Abstract
The active form of vitamin D, 1,25-dihydroxyvitamin D3 (1,25D), plays an important immunomodulatory role, regulating transcription of genes in the innate and adaptive immune system. The present study examines patterns of transcriptome-wide response to 1,25D, and the bacterial lipopolysaccharide (LPS) in primary human monocytes, to elucidate pathways underlying the effects of 1,25D on the immune system. Monocytes obtained from healthy individuals of African-American and European-American ancestry were treated with 1,25D, LPS, or both, simultaneously. The addition of 1,25D during stimulation with LPS induced significant upregulation of genes in the antimicrobial and autophagy pathways, and downregulation of proinflammatory response genes compared to LPS treatment alone. A joint Bayesian analysis enabled clustering of genes into patterns of shared transcriptional response across treatments. The biological pathways enriched within these expression patterns highlighted several mechanisms through which 1,25D could exert its immunomodulatory role. Pathways such as mTOR signaling, EIF2 signaling, IL-8 signaling, and Tec Kinase signaling were enriched among genes with opposite transcriptional responses to 1,25D and LPS, respectively, highlighting the important roles of these pathways in mediating the immunomodulatory activity of 1,25D. Furthermore, a subset of genes with evidence of interethnic differences in transcriptional response was also identified, suggesting that in addition to the well-established interethnic variation in circulating levels of vitamin D, the intensity of transcriptional response to 1,25D and LPS also varies between ethnic groups. We propose that dysregulation of the pathways identified in this study could contribute to immune-mediated disease risk.
Keywords: differential expression profiling, innate immune cells, proinflammatory response, pathway analysis
Vitamin D plays an important immunomodulatory role through a transcriptional mechanism (Liu et al. 2006; Baeke et al. 2010; Aranow 2011; Hewison 2011). In the immune system, the active form of vitamin D, 1,25-dihydroxyvitamin D3 (1,25D), binds the vitamin D receptor (VDR), which translocates into the nucleus, where it modulates the transcription of genes with immune function, such as cathelicidin antimicrobial peptide (CAMP), defensin genes, such as β-defensin 4A (DEFB4A), and autophagy genes, such as autophagy related 5 (ATG5) (Liu et al. 2006, 2009; Adams et al. 2009; Yuk et al. 2009; Baeke et al. 2010; Aranow 2011; Hewison 2011). In monocytes/macrophages, 1,25D can be produced intracellularly from the inactive form, 25-hydroxyvitamin D3 (25D), which is found abundantly in circulation. The circulating levels of 25D vary greatly across individuals and ethnic groups (Rostand 2010; Looker et al. 2011; Murphy et al. 2012). Attesting to the important role of vitamin D in immune response, low levels of 25D have been linked to increased susceptibility to tuberculosis (Tb) (Nnoaham and Clarke 2008; White 2008). Moreover, 25D supplementation in individuals with hypovitaminosis D resulted in an enhanced antimicrobial response (Liu et al. 2006; Martineau et al. 2007; Adams et al. 2009). Although many studies have been conducted on the interindividual and interethnic variation in the circulating inactive 25D levels, with corresponding epidemiological links to immune-related diseases (Hypponen et al. 2001; Kamen et al. 2006; Kamen and Aranow 2008; Correale et al. 2009; Ascherio et al. 2010), little is known about interindividual and interethnic variation in the transcriptional response to active 1,25D.
Previous studies of 1,25D activity in immune cells highlight its complex immunomodulatory role, regulating activities such as enhancement of the response to Mycobacterium tuberculosis (M. tb) in THP-1 macrophage cell lines (Verway et al. 2013), downregulation of immune-related pathways such as interferon signaling in peripheral blood mononuclear cells (PBMCs) (Kupfer et al. 2013), and induction of a tolerogenic phenotype as well as an attenuation of the proinflammatory response in dendritic cells (van Halteren et al. 2004; Ferreira et al. 2012; Ferreira et al. 2015). Though the immunoregulatory role of 1,25D in different innate immune cell types is complex, it generally results in the attenuation of an intense proinflammatory response, which can have toxic consequences, such as sepsis and septic shock (Lehmann et al. 1987; Opal 2007; Zhang et al. 2012).
In this study, we focused on characterizing the transcriptional response to 1,25D in primary monocytes in the presence or absence of a proinflammatory stimulus, bacterial lipopolysaccharide (LPS). Stimulating monocytes with LPS enabled examination of how inflammation modifies the transcriptional response to 1,25D in monocytes. This analysis highlighted several biological pathways that are modulated by 1,25D in the absence of LPS (e.g., oxidative phosphorylation and mitochondrial dysfunction), as well as others that are modulated by LPS and reversed by 1,25D (e.g., proinflammatory cytokine signaling pathways). In addition, we identified some interethnic differential expression patterns, suggesting that the well-established interethnic variations in the vitamin D pathway extend to the intensity of transcriptional response to LPS and 1,25D.
Materials and Methods
Ethics statement
All donors to Research Blood Components (http://researchbloodcomponents.com/) and Sanguine Biosciences (https://www.sanguinebio.com/) sign an Institutional Review Board (IRB)-approved consent form giving permission to collect blood, and use it for research purposes. This study did not require IRB review at the University of Chicago because blood samples were not shipped with individually identifiable information.
Subjects
All subjects were healthy donors collected by Research Blood Components and Sanguine Biosciences. Self-reported ethnicity, age, gender, date, and time of blood drawing were recorded for each donor. Buffy coats from 10 African-American (AA) and 10 European-American (EA) subjects were shipped within 24 hr of collection. We processed samples in multiple batches, balanced by ethnic group. Serum samples from the donors were sent to the Clinical Chemistry Laboratory of the University of Chicago to determine 25-hydroxyvitamin D3 (25D) levels and parathyroid hormone (PTH) levels. Total serum 25D and PTH levels were determined using electrochemiluminescence detection assays (cat. no. 06506780160 and cat. no. 11972103160, respectively, Roche Diagnostics Corporation, Indianapolis, IN).
Monocyte culture and treatment
We isolated peripheral blood mononuclear cells (PBMCs) from the buffy coats of the 20 subjects by density gradient centrifugation using Ficoll-Paque PLUS medium (GE Healthcare Life Sciences, Pittsburgh, PA). We isolated monocytes from the PBMCs by positive selection using magnetic CD14 MicroBeads according to the supplier’s protocol (Miltenyi Biotec, San Diego, CA). We cultured isolated monocytes (1 × 106 cells/mL) in RPMI 1640 medium (Gibco, Life Technologies, Grand Island, NY), 25 mg/mL Gentamicin (Gibco), and 10% charcoal-stripped fetal bovine serum (Gibco) in 24-well plates. Monocytes were cultured in three replicates for 24 hr for each of the following treatments: 1) vehicle solution containing 1% ethanol and 99% culture medium, as a negative control; 2) 100 nM of 1,25D; 3) 10 ng/mL of LPS in the vehicle solution; and 4) 100 nM of 1,25D and 10 ng/mL of LPS (experimental design summarized in Supplemental Material, Figure S1). These four treatments are abbreviated as E, V, L, and V + L, respectively.
Transcriptome analysis
We pooled the three replicates for each treatment, and extracted total RNA from the pool using a Qiagen RNeasy Plus mini kit (Valencia, CA). We extracted RNA from 80 samples consisting of the 20 subjects that each received four treatments, in 10 batches each, balanced by ethnic group. RNA concentration and RNA integrity score (RIN) were recorded for each sample on a 2100 Bioanalyzer instrument (Agilent Technologies, Santa Clara, CA) (average RNA concentration and RIN scores in each ethnic group are summarized in Table S1). Total RNA was reverse transcribed into cDNA, labeled, hybridized to Illumina (San Diego, CA) Human HT-12 v3 Expression Beadchips, and scanned at the University of Chicago Functional Genomics Core facility. The microarrays were hybridized in three batches, and we recorded the array batch number for each sample to be used as a covariate in subsequent analyses. We performed low-level microarray analyses using the Bioconductor software package lumi (Du et al. 2008) in R, as previously described (Maranville et al. 2013). Briefly, we annotated probes by mapping their sequence to RefSeq (GRCh37) transcripts using BLAT. We discarded probes that mapped to multiple genes to avoid ambiguity in the source of a signal due to cross-hybridization of similar RNA molecules. We also discarded probes containing one or more HapMap SNPs to avoid spurious associations between expression measurements and ethnicity due to allele frequency differences between ethnic groups. We applied variance stabilization to all arrays, discarded poor quality probes, and quantile normalized the arrays using the default method implemented in the lumiN function. After these filters, probes mapping to 10,958 genes were used in downstream analyses (data available in File S1).
Differential expression analysis
We tested each gene for differential expression (DE) using a linear mixed-effects model with the R package, lme4 (Bates et al. 2015). The model included fixed effects for ancestry, and the three treatment conditions (V, L, and V + L), as well as interaction effects between ancestry and the treatments. It also included a random effect to model the differences between the individuals. Lastly, the model included covariates for the technical factors with the strongest effects on the expression data (P < 0.05), as determined by their association with the principal components described below, including array batch, age, baseline 25D levels, baseline PTH levels, RNA concentration, and RIN scores. P values were obtained using the R package, lmerTest, which provides a summary function with P values added for the t-test based on the Satterthwaite approximation for denominator degrees of freedom (Kuznetsova et al. 2015). To correct for multiple testing, we estimated the false discovery rate (FDR) using the “qvalue” function in R, based on the Storey method (Storey and Tibshirani 2003). The FDR for DE was set at 1%. To identify genes that were DE between the two ancestries, we tested the significance of the fixed interaction effects between ancestry and the treatments. Here, we used a more relaxed FDR threshold of 10% to determine significance, due to the smaller sample size in the interethnic comparison (10 AA and 10 EA).
We also performed a joint Bayesian analysis using the R package Cormotif (Wei et al. 2015), which jointly models expression data across different experiments, enabling classification of genes into patterns of shared and distinct differential expression. Genes are assigned to correlation motifs, which are the main patterns of differential expression obtained from the shared information across experiments, which in our study are treatments and ethnic groups. We regressed out the technical covariates described above from the expression data using the limma package removeBatchEffect (Ritchie et al. 2015), and used the residuals as input. We used a modified version of Cormotif as described in Blischak et al. (2015), where the original code was modified to return the cluster likelihood for each gene to enable downstream analyses. Also, since Cormotif is nondeterministic, we ran each test 100 times and kept the result with the largest maximum likelihood estimate.
Gene set enrichment analysis
We performed gene set enrichment analyses using the commercially available software Ingenuity Pathway Analysis (IPA). We compared DE genes with curated functional attribution lists organized by canonical pathway function. The magnitude of over-representation of a particular canonical pathway in the gene list from our study was calculated as the ratio of the number of genes from our data set that map to the pathway, divided by the total number of reference genes in that pathway in the IPA database. Statistical significance of the observed enrichment of a particular pathway was determined using Benjamini-Hochberg multiple testing corrected P values provided by IPA (Benjamini and Hochberg 1995).
Identifying vitamin D receptor binding sites near DE genes
We reanalyzed published data sets of VDR ChIP-seq, which used THP-1 monocytic cell lines treated with 1,25D and LPS, or 1,25D alone (Tuoresmaki et al. 2014), and FAIRE-seq, which used THP-1 cells treated with 1,25D (Seuter et al. 2013). First, we aligned sequence reads to the human reference (GRCh37) using BWA backtrack 0.7.5 (Li and Durbin 2009). Second, we kept only sequence reads with phred-scaled mapping quality ≥ 30 using samtools v1.1 (Li et al. 2009). Third, PCR duplicates were removed with Picard v 1.130 (http://broadinstitute.github.io/picard/). For the ChIP-seq data sets, we confirmed the quality of data sets by strand cross-correlation (SCC) analysis (Landt et al. 2012) implemented in the R script “run_spp_nodups.R” packaged in phantompeakqualtools (https://code.google.com/p/phantompeakqualtools/). Statistically significant peaks were identified using MACS version 2 (Zhang et al. 2008) with the following essential command line arguments: macs2 callpeak–bw X -g hs–qvalue = 0.05 -m 5 50, where X is a length of the bandwidth that was defined as a fragment length calculated by SCC for the ChIP-seq data, or as 200 bp for the FAIRE-seq data reported in Seuter et al. (2013).
To identify VDR response elements, we considered peaks that overlapped completely or partially between the ChIP-seq data after 1,25D and LPS treatment and the FAIRE-seq data. We then annotated them using HOMER (Heinz et al. 2010) to find the closest gene to each peak, and, among these genes, we selected those that were DE genes in response to 1,25D (V) and the combined 1,25D + LPS (V + L) treatments from the linear mixed-effects analysis. Enrichment of VDR response elements was determined using Fisher’s exact test, comparing peaks in DE genes to those in nonDE genes.
We also examined the enrichment of VDR binding sites among genes clustered in each expression pattern from the joint Bayesian analysis using Cormotif. To inclusively identify VDR binding sites, we merged the ChIP-seq data from THP-1 cells treated with 1,25D and LPS treatment with data from THP-1 cells treated with 1,25D alone. We examined overlap between the genes that were closest to the peaks, and the genes in each Cormotif pattern. Enrichment of VDR peaks was then determined using Fisher’s exact test, comparing VDR peaks in each Cormotif pattern with peaks in the “No response” Cormotif pattern.
Data availability
The raw microarray data files have been deposited in NCBI’s Gene Expression Omnibus (GEO) (Edgar et al. 2002), and are accessible through GEO Series accession number GSE78083 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE78083). The normalized expression values, the results from the linear mixed-effects model, and the results from Cormotif are provided in File S1.
Results
Sources of transcriptome-wide variation
To evaluate the variation in transcriptional response to 1,25D in the innate immune system in the presence and absence of an inflammatory stimulus, we cultured primary monocytes obtained from 10 African-American and 10 European-American healthy donors in four different conditions in parallel: i. EtOH (i.e., the vehicle control, or E); ii. LPS (L); iii. 1,25 D (V); and iv. 1,25D plus LPS (V + L); the experimental design is illustrated in Figure S1. Transcript levels were measured with gene expression arrays for each treatment condition and each individual, resulting in a total of 80 transcriptome data sets. Relevant covariates, including serum levels of 25D, were measured or recorded and used in downstream analyses (see Materials and Methods). Although there was significant interindividual and interethnic variation in serum 25D levels in our sample of donors (Table S1), this variable was not correlated with the transcriptional response to LPS, 1,25D, or their combination (Figure S5). This suggests that our in vitro system is not affected by 25D levels in vivo.
To evaluate the sources of variation in the overall transcriptome data, we performed principal components analysis (PCA) of the variance-stabilized log2-transformed expression data using the prcomp function in R. Principal component 1 (PC1) separates the samples by LPS treatment, accounting for 22% of the total variation in gene expression, and reflecting the large effect of LPS on the transcriptome (Figure S2, A and C, and Table S2), while PC2 separates the samples by 1,25D treatment, and accounts for 8.6% of the total variation in gene expression (Figure S2, A and D, and Table S2). PC3 and PC4, which account for 6.7% and 5.8% of variation respectively, separate the samples by the three array processing batches (Figure S2, B, E, and F, and Table S2). We also tested for associations between the PCs and the different covariates recorded for each sample (average sample covariates are compared between ancestries in Table S1). PC1 was associated with RNA concentration (P = 1.54 × 10−5, r2 = 0.219), PC2 was weakly associated with the RNA integrity number (RIN) scores (P = 9.0 × 10−3, r2 = 0.086), while PC3 was associated with age (P = 1.36 × 10−6, r2 = 0.266), baseline 25D levels (P = 1.0 × 10−3, r2 = 0.136), baseline PTH levels (P = 6.1 × 10−6, r2 = 0.237), and RIN score (P = 5.0 × 10−3, r2 = 0.099) (Table S2). The effects of array processing batch, RNA concentration, RIN scores, serum 25D, and PTH levels were subsequently included as covariates in the linear mixed-effects model for differential expression.
After regressing out the covariates using the limma package removeBatchEffect, and performing PCA on the residuals of the covariates-corrected expression data, we observed that PC1 and PC2 separated the samples by treatment (Figure S3, A, C, and D, and Table S3), but PC1 was still associated with RNA concentration (P = 3.56 × 10−5, r2 = 0.203) while PC2 was still associated with RIN score (P = 3 × 10−3, r2 = 0.113) (Table S3). PC3, which accounted for 5% of the total variation in gene expression, was associated with sample (P = 4.5 × 10−7, r2 = 0.286), and ancestry (P = 1.04 × 10−5, r2 = 0.227), highlighting the effect of interindividual and interethnic variation on gene expression (Figure S4 and Table S3).
Opposite effects of 1,25D and LPS on the transcriptome
Using the main effects for each treatment from the linear mixed-effects model, we identified genes that were DE in response to the different treatment conditions at a FDR of 1%. A total of 2888 genes were DE in response to 1,25D alone relative to vehicle (V vs. E). Gene set enrichment analysis identified metabolic processes, such as oxidative phosphorylation and the tricarboxylic acid (TCA) cycle, enriched among upregulated genes (Table S4). Pathways that play important roles in regulating translation processes, such as EIF2 signaling and mTOR signaling, were also significantly enriched among upregulated genes, indicating an important role of 1,25D in regulating translation. Immune responses involving chemokine signaling, B and T cell signaling, as well as various proinflammatory signaling cascades, such as Tec kinase signaling, phospholipase C signaling, and integrin signaling, were enriched among the downregulated genes, consistent with the immunomodulatory function of 1,25D.
There was a strong transcriptomic response to LPS treatment relative to vehicle (L vs. E), with 4461 genes DE at a FDR of 1%. Pathways enriched among LPS responsive genes highlight the opposite direction of transcriptional response to 1,25D and LPS, where proinflammatory immune response pathways were enriched among upregulated genes, while oxidative phosphorylation and translational control pathways were enriched among downregulated genes (Table S4), indicating the importance of these pathways in the proinflammatory effects induced by LPS stimulation.
Effects of combined 1,25D + LPS treatment on the transcriptome
The combined treatment of 1,25D + LPS resulted in 4720 genes significantly DE relative to vehicle (V + L vs. E). We also examined the transcriptional response of the combined 1,25D + LPS treatment relative to LPS (V + L vs. L) in an attempt to isolate the effect of 1,25D on the transcriptome in the presence of LPS, and identified 2404 genes significantly DE in this treatment category.
The pattern of response to V + L vs. E followed a similar pattern to LPS treatment alone (L vs. E), with similar pathways enriched among genes in these treatment categories (Table S4), probably because of the overwhelming transcriptional response to LPS. Genes significantly DE in response to V + L vs. L, which effectively subtracts the transcriptional effects of LPS, were similar to the genes significantly DE in response to 1,25D treatment alone (V vs. E), with similar pathways enriched.
We detected additional pathways enriched among genes significantly DE in response to the combined V + L treatments, both relative to vehicle and relative to LPS. These included adipogenesis and insulin receptor signaling pathways, both involved in lipid metabolic processes, which were enriched among upregulated genes. IL-4 signaling, which is associated with allergy and asthma through development of T-cell-mediated immune responses (Chatila 2004; LaPorte et al. 2008), was significantly enriched among downregulated genes (Table S4). Pathways enriched among genes responsive to the combined V + L treatment indicate a regulatory role of 1,25D in these pathways specifically in the context of LPS stimulation.
Bayesian analysis of shared transcriptional response across treatments and ethnic groups
To further dissect the effects of 1,25D and LPS on the transcriptome, we sought to identify the shared and distinct patterns of transcriptional response across treatments, and across ethnic groups. A popular approach to this question is to investigate the overlap of DE genes between conditions at a given FDR threshold. However, this approach fails to account for incomplete power to detect DE genes, thus exaggerating the differences in the transcriptional response between the conditions. In order to identify shared patterns of transcriptional response across treatments and ancestry while accounting for incomplete power, we implemented a joint Bayesian analysis with the R/Bioconductor package Cormotif (Wei et al. 2015). Genes were classified into different response patterns, or correlation motifs, across treatments and ancestry (Figure 1). Since Cormotif does not distinguish the direction of effect across treatments, we used the results of the linear mixed-effects model in conjunction with the Cormotif approach to establish direction of response in the different response patterns (Figure 2).
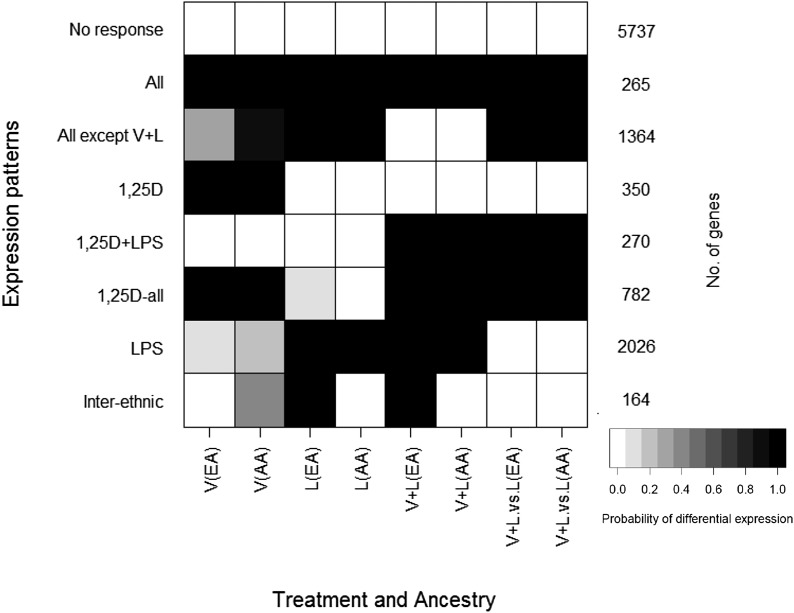
Figure 1.
Transcriptional response patterns shared across the different treatments and ancestries identified by implementing a joint Bayesian analysis using Cormotif. The shading of each box represents the posterior probability that a gene assigned to a given expression pattern (rows) is differentially expressed in individuals from a particular ancestry in response to each treatment (columns). V, response to 1,25D, relative to vehicle; L, response to LPS relative to vehicle; V + L, response to 1,25D + LPS relative to vehicle; V + L.vs.L, response to 1,25D+LPS relative to LPS; EA, European-American; AA, African-American.
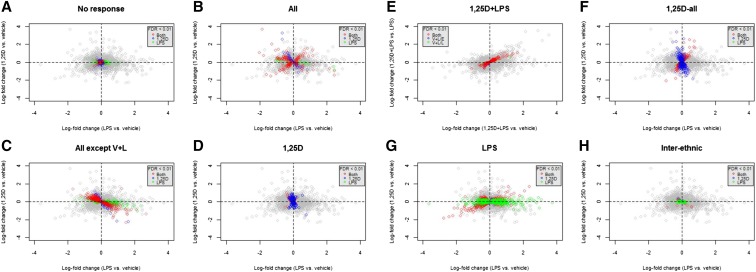
Figure 2.
Direction of response in the different correlation motifs. Patterns of differential response to single treatment with 1,25D (vertical axis) or LPS (horizontal axis) for each correlation motif are shown in (A)–(D) and (F)–(H). (E) Patterns of differential response to the combined treatment with 1,25D and LPS relative to LPS (vertical axis), and 1,25D and LPS relative to vehicle (horizontal axis). Genes are color coded based on q-values < 0.01 from linear mixed-effects analysis as follows: red, DE in response to both 1,25D and LPS; blue, DE in response to 1,25D; green, DE in response to LPS; gray, not DE.
A total of 5737 genes were classified in the “No response” pattern, which includes genes whose expression levels were unchanged across all the treatments (Figure 1 and Figure 2A). This is broadly consistent with the results of the linear mixed-effects model, with 80% of these genes being also classified as nonDE for any treatment in the linear mixed-effects model.
Genes that responded to all the treatments were classified in the “All” pattern, and included 265 genes whose expression levels changed across all treatments and ancestries (Figure 1 and Figure 2B). Genes classified in this Cormotif had response patterns to 1,25D and LPS that were both concordant (i.e., up or downregulated in both treatments) and discordant (i.e., upregulated in one treatment and downregulated in the other). Genes that were upregulated in all treatments (top-right quadrant, Figure 2B) included CD14, which encodes a surface antigen expressed on monocytes that is involved in mediating response to bacterial LPS. Genes that were downregulated in all treatments included chemokine signaling genes such as CCL13 (bottom-left quadrant, Figure 2B). The discordant response patterns included genes that were upregulated by 1,25D and downregulated by LPS (top-left quadrant, Figure 2B), with EIF2 signaling and mTOR signaling pathways significantly enriched among these genes (Figure 3). This is consistent with the opposite transcriptional effects of 1,25D and LPS on genes in these pathways that were highlighted in the linear mixed-effects analysis. Genes that were downregulated by 1,25D and upregulated by LPS (bottom-right quadrant, Figure 2B) included some cytokine receptor genes such as IL7R and IL2RA, which are important components of the proinflammatory signaling cascade.
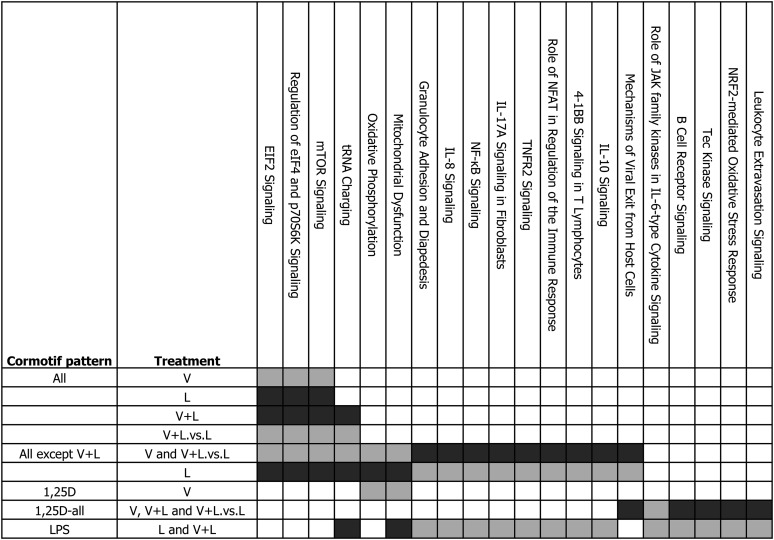
Figure 3.
Sharing of enriched biological pathways across Cormotifs. The table shows the biological pathways that were enriched (FDR < 0.05) in more than one Cormotif subdivided based on the direction of transcriptional response (upregulated genes in light gray, and downregulated genes in dark gray), and the treatment (V, response to 1,25D, relative to vehicle; L, response to LPS relative to vehicle; V + L, response to 1,25D + LPS relative to vehicle; V + L.vs.L, response to 1,25D + LPS relative to LPS).
The “All except V + L” pattern included 1364 genes whose expression levels changed in all treatments, except the combined 1,25D + LPS relative to vehicle (V + L vs. E). All the genes in this Cormotif pattern were discordant in their response to 1,25D and LPS, resulting in a neutral effect in the response to the combined V + L vs. E (Figure 1 and Figure 2C). Genes that were responsive to the combined V + L vs. L followed a similar direction of response to the genes DE in response to the individual V vs. E treatment, suggesting that the response to 1,25D at these genes is not dramatically influenced by LPS. The genes that were upregulated by 1,25D and downregulated by LPS (top-left quadrant, Figure 2C) were enriched for EIF2 and mTOR signaling pathways, similar to the discordant genes in the “All” category (Figure 3). In addition, oxidative phosphorylation and mitochondrial dysfunction pathways were significantly enriched among these genes. On the other hand, genes that were downregulated by 1,25D and upregulated by LPS (bottom-right quadrant, Figure 2C) were enriched for various proinflammatory response pathways, including granulocyte adhesion and diapedesis, IL-8 signaling, NF-kB signaling, TNFR2 signaling, and Role of NFAT in regulation of the immune response (Figure 3).
Genes responsive to 1,25D were divided into three Cormotif patterns: “1,25D”, “1,25D + LPS”, and “1,25D-all”. The “1,25D” pattern included 350 genes that were DE in response to V vs. E alone (Figure 1 and Figure 2D). Oxidative phosphorylation and mitochondrial dysfunction pathways were significantly enriched among upregulated genes (Figure 3). Interestingly, the oxidative phosphorylation pathway genes enriched in this Cormotif pattern responded similarly to the genes in the same pathway classified in the “All except V + L” Cormotif pattern, in that they are significantly induced by 1,25D. However, the oxidative phosphorylation pathway genes in the “1,25D” pattern respond exclusively to 1,25D, while those in the “All except V + L” Cormotif pattern are upregulated by 1,25D, and downregulated by LPS (Figure 3 and Figure S6). This indicates a context-specific response profile among genes in the same pathway, where some genes in the oxidative phosphorylation pathway are uniquely regulated by 1,25D, whereas other genes in the same pathway are regulated by both 1,25D and LPS.
The “1,25D+LPS” pattern included 270 genes that responded to 1,25D only in the presence of LPS. This pattern captured genes that were DE in response to the combined V + L vs. E, and V + L vs. L (Figure 1 and Figure 2E). Although there were no enriched pathways among genes in this Cormotif pattern at an FDR of 5%, some interesting pathways, such as eNOS signaling and cholesterol biosynthesis pathway, were represented among the downregulated genes at a FDR of 27%, suggesting a role for 1,25D in modulating these pathways upon LPS stimulation.
The “1,25D-all” pattern included 782 genes that were DE in response to 1,25D in the both the presence and absence of LPS (Figure 1 and Figure 2F). Genes in the antimicrobial pathway were included in this category, such as the antibacterial peptide gene CAMP, autophagy genes ATG3, ATG5, ATG2A, and ATG9A, and the intracellular pattern recognition receptor gene NOD2. These genes were significantly upregulated in response to 1,25D, alone or in combination with LPS. The role of JAK family kinases in IL-6-type cytokine signaling was the most significantly enriched pathway among the upregulated genes (Figure 3), and included genes such as MAPK14, PTPN11, and STAT5B, all of which could be crucial for triggering antimicrobial responses in monocytes. Biological pathways enriched among downregulated genes in this category included B cell receptor signaling, Tec kinase signaling and leukocyte extravasation signaling (Figure 3 and Figure S7), highlighting the role of 1,25D in repressing proinflammatory response pathways. Interestingly, immunological and inflammatory diseases were among the most enriched disease categories from the IPA analysis among the downregulated genes (Table S6 and Figure S7), suggesting a protective role of 1,25D in immunological diseases. Overall, the “1,25D-all” response pattern illustrates the important dual immunomodulatory role played by 1,25D in monocytes, where antimicrobial pathway genes are upregulated, while proinflammatory pathway genes associated with immunological and inflammatory disease are downregulated by 1,25D in the presence or absence of LPS stimulation.
The “LPS” pattern included 1400 genes whose expression levels changed in response to L vs. E and the combined V + L vs. E (Figure 1 and Figure 2G). Consistent with the results from the linear mixed-effects model, proinflammatory pathways were significantly enriched among the upregulated genes in this category, including IL-8 signaling, NF-kB signaling, IL-17 signaling, and TNFR2 signaling, among others (Figure 3). Among the downregulated genes, tRNA charging, mitochondrial dysfunction, the TCA cycle II, galactose metabolism pathway, and folate transformation pathway were significantly enriched (Figure 3 and Table S5), indicating that LPS modulates transcription of genes in these metabolic pathways.
Genes with interethnic differential response
The “interethnic” pattern was of particular interest, as it identified 164 genes with evidence of differential responses to LPS treatments between AA and EA subjects, with a stronger response in EA compared to AA (Figure 1 and Figure 2H).
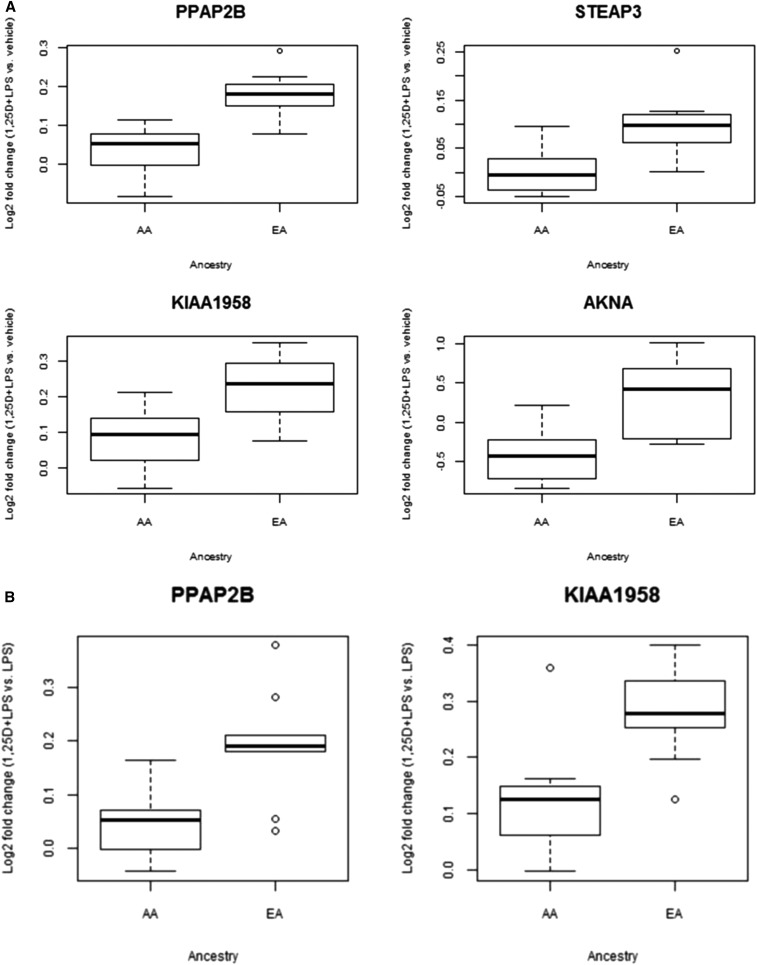
We also interrogated the degree of interethnic differences in transcriptional response using the main interaction term for treatment and ancestry in the linear mixed-effects model. We identified 15 genes with strong interethnic differences in response to V + L vs. E at a FDR < 10%. These genes include PPAP2B, which encodes a member of the phosphatidic acid phosphatase (PAP) family and has been implicated in coronary artery disease risk (Schunkert et al. 2011; Dichgans et al. 2014); STEAP3, which encodes an endosomal ferrireductase required for efficient transferrin-dependent iron uptake; and AKNA, which encodes a transcription factor that specifically activates the expression of the CD40 receptor and its ligand CD40L/CD154 on lymphocyte cell surfaces, which are critical for antigen-dependent-B-cell development (Figure 4A). Interestingly, 13 out of the 15 genes showed more significant differential responses in EA (Figure S8), similar to the pattern observed in the “interethnic” Cormotif pattern.
Figure 4.
Interethnic differential response patterns. Genes with interethnic differences in transcriptional responses to 1,25D + LPS relative to vehicle (A), and relative to LPS (B) were identified using the interaction term for treatment and ancestry in the linear mixed-effects model (FDR < 0.10). The boxplots show examples of these genes with different log-fold change in transcript levels between the two ethnic groups. AA, African-American; EA, European-American.
To account for the effect of LPS, and examine the extent to which the interethnic differential response patterns were modulated by 1,25D, we examined interethnic differential response to the combined V + L vs. L. PPAP2B and KIAA1958 were the only statistically significant genes identified in this category (P = 2.96 × 10−6 and 9.62 × 10−6, respectively), with both of these genes more significantly differentially expressed in EA (Figure 4B).
Regulatory elements near DE genes
We examined the overlap between genes DE in response to 1,25D and the combined 1,25D + LPS treatments in our primary monocytes, and published datasets for VDR ChIP-seq (Tuoresmaki et al. 2014) and FAIRE-seq (Seuter et al. 2013) performed in THP-1 monocytic cells, to examine whether there was enrichment of open chromatin regions and VDR binding sites near the transcription start sites of DE genes. We found a significant enrichment of VDR binding sites among genes DE in response to V vs. E (P = 4.56 × 10−11), V + L vs. E (P = 3.97 × 10−8), and V + L vs. L (P = 1.54 × 10−7) (Table S7). There was an overlap of 201 genes between the DE genes, VDR ChIP-seq, and FAIRE-seq datasets, highlighting genes such as CAMP and CD14, which contain open chromatin regions and VDR binding sites near the transcription start site; these 201 genes are potentially direct VDR targets.
In addition, we examined the enrichment of VDR binding sites across the different Cormotif patterns (Table 1). The genes in the “1,25D-all” and “All” Cormotif patterns had the highest enrichment of VDR binding sites (P = 6.88 × 10−13 and 2.57 × 10−8, respectively), indicating a higher proportion of potentially direct VDR targets represented in these Cormotif patterns. Genes in the “1,25D” Cormotif pattern were not significantly enriched for VDR binding sites, suggesting that the presence of LPS, in addition to 1,25D, is important to enable the 1,25D-VDR transcriptional activity in primary monocytes.
Table 1. Proportion of genes in each Cormotif pattern containing vitamin D receptor (VDR) binding sites.
| Cormotif Pattern | Total No. Genes | No. Genes with VDR Binding Site | Proportion of Genes with VDR Binding Site | Enrichment P Value |
|---|---|---|---|---|
| No response | 5737 | 186 | 0.03 | — |
| All | 265 | 31 | 0.12 | 2.57 × 10−8 |
| All except V + L | 1364 | 65 | 0.05 | 0.01 |
| 1,25D | 350 | 17 | 0.05 | 0.13 |
| 1,25D + LPS | 270 | 23 | 0.09 | 1.49 × 10−4 |
| 1,25D-all | 782 | 76 | 0.10 | 6.88 × 10−13 |
| LPS | 2026 | 96 | 0.05 | 3.79 × 10−3 |
| Interethnic | 164 | 8 | 0.05 | 0.27 |
Enrichment of VDR peaks in each category was calculated using Fisher’s exact test, comparing genes in each Cormotif pattern to those in the “No response” Cormotif pattern.
Discussion
We used a transcriptomic approach to characterize the immunomodulatory role of 1,25D in the presence of a proinflammatory stimulus to identify the mechanisms through which 1,25D exerts its immunomodulatory role. We analyzed differential expression patterns using both a linear mixed-effects analysis, which modeled individual treatment comparisons, and a Bayesian analysis using the Cormotif method, which jointly modeled differential expression across all treatments and ethnic groups, thereby accounting for incomplete power. A similar joint Bayesian framework has been successfully applied to expression quantitative trait loci (eQTL) mapping to distinguish between shared and context-specific eQTLs (Maranville et al. 2011; Flutre et al. 2013). Our joint Bayesian analysis enabled clustering of DE genes into distinct transcriptional response patterns, with pathways enriched within these transcriptional patterns highlighting mechanisms that mediate the immunomodulatory role of 1,25D.
Metabolic pathways involving oxidative phosphorylation were enriched among upregulated genes in the “All except V+L” and “1,25D” Cormotif patterns (Figure 3). We highlight context-specific response pattern of genes within this pathway, where some genes were uniquely induced by 1,25D, while genes in other parts of the pathway were regulated by both 1,25D and LPS. The crucial role played by 1,25D in regulating oxidative phosphorylation was previously reported in PBMCs and dendritic cells (Ferreira et al. 2012, 2015; Kupfer et al. 2013), and this regulation of metabolic reprogramming by 1,25D is thought to be crucial for controlling function, growth, proliferation, and survival of various immune cell subsets (Williams et al. 2013; Kelly and O’Neill 2015). The fact that LPS downregulated genes in the oxidative phosphorylation pathway confirms previous reports indicating that LPS induces a metabolic shift away from oxidative phosphorylation to anaerobic glycolysis in macrophages and dendritic cells to enable ATP production (Kelly and O’Neill 2015). This effect is similar to the Warbug effect in tumor cells, whose high energy demand is met by switching the metabolic profile away from the TCA cycle and the oxidative phosphorylation pathway toward glycolysis, thereby enabling rapid ATP production (Warburg et al. 1927; Kelly and O’Neill 2015). Previous work done in mouse macrophages and dendritic cells (Krawczyk et al. 2010; Freemerman et al. 2014) indicated that a metabolic shift toward glycolysis mediated the proinflammatory response, and this proinflammatory response could be attenuated by pharmacologic inhibition of glycolysis. From our study, this subset of oxidative phosphorylation pathway genes that were downregulated by LPS, were then upregulated by addition of 1,25D in combination with LPS (Figure 3). Therefore, oxidative phosphorylation could be one of the mechanisms through which 1,25D attenuates the proinflammatory response induced by LPS in monocytes, and the subset of genes in this pathway that we identified which were modulated by both LPS and 1,25D could be central to this mechanism.
The mTOR signaling pathway was consistently enriched among genes that were upregulated by 1,25D and downregulated by LPS in the “All” and “All except V + L” patterns. mTOR signaling was previously implicated in inhibition of proinflammatory response in LPS-stimulated monocytes/macrophages and dendritic cells, as well as in the maintenance of a tolerogenic phenotype in dendritic cells (Ohtani et al. 2008; Weichhart et al. 2008; Wang et al. 2011; Ferreira et al. 2015). Inhibition of mTOR resulted in increased proinflammatory cytokine production by LPS-stimulated monocytes/macrophages and dendritic cells (Ohtani et al. 2008; Weichhart et al. 2008), and increased T cell proliferation (Ohtani et al. 2008; Ferreira et al. 2015), implicating a role of mTOR in regulating the proinflammatory response. The genes in this pathway were significantly downregulated by LPS; however, this direction of response was reversed by addition of 1,25D in combination with LPS (Figure 3), implying that 1,25D attenuates the proinflammatory response by upregulating mTOR signaling. In addition, the genes in this pathway play important roles in regulating translation initiation, and include the ribosomal protein gene RPS27, and the eukaryotic translation initiation factor gene EIF2A, which encodes the eukaryotic initiation factor 2 (eIF-2α), which has been shown to be a downstream target of the vitamin D receptor (Wu et al. 2010). Therefore, regulation of translation initiation through targeting the mTOR signaling pathway could be a novel mechanism for the attenuation of the proinflammatory response mediated by 1,25D in monocytes.
Furthermore, the EIF2 signaling pathway was also enriched among genes upregulated by 1,25D and downregulated by LPS in the “All” and “All except V + L” patterns, and this result is consistent with the individual treatment DE analysis using the linear mixed-effects model (Table 1, Table S4, and Table S5). EIF2 signaling plays an important role in regulating translation initiation in response to stress, and was implicated in regulating proinflammatory cytokine production and bacterial invasion in mouse embryonic fibroblast cells (MEFs) (Shrestha et al. 2012). Shrestha et al. (2012) reported that the Yersinia-encoded virulence factor, YopJ, inhibited EIF2 signaling in MEFs. Similarly, in our study, LPS consistently downregulated genes in the EIF2 signaling pathway, in a mechanism that might be similar to that triggered by YopJ. In addition, Shrestha et al. (2012) observed that mutant MEFs with defective EIF2 signaling that were infected with different bacterial pathogens experienced enhanced cytotoxicity compared to wild type, due to increased bacterial invasion, indicating a direct role for EIF2 signaling in the antimicrobial response; 1,25D could hence exert its antimicrobial role in monocytes by upregulating genes in the EIF2 signaling pathway.
The dual immunomodulatory role of 1,25D was also highlighted by the genes clustered in the “1,25D-all” pattern. While 1,25D broadly downregulated genes in the proinflammatory cytokine and signaling cascade pathways, it also played a crucial role in inducing important antimicrobial and autophagy genes in this Cormotif pattern. The most significantly enriched biological pathway among the upregulated genes was the role of JAK family kinases in IL-6-type cytokine signaling, which contained genes such as STAT5B, which regulates signaling in diverse biological processes. Previous reports indicate that the TLR2/1-mediated induction of the vitamin D-dependent antimicrobial pathway requires IL-15 activity (Krutzik et al. 2008), which could be mediated via STAT5 activation, which has been shown to be important for IL-15 signaling (Musikacharoen et al. 2001; Pandiyan et al. 2012). Hence, 1,25D could regulate genes in this pathway to trigger antimicrobial responses in monocytes.
By profiling transcriptional response in monocytes from individuals of African-American and European-American ancestries, we identified some patterns of interethnic variation in response to LPS, and the combined 1,25D + LPS treatment in both the linear mixed-effects analysis and the joint Bayesian analysis, while correcting for interindividual variation in baseline levels of circulating 25D. This raises the intriguing possibility that interethnic variation in the vitamin D pathway is not limited to the well-established differences in circulating levels of 25D (Rostand 2010; Looker et al. 2011; Murphy et al. 2012), but it may extend to the intensity of the transcriptional response to LPS and 1,25D. Interestingly, most of the genes with interethnic differential response showed more significant differential responses in EA. The fact that most of the interethnic transcriptional differences were detected in the response to LPS or to the combined 1,25D + LPS, both relative to vehicle, suggests that these two ethnic groups differ in the proinflammatory transcriptional response. However, two genes had significant interethnic differences in transcriptional response to the combined 1,25D + LPS relative to LPS (Figure 4B), suggesting that they differ more specifically in their response to vitamin D.
We further identified enrichment of VDR ChIP-seq and FAIRE-seq peaks among genes DE in response to the combined 1,25D + LPS treatments. This enrichment was particularly strong for genes in the “1,25D-all” and “All” Cormotif patterns, suggesting that a substantial proportion of these genes are under direct regulation of the 1,25D-VDR transcription factor complex. Intriguingly, we did not detect an enrichment of VDR binding sites near genes in the “1,25D” Cormotif. Different explanations could account for this observation. One is that the combination of both 1,25D and LPS is important for stimulating the transcriptional activity of the 1,25D-VDR transcriptional complex in human monocytes (Liu et al. 2006; Adams et al. 2009; Tuoresmaki et al. 2014). On the other hand, because the genes in the “1,25D” Cormotif are observed to respond only to one treatment condition, it is possible that they are enriched for false positives relative to genes in other Cormotifs that are found to respond to multiple treatment conditions. Another caveat to this analysis is that we examined the overlap of VDR ChIP-seq peaks from published data sets with experimental conditions that were different to ours. While we treated primary monocytes with 100 nM 1,25D and 10 ng/mL LPS for 24 hr, the VDR ChIP-seq data were obtained from THP-1 monocytic cell lines cultured with 100 ng/mL LPS for 24 hr, and then treated with 10 nM 1,25D for 80 min (Tuoresmaki et al. 2014). Future VDR ChIP-seq studies with uniform experimental conditions in primary monocytes will enable better characterization of the regulatory architecture of 1,25D response genes.
Overall, through transcriptomic profiling, our study characterizes the dual immunomodulatory role of 1,25D in primary human monocytes, highlighting the importance of biological pathways such as mTOR signaling and EIF2 signaling in mediating this immunomodulatory role. The pathways highlighted in this study may provide mechanistic clues for the observed associations between insufficient levels of circulating serum 25D and increased disease risk. The interindividual and interethnic variation in intracellular transcriptional response to 1,25D has not been previously characterized, and could serve as an additional contribution to disease risk.
Supplementary Material
Acknowledgments
The authors are grateful to Carole Ober, Marcelo Nobrega, and John Novembre for advice on analysis methods, and Sonia Kupfer, Choongwon Jeong, and the entire Di Rienzo lab for helpful discussions about the project. This work was supported in part by National Institutes of Health (NIH) grant (R01 GM101682) to A.D. S.N.K. was supported by the Howard Hughes Medical Institute Gilliam Fellowship for Advanced Study. J.D.B. was supported by NIH grant AI087658 awarded to Yoav Gilad. This project was supported by the University of Chicago Comprehensive Cancer Center Support Grant (#P30 CA14599), with particular support from the Genomics Core Facility.
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.028712/-/DC1
Communicating editor: J. C. Fay
Literature Cited
- Adams J. S., Ren S., Liu P. T., Chun R. F., Lagishetty V., et al. , 2009. Vitamin D-directed rheostatic regulation of monocyte antibacterial responses. J. Immunol. 182: 4289–4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranow C., 2011. Vitamin D and the immune system. J. Investig. Med. 59: 881–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascherio A., Munger K. L., Simon K. C., 2010. Vitamin D and multiple sclerosis. Lancet Neurol. 9: 599–612. [DOI] [PubMed] [Google Scholar]
- Baeke F., Takiishi T., Korf H., Gysemans C., Mathieu C., 2010. Vitamin D: modulator of the immune system. Curr. Opin. Pharmacol. 10: 482–496. [DOI] [PubMed] [Google Scholar]
- Bates D., Mächler M., Bolker B., Walker S., 2015. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67: 1–48. [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Blischak J. D., Tailleux L., Mitrano A., Barreiro L. B., Gilad Y., 2015. Mycobacterial infection induces a specific human innate immune response. Sci. Rep. 5: 16882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatila T. A., 2004. Interleukin-4 receptor signaling pathways in asthma pathogenesis. Trends Mol. Med. 10: 493–499. [DOI] [PubMed] [Google Scholar]
- Correale J., Ysrraelit M. C., Gaitan M. I., 2009. Immunomodulatory effects of vitamin D in multiple sclerosis. Brain 132: 1146–1160. [DOI] [PubMed] [Google Scholar]
- Dichgans M., Malik R., Konig I. R., Rosand J., Clarke R., et al. , 2014. Shared genetic susceptibility to ischemic stroke and coronary artery disease: a genome-wide analysis of common variants. Stroke 45: 24–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P., Kibbe W. A., Lin S. M., 2008. lumi: a pipeline for processing Illumina microarray. Bioinformatics 24: 1547–1548. [DOI] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A. E., 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira G. B., Kleijwegt F. S., Waelkens E., Lage K., Nikolic T., et al. , 2012. Differential protein pathways in 1,25-dihydroxyvitamin d(3) and dexamethasone modulated tolerogenic human dendritic cells. J. Proteome Res. 11: 941–971. [DOI] [PubMed] [Google Scholar]
- Ferreira G. B., Vanherwegen A. S., Eelen G., Gutierrez A. C., Van Lommel L., et al. , 2015. Vitamin D3 induces tolerance in human dendritic cells by activation of intracellular metabolic pathways. Cell Rep. 10: 711–725. [DOI] [PubMed] [Google Scholar]
- Flutre T., Wen X., Pritchard J., Stephens M., 2013. A statistical framework for joint eQTL analysis in multiple tissues. PLoS Genet. 9: e1003486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freemerman A. J., Johnson A. R., Sacks G. N., Milner J. J., Kirk E. L., et al. , 2014. Metabolic reprogramming of macrophages: glucose transporter 1 (GLUT1)-mediated glucose metabolism drives a proinflammatory phenotype. J. Biol. Chem. 289: 7884–7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., et al. , 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewison M., 2011. Antibacterial effects of vitamin D. Nat. Rev. Endocrinol. 7: 337–345. [DOI] [PubMed] [Google Scholar]
- Hypponen E., Laara E., Reunanen A., Jarvelin M. R., Virtanen S. M., 2001. Intake of vitamin D and risk of type 1 diabetes: a birth-cohort study. Lancet 358: 1500–1503. [DOI] [PubMed] [Google Scholar]
- Kamen D., Aranow C., 2008. Vitamin D in systemic lupus erythematosus. Curr. Opin. Rheumatol. 20: 532–537. [DOI] [PubMed] [Google Scholar]
- Kamen D. L., Cooper G. S., Bouali H., Shaftman S. R., Hollis B. W., et al. , 2006. Vitamin D deficiency in systemic lupus erythematosus. Autoimmun. Rev. 5: 114–117. [DOI] [PubMed] [Google Scholar]
- Kelly B., O’Neill L. A., 2015. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 25: 771–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk C. M., Holowka T., Sun J., Blagih J., Amiel E., et al. , 2010. Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood 115: 4742–4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzik S. R., Hewison M., Liu P. T., Robles J. A., Stenger S., et al. , 2008. IL-15 links TLR2/1-induced macrophage differentiation to the vitamin D-dependent antimicrobial pathway. J. Immunol. 181: 7115–7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupfer S. S., Maranville J. C., Baxter S. S., Huang Y., Di Rienzo A., 2013. Comparison of cellular and transcriptional responses to 1,25-dihydroxyvitamin d3 and glucocorticoids in peripheral blood mononuclear cells. PLoS One 8: e76643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova, A., P. B. Brockhoff, and R. H. B. Christensen, 2015 lmerTest: tests in linear mixed effects models. R package version 2.0–25. Available at: http://CRAN.R-project.org/package=lmerTest. Accessed: October 2, 2015.
- Landt S. G., Marinov G. K., Kundaje A., Kheradpour P., Pauli F., et al. , 2012. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22: 1813–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPorte S. L., Juo Z. S., Vaclavikova J., Colf L. A., Qi X., et al. , 2008. Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell 132: 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann V., Freudenberg M. A., Galanos C., 1987. Lethal toxicity of lipopolysaccharide and tumor necrosis factor in normal and d-galactosamine-treated mice. J. Exp. Med. 165: 657–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P. T., Stenger S., Li H., Wenzel L., Tan B. H., et al. , 2006. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science 311: 1770–1773. [DOI] [PubMed] [Google Scholar]
- Liu P. T., Schenk M., Walker V. P., Dempsey P. W., Kanchanapoomi M., et al. , 2009. Convergence of IL-1beta and VDR activation pathways in human TLR2/1-induced antimicrobial responses. PLoS One 4: e5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looker A. C., Johnson C. L., Lacher D. A., Pfeiffer C. M., Schleicher R. L., et al. , 2011. Vitamin D status: United States, 2001–2006. NCHS Data Brief 59: 1–8. [PubMed] [Google Scholar]
- Maranville J. C., Luca F., Richards A. L., Wen X., Witonsky D. B., et al. , 2011. Interactions between glucocorticoid treatment and cis-regulatory polymorphisms contribute to cellular response phenotypes. PLoS Genet. 7: e1002162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maranville J. C., Baxter S. S., Torres J. M., Di Rienzo A., 2013. Inter-ethnic differences in lymphocyte sensitivity to glucocorticoids reflect variation in transcriptional response. Pharmacogenomics J. 13: 121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martineau A. R., Wilkinson R. J., Wilkinson K. A., Newton S. M., Kampmann B., et al. , 2007. A single dose of vitamin D enhances immunity to mycobacteria. Am. J. Respir. Crit. Care Med. 176: 208–213. [DOI] [PubMed] [Google Scholar]
- Murphy A. B., Kelley B., Nyame Y. A., Martin I. K., Smith D. J., et al. , 2012. Predictors of serum vitamin D levels in African American and European American men in Chicago. Am. J. Men Health 6: 420–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musikacharoen T., Matsuguchi T., Kikuchi T., Yoshikai Y., 2001. NF-kappa B and STAT5 play important roles in the regulation of mouse Toll-like receptor 2 gene expression. J. Immunol. 166: 4516–4524. [DOI] [PubMed] [Google Scholar]
- Nnoaham K. E., Clarke A., 2008. Low serum vitamin D levels and tuberculosis: a systematic review and meta-analysis. Int. J. Epidemiol. 37: 113–119. [DOI] [PubMed] [Google Scholar]
- Ohtani M., Nagai S., Kondo S., Mizuno S., Nakamura K., et al. , 2008. Mammalian target of rapamycin and glycogen synthase kinase 3 differentially regulate lipopolysaccharide-induced interleukin-12 production in dendritic cells. Blood 112: 635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opal S. M., 2007. The host response to endotoxin, antilipopolysaccharide strategies, and the management of severe sepsis. Int. J. Med. Microbiol. 297: 365–377. [DOI] [PubMed] [Google Scholar]
- Pandiyan P., Yang X. P., Saravanamuthu S. S., Zheng L., Ishihara S., et al. , 2012. The role of IL-15 in activating STAT5 and fine-tuning IL-17A production in CD4 T lymphocytes. J. Immunol. 189: 4237–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., et al. , 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostand S. G., 2010. Vitamin D, blood pressure, and African Americans: toward a unifying hypothesis. Clin. J. Am. Soc. Nephrol. 5: 1697–1703. [DOI] [PubMed] [Google Scholar]
- Schunkert H., Konig I. R., Kathiresan S., Reilly M. P., Assimes T. L., et al. , 2011. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 43: 333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seuter S., Heikkinen S., Carlberg C., 2013. Chromatin acetylation at transcription start sites and vitamin D receptor binding regions relates to effects of 1alpha,25-dihydroxyvitamin D3 and histone deacetylase inhibitors on gene expression. Nucleic Acids Res. 41: 110–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha N., Bahnan W., Wiley D. J., Barber G., Fields K. A., et al. , 2012. Eukaryotic initiation factor 2 (eIF2) signaling regulates proinflammatory cytokine expression and bacterial invasion. J. Biol. Chem. 287: 28738–28744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J. D., Tibshirani R., 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuoresmaki P., Vaisanen S., Neme A., Heikkinen S., Carlberg C., 2014. Patterns of genome-wide VDR locations. PLoS One 9: e96105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Halteren A. G., Tysma O. M., van Etten E., Mathieu C., Roep B. O., 2004. 1alpha,25-dihydroxyvitamin D3 or analogue treated dendritic cells modulate human autoreactive T cells via the selective induction of apoptosis. J. Autoimmun. 23: 233–239. [DOI] [PubMed] [Google Scholar]
- Verway M., Bouttier M., Wang T. T., Carrier M., Calderon M., et al. , 2013. Vitamin D induces interleukin-1beta expression: paracrine macrophage epithelial signaling controls M. tuberculosis infection. PLoS Pathog. 9: e1003407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Brown J., Gu Z., Garcia C. A., Liang R., et al. , 2011. Convergence of the mammalian target of rapamycin complex 1- and glycogen synthase kinase 3-beta-signaling pathways regulates the innate inflammatory response. J. Immunol. 186: 5217–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O., Wind F., Negelein E., 1927. The metabolism of tumors in the body. J. Gen. Physiol. 8: 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Tenzen T., Ji H., 2015. Joint analysis of differential gene expression in multiple studies using correlation motifs. Biostatistics 16: 31–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichhart T., Costantino G., Poglitsch M., Rosner M., Zeyda M., et al. , 2008. The TSC-mTOR signaling pathway regulates the innate inflammatory response. Immunity 29: 565–577. [DOI] [PubMed] [Google Scholar]
- White J. H., 2008. Vitamin D signaling, infectious diseases, and regulation of innate immunity. Infect. Immun. 76: 3837–3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A., Henao-Mejia J., Harman C. C., Flavell R. A., 2013. miR-181 and metabolic regulation in the immune system. Cold Spring Harb. Symp. Quant. Biol. 78: 223–230. [DOI] [PubMed] [Google Scholar]
- Wu S., Xia Y., Liu X., Sun J., 2010. Vitamin D receptor deletion leads to reduced level of IkappaBalpha protein through protein translation, protein-protein interaction, and post-translational modification. Int. J. Biochem. Cell Biol. 42: 329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuk J. M., Shin D. M., Lee H. M., Yang C. S., Jin H. S., et al. , 2009. Vitamin D3 induces autophagy in human monocytes/macrophages via cathelicidin. Cell Host Microbe 6: 231–243. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., et al. , 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Leung D. Y., Richers B. N., Liu Y., Remigio L. K., et al. , 2012. Vitamin D inhibits monocyte/macrophage proinflammatory cytokine production by targeting MAPK phosphatase-1. J. Immunol. 188: 2127–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw microarray data files have been deposited in NCBI’s Gene Expression Omnibus (GEO) (Edgar et al. 2002), and are accessible through GEO Series accession number GSE78083 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE78083). The normalized expression values, the results from the linear mixed-effects model, and the results from Cormotif are provided in File S1.