Abstract
Helicobacter pylori (H. pylori) has been recognized as a cause of gastrointestinal diseases and progress of the pathology of gastrointestinal diseases is related to the genotype of H. pylori. Published studies have indicated that the H. pylori vacuolating cytotoxin gene A (vacA) i1/i2 genotype is associated with peptic ulcer disease (PUD) and gastric cancer (GC), but their conclusions are inconsistent. This study aimed to further assess the risk of vacA i gene for PUD and/or GC. A systematic search was conducted across three main electronic databases (PubMed, Web of Science, and CNKI). A meta‐analysis was then performed on the pooled data of the published articles to estimate the overall influence of vacA i polymorphisms on PUD and/or GC by crude odds ratio (OR) with 95% confidence intervals (CI). The reliability of the results were confirmed by publication bias and sensitivity analysis of included studies. A total of 14 studies were selected according to the specific inclusion and exclusion criteria. The pooled results revealed that patients with GC were more vulnerable to infection by H. pylori i1 genotype (OR = 5.12; 95% CI: 2.66–9.85; P < 0.001) than those with chronic gastritis or nonulcer disease. Moreover, the results of subgroup analysis indicated that the i1 genotype of H. pylori was associated with an increased GC risk (OR = 10.89; 95% CI: 4.11–20.88; P < 0.001) in the Middle Asian population. The H. pylori vacA i1 genotype is associated with an increased GC risk, especially in the Middle Asian population.
Keywords: Gastric cancer, Helicobacter pylori, meta‐analysis, vacA i genotype
Abbreviations
- babA2
blood group antigen‐binding adhesion protein A2
- cagA
cytotoxin‐associated gene A
- CG
chronic gastritis
- CI
confidence interval
- DU
duodenal ulcer
- dupA
duodenal ulcer promoting
- E
East Asia
- GC
gastric cancer
- GU
gastric ulcer
- H. pylori
Helicobacter pylori
- χ2
Chi square value
- M
Middle Asia
- NUD
nonulcer disease
- oipA
outer inflammatory protein gene
- OR
odds ratio
- PUD
peptic ulcer disease
- U
Europe
- vacA
the vacuolating cytotoxin gene A
It is estimated that half of the global population have Helicobacter pylori (H. pylori) and this bacteria is proven to be related to gastrointestinal diseases, including chronic gastritis, peptic ulcers, and gastric cancer (GC) 1. Therefore, H. pylori has been categorized as group I carcinogen by the International Agency for Research on Cancer in 1994 2. GC is the fifth most common cancer and the third leading cause of cancer‐related deaths in the world 3. Although the global incidence rate of GC is decreasing, it is still a heavy burden to public health in some developing countries 3.
A previous study suggested that the increased risk of GC was determined by the interaction of H. pylori in the host and was affected by the genetic variation in the patient 4. It is generally accepted that the pathogenicity of H. pylori is determined by genetic variations associated with severe gastroduodenal diseases [including GC and peptic ulcer disease (PUD)], such as the presence of cytotoxin‐associated gene A (cagA), vacuolating cytotoxin gene A (vacA i, s, m, d), duodenal ulcer promoting (dupA), blood group antigen‐binding adhesion protein A2 (babA2), and outer inflammatory protein gene (oipA). Moreover, vacA gene is deduced to have a biological influence on the host gastric epithelial cell, including apoptosis induction, increased permeability of the epithelial monolayer, hexametric pores forming, and suppressed immunity 4, 5. H. pylori vacA gene has two genotypes within its signal (s1, s2), middle (m1, m2), deletion (d1, d2), and intermediate regions (i1, i2). Accumulated evidence suggest that H. pylori strains with s1 lead to stronger cytokine activity than those with s2, indicating the m1 strains to be more virulent than m2 6, 7. In addition, the d1 genotype is found to be involved in the progression of carcinogenesis 8. Among these genotypes, vacA i has been widely investigated since it was first identified by Rhead et al. in 2007 9. In Asia, some studies have reported that the H. pylori vacA i1 genotype may increase the risk of GC in the Iran population 10, 11. In Europe, it has been reported that vacA i1 is associated with GC 10 and PUD 12, respectively. However, vacA i1 is not the only factor to predict the pathological progression of GC 13 and the susceptibility of vacA i1 to severe gastric disease remains unclear. This study aims to evaluate the association of the H. pylori vacA i1 and i2 genotypes with the risk of PUD and GC.
Materials and methods
Study sources and article search
To identify relevant articles, a comprehensive systematic search across three databases (PubMed, Web of Science, and a Chinese digital database CNKI) was performed to identify studies published until June 15, 2015, using the words ‘Helicobacter pylori’, ‘H. pylori’, ‘genotypes’, ‘vacA i1’, ‘intermediate region’, and ‘vacA gene’ with Boolean operators (NOT, AND, OR). Furthermore, a manual search was also performed to obtain substantial relevant studies by reviewing all references in the eligible articles. All analysis methods were in accordance with the preferred reporting items for systematic review and meta‐analyses (PRISMA) statement 14.
Study selection
All included studies must meet the following inclusion criteria: (a) study reported the infection of vacA i1 and/or i2; (b) case–control studies concerned about PUD [including gastric ulcer (GU) and duodenal ulcer (DU)] and/or GC with H. pylori‐positive control group [nonulcer disease (NUD) or chronic gastritis (CG)]; (c) with sufficient original data or information to assess the odds ratio (OR) and with 95% confidence interval (CI); (d) published in English or Chinese. On the other hand, the exclusion criteria include: (a) review articles; (b) case studies; (c) animal or functional experiments; (d) duplicate publications; (e) lack of raw data even after contacting the author; (f) conference proceedings; (g) literatures not written in English or Chinese.
Data extraction
Two independent authors (Xian Liu and Bangshun He) extracted raw data from the eligible studies. For any discrepancy, a final consensus was sought after discussion with the research team. Data concerning the first author's last name, publishing year, the ethnicity or continent of the cases and controls, the amplification primer for detection, adult or children of subjects, and the disease of the control group were extracted from the studies.
Statistical analysis
state software version 11.0 (STATA Corp., College Station, TX, USA) was applied for statistical analysis. The association between the presence of the H. pylori vacA i1 genotype and the risk of PUD or GC was assessed by crude OR with 95% CI. A P value < 0.05 was considered as statistically significant. The Q test (Chi square value, χ2) was used to evaluate heterogeneity (P < 0.05 indicated a significant heterogeneity between studies). The Mantel–Haenszel method was used to calculate the pooled ORs of fixed‐effects model in case if there was no significant heterogeneity among studies (P > 0.05 or χ2 < 50%). On the other hand, the DerSimonian and Laird method was applied for random‐effects model (P < 0.05 or χ2 > 50%) 15. Sensitivity analysis was also conducted to explore the origin of significant heterogeneity. Beside, subgroup analysis was performed to explore the effects of geographical region, ulcer location, and control resources. In addition, publication bias was evaluated by funnel plots and the quantitatively evaluation was conducted by the Egger's test, significant publication bias was reckoned with a P value < 0.1 14.
Results
Included studies
A total of 15 studies 9, 10, 11, 12, 13, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25 were identified, including 13 full papers and two correspondence (a short paper type with available data) articles 22, 25, in which 14 were written in English and one in Chinese (Fig. 1). In the included studies, a total of 2667 patients were infected with the H. pylori vacA i1 or i2 genotype. Among these patients, a total of 85 subjects presented both the vacA i1 and i2 genotypes. These patients were counted in both of the i1 and i2 genotypes to reduce intergroup heterogeneity and to accurately detect i2 pathogenicity. The main characteristics and specific values of each study were shown in Table 1.
Figure 1.
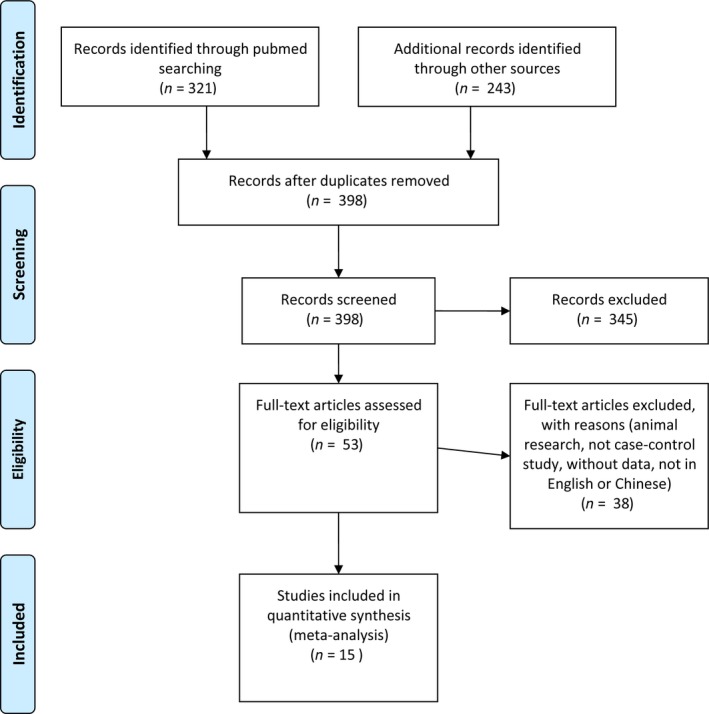
Flow diagram for the studies included in the meta‐analysis.
Table 1.
Characteristics of studies included in the meta‐analysis
| Author/year | Country | Race | CG or NUD (i1/i2) | PUD (i1/i2) | GU (i1/i2) | DU (i1/i2) | GC (i1/i2) |
|---|---|---|---|---|---|---|---|
| Kim/2014 13 | South Korea | E | 75/10 | – | 103/15 | 102/10 | 121/15 |
| Markovska/2011 20 | Bulgarian | U | 14/16 | 27/7 | – | – | – |
| Mottaghi/2014 16 | Iran | M | 19/29 | 6/17 | – | – | 21/3 |
| Memon/2014 11 | Iraq | M | 36/29 | – | – | 27/5 | 15/1 |
| Jiang/2013 17 | China | E | 87/5 | 63/1 | – | – | 21/0 |
| Ferreira/2012 18 | Portugal | E | 24/64 | – | – | – | 45/5 |
| Yordanov/2012 19 | Bulgarian | U | 48/16 | 89/63 | – | – | – |
| Panayotopoulou/2010 21 | Greece | U | 45/34 | 46/19 | – | – | – |
| Douraghi/2010 22 | Iran | M | 75/90 | – | – | – | 42/6 |
| Jang/2010 23 | South Korea | E | 99/4 | – | 41/1 | 46/3 | 30/0 |
| Yakoob/2009 24 | Pakistan | M | 9/113 | – | 27/3 | 27/5 | 35/6 |
| Douraghi/2009 10 | Iran | M | 61/78 | 20/14 | 103 | – | 30/4 |
| Basso/2008 12 | Italy | U | 31/40 | 30/15 | – | – | 35/9 |
| Ogiwara/2008 25 | Japan | E | 118/4 | – | – | 51/4 | 81/2 |
| Rhead/2007 9 | Iran | M | 19/29 | – | – | 102 | 28/16 |
CG, chronic gastritis; DU, duodenal ulcer; E, East Asia; GC, gastric cancer; GU, gastric ulcer; M, Middle Asia; NUD, nonulcer disease; PUD, peptic ulcer disease; U, Europe.
All the included studies used either nonulcer disease or chronic gastritis as the control group. The study conducted by Yakoob et al. 24 had a significant heterogeneity comparing with the other included studies, which caused dramatic publication bias and affected the credibility of the outcome. Therefore, we excluded this study and the remaining 14 studies with 2445 patients were pooled for meta‐analysis. A standard PCR assay was performed by all of the included studies for the detection of the vacA i genotype. The study subjects were divided into East Asian, Middle Asian, and European populations according to geographical location. Considering the difference of geographic location, data from different regions in the same study were retrieved separately for subgroup analysis. The main characteristics and specific values of each subgroup were shown in Table 2.
Table 2.
The main results of pooled analyses in exploring GC and PUD risk with the H. pylori i1/i2 genotype
| Variables | Literatures, n | Patients, n (i1/i2) | Controls, n (i1/i2) | OR | 95% CI | P heterogeneity | χ2 (%) | |
|---|---|---|---|---|---|---|---|---|
| PUD | Total | 11 | 651/174 | 804/289 | 1.38 | 0.87–2.17 | 0.002 | 61.1 |
| GU | 2a | 144/16 | 174/14 | 0.99 | 0.45–2.19 | 0.625 | 0 | |
| DU | 4 | 226/22 | 324/47 | 1.24 | 0.46–3.33 | 0.045 | 62.7 | |
| PU | 7 | 281/136 | 305/228 | 1.52 | 0.79–2.93 | 0.001 | 72.7 | |
| Total | 11 | 651/174 | 807/279 | 1.41 | 0.87–2.29 | 0.001 | 66.6 | |
| E | 4 | 406/34 | 553/37 | 1.00 | 0.60–1.66 | 0.421 | 0 | |
| U | 4 | 192/104 | 138/106 | 1.68 | 0.65–4.37 | 0.000 | 83.2 | |
| M | 3 | 53/36 | 116/136 | 1.64 | 0.57–4.76 | 0.027 | 72.3 | |
| GC | Total | 11 | 504/67 | 653/495 | 5.12 | 2.66–9.85 | 0.001 | 67.4 |
| E | 5 | 298/22 | 403/87 | 3.19 | 0.63–16.21 | 0.000 | 81.9 | |
| M | 5 | 171/36 | 219/368 | 6.64 | 3.57–12.34 | 0.201 | 33.1 | |
| U | 1 | 35/9 | 31/40 | 5.02 | 2.10–11.97 | – | – | |
| NUD | 6 | 317/44 | 384/240 | 3.74 | 1.59–8.81 | 0.004 | 71.3 | |
| CG | 5 | 152/17 | 260/142 | 8.69 | 3.82–19.75 | 0.165 | 38.5 |
CG, chronic gastritis as control source; CI, confidence interval; DU, duodenal ulcer; E, East Asia; GC, gastric cancer; GU, gastric ulcer; χ2, Chi‐square value; M, Middle Asia; NUD, nonulcer disease as control source; OR, odds ratio; PU, peptic ulcer; PUD, peptic ulcer disease; U, Europe. Statistically significant results were in bold.
Two literatures, Kim and Jing, conducted both GU and DU cases resulting in a total of 11 studies.
Association between the vacA i1 genotype and PUD
Meta‐analysis for the pooled results from 11 studies [10, 11, 12, 13, 16, 17, 19, 20, 21, 23, 25] showed no significant association between the H. pylori vacA i1 genotype and PUD (pooled OR = 1.38, 95% CI: 0.87–2.17; P heterogeneity = 0.002, χ2 = 61.1%). In the subgroup analysis for ulcer location (GU and DU), no significant association was observed between the H. pylori i1 genotype and GU (pooled OR = 0.99, 95% CI: 0.45–2.19) or DU (pooled OR = 1.24, 95% CI: 0.46–3.33) (Fig. 2A). When grouping studies according to geographical region (East Asia, Middle Asia, and Europe), the pooled results showed that the H. pylori i1 genotype was not associated with PUD in different region (East Asia: OR = 1.00, 95% CI: 0.60–1.66; Middle Asia: OR = 1.64, 95% CI: 0.57–4.76; Europe: OR = 1.68, 95% CI: 0.65–4.37) as shown in Fig. 2B.
Figure 2.
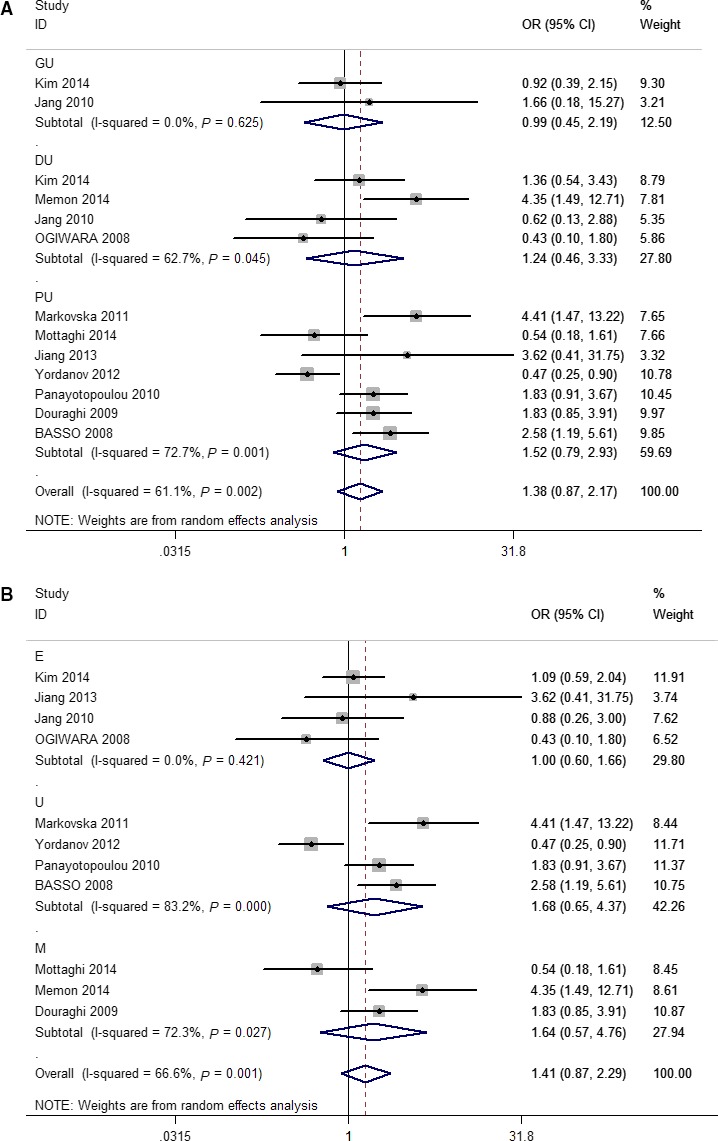
Forest plots of effect estimates for vacA polymorphism and PUD risk (i1 vs i2, overall and subgroup analysis). For each study, the estimation of odds ratio (OR) with 95% confidence interval (CI) is plotted with a box and a horizontal line. Filled diamond means pooled OR with 95% CI. DU, duodenal ulcer; E, East Asia; GU, gastric ulcer; M, Middle Asia; PU, peptic ulcer; U, Europe.
Association between the vacA i1 genotype and GC
The association between the H. pylori vacA i1 genotype and GC was assessed for 11 studies [9, 10, 11, 12, 13, 16, 18, 22, 23, 25] and the results indicated that the H. pylori vacA i1 genotype was associated with an increased risk of GC (pooled OR = 5.12, 95% CI: 2.66–9.85, P heterogeneity = 0.001, χ2 = 67.4%) as shown in Fig. 3A. Furthermore, ethnicity subgroup analysis revealed that the H. pylori vacA i1 genotype was associated with an increased GC risk in the Middle Asian population (OR = 6.64, 95% CI: 3.57–12.34), but not in the East Asian population (OR = 3.19, 95% CI: 0.63–16.21) as shown in Fig. 3B. In addition, the subgroup analysis by the origin of control (NUD and CG) revealed that individuals with infection of H. pylori i1 have an increased GC risk in both the NUD (ORNUD = 3.74, 95% CI: 1.59–8.81) and CG (ORCG = 8.69, 95%CI: 3.82–19.75) control groups as shown in Table 2 and Fig. 3C.
Figure 3.
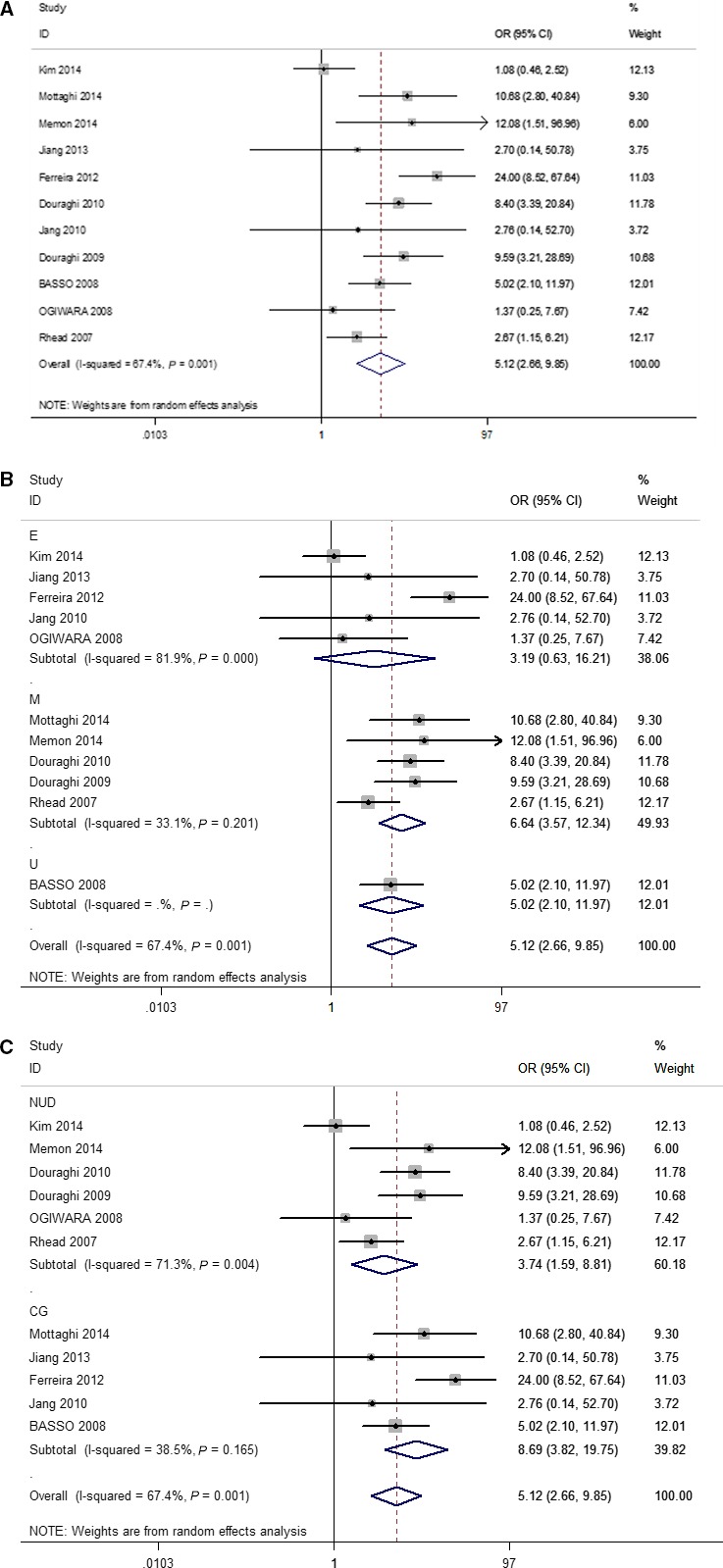
Forest plots of effect estimates for vacA polymorphism and GC risk (i1 vs i2, pooled analysis). For each study, the estimation of odds ratio (OR) with 95% confidence interval (CI) is plotted with a box and a horizontal line. Filled diamond means pooled OR with 95% CI. CG, chronic gastritis; E, East Asia; M, Middle Asia; NUD, nonulcer disease; U, Europe.
Publication bias and sensitivity analysis
Funnel plots were used to evaluate the publication bias of the association between the present of the vacA i1 genotype and GC qualitatively. The shape of the funnel plot did not reveal any evidence of obvious asymmetry (t = 0.048, P = 0.639). The Forest plot demonstrated that no significant publication bias was observed for the studies of GC. Sensitivity analysis revealed that no single study presented a dramatic influence for the final conclusion as shown in Fig. 4A,B.
Figure 4.
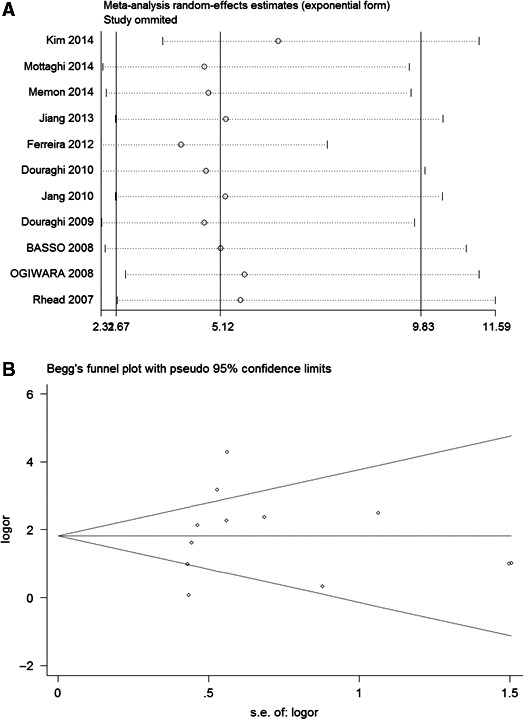
(A) Publication bias test for all included studies concerning GC risk (i1 vs i2). Each circle represents an independent study for the indicated association. Log[odds ratio (OR)]: natural logarithm of OR. Horizontal line shows the mean effect size. SE represents the standard error. (B) Sensitivity analysis of the effect of individual study on the pooled ORs for vacA i1/i2 and GC risk. Each study is represented by one circle. The horizontal line represents the pooled effect estimate.
Discussion
This meta‐analysis has confirmed that the vacA i1 genotype is closely related with an increased risk of GC in the Middle Asian population. Moreover, the pooled analysis also indicates that the vacA i1 genotype has no association with the occurrence of PUD and GC except in Middle Asia. These findings suggest that GC is associated with the infection of H. pylori strains harboring the vacA i1 genotype. The conclusions of four included studies 13, 17, 18, 23 based on Asian population are in agreement with this study. However, this trend does not apply to all of the Asian population 9. The difference in the association between H. pylori vacA i1 and GC among different parts of Asia may be caused by an interaction between the host and other factors.
The results of this study are consistent with previous studies conducted in the Asian population 10, 11, but not in the European population 13, which may be explained by the fact that almost all strains in the East Asian populations are the i1 genotype, whereas only approximately 50% of strains in the European and Middle East populations are the i1 genotype 25. In addition, the dietary habit and host gene polymorphism can partly explain the differences 3.
Although vacA exists in all H. pylori strains, it can hardly induce severe cytoplasmic vacuolation 26. In fact, Rhead et al. 9 first proposed the H. pylori vacA i1 genotype as an independent risk factor for gastric disease. The polymorphic differences in the i‐region may affect vacuolation activity, which present no association with other vacA genotype. Thereafter, many studies tried to find the association among the vacA i1, m1, s1 genotype and diseases of the digestive tract. A similar meta‐analysis has concluded that vacA s1 and m1 exert an increased GC risk than their allele gene s2 (OR 5.32, 95% CI: 2.76–10.26) and m2 (OR 2.50, 95% CI: 1.67–3.750), respectively 27. Interestingly, the data of s1 vs s2 is exactly similar to our result (OR 5.12, 95% CI: 2.66–9.85), which indicates that vacA s1 and i1 may share synergetic effects in the process of carcinogenesis. Argent et al. 28 also found that vacA i1 might not be considered as an independent variable in gastric pathogenesis, they proposed that genetic variation within each virulence factors might affect the function of their own products.
The vacA i1 and i2 genotypes of H. pylori is the target of disease prevention and drug development. Winter et al. 29 found that the strains with vacA i1 remained an increased colonization success rates and planting densities when comparing with the i2 genotype. Moreover, the pathogenesis function of vacA i was further confirmed by Ogiwara et al. 30, who demonstrated that the vacA i1 strains infection induced severer inflammatory cell infiltration in gastric epithelial cell when compared with the i2 genotype. In addition, in the mouse‐colonizing H. pylori strain with cagA negative, vacA demonstrated an independent effect on the induction of inflammation and metaplasia, indicating that s1/i1 type of vacA is the most pathogenic genotype. To a certain extent, H. pylori vacA may drive metaplasia through a different mechanism, which might explain why a different model has obtained a different conclusion. Based on this, the presence of specific i, s, m‐region polymorphisms is identified as a significant risk factor for digestive disease in certain populations.
Currently, it is well known that the infection rate of H. pylori is determined by living environment, dieting habits, and geographic regions. H. pylori is an important cause for PUD and GC. However, the pathogenicity was modified by the genotype of H. pylori. Therefore, it is urgently needed to identify the specific genotype of H. pylori isolated from the PUD and GC patients, which may significantly reduce the costs of screening and prevention for PUD and GC 31, 32. Indeed, the prediction role of the vacA i genotype may be varied in different pathogenic mechanisms. We could not conclude that the vacA i genotype statues are more closely associated with PUD and GC based on the limited samples in this study. Nevertheless, we anticipate that the vacA i1 and i2 genotypes may be a potential indicator for the risk of PUD and GC among patients infected with H. pylori strains.
There are some limitations in this meta‐analysis. Firstly, the languages of the included studies are only limited to English and Chinese, which may contribute to selection bias. Secondly, the control groups are constituted of CG, NUD, and functional dyspepsia. Some studies do not clarify whether the control group has excluded other disease or not, which may result in an underestimate of the effect of H. pylori strains. Thirdly, there is a lack of published reports with a focus on European and American populations, which prevents us from making a generalizable conclusion. Finally, other important data such as age, gender, family history, dieting habits, and other virulent factors are not available to investigate the association between vacA i gene and these factors.
Conclusion
This is the first meta‐analysis evaluating the association between the vacA i genotype and the risk of GC and PUD. Our results demonstrate that H. pylori vacA i1 gene is associated with an increased risk of GC comparing with the CG and PUD controls.
Author contributions
XL extracted the data and wrote the manuscript. BH extracted the data and revised the manuscript. HY and YP searched and screened the eligible articles. FW and KL conducted the statistics. HP prepared Tables and Figures. SW designed the study. SW and WCC reviewed, revised, and approved the manuscript.
Acknowledgements
This study was supported by grants from the National Nature Science Foundation of China (no. 81200401), Nanjing Medical Science and technique Development Foundation to BH (no. JQX13003, QRX11254, QYK11175) and YP (no. QRX11255). We also thank Liu Jian and Xu Tao from Medical Records Division, Nanjing First Hospital, Nanjing Medical University for their consultation and help during the data collection.
References
- 1. McColl KE (2010) Clinical practice. Helicobacter pylori infection. N Engl J Med 362, 1597–1604. [DOI] [PubMed] [Google Scholar]
- 2. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans (1994) Schistosomes, liver flukes and Helicobacter pylori. Lyon, 7–14 June 1994. IARC Monogr Eval Carcinog Risks Hum 61, 1–241. [PMC free article] [PubMed] [Google Scholar]
- 3. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet‐Tieulent J and Jemal A (2015) Global cancer statistics, 2012. CA Cancer J Clin 65, 87–108. [DOI] [PubMed] [Google Scholar]
- 4. Atherton JC (2006) The pathogenesis of Helicobacter pylori‐induced gastro‐duodenal diseases. Annu Rev Pathol 1, 63–96. [DOI] [PubMed] [Google Scholar]
- 5. Iwamoto H, Czajkowsky DM, Cover TL, Szabo G and Shao Z (1999) vacA from Helicobacter pylori: a hexameric chloride channel. FEBS Lett 450, 101–104. [DOI] [PubMed] [Google Scholar]
- 6. McClain MS, Cao P, Iwamoto H, Vinion‐Dubiel AD, Szabo G, Shao Z and Cover TL (2001) A 12‐amino‐acid segment, present in type s2 but not type s1 Helicobacter pylori vacA proteins, abolishes cytotoxin activity and alters membrane channel formation. J Bacteriol 183, 6499–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Van Doorn LJ, Figueiredo C, Megraud F, Pena S, Midolo P, Queiroz DM, Carneiro F, Vanderborght B, Pegado MD, Sanna R et al (1999) Geographic distribution of vacA allelic types of Helicobacter pylori . Gastroenterology 116, 823–830. [DOI] [PubMed] [Google Scholar]
- 8. Basiri Z, Safaralizadeh R, Bonyadi MJ, Somi MH, Mahdavi M and Latifi‐Navid S (2014) Helicobacter pylori vacA d1 genotype predicts risk of gastric adenocarcinoma and peptic ulcers in northwestern Iran. Asian Pac J Cancer Prev 15, 1575–1579. [DOI] [PubMed] [Google Scholar]
- 9. Rhead JL, Letley DP, Mohammadi M, Hussein N, Mohagheghi MA, Eshagh Hosseini M and Atherton JC (2007) A new Helicobacter pylori vacuolating cytotoxin determinant, the intermediate region, is associated with gastric cancer. Gastroenterology 133, 926–936. [DOI] [PubMed] [Google Scholar]
- 10. Douraghi M, Talebkhan Y, Zeraati H, Ebrahimzadeh F, Nahvijoo A, Morakabati A, Ghafarpour M, Esmaili M, Bababeik M, Oghalaie A et al (2009) Multiple gene status in Helicobacter pylori strains and risk of gastric cancer development. Digestion 80, 200–207. [DOI] [PubMed] [Google Scholar]
- 11. Memon AA, Hussein NR, Miendje Deyi VY, Burette A and Atherton JC (2014) Vacuolating cytotoxin genotypes are strong markers of gastric cancer and duodenal ulcer‐associated Helicobacter pylori strains: a matched case‐control study. J Clin Microbiol 52, 2984–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Basso D, Zambon CF, Letley DP, Stranges A, Marchet A, Rhead JL, Schiavon S, Guariso G, Ceroti M, Nitti D et al (2008) Clinical relevance of Helicobacter pylori cagA and vacA gene polymorphisms. Gastroenterology 135, 91–99. [DOI] [PubMed] [Google Scholar]
- 13. Kim JY, Kim N, Nam RH, Suh JH, Chang H, Lee JW, Kim YS, Kim JM, Choi JW, Park JG et al (2014) Association of polymorphisms in virulence factor of Helicobacter pylori and gastroduodenal diseases in South Korea. J Gastroenterol Hepatol 29, 984–991. [DOI] [PubMed] [Google Scholar]
- 14. Welch V, Petticrew M, Tugwell P, Moher D, O'Neill J, Waters E, White H and PRISMA‐Equity Bellagio group (2012) PRISMA‐Equity 2012 extension: reporting guidelines for systematic reviews with a focus on health equity. PLoS Med 9, e1001333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zintzaras E and Ioannidis JP (2005) Heterogeneity testing in meta‐analysis of genome searches. Genet Epidemiol 28, 123–137. [DOI] [PubMed] [Google Scholar]
- 16. Mottaghi B, Safaralizadeh R, Bonyadi M, Latifi‐Navid S and Somi MH (2014) Helicobacter pylori vacA i region polymorphism but not babA2 status associated to gastric cancer risk in northwestern Iran. Clin Exp Med 16, 57–63. [DOI] [PubMed] [Google Scholar]
- 17. Jiang SH, Wu Y and He MJ (2013) Helicobacter pylori vacA i/d genotype features and clinical relevance in Zhenjiang, China. J Jiangsu Univ (Med Ed) 23, 156–160. [Google Scholar]
- 18. Ferreira RM, Machado JC, Letley D, Atherton JC, Pardo ML, Gonzalez CA, Carneiro F and Figueiredo C (2012) A novel method for genotyping the Helicobacter pylori vacA intermediate region directly in gastric biopsy specimens. J Clin Microbiol 50, 3983–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yordanov D, Boyanova L, Markovska R, Gergova G and Mitov I (2012) Significance of Helicobacter pylori vacA intermediate region genotyping‐a Bulgarian study. Diagn Microbiol Infect Dis 74, 253–257. [DOI] [PubMed] [Google Scholar]
- 20. Markovska R, Boyanova L, Yordanov D, Gergova G and Mitov I (2011) Helicobacter pylori oipA genetic diversity and its associations with both disease and cagA, vacA s, m, and i alleles among Bulgarian patients. Diagn Microbiol Infect Dis 71, 335–340. [DOI] [PubMed] [Google Scholar]
- 21. Panayotopoulou EG, Sgouras DN, Papadakos KS, Petraki K, Breurec S, Michopoulos S, Mantzaris G, Papatheodoridis G, Mentis A and Archimandritis A (2010) CagA and vacA polymorphisms are associated with distinct pathological features in Helicobacter pylori‐infected adults with peptic ulcer and non‐peptic ulcer disease. J Clin Microbiol 48, 2237–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Douraghi M, Talebkhan Y, Zeraati H and Mohammadi M (2010) Cooperative genotyping for Helicobacter pylori virulence determinants strengthens the predictive value of gastric cancer risk assessment. Dig Liver Dis 42, 662–663. [DOI] [PubMed] [Google Scholar]
- 23. Jang S, Jones KR, Olsen CH, Joo YM, Yoo YJ, Chung IS, Cha JH and Merrell DS (2010) Epidemiological link between gastric disease and polymorphisms in vacA and CagA. J Clin Microbiol 48, 559–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yakoob J, Abid S, Abbas Z, Jafri W, Ahmad Z, Ahmed R and Islam M (2009) Distribution of Helicobacter pylori virulence markers in patients with gastroduodenal diseases in Pakistan. BMC Gastroenterol 9, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ogiwara H, Graham DY and Yamaoka Y (2008) vacA i‐region subtyping. Gastroenterology 134, 1267; author reply 1268. [DOI] [PubMed] [Google Scholar]
- 26. Tee W, Lambert JR and Dwyer B (1995) Cytotoxin production by Helicobacter pylori from patients with upper gastrointestinal tract diseases. J Clin Microbiol 33, 1203–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matos JI, de Sousa HA, Marcos‐Pinto R and Dinis‐Ribeiro M (2013) Helicobacter pylori CagA and VacA genotypes and gastric phenotype: a meta‐analysis. Eur J Gastroenterol Hepatol 25, 1431–1441. [DOI] [PubMed] [Google Scholar]
- 28. Argent RH, Thomas RJ, Letley DP, Rittig MG, Hardie KR and Atherton JC (2008) Functional association between the Helicobacter pylori virulence factors vacA and CagA. J Med Microbiol 57, 145–150. [DOI] [PubMed] [Google Scholar]
- 29. Winter JA, Letley DP, Cook KW, Rhead JL, Zaitoun AA, Ingram RJ, Amilon KR, Croxall NJ, Kaye PV, Robinson K et al (2014) A role for the vacuolating cytotoxin, vacA, in colonization and Helicobacter pylori‐induced metaplasia in the stomach. J Infect Dis 210, 954–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ogiwara H, Sugimoto M, Ohno T, Vilaichone RK, Mahachai V, Graham DY and Yamaoka Y (2009) Role of deletion located between the intermediate and middle regions of the Helicobacter pylori vacA gene in cases of gastroduodenal diseases. J Clin Microbiol 47, 3493–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yuan Y, Padol IT and Hunt RH (2006) Peptic ulcer disease today. Nat Clin Pract Gastroenterol Hepatol 3, 80–89. [DOI] [PubMed] [Google Scholar]
- 32. Thun MJ, DeLancey JO, Center MM, Jemal A and Ward EM (2010) The global burden of cancer: priorities for prevention. Carcinogenesis 31, 100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]


