Abstract
Epigenetic reprogramming is necessary in somatic cell nuclear transfer (SCNT) embryos in order to erase the differentiation-associated epigenetic marks of donor cells. However, such epigenetic memories often persist throughout the course of clonal development, thus decreasing cloning efficiency. Here, we explored reprogramming-refractory regions in bovine SCNT blastocyst transcriptomes. We observed that histone genes residing in the 1.5 Mb spanning the cow HIST1 cluster were coordinately downregulated in SCNT blastocysts. In contrast, both the nonhistone genes of this cluster, and histone genes elsewhere remained unaffected. This indicated that the downregulation was specific to HIST1 histone genes. We found that, after trichostatin A treatment, HIST1 histone genes were derepressed, and DNA methylation at their promoters was decreased to the level of in vitro fertilization embryos. Therefore, our results indicate that the reduced expression of HIST1 histone genes is a consequence of poor epigenetic reprogramming in SCNT blastocysts.
Keywords: RNA-seq, SCNT, HIST1 cluster, reprogramming, epigenetics, HDAC inhibitor, DNA methylation
Successful cloning by somatic cell nuclear transfer (SCNT) depends on accurate reprogramming processes in which the differentiated state of the donor cell genome drifts to a totipotent, embryonic ground state (Kang et al. 2003). A variety of developmental defects and, consequently, low clonal viability are attributed to incomplete and poor genomic reprogramming in SCNT embryos (Amano et al. 2001; Eggan et al. 2001; Hill et al. 2000; Lanza et al. 2000; Ono et al. 2001). Several efforts have been dedicated to understand the physiogenetics of SCNT embryos, and to tackle the associated problem of low cloning efficiency. In this context, the most direct way to analyze SCNT embryos is to study their whole genome expression profiles. We recently performed RNA-seq in single cow blastocysts, and reported a set of differentially expressed genes (DEGs) between in vitro fertilization (IVF)-derived and SCNT blastocysts (Min et al. 2015). Additionally, we also reported features unique to SCNT blastocysts, including consistently aberrant expression of genes that function in either trophectoderm development or epigenetic modification. As an extension to the study, here we aimed to identify poorly reprogrammed regions where densely packed DEGs are coordinately regulated, and discovered a reprogramming-refractory locus. To the best of our knowledge, this is the first study to report a reprogramming-resistant megabase-sized genomic stretch in SCNT embryos.
Materials and Methods
In this paper, we processed and analyzed preexisting cow blastocyst transcriptomes data obtained from our recent RNA-seq experiment (Min et al. 2015). Therefore, all raw data and related procedures, including in vitro fertilization and culture of bovine oocytes, somatic cell nuclear transfer, isolation and amplification of the whole transcripts, library construction for RNA-seq, differential gene expression analysis, real-time PCR validation, and Circos plot generation were described in the previous report in detail.
In vitro fertilization of bovine oocytes
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Livestock Research Institute of Korea. The protocol was approved by the Committee on the Ethics of Animal Experiments of the Korea Research Institute of Bioscience and Biotechnology. We obtained bovine ovaries from a local slaughterhouse (Daejon-Ojung SH, Korea), and cumulus-oocyte complexes (COC) were extracted from follicles. Oocyte maturation and IVF were performed as previously described (Park et al. 2007). Blastocysts at midexpanded stage were produced after 7–8 d of IVF (Kwon et al. 2015) and used in downstream experiments. Inseminated oocytes were in vitro cultured (IVC) for 20 hr in IVC medium (3 mg/ml BSA in CR1aa), and the embryos were moved to IVC medium containing 25 nM trichostatin A (TSA), and cultured for another 20 hr. Finally, the embryos were transferred back into IVC medium, and cultured to the blastocyst stage.
Somatic cell nuclear transfer
For somatic cell nuclear transfer, bovine oocytes were enucleated, and somatic cells were injected under the zona pellucida using a micromanipulator (Leitz, Wetzlar, Germany). The injected donor and the oocyte were electrofused using an Electro Cell Manipulator 2001 (BTX). The fused eggs were activated and incubated until blastocysts were generated. Ear skin fibroblasts obtained from an adult female cow were passaged three times, and used as donor cells (Kwon et al. 2015). Cumulus cells obtained from COC during denudation were cultured and used as donors on the following day. We counted the cell number of each SCNT blastocyst by Hoechst staining, and picked out only healthy blastocysts comprising 60–80 blastomeres. To generate TSA-treated SCNT blastocysts, the activated oocytes were cultured in IVC medium supplemented with 25 nM TSA, and then removed to regular IVC medium 3.5 hr after. The embryos were further cultured to the blastocyst stage.
RNA-seq of single bovine blastocysts and bioinformatic analyses
Polyadenylated RNAs were isolated from each blastocyst using Dynabeads mRNA DIRECT kit (Invitrogen). cDNAs were amplified by the pico profiling method, and Illumina sequencing libraries were constructed using the amplified dsDNAs using TruSeq DNA Sample Prep kit (Illumina). For bioinformatic analyses, TopHat-HTSeq-DESeq pipeline was followed. Raw read data from HiSeq2500 were aligned on Bos taurus UMD3.1 using TopHat, and gene expression levels were calculated using HTSeq followed by DESeq.
qPCR validation of HIST1 histone gene expression
For quantitative real-time PCR (qPCR) validation of HIST1 histone gene expression, cDNAs were prepared from six male IVF or fSCNT blastocysts. One-twentieth volume of cDNA was mixed with 10 μl TOPreal qPCR 2X PreMIX (Enzynomics), and 10 μM of HIST1 histone gene-specific primers. Quantitative PCR was performed in a 7500 Real-Time PCR System (Applied Biosystems). All experiments were performed in triplicate. The list of PCR primers is as follows: AACAAGCTGCTGGGCAAAGT and CGTTGTTTCCAATCTTGGTTC for HIST1H2AJ; CAAGCTGCTGGGTAAAGTCA and CGAAGTAATCCAGACTTCTA for HIST1H2AG; GCAAAGTCACCATCGCTCAG and CACTGAGATCTAGGTAGGTTCA for LOC618824.
MSRE-PCR
Genomic DNAs were extracted from 20 IVF or fSCNT blastocysts. For this, blastocysts were incubated in embryo lysis buffer (50 mM KCl, 2.5 mM MgCl2, 0.1 μg/ml gelatin, 0.45% NP40, 0.45% Tween20, 10 mM Tris-Cl, and 200 μg/ml proteinase K) at 55° for 1 hr followed by heat inactivation of proteinase K at 85° for 10 min. Whole lysates were reacted with 10 U of HpaII (NEB) or MspI (NEB) at 37° overnight in 40 μl reaction volume (Yeo et al. 2007). Forty cycles of PCR was carried out on 1 μl digestion products using HIST1 histone-promoter-specific primers: TCAGAGCCTGCCAGCCAGG and AACTCGGGTACAAGTGGCAA for HIST1H1C; CACAGAGCAGGCAACCAATCATCA and TGCTTCTGTTTAGCCAGGAAAGA for HIST1H1D; AAGAAGAAGGCGAAGAAATCGG and CAGCCAGAGACACGCCACT for HIST1H1E; TTCACGAACGAATTCATGATGCCC and GAAGAAGGACGGCAAGAAGC for HIST1H2AG; GAGCTACTCCGTGTACGTGTA and AAATGTCGTTGACGAAAGAGTTCATGA for HIST1H2B; TTCACGAACGAATTCATGATGCCC and GCAGAAGAAGGACGGCAAGAA for HIST1H2BB; TACTTGGAGCTGGTGTACTTGG and AAGCGCTCGACCATCACATCTAG for HIST1H2BD; and ATTACAACAAGCGCTCGACCAT and GTTGCTGGACAACTTTACTTGG for HIST1H2BN.
Data availability
Global expression data used in this study is available in our previous report (Min et al. 2015)
Results
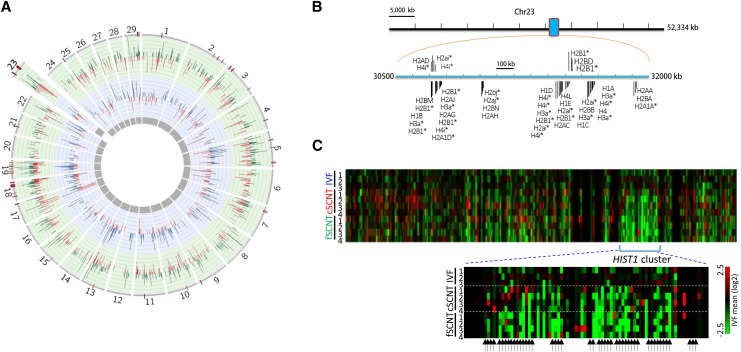
We recently reported RNA-seq data from single bovine blastocysts obtained by either IVF or SCNT. For the latter, we used two different donors, cumulus cells (cSCNT) and ear-skin fibroblasts (fSCNT) (Min et al. 2015). We collected an average of 12.7 million high-quality paired-end reads, and generated an expression profile dataset using TopHat and HTSeq, and Bos taurus UMD3.1 as reference genome. Analysis using DESeq revealed 630 and 1064 DEGs in cSCNT and fSCNT blastocysts, respectively, vs. IVF blastocysts. To identify poorly reprogrammed genomic regions where coordinately regulated DEGs are densely packed, we scanned the entire genome of SCNT blastocysts for erroneous domains by the sliding window analysis. Thirty-eight windows were detected to have six or more DEGs in single windows in the fSCNT blastocysts (Figure 1A). Of these, 13 were common to cSCNT blastocysts.
Figure 1.
The HIST1 cluster is downregulated in somatic cell nuclear transfer (SCNT) blastocysts. (A) Circos plot representing significant differentially expressed genes (DEGs; P < 0.05) in cumulus-cell (cSCNT; blue, inner circular layer), and fibroblast SCNT blastocysts (fSCNT; green, outer circular layer), vs. those in in vitro fertilization (IVF) blastocysts. Their fold-change values are shown (blue, green, and red bars within the circular layers). Red bars on the outermost layer represent DEG-rich regions in cSCNT, and/or fSCNT (five or more DEGs per 2 Mb window). Numbers on the outer layer indicate chromosome numbers; X and Y chromosomes are omitted. (B) Distribution of histone genes within the HIST1 cluster, ranging from 30.5 Mb to 32 Mb, on chromosome 23. Predicted histone genes are marked by asterisks. Only histone-related genes are shown, and other genes in the HIST1 locus are omitted. (C) Heat map of genes in the HIST1 cluster showing relative gene expression of single blastocysts to the mean levels of IVF group along the length of chromosome 23. Below, the enlarged HIST1 cluster. Arrows indicate annotated histone genes including predicted ones.
Among the DEG-rich loci, we were particularly interested in a locus where histone genes are bundled together. In both human (Albig et al. 1997) and mouse (Wang et al. 1996), about 50 histone genes heavily populate this locus, which is called the HIST1 cluster, and are transcriptionally regulated in a coordinated manner (Marzluff et al. 2002; Marzluff and Duronio 2002). The bovine HIST1 cluster on chromosome 23 resembles those of human and mouse. It spans about 1.5 Mb, and contains 47 histone and histone-like genes on Bos taurus UMD3.1 (Figure 1B). A heat map of genes on chromosome 23 unambiguously pinpointed downregulation at the HIST1 cluster in both cSCNT and fSCNT blastocysts (Figure 1C). When the expression of HIST1 histone genes was examined in donor fibroblasts, SCNT blastocysts appeared, overall, to be closer to donor cells than IVF blastocysts (Supplemental Material, Figure S1).
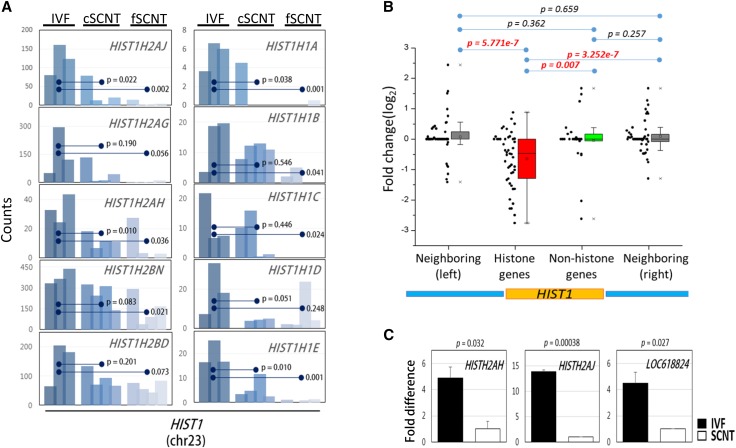
Detailed inspection of the transcriptomes revealed that individual histone genes residing in this cluster were, indeed, weakly expressed in both the fSCNT and cSCNT groups (Figure 2A). We further examined the expression of 47 HIST1 histone genes independently of 26 nonhistone genes in the same cluster. We also examined 47 and 57 genes upstream and downstream of the cluster, respectively (Figure 2B). Interestingly, only the HIST1 histone genes were significantly downregulated in SCNT blastocysts, whereas genes of other subsets, including the nonhistone genes within the HIST1 cluster, were not altered. This result agrees with the previous finding that histone genes in the human HIST1 cluster are coordinately regulated (Marzluff and Duronio 2002). By qPCR using cDNAs from freshly prepared IVF and fSCNT blastocysts, we confirmed that the HIST1H2AJ, HIST1H2AH, and LOC618824 (H2AI-like) genes in the HIST1 cluster were significantly downregulated in the SCNT blastocysts (Figure 2C). Furthermore, we found that bovine SCNT blastocysts also expressed genes, such as YY1, POU2F1, NPAT, and SLBP, known to be involved in coordinated regulation of HIST1 histone genes in human and mouse (Eliassen et al. 1998; Fletcher et al. 1987; Ma et al. 2000; Zhao et al. 2000; Whitfield et al. 2000). However, their expression in SCNT blastocysts was comparable (NPAT and SLBP) to, or even higher (YY1 and POU2F1) than, that in IVF blastocysts (Figure S2).
Figure 2.
Downregulation of histone genes at the HIST1 cluster in somatic cell nuclear transfer (SCNT) blastocysts. (A) Expression profiles of HIST1 histone genes in single blastocysts. Bar graphs represent the expression of representative HIST1 histone genes in individual blastocysts between cSCNT or fSCNT groups vs. IVF embryos. Statistical significance between the groups is denoted (Student t-test). (B) Box plot showing relative expression of HIST1 histone genes in both cSCNT and fSCNT groups vs. those in the IVF group. For comparison, 47 histone and 26 nonhistone genes within the HIST1 cluster (yellow box below), as well as the 57 (left) and 47 (right) upstream and downstream genes, respectively (blue boxes), are included. (C) Validation of the HIST1 histone genes in IVF (solid bar) and fSCNT (blank bar) blastocysts by qPCR. Error bars indicate standard deviation of replicated experiments.
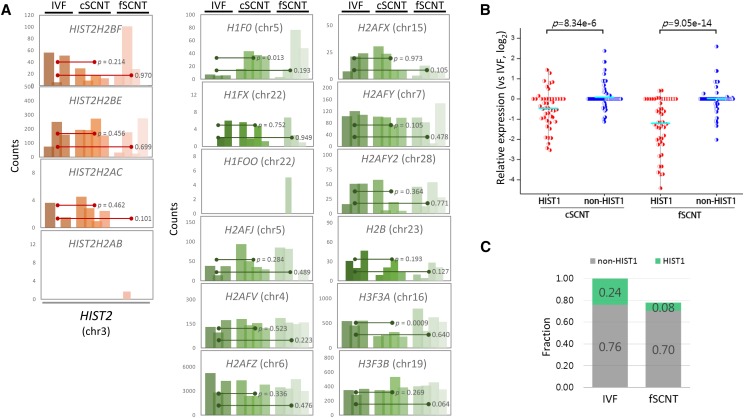
In contrary to HIST1 histones, histone genes such as H2AFV, H2AFY, and H2AFZ on other chromosomes (4, 7, and 6, respectively), and those in the HIST2-like cluster on chromosome 3 (HIST2H2AB, HIST2H2AC, HIST2H2BE, and HIST2H2BF) showed no difference in expression between the blastocyst groups (Figure 3A). When the HIST1 histone genes were compared with ∼100 other histone genes residing elsewhere, their expression levels differed significantly between cSCNT and fSCNT (P = 8.34e–6, and 9.05e–14, respectively; Figure 3B). The sum of expression of all the histone and histone-like genes in the genome revealed that the fSCNT group expressed them at about 80% of what the IVF group did (Figure 3C). Notably, while the HIST1 histone gene expression took ∼24% out of total histone gene expression in the IVF group, their contribution was only 8% in the fSCNT group, which was one-third (0.08/0.24) of the former. This indicates that the difference in total histone gene expression between the IVF and SCNT groups is derived largely from the difference in expression of the HIST1 histone genes.
Figure 3.
NonHIST1 histone gene expression in SCNT blastocysts. (A) Expression profiles of nonHIST1 histone genes including HIST2 histone genes in individual blastocysts. Statistical significance between the groups is denoted (Student t-test). Chromosome location of each histone gene is indicated in parenthesis. (B) Relative expression of HIST1 (red dots) and nonHIST1 (blue dots) histone genes in cSCNT and fSCNT groups vs. the IVF group. Mean expression levels are shown as cyan-colored bars. The statistical significance between HIST1 and nonHIST1 histone gene groups are indicated. (C) Comparison of expression levels of HIST1 and nonHIST1 histone gene groups between IVF and fSCNT blastocysts. Green and gray colors represent the fractions of HIST1 and nonHIST1 histone genes expressed, respectively.
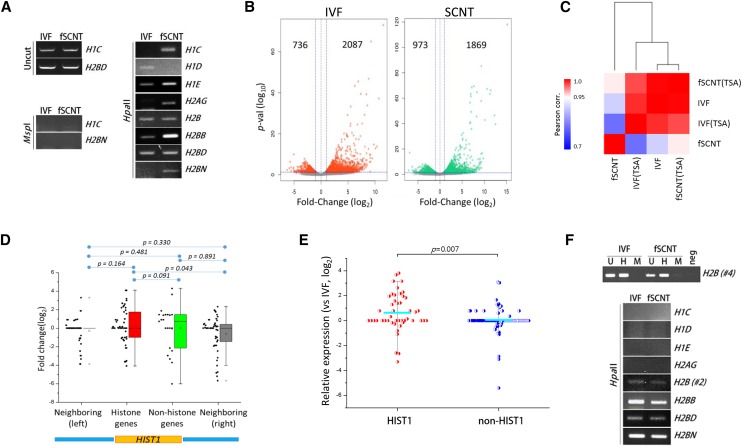
Given the possibility that the HIST1 cluster is epigenetically regulated, we examined the epigenetic state of this region. First, DNA methylation at various HIST1 histone gene promoters was analyzed by methylation-sensitive restriction enzyme polymerase chain reaction (MSRE-PCR), using HpaII-digested genomic DNA from either IVF or fSCNT blastocysts as template. We found that most of the promoters (HIST1H1C, HIST1H1E, HIST1H2AG, HIST1H2BB, and HIST1H2BN) examined were more heavily methylated in the SCNT samples, whereas the rest were either similarly (HIST1H2B and HIST1H2BD) or less (HIST1H1D) methylated (Figure 4A). The results suggest that, in general, gene promoters at the HIST1 locus retain aberrant DNA methylation states in SCNT blastocysts. Next, we addressed whether this abnormal epigenetic state of the HIST1 cluster could be relieved by treatment with TSA, a histone deacetylase (HDAC) inhibitor. TSA positively influences animal cloning by promoting chromatin relaxation and increased global gene expression (Kishigami et al. 2006, 2007; Jee et al. 2012; Cui et al. 2011). RNA-seq was performed in TSA-treated IVF and fSCNT blastocysts. Differential expression analysis showed that TSA treatment upregulates a large number of genes in both blastocyst groups, as expected (Figure 4B). Pearson correlation analysis showed that correlation coefficient was the highest (r = 0.998) between IVF and TSA-treated fSCNT groups, which was a substantial increase considering the relatively low coefficient (r = 0.911) between them in the absence of TSA (Figure 4C). Examination of the genes in the HIST1 cluster revealed that, in contrast to the results shown in Figure 2B, the expression of histone and nonhistone genes within the HIST1 cluster did not significantly differ at P < 0.05 (Figure 4D). Compared with nonHIST1 histone genes, contrary to the result in Figure 2B, HIST1 histones showed increased expression in TSA-treated SCNT blastocysts (Figure 4E). Finally, we analyzed TSA-treated SCNT blastocysts for methylation state using MSRE-PCR. We observed that the methylation states of both SCNT and IVF blastocysts looked similar to each other at the HIST1 histone gene promoters after TSA treatment, suggesting that TSA might directly or indirectly induce demethylation of the HIST1 histone gene promoters (Figure 4F).
Figure 4.
Epigenetic regulation of HIST1 histone gene expression. (A) MSRE-PCR for analysis of DNA methylation at HIST1 histone gene promoters. Genomic DNAs obtained from blastocysts (n = 10) of each group were first digested with either HpaII or MspI. This was followed by PCR using gene-specific primers. Both enzymes equally recognize the 5′-CCGG-3′ sequence, but HpaII cannot function when the second cytosine is methylated. Stronger band intensity in the gel electrophoresis image indicates higher methylation level at the corresponding promoter. PCR was repeated twice for each gene promoter. Uncut, positive control; MspI, negative control. (B) Trichostatin A (TSA)-mediated global gene expression increase in IVF and fSCNT blastocysts. Differentially expressed genes (DEGs; P < 0.05, fold-change > 2) in TSA-treated blastocysts vs. the corresponding untreated groups are shown in either red or green. In TSA-treated IVF and fSCNT blastocysts, 2087 and 1869 genes were upregulated, and 736 and 973 downregulated, respectively. Horizontal blue line indicates P = 0.05, while vertical ones indicate log2(fold change) = ± 1. (C) Heatmap for Pearson correlation between TSA-treated or untreated IVF and fSCNT blastocyst groups. (D) Expressions of HIST1 histone genes in SCNT blastocysts relative to those in IVF blastocysts treated with TSA. The same subsets of genes in Figure 2B were assessed. (E) Expressions of HIST1 (red) and nonHIST1 (blue) histone genes in TSA-treated fSCNT relative to that in TSA-treated IVF blastocysts. Cyan bars indicate mean expression of either HIST1 or nonHIST1 histone genes. Student’s t-test was performed to determine statistical significance. (F) MSRE-PCR analysis of DNA methylation at HIST1 histone gene promoters in TSA-treated IVF and fSCNT blastocysts. Upper panel, control experiment; H, HpaII digested gDNA; U, uncut gDNA for positive control; M, MspI digested gDNA for negative control. Two different promoter regions, H2B #2 and #4, were analyzed for H2B, and the latter is represented as a control experiment.
Discussion
We found that histone genes in the HIST1 cluster were coordinately and epigenetically downregulated in SCNT blastocysts. Our findings suggest that, compared with IVF embryos, SCNT embryos have more repressive chromatin in the HIST1 locus, which hinders transcription factors approaching the region. Histone genes in the human and mouse HIST1 loci are known to be coregulated by YY1, POU2F1, NPAT, and SLBP, but these genes are found normally expressed in cow SCNT blastocysts (Figure S2). This implies that the presence of these regulators is not enough to derepress the HIST1 locus, and that the HIST1 region exists in a transcriptionally nonpermissive chromatin state that is highly resistant to reprogramming in SCNT embryos. This is supported by heavier methylation at the HIST1 histone gene promoters in SCNT (Figure 4A). The low expression of HIST1 histone genes in donor cells (Figure S1) leads to an assumption that the HIST1 histone gene expression anomaly shown in SCNT embryos might have originated in donor cell genomes. Figure 4, D–E shows that TSA can change the refractory chromatin, and relieve the HIST1 histone genes. This gives a molecular mechanistic insight into the low expression of HIST1 histone genes in SCNT embryos. The HIST1 histone genes have lain under an inactive chromatin state in SCNT blastocysts, and were relieved by TSA, which removes repressive chromatin remodelers and recruits various transcription activators to the locus. In support of this view, DNA methylation at HIST1 histone gene promoters was reduced to levels comparable to those in IVF embryos after TSA treatment (Figure 4F). Therefore, our findings indicate that TSA, through modulation of the epigenetic state of the HIST1 locus in SCNT blastocysts, renders the histone genes transcriptionally competent, which is important for proper development of the embryo (see below).
The SCNT blastocyst group expressed lower levels of histone genes compared with the IVF group. This is attributed to the reduced expression of HIST1 histone genes, rather than other histone genes (Figure 3C). The four core histone molecules, H2A, H2B, H3, and H4, comprise a nucleosome, maintaining a ratio of one H3–H4 tetramer, and two H2A–H2B dimers. Unequal histone levels, such as weighing toward a particular molecule, might induce cell cycle arrest in SCNT embryos (Ghule et al. 2014). Since ∼100 histone-like genes with LOC-prefixed names in the cow genome are, as yet, uncharacterized, we were unable to estimate the stoichiometric balance among the four histone molecules from the blastocyst transcriptomes. Therefore, whether the reduced histone expression causes a hazardous imbalance and leads to cell cycle arrest in SCNT embryos, remains to be ascertained. Another hypothesis is that, despite the reduced levels, SCNT blastocysts are able to maintain stable and proportionate histone mRNA levels and, consequently, equilibrate among them. Irrespective of these scenarios, the premise of successful clonal development is that all histone genes, which reside scattered on the chromosomes, are coordinately expressed against genome-wide reprogramming processes.
The effect of reduced HIST1 histone gene expression on SCNT embryos is unresolved. It is known that histone synthesis is tightly coupled to that of DNA, and, likewise, inhibition of the latter during the replication phase of the cell cycle causes rapid decline in the former (Plumb et al. 1983; Sittman et al. 1983; Su et al. 2004; Ghule et al. 2014). Therefore, if the reduced transcription affects protein level, the diminished amount of histones might cause a deregulated cell cycle progression (e.g., delayed S-phase entry) in blastomeres. Unlike normal embryonic cell cycle, which involves rapid successions of DNA replication without the intermittent G1 and G2 phases, that of SCNT embryos is a lengthy process that can be observed in typical differentiated somatic cells (Becker et al. 2006; Boiani et al. 2003; Balbach et al. 2012). Consequently, SCNT blastocysts frequently suffer from low cell counts. If this is true, the reduced expression of HIST1 histone genes is indicative of poor genome reprogramming, which must be overcome for normal clonal development.
Numerous and variable transcriptomic dissimilarities occur unpredictably between SCNT embryos, indicating the stochastic nature of reprogramming processes. Nevertheless, some changes in these embryos are nonrandom and consistent, represented by those in the HIST1 locus. In addition, we recently reported a 400-kb locus harboring zinc-finger (ZNF) protein family genes in chromosome 18 that were coordinately downregulated in fSCNT blastocysts, showing a feature of reprogramming-resistant regions (Min et al. 2015). Therefore, it is not entirely true that each SCNT embryo is unique in terms of its gene expression profile. To the best of our knowledge, this is the first study to report the identification of a reprogramming-resistant megabase-stretch in the genome of bovine SCNT embryos. Extensive efforts to discover such regions, which persistently escape reprogramming, might help establish a reprogramming error-prone genomic map in SCNT embryos. Such a map will shed light on the mechanisms underlying local and global reprogramming events, and help improve the efficiency of such events.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Research Foundation of Korea (2006-2004082 and 2015R1A2A2A01003117), National Research Foundation-Science Research Center program (2011-0030049), and the Korea Research Institute of Bioscience and Biotechnology research initiative program.
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.026666/-/DC1
Communicating editor: D. J. de Koning
Literature Cited
- Albig W., Kioschis P., Poustka A., Meergans K., Doenecke D., 1997. Human histone gene organization: nonregular arrangement within a large cluster. Genomics 40(2): 314–322. [DOI] [PubMed] [Google Scholar]
- Amano T., Tani T., Kato Y., Tsunoda Y., 2001. Mouse cloned from embryonic stem (ES) cells synchronized in metaphase with nocodazole. J. Exp. Zool. 289(2): 139–145. [DOI] [PubMed] [Google Scholar]
- Balbach S. T., Esteves T. C., Houghton F. D., Siatkowski M., Pfeiffer M. J., et al. , 2012. Nuclear reprogramming: kinetics of cell cycle and metabolic progression as determinants of success. PLoS One 7(4): e35322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K. A., Ghule P. N., Therrien J. A., Lian J. B., Stein J. L., et al. , 2006. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J. Cell. Physiol. 209(3): 883–893. [DOI] [PubMed] [Google Scholar]
- Boiani M., Eckardt S., Leu N. A., Scholer H. R., McLaughlin K. J., 2003. Pluripotency deficit in clones overcome by clone-clone aggregation: epigenetic complementation? EMBO J. 22(19): 5304–5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X. S., Xu Y. N., Shen X. H., Zhang L. Q., Zhang J. B., et al. , 2011. Trichostatin A modulates apoptotic-related gene expression and improves embryo viability in cloned bovine embryos. Cell. Reprogram. 13(2): 179–189. [DOI] [PubMed] [Google Scholar]
- Eggan K., Akutsu H., Loring J., Jackson-Grusby L., Klemm M., et al. , 2001. Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc. Natl. Acad. Sci. USA 98(11): 6209–6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliassen K. A., Baldwin A., Sikorski E. M., Hurt M. M., 1998. Role for a YY1-binding element in replication-dependent mouse histone gene expression. Mol. Cell. Biol. 18(12): 7106–7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher C., Heintz N., Roeder R. G., 1987. Purification and characterization of OTF-1, a transcription factor regulating cell cycle expression of a human histone H2b gene. Cell 51(5): 773–781. [DOI] [PubMed] [Google Scholar]
- Ghule P. N., Xie R. L., Medina R., Colby J. L., Jones S. N., et al. , 2014. Fidelity of histone gene regulation is obligatory for genome replication and stability. Mol. Cell. Biol. 34(14): 2650–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J. R., Winger Q. A., Long C. R., Looney C. R., Thompson J. A., et al. , 2000. Development rates of male bovine nuclear transfer embryos derived from adult and fetal cells. Biol. Reprod. 62(5): 1135–1140. [DOI] [PubMed] [Google Scholar]
- Jee B. C., Jo J. W., Lee J. R., Suh C. S., Kim S. H., et al. , 2012. Effect of trichostatin A on fertilization and embryo development during extended culture of mouse oocyte. Zygote 20(1): 27–32. [DOI] [PubMed] [Google Scholar]
- Kang Y. K., Lee K. K., Han Y. M., 2003. Reprogramming DNA methylation in the preimplantation stage: peeping with Dolly’s eyes. Curr. Opin. Cell Biol. 15(3): 290–295. [DOI] [PubMed] [Google Scholar]
- Kishigami S., Mizutani E., Ohta H., Hikichi T., Thuan N. V., et al. , 2006. Significant improvement of mouse cloning technique by treatment with trichostatin A after somatic nuclear transfer. Biochem. Biophys. Res. Commun. 340(1): 183–189. [DOI] [PubMed] [Google Scholar]
- Kishigami S., Bui H. T., Wakayama S., Tokunaga K., Van Thuan N., et al. , 2007. Successful mouse cloning of an outbred strain by trichostatin A treatment after somatic nuclear transfer. J. Reprod. Dev. 53(1): 165–170. [DOI] [PubMed] [Google Scholar]
- Kwon S., Jeong S., Park J. S., Kang Y. K., 2015. Quantifying difference in gene expression profile between bovine blastocysts derived by in vitro fertilization and somatic cell nuclear transfer. Gene Expr. Patterns. 19(1–2): 14–20 [DOI] [PubMed] [Google Scholar]
- Lanza R. P., Cibelli J. B., Blackwell C., Cristofalo V. J., Francis M. K., et al. , 2000. Extension of cell life-span and telomere length in animals cloned from senescent somatic cells. Science 288(5466): 665–669. [DOI] [PubMed] [Google Scholar]
- Ma T., Van Tine B. A., Wei Y., Garrett M. D., Nelson D., et al. , 2000. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 14(18): 2298–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff W. F., Duronio R. J., 2002. Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr. Opin. Cell Biol. 14(6): 692–699. [DOI] [PubMed] [Google Scholar]
- Marzluff W. F., Gongidi P., Woods K. R., Jin J., Maltais L. J., 2002. The human and mouse replication-dependent histone genes. Genomics 80(5): 487–498. [PubMed] [Google Scholar]
- Min, B., S. Cho, J. S. Park, Y. G. Lee, N. Kim et al., 2015 Transcriptomic features of bovine blastocysts derived by somatic cell nuclear transfer. G3 (Bethesda) 5(12): 2527–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y., Shimozawa N., Ito M., Kono T., 2001. Cloned mice from fetal fibroblast cells arrested at metaphase by a serial nuclear transfer. Biol. Reprod. 64(1): 44–50. [DOI] [PubMed] [Google Scholar]
- Park J. S., Jeong Y. S., Shin S. T., Lee K. K., Kang Y. K., 2007. Dynamic DNA methylation reprogramming: active demethylation and immediate remethylation in the male pronucleus of bovine zygotes. Dev. Dyn. 236(9): 2523–2533. [DOI] [PubMed] [Google Scholar]
- Plumb M., Stein J., Stein G., 1983. Influence of DNA synthesis inhibition on the coordinate expression of core human histone genes during S phase. Nucleic Acids Res. 11(22): 7927–7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittman D. B., Graves R. A., Marzluff W. F., 1983. Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc. Natl. Acad. Sci. USA 80(7): 1849–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C., Gao G., Schneider S., Helt C., Weiss C., et al. , 2004. DNA damage induces downregulation of histone gene expression through the G1 checkpoint pathway. EMBO J. 23(5): 1133–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. F., Krasikov T., Frey M. R., Wang J., Matera A. G., et al. , 1996. Characterization of the mouse histone gene cluster on chromosome 13: 45 histone genes in three patches spread over 1Mb. Genome Res. 6(8): 688–701. [DOI] [PubMed] [Google Scholar]
- Whitfield M. L., Zheng L. X., Baldwin A., Ohta T., Hurt M. M., et al. , 2000. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 20(12): 4188–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo S., Jeong S., Kim J., Han J. S., Han Y. M., et al. , 2007. Characterization of DNA methylation change in stem cell marker genes during differentiation of human embryonic stem cells. Biochem. Biophys. Res. Commun. 359(3): 536–542. [DOI] [PubMed] [Google Scholar]
- Zhao J., Kennedy B. K., Lawrence B. D., Barbie D. A., Matera A. G., et al. , 2000. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 14(18): 2283–2297. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Global expression data used in this study is available in our previous report (Min et al. 2015)