Summary
PIWI-interacting RNAs (piRNAs) play a crucial role in transposon silencing in animal germ cells. In piRNA biogenesis, single-stranded piRNA intermediates are loaded into PIWI-clade proteins and cleaved by Zucchini/MitoPLD, yielding precursor piRNAs (pre-piRNAs). Pre-piRNAs that are longer than the mature piRNA length are then trimmed at their 3′ ends. Although recent studies implicated the Tudor domain protein Papi/Tdrkh in pre-piRNA trimming, the identity of Trimmer and its relationship with Papi/Tdrkh remain unknown. Here, we identified PNLDC1, an uncharacterized 3′–5′ exonuclease, as Trimmer in silkworms. Trimmer is enriched in the mitochondrial fraction and binds to Papi/Tdrkh. Depletion of Trimmer and Papi/Tdrkh additively inhibits trimming, causing accumulation of ~35–40-nt pre-piRNAs that are impaired for target cleavage and prone to degradation. Our results highlight the cooperative action of Trimmer and Papi/Tdrkh in piRNA maturation.
Introduction
Small non-coding RNAs form the effector complex with Argonaute (Ago) family proteins and induce post-transcriptional and/or transcriptional silencing of their target genes (Castel and Martienssen, 2013; Ghildiyal and Zamore, 2009; Hutvagner and Simard, 2008; Ketting, 2011; Meister, 2013). In animal germ cells, a class of small RNAs called PIWI-interacting RNAs (piRNAs) are highly expressed and repress the activity of transposable elements together with PIWI-clade Ago family proteins (Aravin et al., 2006; Aravin et al., 2007; Girard et al., 2006; Grivna et al., 2006; Malone and Hannon, 2009; Senti and Brennecke, 2010; Siomi et al., 2011; Vagin et al., 2006; Weick and Miska, 2014). piRNAs are generally 23–30-nt in length with 2′-O-methylation at their 3′ ends (Horwich et al., 2007; Houwing et al., 2007; Kirino and Mourelatos, 2007b; Ohara et al., 2007; Vagin et al., 2006) and are processed from long single-stranded transcripts via two distinct pathways (Beyret et al., 2012; Ghildiyal and Zamore, 2009; Luteijn and Ketting, 2013; Malone and Hannon, 2009; Senti and Brennecke, 2010; Siomi et al., 2011)
Although the initial step of the primary biogenesis pathway still remains elusive, it is thought that long transcripts from discrete genomic regions called piRNA clusters (Brennecke et al., 2007; Li et al., 2013) are first fragmented in an endonucleolytic manner. These RNA fragments or piRNA intermediates are then loaded into PIWI-clade proteins (e.g., Piwi in flies and Mili in mice) and subsequently cleaved by the endonuclease Zucchini in flies (MitoPLD in mice) at a position 3′ downstream of the PIWI-protected region (Han et al., 2015; Ipsaro et al., 2012; Mohn et al., 2015; Nishimasu et al., 2012). In flies, Zucchini cleaves at ~26 nt from the 5′ end, yielding precursor piRNAs (pre-piRNAs) of almost the same size as mature piRNAs (Han et al., 2015; Mohn et al., 2015). In mice, pre-piRNAs produced by MitoPLD-mediated cleavage are ~30–40 nt in length (Han et al., 2015; Mohn et al., 2015) and their 3′ ends need to be trimmed in an exonucleolytic manner before they are matured by the methyltransferase Hen1, which adds a methyl group to the 2′ ribose hydroxyl at the piRNA 3′ end (Horwich et al., 2007; Kamminga et al., 2010; Kirino and Mourelatos, 2007a; Saito et al., 2007). Zucchini/MitoPLD-mediated cleavage not only helps to shape the 3′ end of the upstream piRNA but also triggers the production of another downstream piRNA with the defined 5′ end, consecutively generating “phased” primary piRNAs (Han et al., 2015; Mohn et al., 2015).
In the secondary piRNA biogenesis pathway, a pair of PIWI proteins (e.g., Aubergine and Ago3 in flies and Mili and Miwi2 in mice) loaded with piRNAs of opposite strands reciprocally cleave complementary transcripts at a precise 10-nt offset via their endonucleolytic “slicer” activity (Aravin et al., 2008; Brennecke et al., 2007; De Fazio et al., 2011; Gunawardane et al., 2007). The 3′ cleavage products by one PIWI protein are loaded into the other PIWI protein with the defined 5′ end, serving as piRNA intermediates. Subsequently, Zucchini/MitoPLD cleaves the piRNA intermediates at a position downstream of the PIWI-protected region, followed by 3′-end trimming if the generated pre-piRNAs are longer than the mature length (Han et al., 2015; Mohn et al., 2015). Finally, their 3′ ends are methylated by Hen1 (Horwich et al., 2007; Kamminga et al., 2010; Kirino and Mourelatos, 2007a; Saito et al., 2007). Also in this “ping-pong” piRNA amplification cycle via reciprocal target slicing by PIWI proteins, Zucchini/MitoPLD-mediated cleavage acts both in 3′-end maturation of the upstream piRNA and in production of downstream phased primary piRNAs (Han et al., 2015; Homolka et al., 2015; Mohn et al., 2015), thereby increasing the repertoire of piRNA species. Conversely, the requirement of Zucchini/MitoPLD for secondary piRNA amplification can be bypassed if another neighboring ping-pong site can liberate a pre-piRNA whose length is suitable for 3′-end trimming (Mohn et al., 2015).
We have previously proposed that a putative enzyme named “Trimmer” catalyzes the 3′ end trimming of pre-piRNAs in silkworms (Kawaoka et al., 2011). Although our biochemical analysis strongly suggested that Trimmer is a Mg2+-dependent 3′–5′ exonuclease, the identity of Trimmer remains unknown. Recently, two independent studies reported that depletion of Papi (Tdrkh in mice), a mitochondrially localized Tudor domain protein, causes a 3′ extension of piRNAs (Honda et al., 2013; Saxe et al., 2013), implicating Papi/Tdrkh in pre-piRNA trimming. However, Papi/Tdrkh lacks any recognizable nuclease domains, and the molecular role of Papi/Tdrkh in the trimming reaction is unclear. Here, we identified poly(A)-specific ribonuclease (PARN)-like domain containing 1 (PNLDC1), a previously uncharacterized 3′–5′ exonuclease, as silkworm Trimmer. Trimmer is enriched in the mitochondrial fraction and associates with BmPapi (Bombyx mori Papi). Intriguingly, Trimmer cannot act alone but requires the assistance of BmPapi for trimming. Without trimming, long pre-piRNAs are impaired for target cleavage and prone to degradation. These results highlight the importance of the cooperative action between Trimmer and BmPapi at the 3′-end maturation step in silkworm piRNA biogenesis.
Results
Trimming activity is enriched in the mitochondrial fractions
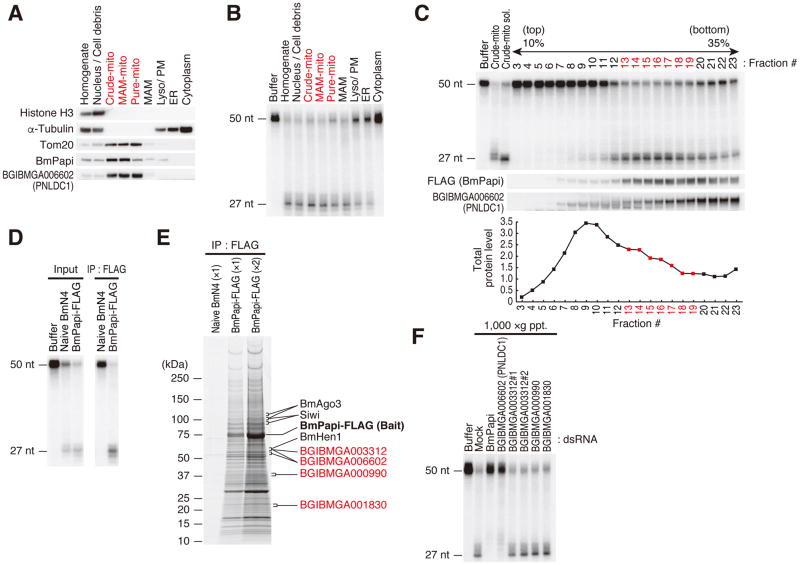
We have previously established a cell-free system that recapitulates pre-piRNA loading into PIWI proteins and 3′ end processing of PIWI-loaded pre-piRNAs by using cell extracts from BmN4, a silkworm ovary-derived cell line (Kawaoka et al., 2011). In this system, single-stranded RNAs (ssRNAs) are first loaded into Siwi (Aubergine ortholog in silkworms) in BmN4 lysate (supernatant of 17,000 × g centrifugation) and then the Siwi-RNA complex is immunopurified. Subsequently, insoluble BmN4 cell pellet from 1,000 × g centrifugation (1,000 × g ppt.) is resuspended and added to the immunoprecipitate, which causes 3′ trimming of the Siwi-loaded RNA to the mature length (27–28 nt) and its 2′-O-methylation (Figure S1A) (Kawaoka et al., 2011). To identify Trimmer biochemically, we first attempted to solubilize the trimming activity from the 1,000 × g pellet fraction with various detergents; however, all attempts were unsuccessful (data not shown). To know where Trimmer is present in cells, we separated BmN4 cells into subcellular fractions (Wieckowski et al., 2009) and examined the trimming activity in each fraction. Except for the cytoplasmic fraction, the trimming activity was broadly detected but was enriched in the mitochondria-related fractions (Figures 1A and 1B). Because the mitochondrial surface is thought to be a site of piRNA biogenesis (Ketting, 2011), we focused on the crude mitochondrial fraction, which contains all mitochondria-related fractions. We again tried various detergents and finally succeeded in partial solubilization of the trimming activity from this fraction (Figure S1B). When the CHAPS-solubilized crude mitochondrial fraction was separated by a sucrose density gradient centrifugation, the trimming activity was detected in relatively heavy fractions (Figure 1C), implying that Trimmer forms a large complex. Intriguingly, the observed trimming activity correlated well with the distribution of BmPapi, one of Tudor domain proteins in silkworms (Figure 1C) (Honda et al., 2013).
Figure 1. Strategy to identify silkworm Trimmer.
(A) Western blot analysis of subcellular fractionated BmN4 cells. Equal amounts of total protein in each fraction were loaded. Histone H3, α-Tubulin and Tom20 were used as fraction markers of nucleus, cytoplasm and mitochondria, respectively. The crude mitochondrial (crude-mito) fraction was further separated into MAM-mito, Pure-mito, and MAM fractions. MAM, mitochondria associated membrane; Lyso/PM, lysosome and plasma membrane; ER, endoplasmic reticulum. Mitochondria enriched fractions were indicated in red. BmPapi and Trimmer are enriched in the mitochondria-containing fractions.
(B) In vitro trimming assay using fractionated BmN4 cell lysate with an equal protein concentration. The mitochondria-containing fractions showed high trimming activity.
(C) CHAPS-solubilized crude mitochondrial (crude-mito) fraction prepared from BmPapi-FLAG stable cells was separated by 10–35% sucrose density gradient centrifugation. In vitro trimming assay was performed using each fraction (top panel). The distribution of BmPapi and Trimmer was analyzed by Western blotting (middle panel). Total protein level in each fraction is shown (bottom panel). The distribution of BmPapi and Trimmer correlated well with the trimming activity. The fractions with high trimming activity are indicated in red.
(D) CHAPS-solubilized crude mitochondrial (crude-mito) fractions were prepared from naive BmN4 or BmPapi-FLAG stable cells and subjected to immunoprecipitation with anti-FLAG antibody. In vitro trimming assay was performed using CHAPS-solubilized crude-mito fraction (IP input, left panel) or FLAG immunoprecipitates (right panel). The BmPapi-FLAG complex showed clear trimming activity.
(E) The immunopurified BmPapi-FLAG complex was resolved by SDS-PAGE and stained with Oriole fluorescent gel stain reagent. Four 3′–5′ exonucleases identified by mass spectrometry in the BmPapi-FLAG complex are shown in red. Details are shown in Table S1.
(F) BmN4 cells were transfected with dsRNAs for Trimmer candidates. In vitro trimming assay was performed using 1,000 ×g pellet fractions. For BGIBMGA003312, two different dsRNAs were used (#1 and #2). Knockdown efficiency is shown in Figure S1C. Mock indicates BmN4 cells transfected with dsRNAs for Renilla luciferase. Only the knockdown of BGIBMGA006602 decreased the trimming activity similarly to BmPapi knockdown.
See also Figures S1, S3 and Table S1.
BmPapi is required for efficient pre-piRNA trimming and associates with Trimmer
Papi was originally identified as an interactor of PIWI proteins in flies (Liu et al., 2011a) and is widely conserved among species (Honda et al., 2013; Saxe et al., 2013). Previous studies reported that BmPapi and Tdrkh are mitochondria-associated proteins and their depletion causes 3′ extension of mature piRNAs, suggesting their involvement in pre-piRNA trimming (Honda et al., 2013; Saxe et al., 2013). Indeed, in our cell-free system, resuspended 1,000 × g pellet from BmPapi knockdown BmN4 cells yielded longer trimming products (29–31 nt) than the mature length (Figure S2A). In addition to the incomplete trimming, BmPapi knockdown reduced the efficiency of the trimming reaction (Figure S2A). Conversely, increased expression of BmPapi accelerated trimming (Figure S2B), suggesting that BmPapi is a limiting factor for trimming in BmN4 cells. These results indicate that BmPapi is required for efficient and complete trimming of pre-piRNAs.
Besides one Tudor domain that directly binds to PIWI proteins (Chen et al., 2011), BmPapi possesses an N-terminal transmembrane domain and subsequent two KH-motifs (Figure S2C) (Honda et al., 2013). To examine the requirement of each functional domain of BmPapi for the activity to promote trimming, we constructed a series of BmPapi mutants (Figure S2C). We overexpressed each BmPapi mutant in BmN4 cells and performed the trimming assay using the 1,000 × g pellet fraction from those cells. Except for BmPapi-ΔC3 (a small C-terminal deletion), all the mutants lost the trimming-accelerating activity; rather they showed dominant negative effect on trimming if they retain the intact Tudor domain (Figures S2D–G). These results suggest that the Tudor domain-mediated interaction of BmPapi with PIWI proteins is important for trimming. Because the Tudor domain of BmPapi binds to symmetrical dimethylarginine (sDMA) modifications of PIWI proteins (Honda et al., 2013), we performed the trimming assay using Siwi-5RK, in which five potential sDMA sites were mutated to substantially weaken its binding to BmPapi (Figure S2H). Consistent with previous reports (Honda et al., 2013; Xiol et al., 2012), Siwi-5RK could load ssRNAs comparable to wild-type Siwi but trimming was severely, if not completely, impaired (Figure S2I). This highlights the importance of the interaction between the Tudor domain of BmPapi and the sDMA modifications of Siwi for efficient trimming.
BmPapi-ΔN, which lacks the N-terminal transmembrane domain and is thus largely soluble (Figures S2D and S2E), failed to promote trimming (Figure S2F), suggesting that only the membrane-anchored form of BmPapi is active. Moreover, mutations in the KH-motifs (Siomi et al., 1993), which generally function in nucleic-acid binding, also abrogated the activity of BmPapi in promoting the trimming reaction (Figure S2F), while the interaction with PIWI proteins was retained (Figure S2G). Together, these data suggest that BmPapi needs to bind both PIWI proteins and PIWI-loaded pre-piRNAs to support pre-piRNA trimming.
Because the distribution of BmPapi correlated well with the trimming activity (Figure 1C), we hypothesized that BmPapi associates with Trimmer to bring pre-piRNA-loaded PIWI proteins and Trimmer together. If so, the purified BmPapi complex should exhibit the trimming activity. To test this, we immunoprecipitated BmPapi from the CHAPS-solubilized crude mitochondrial fraction and performed the trimming assay. As expected, we detected a trimming activity in the immunoprecipitate (Figure 1D), indicating that Trimmer exists in this BmPapi complex. Encouraged by this result, we analyzed proteins contained in the complex by mass spectrometry and identified four 3′–5′ exonucleases, together with two PIWI proteins, Siwi and BmAgo3, and the methyltransferase BmHen1 (Figure 1E and Table S1).
PNLDC1 is Trimmer the pre-piRNA 3′-trimming enzyme in silkworms
To determine which of the four exonucleases is Trimmer, we next knocked down each nuclease by dsRNAs and examined the effects on the trimming activity in vitro. Strikingly, knockdown of BGIBMGA006602 (PNLDC1: poly(A)-specific ribonuclease (PARN)-like domain containing 1) but not others strongly inhibited the trimming activity (Figures 1F and S1C). PNLDC1 is a previously uncharacterized 3′–5′ exonuclease belonging to the CAF1 superfamily. While the nuclease domain of PNLDC1 shows 29% identity to that of BmPARN, it has an additional putative transmembrane domain at the C-terminal end (Figures 2A, S3A and S3B). PNLDC1 seems to be conserved in mice and humans; however, no clear PNLDC1 ortholog is found in flies, worms, and zebrafish (Figure S3; see also Discussion).
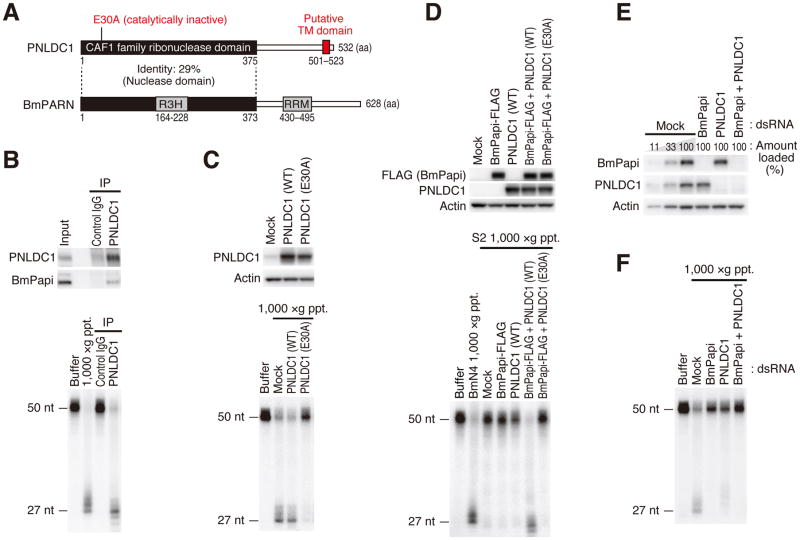
Figure 2. PNLDC1 is the silkworm Trimmer and cooperatively acts with BmPapi in trimming.
(A) Schematic representation of the domain structures of PNLDC1 (Trimmer) and PARN in silkworms. PNLDC1 has a putative transmembrane (TM) domain at the C-terminal end.
(B) PNLDC1 was immunoprecipitated from CHAPS-solubilized crude mitochondrial fraction and analyzed by Western blotting (upper panel). In vitro trimming assay was performed using the immunoprecipitated PNLDC1 complex (lower panel). PNLDC1 co-purified BmPapi and the PNLDC1 complex showed clear trimming activity.
(C) BmN4 cells were transfected with plasmids expressing wild-type or catalytically inactive (E30A) PNLDC1, and the cell lysates were analyzed by Western blotting (upper panel). In vitro trimming assay was performed using 1,000 ×g pellet fraction (lower panel). While overexpression of wild-type PNLDC1 did not increase the cellular trimming activity, overexpression of E30A mutant strongly inhibited trimming.
(D) BmPapi and wild-type or catalytically inactive (E30A) PNLDC1 were expressed in Drosophila S2 cells, and the cell lysates were analyzed by Western blotting (upper panel). In vitro trimming assay was performed using 1,000 ×g pellet fraction from the S2 cells (lower panel). The trimming activity was reconstituted only when BmPapi and wild-type PNLDC1 were co-expressed.
(E, F) BmN4 cells were transfected with dsRNA for BmPapi and/or PNLDC1 and the cell lysates were analyzed by Western blotting (E). In vitro trimming assay was performed using 1,000 ×g pellet fraction (F). Double knockdown of BmPapi and PNLDC1 additively inhibited the trimming reaction.
See also Figure S2.
To confirm that PNLDC1 is Trimmer, we generated an antibody against PNLDC1 and investigated its subcellular distribution. As expected, PNLDC1 was enriched in the mitochondria-related fractions (Figure 1A) and its distribution on sucrose density gradient correlated fairly well with the trimming activity (Figure 1C). Intriguingly, we observed a clear band-shift of PNLDC1 in heavy fractions (Figure 1C), implying its posttranslational modification. For further verification, we immunoprecipitated PNLDC1 from CHAPS-solubilized crude mitochondrial fraction and detected co-precipitation of BmPapi and a clear trimming activity in the immunopurified complex (Figure 2B). Unlike BmPapi (Figure S2B), overexpression of PNLDC1 did not accelerate in vitro trimming (Figure 2C), consistent with the notion that BmPapi, rather than PNLDC1, is a limiting factor for trimming in BmN4 cells. However, overexpression of the catalytically inactive E30A mutant of PNLDC1 strongly inhibited the trimming reaction (Figure 2C), supporting the idea that PNLDC1 is the responsible nuclease for pre-piRNA trimming.
Full-length BmPapi and PNLDC1 were insoluble in E. coli, precluding experiments using purified recombinant proteins. Instead, we ectopically expressed these two silkworm proteins in Drosophila somatic S2 cells, whose naive lysate or 1,000 × g pellet lacks the trimming activity (Figure 2D and data not shown). Importantly, when wild-type PNLDC1 (but not its catalytic mutant) and BmPapi were co-expressed in S2 cells, the trimming activity was reconstituted (Figure 2D). Similarly, co-expression of mouse PNLDC1 and Tdrkh in HEK 293T cells also reconstituted the trimming activity to Mili-loaded 50-nt ssRNA (Figure S2J). Thus, the trimming reaction depends on both PNLDC1 and BmPapi/Tdrkh across species (see below for Discussion).
To further analyze the relationship between BmPapi and PNLDC1, we compared the effects of their single-knockdown and double-knockdown (Figure 2E). Depletion of BmPapi or PNLDC1 alone markedly inhibited the trimming reaction (Figure 2F) without affecting the level of the other protein (Figure 2E). The inhibitory effect on the in vitro trimming reaction was further enhanced by their double knockdown (Figure 2F), indicating their cooperative action in trimming. Based on the above observations, we concluded that PNLDC1 is Trimmer the pre-piRNA trimming enzyme in silkworms (hereinafter we call PNLDC1 “Trimmer”) and that Trimmer requires BmPapi as a partner protein for its function.
Depletion of BmPapi and Trimmer causes 3′ extension and reduction of piRNAs
To confirm that Trimmer functions in pre-piRNA trimming in vivo, we next investigated the effect of Trimmer knockdown on the length of endogenous piRNAs. To this end, we first immunoprecipitated FLAG-Siwi or FLAG-BmAgo3 from stable cells with BmPapi and/or Trimmer knockdown and analyzed Siwi- or BmAgo3-bound RNAs in bulk by dephosphorylating and 32P-radiolabeling their 5′ ends. Consistent with the reduced trimming activity in vitro (Figures 1F and 2F), both Siwi- and BmAgo3-bound piRNAs in Trimmer or BmPapi knockdown were longer than those in the mock knockdown (Figure 3A). In addition, the abundance of piRNAs was decreased in the absence of BmPapi (Figure 3A). Because bulk radiolabeling is limited in resolution, we next performed Northern blotting for piRNA-1 and piRNA-2, which are known to predominantly bind to Siwi and BmAgo3 respectively (Honda et al., 2013). We confirmed that BmPapi knockdown and Trimmer knockdown extend the length of both piRNAs without affecting that of let-7 miRNAs (Figure 3B). Intriguingly, double knockdown of BmPapi and Trimmer resulted in accumulation of longer RNA species around ~35–40 nt for both piRNA-1 and piRNA-2 (Figure 3B, arrowhead). These longer populations most likely correspond to pre-piRNAs generated via cleavage by the endonuclease Zucchini or by slicing at a neighboring ping-pong site (see below and Discussion). Extension of the piRNA length was also caused by simply overexpressing catalytically inactive Trimmer (E30A) (Figures 3C and 3D). These results demonstrated that Trimmer acts in endogenous pre-piRNA trimming together with BmPapi in BmN4 cells.
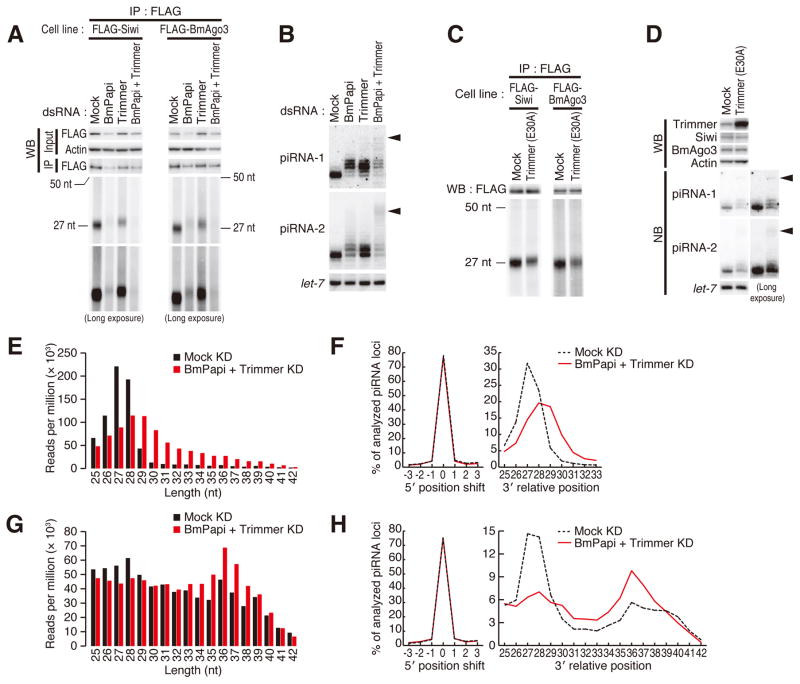
Figure 3. Depletion of BmPapi and Trimmer causes 3′ extension and reduction of piRNAs.
(A) FLAG-Siwi or FLAG-BmAgo3 stable cells were transfected with dsRNAs for BmPapi and/or Trimmer. The cell extracts were subjected to immunoprecipitation by anti-FLAG antibody. The immunoprecipitates were analyzed by Western blotting (upper panel) and bound RNAs were detected by 5′ 32P labeling after dephosphorylation (lower panel). Trimmer or BmPapi knockdown caused an extension of both Siwi- and BmAgo3-bound piRNAs. In addition, BmPapi knockdown decreased the abundance of PIWI proteins and piRNAs.
(B) BmN4 cells were transfected with dsRNA for BmPapi and/or Trimmer. Total RNAs were extracted, and piRNA-1 and piRNA-2 were detected by Northern blotting. let-7 miRNA was used as a loading control. Both piRNAs were elongated by the knockdown of BmPapi and/or Trimmer. Double knockdown of BmPapi and Trimmer accumulated putative pre-piRNAs (arrowhead).
(C) FLAG-Siwi or FLAG-BmAgo3 stable cells were transfected with plasmids expressing catalytically inactive (E30A) Trimmer. The cell lysates were subjected to immunoprecipitation with anti-FLAG antibody. The immunoprecipitates were analyzed by Western blotting (upper panel) and bound RNAs were detected by 5′ labeling with 32P (lower panel). Mock indicates BmN4 cells transfected with an empty plasmid. Overexpression of catalytically inactive (E30A) Trimmer caused an extension of piRNAs.
(D) BmN4 cells were transfected with plasmids expressing catalytically inactive (E30A) Trimmer. Mock indicates BmN4 cells transfected with empty plasmids. The whole cell lysates and total RNAs were extracted and analyzed by Western blotting (upper panel) or Northern blotting (lower panel). let-7 miRNA was used as a loading control for Northern blotting. Overexpression of catalytically inactive (E30A) Trimmer caused an elongation of piRNAs and accumulation of putative pre-piRNAs (arrowhead).
(E, G) The length distribution of piRNAs from ~25–45 nt libraries (E) or ~35–45 nt libraries (G) that mapped to 1,811 transposable elements under double knockdown of BmPapi and Trimmer. Reads were normalized to total mapping reads. Knockdown of BmPapi and Trimmer increased the length distribution of piRNAs.
(F, H) The 5′ and 3′ variation analysis of piRNAs from ~25–45 nt libraries (F) or ~35–45 nt libraries (H) mapped to 1,811 transposable elements under double knockdown of BmPapi and Trimmer. The mode values of 3′- and 5′-end locations at each piRNA locus were calculated and the percentage in each length are indicated (see Figure S4A). Knockdown of BmPapi and Trimmer specifically extended at the 3′ ends of piRNAs.
To further characterize the elongated piRNA species, we deeply sequenced ~25–45-nt small RNAs from cells with BmPapi and/or Trimmer knockdown, and analyzed the obtained reads that were mapped to 1,811 transposons in the silkworm genome (Table S2) (Osanai-Futahashi et al., 2008). As observed in biochemical analysis of bulk and individual piRNAs (Figures 3A–D), double knockdown of BmPapi and Trimmer resulted in a remarkable shift of the RNA length distribution compared to the mock knockdown (Figure 3E). This was due to the extension of the 3′ ends; the 5′ ends were fixed and shared between the two libraries (Figures 3F and S4A). The same trend was also observed in single knockdown of BmPapi or Trimmer (Figures S4B and S4C). To investigate the putative pre-piRNA population detected in the Northern blot analysis (Figures 3B and 3D), we prepared longer RNA libraries (~35–45 nt) from cells with mock knockdown and BmPapi and Trimmer double knockdown. Although there was substantial contamination of mature piRNAs, a longer population peaking at ~36 nt was apparent in the double knockdown library (Figure 3G). Again, their 5′ ends were fixed and shared while the 3′ extended ~36 nt population was markedly increased by the double knockdown (Figure 3H). These results strongly suggest that these longer RNA species correspond to pre-piRNAs before 3′-end trimming.
Mature piRNAs cleave target RNAs more efficiently than pre-piRNAs
To explore the functional significance of pre-piRNA trimming in the silencing activity, we next performed target cleavage assay with or without the trimming reaction. In this assay, we used a 50-nt single-stranded guide RNA to load into Siwi and prepared two target RNAs, 28-perfect and 50-perfect, which harbor a perfectly complementary target site corresponding to 1–28 nt or 1–50 nt of the guide RNA respectively (Figure 4A). To monitor guide trimming and target cleavage simultaneously, both the guide RNA and target RNA were radiolabeled. We first loaded the guide RNA into FLAG-Siwi and incubated with buffer alone (trimming −) or 1,000 ×g pellet suspension for trimming (trimming +). We then compared the target cleavage efficiency between the ~50-nt untrimmed and ~28-nt trimmed guide RNAs loaded in Siwi. For both 28-perfect and 50-prefect targets, the trimmed guide RNA with the mature piRNA length showed ~3–4 fold higher cleavage activity than the untrimmed guide RNA (Figures 4B and 4C). These results indicate that 3′-end trimming is important to exert full silencing activity of piRNAs.
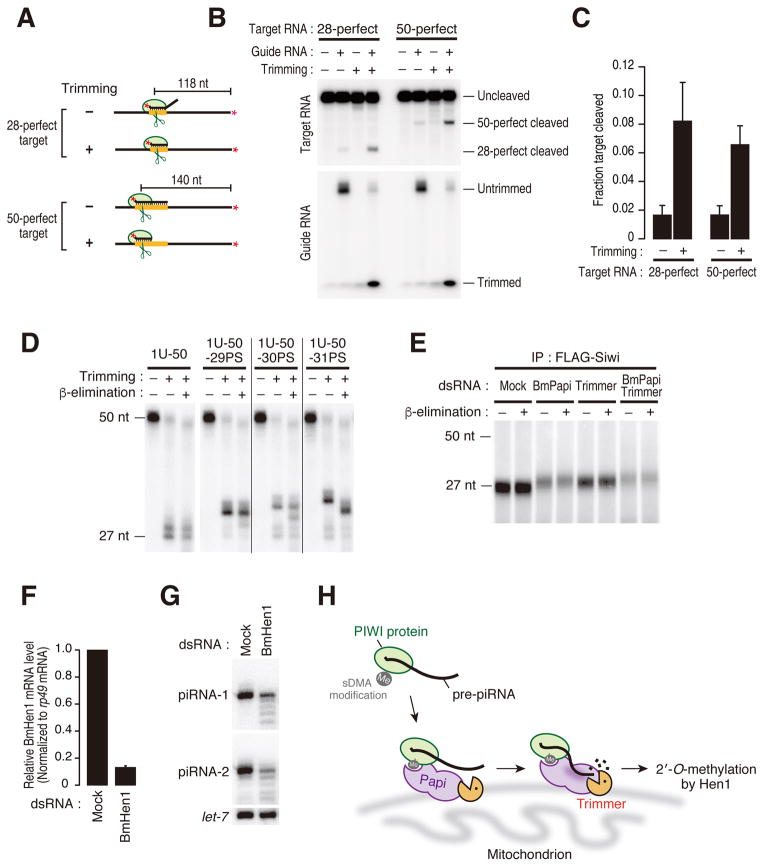
Figure 4. Pre-piRNA trimming is important for the function and stability of piRNAs.
(A) Schematic representation of two target RNAs for cleavage assay. 28-perfect and 50-perfect target RNAs possess a perfectly complementary target site corresponding to 1–28 nt or 1–50 nt of the guide RNA, respectively. The asterisks indicate radiolabeling.
(B) Target cleavage by FLAG-Siwi with or without trimming. After loading 5′-radiolabled guide RNA, FLAG-Siwi was immunoprecipitated and half was subjected to trimming reaction (trimming +). The other half was incubated with buffer (trimming −). After washing, FLAG-Siwi loaded with untrimmed or trimmed guide RNA was incubated with cap-radiolabeled 28-perfect or 50-perfect target RNAs at 25°C for 4 h. The efficiency of target cleavage was increased by the trimming.
(C) Quantification of (B). The graph shows means and standard deviations from three independent cleavage reactions.
(D) In vitro trimming assay was performed using 50-nt ssRNAs with/without a phosphorothioate linkage at different positions from 29 to 31. After trimming reaction, the trimming products were treated with NaIO4 followed by β-elimination. The 3′ end can be 2′-O-methylated only when Siwi-loaded ssRNAs are trimmed down to ≤31 nt.
(E) FLAG-Siwi stable cells were transfected with dsRNAs for BmPapi and/or Trimmer. After immunoprecipitation with anti-FLAG antibody, Siwi-bound RNAs were extracted, treated with NaIO4 followed by β-elimination, and detected by 5′ labeling with 32P. The remaining piRNAs under knockdown of BmPapi and/or Trimmer were completely 2′-O-methylated. Mock indicates BmN4 cells transfected with dsRNAs for Renilla luciferase.
(F, G) BmN4 cells were transfected with dsRNA for BmHen1 and total RNAs were extracted. The mRNA level of BmHen1 was analyzed by quantitative real-time PCR (F). piRNA-1 and piRNA-2 were detected by Northern blotting (G). let-7 miRNA was used as a loading control. BmHen1 knockdown leads to degradation of piRNAs. Mock indicates BmN4 cells transfected with dsRNAs for Renilla luciferase.
(H) A model for piRNA 3′-end maturation in silkworms. BmPapi recruits pre-piRNA loaded PIWI protein to the mitochondrial surface in part via the sDMA modification. BmPapi also binds to pre-piRNAs through its KH motifs and creates an optimal platform for trimming. Trimmer trims the 3′ end of pre-piRNAs to the mature length by the assistance of BmPapi. Coupled to trimming, the piRNA 3′ end is finally 2′-O-methylated by BmHen1.
pre-piRNA trimming stabilizes piRNAs partly via 2′-O-methylation
Although a previous study reported that BmPapi knockdown had no impact on the piRNA level (Honda et al., 2013), we observed severe reduction of piRNAs when BmPapi was depleted (Figures 3A and 3B). We reason that this difference is due to our efficient and long-term knockdown method via multiperiodic repetitive dsRNA transfection (see Experimental Procedures). One possible explanation for the piRNA reduction in BmPapi depletion is that BmPapi stabilizes PIWI proteins and their piRNAs through the protein-protein interaction between BmPapi and PIWI proteins. On the other hand, piRNA levels appeared lower in BmPapi and Trimmer double knockdown than in BmPapi single knockdown (Figures 3A and 3B), suggesting a contribution of Trimmer to piRNA stability. Indeed, deep sequencing analysis of ~25–45-nt small RNAs (Table S2) revealed that transposon-derived piRNA levels were globally decreased in BmPapi and Trimmer double knockdown compared to those in BmPapi single knockdown (Figures S4D and S4E). These observations raise a possibility that insufficient pre-piRNA trimming per se can also cause a reduction of piRNAs.
The methyltransferase Hen1 catalyzes 2′-O-methylation at the 3′ ends of piRNAs, which is generally thought to stabilize piRNAs (Billi et al., 2012; Horwich et al., 2007; Kamminga et al., 2010; Kurth and Mochizuki, 2009; Montgomery et al., 2012), and we have previously shown that 2′-O-methylation is coupled with the trimming reaction in vitro (Kawaoka et al., 2011). Therefore, we speculated that impaired pre-piRNA trimming decreases the efficiency of 2′-O-methylation, which may partly explain destabilization of piRNAs in the absence of BmPapi and Trimmer. To test this idea, we first asked how long piRNAs can be 2′-O-methylated after trimming in vitro. We prepared 50-nt RNA substrates harboring a phosphorothioate linkage at different positions from 29 to 31 to block trimming halfway and examined whether the 3′ end of the trimming products is 2′-O-methylated. Judging from the band shift after treating with NaIO4 followed by β-elimination, the 30-nt trimming product of 1U-50-29PS (phosphorothioate linkage between positions 29 and 30) were fully 2′-O-methylated (Figure 4D), even though they were ~3-nt longer than typical ~27-nt mature piRNAs. In contrast, the 32-nt trimming product of 1U-50-31PS remained completely unmodified (Figure 4D). About half of the 31-nt trimming products of 1U-50-30PS were 2′-O-methylated (Figure 4D). Thus, 31 nt appears to be the border length for efficient 2′-O-methylation coupled to trimming in vitro. This is consistent with the fact that the ≥31 nt population of piRNAs is low in vivo (Figure 3E). In fact, prominent 28–29 nt piRNAs remaining after knockdown of BmPapi and/or Trimmer (Figures 3E and S4B) were fully 2′-O-methylated (Figure 4E), conversely suggesting that longer species are prone to degradation likely due to the lack of 2′-O-methylation. Supporting this idea, blocking 2′-O-methylation by BmHen1 knockdown caused degradation of piRNA-1 and piRNA-2 (Figures 4F and 4G), consistent with previous reports in other animals (Billi et al., 2012; Horwich et al., 2007; Kamminga et al., 2010; Kurth and Mochizuki, 2009; Montgomery et al., 2012). Thus, inhibition of pre-piRNA trimming can cause the reduction of piRNAs at least in part due to the absence of 2′-O-methyl protection in longer RNA species.
Discussion
Trimmer and Papi cooperatively act in 3′-end trimming of pre-piRNAs
We had previously found that the pre-piRNA 3′ trimming activity is enriched in the insoluble fraction of BmN4 cells (Kawaoka et al., 2011). However, its insoluble nature has made it extremely challenging to identify Trimmer using biochemical approaches. In this study, we identified PNLDC1 as Trimmer in silkworms. Several lines of evidence support this conclusion: (1) Knockdown of PNLDC1 or overexpression of its catalytically inactive mutant inhibited trimming and extended 3′ ends of piRNAs in BmN4 cells as well as in our cell-free system. (2) Immunopurified PNLDC1 complex exhibited the trimming activity. (3) Ectopic expression of PNLDC1 together with BmPapi in Drosophila S2 cells reconstituted the trimming activity.
The key to the identification of Trimmer was to focus on the mitochondrial fraction and the mitochondrial Tudor protein Papi. Unfortunately, our Trimmer antibody was unsuitable for immunostaining, and N- or C-terminal tagging of Trimmer hindered its expression in BmN4 cells (data not shown), precluding fluorescent imaging of the intracellular localization. However, the existence of the transmembrane domain and the subcellular fractionation analysis (Figures 1A, 2A, S3A, and S3B) strongly suggest that Trimmer is associated with the mitochondrial surface, similar to other piRNA biogenesis factors including Zucchini/MitoPLD and BmPapi/Tdrkh (Honda et al., 2013; Huang et al., 2011; Saito et al., 2010; Saxe et al., 2013; Watanabe et al., 2011). Therefore, the mitochondrial surface provides the “platform” for at least a part of piRNA biogenesis, presumably by highly concentrating and appropriately arranging relevant factors. This is reminiscent of tRNA biogenesis in yeast, in which enzymes required for pre-tRNA splicing localize on the mitochondrial surface (Yoshihisa et al., 2003). Since overexpression of BmPapi but not that of wild-type Trimmer enhanced the basal efficiency of pre-piRNA trimming in BmN4 cells (Figures 2C and S2B), recruitment of PIWI proteins and their bound pre-piRNAs in proximity to Trimmer on the mitochondrial surface is likely a rate limiting step in trimming. Indeed, many BmPapi mutants showed a dominant negative effect in trimming in vitro, but only if they can bind to PIWI proteins via the intact Tudor domain (Figures S2C and S2F). Moreover, mutating the sDMA modification sites of Siwi (5RK), thereby weakening the interaction between BmPapi and PIWI proteins, inhibited the in vitro trimming reaction (Figures S2H and S2I). This inhibitory effect was not complete, however, as there remained a small amount of the trimming product with the 5RK mutant. This is in line with previous observations that the BmPapi-Siwi interaction is not solely dependent on the Siwi sDMA modifications and that the Siwi sDMAs are not a prerequisite for trimming in BmN4 cells (Honda et al., 2013; Xiol et al., 2012). Perhaps, the sDMA-mediated interaction between BmPapi and PIWI proteins is not essential but facilitates trimming by Trimmer.
The KH motifs (Siomi et al., 1993), which generally function in nucleic acid binding, were required for BmPapi to accelerate trimming in vitro (Figures S2C and S2F), suggesting that BmPapi not only binds to PIWI proteins but also to PIWI-loaded pre-piRNAs. Given that BmPapi interacts with Trimmer (Figures 1E, 2B and S2G), we envision that BmPapi bridges PIWI proteins, their bound pre-piRNAs and Trimmer together, thereby making the pre-piRNA 3′-end accessible to Trimmer (Figure 4H). The fact that reconstitution of the trimming activity in S2 cells requires co-expression of Trimmer and BmPapi (Figure 2D) also supports this model. After trimming, the 3′ end of piRNAs is 2′-O-methylated, which completes piRNA maturation (Billi et al., 2012; Horwich et al., 2007; Kamminga et al., 2010; Kamminga et al., 2012; Kawaoka et al., 2011; Kurth and Mochizuki, 2009; Montgomery et al., 2012; Saito et al., 2007). Indeed, we identified the methyltransferase BmHen1 in the BmPapi complex (Figure 1E). However, it remains to be investigated why 2′-O-methylation occurs in a manner coupled to trimming.
piRNA 3′-end trimming in other animals
We identified PNLDC1, a homolog of PARN, as silkworm Trimmer. Although we could not detect obvious contribution of canonical BmPARN in pre-piRNA trimming in our vitro system (Figure S1D), in worms, PARN-1, one of the two canonical PARN homologs, has been identified as the 3′-end trimming enzyme for worm-specific piRNAs called 21U-RNAs (Tang et al., 2016). Intriguingly, fission yeast possesses a Dicer-independent small RNA biogenesis pathway, in which the 3′ end of Ago1-loaded longer precursor RNAs is trimmed by Triman, a PARN homolog exonuclease (Marasovic et al., 2013). In addition, in vertebrates, canonical PARN participates in trimming of Ago2-cleaved miR-451 precursor hairpin (Yoda et al., 2013). Thus, PARN and PARN-like nucleases seem to act widely in the maturation of small RNAs. However, Trimmer is unique in the sense that it requires the partner protein Papi on the mitochondrial surface for its function.
Curiously, flies lack both PNLDC1 and canonical PARN. However, it was recently reported that, the loss of Nibbler (Nbr), a 3′–5′ exonuclease that was originally characterized as the 3′-end trimming enzyme for Ago1-loaded miRNAs to generate shorter miRNA isoforms (Han et al., 2011; Liu et al., 2011b), causes ~1-nt elongation of a subset of piRNAs in flies (Feltzin et al., 2015), implicating Nbr as the fly pre-piRNA trimming enzyme. In contrast, mice lack the Nbr ortholog EXD3 (exonuclease domain containing 3) but possess PNLDC1 and PARN. According to BioGPS (http://biogps.org/) (Wu et al., 2009), PNLDC1 is highly expressed in mouse testes, suggesting its role in the piRNA pathway. Supporting this, we could reconstitute a trimming activity toward Mili-loaded 50-nt ssRNAs in 293T cells by co-expressing mouse PNLDC1 and Tdrkh (Figure S2J), as is the case for Trimmer and BmPapi in silkworms. Future studies will be needed to confirm the role of PNLDC1 in mouse piRNA biogenesis.
By inhibiting trimming, we detected an accumulation of ~35–40-nt pre-piRNAs, which have extended 3′ ends relative to mature piRNAs (Figures 3B, 3D, 3G, and 3H). Deep sequencing analysis suggested that pre-piRNAs exists at a low level in naive BmN4 cells but become more prominent by the double knockdown of BmPapi and Trimmer (Figures 3G and 3H). In flies and mice, the 3′-end of pre-piRNAs is defined by Zucchini/MitoPLD-mediated cleavage at a downstream U residue or via piRNA-guided slicing at another neighboring ping-pong site (Han et al., 2015; Homolka et al., 2015; Mohn et al., 2015). We have found evidence that at least a part of silkworm pre-piRNAs have a downstream U bias just like fly and mouse pre-piRNAs (K.S., N.I., Y.T., and S.K., unpublished data). In addition, a recent study showed that target cleavage by BmAgo3 can trigger primary piRNA production from the target transcript downstream of the Siwi-loaded ping-pong partner piRNA in BmN4 cells (Homolka et al., 2015), suggesting that the pre-piRNA generating mechanism is conserved among species. However, there is a clear difference in the pre-piRNA lengths; the length of pre-piRNAs in silkworms is similar to that in mice (~30–40 nt) but much longer than that in flies, in which pre-piRNAs are on average only ~0.35-nt longer than mature piRNAs (Han et al., 2015; Homolka et al., 2015; Mohn et al., 2015). Such difference in the pre-piRNA length may be related to which nuclease(s) is mainly used for 3′-end trimming.
Experimental Procedures
Cell culture, transfection and RNAi in BmN4 cells
BmN4 cells were cultured at 27 °C in IPL-41 medium (AppliChem) supplemented with 10% fetal bovine serum. Three microgram of pIZ or pIEx-1 expression vectors or dsRNAs were transfected into BmN4 cells (1 × 106 cells for plasmid DNA, 3 × 105 cells for dsRNA per 6 cm dish) with X-tremeGENE HP DNA Transfection Reagent (Roche). Detailed information is described in Supplemental Experimental Procedures.
Antibodies
Rabbit anti-Trimmer (PNLDC1) and anti-BmPapi antibodies were generated by immunizing N-terminally His-tagged recombinant PNLDC1 (aa 1–223) (MBL) or a synthetic peptide (CGGRSSSVPKDHKDD; Genscript) deriving from BmPapi respectively. Other antibodies are described in Supplemental Experimental Procedures.
In vitro assay for pre-piRNA processing
The sequences of substrate RNAs, in vitro pre-piRNA loading, trimming, NaIO4-mediated oxidation, and β-elimination were described previously (Kawaoka et al., 2011). For the trimming assay, lysates with an equal protein concentration were used in each experimental set.
Preparation of cell lysate, sucrose density gradient centrifugation
Preparation of crude mitochondrial fraction was performed according to previous literature (Wieckowski et al., 2009). The crude mitochondrial pellet was resuspended in two volume of MRB (5 mM Hepes-KOH (pH 7.4), 250 mM mannitol, 0.5 mM EGTA, 0.5 mM DTT, 1× Complete EDTA-free protease inhibitor (Roche)), and extracted proteins by adding CHAPS at the final concentration 0.35%. The soluble fraction was obtained by centrifugation at 14,000 ×g for 30 min at 4°C. For sucrose density gradient centrifugation, the solubilized crude mitochondrial fraction was placed at the top of a 10–35% discontinuous sucrose gradient, and was centrifuged in a Beckman SW-41 rotor at 38,000 rpm for 16.5 h at 4°C.
Immunoprecipitation
For BmPapi-FLAG immunoprecipitation, the solubilized crude mitochondrial fraction was diluted with equal volume of MRB (5 mM Hepes-KOH (pH 7.4), 250 mM mannitol, 0.5 mM EGTA, 0.5 mM DTT, 1× Complete EDTA-free protease inhibitor (Roche)) supplemented with 50 mM NaCl at the final concentration. After incubation with anti-FLAG antibodies bound to Dynabeads protein G (Thermo Fisher) for 2 h at 4°C, the immunoprecipitates were washed with MRB supplemented with 150 mM NaCl and 0.1% CHAPS, and eluted with 3× FLAG peptides (Sigma). Trimmer immunoprecipitation was performed similarly and the immunocomplex was eluted by SDS sample buffer. FLAG-Siwi and FLAG-BmAgo3 immunoprecipitation was described previously (Kawaoka et al., 2009). For immunoprecipitation of BmPapi mutants (Fig. S2G) and Siwi-5RK (Fig. S2H), plasmid-transfected cells were resuspended in NT buffer (20 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1.5 mM MgCl2, 8% glycerol, 0.05% NP-40, 0.1% Triton X-100, 1 mM DTT, 1× Complete EDTA-free protease inhibitor (Roche)) and incubated on ice for 30 min. The soluble fraction was obtained by centrifugation at 17,000 ×g for 30 min at 4°C. After incubation with anti-FLAG antibody conjugated on Dynabeads protein G (Thermo Fisher) at 4°C for 2 h, the immunoprecipitates were washed with NT buffer without 0.1% Triton X-100, and eluted with 3× FLAG peptides (Sigma).
Target cleavage assay
Loading and trimming for cleavage assay were performed essentially as described before (Kawaoka et al., 2011), except that 0.5 μM 5′-32P-radiolabled guide RNA (UCGAAGUAUUCCGCGUACGUUAUGCUAGCUGAUCCUGACCGCUGAGUCG U) was used for loading. After trimming, FLAG-Siwi bound beads were washed with lysis buffer (30 mM Hepes-KOH (pH 7.4), 100 mM KOAc, 2 mM Mg(OAc)2) containing 0.1% (v/v) Empigen (Sigma) five times, rinsed with lysis buffer, and divided into two halves for 28-perfect and 50-perfect target RNAs. Target cleavage was performed with respective 1 nM 32P cap-radiolabeled target RNAs at 25°C for 4 h. Detailed information is described in Supplemental Experimental Procedures.
RNA-seq
Small RNA libraries were prepared from ~25–45 nt or ~35–45 nt total RNAs using Small RNA Library Preparation kit (Illumina) and analyzed by Illumina HiSeq 2500 platform. Detailed informatic analysis is described in Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Wen Tang and Craig Mello for sharing unpublished data, Yuki Okada for providing mouse spermatogonial stem cells, Shinpei Kawaoka for Mili construct and helpful discussion, IMCB Research Center for Epigenetic Disease for sequencing of small RNA libraries. We also thank all of the members of our laboratories for critical comments on the manuscript. This work was in part supported by Grant-in-Aids for Scientific Research on Innovative Areas “Functional machinery for non-coding RNAs” to S.K. and Y.T. and “non-coding RNA neo-taxonomy” to Y.T., and a Grant-in-Aid for Research Activity Start-up to N.I., and NIH grant (GM106047) to Y.K., a JSPS Research Fellowship for Young Researchers to K.S., and a JSPS Postdoctoral Fellowship for Research Abroad to S.H.
Footnotes
Accession Numbers
The accession numbers for the deep sequencing data reported in this paper are DDBJ: DRA003745–DRA003750.
Author Contributions
N.I. and Y.T conceived and designed the experiments and wrote the manuscript. N.I. performed the biochemical experiments and analyzed the data. K.S. and S.K. performed bioinformatics analysis of the deep sequencing data. Y.S. and T.S. performed mass-spectrometric analysis of the immunopurified protein samples. S.H. and Y.K. produced anti-BmPapi antibody. Y.T. supervised the study. All the authors discussed the results and approved the manuscript.
References
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Beyret E, Liu N, Lin H. piRNA biogenesis during adult spermatogenesis in mice is independent of the ping-pong mechanism. Cell Res. 2012;22:1429–1439. doi: 10.1038/cr.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billi AC, Alessi AF, Khivansara V, Han T, Freeberg M, Mitani S, Kim JK. The Caenorhabditis elegans HEN1 ortholog, HENN-1, methylates and stabilizes select subclasses of germline small RNAs. PLoS Genet. 2012;8:e1002617. doi: 10.1371/journal.pgen.1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Nott TJ, Jin J, Pawson T. Deciphering arginine methylation: Tudor tells the tale. Nat Rev Mol Cell Biol. 2011;12:629–642. doi: 10.1038/nrm3185. [DOI] [PubMed] [Google Scholar]
- De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, Funaya C, Antony C, Moreira PN, Enright AJ, O’Carroll D. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature. 2011;480:259–263. doi: 10.1038/nature10547. [DOI] [PubMed] [Google Scholar]
- Feltzin VL, Khaladkar M, Abe M, Parisi M, Hendriks GJ, Kim J, Bonini NM. The exonuclease Nibbler regulates age-associated traits and modulates piRNA length in Drosophila. Aging Cell. 2015;14:443–452. doi: 10.1111/acel.12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Han BW, Hung JH, Weng Z, Zamore PD, Ameres SL. The 3′-to-5′ exoribonuclease Nibbler shapes the 3′ ends of microRNAs bound to Drosophila Argonaute1. Curr Biol. 2011;21:1878–1887. doi: 10.1016/j.cub.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BW, Wang W, Li C, Weng Z, Zamore PD. Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science. 2015;348:817–821. doi: 10.1126/science.aaa1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homolka D, Pandey RR, Goriaux C, Brasset E, Vaury C, Sachidanandam R, Fauvarque MO, Pillai RS. PIWI Slicing and RNA Elements in Precursors Instruct Directional Primary piRNA Biogenesis. Cell Rep. 2015;12:418–428. doi: 10.1016/j.celrep.2015.06.030. [DOI] [PubMed] [Google Scholar]
- Honda S, Kirino Y, Maragkakis M, Alexiou P, Ohtaki A, Murali R, Mourelatos Z. Mitochondrial protein BmPAPI modulates the length of mature piRNAs. RNA. 2013;19:1405–1418. doi: 10.1261/rna.040428.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Huang H, Gao Q, Peng X, Choi SY, Sarma K, Ren H, Morris AJ, Frohman MA. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev Cell. 2011;20:376–387. doi: 10.1016/j.devcel.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature. 2012;491:279–283. doi: 10.1038/nature11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamminga LM, Luteijn MJ, den Broeder MJ, Redl S, Kaaij LJ, Roovers EF, Ladurner P, Berezikov E, Ketting RF. Hen1 is required for oocyte development and piRNA stability in zebrafish. EMBO J. 2010;29:3688–3700. doi: 10.1038/emboj.2010.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamminga LM, van Wolfswinkel JC, Luteijn MJ, Kaaij LJ, Bagijn MP, Sapetschnig A, Miska EA, Berezikov E, Ketting RF. Differential impact of the HEN1 homolog HENN-1 on 21U and 26G RNAs in the germline of Caenorhabditis elegans. PLoS Genet. 2012;8:e1002702. doi: 10.1371/journal.pgen.1002702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka S, Hayashi N, Suzuki Y, Abe H, Sugano S, Tomari Y, Shimada T, Katsuma S. The Bombyx ovary-derived cell line endogenously expresses PIWI/PIWI-interacting RNA complexes. RNA. 2009;15:1258–1264. doi: 10.1261/rna.1452209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka S, Izumi N, Katsuma S, Tomari Y. 3′ end formation of PIWI-interacting RNAs in vitro. Mol Cell. 2011;43:1015–1022. doi: 10.1016/j.molcel.2011.07.029. [DOI] [PubMed] [Google Scholar]
- Ketting RF. The many faces of RNAi. Dev Cell. 2011;20:148–161. doi: 10.1016/j.devcel.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Kirino Y, Mourelatos Z. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA. 2007a;13:1397–1401. doi: 10.1261/rna.659307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Mourelatos Z. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat Struct Mol Biol. 2007b;14:347–348. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- Kurth HM, Mochizuki K. 2′-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA. 2009;15:675–685. doi: 10.1261/rna.1455509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XZ, Roy CK, Dong X, Bolcun-Filas E, Wang J, Han BW, Xu J, Moore MJ, Schimenti JC, Weng Z, et al. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol Cell. 2013;50:67–81. doi: 10.1016/j.molcel.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Qi H, Wang J, Lin H. PAPI, a novel TUDOR-domain protein, complexes with AGO3, ME31B and TRAL in the nuage to silence transposition. Development. 2011a;138:1863–1873. doi: 10.1242/dev.059287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Abe M, Sabin LR, Hendriks GJ, Naqvi AS, Yu Z, Cherry S, Bonini NM. The exoribonuclease Nibbler controls 3′ end processing of microRNAs in Drosophila. Curr Biol. 2011b;21:1888–1893. doi: 10.1016/j.cub.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn MJ, Ketting RF. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nat Rev Genet. 2013;14:523–534. doi: 10.1038/nrg3495. [DOI] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasovic M, Zocco M, Halic M. Argonaute and Triman generate dicer-independent priRNAs and mature siRNAs to initiate heterochromatin formation. Mol Cell. 2013;52:173–183. doi: 10.1016/j.molcel.2013.08.046. [DOI] [PubMed] [Google Scholar]
- Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet. 2013;14:447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- Mohn F, Handler D, Brennecke J. Noncoding RNA. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science. 2015;348:812–817. doi: 10.1126/science.aaa1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery TA, Rim YS, Zhang C, Dowen RH, Phillips CM, Fischer SE, Ruvkun G. PIWI associated siRNAs and piRNAs specifically require the Caenorhabditis elegans HEN1 ortholog henn-1. PLoS Genet. 2012;8:e1002616. doi: 10.1371/journal.pgen.1002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, Bonnefond L, Matsumoto N, Nishizawa T, Nakanaga K, Aoki J, et al. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature. 2012;491:284–287. doi: 10.1038/nature11509. [DOI] [PubMed] [Google Scholar]
- Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat Struct Mol Biol. 2007;14:349–350. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- Osanai-Futahashi M, Suetsugu Y, Mita K, Fujiwara H. Genome-wide screening and characterization of transposable elements and their distribution analysis in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2008;38:1046–1057. doi: 10.1016/j.ibmb.2008.05.012. [DOI] [PubMed] [Google Scholar]
- Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, Siomi MC. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 2010;24:2493–2498. doi: 10.1101/gad.1989510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxe JP, Chen M, Zhao H, Lin H. Tdrkh is essential for spermatogenesis and participates in primary piRNA biogenesis in the germline. EMBO J. 2013;32:1869–1885. doi: 10.1038/emboj.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senti KA, Brennecke J. The piRNA pathway: a fly’s perspective on the guardian of the genome. Trends Genet. 2010;26:499–509. doi: 10.1016/j.tig.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Matunis MJ, Michael WM, Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- Tang W, Tu S, Lee HC, Weng Z, Mello CC. The RNase PARN-1 Trims piRNA 3′ Ends to Promote Transcriptome Surveillance in C. elegans. Cell. 2016;164:974–984. doi: 10.1016/j.cell.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Chuma S, Yamamoto Y, Kuramochi-Miyagawa S, Totoki Y, Toyoda A, Hoki Y, Fujiyama A, Shibata T, Sado T, et al. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev Cell. 2011;20:364–375. doi: 10.1016/j.devcel.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weick EM, Miska EA. piRNAs: from biogenesis to function. Development. 2014;141:3458–3471. doi: 10.1242/dev.094037. [DOI] [PubMed] [Google Scholar]
- Wieckowski MR, Giorgi C, Lebiedzinska M, Duszynski J, Pinton P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat Protoc. 2009;4:1582–1590. doi: 10.1038/nprot.2009.151. [DOI] [PubMed] [Google Scholar]
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, 3rd, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiol J, Cora E, Koglgruber R, Chuma S, Subramanian S, Hosokawa M, Reuter M, Yang Z, Berninger P, Palencia A, et al. A role for Fkbp6 and the chaperone machinery in piRNA amplification and transposon silencing. Mol Cell. 2012;47:970–979. doi: 10.1016/j.molcel.2012.07.019. [DOI] [PubMed] [Google Scholar]
- Yoda M, Cifuentes D, Izumi N, Sakaguchi Y, Suzuki T, Giraldez AJ, Tomari Y. Poly(A)-specific ribonuclease mediates 3′-end trimming of Argonaute2-cleaved precursor microRNAs. Cell Rep. 2013;5:715–726. doi: 10.1016/j.celrep.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihisa T, Yunoki-Esaki K, Ohshima C, Tanaka N, Endo T. Possibility of cytoplasmic pre-tRNA splicing: the yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Mol Biol Cell. 2003;14:3266–3279. doi: 10.1091/mbc.E02-11-0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.