Abstract
Small molecules are known to stabilise membrane proteins and to modulate function and oligomeric state, but their identity is often hard to define. Here we develop and apply a high-resolution, Orbitrap mass spectrometer for intact membrane protein-ligand complexes. Using this platform we resolve the complexity of multiple binding events, quantify small molecule binding and reveal selectivity for endogenous lipids that differ only in acyl chain length.
Keywords: native MS, membrane proteins, lipid binding, drug binding, Orbitrap MS, non-covalent interactions, structural biology
Spanning the membranes within a cell, integral membrane proteins act as biological gatekeepers, controlling the flow of biomolecular substrates across hydrophobic barriers 1. As key protagonists in mediating cellular function, responding to stress and maintaining homeostasis, understanding the interactions of membrane proteins with other biomolecules is essential to develop therapeutic interventions. Membrane proteins however remain challenging targets to study due to their hydrophobic nature and enigmatic interactions with lipid bilayers. Recently non-denaturing or native MS (nMS) has emerged as a powerful technique to probe intact membrane protein assemblies 2. Specifically nano-electrospray quadrupole time-of-flight (Q-ToF) MS has been used to define lipid binding partners and to assess their effects on stability 3. However the lack of spectral resolution, inherent to current instrumentation, restricts interactions that can be measured to those with large mass differences between proteins and substrates (>200-300 Da) 4. High resolution MS has previously been used to interrogate individual membrane proteins but under these conditions quaternary interactions were lost 5. It has therefore not been possible to identify endogenous lipid ensembles, or combinations of similar sized lipids, peptides and drugs bound directly to membrane proteins. Methods exist to quantify and identify lipids in solution 6, 7, primarily those that co-purify, but do not define classes or families of lipids in direct contact with membrane proteins.
To study such interactions higher resolution nMS is required. Modified Orbitrap mass analysers have been shown to provide exceptional mass resolution 8 and transmission of high molecular weight complexes has recently been demonstrated for soluble proteins 9, 10. We set out to develop an Orbitrap platform that enables unambiguous distinction between lipids, detergents, peptides and drugs whilst in complex with membrane protein assemblies.
To maintain interactions micelles or other vehicles are required to protect the membrane protein complex upon transfer into the gas phase 11-13. Previously we used increased backing pressure (1-9 Torr) in the initial stages of a Q-ToF and subsequently removed the micelle using activation in an online collision cell 14. Interestingly we found that with the Q Exactive instrument (Supplementary Figure 1) only nominal pressures were required in the source region (1.4-1.6 mbar). Critical for the Q Exactive was a ‘gentle’ voltage gradient such that the intact membrane protein complex is released from the detergent micelle following collisional activation in either the HCD cell, source region or both (Supplementary Figures 2 and 3 and Supplementary Methods).
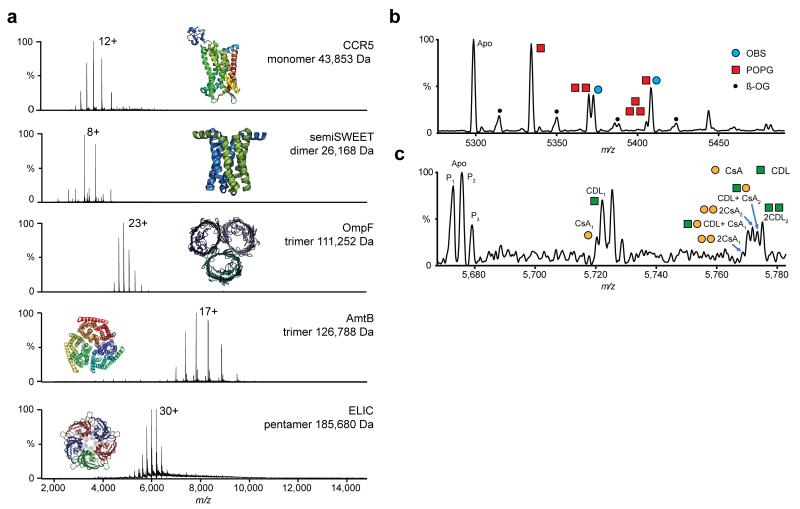
To investigate the applicability of this approach we selected a wide range of membrane proteins with different masses (from 26-186 kDa), structural features and stoichiometries including: a monomeric G-protein coupled receptor (CCR5); dimeric glycan transporter (semiSWEET); trimeric channels, the outer membrane porin OmpF and ammonia transporter AmtB; and pentameric ligand-gated ion channel (ELIC) (Fig 1a). All spectra revealed the anticipated stoichiometry, narrow charge state distributions, and baseline resolution with narrow peak widths (~2-3 m/z) even at modest transient times (64 ms). At increased HCD pressures, and in contrast to previous results 14, the pentameric ion channel ELIC was released from the micelle without appreciable dissociation (185,680 Da) (Supplementary Figure 3). The Orbitrap analyser also allowed greater precision in mass measurement compared with our previous studies (Supplementary Table 1); the measured mass of semiSWEET suggests N-terminal processing and the small molecule adducts, previously convolved with the apo ELIC peak 14 , were identified as bound lipids.
Figure 1.
Non-denaturing Orbitrap mass spectra of purified membrane protein complexes of differing size and topology, bound to multiple lipids, peptides and drugs. (a) High resolution non-denaturing Orbitrap mass spectra of the monomeric GPCR chemokine receptor CCR5, dimeric glycan transporter semiSWEET, trimeric outer membrane porin OmpF, trimeric ammonia transporter AmtB and the pentameric ligand gated ion channel ELIC. Masses and associated errors can be found in Supplementary Table 1. Protein structures have been generated from PDB files 4QND, 4MBS, 2NPE, 2OMF and 3RQU. (b,c) Concomitant small molecule binding. (b) Binding of OBS (blue circle) to OmpF in the presence of POPG lipid (red square). Expansion of the 22+ charge state. The OBS peak can be clearly separated from the 2POPG peak. (c) Binding of CsA drug (yellow circle) to P-gp in the presence of CDL 14:0 (green square). Zoom of two ligand binding events on the 25+ charge state. P-gp is present in three proteoforms (denoted by subscript). Both CsA and CDL binding can be clearly resolved. For the peak corresponding to two ligands bound, fine structure indicates the binding of different numbers of CsA and CDL to distinct proteoforms of P-gp. (For detailed assignment see Supplementary Table 2 and for individual ligand binding Supplementary Figure 7).
To quantify protein-ligand interactions instrument conditions need to be carefully controlled 15. Spectra recorded for the interaction of OmpF with a peptide mimic of colicin OmpF Binding Sequence 1 (OBS1) reveal three successive binding events (Supplementary Figure 3). Intensity values for each state were fit to a multisite binding model using UniDec deconvolution software 16 and yielded a dissociation constant (KD = 0.69 μM) for total OBS1-OmpF binding (Supplementary Figures 4 & 5a) in close agreement with previous measurements (Q-ToF nMS : 1.4 ± 0.1 μM and ITC : 1.0 ± 0.1 μM). Individual microscopic KDs for successive ligand binding events were also extracted highlighting the potential to investigate co-operativity (Supplementary Figures 5b,c) and confirming the suitability of this Orbitrap platform for investigating multiple binding events well separated by mass.
Concomitant drug and lipid binding often involves species of similar mass and has proved challenging, restricting the type of interactions that can be studied to those with large mass differences4, 17. Concomitant binding of the OBS1 peptide to OmpF for example, in the presence of POPG lipids cannot be resolved on current instrumentation due to the small mass difference between OBS1 and 2POPG lipids (Δmass= 55.5 Da). In the case of the ABC transporter P-gp the immunosuppressant drug cyclosporin A (CsA) can affect binding affinity of long chain cardiolipin (CDL) (Δmass= 452 Da). However the lack of spectral resolution abrogated similar studies using shorter chain CDL (CDL14:1 (3)–15:1), (Δmass= 45.0 Da). To investigate if these small mass differences could now be resolved we added OBS1 to OmpF, in the presence of POPG lipid. We found that enhanced resolution allowed separation of OBS and 2POPG binding (Figure 1b). Similarly for P-gp, we observed concomitant binding of CsA and CDL 14:0 (Δmass= 39.0 Da) (Figure 1c, Supplementary Figure 6). Interestingly, the spectrum of P-gp reveals three species separated by 76-80 Da within the major protein peak. These proteoforms are likely post-translationally modified (PTM) forms of the protein. Orbitrap nMS can distinguish multiple binding events of ligands with similar masses.
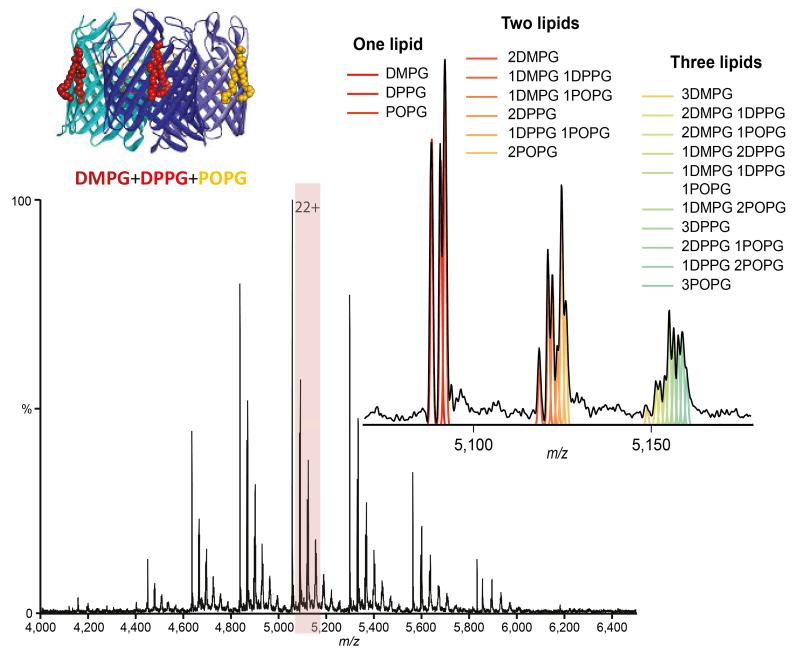
To identify multiple endogenous lipid-binding to membrane protein complexes however requires resolution of smaller mass differences (12-14 Da) that arise from the natural distribution of fatty acid lipid chain lengths. To assess this we created a model system by adding separately phosphoglycerol lipids differing in their acyl chain composition (14:0 - DMPG, 16:0 - DPPG, 18:1 - POPG) to OmpF to confirm lipid binding (Supplementary Figure 7). Next we prepared an equimolar mixture all three lipids, incubated this with OmpF, and observed peaks corresponding to binding of up to six lipids. Closer examination of the singly-bound peak reveals three separate peaks, one each for binding of DMPG, DPPG and POPG (Figure 2). Remarkably, we were able to distinguish binding of multiple lipid combinations with increasingly small mass differences (Figure 2). Intriguingly, the relative abundance of POPG rises in the higher lipid bound states, suggesting that this longer chain, unsaturated lipid binds preferentially. Consequently even when attached to relatively large proteins (>100 kDa) we can distinguish lipids from the same class with different chain lengths and degrees of unsaturation, in line with those in natural membranes. Our method therefore provides a substantial improvement on current nMS measurements that indicate the presence of lipids but have not been able to resolve homologous series directly bound to membrane protein complexes 4, 17, 18.
Figure 2.
Mass spectrum of trimeric OmpF bound to an equimolar ratio of DMPG, DPPG and POPG lipids (cartoon inset) and zoom of the 22+ charge state with peaks showing up to three lipids bound. Theoretical distributions corresponding to different combinations of lipids are shown by coloured lines and correlate with the spectrum.
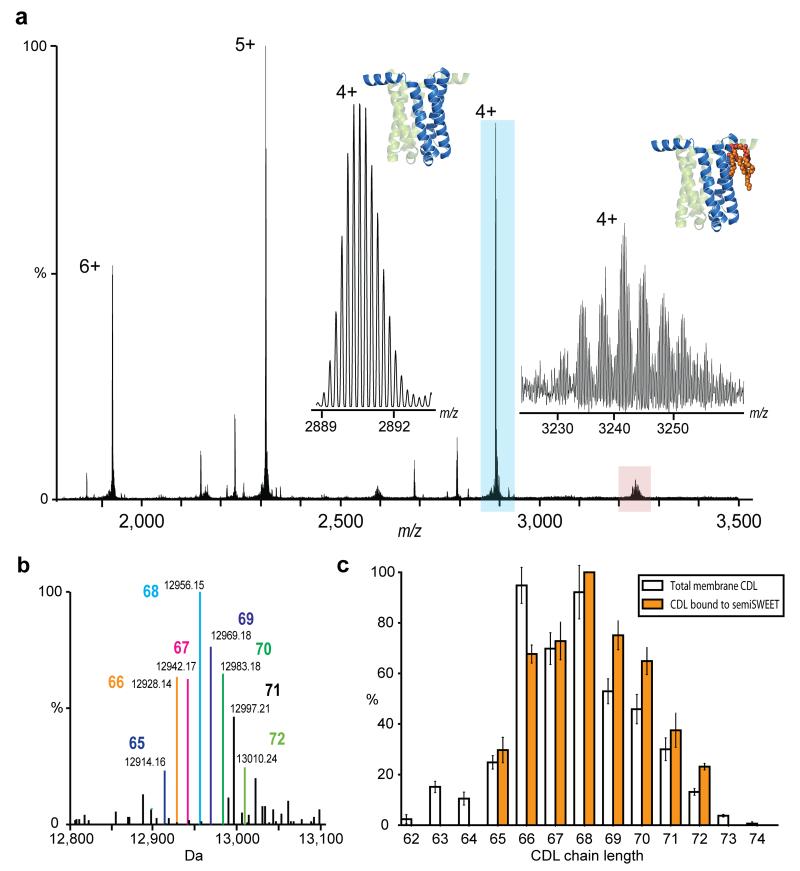
With the ability to resolve unique lipid species, we hypothesized that the Orbitrap approach would allow resolution and identification of endogenous lipids. To test this, the glycan transporter Vibrio species semiSWEET was expressed in E. coli, where the endogenous bilayer lipids essentially comprise of cardiolipin (~5%), phosphoglycerol (~20%) and phosphoethanolamine (~70%) 19. Orbitrap MS of monomeric semiSWEET released from DDM micelles reveals peaks assigned to binding of endogenous CDL (Figure 3a). At increased transient times, isotopic resolution of the protein confirmed the previously unreported N-terminal methionine cleavage (error 0.9 ppm). Acurate mass measurement of the lipid adducts confirmed their identity as different forms of CDL. The lipid acyl chain length was assigned by comparison with a database of reported E. coli lipids 20 (Figure 3b). CDL molecules bound to semiSWEET were then compared with the total distribution of CDL in the E. coli membrane from cells cultured under the same conditions. (Figure 3c, Supplementary Figure 8). Unexpectedly, a marked shift to higher chain length was discerned for CDL in complex with semiSWEET showing clear selectivity for binding longer chain lipids. Such membrane remodelling possibly arises from the need to stabilise the rocking motion associated with the function of this transporter. As a general approach, this direct lipid identification strategy overcomes the shortcomings of “lipid extraction-lipidomics” procedures where typically only co-purified lipids are identified, not necessarily those that are in direct contact with the protein.
Figure 3.
Examination of membrane protein semiSWEET in direct contact with bound lipids. (a) Isotopically resolved mass spectrum of semiSWEET released from DDM micelle obtained at an increased transient time of 256 ms. Monoisotopic mass measurement of the major protein peak (blue) gives an error of 0.9 ppm once N-terminal methionine cleavage had been taken into account. Inset shows a zoom of the apo protein peak (blue) and of the satellite peaks (red) revealing a distribution of homologous series of lipids. (b) Identification of directly bound lipids. Deconvoluted spectrum of semiSWEET with bound CDL adducts showing monoisotopic mass and enabling identification of lipid class as cardiolipin. Acyl chain length assignment is indicated. (c) Comparison of CDL bound to semiSWEET (orange bars) with the distribution of cardiolipin in the total membrane (clear bars). A clear shift of the distribution to longer acyl chain lengths for the lipids bound directly to semiSWEET can be discerned.
In conclusion, we describe the development and application of a general method using a modified Orbitrap platform for high resolution, nMS of membrane protein complexes. We show that careful selection of instrument conditions, particularly the voltage gradient, allows the controlled release of membrane proteins from detergent micelles with retention of stoichiometry and small molecule binding quantitatively similar to both conventional (Q-ToF) nMS and solution measurements. The high resolution afforded by this approach exposed unexpected proteoforms and enables multiple substrates to be distinguished, such as homologous series of endogenous lipids and drug binding in the presence of similarly sized lipids that comprise the natural membrane. Direct identification of individual protein-bound lipid species represents a significant advance on previous methods and showcases the applicability of this technique to more complex membrane proteins, which have undergone multiple ligand binding events or where an abundance of glycoforms and other PTMs are anticipated.
Online Methods
High Resolution Orbitrap Non-Denaturing MS
Experiments were performed using a Q Exactive hybrid quadrupole-Orbitrap mass spectrometer23 (Thermo Fisher Scientific) modified for the transmission and detection of high m/z ions. Hardware alterations include a modified quadrupole RF board that allowed a lower RF frequency to be applied to the selection quadrupole (284 kHz), a modified preamplifier with lower high-pass filter cut-off and several modifications to the HCD gas inlet including replacing the standard PEEKSil capillary from the diverter valve to the HCD cell from 100 to 530 μm internal diameter and replacing the metal restrictor tubing from the manual valve to the diverter valve with 0.02” internal diameter metal tubing. These modifications allowed the selection quadrupole to operate as a mass selector up to 20,000 m/z, allowed the high m/z–low frequency signals to be detected and higher pressures to be achieved in the HCD cell (≈ 1×10−9 mbar rather than the standard operating pressures of 10−11 - 10-10 mbar), respectively. This allows better capture of ions in the HCD cell as described elswhere24. Ions were generated in the positive ion mode from a static nanospray source using gold-coated capillaries prepared in-house, then passed through a temperature controlled transfer tube (40-60 °C), RF-only S-lens, injection flatapole and bent flatapole. The instrument was operated under Tune 2.4 instrument control software in “Native Mode” in which the RF applied to the multipoles (injection flatapole, bent flatapole (950 Vp-p) and C-trap (2950 Vp-p) was increased to the maximum allowed by electronic boards. After traversing the selection quadrupole, which was operated with a wide selection window (2,000-20,000 m/z) in these experiments, ions were trapped in the HCD cell before being transferred into the C-trap and Orbitrap mass analyser for detection.
The key transformative step in getting these experiments to work was to move away from the pressure gradients used previously11, 14, 25 and to apply a gentle voltage gradient throughout the instrument and especially on the ion transfer optics (injection flatapole, inter flatapole lens, bent flatapole, transfer multipole: 8, 7, 6, 4 V respectively) to avoid collisional activation of the ions, while still protected by the detergent micelle and before micelle removal in the HCD cell. Particular care was taken when tuning the C-trap entrance lens as this can act as a mass filter if operated at too high a voltage. To ensure better capture of high-m/z ions in the Orbitrap analyser, the initial setting of central electrode voltage was adjusted from −3.7 kV to −3.2 kV, while the setting during detection remained unchanged (−5 kV). Transient times were 64 ms (corresponding to resolution setting 17,500 at m/z 200) unless otherwise stated and AGC target was 1×106. Spectra were acquired with 10 microscans, and averaged with a noise level parameter of 3 rather than the 4.68 default. Removal of the micelle was achieved through increased acceleration voltages applied in the HCD cell. Additional energy could be applied to the S-lens (in source activation) if required. The collision gas was either nitrogen or argon and pressure in the Orbitrap cell was maintained at around 1×10−9 mbar unless otherwise stated. Calibration was performed up to 11,304 m/z using clusters of CsI in infusion mode. Data was viewed using Xcalibur 2.2 SP1.48 (Thermo Fisher Scientific); no further smoothing or baseline subtraction needed to be applied to the raw data. For non-isotopically resolved data masses were calculated using an in house software tool available online http://benesch.chem.ox.ac.uk/resources.html and based on a minimisation of error calculated from different charge state assignments26, 27. For isotopically resolved data the Xtract algorithm, licenced as part of Qual Browser in Xcalibur 2.2 SP1.48 was used for peak picking and deconvolution. For all proteins presented in this work, once optimal instrument conditions had been established for efficient transmission and release of membrane proteins from the micelle, measurements were found to be entirely reproducible with only minimal variation in parameters and spectral quality. Solutions of purified membrane protein complexes were buffer exchanged into 200 mM ammonium acetate (Sigma Aldrich) supplemented with 2×critical micelle concentration (CMC) detergent (Anatrace, Ohio, USA) using P6 Biospin columns (Biorad) immediately before MS analysis. All proteins were prepared for MS at approximately 10 μM complex concentration before buffer exchange. If the signal intensity was very high (very short ion trap fill time) the concentration was adjusted accordingly.
Protein purification
Samples of CCR5 in n-Dodecyl β-D-maltoside (DDM) were provided by Prof. Beli Wu and were expressed and purified according to the published protocol 28. His-tagged P-gp was expressed in Pichia pastoris and purified in DDM as described previously 29. Vibrio sp. N418 semiSWEET was expressed in a pJexpress411 vector containing sequences for a 3C protease cleavage site and a histidine His-tag (DNA2.0) 30 before exchange into DDM or C8E4 for MS analysis. The ELIC plasmid from I. Zimmerman and R. Dutzler, was transformed into Rosetta 2 (DE3) cells (Novagen, Gibbstown, NJ) and expressed as previously described 14 before exchange into DDM for MS analysis. AmtB was overexpressed as an N-terminal fusion to maltose binding protein (MBP) preceded by a secretion signal peptide (pelB) and 10× His-tag in a modified pET15b vector and purified as previously described3 before exchange into C8E4 for MS. OmpF was purified in β octyl-glucoside (β-OG) according to the published protocol31. Proteins were adjusted to approximately 10 μM complex concentration before buffer exchange and MS.
Measurement and Calculation of OBS1-OmpF Dissociation Constant
Measurement of the dissociation constant for binding of OBS1 to OmpF was performed as described previously 21. Briefly, OBS1 peptide (NH2-2SGGDGRGHNTGAHSTSG18-CONH2) was diluted from a single stock to two fold the desired concentration and mixed with equal volume of OmpF in 25mM ammonium acetate of 1 % (w/v) β-OG immediately prior to mass measurement. Conditions for nanoESI-MS were verified as to generate spectra of sufficient quality to obtain resolved peaks without incurring ligand dissociation. 200V was applied in the HCD cell with no additional in-source activation. Spectra were deconvolved using UniDec software tool 16.
The Data Collector module in Unidec was used to extract and normalize intensities summed over all charge states and for the computation of KD values. For calculation of the overall OmpF·OBS1 KD, peak intensities for all bound states (OmpF·OBS1, OmpF·2OBS1 and OmpF·3OBS1) were combined and data fit to the same binding model as described previously 21, where the [OmpF] is considered as the concentration of binding sites i.e. 3×[OmpF trimer] . For calculation of the individual binding site KDs, extracted intensities for the apo and each separate bound state were fit to KD binding models using either a single fixed KD for all binding sites or free KDs for each individual site. After fitting to both models, an F-test was used to determine the best model.
OmpF-Lipid Binding
Lipid stocks of 1,2-dimyristoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (DMPG), 1,2-dipalmitoyl-snglycero-3-phospho-(1′-rac-glycerol) (DPPG) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (POPG) were prepared from powder (Avanti Polar Lipids Inc., Alabama USA) at concentrations of 10 mg/mL using the previously published procedure 14. Lipid stocks were diluted 50 fold in 1 % (w/v) β-OG, 200 mM ammonium acetate and added to OmpF in an appropriate ratio for multiple lipid species to be bound. For multiple lipid binding an approximately equimolar mixture of all three lipid species was prepared. The spectrum of concomitant binding of DMPG, DPPG and POPG to the OmpF trimer was acquired at an increased transient time of 128 ms (35,000 resolution at m/z 200).
P-gp Binding
Lipid stocks of 1′,3′-bis[1,2-dimyristoyl-sn-glycero-3-phospho]-sn-glycerol (Avanti Polar Lipids Inc., Alabama USA) (CDL 14:0) were prepared at a concentration of 10 mg/mL and diluted 50 fold in 200 mM ammonium acetate containing 2×CMC DDM. Lipid was mixed with P-gp in an appropriate ratio to observe multiply bound species. Binding of cyclosporin A (CsA) (Sigma Aldrich) and concomitant binding was performed in a similar fashion. Spectra of P-gp bound to cardiolipin 14:0, bound to cyclosporin A and to both cardiolipin and cyclosporin were acquired at an increased transient time of 128 ms (35,000 resolution at m/z 200).
Characterisation of Cardiolipin bound to semiSWEET
Three separate preparations of semiSWEET were analysed by native Orbitrap MS as described above but with extended transient times to achieve isotopic resolution (140,000 resolution at m/z 200). Spectra were acquired with 10 microscans for 50-100 scans before being averaged in the Xcalibur software. They were then deconvoluted using the Xtract algorithm and the mass of the lipid adduct calculated by subtracting the mass of the apo protein measured in each case and normalised against the most intense lipid species. By comparison with a previously published list of E. coli cardiolipin species, the lipid adduct was assigned a chain length n (where n is the total number of acyl chain carbon atoms). Averages and standard deviations (error bars are one standard deviation) were then calculated for semiSWEET preparations from three biological replicates (i.e. three separate protein expressions) and represented in Figure 3.
Profiling of Cardiolipin in bulk E. coli membrane
At the moment of cell harvest a 5mL sample of cells was taken from 4 randomly chosen flasks of a 12 L semiSWEET expression. This ensured that the lipid composition of the total membrane extract was representative of the conditions during semiSWEET overexpression. Cells were pelleted, the supernatant removed and replaced with 100 μL lysis buffer (20 mM Tri-HCl, 300 mM NaCl, pH 7.4). After vigorous vortexing, the cell suspension was snap frozen to lyse cells, thawed and the total lipid extracted using a modified Bligh and Dyer procedure32. Briefly, to 100 μL cell suspension, 375 μL chloroform/methanol (2:1) was added, followed by 125 μL chloroform and 125 μL H2O. Samples were vortexed then allowed to phase separate. The bottom (organic) layer was removed and evaporated to dryness before being resuspended in 1000 μL 68% solution A (ACN:H2O 60:40, 10 mM ammonium formate, and 0.1% formic acid), 32% solution B (IPA:ACN 90:10, 10 mM ammonium formate, and 0.1% formic acid) for analysis by reverse phase liquid chromatography tandem MS (RP LC-MS/MS)33.
LC-MS/MS was performed using a Dionex UltiMate 3000 RSLC nano System coupled to an LTQ Orbitrap XL hybrid mass spectrometer (Thermo Fisher Scientific). Total lipid extract resuspended in 32% solution B was loaded into a 1 μL loop using the autosampler and loading pump then transferred onto a C18 column (Acclaim PepMap 100, C18, 75 μm × 15 cm, Thermo Fisher Scientific) at a flow rate of 300 nL/min. After 10 min solvent B was ramped to 65% over 1 min, then 80% over 6 min before being held at 80% for 10min, then ramped to 99% over 6 min and held for 7 min.
The column eluent was delivered to the LTQ Orbitrap XL system operated in negative ion mode via a dynamic nanospray source. Spray voltage was −1.6 kV and capillary temperature 160°C. After a full MS scan acquired in the Orbitrap mass analyser (AGC target 5e5, resolution 60,000 at m/z 400) tandem MS was performed in a data dependant fashion using a top 5 method. Ions were selected, fragmented and detected in the linear ion trap (AGC target 1e4). Collision-induced dissociation (CID) energy was 38% NCE, and activation time 30 ms. Ions were added to a dynamic exclusion list after being fragmented twice.
The proportion of the full scan chromatogram corresponding to cardiolipin (highlighted in Supplementary Figure 8a) was then summed and mass spectra extracted in Xcalibur 2.2 software before being deconvoluted using the Xtract algorithm to give monoisotopic neutral CDL masses. Ions corresponding to the n:2 peaks (where n is the total number of acyl chain carbon atoms and the figure the number of double bind equivalents) were taken as representative of the total lipid population with that chain length. Intensities of these ions were extracted and normalised to the most intense CDL species. The average percentage was then calculated along with the standard deviation and represented in Figure 3.
Supplementary Material
Acknowledgments
The authors would like to thank L Feng and W Frommer (Stanford School of Medicine, USA) for the semiSWEET plasmid. This work has benefited from the expertise of the Protein & Nucleic Acid Chemistry Facility, Department of Biochemistry, University of Cambridge, UK for quantification of the OBS1 peptide. We would also like to thanks A Baldwin for help with dissociation constant calculations. K.G. is a fellow of the Royal Commission for the Exhibition of 1851 and a Junior Research Fellow at St Catherine’s College, Oxford. J.A.C.D. is supported by an Engineering and Physical Sciences Research Council studentship held at the Life Sciences Interface Doctoral Training Centre. M.T.M. & J.T.S.H. are funded by a program grant G1000819 (CVR) from the Medical Research Council. J.G. is funded by a European Research Council Investigator Award (IMPRESSS, grant no. 268851) (CVR). C.V.R. holds a Royal Society Research Professorship and is supported by a Wellcome Trust Senior Investigator Award (grant no. 104633/Z/14/Z) (CVR).
Footnotes
Competing Financial Interests
E.D., M.B. and A.M. are employees of Thermo Fisher Scientific, the manufacturer of the Q Exactive instrument used in this research.
References
- 1.Almén MS, Nordström KJV, Fredriksson R, Schiöth HB. Mapping the human membrane proteome: a majority of the human membrane proteins can be classified according to function and evolutionary origin. BMC Biology. 2009;7:50–50. doi: 10.1186/1741-7007-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marcoux J, Robinson Carol V. Twenty Years of Gas Phase Structural Biology. Structure. 2013;21:1541–1550. doi: 10.1016/j.str.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Laganowsky A, et al. Membrane proteins bind lipids selectively to modulate their structure and function. Nature. 2014;510:172–175. doi: 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marcoux J, et al. MS reveals synergistic effects of nucleotides, lipids, and drugs binding to a multidrug resistance efflux pump. Proceedings of the National Academy of Sciences. 2013;110:9704–9709. doi: 10.1073/pnas.1303888110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitelegge J. Thylakoid membrane proteomics. Photosynthesis Research. 2003;78:265–277. doi: 10.1023/B:PRES.0000006828.65688.0d. [DOI] [PubMed] [Google Scholar]
- 6.Brügger B. Lipidomics: Analysis of the Lipid Composition of Cells and Subcellular Organelles by Electrospray Ionization MS. Annual Review of Biochemistry. 2014;83:79–98. doi: 10.1146/annurev-biochem-060713-035324. [DOI] [PubMed] [Google Scholar]
- 7.Lee AG. How lipids and proteins interact in a membrane: a molecular approach. Molecular BioSystems. 2005;1:203–212. doi: 10.1039/b504527d. [DOI] [PubMed] [Google Scholar]
- 8.Zubarev RA, Makarov A. Orbitrap MS. Analytical Chemistry. 2013;85:5288–5296. doi: 10.1021/ac4001223. [DOI] [PubMed] [Google Scholar]
- 9.Rose RJ, Damoc E, Denisov E, Makarov A, Heck AJR. High-sensitivity Orbitrap mass analysis of intact macromolecular assemblies. Nat Meth. 2012;9:1084–1086. doi: 10.1038/nmeth.2208. [DOI] [PubMed] [Google Scholar]
- 10.Dyachenko A, et al. Tandem Native Mass-Spectrometry on Antibody–Drug Conjugates and Submillion Da Antibody–Antigen Protein Assemblies on an Orbitrap EMR Equipped with a High-Mass Quadrupole Mass Selector. Analytical Chemistry. 2015;87:6095–6102. doi: 10.1021/acs.analchem.5b00788. [DOI] [PubMed] [Google Scholar]
- 11.Barrera NP, Di Bartolo N, Booth PJ, Robinson CV. Micelles Protect Membrane Complexes from Solution to Vacuum. Science. 2008;321:243–246. doi: 10.1126/science.1159292. [DOI] [PubMed] [Google Scholar]
- 12.Reading E, et al. The Role of the Detergent Micelle in Preserving the Structure of Membrane Proteins in the Gas Phase. Angewandte Chemie International Edition. 2015;54:4577–4581. doi: 10.1002/anie.201411622. [DOI] [PubMed] [Google Scholar]
- 13.Hopper JTS, et al. Detergent-free MS of membrane protein complexes. Nat Meth. 2013;10:1206–1208. doi: 10.1038/nmeth.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laganowsky A, Reading E, Hopper JTS, Robinson CV. MS of intact membrane protein complexes. Nat. Protocols. 2013;8:639–651. doi: 10.1038/nprot.2013.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hopper JTS, Robinson CV. MS Quantifies Protein Interactions—From Molecular Chaperones to Membrane Porins. Angewandte Chemie International Edition. 2014;53:14002–14015. doi: 10.1002/anie.201403741. [DOI] [PubMed] [Google Scholar]
- 16.Marty MT, et al. Bayesian Deconvolution of Mass and Ion Mobility Spectra: From Binary Interactions to Polydisperse Ensembles. Analytical Chemistry. 2015;87:4370–4376. doi: 10.1021/acs.analchem.5b00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bechara C, et al. A subset of annular lipids is linked to the flippase activity of an ABC transporter. Nat Chem. 2015;7:255–262. doi: 10.1038/nchem.2172. [DOI] [PubMed] [Google Scholar]
- 18.Zhou M, et al. MS of Intact V-Type ATPases Reveals Bound Lipids and the Effects of Nucleotide Binding. Science. 2011;334:380–385. doi: 10.1126/science.1210148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raetz CR. Enzymology, genetics, and regulation of membrane phospholipid synthesis in Escherichia coli. Microbiol Rev. 1978;42:614–659. doi: 10.1128/mr.42.3.614-659.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garrett TA, O'Neill AC, Hopson ML. Quantification of cardiolipin molecular species in Escherichia coli lipid extracts using liquid chromatography/electrospray ionization MS. Rapid communications in MS : RCM. 2012;26:2267–2274. doi: 10.1002/rcm.6350. [DOI] [PubMed] [Google Scholar]
Methods Only References
- 21.Housden NG, et al. Intrinsically Disordered Protein Threads Through the Bacterial Outer-Membrane Porin OmpF. Science. 2013;340:1570–1574. doi: 10.1126/science.1237864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu F-F, et al. Structural characterization of cardiolipin by tandem quadrupole and multiple-stage quadrupole ion-trap MS with electrospray ionization. Journal of the American Society for MS. 2005;16:491–504. doi: 10.1016/j.jasms.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 23.Michalski A, et al. MS-based Proteomics Using Q Exactive, a High-performance Benchtop Quadrupole Orbitrap Mass Spectrometer. Molecular & Cellular Proteomics. 2011;10 doi: 10.1074/mcp.M111.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snijder J, et al. Defining the Stoichiometry and Cargo Load of Viral and Bacterial Nanoparticles by Orbitrap MS. Journal of the American Chemical Society. 2014;136:7295–7299. doi: 10.1021/ja502616y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barrera NP, et al. MS of membrane transporters reveals subunit stoichiometry and interactions. Nat Meth. 2009;6:585–587. doi: 10.1038/nmeth.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tito MA, Tars K, Valegard K, Hajdu J, Robinson CV. Electrospray Time-of-Flight MS of the Intact MS2 Virus Capsid. Journal of the American Chemical Society. 2000;122:3550–3551. [Google Scholar]
- 27.McKay AR, Ruotolo BT, Ilag LL, Robinson CV. Mass Measurements of Increased Accuracy Resolve Heterogeneous Populations of Intact Ribosomes. Journal of the American Chemical Society. 2006;128:11433–11442. doi: 10.1021/ja061468q. [DOI] [PubMed] [Google Scholar]
- 28.Tan Q, et al. Structure of the CCR5 Chemokine Receptor–HIV Entry Inhibitor Maraviroc Complex. Science. 2013;341:1387–1390. doi: 10.1126/science.1241475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aller SG, et al. Structure of P-Glycoprotein Reveals a Molecular Basis for Poly-Specific Drug Binding. Science. 2009;323:1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Y, et al. Structures of bacterial homologues of SWEET transporters in two distinct conformations. Nature. 2014;515:448–452. doi: 10.1038/nature13670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Housden NG, et al. Directed epitope delivery across the Escherichia coli outer membrane through the porin OmpF. Proceedings of the National Academy of Sciences. 2010;107:21412–21417. doi: 10.1073/pnas.1010780107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bligh EG, Dyer WJ. A Rapid Method of Total Lipid Extraction And Purification. Canadian Journal of Biochemistry and Physiology. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 33.Bird SS, Marur VR, Sniatynski MJ, Greenberg HK, Kristal BS. Lipidomics Profiling by High-Resolution LC–MS and High-Energy Collisional Dissociation Fragmentation: Focus on Characterization of Mitochondrial Cardiolipins and Monolysocardiolipins. Analytical Chemistry. 2011;83:940–949. doi: 10.1021/ac102598u. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.