Abstract
Sex-determining regions (SDRs) or mating-type (MT) loci in two sequenced volvocine algal species, Chlamydomonas reinhardtii and Volvox carteri, exhibit major differences in size, structure, gene content, and gametolog differentiation. Understanding the origin of these differences requires investigation of MT loci from related species. Here, we determined the sequences of the minus and plus MT haplotypes of the isogamous 16-celled volvocine alga, Gonium pectorale, which is more closely related to the multicellular V. carteri than to C. reinhardtii. Compared to C. reinhardtii MT, G. pectorale MT is moderately larger in size, and has a less complex structure, with only two major syntenic blocs of collinear gametologs. However, the gametolog content of G. pectorale MT has more overlap with that of V. carteri MT than with C. reinhardtii MT, while the allelic divergence between gametologs in G. pectorale is even lower than that in C. reinhardtii. Three key sex-related genes are conserved in G. pectorale MT: GpMID and GpMTD1 in MT–, and GpFUS1 in MT+. GpFUS1 protein exhibited specific localization at the plus-gametic mating structure, indicating a conserved function in fertilization. Our results suggest that the G. pectorale–V. carteri common ancestral MT experienced at least one major reformation after the split from C. reinhardtii, and that the V. carteri ancestral MT underwent a subsequent expansion and loss of recombination after the divergence from G. pectorale. These data begin to polarize important changes that occurred in volvocine MT loci, and highlight the potential for discontinuous and dynamic evolution in SDRs.
Keywords: Gonium, Chlamydomonas, Volvox, mating locus, evolution, Genetics of Sex
Sex determining regions (SDRs) can exhibit complex patterns of molecular evolution that are distinct from those of autosomes (Bachtrog et al. 2011). The origins of SDR architecture and function have been studied in diverse species, but our understanding of the evolutionary processes that shape SDRs is still limited. Recombination suppression—a common property of both haploid and diploid SDRs—reinforces linkage between sex-related genes within the SDR, and promotes accumulation of sexually antagonistic alleles linked to the SDR (Bachtrog et al. 2011; Immler and Otto 2015). SDRs often contain genomic rearrangements that may be either the cause, or consequence, of recombination suppression between heteromorphic haplotypes or sex chromosomes. The deleterious effects of suppressed recombination are also seen in SDRs where Y or Z chromosomes in diploid mating systems may undergo degeneration (Charlesworth et al. 2005). Much less is known about the long-term evolutionary histories of SDRs in haploid mating systems where SDRs also undergo degeneration, but where limited data are available (Yamato et al. 2007; Bachtrog et al. 2011; Sun et al. 2012; McDaniel et al. 2013; Ahmed et al. 2014).
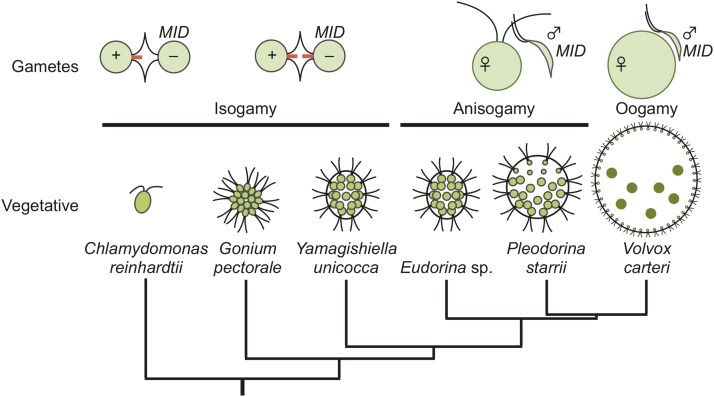
Volvocine algae offer unique advantages as a model for the evolution of SDRs. Like many protists, volvocine algae are haploid and capable of both asexual (vegetative) and sexual reproduction. Environmental or hormonal cues trigger gametic differentiation, which in heterothallic species is controlled by a heteromorphic mating-type (MT) locus with two haplotypes: plus/minus, or male/female (see below). Volvocine algae also encompass a range of morphologies from unicellular genera such as Chlamydomonas, to Volvox, which have thousands of cells, and exhibit germ–soma differentiation and other developmental innovations that result in a functionally integrated multicellular colony (summarized in Figure 1). Sexual reproduction coevolved with colony size in volvocine algae, whose mating strategies include isogamy in unicellular and small colonial genera (Chlamydomonas, Gonium, and Yamagishiella), anisogamy in intermediate-sized genera (Eudorina and Pleodorina), and oogamy in the largest, and most differentiated, genus Volvox. Transitions between heterothallism (genetic sex determination) and homothallism (self-mating) have also occurred within volvocine algae (Coleman 2012) (Figure 1). The well-established phylogenetic relationships between volvocine species enable specific traits and innovations to be ordered and mapped onto a coherent evolutionary framework (Nozaki et al. 2000; Herron and Michod 2008).
Figure 1.
A schematic diagram for phylogenetic relationships of selected volvocine species based on Nozaki et al. (2000) and Herron and Michod (2008). The top row illustrates gamete type and structure. Tubular mating structures in isogamous gametes are indicated with red bars at the flagellar base. The possession of a MID gene is shown next to the minus mating type or male gametes. The lower row of cartoons depicts vegetative morphology (not to scale) for the indicated species.
The full genome sequences, including regions of both haplotypes of MT, were previously described for two heterothallic species: isogamous unicellular C. reinhardtii, and oogamous multicellular Volvox carteri (Figure 2) (Merchant et al. 2007; Ferris et al. 2010; Prochnik et al. 2010). Importantly, the genetic and molecular bases of MT differentiation in volvocine algae, including functions of mating locus genes, have been investigated (Ferris and Goodenough 1994, 1997; Ferris et al. 1996, 2002; De Hoff et al. 2013; Geng et al. 2014). MT haplotypes (plus/minus or male/female) segregate as single Mendelian traits, but the loci themselves are multigenic regions in which recombination is suppressed. The core of the mating locus is the rearranged (R) domain, where gene order and arrangements between the two haplotypes are noncolinear (Umen 2011). In C. reinhardtii, the plus and minus haplotypes of MT reside on chromosome 6 (Ferris and Goodenough 1994; Ferris et al. 2002; De Hoff et al. 2013), while V. carteri MT is located in a region of linkage group I that shares a common origin with C. reinhardtii chromosome 6 (Ferris et al. 2010). Despite their shared chromosomal location, V. carteri MT is much larger in size than C. reinhardtii MT, and shows a far higher degree of differentiation between gametologs (allelic gene pairs residing in MT) (Ferris et al. 2010). This discrepancy raises many questions about why the two MT loci differ so much in genetic divergence and recombination potential (Charlesworth and Charlesworth 2010; Umen 2011). One important feature of C. reinhardtii MT that is not found in V. carteri MT is low-frequency gene conversion between gametologs that acts to reduce allelic differentiation (De Hoff et al. 2013).
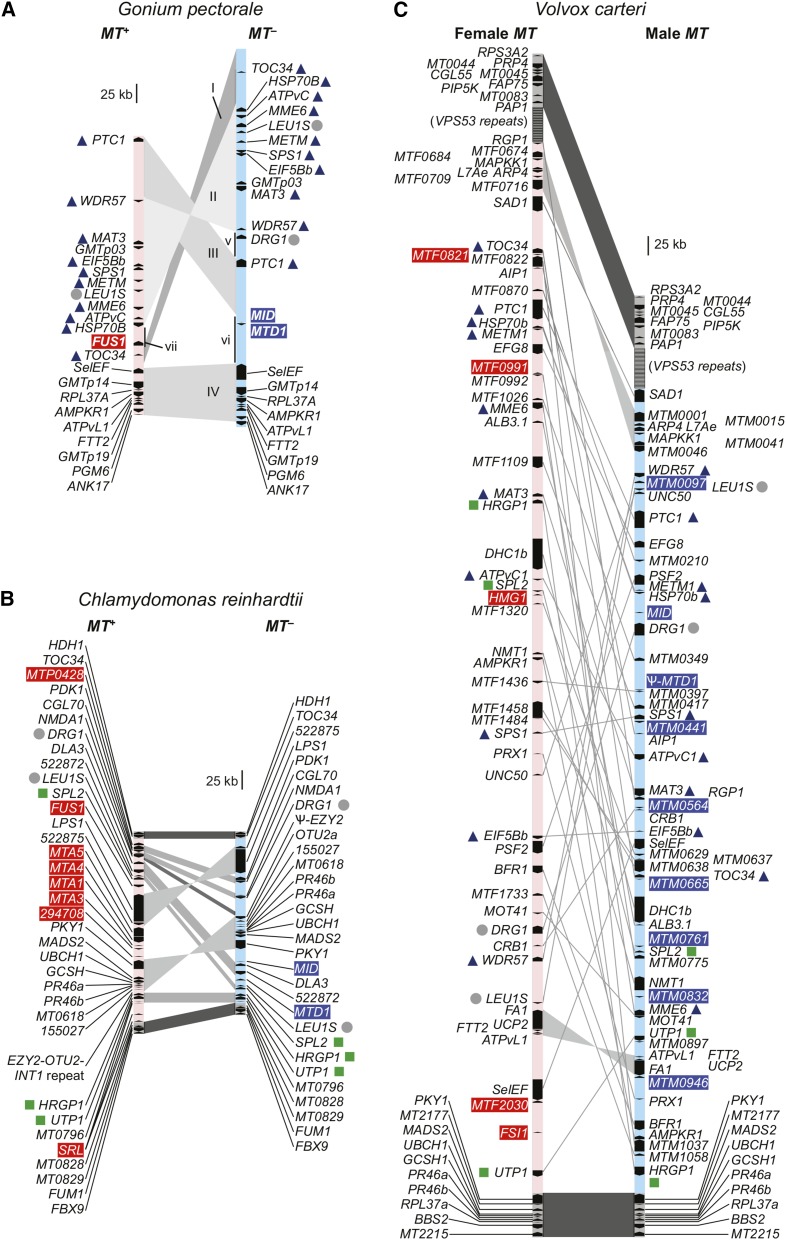
Figure 2.
Schematic of volvocine mating loci for (A) Gonium pectorale, (B) Chlamydomonas reinhardtii (modified from De Hoff et al. 2013), and (C) Volvox carteri (modified from Ferris et al. 2010), with rearranged (R) domains in light blue (minus/male) or pink (plus/female). For G. pectorale MT, syntenic blocs are indicated by gray shading, and labeled with upper case roman numbers (I–IV), while sequences that are unique to one of the two mating haplotypes are indicated with lower case roman numbers (v–vii). Red and blue shading on gene names indicates plus/female and minus/male specific genes, respectively. Gray dots beside gene names indicate those found in the R domains of all three species; dark blue triangles indicate presence of gene in R domain of G. pectorale and V. carteri only; green squares indicate presence of gene in R domain of C. reinhardtii and V. carteri only. No R domain genes are shared exclusively by G. pectorale and C. reinhardtii.
The C. reinhardtii MID (minus-dominance) gene is present only in the MT– haplotype, and plays a critical role in determining mating type (Ferris and Goodenough 1997). Recently, the V. carteri MID gene which is found only in the male MT haplotype (Ferris et al. 2010) was also shown to govern critical aspects of sex determination (Geng et al. 2014). Moreover, MID genes, have been found in the minus or male mating haplotypes of other heterothallic volvocine algae including several Gonium species (Hamaji et al. 2008, 2013a; Setohigashi et al. 2011), and Pleodorina starrii (Nozaki et al. 2006), suggesting that the genetic basis of sex- or mating-type determination is conserved throughout the volvocine lineage (Figure 1). Other than the MID gene, no sex-related genes are conserved between the MT loci of V. carteri and C. reinhardtii. The strikingly different sizes, gametolog contents, gametolog differentiation, and recombination behaviors of C. reinhardtii vs. V. carteri MT loci raise questions about how these two SDRs, which appear to have arisen from a common ancestral SDR region, diverged so markedly from each other. To answer these questions requires information on volvocine algal MT loci from additional species in order to begin reconstructing ancestral states and polarizing changes within the lineage.
Gonium pectorale is a small isogamous colonial volvocine species that is more closely related to V. carteri than it is to C. reinhardtii (Nozaki et al. 2000). As such, it may represent an informative taxon for reconstructing the evolution of MT loci in the volvocine lineage. Previous work on G. pectorale identified MT– homologs of MID and of MTD1, genes that are both also found in the MT– haplotype of C. reinhardtii (Hamaji et al. 2008, 2009). To date, no homologs of MT+ limited genes from C. reinhardtii have been found in other volvocine species, including the gene FUS1, which encodes a membrane-bound protein that localizes to the mating apparatus or “fertilization tubule,” and is required for fusion of plus and minus gametes (Ferris et al. 1996, 1997; Misamore et al. 2003).
Here we report the sequence of G. pectorale MT+ and MT– haplotypes derived from a combination of chromosomal walking, and whole genome sequencing. The overall structure of G. pectorale MT was found to differ from C. reinhardtii MT and V. carteri MT in terms of structural rearrangements between haplotypes. It has fewer rearranged sequence blocs than C. reinhardtii MT, and no large autosomal insertions in either haplotype. G. pectorale shares, in common with C. reinhardtii, the presence of sex-limited genes MID and MTD1 in its MT- haplotype (Hamaji et al. 2008, 2009), and a FUS1 homolog in its MT+ haplotype, whose gene product, GpFUS1, localizes to the fertilization tubule, indicating homologous function with CrFUS1. As in C. reinhardtii, gametolog differentiation in G. pectorale MT is minimal. However, the gametolog gene complement of G. pectorale MT is more similar to that of V. carteri MT than that of C. reinhardtii MT. Taken together, these data enable us to reconstruct a minimal set of changes in the MT region that led to their current states. Our findings suggest a dynamic, and most likely punctuated, history of volvocine MT evolution that involves episodes of structural reconfiguration in which gametolog content and syntenic blocs change, but where mating-related genes remain linked to their ancestral MT haplotypes. The expansion, gametolog differentiation, and loss of MTD1 and FUS1 genes that characterize V. carteri MT likely occurred after the G. pectorale and V. carteri lineages split.
Materials and Methods
Strains used
The genomic DNA sequence data presented here come from previously described strains of G. pectorale: Kaneko3 (minus) and Kaneko4 (plus) (Yamada et al. 2006; Hamaji et al. 2008, 2013b), K3-F3-4 (minus), and K4-F3-4 (plus), which are F3 hybrid progeny from Kaneko3 and Kaneko4 as described previously (Hamaji et al. 2013b).
BAC library construction
DNA plugs were prepared from both mating types (Kaneko4: plus; Kaneko3: minus) in 1% SeaPlaque GTG agarose (Cambrex Bio Science Rockland), treated with Pronase E (Sigma), and washed thoroughly as described (Ferris et al. 2010). BAC library production was performed at the Clemson University Genome Institute. The DNA plugs were partially digested with EcoRI, and then DNA fragments size-fractioned (to ca. 120 kb) by pulse-field gel electrophoresis were ligated into pIndigoBAC536. Single colonies of Escherichia coli strain DH10b transformed with G. pectorale genomic fragment-containing BACs were picked, spotted on nylon membranes (or “BAC filters”), and stored frozen in glycerol at –80°C (total BAC number, plus: 27,648; minus: 18,432, estimated 24× and 16× coverage of G. pectorale plus and minus genome, respectively).
BAC screening and chromosomal walking
BAC filters were hybridized with probes labeled and detected with CDP-Star (GE Healthcare) or DIG-labeling kit (Roche). Positive signals were confirmed by direct PCR with probe-specific primers on the individual clones. BACs of true positives were column-purified from 5 ml cultures, and end-sequenced with M13 Forward and Reverse primers. Each end sequence is designed for a pair of specific primers, which was used to determine the relative locations of overlapping BACs. Primer pairs from both distal ends were selected to label probes for the next round of chromosomal walking. Inverse PCR (Sambrook and Russell 2001) and TAIL-PCR (Liu and Whittier 1995) were performed to identify flanking sequences when no additional BAC clones could be identified. GpMTD1 probes were the same as those used for the GpMTD1 DNA gel-blot analysis (Hamaji et al. 2009). The PCR product of GPLEUFATG and GPLEURTAA was labeled to screen for LEU1S alleles (Supplemental Material, Table S1). Two gametologs (WDR57 and DRG1) and their flanking regions, which were not obtained in the MT+ BAC assembly, were obtained by genomic PCR using specific primers based on the MT– alleles. The linkage of MT+ DRG1 was determined by recombination scores as described below. Abundant repetitive regions flanking the MT assembly prevented further chromosomal walking to connect directly to autosomal sequences.
Shotgun sequencing and assembly
BAC DNA was mechanically sheared either with nebulizers (for 30 sec) in the TOPO shotgun sub-cloning kit (Invitrogen), or with sonication. Blunt-end fragments were subcloned into pCR4, pCRII (Invitrogen), or pUC118 (Takara Bio). Shearing with sonication, and pUC118 subcloning, were performed by the Kazusa DNA Research Institute. Shotgun subclones were sequenced from both ends by the Research Resource Center (BSI, RIKEN), or the Kazusa DNA Research Institute. Raw sequence data were base-called, vector-trimmed for each, end-clipped to remove low-quality regions, and assembled by CodonCode Aligner (CodonCode: http://www.codoncode.com/aligner/). Assembled contigs were queried against C. reinhardtii V4 protein models, or V. carteri protein models (Ver. 2, JGI and male and female MT, NCBI) using BLASTX to identify protein coding genes (Merchant et al. 2007; Ferris et al. 2010).
Annotation
Gene models were generated using Fgenesh (Salamov and Solovyev 2000) (http://linux1.softberry.com/berry.phtml) with the “Chlamydomonas” option, and then manually edited based on similarity to C. reinhardtii (JGI V4 protein models: http://genome.jgi-psf.org/Chlre4/Chlre4.home.html) or V. carteri (male and female MT genes available in GenBank: http://www.ncbi.nlm.nih.gov/genbank/) homologs.
GpFUS1 identification and characterization
BLASTX search of the G. pectorale MT locus contigs using C. reinhardtii proteins (ver. 4) identified a FUS1-like sequence in MT+. RT-PCR fragments of 5′-/3′-RACE were cloned into TOPO TA vector to determine the full-length cDNA sequence. Subsequently, genomic sequences were obtained from plus BAC 71M20. The G. pectorale FUS1 full-length cDNA sequence was determined by the same method as GpMTD1 (specific primers used: GpFUS1AF; GpFUS1AR; GpFUS1BF; GpFUS1BR; GpFUS1CF; GpFUS1CR). The initially gapped genomic sequence of GpFUS1 was filled by genomic PCR. Hydrophobicity with window size of nine, and theoretical protein isoelectric point (pI)/molecular weight (MW) were calculated with ProtScale and Compute pI/Mw in ExPASY (Gasteiger et al. 2003) (http://au.expasy.org/).
Anti-GpFUS1 antibody production, Western blotting, and immunofluorescence
The coding region corresponding to amino acid residues 90–489 out of the predicted 824-aa sequence of GpFUS1 protein was amplified by PCR with primers YM001F and YM001R. The resultant DNA was cloned into pET100/D-TOPO vector (Invitrogen), and expressed as a His-tag fusion protein. The recombinant protein was isolated with a HisTrap HP column (GE Healthcare). Purified protein was injected into two rabbits for antibody production (Riken, Saitama, Japan). The rabbit antisera were affinity-purified with a 6His-GpFUS1-coupled HiTrap NHS-activated HP column (GE Healthcare), according to the manufacturer’s instructions. The final concentration of antibody was ≫0.25 mg/ml.
To stain GpFUS1 protein expressed in gametes, indirect immunofluorescence microscopy assay and Western blot analyses were performed as previously described after dibutyryl-cyclic AMP (db-cAMP) treatment (Mogi et al. 2012).
Linkage mapping
DNA samples of 78 F1 progeny (haploid recombinants from F1 meiosis) derived from G. pectorale Mongolia 1×4 strains (described previously) (Hamaji et al. 2008) were genotyped for markers designed based on polymorphic genome sequences. Primers, restriction enzymes, and accession numbers in PCR-RFLP analyses are listed in Table S1. The PCR conditions were 2 min 94°, 35 cycles of 30 sec at 94°, 30 sec at 53° (for DRG1, 50°), and 30 sec at 72°, followed by 7 min at 72° with the same reaction composition as reported previously (Ferris et al. 2010). Restriction digests of the PCR products were performed (Sambrook and Russell 2001) to identify the two genotypes.
Molecular evolutionary analyses
Previously reported genes linked to, or residing in, MT loci of C. reinhardtii or V. carteri were queried by TBLASTN searching (Altschul et al. 1990) of the G. pectorale de novo draft genome assembly (Hanschen et al. 2016). Genome-wide synteny comparison between C. reinhardtii (v4 assembly) and G. pectorale was done using SyMAP (Soderlund et al. 2006, 2011).
MT shared genes (i.e., gametologs) were analyzed as previously described (Ferris et al. 2010). Coding or genomic sequences were aligned using ClustalX 2.0 (Larkin et al. 2007) and alignments adjusted manually. Divergence scores were calculated using yn00 from the PAML4 package (Yang 2007); nonsynonymous and synonymous site divergence (dN and dS, respectively) of aligned coding sequences of gametologs was estimated according to Yang and Nielsen (2000) with equal weighting between pathways, and the same codon frequency for all pairs. Sliding window analysis of each gametolog was performed using DnaSP v5.10.1 (Librado and Rozas 2009) for Pi (Nei 1987) over the aligned genomic sequences from both haplotypes from start to stop codons in 100-base windows with a 25-base interval.
Data availability
DDBJ/ENA/GenBank accessions: LC062386 (G. pectorale DRG1 plus haplotype); LC062718 (MT- scaffold); LC062719 (MT+ scaffold).
Results and Discussion
Size, structure, and gene content of G. pectorale MT
BAC contigs containing G. pectorale MT sequences comprised approximately 500 kb for MT– and 360 kb for MT+ (Figure 2A, Figure S1, and Table 1). Although these contigs could not be directly connected to autosomal scaffolds, based on their genetic contents and other criteria outlined below, it is likely they contain all or most of the rearranged (R) domain of G. pectorale MT. Contigs from both haplotypes were sequenced and used for BLASTX and TBLASTX queries against the C. reinhardtii proteome, and against the V. carteri MT locus DNA sequence, respectively. These searches detected 24 G. pectorale MT genes, 21 of which are gametolog pairs, meaning that an allele is present in both mating haplotypes. Three additional mating-type-specific genes, MID, MTD1, and FUS1 were also present, with MID and MTD1 in MT–, and FUS1 in MT+ (Figure 2A and Table S2). The gene contents of G. pectorale MT are described in more detail below.
Table 1. Genome and mating locus properties of Gonium pectorale, Chlamydomonas reinhardtii, and Volvox carteri.
| Gonium pectorale | Chlamydomonas reinhardtii | Volvox carteri | |||||||
|---|---|---|---|---|---|---|---|---|---|
| MT+ | MT- | Whole | MT+ | MT– | Whole | Female | Male | Whole | |
| Size (Mb) | 0.366 | 0.499 | 148.8 | 0.310 | 0.204 | 111.1 | 1.51 | 1.13 | 131.1 |
| %GC | 59.7 | 61.0 | 64.5 | 60 | 61 | 64.1 | 52 | 53 | 56.1 |
| Gene density [genes/Mb] | 57.38 | 46.09 | 120.9 | 74.25 | 117.6 | 159.6 | 39 | 54 | 114.1 |
| Introns / gene | 9.62 | 9.09 | 6.50 | 7.1 | 6.37 | 7.46 | 9 | 8 | 6.29 |
| Average intron length (bp) | 175.8 | 278.50 | 349.83 | 354.17 | 358.70 | 420.01 | 618 | 584 | 399.50 |
Three rearranged sequence blocs (designated I–III in Figure 2A and Figure S2) define its R domain of G. pectorale MT with the largest, bloc II, being a > 200-kb inversion that retains collinearity of its 10 resident gametologs, and whose intergenic regions contain a few interspersed repeats (Figure 2A and Figure S2). Bloc III contains a single gene, PTC1 which is flanked by extensive noncoding repeats that comprise the majority of this subregion. While the coding regions of PTC1 are nearly identical between gametologs, the 3′-UTR of the MT– allele is extended compared to the MT+ allele, leading to a larger predicted gene size (Figure 2A). Bloc I contains the TOC34 gene, whose gametolog pairs have low similarity between gametologs compared with bloc II gametologs (Figure 2A; discussed below), and has almost no similarity in intergenic regions (Figure 2A and Figure S2). Bloc IV (Figure 2A and Figure S2) is almost completely collinear between haplotypes, and, as described below, likely represents the centromere-proximal autosomal flank of MT. The only significant difference between the two mating haplotypes in bloc IV is at its proximal end next to the R domain, where a large intron is present in the MT– allele of SelEF at its 3′ end, but absent from the MT+ allele, accounting for their different sizes (Figure 2A and Figure S2). The distal-most gene from the R domain in bloc IV, ANK17, was incompletely assembled in each haplotype, with differences in the length of assembled sequences accounting for the size difference between its two alleles (Figure 2A). On the other side of MT (top end in Figure 2A), no additional sequences distal to TOC34 in MT– and PTC1 in MT+ were found, and it is likely that, beyond this region are repeats and/or telomeric sequences, as is the case in V. carteri (Ferris et al. 2010). Interspersed between the syntenic blocs I–IV are three regions (v–vii) of G. pectorale MT that are unique to one of the two haplotypes. Region vi of MT– contains homologs of two C. reinhardtii sex-determining genes, GpMTD1 and GpMID, which have been previously described (Hamaji et al. 2008, 2009). Region vii of MT+ contains a homolog of FUS1, a C. reinhardtii fertilization protein. Characterization of GpFUS1 is presented below. Region v initially appeared to be unique to MT– and contains a single gene, DRG1, whose orthologs in C. reinhardtii and V. carteri are present as gametolog pairs in their respective MT loci (Ferris et al. 2002, 2010; De Hoff et al. 2013) but which have no known function in the sexual cycle. A MT+ linked allele of DRG1 was subsequently identified, as described below.
Overall, the haplotype bloc structure of G. pectorale MT is notable for being completely different from that of C. reinhardtii or V. carteri MT, with fewer overall rearrangements than in C. reinhardtii (Figure 2) (Ferris et al. 2010; De Hoff et al. 2013). Also, apparently absent from G. pectorale MT are the large autosomal insertions and tandem repeats that are found in C. reinhardtii MT (De Hoff et al. 2013). Although it is larger than C. reinhardtii MT, the structure of G. pectorale MT is simpler, with fewer sequence rearrangements and insertions, consistent with a younger age. It is also very different from V. carteri MT, which is characterized by much more extensive sequence rearrangements, and few collinear blocs of gametologs. Taken together, our findings suggest a nonlinear trajectory of MT evolution in volvocine algae, as discussed further below.
Genomic location and completeness of the G. pectorale MT assembly by linkage mapping
Although we could not connect the G. pectorale MT assembly physically to other scaffolds in the G. pectorale whole genome assembly, we identified a linked 2.8 Mb autosomal scaffold (scaffold00010) that is syntenic to a portion of C. reinhardtii MT-containing chromosome 6 (Table 2 and Figure S3). Recombination was measured between each terminus of G. pectorale scaffold00010 and MT, and revealed that one of the termini did not recombine MT (0/78) but that the other one did, with a genetic distance of 17 cM (13/78 recombinants). Based on physical estimates from two independent mapping intervals (Figure S3 and Table 2), the proximal end of scaffold00010 is less than 100 kb from MT. Ferris et al. (2010) reported a common chromosomal origin for regions containing MT loci of C. reinhardtii (Chr. 6), and V. carteri (Chr. 1). Our findings here further support this idea and show that, despite large changes in structure and gene content, MT loci appear to have remained stably associated with a single chromosomal location throughout the volvocine lineage.
Table 2. Linkage mapping of progeny from Gonium pectorale Mongolia 1 and 4.
| Characterb | RABB1 | MTF1109 | PRX1 | UNC50 | IDA5 | PR46a | ALB3.1 | LEU1S | MID | MT | PGM6 | SpoVS | PLC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Scaffolda | 5 | 3 | 1 | 2 | 97 | 7 | 7 | MT | MT | Phenotyped | MT | 10 | 10 |
| Cre. Chr.c | 2 | 6 | 6 | 6 | 13 | 6 | 6 | 6 | 6 | — | 6 | 6 | 6 |
| RABB1 | 34/78 | 42/78 | 38/77 | 42/78 | 37/77 | 38/78 | 40/78 | 40/78 | 35/69 | 40/78 | 40/78 | 35/78 | |
| MTF1109 | 34/78 | 43/77 | 36/78 | 33/77 | 34/78 | 44/78 | 44/78 | 40/69 | 44/78 | 44/78 | 49/78 | ||
| PRX1 | 34/77 | 40/78 | 32/77 | 26/78 | 38/78 | 38/78 | 36/69 | 38/78 | 38/78 | 41/78 | |||
| UNC50 | 42/77 | 33/76 | 37/77 | 38/77 | 38/77 | 33/68 | 38/77 | 38/77 | 39/77 | ||||
| IDA5 | 39/77 | 36/78 | 48/78 | 48/78 | 42/69 | 48/78 | 48/78 | 49/78 | |||||
| PR46a | 12/77 | 44/77 | 44/77 | 39/68 | 44/77 | 44/77 | 45/77 | ||||||
| ALB3.1 | 40/78 | 40/78 | 38/69 | 40/78 | 40/78 | 41/78 | |||||||
| LEU1S | 0/78 | 0/69 | 0/78 | 0/78 | 13/78 | ||||||||
| MID | 0/69 | 0/78 | 0/78 | 13/78 | |||||||||
| MT | 0/69 | 0/69 | 12/69 | ||||||||||
| PGM6 | 0/78 | 13/78 | |||||||||||
| PLC | 13/78 |
Crossing revised from a former experiment (Hamaji et al. 2008). Values given as ratio of recombinant progeny to total progeny. MT, mating type.
Scaffold in Gonium pectorale genome assembly.
Genotypic or phenotypic characters assayed.
Physical positions in Chlamydomonas reinhardtii chromosome for homologs of genetic markers.
Mating phenotype.
The near completeness of the G. pectorale R domain assembly was supported by our finding gametolog partners in each mating haplotype for every gene other than the three genes described above (MID, MTD1, and FUS1), which are already known to be sex-limited homologs in C. reinhardtii and/or V. carteri. With the exception of DRG1, both gametolog copies are in MT scaffolds. In addition, when we searched for G. pectorale homologs of genes present in, and around, MT from C. reinhardtii and V. carteri, we were able to locate nearly all of them in G. pectorale autosomal scaffolds, or within its MT assembly. Although we could not find a copy of DRG1 in either the MT+ genome scaffolds or in the G. pectorale whole genome assembly (Hanschen et al. 2016), we reasoned that a MT+ allele might be in a region of the MT+ haplotype that was not assembled. Using PCR with primers designed to amplify the MT– copy of GpDRG1, we identified a MT+ DRG1 allele (see Materials and Methods), and found that it is tightly linked to MT (0/78 recombinants). Besides DRG1, there are no other “orphan” gametologs in our MT assembly, and no additional sex-limited genes other than GpMID, GpMTD1, and GpFUS1. Taken together, our data suggest that the MT assembly shown in Figure 2 is nearly complete, as is the inventory of G. pectorale MT-linked genes (Table S3).
Evolution of gene contents and structure of volvocine MT loci
G. pectorale homologs of many V. carteri and C. reinhardtii mating type genes (i.e., gametologs) are autosomal, meaning that, in G. pectorale, these genes reside outside of the MT locus (Table S3). Overall, the gametolog contents of G. pectorale MT show much more overlap with V. carteri gametologs (19 out of 20 in common) than with C. reinhardtii gametologs (two out of 20 in common) showing that G. pectorale MT and V. carteri MT diverged more recently (Figure 1). Besides the sex-limited genes already mentioned above (MTD1, MID, and FUS1 in C. reinhardtii; MID in V. carteri), C. reinhardtii and V. carteri both have additional sex-limited genes that we searched for in G. pectorale (Table S2). Homologs of EZY2 or MTA1 from C. reinhardtii (Ferris and Goodenough 1994; Ferris et al. 2002), or FSI1 and HMG1 from V. carteri (Ferris et al. 2010), could not identified anywhere in the G. pectorale whole genome sequence or MT region. However, a G. pectorale homolog of the C. reinhardtii MT+ gene OTU2 (De Hoff et al. 2013) was found on an autosomal scaffold (scaffold00113). In summary, compared to either C. reinhardtii or V. carteri, G. pectorale MT is the most minimal in terms of structural rearrangements, and in its sex-limited gene content.
Molecular evolution of G. pectorale MT genes
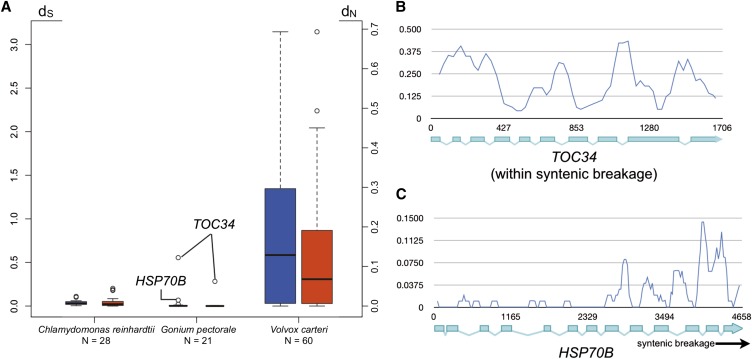
Previous work on C. reinhardtii and V. carteri MT loci revealed very different divergences between gametolog pairs, with high divergences for most V. carteri pairs, and low divergences for C. reinhardtii pairs (Ferris et al. 2010). It was subsequently found that low frequency gene conversion between gametolog sequences plays a role in maintaining sequence homogeneity between the nominally nonrecombining regions of C. reinhardtii MT (De Hoff et al. 2013). We calculated frequencies of synonymous (dS) and nonsynonymous (dN) substitutions between gametologs for G. pectorale MT genes and found, with two exceptions, that they were even lower than in C. reinhardtii (Table S4 and Figure 3A) (Hiraide et al. 2013). These findings suggest either higher rates of gene conversion between G. pectorale gametologs than those in C. reinhardtii, and/or a younger age for its haplotype blocs that may have had less time to diverge. A potentially informative clue about haplotype divergence and the history of G. pectorale MT was revealed from one exceptional region at the distal end of MT– (bloc I in Figure 2A), which encompasses TOC34 and part of neighboring gene HSP70B that resides in bloc II (Figure 2A). The genic regions of TOC34 (dN = 0.0623 and dS = 0.5549; Figure 3B and Table S4), and part of HSP70B (dN = 0.0006 and dS = 0.0678; Figure 3C and Table S4) have up to 10-fold higher divergence values than all other gametolog pairs (Figure S4) (average dN = 0.00333 ± 0.00288 and dS = 0.0331 ± 0.0257), with a boundary in the middle of HSP70B where divergence between gametologs drops and resembles the remainder of bloc II (Figure 3C). High divergences are also evident in the intergenic regions around TOC34 on both its proximal and distal sides relative to HSP70B, none of which can be aligned between the two haplotypes (Figure S2). By contrast, many of the intergenic sequences in blocs II and IV align well between haplotypes (Figure S2).
Figure 3.
Molecular evolutionary analyses of volvocine algal gametologs. (A) Box-whisker plots comparing the distributions of dS (blue) and dN (red) values for Chlamydomonas reinhardtii, Gonium pectorale, and Volvox carteri. Open dots in G. pectorale are values for indicated genes. (B) and (C) Sliding window plots of gametolog similarity (Pi) for TOC34 (B) or HSP70B (C). Position within each gene is indicated on x-axis by bp number beginning with the start codon.
The discontinuity of divergences in the TOC34-HSP70B region has at least two possible explanations. One possibility is that the TOC34-HSP70B region is a “cold-spot” for recombination, and contains a sequence that blocks progression of gene conversion tracts near the middle of HSP70B. There is precedent for interhaplotype gene conversion in C. reinhardtii (De Hoff et al. 2013), though no isolated coldspots were found. A second possibility is that the highly similar regions of G. pectorale MT are relatively young, and that the more diverged region containing TOC34 is a remnant from a previous version of MT with older and more diverged haplotypes. In this case, the low-divergence regions of MT could have arisen as a result of a replacement event, in which bloc II from one of the two MT haplotypes replaced the same region of the opposite mating haplotype (in inverted orientation) with a breakpoint between TOC34 and HSP70B on one end, and between WDR57 and PTC1 on the other end (Figure S5). This resetting event might also have allowed renewed and continuing gene-conversion between gametologs in bloc II, but would leave TOC34 gametologs in bloc I to continue diverging while also restricting the extent of gene conversion for HSP70B that abuts bloc I. The two explanations we propose for the discontinuous divergence patterns of bloc I vs. blocs II–IV of G. pectorale MT are not mutually exclusive, and in both cases they underscore the dynamic, and likely punctuated, nature of volvocine algal MT evolution.
Characterization of GpFUS1
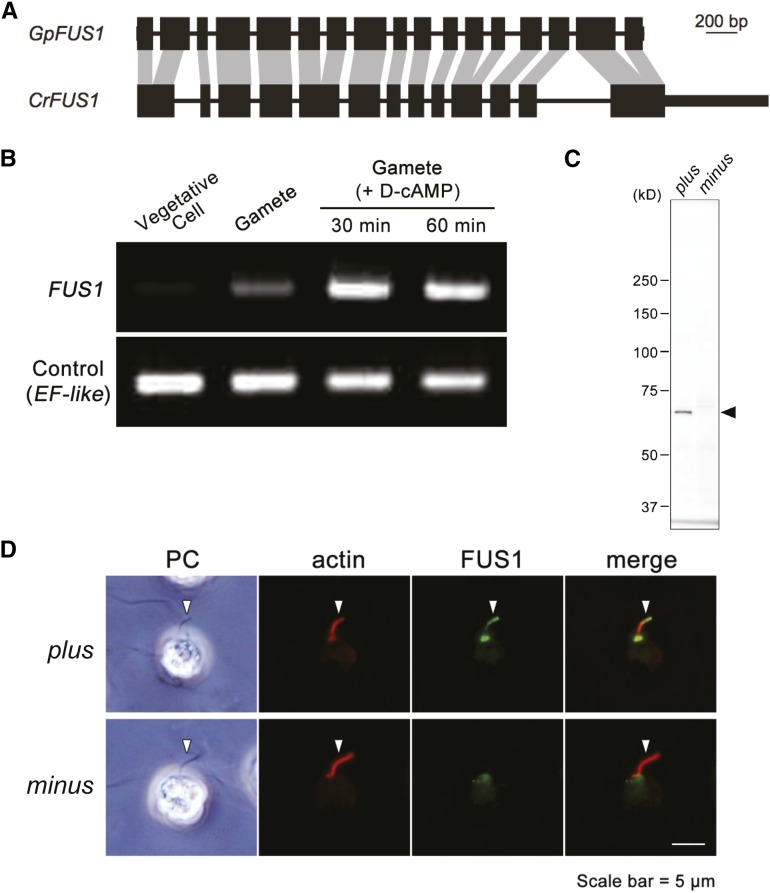
The G. pectorale FUS1 gene (GpFUS1) was identified based on a BLASTX search with the G. pectorale MT region queried against the C. reinhardtii proteome. Overall, GpFUS1 has a similar intron–exon structure as CrFUS1 (Figure 4A), with the addition of four additional introns not present in CrFUS1. The protein sequences of GpFUS1 and CrFUS1 have 31% amino acid identity, but retain the invasion/intimin repeats that may be involved in mediating mate recognition or membrane fusion (Figure S6 and Figure S7) (Misamore et al. 2003), and a signal peptide specifying membrane localization (Figure S8 and Figure S9).
Figure 4.
Identification and characterization of GpFUS1. (A) Comparison of FUS1 intron–exon structures between Chlamydomonas reinhardtii and Gonium pectorale. Homologous CDS (black boxes) sequences are connected by gray shading. Introns are represented by thin lines. UTRs are represented by thick lines. (B) Semiquantitative RT-PCR of GpFUS1 from MT+ G. pectorale strain. mRNAs were isolated from vegetative cells, gametes, and sexually activated gametes with db-cAMP (D-cAMP) treatment. EF-like gene was used as an internal control for RT-PCR. (C) Immunoblot analysis of GpFUS1 expression of activated plus and minus gametes using anti-GpFUS1 antibody. (D) Double immunofluorescence staining of db-cAMP activated mating type plus and minus gametes using anti-GpFUS1 and anti-actin antibody. Arrows indicate fertilization tubule, a mating apparatus that contains actin filament bundles.
GpFUS1 mRNA expression was detected in gametes, but not in vegetative cells, and was upregulated in gametes by treatment with the mating-related signaling agonist db-cAMP (Figure 4B) (Mogi et al. 2012), similar to its ortholog CrFUS1 (Pasquale and Goodenough 1987; Goodenough et al. 2007). Polyclonal antibodies against recombinant GpFUS1 recognized a protein on Western blots from plus but not minus gametes (Figure 4C). The anti-GpFUS1 signal (70 kDa) is smaller than the predicted molecular weight of GpFUS1 (∼90 kDa) in Western blotting analysis, suggesting that GpFUS1 might undergo post-translational processing. Immunofluorescence using the same antibodies localized GpFUS1 to the plus fertilization tubule in G. pectorale (Figure 4D) indicating a probable function in fertilization that is likely to be similar to that for CrFUS1; no signal was observed in the minus fertilization tubule.
Together, our results indicate that the FUS1 gene has been conserved as a mating type plus gene in the volvocine lineage, with an ancestral function in fertilization that has persisted at least since the G. pectorale–C. reinhardtii split, and has been subsequently lost from the lineage leading to V. carteri. Currently, the minus gamete interacting partner for FUS1 is unknown, but our results predict that it will also be conserved between C. reinhardtii and G. pectorale.
A dynamic history of MT locus evolution in the volvocine lineage
Previous comparative studies of volvocine algal MT loci have raised many questions about their evolutionary histories that this current study begins to address (Charlesworth and Charlesworth 2010; Umen 2011). The C. reinhardtii and V. carteri MT occupy a homologous chromosomal location, and share a common ancestral origin, but differ in many respects including gene content, gametolog divergence, locus size, syntenic bloc structure, and repeat content (Ferris et al. 2010). Although the C. reinhardtii MT locus is presumed to more closely reflect the ancestral state of volvocine algal MT loci than the V. carteri MT locus, without additional information about MT loci from other volvocine species, the evolutionary trajectory of the mating type region cannot be unambiguously inferred. The structure and gene content of G. pectorale MT described here begins to clarify the evolutionary history of volvocine algal MT loci. In overall size, structure, and gametolog divergence, G. pectorale MT resembles C. reinhardtii MT: it has just a few syntenic blocs containing multiple genes (Figure 2 and Figure S2), low divergence between most gametolog pairs (Figure 3A), and alignable intergenic regions interrupted in just a few positions with repeats (Figure S2). It also resembles C. reinhardtii in its complement of key mating regulators: MID and MTD1 in MT–, and FUS1 in MT+. However, G. pectorale MT differs from C. reinhardtii MT in its apparent lack of autosomal insertions into the MT region, and in the composition of its gametologs, which have more in common with those in V. carteri MT (Figure 1 and Table S3). Intriguingly, a small region of G. pectorale MT in the TOC34-HSP70B region shows elevated divergence between gametologs, which approaches divergence levels found for V. carteri gametologs, and may represent an early stage of MT differentiation in this lineage (Figure 3 and Figure S5).
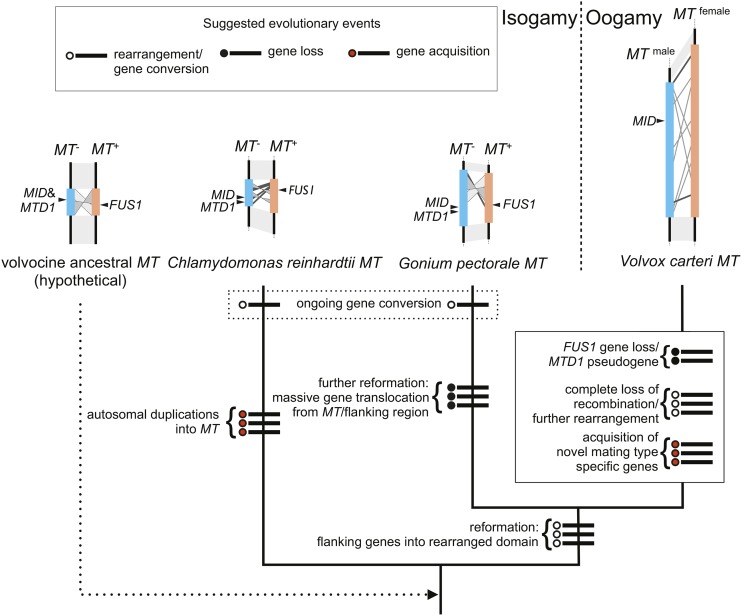
Based on these similarities and differences, we can infer a minimal set of changes that led to the generation of three distinct mating locus regions of C. reinhardtii, G. pectorale, and V. carteri (Figure 5). While the actual history of this region is likely to be more complicated than in our depiction, and awaits validation, our model creates a framework for classifying the types of changes that occurred in volvocine MT loci, and possible ancestral states. The common ancestor of all volvocine algae most likely had a mating locus near its telomere on a chromosome that is syntenic with Chromosome 6 in C. reinhardtii. The ancestral MT locus would have contained MID and MTD1 homologs in its MT– haplotype, and a FUS1 homolog in its MT+ haplotype. Its gametologs were genes found on the current C. reinhardtii Chromosome 6, or V. carteri Linkage Group I. After the G. pectorale and C. reinhardtii lineages split, one or both of their MT loci underwent a major reconfiguration, in which a different group of gametologs became fixed around the rearranged region of MT. During this time, the process of gametolog gene conversion was ongoing, and maintained relative homogeneity between gametolog pairs between plus and minus haplotypes in each lineage. The autosomal insertions in C. reinhardtii MT+ (De Hoff et al. 2013) were probably not lost in the G. pectorale lineage, but gained in the C. reinhardtii lineage, although the latter possibility cannot be excluded (Figure 5). In either case, the appearance of G. pectorale MT is “younger” and less complex than that of C. reinhardtii, which suggests a recent reconfiguration. Supporting the idea of reconfiguration is our finding of a small, and possibly more ancient, region containing TOC34 and part of HSP70B that underwent substantial differentiation (Figure 3 and Figure S2). By the time of the split between the G. pectorale and V. carteri lineages, their common ancestor had acquired many of the gametologs that are still common between these two species. We cannot determine when the FUS1 gene was lost from the V. carteri lineage, but the MTD1 gene may have been lost relatively recently, as a possible MTD1 pseudogene was identified in the male V. carteri MT haplotype (Ferris et al. 2010). In addition, V. carteri MT gained over a dozen male-limited or female-limited MT genes that lack detectable homologs in G. pectorale or elsewhere, suggesting that these additional genes arose after the two lineages split. At some point after the G. pectorale-V. carteri split, gametolog divergence in the V. carteri accelerated, and the R domain of its MT locus underwent a large expansion. Although we do not know when accelerated evolution and expansion began in the V. carteri lineage, there is additional information on gametolog divergence for the MT gene MAT3, whose sequences have been identified in P. starrii, an oogamous colonial species that is more closely related to V. carteri than to G. pectorale (Figure 1). While the male and female MAT3 gametologs of V. carteri are highly diverged (Ferris et al. 2010), those in P. starrii have a low divergence that is comparable to that found in G. pectorale (Hiraide et al. 2013). If the P. starrii MAT3 data are representative of its MT locus as a whole, then the massive divergence observed for V. carteri MT gametologs occurred relatively recently in the lineage.
Figure 5.
Possible evolutionary history for volvocine MT loci based on minimal changes necessary to explain observed results in this study and in previous studies (Ferris et al. 2002, 2010; De Hoff et al. 2013). Each proposed event is indicated by a thick line crossing the node accompanying a circle: open, rearrangement or gene conversion event; filled, gene loss; red, gene acquisition.
Conclusion
While many questions remain about the history, timing, and mechanism of volvocine algal MT evolution, our data begin to elucidate the highly dynamic nature of their diversification, and to establish the trajectory of this process. Overall, there appears to be relative stasis in terms of MT chromosomal location, and in the presence of the sex-determining gene MID. FUS1 and MTD1 appear to have persisted in a large part of the volvocine lineage, but became dispensable in V. carteri, and perhaps its close relatives. While MTD1 is not totally essential for mating-type differentiation in C. reinhardtii, FUS1 is essential for fertilization (Goodenough et al. 2007; Lin and Goodenough 2007). How and why the FUS1 function was dispensed with or replaced in V. carteri (and perhaps other volvocine species), and how this change relates to its sexual cycle are interesting areas for future investigation.
It is unclear why MT remains associated with the same chromosome in the three widely separated volvocine species where it has been characterized, but is suggestive that interchromosomal sequence exchanges are much less frequent than intrachromosomal exchanges and rearrangements. Large changes in MT haplotype structure, gametolog content, and gametolog divergence have all occurred in volvocine algae. In all these areas, there may be episodic upheavals where the MT locus structure is reconfigured, and where the competing effects of autosomal insertions, gametolog gene conversion, and recombination suppression shape haplotype differentiation. Data on haploid mating-type, or sex chromosome evolution, from other lineages suggests that processes similar to those we have documented here for volvocine algae are likely to take place elsewhere. For example, gene conversion between gametologs has been documented in fungal mating loci, as have rearrangements and haplotype bloc formation associated with recombination suppression (Sun et al. 2012). Much less is known about haploid SDR regions outside of fungi, although some data are now available for haploid bryophyte sex chromosomes (Yamato et al. 2007; McDaniel et al. 2013), and for the SDR region of the brown alga Ectocarpus sp. (Ahmed et al. 2014). Future comparative studies in these other groups, and within the volvocine lineage, will help elucidate the mechanisms that contribute to mating locus and sex chromosome evolution, and how these mechanisms impact the evolution of sex and sexual cycles.
Supplementary Material
Acknowledgments
This work was supported by Grants-in-Aid for Scientific Research on Innovative Areas “Genome Science” (grant number 221S0002), Scientific Research (A) (grant number 24247042 to H.N.), National Institutes of Health grant GM 078376 to J.G.U., Japan Society for the Promotion of Sciences Research Fellows (19-7661 and 23-5499 to T.H.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan/ Japan Society for the Promotion of Sciences KAKENHI. T.H. is a Postdoctoral Fellow for Research Abroad (26-495) of the Japan Society for the Promotion of Science.
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.026229/-/DC1
Communicating editor: A. Rokas
Literature Cited
- Ahmed S., Cock J. M., Pessia E., Luthringer R., Cormier A., et al. , 2014. A haploid system of sex determination in the brown alga Ectocarpus sp. Curr. Biol. 24: 1945–1957. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Bachtrog D., Kirkpatrick M., Mank J. E., McDaniel S. F., Pires J. C., et al. , 2011. Are all sex chromosomes created equal? Trends Genet. 27: 350–357. [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., 2010. Evolutionary biology: the origins of two sexes. Curr. Biol. 20: R519–R521. [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., Marais G., 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95: 118–128. [DOI] [PubMed] [Google Scholar]
- Coleman A. W., 2012. A comparative analysis of the Volvocaceae (Chlorophyta). J. Phycol. 48: 491–513. [DOI] [PubMed] [Google Scholar]
- De Hoff P. L., Ferris P., Olson B. J. S. C., Miyagi A., Geng S., et al. , 2013. Species and population level molecular profiling reveals cryptic recombination and emergent asymmetry in the dimorphic mating locus of C. reinhardtii. PLoS Genet. 9: e1003724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris P. J., Goodenough U. W., 1994. The mating-type locus of Chlamydomonas reinhardtii contains highly rearranged DNA sequences. Cell 76: 1135–1145. [DOI] [PubMed] [Google Scholar]
- Ferris P. J., Goodenough U. W., 1997. Mating type in Chlamydomonas is specified by mid, the minus-dominance gene. Genetics 146: 859–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris P., Woessner J., Goodenough U., 1996. A sex recognition glycoprotein is encoded by the plus mating-type gene fus1 of Chlamydomonas reinhardtii. Mol. Biol. Cell 7: 1235–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris P. J., Pavlovic C., Fabry S., Goodenough U. W., 1997. Rapid evolution of sex-related genes in Chlamydomonas. Proc. Natl. Acad. Sci. USA 94: 8634–8639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris P. J., Armbrust E. V., Goodenough U. W., 2002. Genetic structure of the mating-type locus of Chlamydomonas reinhardtii. Genetics 160: 181–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris P., Olson B. J. S. C., De Hoff P. L., Douglass S., Casero D., et al. , 2010. Evolution of an expanded sex-determining locus in Volvox. Science 328: 351–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., et al. , 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31: 3784–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng S., De Hoff P., Umen J. G., 2014. Evolution of sexes from an ancestral mating-type specification pathway. PLoS Biol. 12: e1001904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U., Lin H., Lee J.-H., 2007. Sex determination in Chlamydomonas. Semin. Cell Dev. Biol. 18: 350–361. [DOI] [PubMed] [Google Scholar]
- Hamaji T., Ferris P. J., Coleman A. W., Waffenschmidt S., Takahashi F., et al. , 2008. Identification of the minus-dominance gene ortholog in the mating-type locus of Gonium pectorale. Genetics 178: 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaji T., Ferris P. J., Nishii I., Nozaki H., 2009. Identification of the minus mating-type specific gene MTD1 from Gonium pectorale (Volvocales, Chlorophyta). J. Phycol. 45: 1310–1314. [DOI] [PubMed] [Google Scholar]
- Hamaji T., Ferris P. J., Nishii I., Nishimura Y., Nozaki H., 2013a Distribution of the sex-determining gene MID and molecular correspondence of mating types within the isogamous genus Gonium (Volvocales, Chlorophyta). PLoS One 8: e64385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaji T., Smith D. R., Noguchi H., Toyoda A., Suzuki M., et al. , 2013b Mitochondrial and plastid genomes of the colonial green alga Gonium pectorale give insights into the origins of organelle DNA architecture within the Volvocales. PLoS One 8: e57177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanschen E. R., Marriage T. N., Ferris P. J., Hamaji T., Toyoda A., Fujiyama A., Neme R., Noguchi H., Minakuchi Y., Suzuki M., Kawai-Toyooka H., Smith D. R., Sparks H., Anderson J., Bakarić R., Luria V., Karger A., Kirschner M., Durand P. M., Michod R. E., Nozaki H., Olson B. J. S. C., 2016 Cell cycle regulation, group level adaptation, and the evolution of multicellularity. Nat. Commun. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron M. D., Michod R. E., 2008. Evolution of complexity in the volvocine algae: transitions in individuality through Darwin’s eye. Evolution 62: 436–451. [DOI] [PubMed] [Google Scholar]
- Hiraide R., Kawai-Toyooka H., Hamaji T., Matsuzaki R., Kawafune K., et al. , 2013. The evolution of male–female sexual dimorphism predates the gender-based divergence of the mating locus gene MAT3/RB. Mol. Biol. Evol. 30: 1038–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immler S., Otto S. P., 2015. The evolution of sex chromosomes in organisms with separate haploid sexes. Evolution 69: 694–708. [DOI] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., et al. , 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Librado P., Rozas J., 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- Lin H., Goodenough U. W., 2007. Gametogenesis in the Chlamydomonas reinhardtii minus mating type is controlled by two genes, MID and MTD1. Genetics 176: 913–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.-G., Whittier R. F., 1995. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25: 674–681. [DOI] [PubMed] [Google Scholar]
- McDaniel S. F., Neubig K. M., Payton A. C., Quatrano R. S., Cove D. J., 2013. Recent gene-capture on the UV sex chromosomes of the moss Ceratodon purpureus. Evolution 67: 2811–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S. S., Prochnik S. E., Vallon O., Harris E. H., Karpowicz S. J., et al. , 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misamore M. J., Gupta S., Snell W. J., 2003. The Chlamydomonas Fus1 protein is present on the mating type plus fusion organelle and required for a critical membrane adhesion event during fusion with minus gametes. Mol. Biol. Cell 14: 2530–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogi Y., Hamaji T., Suzuki M., Ferris P., Mori T., et al. , 2012. Evidence for tubular mating structures induced in each mating type of heterothallic Gonium pectorale (Volvocales, Chlorophyta). J. Phycol. 48: 670–674. [DOI] [PubMed] [Google Scholar]
- Nei, M., 1987 Molecular Evolutionary Genetics, Columbia University Press, New York, NY. [Google Scholar]
- Nozaki H., Misawa K., Kajita T., Kato M., Nohara S., et al. , 2000. Origin and evolution of the colonial Volvocales (Chlorophyceae) as inferred from multiple, chloroplast gene sequences. Mol. Phylogenet. Evol. 17: 256–268. [DOI] [PubMed] [Google Scholar]
- Nozaki H., Mori T., Misumi O., Matsunaga S., Kuroiwa T., 2006. Males evolved from the dominant isogametic mating type. Curr. Biol. 16: R1018–R1020. [DOI] [PubMed] [Google Scholar]
- Pasquale S. M., Goodenough U. W., 1987. Cyclic AMP functions as a primary sexual signal in gametes of Chlamydomonas reinhardtii. J. Cell Biol. 105: 2279–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochnik S. E., Umen J., Nedelcu A. M., Hallmann A., Miller S. M., et al. , 2010. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 329: 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamov A. A., Solovyev V. V., 2000. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10: 516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., and D. W. Russell, 2001 Molecular cloning: a laboratory manual, NY Cold Spring Harbor Laboratory Press., Cold Spring Harbor, NY. [Google Scholar]
- Setohigashi Y., Hamaji T., Hayama M., Matsuzaki R., Nozaki H., 2011. Uniparental inheritance of chloroplast DNA is strict in the isogamous Volvocalean Gonium. PLoS One 6: e19545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund C., Nelson W., Shoemaker A., Paterson A., 2006. SyMAP: A system for discovering and viewing syntenic regions of FPC maps. Genome Res. 16: 1159–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund C., Bomhoff M., Nelson W. M., 2011. SyMAP v3.4: a turnkey synteny system with application to plant genomes. Nucleic Acids Res. 39: e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Hsueh Y.-P., Heitman J., 2012. Gene conversion occurs within the mating-type locus of Cryptococcus neoformans during sexual reproduction. PLoS Genet. 8: e1002810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen J. G., 2011. Evolution of sex and mating loci: an expanded view from Volvocine algae. Curr. Opin. Microbiol. 14: 634–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T. K., Nakada T., Miyaji K., Nozaki H., 2006. Morphology and molecular phylogeny of Gonium multicoccum (Volvocales, Chlorophyceae) newly found in Japan. J. Jpn. Bot. 81: 139–147. [Google Scholar]
- Yamato K. T., Ishizaki K., Fujisawa M., Okada S., Nakayama S., et al. , 2007. Gene organization of the liverwort Y chromosome reveals distinct sex chromosome evolution in a haploid system. Proc. Natl. Acad. Sci. USA 104: 6472–6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586. [DOI] [PubMed] [Google Scholar]
- Yang Z., Nielsen R., 2000. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 17: 32–43. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DDBJ/ENA/GenBank accessions: LC062386 (G. pectorale DRG1 plus haplotype); LC062718 (MT- scaffold); LC062719 (MT+ scaffold).