Abstract
Pollen killer genes disable noncarrier pollens, and are responsible for male sterility and segregation distortion in hybrid populations of distantly related plant species. The genetic networks and the molecular mechanisms underlying the pollen killer system remain largely unknown. Two pollen killer genes, S24 and S35, have been found in an intersubspecific cross of Oryza sativa ssp. indica and japonica. The effect of S24 is counteracted by an unlinked locus EFS. Additionally, S35 has been proposed to interact with S24 to induce pollen sterility. These genetic interactions are suggestive of a single S24-centric genetic pathway (EFS–S24–S35) for the pollen killer system. To examine this hypothetical genetic pathway, the S35 and the S24 regions were further characterized and genetically dissected in this study. Our results indicated that S35 causes pollen sterility independently of both the EFS and S24 genes, but is dependent on a novel gene close to the S24 locus, named incentive for killing pollen (INK). We confirmed the phenotypic effect of the INK gene separately from the S24 gene, and identified the INK locus within an interval of less than 0.6 Mb on rice chromosome 5. This study characterized the genetic effect of the two independent genetic pathways of INK–S35 and EFS–S24 in indica–japonica hybrid progeny. Our results provide clear evidence that hybrid male sterility in rice is caused by several pollen killer networks with multiple factors positively and negatively regulating pollen killer genes.
Keywords: pollen killer, Oryza sativa, epistasis, reproductive isolation
Hybrid progeny derived from genetically divergent species frequently suffer reproductive dysfunction. This phenomenon, referred to as hybrid sterility, is assumed to play an important role in the development of speciation by triggering postzygotic reproductive isolation. Segregation distorters, also called transmission ratio distortion factors, are evolutionarily selfish genetic elements that distort Mendelian segregation in their favor at the expense of others. Segregation distorters have been reported in a wide variety of reproductive organisms, including animals, plants, and fungi (Burt and Trivers 2009), and are one of the primary origins of hybrid sterility by causing one-half of the gametes to be dysfunctional. Two well-characterized genetic elements are Segregation distorter (Sd) in Drosophila (Larracuente and Presgraves 2012) and t-complex in mice (Lyon 2003). The role of segregation distorters in speciation of reproductive organisms has been the subject of considerable discussion (Burt and Trivers 2009).
In plants, diverse types of segregation distorters have been identified in many plant species. Some examples include: pollen killer in Nicotiana (Cameron and Moav 1957), gamete eliminator in tomato (Rick 1966), gametocidal factor in wheat (Endo 2015), chromosomal knobs in maize (Buckler et al. 1999; Kanizay et al. 2013) and female meiotic drive in Mimulus (Fishman et al.and Saunders 2008; Finseth et al. 2015). Gamete eliminator (Ge) (both pollen and egg are abortive) was found in a tomato intraspecific cross by plant geneticist Charles M. Rick, who interpreted Ge as a triallelic system at a single locus (Rick 1966). The gamete eliminator (Gep) causes abortion of gametes carrying Gec in the Gep/Gec heterozygote. The third neutral allele, Gen, is compatible with both Gep and Gec in the heterozygous hybrid. Since the neutral allele Gen is widely distributed, Rick proposed that “both Gec and Gep could arise by mutation from Gen without adverse effect on gamete fertility” (Rick 1966). Based on this idea, the killer and the abortive allele were proposed to be derived from a common ancestral allele that is compatible with both of them.
The egg killer locus in rice, S5, is a type of triallelic system (Ikehashi and Araki 1986). The killer allele S5-i from indica causes female gamete abortion in the heterozygous state with the japonica allele, but not with the neutral alleles from other varieties or the wild progenitor. Closer study of the S5 system unveiled the molecular mechanism. The S5 locus is composed of three tightly linked genes, and particular combinations of the alleles lead to endoplasmic reticulum stress, resulting in female gamete abortion (Yang et al. 2012). As a case study of male sterility, two adjacent genes that encode a SUMO E3 ligase-like protein and an F-box protein were identified in an indica/japonica cross (Long et al. 2008). These pioneering studies have provided important insights into the genetic architecture and the evolutionary history of plant gamete killers; namely, the existence of tightly linked multiple genes, and cumulative mutations in these genes lead to the gamete killing phenotype in heterozygotes. In this scenario, not only the ancestral haplotype, but also some of the derived haplotypes can work as compatible (neutral) haplotypes. In this context, several questions about segregation distorters remain to be explored. (1) What is a common or unique aspect of the molecular mechanism? Previous studies have identified highly repetitive and heterochromatic sequences as common causal molecules, e.g., the D locus in Mimulus (Fishman and Saunders 2008), knobs in maize (Buckler et al. 1999), and HSR in mice (Weichenhan et al. 2001). (2) What are the normal functions of these genes within the parental species? (3) Why have a greater number of segregation distorters evolved in plant genomes compared with animal or fungal genomes? (4) Has domestication facilitated the development of segregation distorters? There are many segregation distorters found in crop species, especially in rice, and a previous study proposed an association between reproductive isolation and domestication (Dempewolf et al. 2012). Further studies extended to many more examples are needed to answer these questions.
Asian cultivated rice (Oryza sativa L.) is an autogamous diploid species (2n = 24) and has two major subspecies, indica and japonica, whose genomes have been sequenced (Yu et al. 2002; International Rice Genome Sequencing Project 2005). These subspecies are thought to have originated independently from different subpopulations of the closest wild relative O. rufipogon (Morishima 2001; Zhu and Ge 2005; Londo et al. 2006; Kovach et al. 2007). The intersubspecific cross often exhibits hybrid sterility due to abnormal growth of pollen and/or the embryo sac. Many cases of hybrid sterility in rice are caused by so-called “allelic interaction at a single genetic locus” rather than by epistatic interactions between unlinked loci (Koide et al. 2008; Ouyang et al. 2009). A hybrid male sterility gene, S24, also called f5-Du (Wang et al. 2006) or S-b (Li et al. 2006), is a typical gene showing an allelic interaction effect that is located on rice chromosome 5. S24 causes pollen semisterility when the indica allele is introgressed into the japonica background. The indica allele of (S24-i) acts as a pollen killer and produces maldeveloped male gametes bearing the japonica allele exclusively in the heterozygotes (Kubo et al. 2008; Zhao et al. 2011). Consequently, the S24-i allele is preferentially transmitted to the offspring (90–99%). Zhao et al. (2011) identified an ankyrin protein gene as the primary candidate for S24 by a fine-mapping approach. S24 is under the control of an unlinked dominant suppressor, Epistatic Factor for S24 (EFS), located on chromosome 2 (Kubo et al. 2011). Another hybrid male sterility gene, S35, located on chromosome 1, acts as a pollen killer in a similar fashion to S24. Interestingly, S35 has been proposed to interact genetically with S24 to cause pollen sterility, implying a killer–killer interaction (Kubo et al. 2008). Together with the animal examples, it is plausible that linked and unlinked epistatic factors play key roles in the genetic mechanism of the gamete killer system.
In contrast to the S24 locus, which has been somewhat characterized, many characteristics of the S35 locus remain unknown. For example, the causal gene and molecular mechanism of S35 have not been elucidated, although the basic genetic characteristics, and the approximate position of the S35 locus on chromosome 1 have been identified (Kubo et al. 2008). Also, it remains unclear whether the S24-suppressor gene EFS can restore pollen sterility due to S35, and whether such a killer–killer (S24–S35) interaction really exists as previously proposed. The aim of this study was to characterize the S35 gene and its related genetic network more completely. Using reciprocal near-isogenic lines for S24 and S35, our results indicated that the mechanism for hybrid male sterility in rice involves multiple genes and more complex gene interactions than expected. Here, we report a novel gene that interacts with S35, and evidence for the genetic independency of the S24 and S35 genes.
Materials and Methods
Plant materials
All experimental populations, including mapping populations and near-isogenic lines (NILs), were derived from reciprocal chromosome segment substitution lines (CSSLs), and sister lines that were obtained from a cross between the japonica rice variety Asominori and the indica variety IR24 (Kubo et al. 2002). Both of the parents produced fertile pollen (> 90%) under our growth conditions. An overall view of the breeding procedure used to develop the experimental populations is illustrated in Supplemental Material, Figure S1. The NILs with the Asominori genetic background were developed from crossed/backcrossed and selfed progeny of AIS 86, a CSSL carrying IR24 segments of chromosomes 1 and 5 (Kubo et al. 2002), if not otherwise specified. To evaluate pollen fertility of all 27 genotype classes generated by different combinations of the S24, S35, and EFS alleles, a triple heterozygous plant for S24, S35, and EFS in the Asominori genetic background (NIL-S24+S35+EFS) was developed by crossing CSSL AIS 86 and AB2-6 (a derivative line of the AIS library) (Figure S1A). The NIL S24ILH/I, carrying a very small IR24 segment around the S24 locus in an otherwise uniform Asominori genetic background, was developed by two additional backcrosses of AIS 86 with Asominori (BC5F2) and marker-assisted selection (MAS) (Figure S1B). NILs with the IR24 genetic background were developed from crosses of three different CSSLs, IAS 27/IAS 10//IAS 6 (Figure S1C). Fine mapping of S35 and an S35-activator INK was performed using self-pollinated progeny of AIS 86/Aso and NIL-S24+S35+EFS/Aso. Genotype frequencies were determined using seedling leaves of self-pollinated progeny of a single plant heterozygous for S35 or S24 with different genetic backgrounds. All plant materials for phenotype evaluation were grown under paddy field conditions in 2012–2015 in Mishima, Japan.
DNA analysis
Crude DNA extracts from individual leaves were prepared for genotyping using 0.25 M, NaOH followed by neutralization with 0.1 M Tris-HCl. These DNA extracts (1.0 µl) were used in PCR reactions (10 µl final volume) performed using GoTaq polymerase (Promega, Fitchburg, WI) with the following cycling profile: 94° for 2 min; followed by 30 cycles of 94° for 20 sec, 50−60° for 20 sec, and 72° for 30 sec. The PCR products were resolved on 2.0% agarose gels, and visualized by ethidium bromide staining. PCR-based markers, insertion and deletion (InDel), and simple sequence repeat (SSR) markers were newly designed for mapping of the S35 and INK loci based on DNA sequence polymorphisms between Nipponbare and 93-11 (MSU6, http://rice.plantbiology.msu.edu/; BGI, http://rice.genomics.org.cn/rice/index2.jsp). Also, SSR markers reported by McCouch et al. (2002) were used. The primer sequences for DNA markers are listed in Table S1. The whole-genome genotyping data of the reciprocal CSSLs were obtained from Kubo et al. (2002). For genotyping nontarget chromosome segments retained in the genetic background, PCR markers were used that were evenly distributed over the 12 rice chromosomes (Mizuta et al. 2010; Kubo et al. 2011). Genes and marker loci are shown on the physical chromosome map based on the Nipponbare sequence (MSU7).
Phenotyping and histological experiments
Preflowering panicles from each individual in the population were collected and fixed in 50% ethanol solutions to examine pollen fertility. Pollen grains from three to six anthers at 1 d before anthesis were stained with 1.0% iodine-potassium iodide (I2-KI), and the number of stained/unstained pollen grains were counted using a microscope. More than 400 pollen grains were scored for each individual. In this study, we scored pollen phenotypes with <40%, 40–70%, 70–90%, and >90% pollen fertility as sterile, semisterile, partial sterile, and fertile, respectively. To observe the morphology of mature pollen grains, the ethanol-fixed pollen grains were stained with hematoxylin solution by the method of Kindiger and Beckett (1985). Male gametogenesis was analyzed using young panicles at different developmental stages collected from Asominori and the pollen sterile plants. The panicles were fixed and stored in FAA solution (45% ethanol, 5% formalin, and 5% acetic acid). After fixation, the samples were embedded in paraffin (Paraplast Plus; McMormick Scientific, St. Louis, MO) and cut at a thickness of 8 µm with a Microm HM355 microtome (Microm, Walldorf, Germany). The sections were stained with 0.05% Toluidine blue O. To evaluate seed fertility, three panicles with fully ripened grains were collected from each plant, and the numbers of filled and unfilled spikelets were counted. The seed setting rate was estimated by the formula: the number of filled grains / total number of filled and unfilled grains.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Characteristics and chromosomal localization of S35
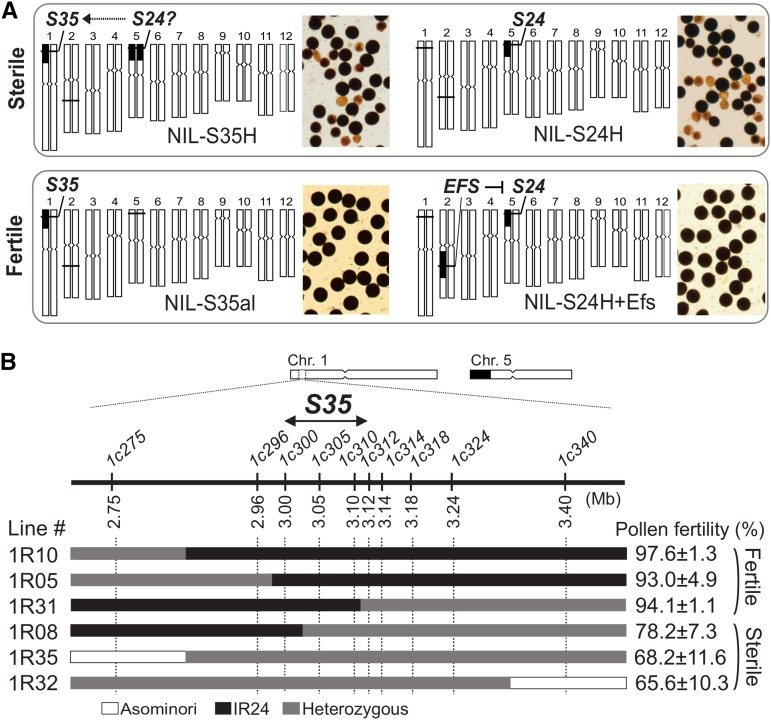
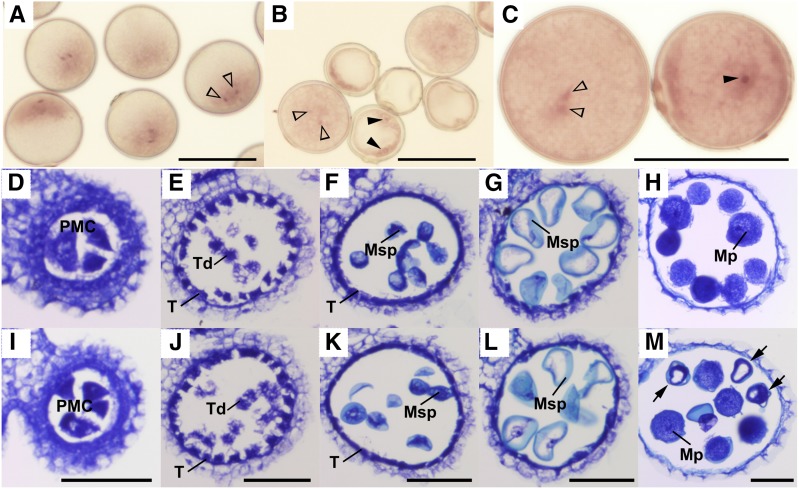
The pollen killer gene S35 has been roughly mapped to the short arm of rice chromosome 1 (Kubo et al. 2008). This previous genetic study indicated that the heterozygous S35 (S35-i/S35-j) caused pollen sterility when the indica segment harboring S24, another pollen killer, was concurrently introgressed into the japonica background (NIL-S35H in Figure 1A) (Kubo et al. 2008). A single introgression of S35-i in the japonica genome does not cause pollen sterility (NIL-S35al in Figure 1A). These observations suggested that S35-i interacts with the dominant indica allele of S24 (S24-i) (Kubo et al. 2008). Reciprocal test crossing of NIL-S35H and Asominori showed that S35-i was inherited at a higher frequency (89%) through the male gamete in the S35 heterozygotes (the expected frequency was 50%) (Table 1), indicating the selective abortion of the male gamete bearing S35-j, and the incomplete penetrance of the S35 gene. Histological observations showed that the mature anthers of the sterile NIL-S35H retained uninucleate and bicellular microspores, suggesting that developmental defects begin from the late uninucleate microspore stage after successful meiosis (Figure 2, A and B). Pollen grains arrested at later developmental stages were also found (Figure 2C). This finding suggested that evaluating some of the large arrested pollen grains as “fertile pollen” might have resulted in an overestimation of pollen fertility in the S35 heterozygotes. No apparent defects were found in the tapetum or anther walls of sterile S35-heterozygotes during male gametogenesis (Figure 2, D–L). We next performed high-resolution mapping to determine the precise position of the S35 locus. Using a large segregating population (N = 2800), the location of the S35 locus was narrowed down within a 122-kb region between marker loci 1c300–1c312 (Figure 1B). The candidate genomic region for S35 encoded 27 putative protein-coding genes and four transposable elements (Table S2). Rice transcriptome data (Fujita et al. 2010) indicated that, out of the 27 candidate genes, three genes (LOC_Os01g06460/Cys-rich domain protein, LOC_Os01g06580/fasciculin-like arabinogalactan-protein, and LOC_Os01g06590/zinc finger C3HC4 type domain protein) had relatively higher expression, or their expression significantly changed around the uninucleate stage (Figure S2), suggesting that these three genes are the most probable candidates for S35 function.
Figure 1.
Genetic basis of hybrid male sterility genes and localization of the S35 locus. (A) Pollen phenotypes of NILs for S35, S24, and EFS genes associated with hybrid male sterility. Graphical genotypes (left) and pollen grains (right) of the NILs stained by I2-KI are shown. Black and white bars denote IR24 (indica) and Asominori (japonica) chromosomes, respectively. The NIL carrying S35 alone (NIL-S35al) were fertile, but cointrogression of the S24 segment from IR24 dominantly activated S35, leading to pollen sterility in the Asominori genetic background (NIL-S35H). The S24-suppressor Efs gene from IR24 dominantly inactivated S24 (NIL-S24H+Efs). (B) Chromosome position of S35. Upper panel: Physical chromosome positions of the S35 locus and DNA markers (1cXXX). Lower panel: Diagram showing the recombination breakpoints and pollen phenotypes of the lines. All recombinant plants carried IR24 homozygous alleles for S24 to activate S35-i. The pollen fertility is shown as the mean (%) ± SD (n = 3–7).
Table 1. Transmission frequency of S35-i-bearing gametes in reciprocal test crosses between NIL-S35H and Asominori.
| Cross Combination | No. of Plantsa | Total | χ2 (1:1) | Frequency of the indica Allele (ki)b | ||
|---|---|---|---|---|---|---|
| ♀ | ♂ | japonica | Heterozygote | |||
| Asominori | NIL-S35H | 17 | 134 | 151 | 45.9*** | 0.89 |
| NIL-S35H | Asominori | 28 | 43 | 71 | 1.63NS | 0.61 |
*** P < 0.001. NS, not significant.
DNA marker 1c350 linked to S35 was used for genotyping.
The expected frequency is 0.5.
Figure 2.
Histological characterization of the sterile S35 heterozygotes. (A)–(C) Mature pollen grains stained with hematoxylin. (A) Pollen grains from Asominori. (B), (C) Pollen grains from the sterile S35 heterozygotes (NIL-S35H). White and black arrowheads indicate normal sperm cells and nuclei of abortive pollen grains, respectively. Pollen grains arrested at the bicellular stage (black arrowheads) and vacuolated pollen at the uninucleate stage are shown in (B). (C) shows that these two pollen grains from NIL-S35H were well filled and similar in size, but the pollen grain on the right has a single bold nucleus instead of two sperm cells as would be observed in normal pollen grains. (D)–(M) Transverse sections showing anther development of Asominori and the S35 heterozygote (NIL-S35H). Five stages of anther development in Asominori and NIL-S35H were compared. Transverse sections were stained with 0.05% Toluidine blue O. (D)–(H) are Asominori, and (I)–(M) are NIL-S35H. (D) and (I) the pollen mother cell stage, (E) and (J) the tetrad stage, (F) and (K) the young microspore stage, (G) and (L) vacuolated pollen stage, (H) and (M) the mature pollen stage. The solid black arrows indicate degenerated pollen grains. PMC, pollen mother cell; T, tapetal layer; Td, tetrad cell; Msp, microspore; Mp, mature pollen. Scale bars = 50 µm for (A)–(M).
Pollen sterility due to S35 is independent of EFS
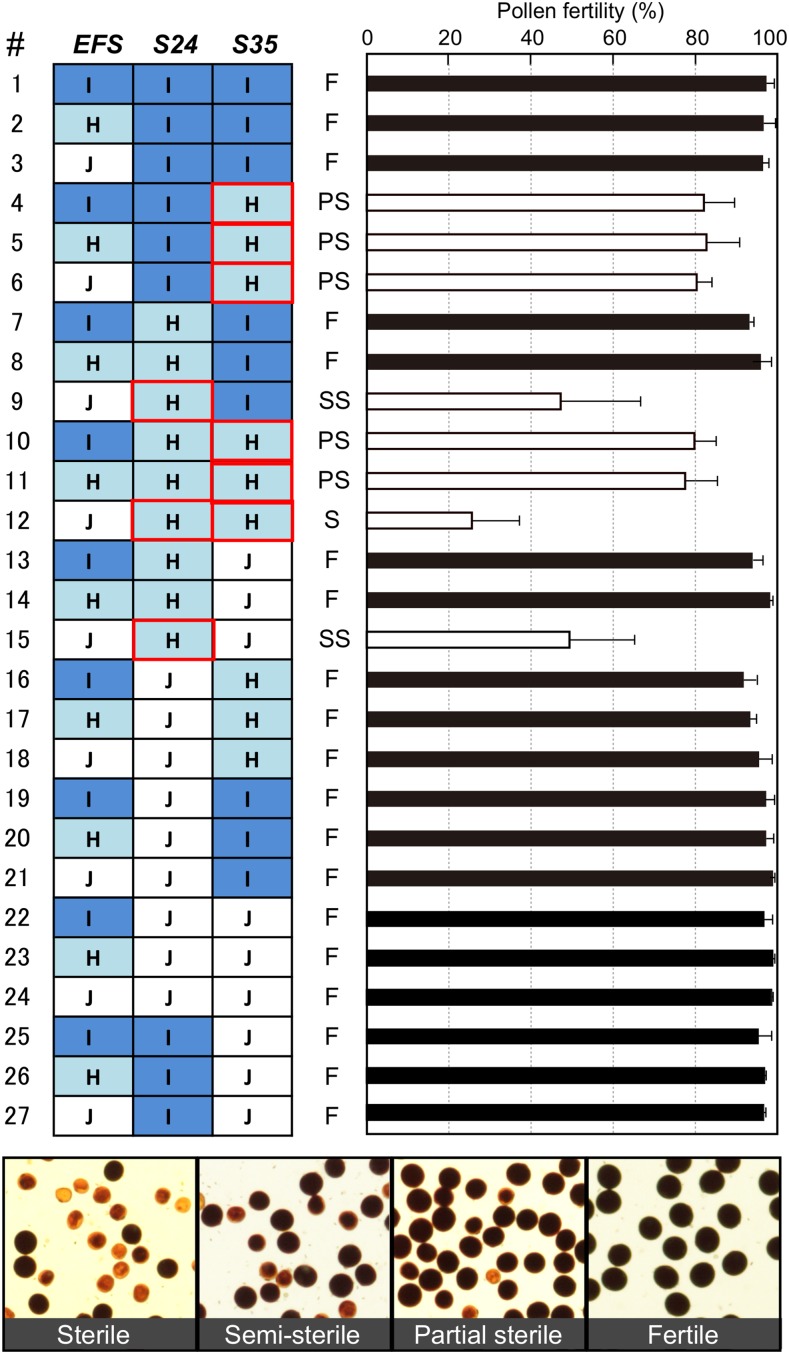
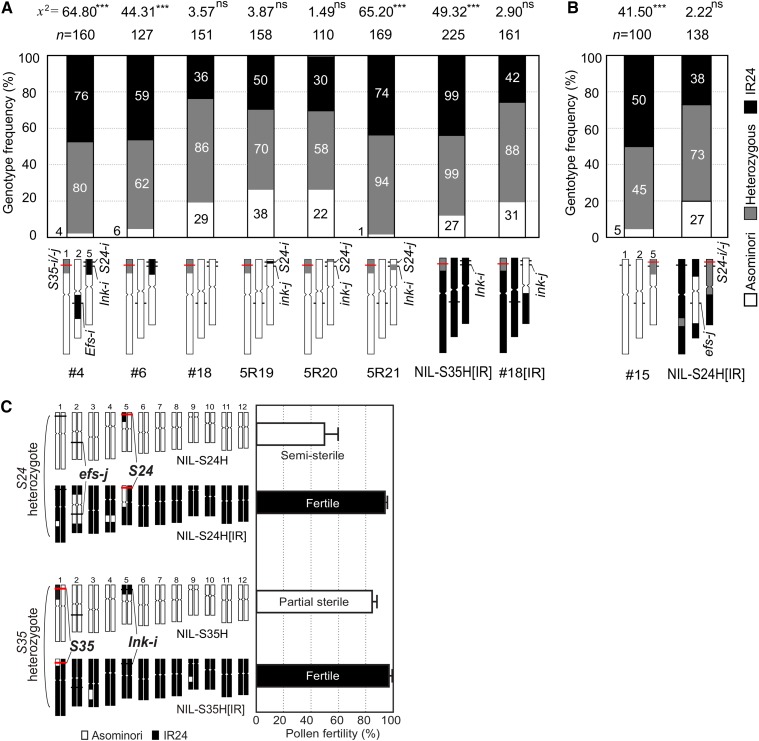
The EFS locus was previously identified as a suppressor of S24; the dominant indica allele Efs-i restores pollen sterility due to S24 (NIL-S24H+Efs in Figure 1A) (Kubo et al. 2011). A direct or indirect interaction between EFS and S35 was presumed based on the previously identified genetic interactions (S24–S35 and EFS–S24) (Kubo et al. 2008, 2011). To examine whether the S35 and EFS genes interact with each other, we phenotyped all 27 allele combinations at the three loci S24, EFS, and S35. The self-pollinated progeny of triple heterozygous plants for S24, EFS, and S35 were used to evaluate the 27 genotypes. Consistent with the previous results (Kubo et al. 2008, 2011), S35 caused pollen sterility only in the presence of the S24-i allele (#6 and #18 in Figure 3), and Efs-i restored the S24-dependent pollen sterility (#13–#15 in Figure 3). However, contrary to our expectations, the heterozygous S35 caused partial pollen sterility regardless of the EFS genotype (#4–#6 in Figure 3). This result indicated that Efs-i is unable to suppress S35, and, therefore, the S24-specific suppressor. Thus, S35 required the IR24 segment harboring S24 to induce pollen sterility, but was independent of EFS. It is worth emphasizing that S24-induced sterility was not affected by the S35 genotype (comparison between #9 and #15 in Figure 3), as previously shown (Kubo et al. 2008).
Figure 3.
Pollen fertility of 27 genotypes determined by three genes S24, S35, and EFS. Pollen fertility (right bar chart) for each genotype class (left panel) is expressed by the mean (%) ± SD (n = 3–10). Effective genes for pollen sterility are marked with red frames on the genotype panel. White and black bars in the bar chart represent the sterile and fertile pollen phenotypes, respectively. Micrographs of the pollen grains for each fertility class are shown in the bottom panel. Plants of each genotype were selected from self-pollinated progeny of NIL-S24+S35+Efs by genotyping using the linked markers mS2 for S24, 1c305 for S35, and 2c2015 for EFS. I: indica (IR24) homozygote. J, japonica (Asominori) homozygote; H, heterozygote; F, fertile; PS, partial-sterile; SS, semisterile; S, sterile.
Genetic dissection of the S24 region and an activator of S35
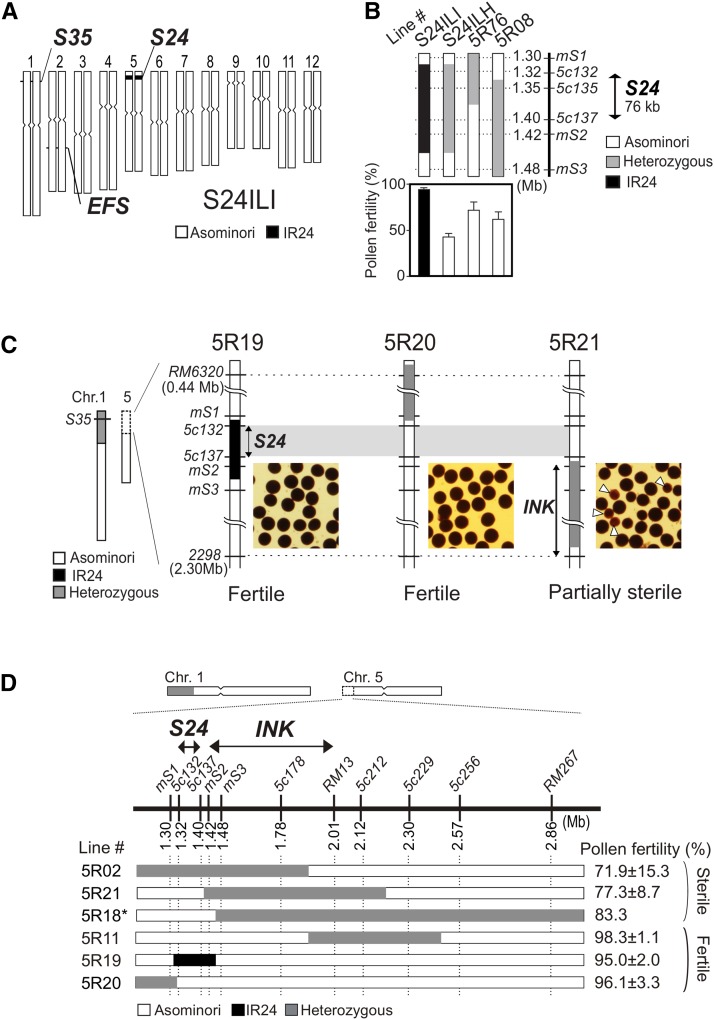
For further characterization of S24 as the pollen killer by itself and as the partner of S35, we genetically dissected the S24 region using a segregating population (n = 485). We obtained a NIL of S24 (called S24ILH/I) carrying a very small segment (181-kb) near the S24 region with an otherwise uniform Asominori genetic background (Figure 4, A and B). The heterozygous segment of S24ILH was able to induce semisterility (43.1%) in the pollen, indicating that the small segment contained the S24 locus (Figure 4B). We next reexamined the S24–S35 interaction by analyzing the F2 and F3 individuals derived from a cross of S24ILI × NIL-S35al. From the self-pollinated progeny of an F1 plant, a plant homozygous for S24-i, and heterozygous for S35, was selected, and the pollen phenotype of its selfed progeny carrying the same parental genotype (5R19) was investigated (Figure S1B). If interaction between S24 and S35 occurred, the 5R19 would have sterile pollen due to the heterozygous S35-i/S35-j genotype. In contrast, the results indicated that 5R19 plants had good pollen fertility (95.0% fertility) (Figure 4, C and D), and the segregation pattern of S35 fit a 1:2:1 ratio in the selfed progeny (Figure 5A). These results indicated that S35 does not interact with S24, but rather with another locus linked to S24. In a similar way, we investigated the effects of other nearby segments on S35-dependent pollen sterility. A distal segment adjacent to the S24 locus had no effect on pollen fertility (5R20 in Figure 4, C and D, and Figure 5A). On the other hand, another centromere-side segment resulted in partial sterility for pollen due to S35 (5R21, 77.3% in Figure 4, C and D) and selective abortion of the S35-j gamete in the selfed progeny (χ21:2:1 = 65.20, P < 0.001, Figure 5A). Thus, we were able to confirm that not S24 but the S24-linked gene is essential for S35 activation. This finding was consistent with the aforementioned result that EFS did not suppress S35 and S24-induced pollen semisterility regardless of the S35 genotype. The newly identified S35-activator locus was designated as incentive for killing pollen (INK). We next genetically dissected INK using 485 individuals. The INK locus was localized within a 592-kb region between marker loci mS2 and RM13 (Figure 4D). These results conclusively showed that the dominant Ink-i gene located close to S24 was required to induce S35-dependent pollen sterility.
Figure 4.
Identification and localization of the S35-activator. (A) Graphical genotype of the near-isogenic line S24ILI. S24ILI was homozygous for S24 and carried a very small IR24 segment of less than 181-kb (mS1–mS3, see also Figure 4B) within a uniform Asominori background. (B) Pollen phenotype of the most informative recombinants and the NILs S24ILI/H. These lines carried Asominori homozygous alleles for both the EFS and S35 loci, representing the pollen sterility dependent exclusively on S24. Bars show the mean (%) with SD (n = 4–9). (C) Pollen fertility of a set of substitution lines covering the S24 region on chromosome 5. All the lines were heterozygous for S35. The white arrows in the photo indicate degenerated pollen grains. (D) Chromosome position of the S35-activator INK locus on chromosome 5. The diagram shows the recombination breakpoints of the obtained plants/lines. All plants/lines carried heterozygous alleles for S35 to evaluate the effect of the recombinant segment on the S35 phenotype. The pollen phenotype of each line was determined using selfed progeny (BC4F3) of the recombinant individuals (BC4F2) excluding 5R18. 5R18 was a single BC5F1 plant derived from a backcross of the recombinant BC4F2 plant carrying homozygous S35-i alleles. The 5R02 line had the Efs-i homozygous genotype that prevented the influence of heterozygous S24 on pollen fertility. The mean (%) ± SD (n = 3–10) are shown on the right.
Figure 5.
Effect of the genetic background on pollen sterility. (A), (B) Frequencies of the S35 and S24 genotypes in the selfed progeny of the NILs with reciprocal genetic backgrounds. The stacked bar chart represents the frequency of the S35 genotype (A) and the S24 genotype (B) in the selfed progeny of the heterozygotes. Population size and chi-square values for the deviation from 1:2:1 are shown on the top. The numbers of plants in each genotype (black, IR24 homozygote; gray, heterozygote; white, Asominori homozygote) are shown inside the bars. The parental genotype is shown at the bottom. DNA markers 1c350 or 1c305 (for S35), and mS2 (for S24) were used for genotyping (the red line on chromosomes). The plant line ID is identical to that shown in Figure 3, Figure 4, and Figure 6. Parental plants #4, #6, #15, and 5R21 had sterile pollen. All other plants had fertile pollen. ** P < 0.01; *** P < 0.001; ns, not significant. (C) Graphical genotype (left) and pollen fertility (right) of the NILs that carried heterozygous segments harboring the S24 or S35 locus in either the Asominori or the IR24 genetic backgrounds. The NILs carried the appropriate genotype for the interacting partner gene (efs-j/efs-j for the S24 activation, or Ink-i/Ink-i for the S35 activation). Bars show the mean (%) with SD (n = 5).
Epistasis involving multiple genes controls pollen killers
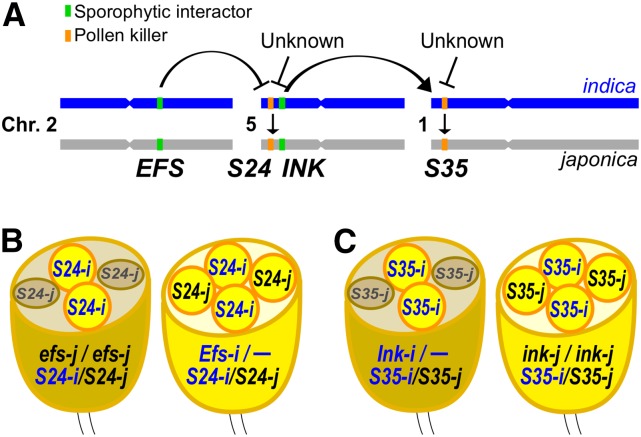
As described above, we genetically dissected the S35 and S24 genes in the japonica genetic background. Through this analysis, we obtained clear evidence for two independent genetic pathways, EFS–S24 and INK–S35. The question then arises as to whether these genes comprise the full set of genes involved in the pollen killer system. To address this question, we examined another set of NILs with an indica genetic background that have a nearly opposite genomic constitution. We developed two NILs, called NIL-S24H[IR] and NIL-S35H[IR] that carried S24 or S35 heterozygous alleles with the indica genetic background, respectively. Unlike the japonica background populations, pollen semisterility due to heterozygous S24 did not occur in the IR24 genetic background (NIL-S24H[IR] in Figure 5C). Consistent with this normal pollen phenotype, a normal Mendelian segregation of S24 was observed in the IR24 genetic background population (χ21:2:1 = 2.22, P = 0.33) (Figure 5B). Also, the pollen fertility of the S35-i/S35-j heterozygotes was not significantly reduced in the IR24 genetic background (NIL-S35H[IR] in Figure 5C). Despite the fertile pollen phenotype, a reduced transmission of the S35-j allele was observed in the selfed progeny of NIL-S35H[IR] (Figure 5A). The frequency of the S35-j homozygote was slightly recovered in the IR24 genetic background (27/225 = 12.0%, in NIL-S35H[IR]) compared with that in the japonica background (2.5–4.7% in #4 and #6). Interestingly, the S35 heterozygote with recessive homozygous ink-j alleles (S35 held inactive) had a higher level of S35-j transmission {the S35-j homozygote emerged at 19.3% (31/161) frequency in the selfed progeny of #18[IR], Figure 5A}. Therefore, we considered that the effect of S35 was slightly diminished, but S35 still remained as a causal agent in the defective fertilization of S35-j pollen in the IR24 genetic background, even though no remarkable visible defects were found in their pollen grains when stained with the I2-KI solution. Genotype frequency data of a recombinant inbred (RI) population reported by Huang et al. (2009) supported these results. In the RI population derived from the cross between Nipponbare (japonica) × 93-11 (indica), the genotype frequency distribution peaked at 3.1 Mb on chromosome 1, where S35 is located (Huang et al. 2009). The japonica homozygotes emerged at a reduced frequency [II:JJ = 115:33 (22%), the expected Mendelian ratio is 1:1 (50%), in Figure S3A), which was slightly higher than our data based on the japonica background populations [the expected value was estimated as II:JJ = 132:16 (11%) when the transmission frequency of the indica S35 allele (ki = 0.89, Table 1) was applied to the RI population]. Equal segregation of S24 in the RI population was also in good agreement with our data obtained from NIL-S24[IR]. In total, these results indicated that S35 was active in the indica background population as well as in the F1 hybrids, and also suggested that some minor modifier(s) existed in the IR24 genome. Furthermore, our results indicated that the action of S24 was repressed by several suppressors encoded by the IR24 genome, including EFS (Figure 6A).
Figure 6.
Diagram showing the genetic interactions between hybrid male sterility loci. (A) Schematic representation of the chromosome position and genetic interactions of hybrid male sterility genes. Arrows and bars represent positive and negative regulation, respectively. Indica has the killer alleles for the S24 and S35 loci (S24-i and S35-i), and these genes kill the male gamete bearing the japonica alleles in heterozygous hybrid progeny. Activation of these pollen killer genes is dependent on the sporophytic factors EFS for S24 (B) and INK for S35 (C). Additional unknown suppressors/modifiers for S24 and S35 exist within the indica genome.
Discussion
Genetic mechanism of the pollen killer system
Although we hypothesized that S35 caused pollen sterility via interaction with the S24 locus (Kubo et al. 2008), the present study revealed that not S24, but a tightly linked locus INK, activates S35 to cause pollen sterility. The indica Ink-i allele dominantly activated S35, and, conversely, S35 had no harmful effect on reproductive or vegetative development in japonica homozygous for ink-j/ink-j. The Ink-i allele did not have a considerable effect on the activity of S24 because S24 caused pollen sterility without Ink-i. The independence of S24 and S35 was consistent with the result that the S24-suppressor Efs-i did not restore pollen sterility caused by S35. Consequently, we concluded that INK–S35 and EFS–S24 are independent pollen killer systems, rather than a single pathway of EFS–S24–S35 (Figure 6B). In addition to these genes, the present study indicated the presence of additional modifier/suppressor(s) in the genetic background. A few minor modifiers for S35, and a major suppressor for S24, were suggested as the causes of phenotypic changes in the indica background. These findings suggest that the plant gamete killer system is regulated by multiple genetic networks involving linked and unlinked multiple genes with major and minor effects.
Candidate genes and putative functions of S35 and INK
In this study, the S35 locus was delimited within a 3.00–3.12 Mb region of chromosome 1 that contained many putative protein-coding genes. Within this chromosomal region (3.04–3.11 Mb), a hybrid male sterility locus S-d was found by using other indica/japonica cross combinations (Li et al. 2008). Since S35 and S-d were located within the same narrow interval, these are hypothesized to be identical genes. Allelic differentiation of S24 and S35 seemed to contribute significantly to pollen sterility in the indica-japonica cross because both loci were found in different cross combinations of the indica and japonica varieties (Wang et al. 2006; Li et al. 2006, 2008). It is of great interest to understand the function of these genes in the parental species, as well as the harmful effects in the heterozygous hybrids. The S35 locus caused developmental defects after the late uninucleate microspore stage (Figure 2A), suggesting that the causal gene is expressed and functions around this stage. The uninucleate stage is followed by two mitotic cell divisions in which many different genes are expressed, and dynamic changes in expression patterns occur, especially during the bicellular–tricellular transition (Fujita et al. 2010). Based on the expression profiles during male gametogenesis in rice, we found three candidate genes for S35 whose expression increased or changed significantly near the uninucleate stage (i.e., Cys-rich domain protein/LOC_Os01g06460, fasciclin-like arabinogalactan-protein/LOC_Os01g06580, and zinc finger C3HC4 type domain protein/LOC_Os01g06590 in Figure S2). Protein sequence polymorphisms were found in the Cys-rich domain protein and the fasciclin-like arabinogalactan-protein based on the indica variety 93-11 (data not shown). Therefore, these two genes are strong candidates for S35/S-d. It is noteworthy that a rice mutant study has revealed that a fasciclin glycoprotein (Microspore and Tapetum Regulator 1, MTR1) is essential for male gametogenesis in rice (Tan et al. 2012). As is the case with the S-d mapping study (N = 2160), no recombination events were found within the 122-kb candidate interval of the S35 locus, suggesting a suppression of recombination within this region (a total of about 5000 individuals were screened for recombination events in the S35/S-d region in these two studies). A large chromosome insertion (∼36 kb) and many SNPs were found in the S35/S-d candidate region of the indica variety 93-11 (data not shown).
The INK locus was localized within a 592-kb region where 72 putative protein-encoding genes, and 20 transposable elements were found based on the Nipponbare genome sequence. No identical or similar genomic fragments were found between the S35 and INK candidate regions. The INK–S35 interaction fits well with an “epistasis-based allelic interaction model” that hypothesizes that the pollen killer system functions by interaction between sporophytic and gametophytic genes, as proposed previously (Kubo et al. 2011). In the case of INK–S35, INK is the sporophytic factor (Figure 6C). Therefore, the INK gene is predicted to be expressed in sporopytic tissues such as tapetal and anther wall layers. Since tapetal cells supply the major components of the pollen wall and nutrition requisite for pollen development, interaction between tapetal cells and microspore cells is essential for normal pollen development (Tan et al. 2012). Thus, the causal molecule encoded by INK may be involved in such a cooperative interaction between sporophytic and gametophytic gene products. This situation resembles the SaF-SaM male sterility system (Long et al. 2008). In that system, two genes, SaF and SaM, interact with each other to cause male sterility in an indica/japonica rice hybrid. The indica SaF+ allele acts sporophytically, whereas the japonica SaM− allele acts as the gametophytic determinant for the selective pollen killing. A physical protein interaction of SaF+ with SaM− but not with SaM+ was demonstrated (Long et al. 2008). Further cloning analyses will provide valuable information on the molecular interaction of INK–S35, and the molecular pathway underlying hybrid male sterility.
Properties of the S24 and S35 gene blocks
In this study, we examined the effect of the S24–INK gene block on chromosome 5, focusing on pollen sterility. Interestingly, a number of studies have detected a major QTL for seed set percentage on the S24–INK region in several indica/japonica populations (Li et al. 1997; Wang et al. 1998; Chin et al. 2011). More recently, Zhao et al. (2007) identified a hybrid female sterility locus S31 at 1.29–1.48 Mb within the S24–INK region by using the same Asominori-IR24 CSSLs. We expect that S31 will facilitate a transmission advantage of the indica S24–INK haplotype through the female gametes. Intriguingly, however, no remarkable phenotype for the S31 gene has been found in our growth conditions (Fukuoka and Mishima in Japan), although S31 caused partial female sterility (about 65%) when the same CSSLs were grown in Nanjing, China (Zhao et al. 2007). This inconsistent gene action may be explained by differences in the environments where the experimental populations were grown. If so, this observation means that S31 is strongly influenced by environment and can enhance the transmission of the S24–S31–INK gene complex under certain conditions. In terms of pollen sterility due to S24 and S35, some variations were found within and among the NIL populations (Figure 1, Figure 3, and Figure 4). Phenotypic variation can result from gene–gene and gene–environment interactions. Linked or unlinked modifier gene(s) located in retained segments in the genetic background is a possible genetic element for phenotypic variation. A straightforward example of this can be found in the difference in pollen fertility among NILs for S24 (Figure 4B). The pollen fertility of S24ILH with the smallest segment harboring the S24 locus was lower than that of other recombinant lines (5R76 and 5R08), implying the presence of linked minor gene(s) for male-fertility restoration. Also, the S35 locus showed some phenotypic differences (65–85%) between the NIL populations (Figure 1B and Figure 3). Environmental effects for pollen sterility due to S35 were seen under different growth conditions [54.3 ± 7.3% in Fukuoka in Kubo et al. (2008); 84.2 ± 4.1% in Mishima in this study]. This environmental susceptibility may be a common feature of genes conferring male sterility in plants (Sakata et al. 2010; Ding et al. 2012; Wang et al. 2013; Kubo et al. 2016).
The impact of pollen killer genes on the genetic characteristics of hybrid offspring
The actions of the S35 and S24 killers were exclusively associated with their heterozygotes. Homozygotes for either alleles did not show any notable phenotype in reproductive development or vegetative growth, excluding phenotypic changes by other linked genes. Both pollen sterile S24-heterozygotes and S35-heterozygotes exhibited nearly full seed set (93.3% ± 3.2, n = 10, in S24ILH; 88.4% ± 0.7, n = 3, in NIL-S35H), suggesting that these pollen killers have a transmission advantage for increasing their frequencies with no or a small effect on individual fecundity, at least in the japonica genetic background. But, as it is, such a strong transmission advantage cannot directly apply to early segregating generations of the indica-japonica hybrid progeny. In view of the results obtained in the RI and the indica background populations, S35-i may weaken its function of eliminating S35-j (japonica allele) but may still keep the killing action to some degree. In contrast, S24 contributed little to the reduction of pollen fertility in the F1 generation. The action of S24 is substantially suppressed by Efs-i and other suppressor(s).
Using indica-japonica hybrid populations, Hiko-Ichi Oka demonstrated that recombination of two independent genes tended to be restricted as if they were linked (referred to as pseudo-linkage or quasi-linkage) (Oka 1988). The same tendency was also observed in O. sativa × Oryza glaberrima (Sano et al. 1980). An example is the inheritance of Phenol reaction (Ph) and Chromogen for anthocyanin (C) located on separate chromosomes. The F2 population of indica (Ph–c) × japonica (ph–C) had excessive numbers of the parental genotypes (Ph–c and ph–C), and reduced numbers of recombined genotypes (Ph–C and ph–c) (Oka 1988). In this study, Oka pointed out that a gene set for duplicate gametic lethals is one possible cause for the pseudo-linkage of the phenotypic markers. Theoretically, the INK–S35 interaction could also partially contribute to pseudo-linkage. The pollen killer S35-i (indica allele) increases its own transmission with the existence of the indica Ink-i allele through the male gamete, resulting in a high frequency of the indica–indica pair for the S35 and INK alleles (S35-i–Ink-i) and a reduction of the recombinant pair of the japonica–indica alleles (S35-j–Ink-i). By this means through male gametes, the parental set of different chromosomes (chromosomal segments) will gradually increase their frequency in the population. Some of this tendency was observed in the two RI populations of Nipponbare × 93-11 (Huang et al. 2009), and Asominori × IR24 (Tsunematsu et al. 1996) (Figure S3B). Thus, several types of hybrid sterility genes may contribute to pseudo-linkage of many chromosomal regions because more than 40 hybrid sterility genes are dispersed throughout the rice genome.
Evolutionary implications of pollen killers
In the traditional triallelic model, both the gamete killer and the sensitive alleles can arise from the original neutral allele without a reduction in fitness, and consequently become fixed in diverging populations. Recent studies of the Sa and S5 loci have afforded tangible evidence of the molecular and the evolutionary mechanisms (Long et al. 2008; Yang et al. 2012). These loci contain multiple linked genes. Presumably, sequential mutations within these genes occur in either or both lineages, and these divergent haplotypes negatively interact, resulting in gamete dysfunction in the heterozygotes (Ouyang and Zhang 2013). This sequential divergence theory could be applicable to the pollen killer systems by S35 and S24, despite the differences in physical positions of the interacting partner genes. Pollen killing can occur only if the pollen killers are paired with particular alleles at other interacting loci in the hybrid offspring. Such a polygenic system allows the accumulation of mutations at related loci without a significant fitness cost during evolution. In fact, an increasing number of studies have suggested that a gradual accumulation of multiple epistatic genes with no, or a small, individual effect may play important roles in the development of reproductive isolation in initial speciation (Cabot et al. 1994; Orr 1995; Kao et al. 2010; Kubo et al. 2016). In addition, the existence of third neutral alleles for S24 and S35 have been reported in studies using numerous cultivars and the wild progenitor Oryza rufipogon (Wang et al. 2006; Shi et al. 2009; Liu et al. 2011). This observation implies that these pollen killer loci may contain tightly linked multiple genes like S5 and Sa, or may be pure trialleles of a single gene. Our study demonstrated the impact of the gene complex S24–S31–INK on hybrid sterility between indica and japonica. Plant species have evolved mainly through repeated duplication of an ancestral genome, chromosome and/or genes. Redundancy of gene sets in plant genomes is important as a prerequisite for diversification of genes adapting to environmental changes. Furthermore, most plants have the ability to reproduce asexually and via autogamy. Therefore, coadapted gene complexes can be maintained over many generations without breakdown by reproductive meiosis and meiotic recombination. These genomic and reproductive characteristics of plants should facilitate the formation of polymorphic gene complexes for reproductive isolation (Dempewolf et al. 2012).
Plant genetic studies have traditionally focused on gamete killer loci. Even if interacting partner genes exist in a genetic background, these partners would have largely gone unnoticed. Though much still remains to be elucidated, it is clear that many genetic factors, and a genetic network, play a pivotal role in the plant gamete killer system. Tracking down the genes one by one can, therefore, shed further light on the molecular and evolutionary mechanisms of gamete killer systems. Knowledge of the chromosomal locations and the nature of gamete killer loci will aid the design of efficient marker-assisted schemes for crop breeding.
Supplementary Material
Acknowledgments
We thank T. Makino, Y. Gonohe, and H. Kondo (National Institute of Genetics, Mishima, Japan) for technical assistance. This study was partly supported by JSPS KAKENHI grant number 25450012 (to T.K.).
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.027573/-/DC1
Communicating editor: K. Dawe
Literature Cited
- Buckler E. S., Phelps-Durr T. L., Buckler C. S., Dawe R. K., Doebley J. F., et al. , 1999. Meiotic drive of chromosomal knobs reshaped the maize genome. Genetics 153: 415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A., Trivers R., 2009. Genes in conflict: the biology of selfish genetic elements, Harvard University Press, Cambridge, MA. [Google Scholar]
- Cabot E. L., Davis A. W., Johnson N. A., Wu C. I., 1994. Genetics of reproductive isolation in the Drosophila simulans clade: complex epistasis underlying hybrid male sterility. Genetics 137: 175–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron D. R., Moav R. M., 1957. Inheritance in Nicotiana tabacum XXVII. Pollen killer, an alien genetic locus inducing abortion of microspores not carrying it. Genetics 42: 326–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin J. H., Chu S.-H., Jiang W., Cho Y.-I., Basyirin R., et al. , 2011. Identification of QTLs for hybrid fertility in inter-subspecific crosses of rice (Oryza sativa L.). Genes Genomics 33: 39–48. [Google Scholar]
- Dempewolf H., Hodgins K. a., Rummell S. E., Ellstrand N. C., Rieseberg L. H., 2012. Reproductive isolation during domestication. Plant Cell 24: 2710–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J., Lu Q., Ouyang Y., Mao H., Zhang P., et al. , 2012. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc. Natl. Acad. Sci. USA 109: 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T. R., 2015. Gametocidal Genes, pp. 121–131 in Alien Introgression in Wheat: Cytogenetics, Molecular Biology, and Genomics, edited by Molnár-Láng M., Ceoloni C., Doležel J. Springer, Berlin. [Google Scholar]
- Finseth F. R., Dong Y., Saunders A., Fishman L., 2015. Duplication and adaptive evolution of a key centromeric protein in Mimulus, a genus with female meiotic drive. Mol. Biol. Evol. 32: 2694–2706. [DOI] [PubMed] [Google Scholar]
- Fishman L., and Saunders A., 2008 Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science 322: 1559–1562. [DOI] [PubMed]
- Fujita M., Horiuchi Y., Ueda Y., Mizuta Y., Kubo T., et al. , 2010. Rice expression atlas in reproductive development. Plant Cell Physiol. 51: 2060–2081. [DOI] [PubMed] [Google Scholar]
- Huang X., Feng Q., Qian Q., Zhao Q., Wang L., et al. , 2009. High-throughput genotyping by whole-genome resequencing. Genome Res. 19: 1068–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikehashi H., Araki H., 1986. Genetics of F1 sterility in remote crosses of rice, pp. 119–130 in Rice Genetics, edited by Khush G. S.International Rice Research Institute, Los Baños, Philippines. [Google Scholar]
- International Rice Genome Sequencing Project , 2005. The map-based sequence of the rice genome. Nature 436: 793–800. [DOI] [PubMed] [Google Scholar]
- Kanizay L. B., Albert P. S., Birchler J. A., Dawe R. K., 2013. Intragenomic conflict between the two major knob repeats of maize. Genetics 194: 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao K. C., Schwartz K., Sherlock G., 2010. A genome-wide analysis reveals no nuclear Dobzhansky-Muller pairs of determinants of speciation between S. cerevisiae and S. paradoxus, but suggests more complex incompatibilities. PLoS Genet. 6: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindiger B., Beckett J. B., 1985. A hematoxylin staining procedure for maize pollen grain chromosomes. Biotech. Histochem. 60: 265–269. [DOI] [PubMed] [Google Scholar]
- Koide Y., Onishi K., Kanazawa A., Sano Y., 2008. Genetics of Speciation in Rice, pp. 247–259 in Rice Biology in the Genomics Era. Biotechnology in Agriculture and Forestry, edited by Hirano H., Sano Y., Hirai A., Sasaki T. Springer-Verlag, Berlin, Heidelberg. [Google Scholar]
- Kovach M. J., Sweeney M. T., McCouch S. R., 2007. New insights into the history of rice domestication. Trends Genet. 23: 578–587. [DOI] [PubMed] [Google Scholar]
- Kubo T., Aida Y., Nakamura K., Tsunematsu H., Doi K., et al. , 2002. Reciprocal chromosome segment substitution series derived from Japonica and Indica cross of rice (Oryza sativa L.). Breed. Sci. 52: 319–325. [Google Scholar]
- Kubo T., Yamagata Y., Eguchi M., Yoshimura A., 2008. A novel epistatic interaction at two loci causing hybrid male sterility in an inter-subspecific cross of rice (Oryza sativa L.). Genes Genet. Syst. 83: 443–453. [DOI] [PubMed] [Google Scholar]
- Kubo T., Yoshimura A., Kurata N., 2011. Hybrid male sterility in rice is due to epistatic interactions with a pollen killer locus. Genetics 189: 1083–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T., Takashi T., Ashikari M., Yoshimura A., Kurata N., 2016. Two tightly linked genes at the hsa1 locus cause both F1 and F2 hybrid sterility in rice. Mol. Plant 9: 221–232. [DOI] [PubMed] [Google Scholar]
- Larracuente A. M., Presgraves D. C., 2012. The selfish segregation distorter gene complex of Drosophila melanogaster. Genetics 192: 33–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zeng R., Zhang Z., Ding X., Zhang G., 2006. Fine mapping of locus S-b for F1 pollen sterility in rice (Oryza sativa L.). Chin. Sci. Bull. 51: 675–680. [Google Scholar]
- Li W., Zeng R., Zhang Z., Ding X., Zhang G., 2008. Identification and fine mapping of S-d, a new locus conferring the partial pollen sterility of intersubspecific F1 hybrids in rice (Oryza sativa L.). Theor. Appl. Genet. 116: 915–922. [DOI] [PubMed] [Google Scholar]
- Li Z., Pinson S. R. M., Paterson A. H., Park W. D., Stancel J. W., 1997. Genetics of hybrid sterility and hybrid breakdown in an intersubspecific rice (Oryza sativa L.) populations. Genetics 145: 1139–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Li J. Q., Liu X. D., Shahid M. Q., Shi L. G., et al. , 2011. Identification of neutral genes at pollen sterility loci Sd and Se of cultivated rice (Oryza sativa) with wild rice (O. rufipogon) origin. Genet. Mol. Res. 10: 3435–3445. [DOI] [PubMed] [Google Scholar]
- Londo J. P., Chiang Y.-C., Hung K.-H., Chiang T.-Y., Schaal B. A., 2006. Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc. Natl. Acad. Sci. USA 103: 9578–9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Y., Zhao L., Niu B., Su J., Wu H., et al. , 2008. Hybrid male sterility in rice controlled by interaction between divergent alleles of two adjacent genes. Proc. Natl. Acad. Sci. USA 105: 18871–18876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon M. F., 2003. Transmission ratio distortion in mice. Annu. Rev. Genet. 37: 393–408. [DOI] [PubMed] [Google Scholar]
- McCouch S. R., Teytelman L., Xu Y., Lobos K. B., Clare K., et al. , 2002. Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res. 9: 199–207. [DOI] [PubMed] [Google Scholar]
- Mizuta Y., Harushima Y., Kurata N., 2010. Rice pollen hybrid incompatibility caused by reciprocal gene loss of duplicated genes. Proc. Natl. Acad. Sci. USA 107: 20417–20422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishima H., 2001. Evolution and domestication of rice, pp. 63–77 in Rice Genetics IV, edited by Khushu G. S., Brar D. S., Hardy B. International Rice Research Institute, Los Baños, Philippines. [Google Scholar]
- Oka H., 1988. Origin of cultivated rice, Japan Scientific Societies Press, Tokyo, Japan. [Google Scholar]
- Orr H. A., 1995. The population genetics of speciation: the evolution of hybrid incompatibilities. Genetics 139: 1805–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Y., Zhang Q., 2013. Understanding reproductive isolation based on the rice model. Annu. Rev. Plant Biol. 64: 111–135. [DOI] [PubMed] [Google Scholar]
- Ouyang Y., Chen J., Ding J., Zhang Q., 2009. Advances in the understanding of inter-subspecific hybrid sterility and wide-compatibility in rice. Chin. Sci. Bull. 54: 2332–2341. [Google Scholar]
- Rick C. M., 1966. Abortion of male and female gametes in the tomato determined by allelic interaction. Genetics 53: 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata T., Oshino T., Miura S., Tomabechi M., Tsunaga Y., et al. , 2010. Auxins reverse plant male sterility caused by high temperatures. Proc. Natl. Acad. Sci. USA 107: 8569–8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano Y., Chu Y., Oka H., 1980. Genetic studies of speciation in cultivated rice. 2. Character variations in backcross derivatives between Oryza sativa and O. graberrima: MV linkage and key. Jpn. J. Genet. 55: 19–39. [Google Scholar]
- Shi L., Liu X., Liu B., Zhao X., Wang L., et al. , 2009. Identifying neutral allele Sb at pollen-sterility loci in cultivated rice with Oryza rufipogon origin. Chin. Sci. Bull. 54: 3813–3821. [Google Scholar]
- Tan H., Liang W., Hu J., Zhang D., 2012. MTR1 encodes a secretory fasciclin glycoprotein required for male reproductive development in rice. Dev. Cell 22: 1127–1137. [DOI] [PubMed] [Google Scholar]
- Tsunematsu H., Yoshimura A., Harushima Y., Nagamura Y., Kurata N., et al. , 1996. RFLP framework map using recombinant inbred lines in rice. Breed. Sci. 46: 279–284. [Google Scholar]
- Wang G. W., He Y. Q., Xu C. G., Zhang Q., 2006. Fine mapping of f5-Du, a gene conferring wide-compatibility for pollen fertility in inter-subspecific hybrids of rice (Oryza sativa L.). Theor. Appl. Genet. 112: 382–387. [DOI] [PubMed] [Google Scholar]
- Wang H., Lu Y., Jiang T., Berg H., Li C., et al. , 2013. The Arabidopsis U-box/ARM repeat E3 ligase AtPUB4 influences growth and degeneration of tapetal cells, and its mutation leads to conditional male sterility. Plant J. 74: 511–523. [DOI] [PubMed] [Google Scholar]
- Wang J., Liu K. D., Xu C. G., Li X. H., Zhang Q., 1998. The high level of wide-compatibility of variety “Dular” has a complex genetic basis. Theor. Appl. Genet. 97: 407–412. [Google Scholar]
- Weichenhan D., Kunze B., Winking H., van Geel M., Osoegawa K., et al. , 2001. Source and component genes of a 6–200 Mb gene cluster in the house mouse. Mamm. Genome 12: 590–594. [DOI] [PubMed] [Google Scholar]
- Yang J., X. Zhao, K. Cheng, H. Du, Y. Ouyang, et al., 2012 A killer-protector system regulates both hybrid sterility and segregation distortion in rice. Science 337: 1336–1340. [DOI] [PubMed]
- Yu J., Hu S., Wang J., Wong G. K., Li S., et al. , 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92. [DOI] [PubMed] [Google Scholar]
- Zhao Z. G., Jiang L., Zhang W. W., Yu C. Y., Zhu S. S., et al. , 2007. Fine mapping of S31, a gene responsible for hybrid embryo-sac abortion in rice (Oryza sativa L.). Planta 226: 1087–1096. [DOI] [PubMed] [Google Scholar]
- Zhao Z. G., Zhu S. S., Zhang Y. H., Bian X. F., Wang Y., et al. , 2011. Molecular analysis of an additional case of hybrid sterility in rice (Oryza sativa L.). Planta 233: 485–494. [DOI] [PubMed] [Google Scholar]
- Zhu Q., Ge S., 2005. Phylogenetic relationships among A-genome species of the genus Oryza revealed by intron sequences of four nuclear genes. New Phytol. 167: 249–265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.