ABSTRACT
Inflammatory bowel disease (IBD), comprised of Crohn's disease and ulcerative colitis, is a chronic inflammatory condition of multifactorial etiology and risk factors. Currently, one of the most effective treatments for IBD is the use of Tumor Necrosis Factor (TNF) functional inhibitor drugs, however, this treatment can cause adverse reactions and has a relatively large percentage of incomplete or non-responders. This lack of response may be related to differences in patients’ gut microbiomes prior to and after disease initiation or treatment. Recent observations in our lab using a rodent model of IBD support the theory that TNF drives acute colitis, but also that the microbiome differs in association with TNF production and colitis severity. Studies such as this and others provide new insights into host-microbiome interactions associated with colitis that can lead to new therapies to prevent or treat the disease.
KEYWORDS: colitis, inflammatory bowel disease, microbiota, mouse model, TNBS, tumor necrosis factor
Introduction
Inflammatory bowel diseases (IBD), comprised of Crohn's Disease (CD) and Ulcerative Colitis (UC), are chronic inflammatory disorders, primarily in the gastrointestinal tract. The exact pathogenesis of these diseases is unclear, however environmental risk factors, genetic predisposition and aberrant immune responses to the patient's normal intestinal flora are all involved.1-3 Recent studies exploring the gut microbiome function and makeup indicate that, under normal conditions, the commensal gut microbiota stimulates regulatory T cells (Tregs) that, in turn, are responsible for maintaining gut homeostasis (reviewed:4). Commensal bacteria are thought to be the primary instigators of intestinal inflammation in IBD patients; however the relationship between IBD pathogenesis and commensal bacteria is not completely understood. Several theories exist, all with the common endpoint—increased exposure of mucosal T cells to bacteria resulting in chronic inflammation (reviewed:5). We recently provided novel insights into the interplay of tumor necrosis factor (TNF), IBD and the microbiome in a mouse model of IBD.6 Using these data as a framework, here we provide a brief overview of some of the recent highlights in the field of IBD and the microbiome and how TNF influences inflammation and microbiota.
Tumor necrosis factor and the inflammation of IBD
CD, manifests as discontinuous, transmural and granulomatous inflammation, affecting any part of the gastrointestinal tract, but preferentially affecting the ileocecal section of the gut. As the name implies, UC is confined to the large colon, and results in continuous, superficial ulceration and mucosal inflammation of the colon and/or rectum.7 Dysregulation of mucosal immunity is a key component of IBD development and progression, with mucosal CD4+ T cells, including T helper (Th)-1, Th-17 and Th-2, cells producing the preponderance of pro-inflammatory cytokines. Th-1 cytokines (e.g. TNF, interleukin (IL)-12, and interferon (IFN)-γ) and Th-17 cytokines (e.g., IL-17 and IL-23) are elevated in the mucosa of CD patients. Th-2 (e.g. IL-4 and IL-13) and Th-17 cytokines are elevated in areas of active disease in the mucosa of UC patients.8,9 Under homeostatic conditions, a balance exists between the production of pro-inflammatory cytokines by CD4+ T cells and anti-inflammatory cytokines (e.g., IL-10) by Tregs. Evidence (in mice and humans) suggests that lacking Tregs can lead to IBD; however a clear association between a local deficit of Tregs or the presence of defective Tregs and IBD development has not been established.10
TNF is chronically elevated, locally and systemically, in patients with IBD.11-13 Currently, one of the most effective therapies for treating refractory IBD is suppression of TNF function, but adverse side effects do occur and some patients do not respond to this therapy (reviewed:14). Supporting the importance of TNF in CD, mice with deletion of the 3′ regulatory element from the TNF transcript have increased, sustained production of TNF, resulting in CD-like inflammation and immune profile.15 Similarly, TNF has been shown to be pathogenic, both in a variety of mouse models of IBD16-18 and colitis associated colon cancer.19 We recently confirmed these results using the trinitrobenzene (TNBS) mouse model of acute colitis.6 TNBS is a haptenating agent and when given intrarectally, induces an immunopathology in the colon of mice that is similar to that seen in CD associated colitis.20 As described in this study, the TNBS given intra-rectally is dissolved in ethanol. Similarly, SHAM treated mice receive and intra-rectal installation of an ethanol/PBS solution. The ethanol is needed to cause microabrasions and irritation in the colon mucosa to allow for absorption of the TNBS into the submucosa, and therefore itself initiates a low level of colitis. In our published study, we showed that TNBS treated mice with ablation of the TNF gene (Tnf −/− mice) had less severe colonic inflammation than similarly treated wild type (WT) mice.6 Additional studies (unpublished and preliminary), from our lab examining male and female mice subjected to acute TNBS colitis confirm that Tnf −/− mice had significantly less colitis than WT mice (Fig. 1). Colitis scores evaluation and study design for these studies were as previously described.6
Figure 1.
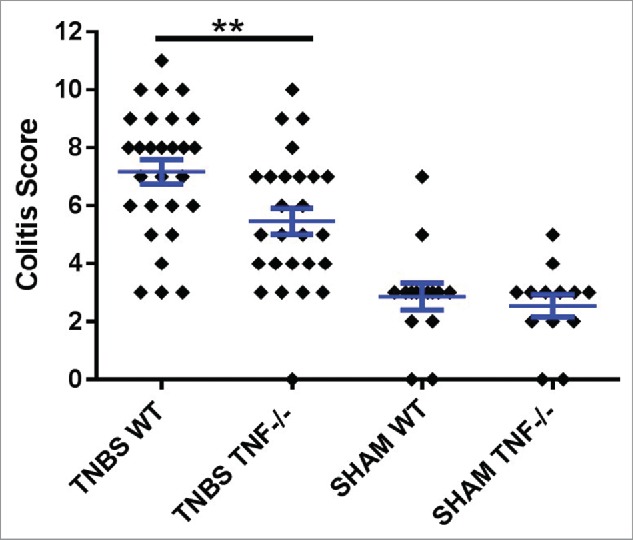
TNBS treatment in WT mice induces significantly more inflammation than TNBS treated Tnf −/− mice. Histopathological semi-quantitative scores are presented as the mean score ± SE. **= p.01. Data represents 5 independent experiments using male and female, 6–8 week old mice. Experiment 1 used 20 mice; 2 used 10 mice; 3 used 20 mice; 4 used 8 mice; and 5 used 23 mice, for a total of 81 mice.
The intersection of the microbiome, TNF and IBD
The phyla Bacteroidetes and Firmicutes dominate the gut microbiota of healthy humans.21 CD and UC patients exhibit dysbiosis consisting of decreased proportions of Firmicutes and Bacteroidetes and increased proportions of microbial commensal communities, such as those in the phyla Actinobacteria and Proteobacteria, or elevation of specific pathogenic organisms, such as adherent-evasive E. coli.4,22,23,24 A study by Morgan et al., correlated variation between the gut microbiome of IBD patients and a number of environmental factors, including treatment with anti-TNF drugs.23 Anti-TNF therapeutics dampens inflammation in IBD, at least in part, by modulating the gut microbiota, inducing T cell apoptosis, and by inhibiting vasculitis.25,26 With respect to the microbiota, this decreased inflammation is associated with modulation of the gut microbiota tending toward eubiosis. The microbiome of treated individuals was characterized by reduced Enterobacteriaceae (specifically E. coli) and Ruminococcus, and increased proportions of Bacteroidetes and Firmicutes, restoring the microbiome to a composition more reflective of healthy individuals.26
Recently, we compared the microbiome of WT and Tnf−/− mice in the TNBS model and found that prior to and after colitis, the predominant phyla in gut were Bacteroidetes and Firmicutes. Specifically, prior to treatment (F0) proportions of Bacteroidetes were significantly (Mann Whitney, p < 0.05) greater and Firmicutes lesser in Tnf −/− mice compared to WT mice. Comparing gut communities post colitis to pre colitis, Bacteroidetes significantly (Mann Whitney, p < 0.05) increased and Firmicutes decreased in WT mice (Fig. 2). A number of human studies have also noted a similar trend with greater proportions of Bacteroidetes and lesser Firmicutes in IBD patients compared to healthy subjects (reviewed:24). In contrast, Rooks et al. showed an increase in proportions of Firmicutes and decrease in proportions of Bacteroidetes in TRUC mice (mice that develop an UC-type disease) treated with anti-TNF antibodies post colitis when compared to untreated and antibiotic treated mice.27 These varied results may be simply dependent on the mouse model used. However; given the variety of methods of action of the different anti-TNF therapies, these contrasting results also highlight potential differences in microbial responses and potentially, patient responses to anti-TNF treatment. This is further supported by a preliminary study in which the sub-mucosa microbial communities of patients with CD appear to fall into 2 biotypes that suggest CD may not have single etiology, but represent a spectrum of disease.28
Figure 2.

Phyla level comparisons of relative proportion of bacterial in fecal sample of WT and Tnf −/− mice at the beginning (F0) versus end (F10) of acute TNBS treatment. Phyla SR1 and other Bacteria not illustrated because of low proportions (<0.04%).
It is important to note that these 2 dominant phyla contain both colitis- and “health”-associated genera. Recently, Firmicutes were shown to be the prevalent phyla recovered from CD patients’ mucosa that were treated with Adalimumab (an anti-TNF drug) for 3 months. At the genus level, however the predominant bacteria that increased in this phylum were Clostridium spp., whereas the Faecalibacterium, also in this phyla, were more abundant in healthy controls.26 We showed that Firmicutes decreased post colitis in Tnf −/− mice, and within this phylum, Ruminococcus and Clostridiaceae (colitis associated bacteria) decreased.6 Another important finding is that in CD patients, E. coli (a member of the phylum Proteobacteria) often increases and E. coli exhibiting pathogenic features adhesion and invasion are frequently found in patients with CD.26,29,30 However, in our acute study this genus was not found. Our current studies have been conducted under “acute” conditions and perhaps because disease associated taxa are more of an indicator of chronic infection. Future studies will use the chronic model of TNBS colitis to evaluate the same parameters that were evaluated in the acute studies. This is also illustrates the limitation of using a 16S rRNA gene sequencing of fecal samples. Disease associated microbes typically require sufficient time to multiply to be detected in sufficient quantities in fecal samples (a mixture of microbes from the entire GI tract) that likely will not occur in an acute study. In animal studies this can be overcome by directly sampling mucosa from inflamed regions but this is not as easily done in humans. It is also possible that microbes actively associated with disease only make up a small fraction of the community and methods to preferentially select this sub-group are needed, such as, using flow cytometry to sort IgA coated cells that can then be sequenced.31,32
Conclusion
The inclusion of microbiome analysis in IBD studies provides insights into the underlying reasons for the lack of consistent findings among clinical and animal studies and, potentially, the differential responses to anti-TNF therapy. A single genotypic change to this mouse model was clearly reflected in a distinctly different microbiome when compared to the WT. This difference also was reflected in the response of the microbiome to IBD induced by TNBS in which there was a significant change of microbiome composition in WT that was not as evident in the Tnf −/− mice. This demonstrates the importance of TNF production in potential severity of IBD and also identifies some microorganisms that could potentially be used as probiotics to reduce disease. Just as important as alterations in microbiota composition, however, is the dysfunction of the gut microbiota. Microbial dysfunction is consistently seen in both UC and CD patients and likely plays a key role in the pathogenesis of IBD.23 Limited work has been done to investigate the microbiome function in IBD,33 and these studies did not consider the influences of treatment and other environmental factors. Additionally, it has yet to be determined if microbial composition and function alterations cause or are the result of IBD and how anti-TNF therapeutics (and others) potentially contribute to or help correct microbial dysfunctions.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature 2007; 448:427-34; PMID:17653185; http://dx.doi.org/ 10.1038/nature06005 [DOI] [PubMed] [Google Scholar]
- 2.Rubin DC, Shaker A, Levin MS. Chronic intestinal inflammation: inflammatory bowel disease and colitis-associated colon cancer. Front Immunol 2012; 3:107; PMID:22586430; http://dx.doi.org/ 10.3389/fimmu.2012.00107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Latella G, Papi C. Crucial steps in the natural history of inflammatory bowel disease. World J Gastroenterol 2012; 18:3790-9; PMID:22876029; http://dx.doi.org/ 10.3748/wjg.v18.i29.3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strober W. Impact of the gut microbiome on mucosal inflammation. Trends Immunol 2013; 34:423-30; PMID:23957963; http://dx.doi.org/ 10.1016/j.it.2013.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sartor RB, Mazmanian SK. Intestinal microbes in inflammatory bowel diseases. Am J Gastroenterol Suppl 2012; 1:15-21; http://dx.doi.org/ 10.1038/ajgsup.2012.4 [DOI] [Google Scholar]
- 6.Jones-Hall YL, Kozik A, Nakatsu C. Ablation of tumor necrosis factor is associated with decreased inflammation and alterations of the microbiota in a mouse model of inflammatory bowel disease. PLoS One 2015; 10:e0119441; PMID:25775453; http://dx.doi.org/ 10.1371/journal.pone.0119441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med 2009; 361:2066-78; PMID:19923578; http://dx.doi.org/ 10.1056/NEJMra0804647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu XR, Liu CQ, Feng BS, Liu ZJ. Dysregulation of mucosal immune response in pathogenesis of inflammatory bowel disease. World J Gastroenterol 2014; 20:3255-64; PMID:24695798; http://dx.doi.org/ 10.3748/wjg.v20.i12.3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monteleone I, Pallone F, Monteleone G. Th17-related cytokines: new players in the control of chronic intestinal inflammation. BMC Med 2011; 9:122; PMID:22082127; http://dx.doi.org/ 10.1186/1741-7015-9-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Himmel ME, Yao Y, Orban PC, Steiner TS, Levings MK. Regulatory T-cell therapy for inflammatory bowel disease: more questions than answers. Immunology 2012; 136:115-22; PMID:22348589; http://dx.doi.org/ 10.1111/j.1365-2567.2012.03572.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braegger CP, Nicholls S, Murch SH, Stephens S, MacDonald TT. Tumour necrosis factor alpha in stool as a marker of intestinal inflammation. Lancet 1992; 339:89-91; PMID:1345871; http://dx.doi.org/ 10.1016/0140-6736(92)90999-J [DOI] [PubMed] [Google Scholar]
- 12.Reimund JM, Wittersheim C, Dumont S, Muller CD, Kenney JS, Baumann R, Poindron P, Duclos B. Increased production of tumour necrosis factor-alpha interleukin-1 beta, and interleukin-6 by morphologically normal intestinal biopsies from patients with Crohn's disease. Gut 1996; 39:684-9; PMID:9026483; http://dx.doi.org/ 10.1136/gut.39.5.684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reimund JM, Wittersheim C, Dumont S, Muller CD, Baumann R, Poindron P, Duclos B. Mucosal inflammatory cytokine production by intestinal biopsies in patients with ulcerative colitis and Crohn's disease. J Clin Immunol 1996; 16:144-50; PMID:8734357; http://dx.doi.org/ 10.1007/BF01540912 [DOI] [PubMed] [Google Scholar]
- 14.Lichtenstein GR. Comprehensive review: antitumor necrosis factor agents in inflammatory bowel disease and factors implicated in treatment response. Therap Adv Gastroenterol 2013; 6:269-93; PMID:23814608; http://dx.doi.org/ 10.1177/1756283X13479826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity 1999; 10:387-98; PMID:10204494; http://dx.doi.org/ 10.1016/S1074-7613(00)80038-2 [DOI] [PubMed] [Google Scholar]
- 16.Neurath MF, Fuss I, Pasparakis M, Alexopoulou L, Haralambous S, Meyer zum Buschenfelde KH, Strober W, Kollias G. Predominant pathogenic role of tumor necrosis factor in experimental colitis in mice. Eur J Immunol 1997; 27:1743-50; PMID:9247586; http://dx.doi.org/ 10.1002/eji.1830270722 [DOI] [PubMed] [Google Scholar]
- 17.Mueller C. Tumour necrosis factor in mouse models of chronic intestinal inflammation. Immunology 2002; 105:1-8; PMID:11849309; http://dx.doi.org/ 10.1046/j.1365-2567.2002.01329.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindebo Holm T, Poulsen SS, Markholst H, Reedtz-Runge S. Pharmacological evaluation of the SCID T cell transfer model of colitis: as a model of crohn's disease. Int J Inflam 2012; 412178:19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popivanova BK, Kitamura K, Wu Y, Kondo T, Kagaya T, Kaneko S, Oshima M, Fujii C, Mukaida N. Blocking TNF-alpha in mice reduces colorectal carcinogenesis associated with chronic colitis. J Clin Invest 2008; 118:560-70; PMID:18219394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strober W, Fuss IJ, Blumberg RS. The immunology of mucosal models of inflammation. Annu Rev Immunol 2002; 20:495-549; PMID:11861611; http://dx.doi.org/ 10.1146/annurev.immunol.20.100301.064816 [DOI] [PubMed] [Google Scholar]
- 21.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science 2005; 308:1635-8; PMID:15831718; http://dx.doi.org/ 10.1126/science.1110591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walters WA, Xu Z, Knight R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett 2014; 588:4223-33; PMID:25307765; http://dx.doi.org/ 10.1016/j.febslet.2014.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, Bousvaros A, Korzenik J, Sands BE, Xavier RJ, Huttenhower C. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 2012; 13:R79; PMID:23013615; http://dx.doi.org/ 10.1186/gb-2012-13-9-r79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright EK, Kamm MA, Teo SM, Inouye M, Wagner J, Kirkwood CD. Recent advances in characterizing the gastrointestinal microbiome in Crohn's disease: a systematic review. Inflamm Bowel Dis 2015; 21:1219-28; PMID:25844959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Danese S, Sans M, Scaldaferri F, Sgambato A, Rutella S, Cittadini A, Pique JM, Panes J, Katz JA, Gasbarrini A, Fiocchi C. TNF-alpha blockade down-regulates the CD40/CD40L pathway in the mucosal microcirculation: a novel anti-inflammatory mechanism of infliximab in Crohn's disease. J Immunol 2006; 176:2617-24; PMID:16456024; http://dx.doi.org/ 10.4049/jimmunol.176.4.2617 [DOI] [PubMed] [Google Scholar]
- 26.Busquets D, Mas-de-Xaxars T, Lopez-Silas M, Martinez-Medina M, Bahi A, Sabat M, Louvriex R, Miguel-Cusachs O, Garcia-Gil J, Aldeguer X. Anti-TNF treatment with adalimumab induces changes in the microbiota of Crohn's disease. J Crohns Colitis 2015; PMID:26142465 [DOI] [PubMed] [Google Scholar]
- 27.Rooks MG, Veiga P, Wardwell-Scott LH, Tickle T, Segata N, Michaud M, Gallini CA, Beal C, van Hylckama-Vlieg JE, Ballal SA, et al.. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J 2014; 8:1403-17; PMID:24500617; http://dx.doi.org/ 10.1038/ismej.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiodini RJ, Dowd SE, Davis B, Galandiuk S, Chamberlin WM, Kuenstner JT, McCallum RW, Zhang J. Crohn's disease may be differentiated into 2 distinct biotypes based on the detection of bacterial genomic sequences and virulence genes within submucosal tissues. J Clin Gastroenterol 2013; 47:612-20; PMID:23426447; http://dx.doi.org/ 10.1097/MCG.0b013e31827b4f94 [DOI] [PubMed] [Google Scholar]
- 29.Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R, Orsi RH, Wiedmann M, McDonough P, Kim SG, et al.. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. ISME J 2007; 1:403-18; PMID:18043660; http://dx.doi.org/ 10.1038/ismej.2007.52 [DOI] [PubMed] [Google Scholar]
- 30.Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, Bringer MA, Swidsinski A, Beaugerie L, Colombel JF. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology 2004; 127:412-21; PMID:15300573; http://dx.doi.org/ 10.1053/j.gastro.2004.04.061 [DOI] [PubMed] [Google Scholar]
- 31.D'Auria G, Peris-Bondia F, Dzunkova M, Mira A, Collado MC, Latorre A, Moya A. Active and secreted IgA-coated bacterial fractions from the human gut reveal an under-represented microbiota core. Sci Rep 2013; 3:3515; PMID:24343271; http://dx.doi.org/ 10.1038/srep03515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, et al.. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 2014; 158:1000-10; PMID:25171403; http://dx.doi.org/ 10.1016/j.cell.2014.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greenblum S, Turnbaugh PJ, Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci U S A 2012; 109:594-9; PMID:22184244; http://dx.doi.org/ 10.1073/pnas.1116053109 [DOI] [PMC free article] [PubMed] [Google Scholar]


