Abstract
Microbiota has been shown to promote tolerogenic differentiation of T lymphocytes. It remains unclear to what extent microbiota triggers de novo re-programming or amplify pre-existing plasticity intrinsic to T cells. In a study with mouse models to track the clonal fate of CD4 and CD8 T cells, we discovered that CD8 T cells converted to MHC class I-restricted CD4 T cells without regard to selfness of their antigen specificity. In mesenteric lymph nodes (MLN), CD8 T cells converted to CD4+Foxp3+ regulatory T (Treg) cells which were enriched in the large intestine lamina propria (LILP) and suppressed chemical- or immune-mediated inflammatory damage. In germ-free conditions, the converted CD4 populations were present in MLN, but absent in LILP. Therefore, an intrinsic plasticity in the host was amplified by the gut microbiota, leading to selfless tolerance induction in the intestinal mucosa. The findings may be relevant to HIV infection, cancer and autoimmune disorders.
Keywords: intrinsic, immune tolerance, lineage plasticity, lamina propria, microbiota, mesenteric lymph nodes, selfless, T cells
Harmony between host adaptive immune system and microbiota (“friendly” bacteria)
A symbiosis between the gut microbiota and the host requires a harmonious relationship between the adaptive immune system and the microbiota. In exchange for refuge and nutrients, microbial symbionts offer a number of advantages to their host, including essential vitamins to the host and metabolic assistance. The microbiome can also protect against colonization or invasion of the host by pathogenic bacteria through occupation of available gut niches or production of toxins that target their pathogenic counterparts.1,2 Vertebrate animals are equipped with a large repertoire of lymphocytes with fine specificities, which have evolved to combat harmful infectious microbes. Recent studies have gathered a substantial body of evidence for the mutualistic relationship between the adaptive immune system and the “friendly” microbiota. Microbiota can boost adaptive immune responses against infectious agents.3,4 On the other hand, adaptive immunity can regulate the diversity of the microbiota. 5,6 Gut microbiota is instrumental in shaping the adaptive immune repertoire. For instance, segmented filamentous bacteria in the small intestine are known to induce Th17 cells.7 In the colon, microbiota can induce CD4+Foxp3+ regulatory T (Treg) cells to promote peripheral tolerance.8 Therefore, understanding the immunological basis of the mutualistic relationship between host adaptive immune system and gut microbiota will provide insight into various types of immune-mediated diseases in humans and potential novel strategies for therapeutic and preventive interventions.
Dilemma of “selfhood”-based tolerance: How to tolerate “friends” if “friends” look like “enemies”
Immune tolerance induction is largely characterized by discrimination of self-antigens in the host against nonself-antigens in foreign microbes. In the adaptive immune system, tolerance is achieved by clonal selection of lymphocytes with fine specificities. In the innate immune system, the discrimination against harmful microbes is based on molecular pattern recognition of microbial agents.9 How, then, does the immune system recognize the microbiota, which share most molecular patterns with harmful microbes? Studies have documented peripheral generation of Treg cells specific to the bacteria in the gut microbiota.10 However, one might argue that although overall tolerance to the gut microbiota is necessary for preservation of their symbiotic presence, specific tolerance to a broad range of bacteria can be harmful due to the minimal difference between pathogenic bacteria and their microbiota. Given this dilemma, how does the immune system tolerate the microbiome even though these “friendly” microbes appear immunologically like the harmful infectious microbes?
Evidence for selfless tolerance induction at the interface with gut microbiota
We explored how T lymphocytes, major players of the adaptive immune system, tolerate the microbiome in the gut. We followed the clonal fate of CD4 and CD8 T cells in vivo, using some of the commonly used T-cell receptor (TCR)-transgenic models. We chose T cell clones (OT1 and OTII) that recognize a nonself-antigen. In our studies, the animal models harboring those T cell clones did not carry the cognate antigens. We also chose 2 clones (BDC2.5 and 8.3) that recognize a self-antigen that is present mainly in a gut-distal organ (the pancreas) rather than in the gut.11-14 These TCR transgenic models were bred onto the recombination activating gene (Rag)-deficient background 15 to create mouse models with a single clone of T cells. We made a number of surprising findings from these models.16
First, we discovered the presence of CD4 T cell populations in Rag-deficient OT1 mice, a commonly used model which has been assumed to harbor a monoclonal repertoire of CD8 T cells. The converted CD4 T cells were particularly enriched in the gut-associated environment and expressed the same clonotypic TCR as in OT1 CD8 T cells. Indeed, they recognized the MHC Class I (MHCI)-restricted SIINFEKL peptide of the OT1 TCR. The CD8-to-CD4 lineage conversion was also found in the other CD8 clone we analyzed, 8.3. Using a lineage-tracking model we determined that mature peripheral CD8 T cells from a natural polyclonal repertoire could also convert to CD4 T cells, thus establishing the generality of CD8-to-CD4 lineage conversion in the periphery.
Second, the converted CD4 T cells were highly enriched in the LILP, with the MHCI-restricted CD4+Foxp3+ Treg (CI-Treg) accounting for the vast majority of CD4 T cells. Interestingly, in such a steady state of CD4 versus CD8 imbalance, we did not detect conversion from the CD4 lineage to CD8 T cells. Furthermore, we did not detect gut-associated generation of CD4+Foxp3+ T cells from BDC2.5 and OTII CD4 T cell clones,16 even though the potential of the conventional CD4 T cells to convert to the CD4+Foxp3+ Treg cell lineage has been shown in pharmacological interventions.17
Third, the process of cross-differentiation to CI-Treg cells required the MLN. Surgical removal of the MLN at 2 weeks of age eliminated the cross-differentiation of CD8 T cells to CI-Treg cells. Of note, the converted CD4+Foxp3− T cells were still detectable in the animals which had their MLN removed at 2 weeks of age. Technical limits in survival surgeries precluded us from determining whether or not removing the MLN of the animals at an earlier age would completely eliminate CD8-to-CD4 lineage conversion, or if the conversion from the CD8 lineage to CD4+Foxp3− cells does not require the MLN.
Fourth, despite the TCR recognition of cognate antigens presented by MHCI, cross-differentiation from the CD8 lineage to CD4 T cells required MHC Class II (MHCII). However, we did not detect any pre-requisite of MHCII-based thymic selection, or “mis-selection,” of the CD8 T cells that would be needed to potentiate the peripheral conversion from the CD8 lineage to CD4 T cells.
Lastly, the CD8-to-CD4 lineage conversion occurred regardless of the types of diet and housing facilities we tested. Indeed, the cross-differentiation from the CD8 T cell lineage to CD4 T cells occurred even in the absence of microbiota. As illustrated in Figure. 1, in germ-free conditions, the converted CD4 T cells were present in the MLN, but absent in the LILP.
Figure 1.
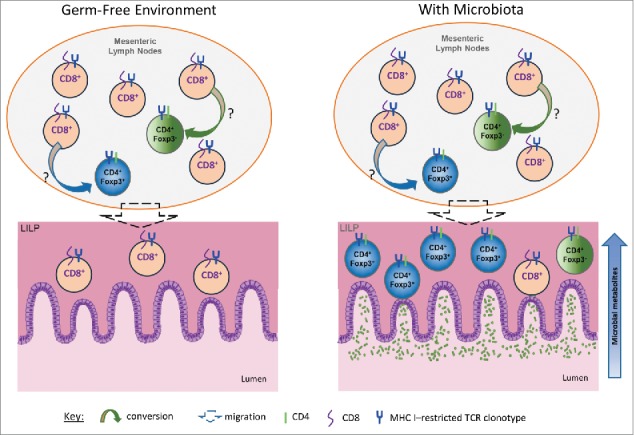
Intrinsic plasticity of CD8 T cell lineage amplified by microbiota. A hypothetical model is presented to show the plasticity of the CD8 T cell lineage spontaneously converting to CD4 T cells in the periphery. In this model, the presence of the gut microbiota is not required to trigger the original generation of CD4+Foxp3+ CI-Treg cells from CD8 T cells. However, the gut microbiota greatly amplifies the converted population, especially in the LILP. The yet-to-be-identified, host-intrinsic mechanisms initiate the conversion from the CD8 lineage to CD4 T cells including CD4+Foxp3+ T cells. The gut microbiota, or more likely their metabolites, may promote the homing and expansion of the converted population in the LILP. The enrichment of CI-Treg cells in the LILP suggests that food antigens are unlikely involved since they are present primarily in the small intestine. The converted CD4 CI-Treg cells may lead to the creation of a “friendly zone” at the interface of the adaptive immune system and the gut microbiota that facilitates immune tolerance induction in a “selfless” mode.
These data, collectively, suggest that there is a host-intrinsic plasticity in the CD8 lineage for cross-differentiation to CI-Treg cells. Microbiota is not the original trigger for CD8-to-CD4 lineage conversion, but they may play a critical role in attracting and expanding the converted population in the LILP to facilitate mucosal tolerance induction (Fig. 1).16
Since bacterial lipopolysaccharides (LPS) exist in abundance, and are even suspected to be present in the sterile feed in germ-free conditions, we examined if signals from LPS could be the trigger for the CD8 lineage to cross-differentiate to CD4 T cells. Given that the Toll-like receptor 4 (TLR4) is the mammalian cell receptor for LPS,18 we crossed TLR4 knockout mutations into the OT1 mouse model on the Rag-deficient background. As shown in Figure. 2A-B, TLR4-deficiencies did not diminish the percentages and total numbers of CD4+ T cells or the CD4+Foxp3+ Treg subset in the MLN or LILP. Therefore, TLR4 was not required for conversion from the CD8 T cell lineage to CD4 T cells, nor was it required for the homing and expansion of the converted population in the LILP. It remains to be determined if and how other types of bacterial components or metabolites from gut microbes play a role in this novel pathway of T cell differentiation and tolerance induction in the intestinal mucosa. For example, short-chain fatty acids were shown to promote the population size and function of colonic Treg cells through G protein–coupled receptors.19 These metabolites could similarly expand the CI-Treg cell population in the LILP.
Figure 2.
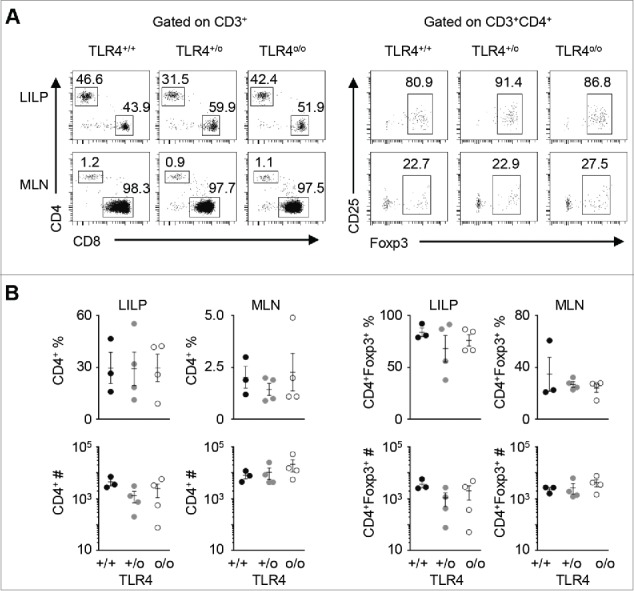
TLR4, the mammalian cell receptor for LPS, one of the most abundant products of microbial agents, is not required for gut-associated cross-differentiation from the CD8 lineage to CD4 T cells. OT1+Rago mice were crossed with the TLR4 knockout line to generate TLR4-deficient OT1+Rago mice and controls. A) Representative flow cytometry plots of OT1+RagoTLR4+/+ (n = 3), OT1+RagoTLR4+/o (n = 4), and OT1+RagoTLR4o/o (n = 4) mice analyzed at 3-5 weeks of age. Numbers in each plot represent the percentage of the gated population. B) Summary of CD4+ and CD4+Foxp3+ T cell percentages and counts. Each data point represents one animal (mean ± SEM; no statistical significance was detected among the groups).
If not microbiota, what else distinguishes the gut environment?
The germ-free experiment clearly demonstrated that microorganisms were not required as an original trigger of the CD8-to-CD4 lineage conversion. What else then is unique in the gut-associated environment, especially in the MLN? We tested a few commercially available diets for the animals, and did not detect substantial differences.16 Nevertheless, we could not exclude the role of food antigens since we do not have a model to test the effect of its absence. However, we did not detect a substantial population of converted CD4 T cells in the small intestine lamina propria. This suggests that it is unlikely the food antigens are causing the conversion, as food antigens are mostly found in the small intestine,20 whereas the converted CD4 T cells were found primarily in the large intestine.16
Could the cytokine milieu in the gut-associated environment be involved in cross-differentiation from the CD8 lineage to CD4 T cells? TGFβ is the usual suspect. Indeed, it has been well recognized for its role in the conversion of conventional CD4 T cells into CD4 Treg cells.21 We reported in our study that blocking TGFβ signaling in T cells did not prevent the CD8-to-CD4 cross-differentiation.16 The role of other cytokines has yet to be tested. Perhaps, anatomic and / or physiological characteristics in the gut-associated environment 20 also play an instrumental role for the host-intrinsic plasticity of the CD8 T cell lineage. Of note, previous studies showed that in the MLN, both dendritic cells and stromal cells presented antigens to stimulate OT1 cell responses.22 Recent studies have also demonstrated that lymph node stromal cells not only express MHCI molecules, but can also express MHCII molecules and thus play a role in CD4 T cell-mediated immunity vs. tolerance.23-26
Functions in physiological niches of gut mucosa and cell transfer settings for protection from colitis
To examine the function of CI-Treg cells in their physiological niches in LILP, we resorted to a genetic approach to test the impact of their absence. We compared Rag-deficient OT1 mice with or without a null-mutation of the Foxp3 gene. If one assumes Rag-deficient OT1 mice indeed harbor a monoclonal repertoire of CD8 T cells as generally thought, and minor populations of CD4 T cells in the model are mere “noise” and biologically non-consequential, then the Foxp3 null mutation should not have any effect. After all, Foxp3 expression in mice is restricted to the CD4+Foxp3+ Treg cell lineage. Foxp3 mutant Rag-deficient OT1 mice were apparently healthy. However, an age-associated accumulation of activated CD8 T cells did occur in the immune system. When the animals were challenged with a chemical—dextran sodium sulfate, which induces inflammatory damage to the colon—the Foxp3 mutant animals developed moribund inflammatory pathology in the intestine even after the inflammatory trigger was discontinued, whereas the Foxp3 wildtype Rag-deficient OT1 mice recovered from the colitis. It remains unknown how exactly the CI-Treg cells facilitate the repair of intestinal tissue from inflammatory destruction. Along this line, it should be noted that a previous study showed that CD4 Treg cells in the muscle could induce tissue repair by secreting amphiregulin.27
We also tested the potential of CI-Treg cells in adoptive cell transfer settings. Consistent with the observations from the Foxp3 mutant Rag-deficient OT1 model, adoptively transferred CI-Treg cells suppressed homeostatic activation of CD8 T cells. Importantly, CI-Treg cells isolated from Rag-deficient OT1 mice suppressed autoimmune colitis induced by polyclonal CD8 T cells, even in the absence of the cognate antigen for the specific OT1 CI-Treg cells. They were suppressive even in a setting and dose in which standard CD4+Foxp3+ Treg cells from C57BL/6 mice were not effective.16 Therefore, future studies are warranted to test whether CI-Treg cells are indeed more potent than standard MHCII-restricted CD4 Treg cells in various settings of autoimmune or inflammatory pathology.
Implications to other conditions of immune-related disorders
Treg cells need to be activated for functioning. Presumably, Treg cells which recognize antigens presented by MHCI are more likely to encounter their antigens, given the ubiquitous expression of MHCI on all nucleated cells, as opposed to the more restricted expression of MHCII on antigen-presenting cells. In vivo, Treg cells can suppress autoimmune damage through a number of mechanisms, including contact-dependent interaction with pathogenic T cells.28 It is conceivable, then, that MHCI-based interaction would further facilitate the interaction of Treg cells with their targets. Furthermore, the likelihood of MHCI-based antigenic recognition would lead to a higher probability of memory cell formation, which may perpetuate the potency of Treg cells' suppressive activities.29-31
The overall suppressive nature of the gut-associated environment is shared in a pathological setting, the tumor microenvironment. It too presents a challenge to immunological discrimination based on self and nonself. On one hand, tumors present an immunoprivileged self that can suppress potent autoimmune damage.32-34 On the other hand, the genome instabilities of tumor cells also produce mutations that generate potential neoantigens.35 It would be interesting to examine whether conversion from CD8 T cells to CD4 CI-Treg cells occurs in the tumor microenvironment, not only in gastrointestinal cancer, but also in cancer in general, especially in settings of adoptive CD8 T cell therapies.
One of the striking findings from our studies is that the 8.3 clone of the CD8 lineage exhibited conversion to CD4 T cells in the gut-associated environment, rather than in the draining lymph node of the pancreas, where the specific self-antigen of the 8.3 clone is expressed. This finding suggests a selfless mode of tolerance induction in the gut environment that can protect gut-distal organs from autoimmune damage. A particularly interesting result is that the conversion appeared to depend on the genetic background. It was absent in the autoimmune-prone NOD genetic background, but occurred on the NOD/B6 mixed background. Our preliminary tests found that this was not due to an association with the MHC locus of NOD, but a yet-to-be-identified genetic element(s) not linked to the MHC locus.16
Perhaps, the most obvious implication of these findings is to HIV infection. In this setting, CD4 T cells are depleted, leaving an imbalance of CD8 versus CD4 lineage analogous to that in the Rag-deficient OT1 model. Of note, CD4 T cells are severely depleted in the gut-associated lymphoid tissue (GALT) in HIV-infected patients, even though some of these individuals have relatively normal CD4 T cell frequencies in the peripheral blood. Effective long term antiretroviral therapies (9 years) did not completely restore the CD4 T cell population in the GALT.36,37 Then would one expect the conversion from the CD8 lineage to CD4 T cells and an enrichment of potent CI-Treg cells in the LILP? Could those converted CD4 T cells become a new target for HIV infection? The availability of MHC-tetramer reagents and matched clinical samples can help future studies examine the potential existence of MHCI-restricted CD4 T cells in the gut-associated environment in settings of HIV infection. However, it may not be feasible to determine the origin of such cells, if they are detected, until the successful development of an in vitro organ culture model and / or a humanized mouse model that enables lineage tracing of human CD8 T cell development.
MHC class I-restricted CD4 T cells in humans
The existence of MHCI-restricted CD4 T cells at the clonal and population levels in healthy humans was discovered by Strassman and Bach more than 30 years ago.38 Later studies implicated this type of cell in cancer and autoimmune diseases.39-45 In human ankylosing spondylitis, its strong association with HLA-B2746,47 contrasts with the MHCII association of most autoimmune diseases. HLA-B27-restricted cells in the patients were found in both CD8 and CD4 lineages.45,48 As in studies of other types of human cells, it is difficult to pinpoint the origin of MHCI-restricted CD4 T cells in humans, e.g., from peripheral conversion, or due to “mis-selection” by MHCII in the thymus. In the mouse model, we showed strong evidence that CD8-to-CD4 cross-differentiation was not due to “imprinting” by thymic MHCII.16 Rather, it was generated in the periphery in the gut-associated environment, especially the MLN.
Although it is not feasible to determine the origin of the MHCI-restricted CD4 T cell clones in humans, the interventions based on CI-Treg cells, and for that matter, MHCI-restricted CD4 T helper cells, may hold major potential for clinical translation. The gap of knowledge presents a substantial obstacle, though. For example, in human inflammatory bowel diseases (IBD) and other disorders, if and how CI-Treg cells are involved awaits tools for effective detection of this type of cells. For that purpose, the molecular signature of CI-Treg cells need to be uncovered. Their antigen specificity needs to be determined, so new MHCI-tetramer reagents can be designed to identify and isolate those cells. Efforts have been made to engineer MHC class-I restricted CD4 Treg cells,49,50 but it is not known how adding an exogenous MHCI-restricted TCR to the native MHCII-restricted TCR in the same CD4 T cell may confound the antigen specificity of the T cell. Future studies with animal models are needed to understand the cellular and molecular signals that trigger the spontaneous conversion from the CD8 T cell lineage to CD4 T cell clones in the gut-associated environment. The knowledge from those studies will be useful for designing strategies to reliably and efficiently generate human CD4 CI-Treg cells as well as MHCI-restricted cytotoxic and helper CD4 T cells in vitro, for potential applications in cell therapies.
Overall, the finding of CD8-to-CD4 lineage plasticity and the major role of the gut microbiota in amplification of this host-intrinsic plasticity may offer new potential interventions to IBD and autoimmune diseases in general. The potential of CD8-to-CD4 T cell lineage conversion also has critical implications to HIV infection and cancer immunotherapies.
Abbreviations
- CI-Treg
MHC class I (MHCI)-restricted CD4+Foxp3+ regulatory T cells
- LILP
large intestine lamina propria
- MLN
mesenteric lymph nodes
Disclosure of potential conflict of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We apologize to all colleagues who contribute important knowledge to the surveyed topics but are not cited due to the limit of citation number and the scope of our literature knowledge.
Funding
Z.C. received support from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK/NIH) (DP3DK085696) and the University of Miami Miller School of Medicine Dean's Bridge funding. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agency.
References
- 1.Kamada N, Chen GY, Inohara N, Nunez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol 2013; 14:685-90; PMID:23778796; http://dx.doi.org/ 10.1038/ni.2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hammami R, Fernandez B, Lacroix C, Fliss I. Anti-infective properties of bacteriocins: an update. Cell Mol Life Sci 2013; 70:2947-67; PMID:23109101; http://dx.doi.org/ 10.1007/s00018-012-1202-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, Paley MA, Antenus M, Williams KL, Erikson J, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 2012; 37:158-70; PMID:22705104; http://dx.doi.org/ 10.1016/j.immuni.2012.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geuking MB, Koller Y, Rupp S, McCoy KD. The interplay between the gut microbiota and the immune system. Gut Microbes 2014; 5:411-8; PMID:24922519; http://dx.doi.org/ 10.4161/gmic.29330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang H, Sparks JB, Karyala SV, Settlage R, Luo XM. Host adaptive immunity alters gut microbiota. ISME J 2015; 9:770-81; PMID:25216087; http://dx.doi.org/ 10.1038/ismej.2014.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H, Luo XM. Control of commensal microbiota by the adaptive immune system. Gut Microbes 2015; 6:156-60; PMID:25901893; http://dx.doi.org/ 10.1080/19490976.2015.1031946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009; 139:485-98; PMID:19836068; http://dx.doi.org/ 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011; 331:337-41; PMID:21205640; http://dx.doi.org/ 10.1126/science.1198469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janeway CA Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol 2002; 20:197-216; PMID:11861602; http://dx.doi.org/ 10.1146/annurev.immunol.20.083001.084359 [DOI] [PubMed] [Google Scholar]
- 10.Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, Peterson DA, Stappenbeck TS, Hsieh CS. Peripheral education of the immune system by colonic commensal microbiota. Nature 2011; 478:250-4; PMID:21937990; http://dx.doi.org/ 10.1038/nature10434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell 1994; 76:17-27; PMID:8287475; http://dx.doi.org/ 10.1016/0092-8674(94)90169-4 [DOI] [PubMed] [Google Scholar]
- 12.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based α- and β-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol 1998; 76:34-40; PMID:9553774; http://dx.doi.org/ 10.1046/j.1440-1711.1998.00709.x [DOI] [PubMed] [Google Scholar]
- 13.Verdaguer J, Schmidt D, Amrani A, Anderson B, Averill N, Santamaria P. Spontaneous autoimmune diabetes in monoclonal T cell nonobese diabetic mice. J Exp Med 1997; 186:1663-76; PMID:9362527; http://dx.doi.org/ 10.1084/jem.186.10.1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katz JD, Wang B, Haskins K, Benoist C, Mathis D. Following a diabetogenic T cell from genesis through pathogenesis. Cell 1993; 74:1089-100; PMID:8402882; http://dx.doi.org/ 10.1016/0092-8674(93)90730-E [DOI] [PubMed] [Google Scholar]
- 15.Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 1992; 68:869-77; PMID:1547488; http://dx.doi.org/ 10.1016/0092-8674(92)90030-G [DOI] [PubMed] [Google Scholar]
- 16.Lui JB, Devarajan P, Teplicki SA, Chen Z. Cross-Differentiation from the CD8 Lineage to CD4 T Cells in the Gut-Associated Microenvironment with a Nonessential Role of Microbiota. Cell Rep 2015; 10:574-85; PMID:25640181; http://dx.doi.org/ 10.1016/j.celrep.2014.12.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu Y, Suzuki J, Guillioli M, Umland O, Chen Z. Induction of self-antigen-specific Foxp3+ regulatory T cells in the periphery by lymphodepletion treatment with anti-mouse thymocyte globulin in mice. Immunology 2011; 134:50-9; PMID:21711461; http://dx.doi.org/ 10.1111/j.1365-2567.2011.03466.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 1998; 282:2085-8; PMID:9851930; http://dx.doi.org/ 10.1126/science.282.5396.2085 [DOI] [PubMed] [Google Scholar]
- 19.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013; 341:569-73; PMID:23828891; http://dx.doi.org/ 10.1126/science.1241165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol 2014; 14:667-85; PMID:25234148; http://dx.doi.org/ 10.1038/nri3738 [DOI] [PubMed] [Google Scholar]
- 21.Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, McGrady G, Wahl SM. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-β induction of transcription factor Foxp3. J Exp Med 2003; 198:1875-86; PMID:14676299; http://dx.doi.org/ 10.1084/jem.20030152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JW, Epardaud M, Sun J, Becker JE, Cheng AC, Yonekura AR, Heath JK, Turley SJ. Peripheral antigen display by lymph node stroma promotes T cell tolerance to intestinal self. Nat Immunol 2007; 8:181-90; PMID:17195844; http://dx.doi.org/ 10.1038/ni1427 [DOI] [PubMed] [Google Scholar]
- 23.Roozendaal R, Mebius RE. Stromal cell-immune cell interactions. Annu Rev Immunol 2011; 29:23-43; PMID:21073333; http://dx.doi.org/ 10.1146/annurev-immunol-031210-101357 [DOI] [PubMed] [Google Scholar]
- 24.Malhotra D, Fletcher AL, Astarita J, Lukacs-Kornek V, Tayalia P, Gonzalez SF, Elpek KG, Chang SK, Knoblich K, Hemler ME, et al. Transcriptional profiling of stroma from inflamed and resting lymph nodes defines immunological hallmarks. Nat Immunol 2012; 13:499-510; PMID:22466668; http://dx.doi.org/ 10.1038/ni.2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng CT, Nayak BP, Schmedt C, Oldstone MB. Immortalized clones of fibroblastic reticular cells activate virus-specific T cells during virus infection. Proc Natl Acad Sci U S A 2012; 109:7823-8; PMID:22550183; http://dx.doi.org/ 10.1073/pnas.1205850109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dubrot J, Duraes FV, Potin L, Capotosti F, Brighouse D, Suter T, LeibundGut-Landmann S, Garbi N, Reith W, Swartz MA, et al. Lymph node stromal cells acquire peptide-MHCII complexes from dendritic cells and induce antigen-specific CD4+ T cell tolerance. J Exp Med 2014; 211(6):1153-66; PMID:24842370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burzyn D, Kuswanto W, Kolodin D, Shadrach JL, Cerletti M, Jang Y, Sefik E, Tan TG, Wagers AJ, Benoist C, et al. A special population of regulatory T cells potentiates muscle repair. Cell 2013; 155:1282-95; PMID:24315098; http://dx.doi.org/ 10.1016/j.cell.2013.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miska J, Abdulreda MH, Devarajan P, Lui JB, Suzuki J, Pileggi A, Berggren PO, Chen Z. Real-time immune cell interactions in target tissue during autoimmune-induced damage and graft tolerance. J Exp Med 2014; 211:441-56; PMID:24567447; http://dx.doi.org/ 10.1084/jem.20130785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenblum MD, Gratz IK, Paw JS, Lee K, Marshak-Rothstein A, Abbas AK. Response to self antigen imprints regulatory memory in tissues. Nature 2011; 480:538-42; PMID:22121024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Devarajan P, Miska J, Lui JB, Swieboda D, Chen Z. Opposing effects of CTLA4 insufficiency on regulatory vs. conventional T cells in autoimmunity converge on effector memory in target tissue. J Immunol 2014; 193:4368-80; PMID:25246499; http://dx.doi.org/ 10.4049/jimmunol.1400876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Devarajan P, Chen Z. Autoimmune effector memory T cells: the bad and the good. Immunol Res 2013; 57:12-22; PMID:24203440; http://dx.doi.org/ 10.1007/s12026-013-8448-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miska J, Bas E, Devarajan P, Chen Z. Autoimmunity-mediated antitumor immunity: tumor as an immunoprivileged self. Eur J Immunol 2012; 42:2584-96; PMID:22777737; http://dx.doi.org/ 10.1002/eji.201242590 [DOI] [PubMed] [Google Scholar]
- 33.Miska J, Devarajan P, Chen Z. The immunological identity of tumor: Self implications. Oncoimmunology 2013; 2:e23794; PMID:23734327; http://dx.doi.org/ 10.4161/onci.23794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toomer KH, Chen Z. Autoimmunity as a double agent in tumor killing and cancer promotion. Front Immunol 2014; 5:116; PMID:24672527; http://dx.doi.org/ 10.3389/fimmu.2014.00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science 2015; 348:69-74; PMID:25838375; http://dx.doi.org/ 10.1126/science.aaa4971 [DOI] [PubMed] [Google Scholar]
- 36.Guadalupe M, Reay E, Sankaran S, Prindiville T, Flamm J, McNeil A, Dandekar S. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J Virol 2003; 77:11708-17; PMID:14557656; http://dx.doi.org/ 10.1128/JVI.77.21.11708-11717.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chun TW, Nickle DC, Justement JS, Meyers JH, Roby G, Hallahan CW, Kottilil S, Moir S, Mican JM, Mullins JI, et al. Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. J Infect Dis 2008; 197:714-20; PMID:18260759; http://dx.doi.org/ 10.1086/527324 [DOI] [PubMed] [Google Scholar]
- 38.Strassman G, Bach FH. OKT4+ cytotoxic T cells can lyse targets via class I molecules and can be blocked by monoclonal antibody against T4 molecules. J Immunol 1984; 133:1705-9; PMID:6332131 [PubMed] [Google Scholar]
- 39.Wang P, Vanky F, Klein E. MHC class-I-restricted auto-tumor-specific CD4+CD8- T-cell clones established from autologous mixed lymphocyte-tumor-cell culture (MLTC). Int J Cancer 1992; 51:962-7; PMID:1386348; http://dx.doi.org/ 10.1002/ijc.2910510621 [DOI] [PubMed] [Google Scholar]
- 40. De Bueger M, Bakker A, Goulmy E. Existence of mature human CD4+ T cells with genuine class I restriction. Eur J Immunol 1992; 22:875-8; PMID:1372262; http://dx.doi.org/ 10.1002/eji.1830220338 [DOI] [PubMed] [Google Scholar]
- 41.LeMay LG, Kan-Mitchell J, Goedegebuure P, Harel W, Mitchell MS. Detection of melanoma-reactive CD4+ HLA-class I-restricted cytotoxic T cell clones with long-term assay and pretreatment of targets with interferon-gamma. Cancer Immunol Immunother 1993; 37:187-94; PMID:8101473; http://dx.doi.org/ 10.1007/BF01525434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Darrow TL, Abdel-Wahab Z, Quinn-Allen MA, Seigler HF. Recognition and lysis of human melanoma by a CD3+, CD4+, CD8- T-cell clone restricted by HLA-A2. Cell Immunol 1996; 172:52-9; PMID:8806806; http://dx.doi.org/ 10.1006/cimm.1996.0214 [DOI] [PubMed] [Google Scholar]
- 43.Nishimura MI, Avichezer D, Custer MC, Lee CS, Chen C, Parkhurst MR, Diamond RA, Robbins PF, Schwartzentruber DJ, Rosenberg SA. MHC class I-restricted recognition of a melanoma antigen by a human CD4+ tumor infiltrating lymphocyte. Cancer Res 1999; 59:6230-8; PMID:10626817 [PubMed] [Google Scholar]
- 44.Somasundaram R, Robbins P, Moonka D, Loh E, Marincola F, Patel A, Guerry D, Herlyn D. CD4(+), HLA class I-restricted, cytolytic T-lymphocyte clone against primary malignant melanoma cells. Int J Cancer 2000; 85:253-9; PMID:10629086; http://dx.doi.org/ 10.1002/(SICI)1097-0215(20000115)85:2%3C253::AID-IJC17%3E3.0.CO;2-U [DOI] [PubMed] [Google Scholar]
- 45.Boyle LH, Goodall JC, Opat SS, Gaston JS. The recognition of HLA-B27 by human CD4(+) T lymphocytes. J Immunol 2001; 167:2619-24; PMID:11509603; http://dx.doi.org/ 10.4049/jimmunol.167.5.2619 [DOI] [PubMed] [Google Scholar]
- 46.Brewerton DA, Hart FD, Nicholls A, Caffrey M, James DC, Sturrock RD. Ankylosing spondylitis and HL-A 27. Lancet 1973; 1:904-7; PMID:4123836; http://dx.doi.org/ 10.1016/S0140-6736(73)91360-3 [DOI] [PubMed] [Google Scholar]
- 47.Brown MA, Pile KD, Kennedy LG, Calin A, Darke C, Bell J, Wordsworth BP, Cornelis F. HLA class I associations of ankylosing spondylitis in the white population in the United Kingdom. Ann Rheum Dis 1996; 55:268-70; PMID:8733445; http://dx.doi.org/ 10.1136/ard.55.4.268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hermann E, Yu DT, Meyer zum Buschenfelde KH, Fleischer B. HLA-B27-restricted CD8 T cells derived from synovial fluids of patients with reactive arthritis and ankylosing spondylitis. Lancet 1993; 342:646-50; PMID:8103147; http://dx.doi.org/ 10.1016/0140-6736(93)91760-J [DOI] [PubMed] [Google Scholar]
- 49.Plesa G, Zheng L, Medvec A, Wilson CB, Robles-Oteiza C, Liddy N, Bennett AD, Gavarret J, Vuidepot A, Zhao Y, et al. TCR affinity and specificity requirements for human regulatory T-cell function. Blood 2012; 119:3420-30; PMID:22318202; http://dx.doi.org/ 10.1182/blood-2011-09-377051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Geiger TL. Turning Tregs into class I suppressors. Blood 2012; 119:3373-4; PMID:22500048; http://dx.doi.org/ 10.1182/blood-2012-02-410969 [DOI] [PubMed] [Google Scholar]


