. RINGO C is required to sustain the spindle-assembly checkpoint. J Cell Sci. 2010;123:2586–2595. doi: 10.1242/jcs.059964. [DOI] [PubMed] [Google Scholar]
- Mui MZ, Roopchand DE, Gentry MS, Hallberg RL, Vogel J, Branton PE. Adenovirus protein E4orf4 induces premature APCCdc20 activation in Saccharomyces cerevisiae by a protein phosphatase 2A–dependent mechanism. J Virol. 2010;84:4798–4809. doi: 10.1128/JVI.02434-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Niault T, Hached K, Sotillo R, Sorger PK, Maro B, Benezra R, Wassmann K. Changing Mad2 levels affects chromosome segregation and spindle assembly checkpoint control in female mouse meiosis I. PLoS ONE. 2007;2:e1165. doi: 10.1371/journal.pone.0001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson J, Yekezare M, Minshull J, Pines J. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat Cell Biol. 2008;10:1411–1420. doi: 10.1038/ncb1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelschlaegel T, Schwickart M, Matos J, Bogdanova A, Camasses A, Havlis J, Shevchenko A, Zachariae W. The yeast APC/C subunit Mnd2 prevents premature sister chromatid separation triggered by the meiosis-specific APC/C-Ama1. Cell. 2005;120:773–788. doi: 10.1016/j.cell.2005.01.032. [DOI] [PubMed] [Google Scholar]
- Ohe M, Kawamura Y, Ueno H, Inoue D, Kanemori Y, Senoo C, Isoda M, Nakajo N, Sagata N. Emi2 inhibition of the anaphase-promoting complex/cyclosome absolutely requires Emi2 binding via the C-terminal RL tail. Mol Biol Cell. 2010;21:905–913. doi: 10.1091/mbc.E09-11-0974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi MD, Feoktistova A, Ren L, Yip C, Cheng Y, Chen JS, Yoon HJ, Wall JS, Huang Z, Penczek PA, Gould KL, Walz T. Structural organization of the anaphase-promoting complex bound to the mitotic activator Slp1. Mol Cell. 2007;28:871–885. doi: 10.1016/j.molcel.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi K, Koyanagi A, Yamamoto TM, Gotoh T, Kishimoto T. Emi1-mediated M-phase arrest in Xenopus eggs is distinct from cytostatic factor arrest. Proc Natl Acad Sci USA. 2004;101:12531–12536. doi: 10.1073/pnas.0405300101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto Y, Ozaki T, Miyazaki K, Aoyama M, Miyazaki M, Nakagawara A. UbcH10 is the cancer-related E2 ubiquitin-conjugating enzyme. Cancer Res. 2003;63:4167–4173. [PubMed] [Google Scholar]
- Osaka F, Seino H, Seno T, Yamao F. A ubiquitin-conjugating enzyme in fission yeast that is essential for the onset of anaphase in mitosis. Mol Cell Biol. 1997;17:3388–3397. doi: 10.1128/mcb.17.6.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Chen RH. Spindle checkpoint regulates Cdc20p stability in Saccharomyces cerevisiae. Genes Dev. 2004;18:1439–1451. doi: 10.1101/gad.1184204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, McCormack EA, Au SW, Paul A, Willison KR, Harper JW, Barford D. Doc1 mediates the activity of the anaphase-promoting complex by contributing to substrate recognition. EMBO J. 2003;22:786–796. doi: 10.1093/emboj/cdg084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, Booth CR, Vénien-Bryan C, Ludtke SJ, Fioretto C, Johnson LN, Chiu W, Barford D. Structural analysis of the anaphase-promoting complex reveals multiple active sites and insights into polyubiquitylation. Mol Cell. 2005;20:855–866. doi: 10.1016/j.molcel.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Patra D, Dunphy WG. Xe-p9, a Xenopus Suc1/Cks protein, is essential for the Cdc2-dependent phosphorylation of the anaphase- promoting complex at mitosis. Genes Dev. 1998;12:2549–2559. doi: 10.1101/gad.12.16.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkner AM, Prinz S, Ferscha S, Klein F. Mnd2, an essential antagonist of the anaphase-promoting complex during meiotic prophase. Cell. 2005;120:789–801. doi: 10.1016/j.cell.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Pesin JA, Orr-Weaver TL. Developmental role and regulation of cortex, a meiosis-specific anaphase-promoting complex/cyclosome activator. PLoS Genet. 2007;3:e202. doi: 10.1371/journal.pgen.0030202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesin JA, Orr-Weaver TL. Regulation of APC/C activators in mitosis and meiosis. Annu Rev Cell Dev Biol. 2008;24:475–499. doi: 10.1146/annurev.cellbio.041408.115949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, King RW, Höög C, Kirschner MW. Identification of BIME as a subunit of the anaphase-promoting complex. Science. 1996;274:1199–1201. doi: 10.1126/science.274.5290.1199. [DOI] [PubMed] [Google Scholar]
- Peters JM. SCF and APC: the Yin and Yang of cell cycle regulated proteolysis. Curr Opin Cell Biol. 1998;10:759–768. doi: 10.1016/s0955-0674(98)80119-1. [DOI] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Pfleger CM, Kirschner MW. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- Pfleger CM, Lee E, Kirschner MW. Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes Dev. 2001;15:2396–2407. doi: 10.1101/gad.918201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Pinsky BA, Nelson CR, Biggins S. Protein phosphatase 1 regulates exit from the spindle checkpoint in budding yeast. Curr Biol. 2009;19:1182–1187. doi: 10.1016/j.cub.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz S, Hwang ES, Visintin R, Amon A. The regulation of Cdc20 proteolysis reveals a role for APC components Cdc23 and Cdc27 during S phase and early mitosis. Curr Biol. 1998;8:750–760. doi: 10.1016/s0960-9822(98)70298-2. [DOI] [PubMed] [Google Scholar]
- Rabitsch KP, Gregan J, Schleiffer A, Javerzat JP, Eisenhaber F, Nasmyth K. Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis I and II. Curr Biol. 2004;14:287–301. doi: 10.1016/j.cub.2004.01.051. [DOI] [PubMed] [Google Scholar]
- Rape M, Kirschner MW. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature. 2004;432:588–595. doi: 10.1038/nature03023. [DOI] [PubMed] [Google Scholar]
- Rape M, Reddy SK, Kirschner MW. The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell. 2006;124:89–103. doi: 10.1016/j.cell.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Rauh NR, Schmidt A, Bormann J, Nigg EA, Mayer TU. Calcium triggers exit from meiosis II by targeting the APC/C inhibitor XErp1 for degradation. Nature. 2005;437:1048–1052. doi: 10.1038/nature04093. [DOI] [PubMed] [Google Scholar]
- Reddy SK, Rape M, Margansky WA, Kirschner MW. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446:921–925. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- Reimann JD, Freed E, Hsu JY, Kramer ER, Peters JM, Jackson PK. Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the anaphase promoting complex. Cell. 2001;105:645–655. doi: 10.1016/s0092-8674(01)00361-0. [DOI] [PubMed] [Google Scholar]
- Reimann JD, Jackson PK. Emi1 is required for cytostatic factor arrest in vertebrate eggs. Nature. 2002;416:850–854. doi: 10.1038/416850a. [DOI] [PubMed] [Google Scholar]
- Reis A, Levasseur M, Chang HY, Elliott DJ, Jones KT. The CRY box: a second APCcdh1-dependent degron in mammalian cdc20. EMBO Rep. 2006;7:1040–1045. doi: 10.1038/sj.embor.7400772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenkova E, Herrmann K, Adelfalk C, Jessberger R. Oocyte cohesin expression restricted to predictyate stages provides full fertility and prevents aneuploidy. Curr Biol. 2010;20:1529–1533. doi: 10.1016/j.cub.2010.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Brenni MC, Morgan DO. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130:127–139. doi: 10.1016/j.cell.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Rudner AD, Murray AW. Phosphorylation by Cdc28 activates the Cdc20-dependent activity of the anaphase-promoting complex. J Cell Biol. 2000;149:1377–1390. doi: 10.1083/jcb.149.7.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salah SM, Nasmyth K. Destruction of the securin Pds1p occurs at the onset of anaphase during both meiotic divisions in yeast. Chromosoma. 2000;109:27–34. doi: 10.1007/s004120050409. [DOI] [PubMed] [Google Scholar]
- Salic A, Waters JC, Mitchison TJ. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell. 2004;118:567–578. doi: 10.1016/j.cell.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Duncan PI, Rauh NR, Sauer G, Fry AM, Nigg EA, Mayer TU. Xenopus polo-like kinase Plx1 regulates XErp1, a novel inhibitor of APC/C activity. Genes Dev. 2005;19:502–513. doi: 10.1101/gad.320705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Rauh NR, Nigg EA, Mayer TU. Cytostatic factor: an activity that puts the cell cycle on hold. J Cell Sci. 2006;119:1213–1218. doi: 10.1242/jcs.02919. [DOI] [PubMed] [Google Scholar]
- Schwab M, Neutzner M, Möcker D, Seufert W. Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. EMBO J. 2001;20:5165–5175. doi: 10.1093/emboj/20.18.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sczaniecka M, Feoktistova A, May KM, Chen JS, Blyth J, Gould KL, Hardwick KG. The spindle checkpoint functions of Mad3 and Mad2 depend on a Mad3 KEN box-mediated interaction with Cdc20-anaphase-promoting complex (APC/C) J Biol Chem. 2008;283:23039–23047. doi: 10.1074/jbc.M803594200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seino H, Kishi T, Nishitani H, Yamao F. Two ubiquitin-conjugating enzymes, UbcP1/Ubc4 and UbcP4/Ubc11, have distinct functions for ubiquitination of mitotic cyclin. Mol Cell Biol. 2003;23:3497–3505. doi: 10.1128/MCB.23.10.3497-3505.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Zachariae W, Ciosk R, Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shonn MA, McCarroll R, Murray AW. Requirement of the spindle checkpoint for proper chromosome segregation in budding yeast meiosis. Science. 2000;289:300–303. doi: 10.1126/science.289.5477.300. [DOI] [PubMed] [Google Scholar]
- Shteinberg M, Protopopov Y, Listovsky T, Brandeis M, Hershko A. Phosphorylation of the cyclosome is required for its stimulation by Fizzy/cdc20. Biochem Biophys Res Commun. 1999;260:193–198. doi: 10.1006/bbrc.1999.0884. [DOI] [PubMed] [Google Scholar]
- Simpson-Lavy KJ, Sajman J, Zenvirth D, Brandeis M. APC/CCdh1 specific degradation of Hsl1 and Clb2 is required for proper stress responses of S. cerevisiae. Cell Cycle. 2009;8:3003–3009. [PubMed] [Google Scholar]
- Sironi L, Melixetian M, Faretta M, Prosperini E, Helin K, Musacchio A. Mad2 binding to Mad1 and Cdc20, rather than oligomerization, is required for the spindle checkpoint. EMBO J. 2001;20:6371–6382. doi: 10.1093/emboj/20.22.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar JR, Pagano M. Cdh1: a master G0/G1 regulator. Nat Cell Biol. 2008;10:755–757. doi: 10.1038/ncb0708-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar JR, Pagano M. Control of cell growth by the SCF and APC/C ubiquitin ligases. Curr Opin Cell Biol. 2009;21:816–824. doi: 10.1016/j.ceb.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MS, Lim DS. Control of APC-Cdc20 by the tumor suppressor RASSF1A. Cell Cycle. 2004;3:574–576. [PubMed] [Google Scholar]
- Song MS, Song SJ, Ayad NG, Chang JS, Lee JH, Hong HK, Lee H, Choi N, Kim J, Kim H, Kim JW, Choi EJ, Kirschner MW, Lim DS. The tumour suppressor RASSF1A regulates mitosis by inhibiting the APC-Cdc20 complex. Nat Cell Biol. 2004;6:129–137. doi: 10.1038/ncb1091. [DOI] [PubMed] [Google Scholar]
- Steen JA, Steen H, Georgi A, Parker K, Springer M, Kirchner M, Hamprecht F, Kirschner MW. Different phosphorylation states of the anaphase promoting complex in response to antimitotic drugs: a quantitative proteomic analysis. Proc Natl Acad Sci USA. 2008;105:6069–6074. doi: 10.1073/pnas.0709807104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Rape M, Draviam VM, Nalepa G, Sowa ME, Ang XL, McDonald ER, 3rd, Li MZ, Hannon GJ, Sorger PK, Kirschner MW, Harper JW, Elledge SJ. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature. 2007;446:876–881. doi: 10.1038/nature05694. [DOI] [PubMed] [Google Scholar]
- Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca FC, Ruderman JV, Hershko A. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6:185–197. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V, Shteinberg M, Ganoth D, Hershko J, Hershko A. Binding of activated cyclosome to p13(suc1). Use for affinity purification. J Biol Chem. 1997;272:18051–18059. doi: 10.1074/jbc.272.29.18051. [DOI] [PubMed] [Google Scholar]
- Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M, Morgan DO. A novel destruction sequence targets the meiotic regulator Spo13 for anaphase-promoting complex-dependent degradation in anaphase I. J Biol Chem. 2007;282:19710–19715. doi: 10.1074/jbc.M701507200. [DOI] [PubMed] [Google Scholar]
- Summers MK, Pan B, Mukhyala K, Jackson PK. The unique N terminus of the UbcH10 E2 enzyme controls the threshold for APC activation and enhances checkpoint regulation of the APC. Mol Cell. 2008;31:544–556. doi: 10.1016/j.molcel.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan A, Schüpbach T. The Cdc20 (Fzy)/Cdh1-related protein, Cort, cooperates with Fzy in cyclin destruction and anaphase progression in meiosis I and II in Drosophila. Development. 2007;134:891–899. doi: 10.1242/dev.02784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K. The proteasome: overview of structure and functions. Proc Jpn Acad, Ser B, Phys Biol Sci. 2009;85:12–36. doi: 10.2183/pjab.85.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Wu JQ, Chen C, Yang CS, Guo JY, Freel CD, Kornbluth S. Emi2-mediated inhibition of E2-substrate ubiquitin transfer by the anaphase-promoting complex/cyclosome through a D-box-independent mechanism. Mol Biol Cell. 2010;21:2589–2597. doi: 10.1091/mbc.E09-08-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Bharadwaj R, Li B, Yu H. Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev Cell. 2001a;1:227–237. doi: 10.1016/s1534-5807(01)00019-3. [DOI] [PubMed] [Google Scholar]
- Tang Z, Li B, Bharadwaj R, Zhu H, Ozkan E, Hakala K, Deisenhofer J, Yu H. APC2 Cullin protein and APC11 RING protein comprise the minimal ubiquitin ligase module of the anaphase-promoting complex. Mol Biol Cell. 2001b;12:3839–3851. doi: 10.1091/mbc.12.12.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Shu H, Oncel D, Chen S, Yu H. Phosphorylation of Cdc20 by Bub1 provides a catalytic mechanism for APC/C inhibition by the spindle checkpoint. Mol Cell. 2004;16:387–397. doi: 10.1016/j.molcel.2004.09.031. [DOI] [PubMed] [Google Scholar]
- Tedesco D, Zhang J, Trinh L, Lalehzadeh G, Meisner R, Yamaguchi KD, Ruderman DL, Dinter H, Zajchowski DA. The ubiquitin-conjugating enzyme E2-EPF is overexpressed in primary breast cancer and modulates sensitivity to topoisomerase II inhibition. Neoplasia. 2007;9:601–613. doi: 10.1593/neo.07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teodoro JG, Heilman DW, Parker AE, Green MR. The viral protein Apoptin associates with the anaphase-promoting complex to induce G2/M arrest and apoptosis in the absence of p53. Genes Dev. 2004;18:1952–1957. doi: 10.1101/gad.1198404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirthagiri E, Robinson CM, Huntley S, Davies M, Yap LF, Prime SS, Paterson IC. Spindle assembly checkpoint and centrosome abnormalities in oral cancer. Cancer Lett. 2007;258:276–285. doi: 10.1016/j.canlet.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Thornton BR, Toczyski DP. Securin and B-cyclin/CDK are the only essential targets of the APC. Nat Cell Biol. 2003;5:1090–1094. doi: 10.1038/ncb1066. [DOI] [PubMed] [Google Scholar]
- Thornton BR, Ng TM, Matyskiela ME, Carroll CW, Morgan DO, Toczyski DP. An architectural map of the anaphase-promoting complex. Genes Dev. 2006;20:449–460. doi: 10.1101/gad.1396906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton BR, Toczyski DP. Precise destruction: an emerging picture of the APC. Genes Dev. 2006;20:3069–3078. doi: 10.1101/gad.1478306. [DOI] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres JZ, Ban KH, Jackson PK. A specific form of phospho protein phosphatase 2 regulates anaphase-promoting complex/cyclosome association with spindle poles. Mol Biol Cell. 2010;21:897–904. doi: 10.1091/mbc.E09-07-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MP, Borchers CH. Mitotic phosphorylation of the anaphase-promoting complex inhibitory subunit Mnd2 is necessary for efficient progression through meiosis i. J Biol Chem. 2007;282:17351–17362. doi: 10.1074/jbc.M610841200. [DOI] [PubMed] [Google Scholar]
- Townsley FM, Aristarkhov A, Beck S, Hershko A, Ruderman JV. Dominant-negative cyclin-selective ubiquitin carrier protein E2-C/UbcH10 blocks cells in metaphase. Proc Natl Acad Sci USA. 1997;94:2362–2367. doi: 10.1073/pnas.94.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran K, Mahr JA, Choi J, Teodoro JG, Green MR, Spector DH. Accumulation of substrates of the anaphase-promoting complex (APC) during human cytomegalovirus infection is associated with the phosphorylation of Cdh1 and the dissociation and relocalization of APC subunits. J Virol. 2008;82:529–537. doi: 10.1128/JVI.02010-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran K, Kamil JP, Coen DM, Spector DH. Inactivation and disassembly of the anaphase-promoting complex during human cytomegalovirus infection is associated with degradation of the APC5 and APC4 subunits and does not require UL97-mediated phosphorylation of Cdh1. J Virol. 2010;84:10832–10843. doi: 10.1128/JVI.01260-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurumi C, Hoffmann S, Geley S, Graeser R, Polanski Z. The spindle assembly checkpoint is not essential for CSF arrest of mouse oocytes. J Cell Biol. 2004;167:1037–1050. doi: 10.1083/jcb.200405165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugendreich S, Tomkiel J, Earnshaw W, Hieter P. CDC27Hs colo-calizes with CDC16Hs to the centrosome and mitotic spindle and is essential for the metaphase to anaphase transition. Cell. 1995;81:261–268. doi: 10.1016/0092-8674(95)90336-4. [DOI] [PubMed] [Google Scholar]
- Tung JJ, Hansen DV, Ban KH, Loktev AV, Summers MK, Adler JR, 3rd, Jackson PK. A role for the anaphase-promoting complex inhibitor Emi2/XErp1, a homolog of early mitotic inhibitor 1, in cytostatic factor arrest of Xenopus eggs. Proc Natl Acad Sci USA. 2005;102:4318–4323. doi: 10.1073/pnas.0501108102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung JJ, Jackson PK. Emi1 class of proteins regulate entry into meiosis and the meiosis I to meiosis II transition in Xenopus oocytes. Cell Cycle. 2005;4:478–482. doi: 10.4161/cc.4.3.1532. [DOI] [PubMed] [Google Scholar]
- van Leuken R, Clijsters L, Wolthuis R. To cell cycle, swing the APC/C. Biochim Biophys Acta. 2008;1786:49–59. doi: 10.1016/j.bbcan.2008.05.002. [DOI] [PubMed] [Google Scholar]
- van Ree JH, Jeganathan KB, Malureanu L, van Deursen JM. Overexpression of the E2 ubiquitin-conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. J Cell Biol. 2010;188:83–100. doi: 10.1083/jcb.200906147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zon W, Ogink J, ter Riet B, Medema RH, te Riele H, Wolthuis RM. The APC/C recruits cyclin B1-Cdk1-Cks in promet-aphase before D box recognition to control mitotic exit. J Cell Biol. 2010;190:587–602. doi: 10.1083/jcb.200912084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoosthuyse V, Hardwick KG. A novel protein phosphatase 1-dependent spindle checkpoint silencing mechanism. Curr Biol. 2009;19:1176–1181. doi: 10.1016/j.cub.2009.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- Vodermaier HC, Gieffers C, Maurer-Stroh S, Eisenhaber F, Peters JM. TPR subunits of the anaphase-promoting complex mediate binding to the activator protein CDH1. Curr Biol. 2003;13:1459–1468. doi: 10.1016/s0960-9822(03)00581-5. [DOI] [PubMed] [Google Scholar]
- Vodermaier HC. APC/C and SCF: controlling each other and the cell cycle. Curr Biol. 2004;14:R787–R796. doi: 10.1016/j.cub.2004.09.020. [DOI] [PubMed] [Google Scholar]
- Wai P, Truong B, Bhat KM. Cell division genes promote asymmetric interaction between Numb and Notch in the Drosophila CNS. Development. 1999;126:2759–2770. doi: 10.1242/dev.126.12.2759. [DOI] [PubMed] [Google Scholar]
- Walker A, Acquaviva C, Matsusaka T, Koop L, Pines J. UbcH10 has a rate-limiting role in G1 phase but might not act in the spindle checkpoint or as part of an autonomous oscillator. J Cell Sci. 2008;121:2319–2326. doi: 10.1242/jcs.031591. [DOI] [PubMed] [Google Scholar]
- Wang J, Dye BT, Rajashankar KR, Kurinov I, Schulman BA. Insights into anaphase promoting complex TPR subdomain assembly from a CDC26-APC6 structure. Nat Struct Mol Biol. 2009;16:987–989. doi: 10.1038/nsmb.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Moyret-Lalle C, Couzon F, Surbiguet-Clippe C, Saurin JC, Lorca T, Navarro C, Puisieux A. Alterations of anaphase-promoting complex genes in human colon cancer cells. Oncogene. 2003;22:1486–1490. doi: 10.1038/sj.onc.1206224. [DOI] [PubMed] [Google Scholar]
- Wäsch R, Robbins JA, Cross FR. The emerging role of APC/CCdh1 in controlling differentiation, genomic stability and tumor suppression. Oncogene. 2010;29:1–10. doi: 10.1038/onc.2009.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassmann K, Liberal V, Benezra R. Mad2 phosphorylation regulates its association with Mad1 and the APC/C. EMBO J. 2003a;22:797–806. doi: 10.1093/emboj/cdg071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassmann K, Niault T, Maro B. Metaphase I arrest upon activation of the Mad2-dependent spindle checkpoint in mouse oocytes. Curr Biol. 2003b;13:1596–1608. doi: 10.1016/j.cub.2003.08.052. [DOI] [PubMed] [Google Scholar]
- Wei L, Liang XW, Zhang QH, Li M, Yuan J, Li S, Sun SC, Ouyang YC, Schatten H, Sun QY. BubR1 is a spindle assembly checkpoint protein regulating meiotic cell cycle progression of mouse oocyte. Cell Cycle. 2010;9:1112–1121. doi: 10.4161/cc.9.6.10957. [DOI] [PubMed] [Google Scholar]
- Wendt KS, Vodermaier HC, Jacob U, Gieffers C, Gmachl M, Peters JM, Huber R, Sondermann P. Crystal structure of the APC10/DOC1 subunit of the human anaphase-promoting complex. Nat Struct Biol. 2001;8:784–788. doi: 10.1038/nsb0901-784. [DOI] [PubMed] [Google Scholar]
- Whitehurst AW, Ram R, Shivakumar L, Gao B, Minna JD, White MA. The RASSF1A tumor suppressor restrains anaphase-promoting complex/cyclosome activity during the G1/S phase transition to promote cell cycle progression in human epithelial cells. Mol Cell Biol. 2008;28:3190–3197. doi: 10.1128/MCB.02291-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebusch L, Bach M, Uecker R, Hagemeier C. Human cytome-galovirus inactivates the G0/G1-APC/C ubiquitin ligase by Cdh1 dissociation. Cell Cycle. 2005;4:1435–1439. doi: 10.4161/cc.4.10.2077. [DOI] [PubMed] [Google Scholar]
- Williamson A, Wicklife KE, Mellone BG, Song L, Karpen GH, Rape M. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc Natl Acad Sci USA. 2009;106:18213–18218. doi: 10.1073/pnas.0907887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Grady JT, Villén J, Gygi SP. Phosphoproteome analysis of fission yeast. J Proteome Res. 2008;7:1088–1097. doi: 10.1021/pr7006335. [DOI] [PubMed] [Google Scholar]
- Wirth KG, Ricci R, Giménez-Abián JF, Taghybeeglu S, Kudo NR, Jochum W, Vasseur-Cognet M, Nasmyth K. Loss of the anaphase-promoting complex in quiescent cells causes unscheduled hepatocyte proliferation. Genes Dev. 2004;18:88–98. doi: 10.1101/gad.285404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolthuis R, Clay-Farrace L, van Zon W, Yekezare M, Koop L, Ogink J, Medema R, Pines J. Cdc20 and Cks direct the spindle checkpoint-independent destruction of cyclin A. Mol Cell. 2008;30:290–302. doi: 10.1016/j.molcel.2008.02.027. [DOI] [PubMed] [Google Scholar]
- Wu T, Merbl Y, Huo Y, Gallop JL, Tzur A, Kirschner MW. UBE2S drives elongation of K11-linked ubiquitin chains by the anaphase-promoting complex. Proc Natl Acad Sci USA. 2010;107:1355–1360. doi: 10.1073/pnas.0912802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia G, Luo X, Habu T, Rizo J, Matsumoto T, Yu H. Conformation-specific binding of p31(comet) antagonizes the function of Mad2 in the spindle checkpoint. EMBO J. 2004;23:3133–3143. doi: 10.1038/sj.emboj.7600322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H, Kumada K, Yanagida M. Distinct subunit functions and cell cycle regulated phosphorylation of 20S APC/cyclosome required for anaphase in fssion yeast. Cell Sci. 1997;110(Pt 15):1793–1804. doi: 10.1242/jcs.110.15.1793. [DOI] [PubMed] [Google Scholar]
- Yamada HY, Matsumoto S, Matsumoto T. High dosage expression of a zinc finger protein, Grt1, suppresses a mutant of fission yeast slp1(+), a homolog of CDC20/p55CDC/Fizzy. J Cell Sci. 2000;113(Pt 22):3989–3999. doi: 10.1242/jcs.113.22.3989. [DOI] [PubMed] [Google Scholar]
- Yamano H, Tsurumi C, Gannon J, Hunt T. The role of the destruction box and its neighbouring lysine residues in cyclin B for anaphase ubiquitin-dependent proteolysis in fission yeast: defining the D-box receptor. EMBO J. 1998;17:5670–5678. doi: 10.1093/emboj/17.19.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita YM, Nakaseko Y, Samejima I, Kumada K, Yamada H, Michaelson D, Yanagida M. 20S cyclosome complex formation and proteolytic activity inhibited by the cAMP/PKA pathway. Nature. 1996;384:276–279. doi: 10.1038/384276a0. [DOI] [PubMed] [Google Scholar]
- Yang M, Li B, Tomchick DR, Machius M, Rizo J, Yu H, Luo X. p31comet blocks Mad2 activation through structural mimicry. Cell. 2007;131:744–755. doi: 10.1016/j.cell.2007.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Kim AH, Bonni A. The dynamic ubiquitin ligase duo: Cdh1-APC and Cdc20-APC regulate neuronal morphogenesis and connectivity. Curr Opin Neurobiol. 2010;20:92–99. doi: 10.1016/j.conb.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HJ, Feoktistova A, Chen JS, Jennings JL, Link AJ, Gould KL. Role of Hcn1 and its phosphorylation in fission yeast anaphase-promoting complex/cyclosome function. J Biol Chem. 2006;281:32284–32293. doi: 10.1074/jbc.M603867200. [DOI] [PubMed] [Google Scholar]
- Yu H, King RW, Peters JM, Kirschner MW. Identification of a novel ubiquitin-conjugating enzyme involved in mitotic cyclin degradation. Curr Biol. 1996;6:455–466. doi: 10.1016/s0960-9822(02)00513-4. [DOI] [PubMed] [Google Scholar]
- Yu H. Cdc20: a WD40 activator for a cell cycle degradation machine. Mol Cell. 2007;27:3–16. doi: 10.1016/j.molcel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Yuan B, Xu Y, Woo JH, Wang Y, Bae YK, Yoon DS, Wersto RP, Tully E, Wilsbach K, Gabrielson E. Increased expression of mitotic checkpoint genes in breast cancer cells with chromosomal instability. Clin Cancer Res. 2006;12:405–410. doi: 10.1158/1078-0432.CCR-05-0903. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Schwab M, Nasmyth K, Seufert W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 1998;282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Kulkarni K, Hanrahan SJ, Thompson AJ, Barford D. The APC/C subunit Cdc16/Cut9 is a contiguous tetratricopeptide repeat superhelix with a homo-dimer interface similar to Cdc27. EMBO J. 2010a;29:3733–3744. doi: 10.1038/emboj.2010.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Roe SM, Diogon M, Kong E, El Alaoui H, Barford D. Molecular structure of the N-terminal domain of the APC/C subunit Cdc27 reveals a homo-dimeric tetratricopeptide repeat architecture. J Mol Biol. 2010b;397:1316–1328. doi: 10.1016/j.jmb.2010.02.045. [DOI] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- Zielke N, Querings S, Rottig C, Lehner C, Sprenger F. The anaphase-promoting complex/cyclosome (APC/C) is required for rereplication control in endoreplication cycles. Genes Dev. 2008;22:1690–1703. doi: 10.1101/gad.469108. [DOI] [PMC free article] [PubMed] [Google Scholar]