Abstract
The double-stranded RNA-binding protein Staufen has been implicated in various posttranscriptional gene regulatory processes. Here, we demonstrate that the Caenorhabditis elegans homolog of Staufen, STAU-1, functionally interacts with microRNAs. Loss-of-function mutations of stau-1 significantly suppress phenotypes of let-7 family microRNA mutants, a hypomorphic allele of dicer, and a lsy-6 microRNA partial loss-of-function mutant. Furthermore, STAU-1 modulates the activity of lin-14, a target of lin-4 and let-7 family microRNAs, and this modulation is abolished when the 3′ untranslated region of lin-14 is removed. Deep sequencing of small RNA cDNA libraries reveals no dramatic change in the levels of microRNAs or other small RNA populations between wild-type and stau-1 mutants, with the exception of certain endogenous siRNAs in the WAGO pathway. The modulation of microRNA activity by STAU-1 does not seem to be associated with the previously reported enhanced exogenous RNAi (Eri) phenotype of stau-1 mutants, since eri-1 exhibits the opposite effect on microRNA activity. Altogether, our results suggest that STAU-1 negatively modulates microRNA activity downstream of microRNA biogenesis, possibly by competing with microRNAs for binding on the 3′ untranslated region of target mRNAs.
Keywords: microRNA, stau-1, RNAi, RNA-binding protein, 3′UTR
MicroRNAs (miRNAs) are a class of endogenous noncoding small RNAs that posttranscriptionally regulate gene expression, primarily through binding to the 3′ untranslated region (3′UTR) of target mRNAs and inhibiting translation and/or mRNA stability (He and Hannon 2004). miRNAs are usually transcribed into primary transcripts (pri-miRNAs) from independent genes by RNA polymerase II. Pri-miRNAs are processed into hairpin structured precursor miRNAs (pre-miRNAs) by the Drosha-DGCR8 complex in the nucleus, and then pre-miRNAs are exported into the cytoplasm and further processed by Dicer to generate the ∼21 nucleotide-long mature miRNAs (Krol et al. 2010). The seed sequence (nucleotides 2–7) of a mature miRNA dictates the specificity of a miRNA’s recognition of target mRNAs. Therefore, miRNAs with the same seed sequence are grouped into a family and are predicated to potentially share the same set of target mRNAs (Bartel 2009).
MiRNAs exert their repression on mRNAs through the assembly of the miRNA-induced silencing complex (miRISC) on the 3′UTR of target mRNAs. MiRISC is a ribonucleoprotein complex with a miRNA-specific Argonaute (AGO) protein loaded with a mature miRNA, and an AGO binding partner GW182 protein (Fabian and Sonenberg 2012). Besides AGO and GW182, other RNA-binding proteins have been shown to affect miRNA activity through biogenesis, such as the case of LIN-28 (Viswanathan and Daley 2010), miRISC activity, as for NHL-2 (Hammell et al. 2009), and target site accessibility, as shown for Pumilio, HuR, and Dnd1 (Nolde et al. 2007; Kedde et al. 2010, 2007; Bhattacharyya et al. 2006; Tominaga et al. 2011; Kundu et al. 2012; Young et al. 2012).
Staufen is a conserved double-stranded RNA-binding protein that contains five double-stranded RNA-binding domains, and was first identified in Drosophila to regulate mRNA localization and translation (oskar in oocytes, bicoid in embryos, and prospero in neuroblasts) (St Johnston et al. 1991; Li et al. 1997; Broadus et al. 1998). For example, Staufen binding to the 3′UTR of bicoid and prospero mRNAs is required for their localization (Ferrandon et al. 1994, 1997; Shen et al. 1997). In mammalian neurons, Staufen homologs (Staufen1 and Staufen2) are also known to regulate mRNA transport and the activation of localized mRNA translation (Köhrmann et al. 1999; Kiebler et al. 1999). Two groups have shown that Staufen1 can bind to long-range duplexes in the 3′UTR of mRNAs (Ricci et al. 2014; Sugimoto et al. 2015). Besides regulating mRNA localization and translation, mammalian Staufen can also mediate mRNA decay through interaction with the nonsense medicated decay regulator Upf1 (Park and Maquat 2013). In Caenorhabditis elegans, there is only one Staufen homolog, stau-1, and it has been shown to have high binding affinity for double-stranded RNA in vitro. STAU-1 is expressed at all developmental stages in C. elegans, and partial loss-of-function mutants [stau-1(tm2266) and stau-1(q798)] exhibit phenotypes that include enhanced transgene silencing, enhanced exogenous RNAi, and mild germline defects (LeGendre et al. 2013).
Previous studies have demonstrated that miRISC components and miRNAs are present in Staufen-containing RNA granules (Barbee et al. 2006; Peredo et al. 2014), which indicates that Staufen might affect the miRNA pathway, perhaps by influencing miRNA biogenesis and/or function. Here, we report genetic evidence that C. elegans STAU-1 exerts activity in opposition to certain miRNAs; we show that loss of function for stau-1 genetically suppresses the phenotypes of mutants in several distinct miRNA genes and of a Dicer (dcr-1) mutant. Further, our small RNA sequencing data show that STAU-1 does not have any significant effect on the levels of mature miRNAs, indicating that Staufen can inhibit the activity of miRNAs downstream of miRNA biogenesis. Finally, we report data suggesting that Staufen-mediated opposition of miRNA activity acts via the 3′UTR of miRNA target mRNAs.
Materials and Methods
Nematode methods and phenotypic analysis
C. elegans were cultured on nematode growth media (NGM) (Brenner 1974) and fed with Escherichia coli HB101. All the C. elegans strains used in this study are listed in Supplemental Material, Table S11. Synchronized populations of developmentally staged worms were obtained by standard methods (Stiernagle 2006).
For heterochronic phenotype analysis, gravid adult animals raised at 20° were placed on NGM plates seeded with E. coli HB101 at 20°, unless otherwise noted, and their progeny were scored at the young adult stage for adult lateral alae formation and seam cell number. Nomarski DIC microscopy was used to score alae formation, and fluorescence microscopy with the maIs105 [col-19::gfp] or wIs51 [scm::gfp] transgenes to mark lateral hypodermal cell nuclei was used to score seam cell number.
The ASE neuron phenotype was scored by the expression of otIs114 [lim-6::gfp] (ASEL marker) in the larvae of each genotype using a Zeiss SteREO Discovery.V12 microscope (Hammell et al. 2009).
Targeted genome editing by CRISPR/Cas9
In order to generate stau-1 null mutants, we adapted previously described co-CRISPR strategies (Kim et al. 2014; Arribere et al. 2014) with our modifications. Wild type animals (N2) were injected with a mixture containing 40 ng/μl eft-3::cas9 vector, 35 ng/μl unc-22 sgRNA vector, 35 ng/μl dpy-10 sgRNA vector, 35 ng/μl stau-1 sgRNA-1 vector, 35 ng/μl stau-1 sgRNA-2 vector, and 15 ng/μl sur-5::gfp vector. The sequences for stau-1 sgRNAs were: 5′-GGATGGAGTGATGATAGTAC-3′ (sgRNA-1) and 5′-TACGGATCTGGCAGATACTT-3′ (sgRNA-2). F1 worms exhibiting any of the dumpy and/or twitching phenotypes, and/or sur-5::gfp expression, were picked individually to plates and allowed them to produce F2 progeny. These F1 animals were lysed in 10 μl single-worm lysis buffer (50 mM KCl, 10 mM Tris-HCl pH 8.2, 2.5 mM MgCl2, 0.45% NP-40, 0.45% Tween-20, 0.01% Gelatin, and 60 ng/μl proteinase K) at 60° for 1 hr. PCR reactions were performed with primers (5′-TCCTTCAATCGATGTGGCCAA-3′ and 5′-TGGCTCACATTTTGTTAAACGACA-3′) and the sequence of PCR products was determined using Sanger sequencing. Both of the two mutations recovered were from CRISPR/Cas9 editing events by sgRNA-1.
Western blot analysis
For STAU-1 western blots, samples were prepared from populations of mixed stage embryos and synchronized L4 stage larvae grown on E. coli HB101 at 20°. Animals were washed off plates with M9 buffer and flash frozen in liquid nitrogen. Lysates were prepared by resuspending samples in lysis buffer [25 mM HEPES pH 7.5, 100 mM NaCl, 0.25 mM EDTA, 0.1% NP-40, 2 mM DTT, PhosSTOP (Roche), and Protease inhibitor (Roche)] and homogenized with a Branson SLPe sonicator. Lysates were centrifuged at 164,000 rpm for 15 min at 4° and the supernatants were collected. BioRad Protein Assay Dye Reagent Concentrate (Cat# 500-0006) was used to measure the protein concentration. 80 μg of protein were used for the immunoblot analysis. STAU-1 was recognized by an antiserum generated in the laboratory of Dr. Marvin Wickens (LeGendre et al. 2013) at 1:1000 dilution, gel loading was calibrated by reprobing blots with anti-α-tubulin antibody (Sigma-Aldrich Cat# T6074) at 1:20,000 dilution.
RNA extraction and small RNA cDNA cloning
Wild type (N2) and stau-1(tm2266) young adults were collected and flash frozen in liquid nitrogen. Three biological replicates were analyzed for each strain. Total RNA was extracted using Trizol reagent (Invitrogen). 20 μg of total RNA for each sample was used to isolate small RNA populations. RNA samples were run on a 15% PAGE/urea gel and small RNA populations were isolated from the gel with sizes ranging from 18–26 nucleotides (nt). A previously published small RNA cloning protocol (Sterling et al. 2015) was used to generate cDNA libraries with the following modifications: 1) 3′ ligation reactions were performed at 4° overnight; 2) 100 units of Superscript III Reverse Transcriptase were used for first-strand cDNA synthesis for each sample and the Reverse Transcriptase reaction was performed at 42° for 90 min. The Sterling et al. (2015) protocol involves only one RNA ligation to the 3′ end of the RNA, and hence recovers RNA species regardless of 5′ end structure.
Computational analysis of small RNA libraries
cDNA libraries were sequenced on the Ion Torrent (Proton) instrument according to manufacturer’s protocols. Sequencing files in FastQ formats were processed using the Cutadapt method (version 1.2.1) (Martin 2011) to remove the adapter sequences with the following options -e 0.25 -g 5′-ATTGATGGTGCCTACAG-3′ -a 5′-GATCGTTCGGACTGTAGATC-3′.
Sequence files were split into libraries according to barcode sequences, and reads shorter than 16 nt were removed. For each library, reads with identical sequences were combined and the combined count was saved in Fasta files. Reads were then aligned to the C. elegans genome (WormBase release WS215) using bowtie (Langmead et al. 2009) with arguments, -v 3 -f -B 1 -a–best –strata. Alignments were then filtered based on the length of the reads and the number of mismatches as follows: for sequence lengths 16–17, 18–19, 20–24, or >24: zero, one, two, or three mismatches were allowed, respectively.
Annotations of coding genes, transposons, tRNAs, rRNAs, piRNAs, and miRNAs were obtained from WormBase (release WS215) and miRBase (Griffiths-Jones et al. 2008) (release 20). An in-house developed code was used to analyze the mapping results. To assign read counts to the miRNA sequences, we considered all reads that mapped to the miRNA genomic loci starting within –5 to +5 nt of the annotated 5′ end of mature miRNAs. For all the other small RNA species and genomic features (e.g., coding genes and transposons), we counted all reads that mapped within the annotated region in sense and antisense orientations separately.
For endo-siRNA analysis, we considered all reads that mapped antisense to the 5′UTR, coding exons, and 3′UTR regions of each annotated gene. Annotations of target genes in the CSR-1, WAGO, ALG-3/4, and ERGO-1 pathways were downloaded from (Lee et al. 2012).
Differential expression analysis was performed using the edgeR package in R (Robinson et al. 2010).
Data availability
Sequence data files are available in the GEO database under the accession number GSE79217. Additional methods are described in File S1.
Results
STAU-1 functionally modulates the activity of several miRNAs
In order to identify modulators of miRNA activity, we established a panel of worm strains containing mutations designed to produce sensitized genetic backgrounds with compromised activity of specific miRNA families or miRNA biogenesis factors. There are three categories of mutations in these sensitized genetic backgrounds: 1) null mutations of a subset of genes encoding a miRNA family; 2) hypomorphic (nonnull partial loss-of-function) mutations of a particular miRNA; 3) hypomorphic mutations of a miRNA biogenesis factor or miRISC component. One essential feature of the sensitized genetic backgrounds is that these mutants all have partially penetrant phenotypes. This feature allows the identification of either positive or negative modulators of miRNA activity by testing for enhancement or suppression, respectively, of these sensitized phenotypes after genetic or RNAi knockdown of candidate gene activity.
The first sensitized genetic backgrounds we investigated were the let-7 family miRNA mutants. C. elegans let-7 family miRNAs (including let-7, mir-48, mir-84, and mir-241) function semiredundantly in controlling the developmental timing of certain stage-specific hypodermal seam cell fates. Loss of let-7 family miRNAs results in reiterations of early larval seam cell division patterns at later stages, and seam cells in these mutants also fail to properly differentiate adult specific cuticular structures (called adult alae) at the young adult stage (Figure 1A). Three of the let-7 family miRNAs (mir-48, mir-84, and mir-241) are expressed starting at the L2 stage and function to regulate the L2 stage proliferative seam cell division, while the let-7 miRNA is strongly upregulated from the L3 stage to control the larval-to-adult transition of seam cells (Reinhart et al. 2000; Abbott et al. 2005). These heterochronic let-7 family miRNA mutant phenotypes are easily quantified by using microscopy to measure the formation of adult alae and to score the number of seam cells in young adults.
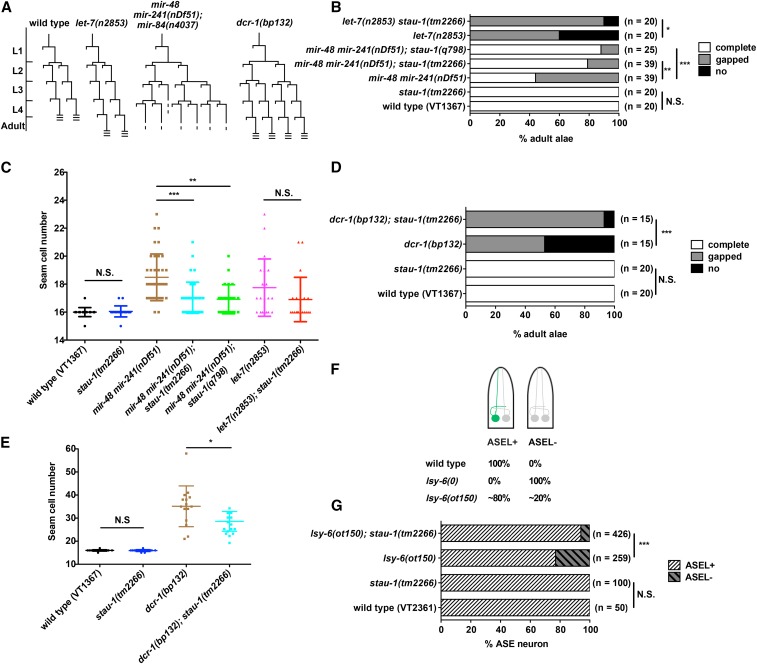
Figure 1.
Loss of function for stau-1 suppresses the phenotypes associated with mutations of genes encoding miRNAs, or DCR-1, a miRNA biogenesis factor. (A) Diagrams of seam cell V lineage in wild type (N2), let-7(n2853), mir-48 mir-241(nDf51); mir-84(n4037), and dcr-1(bp132) animals. L1 to L4 are the four larval stages in C. elegans postembryonic development. The three horizontal lines indicate adult alae formation. (B) Adult alae and (C) seam cell phenotypes of stau-1 mutants in wild type and let-7 family miRNA mutant backgrounds. (D) Adult alae and (E) seam cell phenotypes of stau-1(tm2266) in combination with dcr-1(bp132). The experiment with dcr-1(bp132) was carried out at 15°. (F) A diagram of C. elegans larvae illustrating the ASE (amphid sensilla) neuron phenotype of miRNA lsy-6 mutants. The green fluorescent protein (GFP) expression in ASEL (amphid sensilla, left) neuron is driven by lim-6 promoter. lsy-6(0) indicates the null allele of lsy-6; lsy-6(ot150) is a partial loss-of-function mutation. (G) ASE neuron phenotype of stau-1(tm2266) and double mutant of stau-1(tm2266); lsy-6(ot150). miRNA, microRNA; N.S., not significant (chi-square test for adult alae phenotype and ASE neuron phenotype, two-tailed t-test for seam cell phenotype); * P < 0.05; ** P < 0.01; *** P < 0.001.
To test if STAU-1 modulates let-7 family miRNA activity, we used two mutant strains [mir-48mir-241(nDf51) and let-7(n2853)], which both have partially penetrant heterochronic phenotypes with gaps in adult alae and increased number of seam cells at the young adult stage. mir-48mir-241(nDf51) mutant has two let-7 family miRNAs removed while let-7(n2853) is a strong loss-of-function mutation at the seed region of let-7 mature miRNA (Reinhart et al. 2000). Although the stau-1 loss-of-function mutant does not exhibit any developmental timing defects in an otherwise wild-type genetic background, we observed that both stau-1(tm2266) and stau-1(q798) significantly suppresses the heterochronic phenotypes of the mir-48mir-241(nDf51) mutant (Figure 1, B and C). Since stau-1(tm2266) and stau-1(q798) have similar effects on the phenotypes of mir-48mir-241(nDf51) animals, we focused only on stau-1(tm2266) for further analysis. Besides suppressing the heterochronic phenotypes of mir-48mir-241(nDf51) animals, stau-1(tm2266) also exerts significant suppression of the heterochronic adult alae phenotype of let-7(n2853) animals (Figure 1B). We interpret this suppression of heterochronic phenotypes of let-7 family miRNA mutants by stau-1 loss of function to suggest that loss of stau-1 function causes an increase in the activity of the remaining let-7 family miRNAs. These results indicate that STAU-1 acts as a negative modulator of let-7 family miRNA biogenesis or activity.
The second sensitized genetic background we tested is a dicer (dcr-1) hypomorphic allele, bp132. This mutation causes a single amino acid change in the RNase III domain of DCR-1 and has been previously shown to cause developmental timing defects, as indicated by an increased number of seam cells and failure to form complete adult alae at the young adult stage (Figure 1A). The phenotypes of this partial loss-of-function mutant dcr-1(bp132) are stronger at 15° and can be suppressed by a point mutation that alters the sequence of the lin-4 miRNA precursor (Lee et al. 1993; Wightman et al. 1993; Ren and Zhang 2010), indicating that the dcr-1(bp132) phenotypes reflect partially compromised lin-4 biogenesis. We found that stau-1(tm2266) suppresses both adult alae and seam cell phenotypes of this dcr-1(bp132) mutant at 15° (Figure 1, D and E), suggesting that STAU-1 negatively modulates lin-4 biogenesis or activity.
The third sensitized genetic background that we employed is a lsy-6 miRNA hypomorphic mutant. lsy-6 is known to regulate the asymmetric cell fate decision in ASE neurons (Johnston and Hobert 2003). The null allele of lsy-6 causes a highly penetrant cell fate transformation phenotype, where the ASEL neuron adopts the cell fate of the ASER neuron, which is detected by loss of expression of the ASEL marker lim-6. The lsy-6(ot150) allele is a nonnull (hypomorphic) point mutation 111 nt upstream of the lsy-6 hairpin, which disrupts a cis-regulatory element required for lsy-6 expression. The lsy-6(ot150) animals exhibit a weak phenotype with ∼20% penetrance (Sarin et al. 2007) (Figure 1F). stau-1(tm2266) animals do not exhibit any ASE neuron cell fate defects since all the animals have lim-6 expression only in the ASEL neurons, yet the phenotype of lsy-6(ot150) animals is significantly suppressed by stau-1(tm2266) (Figure 1G). These results indicate that loss of function of stau-1 can potentiate the activity of lsy-6, suggesting that the role of STAU-1 as a negative modulator of miRNA activity is not restricted to miRNAs of the heterochronic pathway.
stau-1 null mutants have an effect on miRNA activity similar to that of partial loss-of-function mutants
The stau-1 mutants available so far are partial loss-of-function mutants that remove either the second (tm2266) or the fourth (q798) double-stranded RNA-binding domain (Figure 2A). Therefore, to test the effect of stau-1 null mutants on miRNA activity and to determine if stau-1 may have additional functions, we carried out CRISPR/Cas9 experiments to generate null mutants of stau-1. The guide RNA was designed to target the first exon of stau-1 and we screened for frameshift mutations that lead to premature stop codons. Two independent mutations were isolated: ma327, an 11 bp insertion, and ma346, a 5 bp deletion, both of which generate early premature stop codons (Figure 2, A and B).
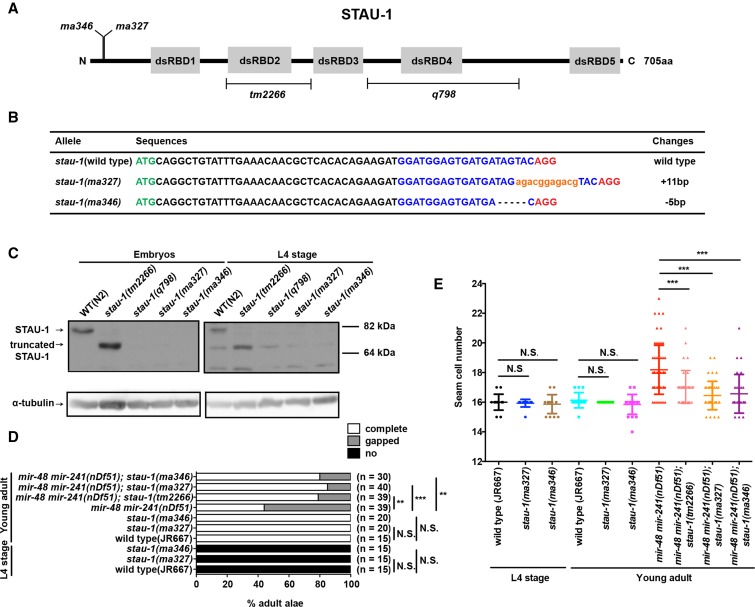
Figure 2.
Characterization of stau-1 null alleles. (A) A diagram of protein domains and mutations of STAU-1 used in this study. The five double-stranded RNA-binding domains (dsRBDs) are illustrated as gray boxes. The deletions in stau-1(tm2266) and stau-1(q798) (LeGendre et al. 2013) are shown by brackets. The positions of ma327 and ma346 mutations are indicated. (B) Nucleotide changes of stau-1(ma327) and stau-1(ma346) mutants. All the sequences shown here are in the beginning of stau-1’s first exon. The ATG start codon is in green. The sgRNA sequence is highlighted in blue and the PAM sequence is in red. The inserted sequence in ma327 is in orange and lower case. The premature termination codons in ma327 and ma346 are 89 and 65 amino acids downstream from the N-terminus respectively. (C) Western blots of STAU-1 and α-tubulin in wild type and stau-1 mutants. Both mix stage embryos and L4 stage animals were used for this experiment. There are nonspecific bands in the L4 stage blot. (D) Adult alae and (E) seam cell phenotypes of the stau-1 null mutants at the L4 and the young adult stage in wild type and mir-48 mir-241(nDf51) backgrounds. ** P < 0.01; *** P < 0.001; N.S., not significant (chi-square test for adult alae phenotype and two-tailed t-test for seam cell phenotype); sgRNA, single guide RNA.
To confirm that ma327 and ma346 mutations are null alleles, we tested for expression of STAU-1 protein in these mutant animals. We performed western blot analysis of wild type and stau-1 mutant embryos and L4 stage larvae using an antiserum generated against the fourth double-stranded RNA-binding domain of STAU-1 (LeGendre et al. 2013) (Figure 2C). stau-1(q798) animals lacking the fourth double-stranded RNA-binding domain were used as negative controls. As expected, the stau-1(tm2266) mutation resulted in a truncated STAU-1 protein recognizable by the antiserum. In the embryos, we only observed full-length and truncated STAU-1 in wild type animals and stau-1(tm2266) mutants, respectively, whereas neither stau-1(ma327) nor stau-1(ma346) embryos contained any detectable STAU-1 protein. In the L4 stage samples, there were nonspecific bands close to the size of the truncated STAU-1, but the stau-1(ma327) and stau-1(ma346) mutants had the same band pattern as the negative control. Therefore, the stau-1 alleles ma327 and ma346 generated in this study appear to be null alleles.
Next, we sought to characterize the phenotypes of these stau-1 null alleles. Homozygous mutants of either ma327 or ma346 are viable. However, similar to tm2266 and q798, these animals exhibit a 4 hr delay in larval development at 20° and they are smaller in size compared to wild type animals at the young adult stage (Figure S1). Furthermore, to test if the stau-1 null mutants also suppress phenotypes of miRNA mutants, we crossed the null alleles into the let-7 family mutant mir-48mir-241(nDf51). As expected, both of these null alleles significantly suppressed the adult alae and seam cell phenotypes of mir-48mir-241(nDf51) animals (Figure 2, D and E). Interestingly, neither stau-1(ma327) nor stau-1(ma346) animals exhibited any heterochronic defects in an otherwise wild-type genetic background (Figure 2, D and E), indicating that the let-7 family hyperactivity that would result from the loss of STAU-1 was below the threshold required to elicit a precocious developmental timing phenotype.
Since the effects of these stau-1 null alleles on let-7 family miRNA activity are similar to the effect of stau-1(tm2266), subsequent studies were conducted using stau-1(tm2266).
STAU-1 does not dramatically affect mature miRNA levels
To investigate the mechanism of how stau-1 modulates miRNA activity, we first performed small RNA high-throughput sequencing analysis of wild type and stau-1(tm2266) animals at the young adult stage. Three replicate samples were sequenced for each genotype and all the replicates yielded more than 5,000,000 reads, with ∼80% of the reads mapping to the C. elegans genome. Since the small RNA cloning technique we used is not dependent on the structure of the 5′ nucleotide of the RNA (Sterling et al. 2015), we were able to examine STAU-1’s effect on diverse small RNA populations, including miRNAs, piRNAs, and endogenous siRNAs. Based on the reads distribution data of small RNA populations, we did not observe any dramatic difference between wild type and stau-1(tm2266) animals (Figure 3A).
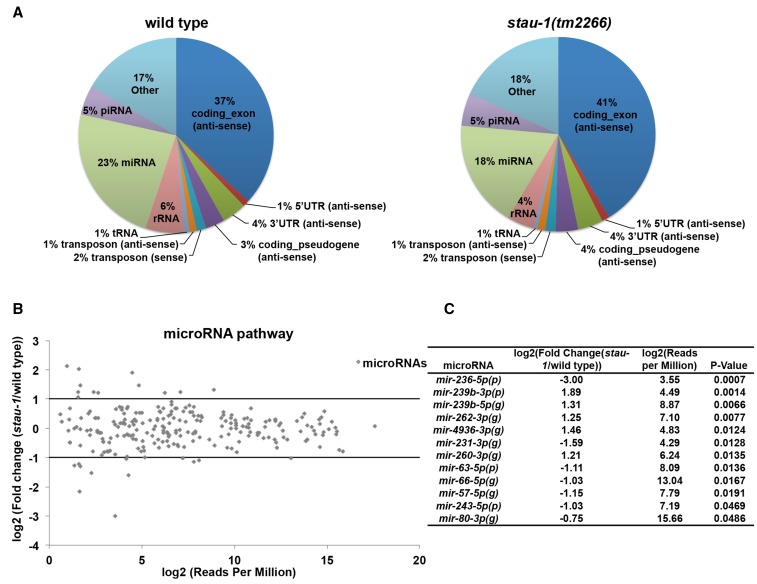
Figure 3.
STAU-1 does not significantly affect mature miRNA levels. (A) Read distribution of small RNAs mapped to C. elegans genome in wild type and stau-1(tm2266) animals. All the antisense reads are endo-siRNA reads since C. elegans endo-siRNAs are mapped antisense to various regions of different gene transcripts (Gu et al. 2009). Most of reads in “other” were mapped to the sense strand of protein coding genes. (B) Differential gene expression analysis of miRNAs between wild type and stau-1(tm2266) animals. (C) List of miRNAs whose levels are significantly affected in stau-1(tm2266) animals. (g), miRNA guide strand; (p), miRNA passenger strand.
Next, we focused our analysis on the levels of mature miRNAs. In our sequencing data, we identified a total of 239 miRNAs. However, only 12 miRNAs were significantly changed in stau-1(tm2266) compared to wild type animals (five upregulated and seven downregulated) (Figure 3, B and C, Table S1, and Table S2). None of the miRNAs that are differentially expressed in stau-1(tm2266) animals are known to be able to contribute to the phenotypes suppressed by stau-1(tm2266) (Figure 1). Additionally, since the precocious expression of let-7 at the L2 stage, rather than the overexpression at the young adult stage, could affect the developmental timing phenotypes (Vadla et al. 2012), we examined the mature miRNA levels at the L2 stage. No significant change in mature miRNA levels was detected in either stau-1(tm2266) or stau-1(q798) animals compared to wild type animals (Figure S2). Consistent with these results, we did not observe any significant change in the protein levels of two miRNA biogenesis factors [DCR-1 and ALG-1 (a C. elegans AGO)] in several stau-1 mutants (Figure S3). Therefore, we concluded that the modulation by STAU-1 of miRNA activity occurs downstream of miRNA biogenesis.
STAU-1 does not dramatically affect other small RNA populations
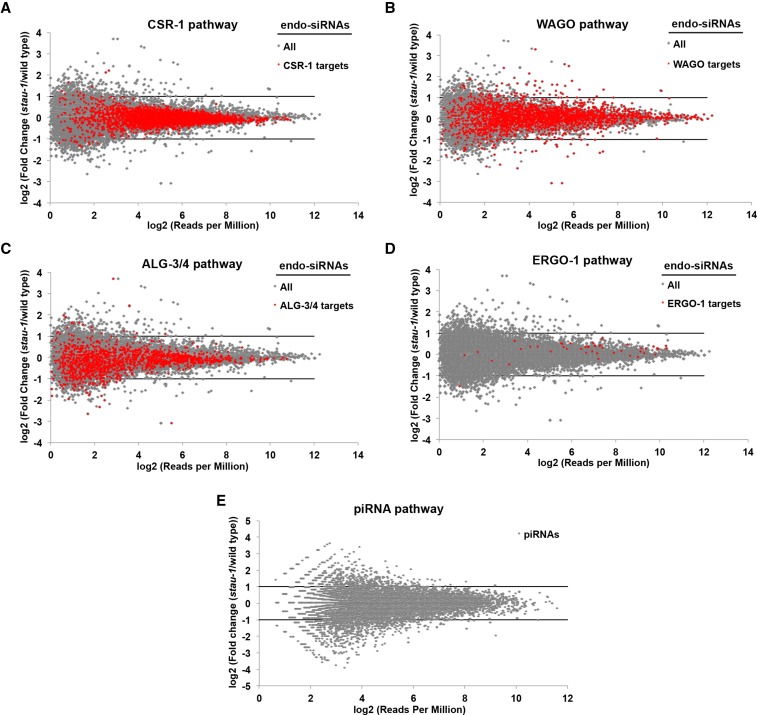
Since stau-1 mutants exhibit enhanced RNAi and transgene silencing phenotypes (LeGendre et al. 2013), we examined whether there is any change in other small RNA populations between wild type and stau-1(tm2266) animals. Besides miRNAs, C. elegans also possesses several classes of endogenous siRNAs (endo-siRNAs) and piRNAs. Since different functional classes of endo-siRNAs are loaded into the distinct Argonaute proteins, CSR-1, WAGO, ALG-3/4, and ERGO-1 (Lee et al. 2012), we mapped our sequencing reads to these annotations and carried out differential gene expression analysis (Figure 4, A–D, Table S3, Table S4, Table S5, Table S6, Table S7, and Table S8). Of all these endo-siRNA categories, the only noteworthy changes we observed were for a subset of WAGO endo-siRNAs, which may function to maintain the silencing of “nonself” transcripts in C. elegans germline (Shirayama et al. 2012) (Figure 4B and Table S6). STAU-1 does not seem to simply promote biogenesis of endo-siRNAs in the WAGO pathway since cases of upregulation and downregulation of endo-siRNAs were evident in stau-1(tm2266) animals (Figure 4B). The final class of small RNAs that we analyzed was piRNAs, and there was no dramatic change in the piRNA levels between wild type and stau-1(tm2266) animals (Figure 4E, Table S9, and Table S10).
Figure 4.
The effects of STAU-1 on small RNA pathways other than miRNAs. Comparisons between wild type and stau-1(tm2266) animals regarding expression of endo-siRNAs (A–D) associated with the CSR-1 (A), WAGO (B), ALG-3/4 (C), and ERGO-1 pathways (D), and piRNAs (PIWI-interacting RNAs) (E).
The modulation of miRNA activity by stau-1 mutations is likely independent of their enhanced RNAi phenotype
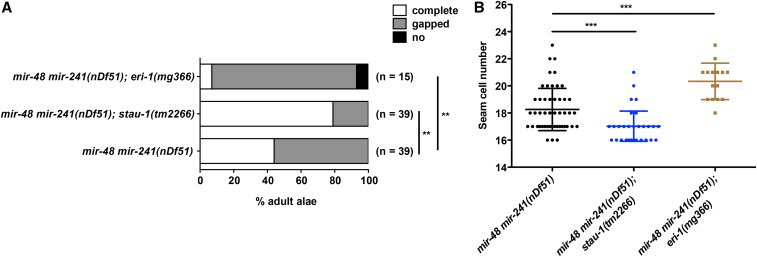
C. elegans stau-1 mutants have been shown to exhibit an enhanced RNAi (Eri) phenotype, indicating that STAU-1 negatively modulates one or more RNAi pathways. This suggests that STAU-1’s negative modulation of miRNA activity that we have shown here could reflect a common underlying effect of STAU-1 on small RNA silencing more broadly. In that case, one might expect that other Eri mutants may also exhibit enhanced miRNA activity. Because stau-1 mutants had been shown to interact genetically with eri-1 (LeGendre et al. 2013), we tested whether eri-1 loss-of-function could affect the heterochronic phenotypes of mir-48mir-241(nDf51) animals. Interestingly, loss of function of eri-1 enhanced the adult alae and seam cell defects of mir-48mir-241(nDf51) animals (Figure 5), which is opposite to the suppression caused by stau-1 mutation. Therefore, the modulation of miRNA activity by STAU-1 is unlikely to be simply the result of a general enhancement of RNA interference.
Figure 5.
ERI-1 positively modulates let-7 family miRNA activity. (A) Adult alae and (B) seam cell phenotype of eri-1(mg366) in combination with mir-48 mir-241(nDf51). N.S., not significant ** P < 0.01; *** P < 0.001 (chi-square test for adult alae phenotype and two-tailed t-test for seam cell phenotype).
STAU-1 may act through the 3′UTR of miRNA targets to modulate miRNA activity
Staufen has been shown to promote the translation of its target mRNAs (Micklem et al. 2000; Dugré-Brisson et al. 2005; Ricci et al. 2014), suggesting that the negative effect of STAU-1 on miRNA activity shown here could reflect its role in promoting the translation of miRNA target mRNAs, either by binding to 3′UTR sequences (perhaps at or near miRNA binding sites), or to other regions of the mRNAs (such as 5′UTR and/or coding sequences). To investigate whether STAU-1 could oppose miRNA activity relatively directly, via the 3′UTR sequences of the target mRNAs, we utilized mutants of the heterochronic miRNA lin-4 and its primary target lin-14 (Lee et al. 1993; Wightman et al. 1993).
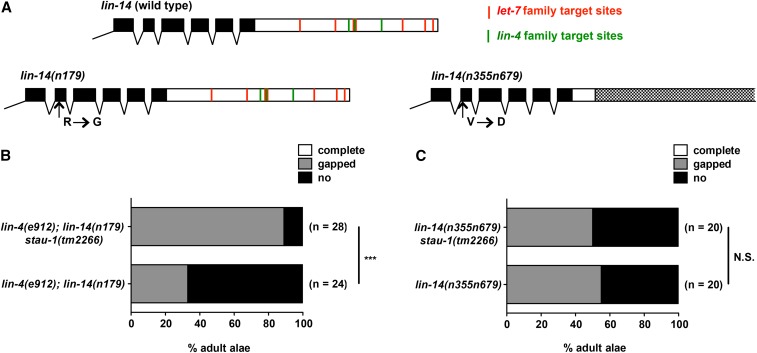
The 3′UTR of lin-14 possesses several lin-4 and let-7 family miRNA target sites (Figure 6A). The first strain we tested was lin-4(e912); lin-14(n179), which has a null mutation of lin-4 and a point mutation in lin-14, and this double mutant exhibits a temperature sensitive heterochronic phenotype. At permissive temperature (15°), lin-4(e912); lin-14(n179) animals exhibit a partially penetrant phenotype (∼30% animals have gaps in the adult alae) and can be considered a sensitized genetic background. stau-1(tm2266) significantly suppresses the adult alae phenotype of lin-4(e912); lin-14(n179) animals (Figure 6B). This indicates that STAU-1 could modulate the activity of lin-14, possibly through let-7 family miRNAs or mir-237 (the other member of lin-4 family miRNAs in C. elegans).
Figure 6.
The 3′UTR of lin-14 is required for modulation of lin-14 gain-of-function phenotypes by stau-1 mutation. (A) Diagrams of C-terminal end of lin-14 gene in wild type, lin-14(n719), and lin-14(n355n679). The black boxes represent exons and the white boxes represent 3′UTR. The dotted box indicates the region of 3′UTR deleted in lin-14(n355n679). The predicated lin-4 family (green lines) and let-7 family (red lines) target sites are indicated; the target site predication was obtained from TargetScan (Lewis et al. 2005; Jan et al. 2011). (B) Adult alae phenotype of lin-4(e912); lin-14(n179) and lin-4(e912); lin-14(n179) stau-1(tm2266) animals. (C) Adult alae phenotype of lin-14(n355n679) and lin-14(n355n679) stau-1(tm2266) animals. *** P < 0.001; N.S., not significant (chi-square test for adult alae phenotype and two-tailed t-test for seam cell phenotype).
Next, we used another lin-14 mutant strain lin-14(n355n679). n355 is a breakpoint mutation in the 3′UTR of lin-14, which results in the removal of most of the lin-14 3′UTR, including all the lin-4 and let-7 family miRNA binding sites (Shi et al. 2013). In combination with another point mutation (n679) that partially compromises LIN-14 function, the phenotype of lin-14(n355n679) is similar to lin-4(e912); lin-14(n179) animals. However, we failed to observe any suppression by stau-1(tm2266) on the adult alae phenotype on lin-14(n355n679) animals (Figure 6C). These data suggest that STAU-1 may modulate miRNA activity through the 3′UTR of miRNA targets.
Discussion
The double-stranded RNA-binding protein Staufen has been characterized as a master regulator of mRNA localization and translation in many metazoan species (Roegiers and Jan 2000). Staufen is known to positively regulate translation upon localization through interactions with structured regions of mRNAs (3′UTRs, coding sequences, and 5′UTRs) and various partner proteins and/or ribosomes (Ferrandon et al. 1994; Micklem et al. 2000; Dugré-Brisson et al. 2005; Ricci et al. 2014). Besides such positive roles in gene expression, Staufen can also negatively regulate gene expression by recruiting the nonsense-mediated decay factor Upf1 to the 3′UTR of mRNAs to trigger mRNA degradation (Park and Maquat 2013). Because of these alternative positive or negative roles in posttranscriptional regulation of mRNA activity, we predicted that Staufen could be expected to interact functionally with miRNA-mediated repression of mRNA targets, and could exert either promotion or inhibition of miRNA activity.
In this study, we found that loss-of-function Staufen (stau-1) mutations in C. elegans can suppress the phenotypes of miRNA partial loss of function, indicating that STAU-1 inhibits miRNA activity. This suggests that, at least with respect to the miRNAs whose functions we examined here, STAU-1 engages its translational enhancer function, rather than its mRNA decay activity. We show that stau-1 loss-of-function mutation does not appreciably affect the levels of mature miRNAs; in particular, there was no detectable change, in stau-1 mutants, of the levels of the lin-4, let-7 family, and lsy-6 miRNAs whose function we monitored phenotypically in our genetic interaction experiments. This strongly suggests that STAU-1 likely opposes the activity of these miRNAs independently of their biogenesis or turnover, and perhaps may act by binding to their target mRNAs. Consistent with a model where STAU-1 can modulate miRNA activity by binding to the 3′UTR of miRNA targets, our data show that the 3′UTR of a miRNA target, lin-14, is required for STAU-1-mediated modulation of lin-14 heterochronic phenotypes.
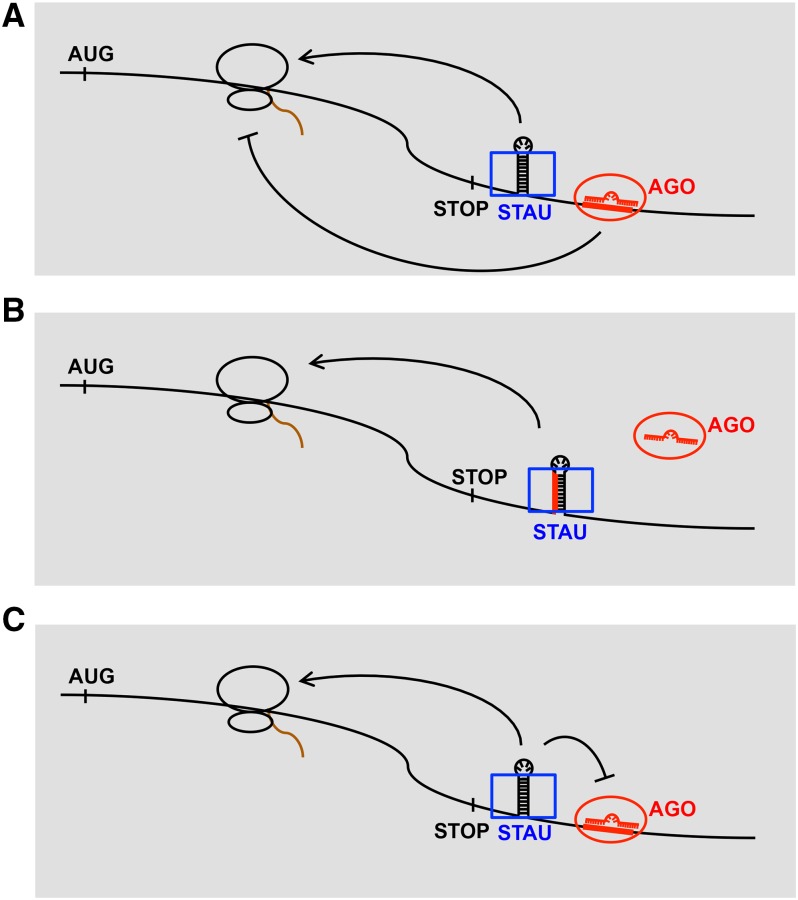
Various possible molecular mechanisms could be the basis for an opposition of miRNA repression by STAU-1. STAU-1 bound to target mRNA could oppose miRNA activity by exerting an independent translational activation (Figure 7A), or perhaps also by directly inhibiting the binding (Figure 7B) or efficacy (Figure 7C) of miRISC. It should be noted that a target site occlusion model for STAU-1 (Figure 7B) would be similar to the action attributed to other 3′UTR binding proteins (Pumilio, HuR, and Dnd1) that can apparently affect miRNA target accessibility by binding at or near miRNA sites (Fabian and Sonenberg 2012).
Figure 7.
Alternative models for STAU-1-mediated modulation of miRNA activity wherein STAU-1 is proposed to bind to miRNA targets, for example via 3′UTR double-stranded RNA secondary structure. The previously-described translational activation function of Staufen (STAU) could oppose, and hence inhibit the net potency of, miRNA-based translational repression (A); STAU-1 could also affect miRNA activity by binding at (or close to) the miRNA binding site and hence inhibit miRISC binding (B) or activity (C). Blue rectangles are STAU-1 proteins and red ovals are AGO proteins. The red single stranded nucleic acids in AGO proteins represent miRNAs and red lines on mRNAs are miRNA binding sites. Brown curves are newly synthesized protein peptides from the mRNAs.
All the models proposed here (Figure 7) involve a hypothetical physical interaction of STAU-1 with miRNA targets. Accordingly, one would predict that the miRNA target mRNAs that likely contribute to the phenotypes we observed here (lin-14, hbl-1, lin-41, and cog-1) should be recoverable associated with STAU-1 immunoprecipitated from worms of the appropriate developmental stage (embryo for cog-1; L1 for lin-14, L2 for hbl-1, and L4/adult for lin-41). We have not tested for these mRNA associations by immunoprecipitation, but a previous study (LeGendre et al. 2013) did recover the let-7 target lin-41 as enriched in a STAU-1 RNA Immunoprecipitation (RIP) experiment using extracts of C. elegans adults, supporting the model that STAU-1 could interact with miRNA targets. LeGendre et al. (2013) did not recover cog-1, lin-14, or hbl-1 in their RIP experiments, but this is perhaps not unexpected since larval stage extracts were not tested. Consistent with the hypothesis that STAU-1 could interact with the 3′UTRs of these genes by binding to regions of double-stranded RNA (dsRNA) secondary structure to potentiate miRNA activity, these 3′UTRs contain numerous regions predicted to form dsRNA structures, and many of the miRNA complementary sites reside at or near the structured regions (Figure S4). However, the identification of STAU-1 binding sites in C. elegans is needed to fully test this hypothesis.
Our small RNA sequencing data indicate that stau-1 loss-of-function mutation does not affect miRNA biogenesis in general, despite having clear effects on the developmental phenotypes of certain miRNA mutants. Importantly, the levels of the particular miRNAs responsible for those phenotypes were unchanged in stau-1 mutants. However, we did not examine levels of these miRNAs in specific cell types, therefore, it is possible that STAU-1 could modulate miRNA biogenesis or stability cell type-specifically, and we might not have detected cell type-specific changes of those miRNAs in our RNA samples extracted from whole animals. Indeed, our sequencing data contain a hint that the abundance of some miRNAs could be affected by STAU-1; 11 miRNAs exhibited at least twofold change in levels in the stau-1 mutant compared to wild type animals (Figure 3C). In such cases, perhaps STAU-1, through its double-stranded RNA-binding activity, can associate with secondary structure elements in miRNA primary transcripts and/or precursors and modulate their processing into mature miRNAs.
Prompted by the finding from a previous study that C. elegans stau-1 mutants exhibit an enhanced RNAi (Eri) phenotype, and interact genetically with eri-1 mutation (LeGendre et al. 2013), we tested whether an eri-1 mutation, similarly to stau-1, could suppress let-7 family miRNA mutant’s heterochronic phenotypes. Surprisingly, the eri-1(mg366); mir-48mir-241(nDf51) mutant exhibited enhanced heterochronic phenotypes, which is the opposite from the effect of stau-1. First of all, this finding indicates that the modulation of miRNA activity by STAU-1 is unlikely to stem simply from an enhanced exogenous RNAi pathway; otherwise, we would have expected that eri-1(mg366), like stau-1(loss-of-function), should suppress mir-48mir-241(nDf51) mutant phenotypes. Rather, these findings, particularly the opposite effects of different Eri loci on let-7 family miRNA phenotypes, suggest important but as yet uncharacterized interactions among RNAi and miRNA pathways in C. elegans. ERI-1 is known to be an exonuclease and important for the production of siRNAs in C. elegans (Kennedy et al. 2004), and a few studies have examined miRNA levels in an eri-1 loss-of-function context with mixed results, perhaps reflecting differences among experimental systems and/or the particular miRNAs assayed (Lee et al. 2006; Duchaine et al. 2006; Pavelec et al. 2009; Thomas et al. 2012). It is clear that further studies are needed to characterize the mechanisms by which ERI-1 affects miRNA activity.
Interestingly, we did not observe any overt miRNA gain-of-function phenotypes for stau-1 mutations in an otherwise wild-type genetic background, as might be expected for loss of a potent miRNA inhibitor. Rather, the stau-1 mutants’ miRNA phenotypes were only detected in sensitized genetic backgrounds with compromised miRNA activity. These findings suggest a modulatory effect of STAU-1 on miRNA activity and underscore the importance of miRNA pathways in conferring robustness to biological systems. The modulatory role of STAU-1 on miRNA activity could perhaps be important in refining the posttranscriptional regulation of important miRNA targets and to modulate the efficacy of miRNAs in response to physiological and environmental signals.
In conclusion, our study demonstrates that the RNA-binding protein STAU-1 negatively modulates miRNA activity downstream of miRNA biogenesis, possibly by interacting with the 3′UTR of miRNA targets. These findings reveal an expanded suite of RNA regulatory roles for STAU-1; besides regulating mRNA localization, translation and decay, Staufen can also exert posttranscriptional gene regulation through its engagement with miRNA targets. It should be noted that our results to date indicate that STAU-1 can inhibit the activity of let-7 family, lsy-6, and perhaps lin-4 miRNAs, but further studies are required to test for similar roles of STAU-1 in opposing the activity of other miRNAs. Moreover, we should not rule out the possibility that STAU-1 could promote the activity of certain other miRNAs, for example through its known role in mediating mRNA decay (Park and Maquat 2013).
Supplementary Material
Acknowledgments
We thank members of the Ambros lab for helpful discussions, and the laboratories of Marvin Wickens, Craig Mello, Amy Walker, and Roger Davis for reagents and technical assistance. Several nematode strains and reagents used in this study were kindly provided by the Caenorhabditis Genetics Center (which is funded by the National Institutes of Health [NIH] Office of Research Infrastructure Programs [P40 OD010440]). Firefly Bioworks Inc. developed the firefly miRNA assay for C. elegans-specific miRNAs. This work was funded by a NIH grant (R01 GM34028) to V.A. and a Leukemia and Lymphoma society postdoctoral fellowship to I.V-L.
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.027300/-/DC1
Communicating editor: S. Lee
Literature Cited
- Abbott A. L., Alvarez-Saavedra E., Miska E. A., Lau N. C., Bartel D. P., et al. , 2005. The let-7 microRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev. Cell 9: 403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere J. A., Bell R. T., Fu B. X. H., Artiles K. L., Hartman P. S., et al. , 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198: 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbee S. A., Estes P. S., Cziko A.-M., Hillebrand J., Luedeman R. A., et al. , 2006. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron 52: 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P., 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S. N., Habermacher R., Martine U., Closs E. I., Filipowicz W., 2006. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125: 1111–1124. [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadus J., Fuerstenberg S., Doe C. Q., 1998. Staufen-dependent localization of prospero mRNA contributes to neuroblast daughter-cell fate. Nature 391: 792–795. [DOI] [PubMed] [Google Scholar]
- Duchaine T. F., Wohlschlegel J. A., Kennedy S., Bei Y., Conte D., Jr, et al. , 2006. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124: 343–354. [DOI] [PubMed] [Google Scholar]
- Dugré-Brisson S., Elvira G., Boulay K., Chatel-Chaix L., Mouland A. J., et al. , 2005. Interaction of Staufen1 with the 5′ end of mRNA facilitates translation of these RNAs. Nucleic Acids Res. 33: 4797–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian M. R., Sonenberg N., 2012. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat. Struc.t Mol. Biol. 19: 586–593. [DOI] [PubMed] [Google Scholar]
- Ferrandon D., Elphick L., Nüsslein-Volhard C., St Johnston D., 1994. Staufen protein associates with the 3′UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell 79: 1221–1232. [DOI] [PubMed] [Google Scholar]
- Ferrandon D., Koch I., Westhof E., Nüsslein-Volhard C., 1997. RNA-RNA interaction is required for the formation of specific bicoid mRNA 3′ UTR-STAUFEN ribonucleoprotein particles. EMBO J. 16: 1751–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Saini H. K., van Dongen S., Enright A. J., 2008. miRBase: tools for microRNA genomics. Nucleic Acids Res. 36: D154–D158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Shirayama M., Conte D., Vasale J., Batista P. J., et al. , 2009. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell 36: 231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell C. M., Lubin I., Boag P. R., Blackwell T. K., Ambros V., 2009. nhl-2 modulates microRNA activity in Caenorhabditis elegans. Cell 136: 926–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Hannon G. J., 2004. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5: 522–531. [DOI] [PubMed] [Google Scholar]
- Jan C. H., Friedman R. C., Ruby J. G., Bartel D. P., 2011. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature 469: 97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R. J., Hobert O., 2003. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426: 845–849. [DOI] [PubMed] [Google Scholar]
- Kedde M., Strasser M. J., Boldajipour B., Oude Vrielink J. A. F., Slanchev K., et al. , 2007. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 131: 1273–1286. [DOI] [PubMed] [Google Scholar]
- Kedde M., van Kouwenhove M., Zwart W., Oude Vrielink J. A. F., Elkon R., et al. , 2010. A Pumilio-induced RNA structure switch in p27–3′ UTR controls miR-221 and miR-222 accessibility. Nature Publishing Group 12: 1014–1020. [DOI] [PubMed] [Google Scholar]
- Kennedy S., Wang D., Ruvkun G., 2004. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 427: 645–649. [DOI] [PubMed] [Google Scholar]
- Kiebler M. A., Hemraj I., Verkade P., Köhrmann M., Fortes P., et al. , 1999. The mammalian staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: implications for its involvement in mRNA transport. J. Neurosci. 19: 288–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Ishidate T., Ghanta K. S., Seth M., Conte D., et al. , 2014. A co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans. Genetics 197: 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhrmann M., Luo M., Kaether C., DesGroseillers L., Dotti C. G., et al. , 1999. Microtubule-dependent recruitment of Staufen-green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol. Biol. Cell 10: 2945–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J., Loedige I., Filipowicz W., 2010. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11: 597–610. [DOI] [PubMed] [Google Scholar]
- Kundu P., Fabian M. R., Sonenberg N., Bhattacharyya S. N., Filipowicz W., 2012. HuR protein attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA. Nucleic Acids Res. 40: 5088–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.-C., Gu W., Shirayama M., Youngman E., Conte D., et al. , 2012. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell 150: 78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L., Ambros V., 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854. [DOI] [PubMed] [Google Scholar]
- Lee R. C., Hammell C. M., Ambros V., 2006. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA 12: 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeGendre J. B., Campbell Z. T., Kroll-Conner P., Anderson P., Kimble J., et al. , 2013. RNA targets and specificity of Staufen, a double-stranded RNA-binding protein in Caenorhabditis elegans. J. Biol. Chem. 288: 2532–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B. P., Burge C. B., Bartel D. P., 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. [DOI] [PubMed] [Google Scholar]
- Li P., Yang X., Wasser M., Cai Y., Chia W., 1997. Inscuteable and Staufen mediate asymmetric localization and segregation of prospero RNA during Drosophila neuroblast cell divisions. Cell 90: 437–447. [DOI] [PubMed] [Google Scholar]
- Martin M., 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12. [Google Scholar]
- Micklem D. R., Adams J., Grünert S., St Johnston D., 2000. Distinct roles of two conserved Staufen domains in oskar mRNA localization and translation. EMBO J. 19: 1366–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolde M. J., Saka N., Reinert K. L., Slack F. J., 2007. The Caenorhabditis elegans pumilio homolog, puf-9, is required for the 3′UTR-mediated repression of the let-7 microRNA target gene, hbl-1. Dev. Biol. 305: 551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E., Maquat L. E., 2013. Staufen-mediated mRNA decay. Wiley Interdiscip. Rev. RNA 4: 423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelec D. M., Lachowiec J., Duchaine T. F., Smith H. E., Kennedy S., 2009. Requirement for the ERI/DICER complex in endogenous RNA interference and sperm development in Caenorhabditis elegans. Genetics 183: 1283–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peredo J., Villacé P., Ortín J., de Lucas S., 2014. Human Staufen1 associates to miRNAs involved in neuronal cell differentiation and is required for correct dendritic formation. PLoS One 9: e113704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B. J., Slack F. J., Basson M., Pasquinelli A. E., Bettinger J. C., et al. , 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906. [DOI] [PubMed] [Google Scholar]
- Ren H., Zhang H., 2010. Wnt signaling controls temporal identities of seam cells in Caenorhabditis elegans. Dev. Biol. 345: 144–155. [DOI] [PubMed] [Google Scholar]
- Ricci E. P., Kucukural A., Cenik C., Mercier B. C., Singh G., et al. , 2014. Staufen1 senses overall transcript secondary structure to regulate translation. Nat. Struct. Mol. Biol. 21: 26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K., 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roegiers F., Jan Y. N., 2000. Staufen: a common component of mRNA transport in oocytes and neurons? Trends Cell Biol. 10: 220–224. [DOI] [PubMed] [Google Scholar]
- Sarin S., O’Meara M. M., Flowers E. B., Antonio C., Poole R. J., et al. , 2007. Genetic screens for Caenorhabditis elegans mutants defective in left/right asymmetric neuronal fate specification. Genetics 176: 2109–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C. P., Jan L. Y., Jan Y. N., 1997. Miranda is required for the asymmetric localization of Prospero during mitosis in Drosophila. Cell 90: 449–458. [DOI] [PubMed] [Google Scholar]
- Shi Z., Hayes G., Ruvkun G., 2013. Dual regulation of the lin-14 target mRNA by the lin-4 miRNA. PLoS One 8: e75475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M., Seth M., Lee H.-C., Gu W., Ishidate T., et al. , 2012. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150: 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D., Beuchle D., Nüsslein-Volhard C., 1991. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell 66: 51–63. [DOI] [PubMed] [Google Scholar]
- Sterling C. H., Veksler-Lublinsky I., Ambros V., 2015. An efficient and sensitive method for preparing cDNA libraries from scarce biological samples. Nucleic Acids Res. 43: e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle, T., 2006 Maintenance of C. elegans (February 11, 2006), WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.101.1, http://www.wormbook.org.
- Sugimoto Y., Vigilante A., Darbo E., Zirra A., Militti C., et al. , 2015. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature 519: 491–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M. F., Abdul-Wajid S., Panduro M., Babiarz J. E., Rajaram M., et al. , 2012. Eri1 regulates microRNA homeostasis and mouse lymphocyte development and antiviral function. Blood 120: 130–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga K., Srikantan S., Lee E. K., Subaran S. S., Martindale J. L., et al. , 2011. Competitive regulation of nucleolin expression by HuR and miR-494. Mol. Cell. Biol. 31: 4219–4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadla B., Kemper K., Alaimo J., Heine C., Moss E. G., 2012. lin-28 controls the succession of cell fate choices via two distinct activities. PLoS Genet. 8: e1002588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan S. R., Daley G. Q., 2010. Lin28: A microRNA regulator with a macro role. Cell 140: 445–449. [DOI] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G., 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75: 855–862. [DOI] [PubMed] [Google Scholar]
- Young L. E., Moore A. E., Sokol L., Meisner-Kober N., Dixon D. A., 2012. The mRNA stability factor HuR inhibits microRNA-16 targeting of COX-2. Mol. Cancer Res. 10: 167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data files are available in the GEO database under the accession number GSE79217. Additional methods are described in File S1.