Abstract
Development of precision therapeutics is of immense interest, particularly as applied to the treatment of cancer. By analyzing the preferred cellular RNA targets of small molecules, we discovered that 5″-azido neomycin B binds the Drosha processing site in the microRNA (miR)-525 precursor. MiR-525 confers invasive properties to hepatocellular carcinoma (HCC) cells. Although HCC is one of the most common cancers, treatment options are limited, making the disease often fatal. Herein, we find that addition of 5″-azido neomycin B and its FDA-approved precursor, neomycin B, to an HCC cell line selectively inhibits production of the mature miRNA, boosts a downstream protein, and inhibits invasion. Interestingly, neomycin B is a second-line agent for hepatic encephalopathy (HE) and bacterial infections due to cirrhosis. Our results provocatively suggest that neomycin B, or second-generation derivatives, may be dual functioning molecules to treat both HE and HCC. Collectively, these studies show that rational design approaches can be tailored to disease-associated RNAs to afford potential lead therapeutics.
Keywords: Chemical Biology, Precision Medicine, RNA, Nucleic Acids, Medicinal Chemistry
MicroRNAs (miRNAs) are small noncoding RNAs that regulate gene expression by binding to the 3′ untranslated regions (UTRs) of mRNAs.(1) Although miRNAs comprise only ~1% of the human genome, they regulate expression of up to 50% of all genes.(2) Not surprisingly, miRNA expression is tightly controlled,(3, 4) and dysregulation of miRNA abundance has been associated with many diseases including cancers,(4, 5) Alzheimer’s disease,(6) and cardiovascular disease,(7) among others.(8) Indeed, many diseases and their prognoses have signature miRNA profiles.(9)
Like many cellular RNAs, miRNAs are synthesized as precursors, often as clusters of multiple miRNAs.(10) They are processed in two steps, the first in the nucleus by the nuclease Drosha.(11) After Drosha cleavage, the pre-miRNA is ~70 nucleotides and is exported to the cytoplasm where final processing is completed by the nuclease Dicer.(12) The mature miRNA is 21–23 nucleotide dsRNA and binds to the 3′ untranslated regions (UTRs) of mRNA to either repress its translation or induce its degradation.(1, 13)
MiRNAs are perhaps an ideal class for the development of precision therapeutics as aberrantly expressed miRNAs in disease-affected cells, especially various cancers, have been widely reported. One manner to selectivity drug miRNAs is to identify compounds that bind specifically to Drosha or Dicer sites in a miRNA hairpin precursor, thereby inhibiting production of the mature miRNA. In our previous study, we identified lead small molecules that bind to Drosha and Dicer processing sites of human miRNAs that are disease-associated using a computational approach developed in our lab named Inforna.(14) Inforna compares the secondary structural elements (motifs) that comprise a target RNA’s structure to known interactions between small molecules and RNA motifs. Our hypothesis was that Drosha and Dicer processing sites represent functional sites; thus, small molecule binding to these sites could inhibit miRNA biogenesis. Indeed, 44% of our lead small molecules decreased levels of the desired miRNA target in cells.(14)
One of the lead RNA–small molecule interactions identified in those studies was between the Drosha site of microRNA (miR)-525 and the aminoglycoside derivative 5″-azido-neomycin B (Neo-N3; Figure 1).(14) Indeed, Neo-N3 decreases levels of the mature miRNA by ~50% when MCF7 (breast cancer) cells were treated with 10 µM compound. Interestingly, miR-525 is overexpressed in ~60% of liver cancer tissues and indicates poor prognosis.(15) Overexpression of miR-525 enhances migratory and invasive properties via down-regulation of zinc finger protein 395 (ZNF395; also known as HDBP2).(15) Herein, we report that Neo-N3 decreases levels of mature miR-525, up-regulates ZNF395, and inhibits the invasive properties of a hepatocellular carcinoma cell line. Interestingly, other miRNAs predicted to bind the ZNF395 UTR are not affected by Neo-N3, showing that the compound’s effect can be traced to inhibition of miR-525. Importantly, the functional outcomes of inhibiting miR-525 biogenesis are reversed upon forced expression of pri-miR-525. Collectively, these results demonstrate that the compound is pathway-selective.
Figure 1.
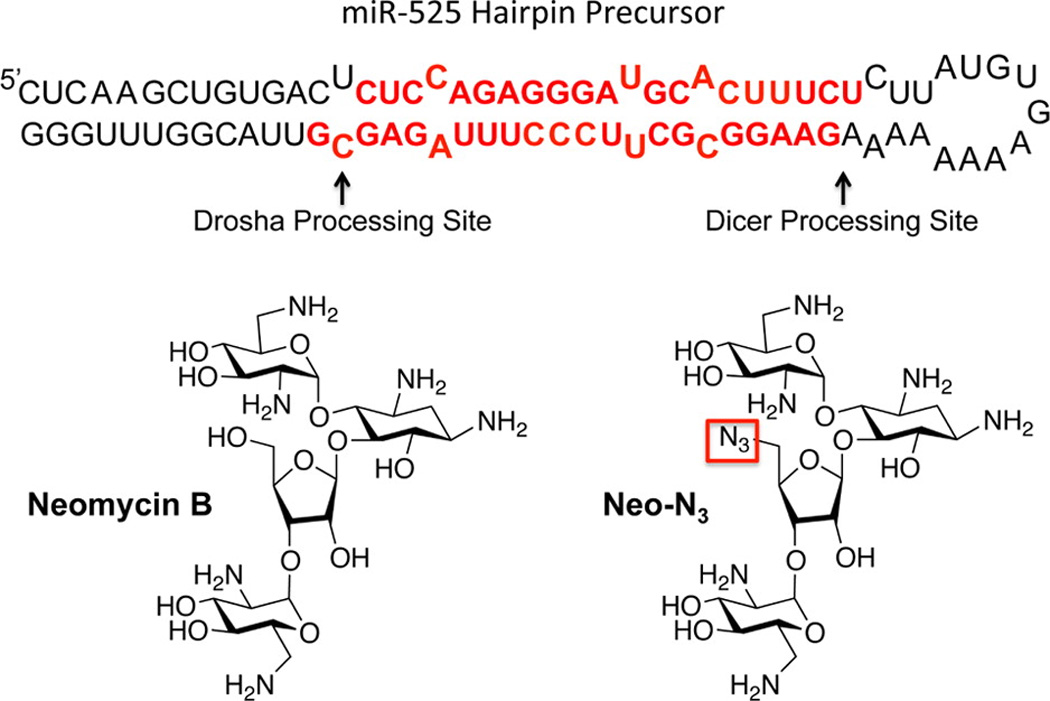
Secondary structure of the miR-525 hairpin precursor and structures of the compounds used in these studies. Drosha and Dicer processing sites are indicated with arrows. The mature miRNA sequences, miR-525-3p and miR-525-5p, are indicated with red lettering. The azide functional group in Neo-N3 is boxed in red.
Results and Discussion
Neo-N3 Inhibits miR-525 Biogenesis in HepG2 Cells
We first confirmed that Neo-N3 inhibits miR-525 biogenesis in the hepatocellular carcinoma (HCC) cell line HepG2 by using qRT-PCR. In agreement with our previous studies in MCF7 cells,(14) Neo-N3 decreases levels of miR-525–3p by ~40% when HepG2 cells are treated with 6.25 µM or 25 µM compound as compared to untreated cells (Figure 2A, black bars). To gain insight into if the observed decrease in miR-525–3p levels were due to inhibition of biogenesis and not transcriptional inhibition, we also determined how Neo-N3 affected levels of pri- and pre-miR-525. If Neo-N3 binds to the Drosha site and inhibits processing, an increase of pri-miR-525 levels and a decrease in pre-miR-525 levels are expected. Indeed, pri-miR-525 levels increase by ~2-fold upon treatment with 25 µM Neo-N3 (Figure 2A, dark gray bars), while pre-miR-525 levels were decreased by ~40% (Figure 2A, light gray bars); more modest effects were observed with treatment of 6.25 µM compound. Interestingly, Neo-N3’s, parent compound neomycin B, inhibits levels of mature miR-525, but kanamycin A, tobramycin, and streptomycin do not (Figure 2B). In good agreement with our qRT-PCR studies, Neo-N3 inhibits Drosha processing of pri-miR-525 in vitro in a dose-dependent manner, inhibiting ~40% of processing at 25 µM (Figure 3). As we previously defined the preferred motifs that bind Neo-N3 and showed that it inhibited processing of miRNAs with those motifs, we focused the remainder of our studies on characterizing Neo-N3 rather than neocymin B.
Figure 2.
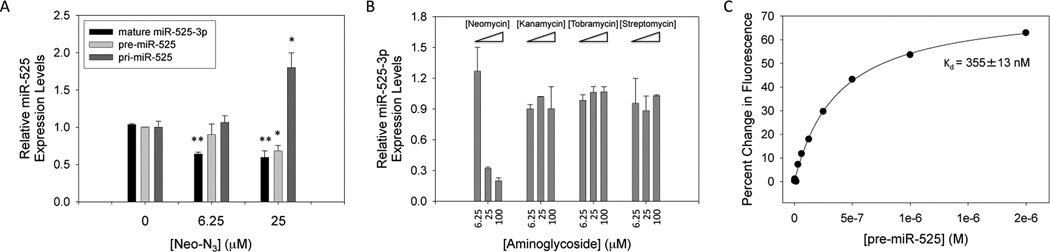
Effect of Neo-N3 and other aminoglycosides on pri-, pre-, and mature miR-525 expression levels in HepG2 cells and binding affinity of Neo-N3 for miR-525 hairpin precursor. (a) Quantification of the effect of Neo-N3 on pri-miR-525, pre-miR-525, and mature miR-525-3p expression levels in HepG2 cells as determined by qRT-PCR. (b) Quantification of the effects of neomycin B, kanamycin A, tobramycin, and streptomycin on mature miR-525 levels in HepG2 as determined by qRT-PCR. (c) Representative binding curve for Neo-N3 and the miR-525 hairpin precursor. The Kd, 355 ± 13 nM, is the average of individual curve fits ± standard deviation. *p < 0.05 and **p < 0.01, as determined by a Student t test.
Figure 3.
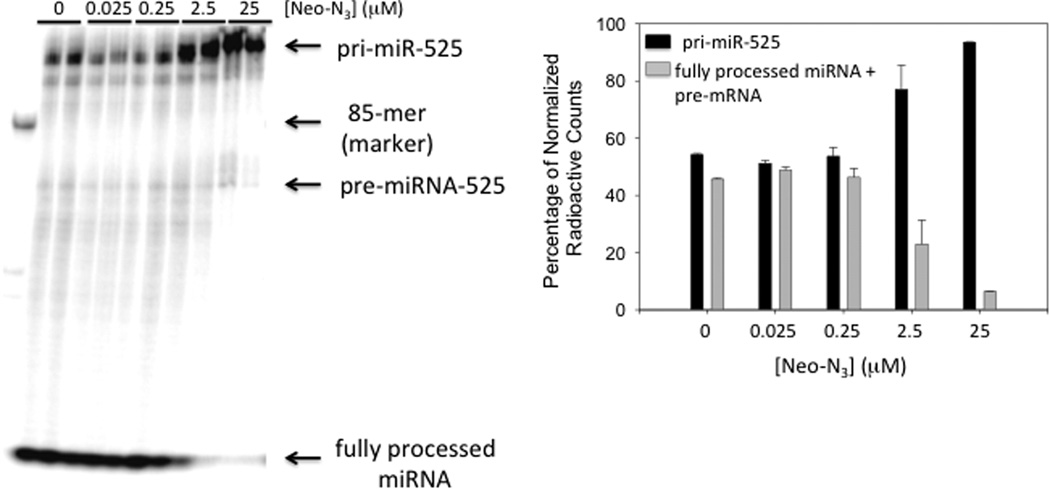
Neo-N3 inhibits Drosha processing of pri-miR-525 in cell extracts. Note: cell extracts have endogenous Dicer activity, enabling processing to the mature miRNA. Left, representative gel image of pri-miR-525 processing in the presence and absence of Neo-N3. Right, quantification of processing as a function of Neo-N3 concentration. Data were normalized for the number of A residues in each product, as the substrate was internally labeled.
Next, we measured the affinity of Neo-N3 for the miR-525 precursor hairpin using a previously described in solution binding assay using a fluorescently labeled derivative of Neo-N3, or Neo-FL. Neo-FL binds the miRNA hairpin with a Kd of 355 ± 13 nM (Figure 2C). The difference between the measured Kd and the ability to inhibit processing in vitro and in cells could be due to several factors. For example, there could be additional binding sites for Neo-N3 in other cellular RNAs (also present in cell lysate used for in vitro Drosha processing studies), although they may not be functional sites that elicit a downstream effect. Alternatively, the relative association and dissociation rates of Neo-N3 and Drosha as they compete for binding to pri-miR-525 could affect potency. Of note, neomycin does not bind to human serum proteins.(16)
Since aminoglycosides, such as neomycin B, target the bacterial and human ribosome and affect translation, we tested if Neo-N3 affects translation in mammalian cells. Studies were completed by transfecting HepG2 cells with a luciferase reporter plasmid followed by compound treatment. No effect on translation was observed at the concentrations of Neo-N3 that inhibit miR-525 biogenesis (Supporting Information Figure 1). Thus, the translational machinery is not a significant off-target. Although it is widely accepted that aminoglycosides used as antibacterials also bind the human A-site, it should be noted that kasugamycin and streptomycin (up to 1.6 mM, the highest concentration tested) do not affect mitochondrial translation in liver or heart cells.(17)
Neo-N3 Upregulates ZNF395 and Inhibits Invasion
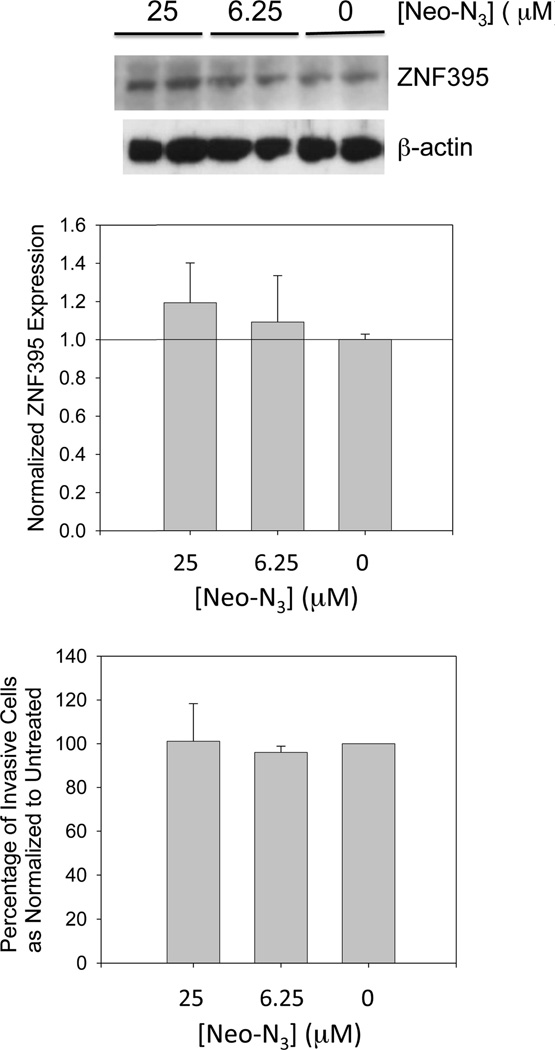
One of miR-525’s downstream targets is ZNF395. ZNF395 protein functions as a transcription factor and was originally identified in a yeast one-hybrid system for regulating transcription of the HTT gene in Huntington’s disease (HD).(18) Importantly, Pang et al. discovered that miR-525 is up-regulated in ~60% of liver cancer tissues and that these increased levels promote migration and invasion via down-regulation of ZNF395.(15) Because of the urgent need for compounds to treat HCC, we studied if inhibition of miR-525 biogenesis by Neo-N3 is sufficient to up-regulate ZNF395 levels. Indeed, Western blotting revealed a dose-dependent increase in ZNF-395 levels, with an ~1.7- and ~2-fold increase observed when cells were treated with 6.25 or 25 µM compound, respectively (Figure 4A). We next determined if up-regulation of ZNF395 was sufficient to inhibit the invasive properties of HepG2 cells using a Boyden chamber assay.(19) Neo-N3 decreases invasion by ~40% and ~80% when cells are treated with 6.25 and 25 µM compound, respectively (Figure 4B).
Figure 4.
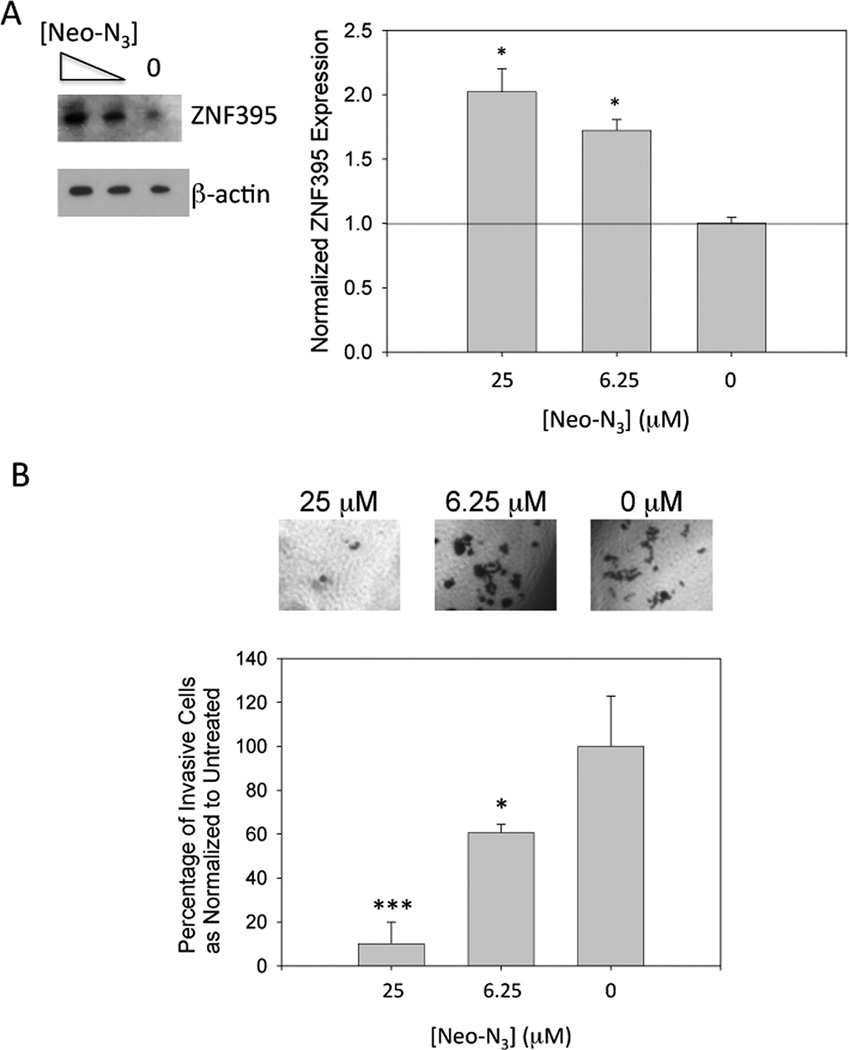
Treatment with Neo-N3 derepresses inhibition of ZNF395 expression by miR-525 and attenuates invasion of HepG2 cells. (a) Representative Western blot of ZNF395 expression and quantification as normalized to β-actin. (b) Top, representative microscopic images of invasive cells as a function of Neo-N3 concentration. Bottom, quantification of the number of invasive cells as a function of Neo-N3 concentration. ***p < 0.001; *p < 0.05 as determined by a Student t test.
Effect of Neo-N3 on Levels of Other miRNAs
Our previous studies showed that treatment of MCF7 cells with 100 µM Neo-N3 decreased levels of miR-517c (~20% reduction), miR-518e (~50% reduction), miR-519-d (~50% reduction), and miR-525 (~60% reduction).(14) In HepG2 cells, miR-517c, miR-518e, and miR-519d are all expressed at low levels and could not be assayed (Ct values >35). We therefore profiled the effect of Neo-N3 on conserved miRNAs that are predicted by both TargetScan(13) and miRanda(2, 20, 21) to bind ZNF395 3′ UTR (Ct values ≤ 30; n = 19). No statistically significant changes were observed (Figure 5). Notably, miR-126, which is significantly down-regulated in hepatocellular carcinoma as compared to healthy hepatocytes,(22–24) was unaffected by compound treatment.
Figure 5.
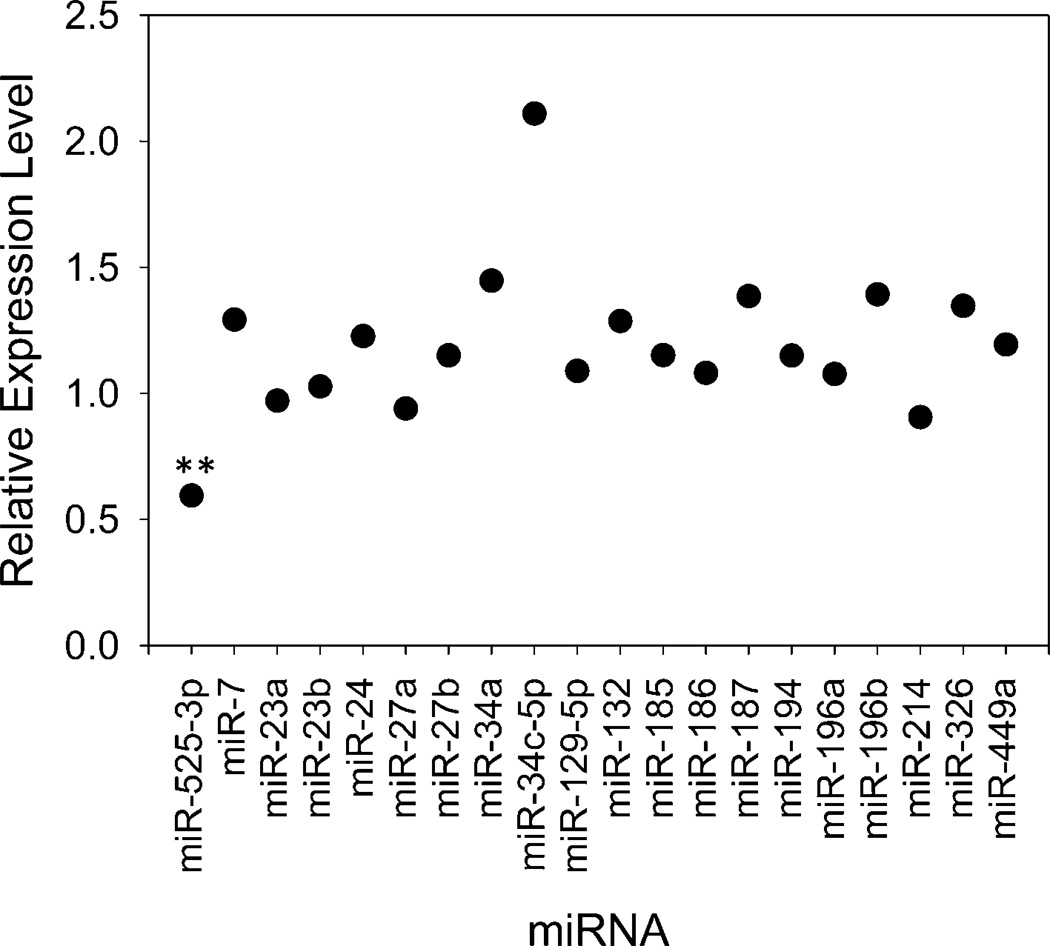
Neo-N3 does not statistically significantly affect levels of other mature miRNAs predicted to bind ZNF395 3′ UTR when HepG2 cells were treated with 25 µM compound. **p < 0.01, as determined by a Student t test.
Although Neo-N3 could bind to various RNAs, binding events likely occur in nonfunctional sites; that is, binding to these sites has no downstream effect. For example, a recent report screened the binding of various aminoglycosides, anticancer therapeutics, and RNA intercalators to pre-miR-155.(25) Although neomycin B binds with moderate affinity to pre-miR-155 (11 µM), it only inhibits in vitro Dicer processing by ~10% at 1 mM concentration.(25) Not surprisingly, miR-155 abundance levels were unaffected by treatment with Neo-N3.
Pri-miR-525 Overexpression Decreases Potency
To further probe the selectivity of Neo-N3 on the miR-525-ZNF395 circuit, we studied if overexpression of pri-miR-525 attenuates compound potency. Indeed, increased levels of pri-miR-525 attenuate derepression of ZNF395 as assessed by Western blotting and decreases Neo-N3’s anti-invasion effect (Figure 6). Thus, Neo-N3 is a selective modulator of important downstream processes by targeting miR-525.
Figure 6.
Forced overexpression of pri-miR-525 decreases the potency of Neo-N3, as assessed by derepression of ZNF395 and invasion of HepG2 cells. Top, representative Western blot showing the reduced effect of Neo-N3 on ZNF395 levels when miR-525 is overexpressed. Middle, quantification of ZNF395 levels as determined by Western blotting. Bottom, overexpression of miR-525 also decreases the potency of Neo-N3 to attenuate invasion of HepG2 cells.
Potential Therapeutic Use of Neo-N3
Neomycin B is used clinically to treat hepatic encephalopathy (by reducing ammonium levels in the gut) and enteropathogenic Escherichia coli infections.(26) Dosage for the latter in adults is 6 g per day spread out over four doses. For a 100 kg adult, assuming an 8 L blood volume and 3% absorption,(27) this correlates to a potential blood concentration of 7.5 µM. It should be noted, however, that systemic absorption and tissue accumulation increases cumulatively with each dose.(28, 29) Although neomycin B can cause nephro- and ototoxicity,(30) it is possible that Neo-N3 could be administered at low doses to work synergistically with HCC therapeutics. Interestingly, neomycin B decreases mature miR-525 abundance to a similar extent as Neo-N3 in HepG2 cells (Figures 2A,B).
Conclusions
Using the privileged RNA targets of small molecules, Inforna, our lead identification strategy,(14) identified that a derivative of the well-studied RNA-binding compound neomycin B binds the Drosha site in miR-525. Fortuitously, this drug has been used to treat bacterial infections that occur during cirrhosis and HE, which are underlying conditions associated with HCC. Further, our approach could enable precision therapeutics once a tumor is known to overexpress miR-525. Although there may be many cellular RNAs to which this compound binds, only binding to functional sites, such as the miR-525’s Drosha site, elicited a biological response. Thus, perhaps one advantage that RNA may have as a drug target is that functional responses are less sensitive to simple binding that plagues targeting of DNA. RNA may therefore be a more attractive drug target than DNA. These studies also suggest that identification of the privileged targets of known drugs may inform off-label uses in a predictable way.
Methods
Synthesis of Neo-N3 and Neo-FL
Neo-N3 and Neo-FL were synthesized as previously described.(31)
RNA Constructs and in Vitro Transcription
A DNA template encoding the miR-525 hairpin precursor was purchased from Integrated DNA Technologies, Inc. (IDT) and amplified using the following primers to install a T7 promoter (underlined): forward, 5′-GGCCGGATCCTAATACGACTCACTATACTCAAGCTGTGACTCTCC; reverse, 5′-CCCAAACCGTAACGCTC. The template for pri-miR-525 was produced from the miR-525 hairpin precursor PCR product in two steps. First, 25 nucleotides upstream and downstream of the miR-525 precursor hairpin were added: forward, 5′-ACCCACGGTGCTGGAGCAAGAAGATCTCAAGCTGTGACTCTCC, and reverse, 5′-CAGCATCAACTTCAACGTTGCTTTACCCAAACCGTAACGCTC. The resulting PCR product was then amplified to install a T7 promoter (underlined): forward, 5′-GGCCGGATCCTAATACGACTCACTATAGGACCCACGGTGCTGGAGC, and reverse, 5′-CAGCATCAACTTCAACGTTGCTTTACCCAAACCGTAACGCTC.
PCR amplification was carried out in 1× PCR Buffer (10 mM Tris, pH 9, 50 mM KCl, and 0.1% (v/v) Triton X-100), 1 µM forward primer, 1 µM reverse primer, 4.25 mM MgCl20.33 mM each dNTP, and 1 µL of Taq DNA polymerase. PCR cycling conditions were 95 °C for 30 s, 50 °C for 30 s, and 72 °C for 1 min.
Run-off transcription of the miR-525 hairpin precursor was completed using the PCR-amplified DNA template (600 µL) in 1× Transcription Buffer (40 mM Tris HCl, pH 8.1, 1 mM spermidine, 0.001% (v/v) Triton X-100 and 10 mM DTT(32)) containing 2.25 mM each NTP, 10 mM MgCl2, and 40 µL of T7 RNA polymerase in a total volume of 2 mL. The RNA was purified on a denaturing 10% (w/v) polyacrylamide gel and extracted into 300 mM NaCl by tumbling overnight at 4 °C. After concentrating with 2-butanol and precipitating with ethanol, the concentration of RNA was determined by UV absorbance at 80 °C and the corresponding extinction coefficient.(33–35)
Radioactively labeled pri-miR-525 was produced by in vitro transcription using an RNAMaxx transcription kit (Stratagene) per the manufacturer’s recommended protocol except that the concentration of ATP was reduced by half and supplemented with 4 µL of α-32P-ATP (3000 Ci mmol−1; 10 mCi mL−1). The RNA was purified by gel electrophoresis as described above.
Binding Assays
The affinity of Neo-N3 for the miR-525 hairpin precursor was completed as previously described.(31) Briefly, the RNA was folded in 1× Binding Buffer (20 mM N-(2-hydroxyethyl)piperazine-N′-2-ethanesulfonic acid (Hepes), pH 7.5, 140 mM NaCl, and 5 mM KCl, 1 mM MgCl2, and 40 µg mL−1 BSA) lacking MgCl2 by heating to 60 °C for 5 min. After slowly cooling to RT on the benchtop, MgCl2 and Neo-FL were added to final concentrations of 1 mM and 10 nM, respectively. Serial dilutions were then completed with 1× Binding Buffer containing 10 nM Neo-FL. Samples were incubated for 1 h at RT, after which fluorescence intensity was measured using a Molecular Devices SpectraMax M5 plate reader. Dissociation constants were determined by fitting curves of fluorescence intensity as a function of RNA concentration to eq 1:(36)
| (1) |
where I is the observed fluorescence intensity, I0 is the fluorescence intensity in the absence of RNA, Δε is the difference between the fluorescence intensity in the absence of RNA and in the presence of infinite RNA concentration, [FL]0 is the concentration of fluorescently labeled Neo-FL, [RNA]0 is the concentration of the RNA, and Kd is the dissociation constant. The reported Kd is the average of three measurements ± standard deviation.
Cell Culture
The HepG2 cell line was maintained as monolayers in growth medium (1× DMEM supplemented with 10% (v/v) FBS, 2 mM Glutagro (Corning)) containing 10 IU mL−1 penicillin, and 10 µg mL−1 streptomycin (MP Biomedical). Cells were plated in six-well plates to assess levels of miRNAs and ZNF395 protein. Total RNA was harvested using a Zymo Quick-RNA MiniPrep kit, including an on-column DNase treatment, per the manufacturer’s recommended protocol. Total protein was isolated by adding 100 µL of M-PER Mammalian Protein Extraction Reagent (Pierce Biotechnology/ThermoFisher) to the cells after washing with 1× DPBS.
Reverse Transcription and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Levels of pri-, pre-, and mature miR-525 were measured by qRT-PCR. Reverse transcription was completed by using a miScript II kit (Qiagen) per the manufacturer’s standard protocol (2 µg of total RNA in a 20 µL reaction). qRT-PCR samples (10 µL) were prepared in 1× Power SYBR Green Master Mix (Life Technologies) containing ~50 ng of cDNA, and 500 nM of each primer. qRT-PCR was completed using an ABI 7900 HT Real-Time PCR System (Applied Biosystems). Data were analyzed using the ΔΔCt method and normalized to RNU6. Please see Supporting Information Table 1 for a list of primers and cycling conditions.
Inhibition of Drosha Processing in Vitro
Drosha was overexpressed in HEK293 cells as previously described.(14) Radioactively labeled pri-miR-525 was folded by heating at 70 °C for 5 min followed by slowly cooling to RT on the benchtop. MgCl2 was then added to a final concentration of 6.5 mM, followed by various concentrations of Neo-N3. After incubation at RT for 15 min, 1 µL of cell extract containing overexpressed Drosha was added to each sample (25 µL total volume), and the sample was incubated at RT for an additional 30 min. The reaction was quenched by the addition of an equal volume of 2× Loading Buffer (8 M urea, 2 mM Tris, pH 7.5, and 20 mM EDTA). Reaction products were separated on a denaturing 10% (w/v) polyacrylamide gel. Following exposure to a phosphorimager plate, the gel was imaged using a Molecular Dynamics Typhoon phosphorimager and quantified using QuantityOne software (Bio-Rad).
Effect of Neo-N3 on Translation
HepG2 cells were plated into 96-well plates and transfected with a plasmid encoding firefly luciferase with jetPRIME transfection reagent per the manufacturer’s standard protocol. The transfection cocktail was removed and replaced with growth medium with or without Neo-N3. After 24 h, cell viability was measured using WST-1 reagent (used for normalizing luciferase activity). The cells were then lysed, and luciferase activity was measured using a luciferase assay system (Promega).
Prediction of MiRNAs that Regulate ZNF395
The miRNAs that bind ZNF395’s 3′ UTR were predicted using miRanda(2, 20, 21) and TargetScan.(13) The effect of Neo-N3 on other miRNAs was assessed by qRT-PCR as described above using SNORD44, SNORD47, SNORD48, and RNU6 for normalization. Supporting Information Table 2 provides a list of all miRNAs profiled and the corresponding qPCR primers. Cycling conditions were 95 °C for 15 s and 60 °C for 60 s.
Overexpression of pri-miR-525
A plasmid that allows for the overexpression of pri-miR-525 via a CMV promoter was purchased from Genecopoeia. HepG2 cells were plated in six-well plates and grown to ~70% confluency. They were then transfected with 1 µg of plasmid using Lipofectamine 2000 per the manufacturer’s recommended protocol. After 5 h, the transfection cocktail was removed and replaced with growth medium (no antibiotics). The cells were grown for 48 h to allow for miR-525 overexpression before treatment with Neo-N3. For Western blotting, cells were then treated with various concentrations of Neo-N3 for 18 h followed by harvesting of total protein.
Western Blotting
Total protein (~15 µg) was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by transfer to a PVDF membrane. The membrane was blocked with 5% (w/v) bovine serum albumin (BSA) in 1× Tris-buffered saline (TBS) containing 0.05% (v/v) Tween-20 (1× TBST). ZNF395 was detected by using a 1:1000 dilution of anti-HDBP2 (Thermo Fisher) and a 1:2000 dilution of rabbit anti-IgG-HRP conjugate, both diluted in 1× TBST containing 3% (w/v) BSA. SuperSignal West Pico chemiluminescent substrate (Pierce Biotechnology) was used for imaging. After detection of ZNF395 levels, the blot was stripped and reprobed to measure levels of β-actin, which were used for normalization of Western blot signals (1:2000 dilution of anti-β-actin (Cell Signaling) and 1:2000 dilution of rabbit anti-IgG-HRP conjugate in 1× TBST containing 5% (w/v) milk). A luminescent signal was generated using SuperSignal West Pico Chemiluminscent Substrate (Thermo), imaged with film, and quantified with ImageJ software.
Cell Invasion
Invasion assays were completed using the Boyden (chamber assay) method.(19) Briefly, ~100 µL of diluted Matrigel (Corning) was added to a Millicell hanging insert (Millipore) containing a polyethylene terephthalate membrane with 8 µm pores. The inserts were incubated at 37 °C for 2 h and then placed at 4 °C overnight. Prior to adding cells, growth medium (no penicillin/streptomycin) was added to the wells of the plate, and the Matrigel and medium were prewarmed to 37 °C. HepG2 cells were harvested from a 75 cm2 plate using accutase and resuspended in 1× DMEM supplemented with 1% (v/v) FBS and 2 mM Glutagro (no penicillin/streptomycin) containing various concentrations of Neo-N3.
To assess the effect of miR-525 overexpression on the potency of Neo-N3, cells were transfected with plasmid that encodes pri-miR-525 (described above). Approximately 5 h post-transfection, the transfection cocktail was replaced with growth medium. After 48 h, the cells were removed from the surface with accutase and resuspended in 1× DMEM supplemented with 1% (v/v) FBS and 2 mM Glutagro (no penicillin/streptomycin) containing various concentrations of Neo-N3.
Approximately 50 000 cells (treated or untreated) were then applied to the top of the Matrigel. After 48 h, cells were fixed with 1% (w/v) paraformaldehyde in 1× DPBS and stained with 0.5% (w/v) crystal violet in water. Noninvasive cells were removed with a cotton swab, and invasive cells were imaged by microscopy and counted.
Supplementary Material
Acknowledgments
This work was funded by the National Institutes of Health R01GM097455 and The Scripps Research Institute. M.D.D. is a Camille and Henry Dreyfus Teacher–Scholar.
Footnotes
The authors declare no competing financial interest.
References
- 1.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 2.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Visone R, Croce CM. MiRNAs and cancer. Am. J. Pathol. 2009;174:1131–1138. doi: 10.2353/ajpath.2009.080794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 5.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 6.Dickson JR, Kruse C, Montagna DR, Finsen B, Wolfe MS. Alternative polyadenylation and miR-34 family members regulate tau expression. J. Neurochem. 2013;127:739–749. doi: 10.1111/jnc.12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han M, Toli J, Abdellatif M. MicroRNAs in the cardiovascular system. Curr. Opin. Cardiol. 2011;26:181–189. doi: 10.1097/HCO.0b013e328345983d. [DOI] [PubMed] [Google Scholar]
- 8.Garofalo M, Condorelli G, Croce CM. MicroRNAs in diseases and drug response. Curr. Opin. Pharmacol. 2008;8:661–667. doi: 10.1016/j.coph.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 9.Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37:D98–D104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 11.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 12.Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 14.Velagapudi SP, Gallo SM, Disney MD. Sequence-based design of bioactive small molecules that target precursor microRNAs. Nat. Chem. Biol. 2014;10:291–297. doi: 10.1038/nchembio.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pang F, Zha R, Zhao Y, Wang Q, Chen D, Zhang Z, Chen T, Yao M, Gu J, He X. MiR-525-3p enhances the migration and invasion of liver cancer cells by downregulating. PLoS One. 2014;9:e90867. doi: 10.1371/journal.pone.0090867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenkranz H, Scheer M, Scholtan W. Binding of aminoglycoside antibiotics to human serum proteins. III. Effect of experimental conditions Infection. 1978;6:57–64. doi: 10.1007/BF01642159. [DOI] [PubMed] [Google Scholar]
- 17.McKee EE, Ferguson M, Bentley AT, Marks TA. Inhibition of mammalian mitochondrial protein synthesis by oxazolidinones. Antimicrob. Agents Chemother. 2006;50:2042–2049. doi: 10.1128/AAC.01411-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka K, Shouguchi-Miyata J, Miyamoto N, Ikeda JE. Novel nuclear shuttle proteins, HDBP1 and HDBP2, bind to neuronal cell-specific cis-regulatory element in the promoter for the human Huntington’s disease gene. J. Biol. Chem. 2004;279:7275–7286. doi: 10.1074/jbc.M310726200. [DOI] [PubMed] [Google Scholar]
- 19.Boyden S. The chemotactic effect of mixtures of antibody and antigen on polymorphonuclear leucocytes. J. Exp. Med. 1962;115:453–466. doi: 10.1084/jem.115.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen H, Miao R, Fan J, Han Z, Wu J, Qiu G, Tang H, Peng Z. Decreased expression of miR-126 correlates with metastatic recurrence of hepatocellular carcinoma. Clin. Exp. Metastasis. 2013;30:651–658. doi: 10.1007/s10585-013-9569-6. [DOI] [PubMed] [Google Scholar]
- 23.Du C, Lv Z, Cao L, Ding C, Gyabaah OA, Xie H, Zhou L, Wu J, Zheng S. MiR-126–3p suppresses tumor metastasis and angiogenesis of hepatocellular carcinoma by targeting LRP6 and PIK3R2. J. Transl. Med. 2014;12:259. doi: 10.1186/s12967-014-0259-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao C, Li Y, Zhang M, Yang Y, Chang L. miR-126 inhibits cell proliferation and induces cell apoptosis of hepatocellular carcinoma cells partially by targeting Sox2 Hum. Hum. Cell. 2015;28:91–99. doi: 10.1007/s13577-014-0105-z. [DOI] [PubMed] [Google Scholar]
- 25.Maiti M, Nauwelaerts K, Herdewijn P. Pre-microRNA binding aminoglycosides and antitumor drugs as inhibitors of Dicer catalyzed microRNA processing. Bioorg. Med. Chem. Lett. 2012;22:1709–1711. doi: 10.1016/j.bmcl.2011.12.103. [DOI] [PubMed] [Google Scholar]
- 26.Al Sibae MR, McGuire BM. Current trends in the treatment of hepatic encephalopathy. Ther. Clin. Risk Manage. 2009;5:617–626. doi: 10.2147/tcrm.s4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neomycin Sulfate - FDA Prescribing Information, Side Effects and Uses. http://www.drugs.com/pro/neomycin-sulfate.html.
- 28.Blei AT, Cordoba J. Hepatic encephalopathy. Am. J. Gastroenterol. 2001;96:1968–1976. doi: 10.1111/j.1572-0241.2001.03964.x. [DOI] [PubMed] [Google Scholar]
- 29.Manuel MA, Kurtz I, Saiphoo CS, Nedzelski JM. Nephrotoxicity and ototoxicity following irrigation of wounds with neomycin. Can. J. Surg. 1979;22:274–277. [PubMed] [Google Scholar]
- 30.Mingeot-Leclercq MP, Glupczynski Y, Tulkens PM. Aminoglycosides: activity and resistance. Antimicrob. Agents Chemother. 1999;43:727–737. doi: 10.1128/aac.43.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Disney MD, Labuda LP, Paul DJ, Poplawski SG, Pushechnikov A, Tran T, Velagapudi SP, Wu M, Childs-Disney JL. Two-dimensional combinatorial screening identifies specific aminoglycoside-RNA internal loop partners. J. Am. Chem. Soc. 2008;130:11185–11194. doi: 10.1021/ja803234t. [DOI] [PubMed] [Google Scholar]
- 32.McKenna SA, Kim I, Puglisi EV, Lindhout DA, Aitken CE, Marshall RA, Puglisi JD. Purification and characterization of transcribed RNAs using gel filtration chromatography. Nat. Protoc. 2007;2:3270–3277. doi: 10.1038/nprot.2007.480. [DOI] [PubMed] [Google Scholar]
- 33.Peyret N, Seneviratne PA, Allawi HT, SantaLucia J., Jr Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A.A, C.C, G.G, and T.T mismatches. Biochemistry. 1999;38:3468–3477. doi: 10.1021/bi9825091. [DOI] [PubMed] [Google Scholar]
- 34.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. U.S.A. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puglisi JD, Tinoco I., Jr Absorbance melting curves of RNA. Methods Enzymol. 1989;180:304–325. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Rando RR. Specific binding of aminoglycoside antibiotics to RNA. Chem. Biol. 1995;2:281–290. doi: 10.1016/1074-5521(95)90047-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


