Abstract
Pseudomonas aeruginosa strain 1001 produces an esterase (EstA) that can hydrolyse the racemic methyl ester of β‐acetylthioisobutyrate to produce the (D)‐enantiomer, which serves as a precursor of captopril, a drug used for treatment of hypertension. We show here that PA2949 from P. aeruginosa PA01, a homologue of EstA, can efficiently be expressed in an enzymatically active form in E. coli. The enzyme is membrane‐associated as demonstrated by cell fractionation studies. PA2949 was purified to homogeneity after solubilisation with the nonionic detergent, Triton X‐100, and was shown to possess a conserved esterase catalytic triad consisting of Ser137–His258–Asp286. Our results should allow the development of an expression and purification strategy to produce this biotechnologically relevant esterase in a pure form with a high yield.
Keywords: (D)‐β‐acetylthioisobutyric acid, Escherichia coli, esterase, inhibition, membrane protein, Pseudomonas aeruginosa
Abbreviations
- DAT
(D)‐β‐acetylthioisobutyric acid
- EDTA
ethylenediaminetetraacetic acid
- IMAC
immobilised metal affinity chromatography
- MUB
4‐methylumbelliferyl butyrate
- PMSF
phenylmethylsulfonyl fluoride
- pNPC6
p‐nitrophenyl hexanoate
- pNPP
p‐nitrophenyl palmitate
- SDS/PAGE
sodium dodecyl sulphate polyacrylamide gel electrophoresis
- THL
tetrahydrolipstatine
Carboxyl esterases (EC 3.1.1.1) and lipases (EC 3.1.1.3), usually referred to as lipolytic enzymes, hydrolyse ester bonds of a wide range of lipidic substrates 1. They exert various cellular functions in animals, plants and microorganisms 2, 3, 4. The present classification scheme consists of 15 bacterial lipase families, which contain several hundreds of protein sequences and three dimensional structures 5, 6. Despite relatively low amino acid sequence similarity (typically below 20%) lipolytic enzymes have a common α/β‐hydrolase fold, characterised by a central hydrophobic sheet composed of mostly eight β‐strands, which are connected by α‐helices 7. These enzymes are characterised by a typical GXSXG‐consensus motif containing the catalytic serine, which forms the catalytic triad with histidine and aspartate residues 8. Glycine residues of the GXSXG‐motif account for the localisation of the catalytic serine on top of a sharp turn preceded by an α‐helix followed by a β‐strand. This structural motif, named nucleophilic elbow, is essential for substrate hydrolysis and represents one of the best conserved structural motifs among α/β‐hydrolases. The catalytic triad and the residues forming the oxyanion hole, which stabilises the tetrahedral intermediates formed during the hydrolysis, are structurally strongly conserved in all α/β‐hydrolases 9.
Although many structural features of α/β‐hydrolases are similar, these enzymes show a wide range of functionalities 10. In recent years, the number of biochemical and structural data about lipolytic enzymes are rapidly increased, however, the understanding of structure‐function relationship is still limited 11. Nowadays, lipolytic enzymes are broadly used as industrial biocatalysts, since many of them are fairly stable in harsh conditions that are often needed for industrial bioprocess 12, 13, for example, high temperatures, presence of organic solvents or high ionic strength buffers. Moreover, their enantio‐, stereo‐ and regioselectivity allow controlled organic synthesis reactions using complex unnatural substrates, as, for example, precursors for the biopharmaceuticals paclitaxel 14, naproxen 15 and captopril 16. The latter is the first drug used for the treatment of hypertension, congestive heart failure and diabetic nephropathy, which acts by inhibiting angiotensin‐converting enzyme 16. Presently, captopril is still prescribed and distributed under the trade name Capoten®. Its chemical synthesis requires the optically pure key intermediate, (D)‐β‐acetylthioisobutyric acid (DAT) as the inhibiting potency of the (D)‐enantiomer is 100‐fold higher than of the (L)‐enantiomer 16, 17. To overcome the laborious and expensive chemical synthesis of optically active DAT, Sakimae et al. 18 screened for microorganisms producing esterases, which can stereoselectively hydrolyse the racemic methyl ester of β‐acetylthioisobutyrate 18. The functional screening revealed several strains capable of DAT synthesis with Pseudomonas strains producing DAT of highest optical purity 18.
The esterase EstA from P. aeruginosa 1001 was cloned, overexpressed and shown to produce DAT 19, 20. However, expression of EstA in E. coli resulted in low enzymatic activity presumably caused by formation of protein aggregates, which could not be completely abolished by fusion with maltose‐binding protein 19, 20. Here, we report cloning, expression and purification of the protein encoded by open reading frame pa2949 from P. aeruginosa PA01, which is homologous to EstA from P. aeruginosa 1001. We have developed a system for efficient expression of highly active PA2949 in common laboratory strain E. coli BL21(DE3). Biochemical analysis showed esterase but no lipase activity of PA2949, and enzyme activity could be inhibited by serine‐hydrolase inhibitors. Furthermore, we showed that PA2949 is localised in the membrane of E. coli prompting us to develop a detergent‐based purification method, which yielded mg amounts of enzymatically active protein. Our data demonstrates that PA2949 can be functionally expressed, easily purified and adequately stabilised thus making it available for a range of different biotechnological applications.
Materials and methods
Bioinformatic analysis
Amino acid sequences were analysed and aligned using the blast search and alignment tool of the Universal Protein Knowledge Base (www.uniprot.org) 21. Signal peptide cleavage sites were predicted by two different methods, namely the Hidden Markov Model (Signal P‐HMM) 22 and neural network (Signal P‐NN) 22. Signal peptides were distinguished from nonsignal peptides by a threshold D‐score of SignalP‐NN higher than 0.5 and by a threshold C‐score of SignalP‐HMM higher than 0.95. The transmembrane helix was predicted using the Toppred 23 online tool with a score higher than 0.8.
Cloning, site‐directed mutagenesis, expression and purification of PA2949
Restriction endonucleases, Pfu DNA polymerase and bacteriophage T4 DNA ligase (Thermo Scientific, Darmstadt, Germany) reactions were carried out as recommended by the manufacturers. DNA fragments were analysed on 1% (w/v) agarose gels. Plasmid DNA was purified using the InnuPREP DOUBLE pure kit (Analytik Jena, Jena, Germany) or, for genomic DNA from P. aeruginosa PA01 using the DNeasy tissue kit (Qiagen, Hilden, Germany). Used strains and plasmids are listed in Table 1 24, 25, 26, 27.
Table 1.
Strains and plasmids used in this study
| Strains | Genotype | Source |
|---|---|---|
| E. coli DH5α | supE44 Δ(lacZYA‐argF)U196 (Φ80ΔlacZM15) hsdR17 recA1 endA1 gyrA96 thi‐1 relA1 | 24 |
| E. coli BL21(DE3) | F− ompT hsdSB(rB −mB −) gal dcm (λIts857 indI sam7 nin5 lacUV5‐T7gene1) | 25 |
| P. aeruginosa PA01 | Wild‐type, originating from Dieter Haas laboratory (Lausanne, CH) | 26 |
| Plasmids | Description | Source |
|---|---|---|
| pET22b+ | ColE1 PT7Ф10 pelB Apr C‐His6‐‘Tag’ lacIq | Novagen |
| pBBR1mcs3 | Cmr mob lacZα Plac PT7 Tcr | 27 |
| pET22b‐pa2949 | pa2949H6 gene inserted in NdeI/SacI of pET22b(+) | This study |
| pET22b‐pa2949_S137A | pa2949 gene with substitutions of Ser137 with Ala inserted in NdeI/SacI sites of pET22b(+) | This study |
| pBBR1mcs3‐pa2949 | XbaI/SacI fragment of pET22b‐pa2949 inserted in pBBR1mcs‐3 | This study |
The pa2949 gene was amplified by standard PCR using Pfu DNA polymerase, the genomic DNA of P. aeruginosa PA01 as a template and oligonucleotide pair 5′‐AAACATATGAAACGATTCCTC‐3′/5′‐TCAGAGCTC CACCACCACCACCACCACGCGACCGGCCAC‐3′ encoding NdeI and SacI restriction sites (underlined) and a C‐terminal His6‐tag (bold). Primers were synthesised by MWG Biotech. The pa2949 gene was cloned into NdeI and SacI restriction sites of pET22b yielding expression plasmid pET‐pa2949 (Table 1) allowing for bacteriophage T7‐RNA polymerase‐dependent expression from the T7 promoter. The mutation of Ser137Ala in PA2949 was performed by the Quik‐change PCR method using Pfu DNA polymerase (Invitrogen, Darmstadt, Germany), pET‐pa2949 plasmid and the complementary mutagenic oligonucleotide pair 5′‐TGGCCGGCAACG T C C C CATGGGCGGG‐3′/5′‐CCCGCCCATG G G G C AGTTGCCGGCCA‐3′ (mutated codons are underlined and nucleotides of the wild‐type gene are indicated in the subscript). Correctness of plasmids pET‐pa2949 and pET‐pa2949_S137A was confirmed by DNA sequencing (MWG Biotech, Ebersberg, Germany).
For the expression of PA2949 and PA2949 S137A, E. coli BL21(DE3) cells transformed respectively with pET‐pa2949 and pET‐pa2949_S137A were grown overnight at 37 °C in a Luria–Bertani (LB) medium supplemented with ampicillin (100 μg·mL−1). These cultures were used to inoculate an expression culture in LB medium supplemented with ampicillin (100 μg·mL−1) to an initial OD580 nm = 0.05. The cultures were grown at 37 °C until they reached logarithmic phase (OD580 nm = 0.5–0.8) and gene expression was induced by addition of isopropyl‐β‐D‐thiogalactosid (IPTG) to a final concentration of 0.4 mm. After 2 h, cells were harvested by centrifugation at 4000 g and 4 °C for 20 min and stored at −20 °C.
SDS/PAGE, zymography and immunodetection
Proteins were analysed by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS/PAGE) under denaturation conditions on 12% (w/v) gels as described by Laemmli 28. Esterase activity in SDS/PAGE gels was detected by zymography using the fluorescent substrate 4‐methylumbelliferyl butyrate (MUB) 29. The proteins transferred from SDS/PAGE gel to the polyvinylidene difluoride membranes by western blotting 30 were detected using anti‐His(C‐term)‐HRP antibodies (Invitrogen) according the manufacturers' instructions. The protein concentration was determined by the method of Bradford with bovine serum albumin as a standard 31.
Cellular localisation, purification and biochemical characterisation
Cellular localisation
Gene pa2949 was subcloned from pET‐pa2949 into pBBR1mcs‐3 behind the lac promoter using XbaI and SacI restriction sites yielding plasmid pBBR‐pa2949. E. coli DH5α cells harbouring pBBR‐pa2949 were cultivated overnight in LB medium supplemented with tetracycline (10 μg·mL−1) at 37 °C. The cells were harvested by centrifugation (1 min, 19 000 g, and 4 °C), resuspended in 100 mm Tris‐HCl buffer (pH 8), disrupted by sonication and total cell membranes were isolated by ultracentrifugation (30 min, 180 000 g, 4 °C) 29, 32.
Purification
The total membrane fraction containing PA2949 with a C‐terminal His6‐tag was solubilised with 1% (w/v) Triton X‐100 by gentle agitation overnight at 4 °C. Solubilised membranes were subjected to ultracentrifugation (30 min, 180 000 g, 4 °C) and PA2949 was purified from the supernatant by immobilised metal affinity chromatography (IMAC) using Ni‐NTA agarose (Qiagen) 33. All buffers used were supplemented with (1% w/v) Triton X‐100 to keep PA2949 in soluble form. Purified PA2949 samples eluted from the Ni‐NTA column were transferred into Tris‐HCl buffer (100 mm, pH 8) containing 1% (v/v) Triton X‐100 by gel filtration using PD‐10 column (GE Healthcare, Solingen, Germany) according to the manufacturers' protocol. The purified protein was then stored at room temperature.
Enzyme activity assays
Enzymatic activities were determined in 96‐well microplates by adding 5 μL of enzyme sample to 200 μL of substrate with p‐nitrophenyl hexanoate (pNPC6) for esterase and p‐nitrophenyl palmitate (pNPP) for lipase activity 1.
Temperature optimum
Esterase activities were measured over a range of temperatures from 10 °C to 70 °C as described previously 34. Assays were performed in a 96‐well microplate by adding 2 μL of enzyme sample to 200 μL of pNPC6 substrate.
pH and organic solvents stability
Esterase activities of PA2949 incubated for 1 h with buffers of a pH range from 3 to 10.5 34 or for 3 h with various organic solvents (dimethyl sulfoxide, N, N‐dimethyl formamide, methanol, acetonitrile, ethanol, acetone, propan‐2‐ol, diethyl ether, hexane, toluene) 34 were measured in a 96‐well microplate by adding 2 μL of enzyme sample to 200 μL of pNPC6 substrate.
Inhibition
The inhibition of PA2949 was tested according Asler et al. 35 using THL, PMSF, paraoxon (all dissolved in propan‐2‐ol) and the EDTA (dissolved in H2O). Inhibition of PA2949 was performed by incubating enzyme aliquots with the inhibitors for 3 h at the room temperature and subsequent determination of enzyme activity using pNPC6 as the substrate.
Results and Discussion
Open reading frame PA2949 of P. aeruginosa PA01 encodes a putative lipase
By searching the Pseudomonas genome database (www.pseudomonas.com) 36 we have identified about hundred genes of P. aeruginosa PA01 encoding putative lipolytic enzymes, among them ORF pa2949. This gene of 948 bp length encodes a protein of Mr 34.8 kDa with a predicted Abhydrolase_6 Pfam domain (PF12697) spanning residues 65–299 (Fig. 1A). A blast search revealed homology of PA2949 with known esterases and lipases, namely EstA from P. aeruginosa 1001 19, 20, and two lipases from psycrophilic bacteria, Moraxella sp. lipase 37 and Psychrobacter immobilis lipase 38 (Fig. 1B). The genes encoding pa2949 from P. aeruginosa PA01 and estA from P. aeruginosa 1001 share 99% identity (data not shown), and the protein sequences are identical (Fig. 1). The amino acid sequences of Moraxella sp. and P. immobilis lipases are ~ 50% similar to the one of PA2949. The sequence alignment revealed the strictly conserved amino acid Ser137 embedded in the conserved GXSXG‐lipase motif, as well as Asp258 and His286 predicted to form the catalytic triad of PA2949.
Figure 1.

Sequence analysis of PA2949. (A) Molecular organisation of PA2949 from P. aeruginosa PA01 showing putative transmembrane domain (TM, amino acids 4‐24) and catalytic α/β‐hydrolase domain (amino acids 65–299). The cleavage site for the predicted signal peptide (LLA) and the putative active site residues (Ser, Asp, His) are marked by the grey pin and the red diamonds respectively. (B) Sequence alignment of PA2949 with EstA from P. aeruginosa 1001 19, 20, Moraxella sp. lipase MsLip 37 and Psychrobacter immobilis lipase PiLip 50. Residues identical and similar in at least three sequences were shaded in black and yellow respectively.
Expression of PA2949 in E. coli BL21(DE3)
In order to obtain enzymatically active and soluble PA2949, we constructed a heterologous expression system using E. coli BL21(DE3) carrying plasmid pET‐pa2949. Bacteria were grown at 37 °C and expression of pa2949 was induced by addition of 0.4 mm IPTG. SDS/PAGE and western blot analyses revealed expression of a protein with an estimated molecular weight of 35 kDa. In parallel, we measured significantly increased esterase activity in the cell lysate of the expression strain compared with the strain carrying the empty vector (Fig. 2). Additionally, esterase activity of the 35 kDa protein was detected by zymographic analysis. Interestingly, PA2949 did not show activity with palmitic acid p‐nitrophenyl ester considered as a typical lipase substrate. In conclusion, we could demonstrate that PA2949 of P. aeruginosa can be functionally expressed in E. coli BL21(DE3) and displays esterase but not lipase activity.
Figure 2.

Expression and esterase activity of PA2949. (A) Coomassie Blue‐stained SDS gel (12%) after separation of extracts from E. coli BL21(DE3) cells carrying empty vector (EV, pET22b) or PA2949 expression vector (pET‐pa2949). The gel was loaded with equal amount of cell extracts collected before induction (0 h) and 1 or 2 h after induction with IPTG. Molecular weights of standard proteins (St) are indicated on the left; the black arrow indicates overexpressed PA2949. (B) Western blot of samples shown in A using anti‐His‐tag antibodies. (C) Esterase activity of cell extracts from samples shown in Fig. 2A. The results ± standard deviations are means of three independent experiments, each set in triplicate.
PA2949 is localised in the membrane of E. coli
The subcellular localisation of a protein must be considered to develop an efficient purification protocol 39. As cellular localisation was unknown for PA2949 and its homologues, we first performed a bioinformatic analysis revealing either a putative N‐terminal type I signal peptide spanning amino acids 1–28 or a putative transmembrane helix spanning amino acids 4–24 suggesting a periplasmic, extracellular or membrane localisation. Many membrane proteins containing transmembrane helices display anomalous migration in SDS/PAGE 40 that is often heat inducible 41, and caused by differences in binding of SDS to heat‐treated and untreated forms of the protein. Hence, we have tested the effect of temperature on the electrophoretic mobility of PA2949 and have shown that PA2949 migrated faster after incubation at 99 °C prior to electrophoresis compared to the sample incubated at room temperature indicating that it is a membrane protein (data not shown).
Therefore, we have experimentally separated membranes and soluble fraction of E. coli cells expressing PA2949. For this experiment, we expressed PA2949 in E. coli DH5α under the control of the weak lac promoter rather than the strong T7 promoter to avoid overloading of membranes and prevent mislocalisation 42. The gene pa2949 was subcloned from plasmid pET‐pa2949 into the broad host range vector pBBR1mcs‐3 43 and expression was performed in E. coli DH5α without induction. Cells were disrupted by sonication, membranes were isolated by ultracentrifugation, analysed by SDS/PAGE and western blotting and PA2949 was detected solely in the membrane fraction (Fig. 3). Thus, recombinant PA2949 expressed in E. coli is localised in the cell membrane, however, the localisation of PA2949 in the homologous host P. aeruginosa is still unknown. As E. coli and P. aeruginosa belong to the same class of Gamma‐proteobacteria and share evolutionarily conserved signal recognition particles and Sec‐translocation systems for targeting membrane proteins 44 we predict membrane localisation of PA2949 in P. aeruginosa as well.
Figure 3.

Subcellular localisation of recombinant PA2949 in E. coli DH5α. The membranes (M) and soluble fraction (SF) of E. coli DH5α carrying pBBR‐pa2949 or pBBR1mcs‐3 (empty vector, EV) were separated by ultracentrifugation and equal amount of proteins from each cell fraction were analysed by western blotting using anti‐His‐tag antibodies (upper panel) and by SDS/PAGE (lower panel). Molecular weights of protein standard (St) are indicated on the left.
Purification and biochemical characterisation of PA2949
Purification of membrane proteins requires the usage of detergents for solubilisation as well as to prevent subsequent protein aggregation by stabilising hydrophobic domains, which are naturally embedded in the membrane 45. Here, we have selected the nonionic detergent Triton X‐100 commonly used for purification of membrane proteins from E. coli 46 to extract PA2949 from the membranes. Initially, the total membrane fraction of E. coli BL21(DE3) expressing PA2949 was incubated for 1 h at room temperature with Triton X‐100 at concentrations exceeding the critical micellar concentration. Although, Triton X‐100 in a concentration range from 0.5 to 2% (w/v) did not reduce the esterase activity of PA2949, the protein was not quantitatively solubilised from the membranes (results not shown). Almost quantitative extraction of PA2949 was achieved after overnight incubation of membranes with detergent without losing esterase activity. Solubilisation of membranes with Triton X‐100 and subsequent purification using immobilised metal affinity chromatography yielded ~ 1 mg/L culture/OD580 nm of pure PA2949 (Fig. 4). Purified PA2949 preparation had 198.8 ± 5.1 U·mg−1‐specific esterase activity measured with p‐nitrophenyl hexanoate at 30 °C that corresponds to approximately 60–70% of total esterase activity of membrane fraction of E. coli BL21(DE3)‐expressing PA2949. Incubation of PA2949 with various buffers (pH 3.0–10.5) revealed that activity was best retained in Tris‐HCl buffer pH 8.0 at room temperature (Table 2), whereas storage at 4 °C or freezing (also in the presence of 30% glycerol) lead to the precipitation and inactivation of PA2949.
Figure 4.
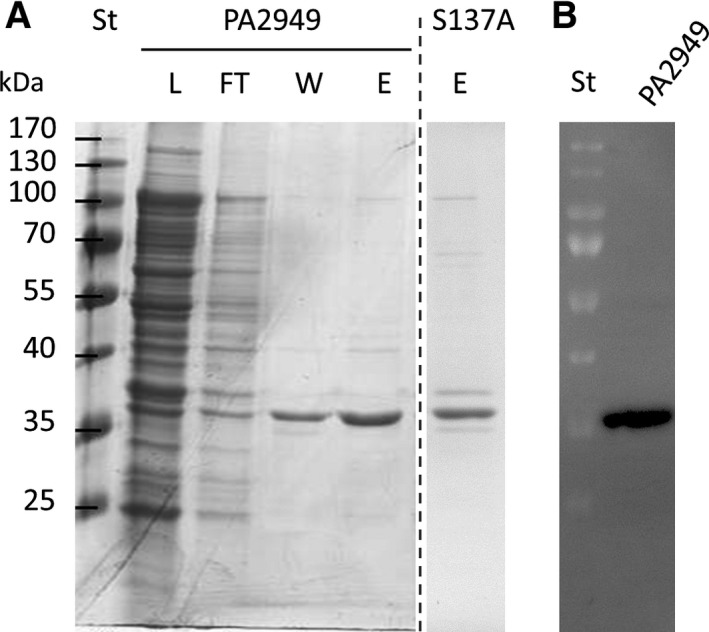
Purification of recombinant PA2949 from E. coli BL21(DE3). (A) Coomassie Blue‐stained SDS‐gel (12%) showing fractions obtained by purification of PA2949 and its catalytic inactive variant PA2949_S137A using a Ni‐NTA column. L, load fraction containing membranes of E. coli BL21(DE3) expressing PA2949 solubilised with Triton X‐100, FT, flow through; W, fraction after washing with buffer containing 60 mm imidazole and E, fraction eluted from the column with buffer containing 250 mm imidazole. Molecular weights of protein standard (St) are indicated on the left. The fraction containing PA2949_S137A is shown on a separate SDS gel (indicated by a dashed line). (B) Zymogram indicating esterase activity of purified PA2949 (elution fraction shown in A) monitored under UV light using the fluorescent substrate 4‐methylumbelliferyl butyrate.
Table 2.
pH stability of PA2949
| pH | Activity ± SD (%) |
|---|---|
| 3.0 | 40.1 ± 2.0 |
| 4.5 | 60.2 ± 4.3 |
| 6.0 | 80.4 ± 3.7 |
| 7.5 | 88.7 ± 7.8 |
| 8.0 | 100.0 ± 6.7 |
| 8.5 | 87.4 ± 8.7 |
| 9.0 | 81.1 ± 6.7 |
| 9.5 | 62.5 ± 12.6 |
| 10.5 | 42.3 ± 9.1 |
Esterase activities ± standard deviations are means of three independent experiments, each set in triplicate.
The similarity of the PA2949 amino acid sequence with psychrophilic esterases (Fig. 1) prompted us to test if PA2949 retained its activity also at low temperatures. Determination of esterase activities at temperatures ranging from 10 °C to 70 °C revealed 30 °C as the optimal temperature (Fig. 5). Interestingly, even at a temperature of 10 °C PA2949 retained 47% of its activity measured at 30 °C. This data indicate that PA2949 behaves similar to psychrophilic rather than mesophilic enzymes that are usually inactive at low temperatures 47.
Figure 5.

Optimum temperature of PA2949. Esterase activities of purified PA2949 measured at respective temperatures are shown as relative activities compared with the highest activity measured at 30 °C with 100% activity corresponding to the specific activity of the wild‐type (198.8 ± 5.1 U·mg−1). The results ± standard deviations are means of three independent experiments, each set in triplicate.
Stability in the presence of organic solvents is an essential property of enzymes used as biocatalysts for organic synthesis 13. Membrane proteins usually contain membrane‐embedded domains and soluble domains protruding into the surrounding water milieu, which are differently resistant to hydrophobic (high log P) and hydrophilic (low log P) solvents 48. Therefore, we have tested the stability of PA2949 after incubation with various solvents (Table 3). Notably, PA2949 was fairly stable in most of the tested solvents, some of them, for example, diethyl ether and methanol even enhanced its activity, but some hydrophilic solvents (acetonitrile, ethanol and propan‐2‐ol) severely reduced esterase activity.
Table 3.
Stability of PA2949 in the presence of various organic solvents
| Organic solvent | Concentration (%, v/v) | Log P | Residual activity ± SD (%)a |
|---|---|---|---|
| DMSO | 30 | −1.300 | 82.2 ± 4.7 |
| N,N‐Dimethyl formamide | 30 | −1.000 | 26.0 ± 2.7 |
| Methanol | 30 | −0.760 | 111.0 ± 11.1 |
| Acetonitrile | 30 | −0.330 | 0.0 ± 0.0 |
| Ethanol | 30 | −0.240 | 5.4 ± 0.2 |
| Acetone | 30 | −0.230 | 79.1 ± 5.6 |
| Propan‐2‐ol | 30 | 0.074 | 8.1 ± 0.4 |
| Diethyl ether | 30 | 0.850 | 125.6 ± 8.8 |
| Hexane | 5 | 3.500 | 106.9 ± 5.8 |
| Toluene | 5 | 2.500 | 65.1 ± 2.7 |
Residual esterase activities are expressed as a percentage of PA2949 activity in buffer without organic solvent. ± standard deviations are means of three independent experiments, each set in triplicate.
Binding of substrate‐mimicking inhibitors to the enzyme active site can provide valuable data for understanding the catalytic mechanism 49 as well as regio‐ and enantio preference 50 of lipases. We thus determined the inhibition kinetics of PA2949 using typical lipase inhibitors containing long hydrophobic acyl chains, namely tetrahydrolipstatine (THL) 51, the short acyl chain arylesterase inhibitors paraoxon 52, phenylmethylsulfonyl fluoride (PMSF) 53 and an inhibitor of metal‐dependent enzymes, ethylenediaminetetraacetic acid (EDTA) 54. Resistance towards EDTA indicated that PA2949 is not a metalloenzyme (Fig. 6). All three inhibitors targeting the catalytic serine residue (THL, PMSF and paraoxon) inhibited PA2949 activity, although to a different degree (Fig. 6). Incubation of PA2949 with THL for 3 h resulted in 28% of residual activity, whereas THL, PMSF and paraoxon completely abolished PA2949 activity indicating irreversible inhibition. Phosphonate or sulphonate inhibitors covalently linked to the catalytic serine, mimic the first tetrahedral intermediate (before dissociation of alcohol moiety) and the second tetrahedral intermediate (after dissociation of alcohol moiety) formed during ester hydrolysis respectively 10. These results provided further evidence that PA2949 contains in its active site a nucleophlic serine. Using site‐directed mutagenesis, we constructed variant PA2949 S137A, which was purified (Fig. 4), but did not show any esterase activity. These results are in agreement with the bioinformatic prediction of Ser137 as the catalytic residue. Additionally, our data suggest a rather narrow active site of PA2949 because the bulky inhibitor molecule THL apparently could not efficiently bind to the active site. The preparation of stable PA2949‐PMSF and PA2949‐paraoxon complexes we have described here will be used for further crystallographic and kinetic studies.
Figure 6.

Inhibition of PA2949. Residual esterase activities were determined after preincubation for 3 h of 1.7 μm of PA2949 with 10 mm EDTA, 2 mm THL, 1 mm paraoxon and 1 mm PMSF at room temperature. Inhibited PA2949 samples (15 μg) were incubated with 100 μL of substrate at 30 °C and residual esterase activity was measured. PA2949 samples treated with propan‐2‐ol and water represent noninhibited controls. On the right, the chemical structures of inhibitors THL, paraoxon and PMSF are shown with the bonds to be hydrolysed indicated in red. The results ± standard deviations are means of three independent experiments, each set in triplicate. The standard deviations were below 8%.
Biotechnological potential of PA2949
PA2949 from P. aeruginosa PA01 is homologous to the esterase EstA from P. aeruginosa 1001, which was previously shown to hydrolyse racemic β‐acetylthioisobutyrate methyl ester to form enantiopure DAT 19, 20, an important intermediate in the synthesis of pharmaceuticals 16. However, heterologous expression of enzymatically active EstA turned out to be difficult because of poor solubility and low production yield 20. EstA was predicted to be a soluble protein with an N‐terminal signal sequence instead of a transmembrane helix 19, 20. This misleading assumption resulted in the construction of an expression system where maltose‐binding protein (which was shown to enhance the solubility of proteins) was fused to the N‐terminus of EstA resulting in blocking of EstA secretion and expression of only poorly active enzyme 20. Using the pa2949 gene encoding the full‐length protein with C‐terminal His6‐tag we have successfully expressed mg/Lculture quantities of active PA2949 located in the membrane of the common laboratory strain E. coli. Subsequent extraction with the nonionic detergent Triton X‐100 allowed for the efficient purification of up to 70% of PA2949 from membranes and enabled storage at room temperature without loss of activity. The biochemical characterisation of PA2949 revealed 30 °C as the optimal temperature for catalysis (Fig. 5), high stability at pH 7.5–8.5 (Table 2), and notable resistance to various organic solvents (Table 3). Interestingly, a study of DAT production with whole cells of P. putida MR‐2068, which expressed a PA2949 homologue revealed pH 7.5 and 45 °C as the best conditions 55. At higher temperatures and in alkaline pH spontaneous hydrolysis of the racemic β‐acetylthioisobutyrate methyl ester was observed, thereby reducing the optical purity of DAT 55. In summary, PA2949 possesses biochemical properties, which match the requirements for the enzymatic synthesis of DAT as well as its subsequent extraction with organic solvent.
Author contributions
FK, KEJ, RBS and JG conceived and supervised the study; FK, SW and KEJ designed experiments; FK, FB and MC performed experiments; FK, RBS and JG analysed data; FK, KEJ and RBS wrote the manuscript.
Acknowledgements
This work was mainly supported by a research grant from the European Union (FP6 Marie Curie EST project ANTIBIOTARGET; MEST‐CT‐2005‐020278). This work was partially supported by a research grant from the German Research Foundation (DFG) within the Collaborative Research Center 1208 ‘Identity and Dynamics of Membrane Systems’. We thank Frank Rosenau (Zentrum für Peptidpharmazeutika, Universität Ulm, Germany) for helpful discussions during planning of this project and Peter Dollinger for critical reading of the manuscript.
References
- 1. Jaeger K‐E and Kovacic F (2014) Determination of lipolytic enzyme activities. Methods Mol Biol 1149, 111–134. [DOI] [PubMed] [Google Scholar]
- 2. Huang C and Freter C (2015) Lipid metabolism, apoptosis and cancer therapy. Int J Mol Sci 16, 924–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Seth S, Chakravorty D, Dubey VK and Patra S (2014) An insight into plant lipase research – challenges encountered. Protein Expr Purif 95, 13–21. [DOI] [PubMed] [Google Scholar]
- 4. Wilhelm S, Gdynia A, Tielen P, Rosenau F and Jaeger KE (2007) The autotransporter esterase EstA of Pseudomonas aeruginosa is required for rhamnolipid production, cell motility, and biofilm formation. J Bacteriol 189, 6695–6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arpigny JL and Jaeger KE (1999) Bacterial lipolytic enzymes: classification and properties. Biochem J 343, 177–183. [PMC free article] [PubMed] [Google Scholar]
- 6. Charbonneau DM and Beauregard M (2013) Role of key salt bridges in thermostability of G. thermodenitrificans EstGtA2: distinctive patterns within the new bacterial lipolytic enzyme family XV. PLoS One 8, e76675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Heikinheimo P, Goldman A, Jeffries C and Ollis DL (1999) Of barn owls and bankers: a lush variety of α/β hydrolases. Structure 7, 141–146. [DOI] [PubMed] [Google Scholar]
- 8. Upton C and Buckley JT (1995) A new family of lipolytic enzymes? Trends Biochem Sci 20, 178–179. [DOI] [PubMed] [Google Scholar]
- 9. Carr PD and Ollis DL (2009) Alpha/beta hydrolase fold: an update. Protein Pept Lett 16, 1137–1148. [DOI] [PubMed] [Google Scholar]
- 10. Rauwerdink A and Kazlauskas RJ (2015) How the same core catalytic machinery catalyzes 17 different reactions: the serine‐histidine‐aspartate catalytic triad of α/β‐hydrolase fold enzymes. ACS Catal 5, 6153–6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ali YB, Verger R and Abousalham A (2012) Lipases or esterases: does it really matter? Toward a new bio‐physico‐chemical classification. Methods Mol Biol 861, 31–51. [DOI] [PubMed] [Google Scholar]
- 12. Daiha KDG, Angeli R, de Oliveira SD and Almeida RV (2015) Are lipases still important biocatalysts? A study of scientific publications and patents for technological forecasting PLoS One 10, e0131624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jaeger KE and Eggert T (2002) Lipases for biotechnology. Curr Opin Biotechnol 13, 390–397. [DOI] [PubMed] [Google Scholar]
- 14. Schmid A, Dordick JS, Hauer B, Kiener A, Wubbolts M and Witholt B (2001) Industrial biocatalysis today and tomorrow. Nature 409, 258–268. [DOI] [PubMed] [Google Scholar]
- 15. Kwon CH, Lee JH, Kim SW and Kang JW (2009) Lipase‐catalyzed esterification of (S)‐naproxen ethyl ester in supercritical carbon dioxide. J Microbiol Biotechnol 19, 1596–1602. [DOI] [PubMed] [Google Scholar]
- 16. Ondetti MA, Rubin B and Cushman DW (1977) Design of specific inhibitors of angiotensin‐converting enzyme: new class of orally active antihypertensive agents. Science 196, 441–444. [DOI] [PubMed] [Google Scholar]
- 17. Shimazaki M, Hasegawa J, Kan K, Nomura K, Nose Y, Kondo H, Ohashi T and Watanabe K (1982) Synthesis of captopril starting from an optically active β‐hydroxy acid. Chem Pharm Bull 30, 3139–3146. [Google Scholar]
- 18. Sakimae A, Hosoi A, Kobayashi E, Ohsuga N, Numazawa R, Watanabe I and Ohnishi H (1992) Screening of microorganisms producing (D)‐β‐acetylthioisobutyric acid from methyl (DL)‐β‐acetylthioisobutyrate. Biosci Biotechnol Biochem 56, 1252–1256. [Google Scholar]
- 19. Lee J, Boyapati G, Song K, Rhee S and Kim C (2000) Cloning and sequence analysis of the estA gene encoding enzyme for producing (R)‐beta‐acetylmercaptoisobutyric acid from Pseudomonas aeruginosa 1001. J Biosci Bioeng 90, 684–687. [DOI] [PubMed] [Google Scholar]
- 20. Lee JH, Rhee SK and Kim CH (2004) Expression and activation of an esterase from Pseudomonas aeruginosa 1001 in Escherichia coli . Enzyme Microb Technol 35, 563–567. [Google Scholar]
- 21. Magrane M and Consortium U (2011) UniProt knowledgebase: a hub of integrated protein data. Database (Oxford) 2011, bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Emanuelsson O, Brunak S, von Heijne G and Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2, 953–971. [DOI] [PubMed] [Google Scholar]
- 23. von Heijne G (1992) Membrane protein structure prediction: hydrophobicity analysis and the positive‐inside rule. J Mol Biol 225, 487–494. [DOI] [PubMed] [Google Scholar]
- 24. Woodcock DM, Crowther PJ, Doherty J, Jefferson S, DeCruz E, Noyer‐Weidner M, Smith SS, Michael MZ and Graham MW (1989) Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res 17, 3469–3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Studier FW and Moffatt BA (1986) Use of bacteriophage T7 RNA polymerase to direct selective high‐level expression of cloned genes. J Mol Biol 189, 113–130. [DOI] [PubMed] [Google Scholar]
- 26. Holloway BW, Krishnapillai V and Morgan AF (1979) Chromosomal genetics of Pseudomonas . Microbiol Rev 43, 73–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kovach ME, Phillips RW, Elzer PH, Roop RM and Peterson KM (1994) pBBR1MCS: a broad‐host‐range cloning vector. Biotechniques 16, 800–802. [PubMed] [Google Scholar]
- 28. Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 29. Kovacic F, Granzin J, Wilhelm S, Kojic‐Prodic B, Batra‐Safferling R and Jaeger KE (2013) Structural and functional characterisation of TesA – a novel lysophospholipase A from Pseudomonas aeruginosa . PLoS One 8, e69125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yuen SW, Chui AH, Wilson KJ and Yuan PM (1989) Microanalysis of SDS‐PAGE electroblotted proteins. Biotechniques 7, 74–83. [PubMed] [Google Scholar]
- 31. Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Anal Biochem 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 32. Witholt B, Boekhout M, Brock M, Kingma J, Heerikhuizen HV and Leij LD (1976) An efficient and reproducible procedure for the formation of spheroplasts from variously grown Escherichia coli . Anal Biochem 74, 160–170. [DOI] [PubMed] [Google Scholar]
- 33. Lescic Asler I, Ivic N, Kovacic F, Schell S, Knorr J, Krauss U, Wilhelm S, Kojic‐Prodic B and Jaeger KE (2010) Probing enzyme promiscuity of SGNH hydrolases. ChemBioChem 11, 2158–2167. [DOI] [PubMed] [Google Scholar]
- 34. Perez D, Kovacic F, Wilhelm S, Jaeger KE, Garcia MT, Ventosa A and Mellado E (2012) Identification of amino acids involved in the hydrolytic activity of lipase LipBL from Marinobacter lipolyticus . Microbiology 158, 2192–2203. [DOI] [PubMed] [Google Scholar]
- 35. Asler IL, Zehl M, Kovacic F, Muller R, Abramic M, Allmaier G and Kojic‐Prodic B (2007) Mass spectrometric evidence of covalently‐bound tetrahydrolipstatin at the catalytic serine of Streptomyces rimosus lipase. Biochim Biophys Acta 1770, 163–170. [DOI] [PubMed] [Google Scholar]
- 36. Winsor GL, Lam DK, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock RE and Brinkman FS (2011) Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res 39, D596–D600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Feller G, Thiry M and Gerday C (1991) Nucleotide sequence of the lipase gene lip3 from the antarctic psychotroph Moraxella TA144. Biochim Biophys Acta 1088, 323–324. [DOI] [PubMed] [Google Scholar]
- 38. Arpigny JL, Feller G and Gerday C (1993) Cloning, sequence and structural features of a lipase from the antarctic facultative psychrophile Psychrobacter immobilis B10. Biochim Biophys Acta 1171, 331–333. [DOI] [PubMed] [Google Scholar]
- 39. Kubicek J, Block H, Maertens B, Spriestersbach A and Labahn J (2014) Expression and purification of membrane proteins. Methods Enzymol 541, 117–140. [DOI] [PubMed] [Google Scholar]
- 40. Rath A, Glibowicka M, Nadeau VG, Chen G and Deber CM (2009) Detergent binding explains anomalous SDS‐PAGE migration of membrane proteins. Proc Natl Acad Sci USA 106, 1760–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oomen CJ, van Ulsen P, Van Gelder P, Feijen M, Tommassen J and Gros P (2004) Structure of the translocator domain of a bacterial autotransporter. EMBO J 23, 1257–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wagner S, Klepsch MM, Schlegel S, Appel A, Draheim R, Tarry M, Högbom M, van Wijk KJ, Slotboom DJ, Persson JO et al (2008) Tuning Escherichia coli for membrane protein overexpression. Proc Natl Acad Sci USA 105, 14371–14376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM and Peterson KM (1995) Four new derivatives of the broad‐host‐range cloning vector pBBR1MCS, carrying different antibiotic‐resistance cassettes. Gene 166, 175–176. [DOI] [PubMed] [Google Scholar]
- 44. Facey SJ and Kuhn A (2010) Biogenesis of bacterial inner‐membrane proteins. Cell Mol Life Sci 67, 2343–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Arnold T and Linke D (2008) The use of detergents to purify membrane proteins. Curr Protoc Protein Sci 4, 1–4. [DOI] [PubMed] [Google Scholar]
- 46. Schnaitman CA (1971) Solubilization of the cytoplasmic membrane of Escherichia coli by Triton X‐100. J Bacteriol 108, 545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kovacic F, Mandrysch A, Poojari C, Strodel B and Jaeger K‐E (2016) Structural features determining thermal adaptation of esterases. Protein Eng Des Sel 29, 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Laane C, Boeren S, Vos K and Veeger C (1987) Rules for optimization of biocatalysis in organic solvents. Biotechnol Bioeng 30, 81–87. [DOI] [PubMed] [Google Scholar]
- 49. Kim KK, Song HK, Shin DH, Hwang KY and Suh SW (1997) The crystal structure of a triacylglycerol lipase from Pseudomonas cepacia reveals a highly open conformation in the absence of a bound inhibitor. Structure 5, 173–185. [DOI] [PubMed] [Google Scholar]
- 50. Nardini M, Lang DA, Liebeton K, Jaeger KE and Dijkstra BW (2000) Crystal structure of Pseudomonas aeruginosa lipase in the open conformation. The prototype for family I.1 of bacterial lipases. J Biol Chem 275, 31219–31225. [DOI] [PubMed] [Google Scholar]
- 51. Hadvary P, Sidler W, Meister W, Vetter W and Wolfer H (1991) The lipase inhibitor tetrahydrolipstatin binds covalently to the putative active site serine of pancreatic lipase. J Biol Chem 266, 2021–2027. [PubMed] [Google Scholar]
- 52. Crow JA, Bittles V, Herring KL, Borazjani A, Potter PM and Ross MK (2012) Inhibition of recombinant human carboxylesterase 1 and 2 and monoacylglycerol lipase by chlorpyrifos oxon, paraoxon and methyl paraoxon. Toxicol Appl Pharmacol 258, 145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nam KH, Kim SJ, Priyadarshi A, Kim HS and Hwang KY (2009) The crystal structure of an HSL‐homolog EstE5 complex with PMSF reveals a unique configuration that inhibits the nucleophile Ser144 in catalytic triads. Biochem Biophys Res Commun 389, 247–250. [DOI] [PubMed] [Google Scholar]
- 54. Borkar PS, Bodade RG, Rao SR and Khobragade CN (2009) Purification and characterization of extracellular lipase from a new strain: Pseudomonas aeruginosa SRT 9. Braz J Microbiol 40, 358–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sakimae A, Ozaki E, Toyama H, Ohsuga N, Numazawa R, Muraoka I, Hamada E and Ohnishi H (1993) Process conditions for production of D‐β‐acetylthioisobutyric acid from methyl DL‐β‐acetylthioisobutyrate with the cells of Pseudomonas putida MR‐2068. Biosci Biotechnol Biochem 57, 782–786. [Google Scholar]


