ABSTRACT
The gut microbiota is a complex, densely populated community, home to many different species that collectively provide huge benefits for host health. Disruptions to this community, as can result from recurrent antibiotic exposure, alter the existing network of interactions between bacteria and can render this community susceptible to invading pathogens. Recent findings show that direct antagonistic and metabolic interactions play a critical role in shaping the microbiota. However, the part played by quorum sensing, a means of regulating bacterial behavior through secreted chemical signals, remains largely unknown. We have recently shown that the interspecies signal, autoinducer-2 (AI-2), can modulate the structure of the gut microbiota by using Escherichia coli to manipulate signal levels. Here, we discuss how AI-2 could influence bacterial behaviors to restore the balance between the 2 major bacteria phyla, the Bacteroidetes and Firmicutes, following antibiotic treatment. We explore how this may impact on host physiology, community susceptibility or resistance to pathogens, and the broader potential of AI-2 as a means to redress the imbalances in microbiota composition that feature in many infectious and non-infectious diseases.
KEYWORDS: AI-2; antibiotics, autoinducer-2, dysbiosis, lsr, luxS, microbiota, quorum sensing
The gut microbiota - a multispecies consortium
The human body is home to a multitude of microorganisms, collectively known as the microbiota. These organisms live in intimate association with their host, often providing many gains for human health. This is particularly evident in the large intestine, which harbors the most densely populated and diverse bacterial community in the body.1 The massive genetic capacity of these organisms encodes for many functions, supplementing the host's existing metabolic repertoire to provide sources of vitamins B and K, break down complex polysaccharides and produce a plethora of important molecules such as short chain fatty acids (SCFAs).2–4 These interactions promote host homeostasis and protection from insults associated with intestinal injury,4–6 as well as conferring resistance to both intestinal and systemic infection.7,8
The need to understand how interactions within this microbial community influence human health stems from the ever-increasing spectrum of infectious and non-infectious diseases associated with imbalances or disturbances in the microbiota (known as dysbiosis). One of the more frequent causes of dysbiosis in humans is the use of broad spectrum antibiotics, which exert a significant impact upon gut commensal bacteria. Antibiotic-induced changes in bacterial population density and diversity, coincident with declines and expansions in certain taxa,9 frequently leave the host at increased risk of infection by specific pathogens such as vancomycin-resistant Enterococcus and Clostridium difficile.10,11 This link between particular changes to the microbiota and increased infection by certain pathogens illustrates the protection, referred to as colonization resistance, provided by the specific interactions between bacteria within the microbiota that limit the ability of invading organisms to become established within this community.12
Many of these interactions have a basis in metabolism and nutrition. Intricate cross-feeding networks exist, which limit nutrient availability and thus population expansion by potential colonizers.13 By-products from metabolism that have antagonistic effects including SCFAs and secondary bile acids can further inhibit growth and in some cases also the virulence of bacteria such as Salmonella and C. difficile.13–16 Non-nutritional antagonism between bacteria such as the removal of nearby competitors using secreted bacteriocins or direct cell contact-dependent delivery of toxins by specialized machineries such as Type 6 Secretion Systems, likely also influence community composition and its ability to provide colonization resistance.17,18 Motility, adherence and biofilm formation are further non-nutrititive behaviors that can also contribute to the ability of both commensal and pathogenic bacteria alike to expand in the gut. These behaviors are often regulated through a form of cell-to-cell communication called quorum sensing, in which small chemical signals known as autoinducers are secreted and accumulate as bacteria multiply. Subsequent signal detection enables bacteria to determine cell density, regulate gene expression at the population level and engage in group behaviors.19 Though such regulation has proved integral to the success of many host-microbe interactions, its precise role within the gut microbiota is not yet known. Given the polyspecies nature of this complex community, communication between species must be required to enable bacteria to optimally fine-tune behavior to their given social and environmental context. One quorum sensing molecule, Autoinducer-2 (AI-2), is produced by multiple bacterial species found in the gastrointestinal tract, including Bacteroides spp., Ruminococcus spp, Eubacterium rectale and Lactobacillus spp.,20–24 while AI-2-dependent signaling and subsequent modulation of bacterial behaviors across the species barrier has been shown in several other multispecies settings.25–27 These observations led to the hypothesis that AI-2 is one of the signaling molecules which might regulate bacterial behavior and community dynamics in the microbiota.28
A toolkit to investigate AI-2 quorum sensing in the gut microbiota
To determine if AI-2 signaling influences the gut microbiota, a means of manipulating its levels in vivo was required. This small, 5 carbon molecule can be chemically synthesized, but its relative instability makes production of large quantities difficult and raised questions over its longevity following oral administration to mice.29 This approach also gave no means of lowering signal concentration in vivo. To overcome these technical hurdles, we harnessed the natural properties of the AI-2-producing and -importing bacterial strain, Escherichia coli K-12 MG1655. This bacterium synthesizes and releases AI-2 into the environment as it grows. Once a threshold concentration of signal is reached, binding and internalization ensues through the activity of a high affinity periplasmic binding protein and an ABC-type transporter, collectively known as the Lsr transporter (Fig 1A). AI-2 is then sequestered inside the cell following phosphorylation by a kinase, known as LsrK. This phospho-AI-2 then binds and inhibits a repressor, LsrR, which leads to de-repression of expression of its target, the Lsr transporter. Thus AI-2 uptake creates a positive feedback loop, whereby signal import drives increased expression of the transporter, the subsequent uptake of more signal and rapid depletion of extracellular AI-230. E. coli mutants affected in certain components of this Lsr system consequently have altered abilities to accumulate or deplete extracellular AI-2 levels in vitro, with consequent effects upon the AI-2-dependent regulation of behaviors in other species upon co-culture.25,30 No AI-2-dependent transcriptional responses other than induction of the Lsr system have been described for this MG1655 K-12 strain of E. coli, indicating that the effects upon neighboring bacteria are likely due to changes in AI-2 availability and not the result of altered E. coli physiology.
Figure 1.
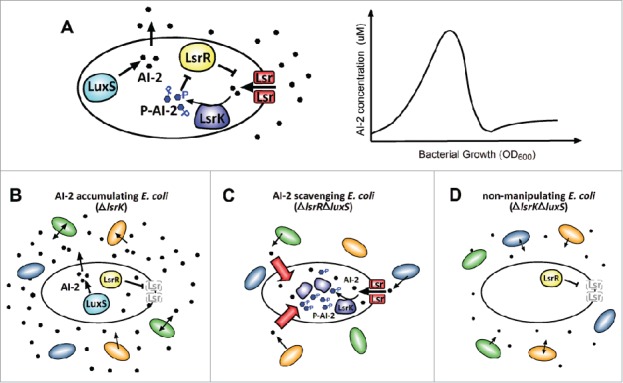
Using the Lsr transporter system to manipulate AI-2 levels. A. Wild-type E. coli synthesize AI-2 using the LuxS synthase (left panel). Signal is secreted to the extracellular environment where it accumulates with cell number. Once a threshold concentration is reached, signal binds and is internalised by the Lsr transporter. Imported AI-2 is then phosphorylated by LsrK, which sequesters the signal intracellularly. Phospho-AI-2 binds and inhibits the repressor, LsrR, which enables the expression of more Lsr transporter and an amplification loop that leads to further signal uptake and the rapid removal of AI-2 from the environment. A graphical representation of how this affects extracellular signal concentration during growth in vitro is shown in the right panel. B. ΔlsrK mutant E. coli accumulate AI-2 extracellularly. Deletion of lsrK means phospho-AI-2 cannot be generated, transporter expression is not induced and signal cannot be sequestered intracellularly. This mutant does not degrade AI-2, so AI-2 produced by E. coli or its neighbors accumulates. C. ΔlsrRΔluxS mutant E. coli scavenge extracellular AI-2. Deletion of the signal synthase, LuxS, abolishes signal production, while removal of the repressor, LsrR, results in over-expression of the transporter. The resulting mutant E. coli efficiently imports AI-2 produced by bacteria in its vicinity. D. ΔlsrKΔluxS mutant E. coli do not manipulate exogenous signal levels. Deletion of these 2 genes results in a mutant which does not produce nor import and degrade signal, so it does not affect ambient levels of AI-2 in polyspecies communities.
With this knowledge, we engineered fluorescent, streptomycin-resistant strains for use in vivo, targeting LsrK to produce E. coli which do not sequester AI-2 intracellularly, nor induce transporter expression (Fig. 1B). This mutant successfully accumulated AI-2 in the gastrointestinal tract of mono-colonized gnotobiotic mice.28 To deplete extracellular signal levels, we deleted both luxS, so that bacteria cannot produce AI-2, and lsrR, to induce constitutive Lsr transporter expression and signal import. The resulting E. coli mutant strain efficiently scavenged signal produced by neighboring LuxS-positive bacteria in vivo (Fig. 1C). To distinguish between the effects of introducing E. coli into the microbiota and of modulating AI-2 availability, we constructed a ΔluxS and ΔlsrK double mutant strain. These bacteria neither produced nor sequestered signal upon co-colonization of gnotobiotic mice, validating their use as a control that does not manipulate ambient signal levels (Fig. 1D).28 The engineered mutant strains all efficiently colonized the mice to the same extent, making them effective tools with which to investigate the role of AI-2 within the gut microbiota.
Chemical communication between bacteria alters antibiotic-induced gut dysbiosis
To determine whether and how changes in AI-2 availability influenced the microbiota, mice were first treated with streptomycin to create conditions of dysbiosis that are typically associated with expansion of enteric bacteria like E. coli.31 Streptomycin affected the microbiota hugely, causing transient decreases in bacterial load and progressive changes in the bacterial community structure during the month-long treatment.28 Huge losses in the Firmicutes occurred, while abundance of just a few members of the Bacteroidetes increased to such an extent that they almost completely dominated the community.28 Despite the strong influence of streptomycin, changes in the microbiota were observed in mice colonized by ΔlsrK mutant E. coli (which increase signal availability) compared to that in mice colonized by either of the other 2 mutant strains.28 Increased AI-2 levels led to the decreased abundance of 11 members of the Bacteroidetes phylum and significant increases in abundance of 2 members of the Lachnospiraceae. Multiple changes of this nature combined to give an increase in the ratio of Firmicutes to Bacteroidetes, which opposed the huge imbalance imposed by streptomycin (Fig. 2A).28 In promoting the Firmicutes, AI-2 also promoted the phylum with the greatest proportion of signal producers: 83% of currently available sequenced genomes belonging to the Firmicutes encode a LuxS homolog compared to only 17% of those pertaining to the Bacteroidetes. These results suggest that positive feedback (a common feature of quorum sensing systems) might exist within the microbiota, whereby AI-2 signaling and downstream responses drive increases in abundance of the AI-2 producers which then further increases signal levels and amplifies the response throughout the community.
Figure 2.
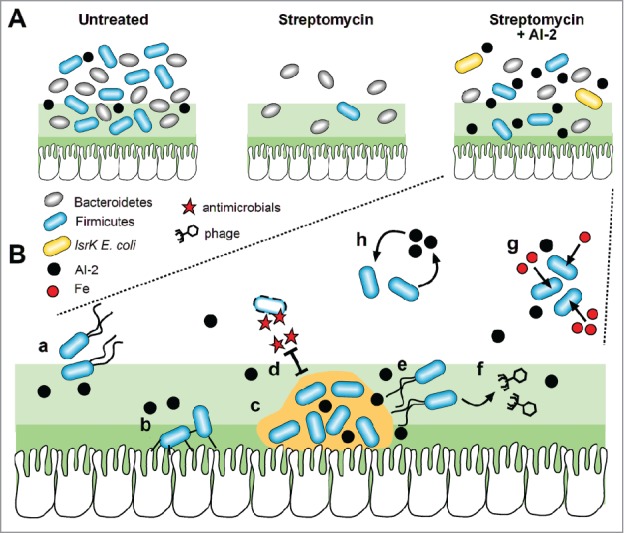
AI-2-dependent effects within the gut microbiota A. Approximately equal ratios of the 2 major phyla, the Bacteroidetes and Firmicutes, were found in the gut microbiota of untreated mice (left panel). Though streptomycin caused a depletion of the Firmicutes and enabled the Bacteroidetes to dominate the microbiota (middle panel), colonisation by AI-2-accumulating ΔlsrK mutant E. coli promoted an increase in abundance of the Firmicutes (right panel). B. In vitro and in vivo studies show that AI-2 regulates multiple phenotypes in different bacteria which might promote colonisation if also induced in the Firmicutes. AI-2 induces motility which could help bacteria find nutrients under conditions of starvation (a). This signal can also upregulate attachment to epithelial cells (b) and biofilm formation (c) which could also increase resistance to antimicrobials such as streptomycin (d). In some bacteria, AI-2 acts as a signal for biofilm dispersal (e), and can trigger the release of phage and transfer of genetic material between bacteria within the gut microbiota (f). Expression of iron uptake mechanisms in response to AI-2 could help bacteria to outcompete their neighbors when this important metal ion is limiting (g). The discovery that a much greater proportion of species of Firmicutes than Bacteroidetes encode the AI-2 synthase, LuxS, suggests positive feedback might also occur: the bacteria which respond positively to this signal also produce it (h), leading to increasing amounts of AI-2, further induction of the above responses and greater expansion of the Firmicutes.
Questions now arise as to how these AI-2-dependent changes in the microbiota are brought about, not to mention the effects they have upon the community itself and the host. It is difficult to know at this stage whether these changes are the direct result of positive effects of AI-2 upon the Firmicutes, inhibitory effects upon the Bacteroidetes, or a mix of both. AI-2 signaling could also have some indirect effect, as behavioral changes in one species are likely to affect neighboring bacteria, through altered nutrient and attachment site availability, incorporation into developing biofilms or changes in production of inhibitory metabolites, for example. One thing is clear: at least some of the bacteria involved must detect AI-2 (whether originating from E. coli or other AI-2-producing members of the microbiota) in order to respond to it. Two classes of AI-2 receptors have been identified so far: LuxP, restricted to the Vibrionales; and LsrB, found more widely across members of the Proteobacteria and in several species of Bacilli.32–34 Six residues are critical for AI-2 binding by the LsrB receptor, which are conserved across all functional homologues so far identified. This, alongside an overall sequence identity of more than 60% and the presence of all necessary accessory proteins for signal transport and processing, have already provided criteria through which LsrB homologues were identified in Bacillus cereus and B. anthracis.32 As these bacteria internalize AI-2 and have an Lsr system that responds to the same AI-2 adduct as E. coli (personal communication from K. Xavier32), it is likely that other Firmicutes, more relevant in the gut environment, will also respond to AI-2 using receptors similar to LsrB. Interestingly, bacteria such as Streptococcus mutans and Staphylococcus epidermidis respond to AI-2 without having either of the known receptors.23 Novel, as yet uncharacterised, receptor classes must exist in these organisms; this may also be the case for some or all of the AI-2-responsive bacteria inhabiting the gut. Identifying new receptor classes remains a major challenge: despite overall structural similarity, the known receptors, LsrB and LuxP, share only 11% sequence identity, suggesting that sequence analysis alone is unlikely to find new candidates effectively. Novel strategies are clearly required to make this essential step forward in determining the molecular mechanisms through which microbiota members respond to AI-2.
The phenotypes regulated by AI-2 in vitro can provide some clues as to how signaling might drive the community shifts seen in vivo. Adherence to epithelial cells is upregulated in an AI-2-dependent manner in Actinobacillus pleuropneumoniae,35 while increased expression of motility genes and regulation of chemotaxis in response to AI-2 is reported for several strains of E. coli.36,37 If such behaviors are also induced among the Firmicutes, they could promote nutrient scavenging and provide access to new niches that confer a competitive edge over non-responding bacteria in this environment (Fig. 2B). Flagellar structures and motility also promote attachment and influence the development and structure of bacterial biofilms. These multicellular aggregates of bacteria are surrounded in an extracellular matrix comprised of polysaccharides, proteins and DNA, all of which provides protection from both immune attack and antimicrobial exposure.38 AI-2 can promote the formation of such biofilms, not only between bacteria of the nasopharyngeal and oral cavities,26,27,39 but also by certain gut commensal bacteria including Bifidobacterium longum.40 This behavior might further promote colonization and persistence within the intestinal tract, potentially also contributing to the AI-2-dependent expansions that we observed for some members of the community.28 Conversely, AI-2 stimulates biofilm dispersal in some bacteria, acting as a chemorepellent for Helicobacter pylori, for example.41 Such a response could be of benefit in promoting clearance of biofilm-forming pathogens from the host, and if induced in the Bacteroidetes, might explain their decreased abundance when AI-2 levels increased.
AI-2-associated changes in the gut microbiota might also result from interactions with the host, which also responds to and exerts some influence over the bacteria residing within the intestinal tract. Transient proinflammatory responses induced upon exposure to AI-2 were detected in epithelial cells in vitro,42 suggesting that detection of this signal in vivo may alter inflammatory status within the intestinal epithelium. Changes in the metabolite pool caused by bacterial responses to AI-2 are also likely to affect host physiology: AI-2-induced shifts in the proportions of Firmicutes and Bacteroidetes will alter the relative concentrations of SCFAs such as butyrate, propionate and acetate. These molecules influence gene expression, cytokine secretion and regulatory T cell induction, ultimately also modulating host inflammatory responses.2 Antibiotic-induced losses in such metabolites, and the changes in inflammation and physiology that result, can lead to changes in the microbiota and generate conditions conducive to pathobiont outgrowth or pathogen invasion. Thus AI-2, by promoting the expansion of the Firmicutes, which are the major source of many of these compounds, might restore the intestinal metabolite pool and thereby indirectly inhibit such events.
Alongside the possibility of restoring community structure, function and its associated metabolome, AI-2 signaling by commensals might also contribute more directly to host health and protection from pathogens. This molecule can promote probiotic capabilities in some bacteria and reduce virulence in others: AI-2 regulates iron acquisition and the ability to inhibit Salmonella infection by Bifidobacteria breve,43 while signal produced by Ruminococcus obeum downregulates virulence and reduces colonization by Vibrio cholerae.22,44 Although signaling through AI-2 can also upregulate virulence in a few bacteria: it triggers phage release from the pathobiont, Enterococcus faecalis, which is associated with increased virulence and transfer of genetic material;45 this simply highlights how individually tailored approaches to either quench or enhance signaling will be required if using AI-2 to manipulate the microbiota.
Though the levels of AI-2 within a normal healthy microbiota are not yet known, the high incidence of putative LuxS homologues among the Firmicutes, a prevalent group within the normal gut microbiota, strongly suggests that AI-2 is present. It is not yet clear however, how this signal will influence the balance between Bacteroidetes and Firmicutes under conditions of homeostasis. Our finding that AI-2 promotes Firmicutes, the group most affected during a streptomycin-induced dysbiosis, provides a strong case for exploring AI-2 signaling as a means to shape community composition and function for the benefit of human health. Increasing AI-2 availability during antibiotic treatment moved the microbiota toward re-establishment of the balance between the Bacteroidetes and the Firmicutes.28 This shift, alongside the positive effect of AI-2 upon AI-2 producers, argues for the possibility of using signal supplementation to accelerate microbiota recovery and restore colonisation resistance following antibiotic treatment. This ability of AI-2 signaling to shape the microbiota is unlikely to be specific to streptomycin-induced dysbiosis, but as different antibiotics have distinct effects on the microbiota, increasing AI-2 levels might not always be beneficial. For example, some antibiotics favor the expansion of the Firmicutes, which should per se lead to an increase in AI-2 levels. In these cases, signal depletion may be of more benefit in restoring communities to their pre-treatment state.
As many non-infectious diseases including obesity, irritable bowel disease, autism and stress are also associated with abnormal abundances of the Bacteroidetes and Firmicutes, AI-2 could perhaps provide a tool with which to investigate and perhaps redress the balance between these 2 phyla in a whole range of microbiota-associated pathologies. Multiple strategies can be envisioned, either involving signal supplementation to restore certain commensal populations, quorum quenching using antagonistic signal analogs to block species-specific receptors, or engineered AI-2-manipulating bacteria as probiotics to inhibit specific pathogen virulence or growth. Increased understanding of the mechanisms and consequences of interspecies chemical communication working within the gut microbiota is clearly required: such knowledge could yield much-needed alternatives to antibiotics and simultaneously enable us to manipulate community composition and bacterial functions to the great benefit of human health.
Abbreviations
- AI-2
Autoinducer-2
- SCFAs
Short chain fatty acids
- ABC
ATP-binding cassette
- Lsr
LuxS-regulated
Funding
The work discussed in this addendum was supported by Howard Hughes Medical Institute (International Early Career Scientist, HHMI 55007436) and Fundaçao para a Ciência e Tecnologia (PTDC/BIA-EVF/118075/2010 and RECI/IMI-IMU/0038/2012).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Miguel P. Soares and Inês Torcato for critically reading the manuscript.
References
- [1].The Human Microbiome Project Consortium . Structure, function and diversity of the healthy human microbiome. Nature 2012; 486:207-14; PMID:22699609; http://dx.doi.org/ 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Micro 2014; 12:661-72; PMID:25198138; http://dx.doi.org/ 10.1038/nrmicro3344 [DOI] [PubMed] [Google Scholar]
- [3].Savage DC. Microbial Ecology of the Gastrointestinal Tract. Annu Rev Microbiol 1977; 31:107-33; PMID:334036; http://dx.doi.org/ 10.1146/annurev.mi.31.100177.000543 [DOI] [PubMed] [Google Scholar]
- [4].Berg RD. The indigenous gastrointestinal microflora. Trends Microbiol 1996; 4:430-5; PMID:8950812; http://dx.doi.org/ 10.1016/0966-842X(96)10057-3 [DOI] [PubMed] [Google Scholar]
- [5].Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of Commensal Microflora by Toll-Like Receptors Is Required for Intestinal Homeostasis. Cell 2004; 118:229-41; PMID:15260992; http://dx.doi.org/ 10.1016/j.cell.2004.07.002 [DOI] [PubMed] [Google Scholar]
- [6].Hooper LV, Littman DR, Macpherson AJ. Interactions Between the Microbiota and the Immune System. Science 2012; 336:1268-73; PMID:22674334; http://dx.doi.org/ 10.1126/science.1223490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yilmaz B, Portugal S, Tran TM, Gozzelino R, Ramos S, Gomes J, Regalado A, Cowan PJ, d'Apice AJF, Chong AS, et al.. Gut Microbiota Elicits a Protective Immune Response against Malaria Transmission. Cell 2014; 159:1277-89; PMID:25480293; http://dx.doi.org/ 10.1016/j.cell.2014.10.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Schieber AMP, Lee YM, Chang MW, Leblanc M, Collins B, Downes M, Evans RM, Ayres JS. Disease tolerance mediated by microbiome E. coli involves inflammasome and IGF-1 signaling. Science 2015; 350:558-63; PMID:26516283; http://dx.doi.org/ 10.1126/science.aac6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dethlefsen L, Huse S, Sogin ML, Relman DA. The Pervasive Effects of an Antibiotic on the Human Gut Microbiota, as Revealed by Deep 16S rRNA Sequencing. PLoS Biol 2008; 6:e280; PMID:19018661; http://dx.doi.org/ 10.1371/journal.pbio.0060280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Buffie CG, Jarchum I, Equinda M, Lipuma L, Gobourne A, Viale A, Ubeda C, Xavier J, Pamer EG. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect Immun 2012; 80:62-73; PMID:23090957; http://dx.doi.org/ 10.1128/IAI.05496-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, Viale A, Socci ND, van den Brink MRM, Kamboj M, et al.. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest 2010; 120:4332-41; PMID:21099116; http://dx.doi.org/ 10.1172/JCI43918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lawley TD, Walker AW. Intestinal colonization resistance. Immunology 2013; 138:1-11; PMID:23240815; http://dx.doi.org/ 10.1111/j.1365-2567.2012.03616.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fischbach MA, Sonnenburg JL. Eating For Two: How Metabolism Establishes Interspecies Interactions in the Gut. Cell Host Microbe 2011; 10:336-47; PMID:22018234; http://dx.doi.org/ 10.1016/j.chom.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kamada N, Kim Y-G, Sham HP, Vallance BA, Puente JL, Martens EC, Núñez G. Regulated Virulence Controls the Ability of a Pathogen to Compete with the Gut Microbiota. Science 2012; 336:1325-9; PMID:22582016; http://dx.doi.org/ 10.1126/science.1222195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lawhon SD, Maurer R, Suyemoto M, Altier C. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol Microbiol 2002; 46:1451-64; PMID:12453229; http://dx.doi.org/ 10.1046/j.1365-2958.2002.03268.x [DOI] [PubMed] [Google Scholar]
- [16].Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, et al.. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015; 517:205-8; PMID:25337874; http://dx.doi.org/ 10.1038/nature13828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].McKenney PT, Pamer EG. From Hype to Hope: The Gut Microbiota in Enteric Infectious Disease. Cell 2015; 163:1326-32; PMID:26638069; http://dx.doi.org/ 10.1016/j.cell.2015.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Russell AB, Wexler AG, Harding BN, Whitney JC, Bohn AJ, Goo YA, Tran BQ, Barry NA, Zheng H, Peterson SB, et al.. A Type VI Secretion-Related Pathway in Bacteroidetes Mediates Interbacterial Antagonism. Cell Host Microbe 2014; 16:227-36; PMID:25070807; http://dx.doi.org/ 10.1016/j.chom.2014.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rutherford ST, Bassler BL. Bacterial Quorum Sensing: Its Role in Virulence and Possibilities for Its Control. Cold Spring Harb Perspect Med [Internet] 2012; 2:1-25; PMID:23125205; http://dx.doi.org/ 10.1101/cshperspect.a012427 Available from: http://perspectivesinmedicine.cshlp.org/content/2/11/a012427.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tannock GW, Ghazally S, Walter J, Loach D, Brooks H, Cook G, Surette M, Simmers C, Bremer P, Dal Bello F, et al.. Ecological Behavior of Lactobacillus reuteri 100-23 Is Affected by Mutation of the luxS Gene. Appl Environ Microbiol 2005; 71:8419-25; PMID:16332830; http://dx.doi.org/ 10.1128/AEM.71.12.8419-8425.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Antunes LCM, Ferreira LQ, Ferreira EO, Miranda KR, Avelar KES, Domingues RMCP, Ferreira MC de S. Bacteroides species produce Vibrio harveyi autoinducer 2-related molecules. Anaerobe 2005; 11:295-301; PMID:16701587; http://dx.doi.org/ 10.1016/j.anaerobe.2005.03.003 [DOI] [PubMed] [Google Scholar]
- [22].Hsiao A, Ahmed AMS, Subramanian S, Griffin NW, Drewry LL, Petri WA, Haque R, Ahmed T, Gordon JI. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature 2014; 515:423-6; PMID:25231861; http://dx.doi.org/ 10.1038/nature13738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Pereira CS, Thompson JA, Xavier KB. AI-2-mediated signalling in bacteria. FEMS Microbiol Rev 2013; 37:156-81; PMID:22712853; http://dx.doi.org/ 10.1111/j.1574-6976.2012.00345.x [DOI] [PubMed] [Google Scholar]
- [24].Lukas F, Gorenc G, Kopecny J. Detection of possible AI-2-mediated quorum sensing system in commensal intestinal bacteria. Folia Microbiol (Praha) 2008; 53:221-4; PMID:18661296; http://dx.doi.org/ 10.1007/s12223-008-0030-1 [DOI] [PubMed] [Google Scholar]
- [25].Xavier KB, Bassler BL. Interference with AI-2-mediated bacterial cell-cell communication. Nature 2005; 437:750-3; PMID:16193054; http://dx.doi.org/ 10.1038/nature03960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Armbruster CE, Hong W, Pang B, Weimer KED, Juneau RA, Turner J, Swords WE. Indirect pathogenicity of Haemophilus influenzae and Moraxella catarrhalis in polymicrobial otitis media occurs via interspecies quorum signaling. mBio 2010; 1:e00102-10; PMID:20802829; http://dx.doi.org/ 10.1128/mBio.00102-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Cuadra-Saenz G, Rao DL, Underwood AJ, Belapure SA, Campagna SR, Sun Z, Tammariello S, Rickard AH. Autoinducer-2 influences interactions amongst pioneer colonizing streptococci in oral biofilms. Microbiol Read Engl 2012; 158:1783-95; PMID:22493304; http://dx.doi.org/ 10.1099/mic.0.057182-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Thompson JA, Oliveira RA, Djukovic A, Ubeda C, Xavier KB. Manipulation of the Quorum Sensing Signal AI-2 Affects the Antibiotic-Treated Gut Microbiota. Cell Rep 2015; 10:1861-71; PMID:25801025; http://dx.doi.org/ 10.1016/j.celrep.2015.02.049 [DOI] [PubMed] [Google Scholar]
- [29].Ascenso OS, Marques JC, Santos AR, Xavier KB, Ventura MR, Maycock CD. An efficient synthesis of the precursor of AI-2, the signalling molecule for inter-species quorum sensing. Bioorg Med Chem 2011; 19:1236-41; PMID:21216605; http://dx.doi.org/ 10.1016/j.bmc.2010.12.036 [DOI] [PubMed] [Google Scholar]
- [30].Xavier KB, Bassler BL. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J Bacteriol 2005; 187:238-48; PMID:15601708; http://dx.doi.org/ 10.1128/JB.187.1.238-248.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Barthel M, Hapfelmeier S, Quintanilla-Martínez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Rüssmann H, Hardt W-D. Pretreatment of Mice with Streptomycin Provides a Salmonella enterica Serovar Typhimurium Colitis Model That Allows Analysis of Both Pathogen and Host. Infect Immun 2003; 71:2839-58; PMID:12704158; http://dx.doi.org/ 10.1128/IAI.71.5.2839-2858.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Pereira CS, de Regt AK, Brito PH, Miller ST, Xavier KB. Identification of functional LsrB-like autoinducer-2 receptors. J Bacteriol 2009; 191:6975-87; PMID:19749048; http://dx.doi.org/ 10.1128/JB.00976-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 2002; 415:545-9; PMID:11823863; http://dx.doi.org/ 10.1038/415545a [DOI] [PubMed] [Google Scholar]
- [34].Miller ST, Xavier KB, Campagna SR, Taga ME, Semmelhack MF, Bassler BL, Hughson FM. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol Cell 2004; 15:677-87; PMID:15350213; http://dx.doi.org/ 10.1016/j.molcel.2004.07.020 [DOI] [PubMed] [Google Scholar]
- [35].Li L, Xu Z, Zhou Y, Li T, Sun L, Chen H, Zhou R. Analysis on Actinobacillus pleuropneumoniae LuxS regulated genes reveals pleiotropic roles of LuxS/AI-2 on biofilm formation, adhesion ability and iron metabolism. Microb Pathog 2011; 50:293-302; PMID:21320583; http://dx.doi.org/ 10.1016/j.micpath.2011.02.002 [DOI] [PubMed] [Google Scholar]
- [36].Hegde M, Englert DL, Schrock S, Cohn WB, Vogt C, Wood TK, Manson MD, Jayaraman A. Chemotaxis to the quorum-sensing signal AI-2 requires the Tsr chemoreceptor and the periplasmic LsrB AI-2-binding protein. J Bacteriol 2011; 193:768-73; PMID:21097621; http://dx.doi.org/ 10.1128/JB.01196-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gonzalez Barrios AF, Zuo R, Hashimoto Y, Yang L, Bentley WE, Wood TK. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J Bacteriol 2006; 188:305-16; PMID:16352847; http://dx.doi.org/ 10.1128/JB.188.1.305-316.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hobley L, Harkins C, MacPhee CE, Stanley-Wall NR. Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes. FEMS Microbiol Rev 2015; 39:649-69; PMID:25907113; http://dx.doi.org/ 10.1093/femsre/fuv015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Rickard AH, Palmer RJJ, Blehert DS, Campagna SR, Semmelhack MF, Egland PG, Bassler BL, Kolenbrander PE. Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol Microbiol 2006; 60:1446-56; PMID:16796680; http://dx.doi.org/ 10.1111/j.1365-2958.2006.05202.x [DOI] [PubMed] [Google Scholar]
- [40].Sun Z, He X, Brancaccio VF, Yuan J, Riedel CU. Bifidobacteria exhibit LuxS-dependent autoinducer 2 activity and biofilm formation. PloS One 2014; 9:e88260; PMID:24505453; http://dx.doi.org/ 10.1371/journal.pone.0088260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Anderson JK, Huang JY, Wreden C, Sweeney EG, Goers J, Remington SJ, Guillemin K. Chemorepulsion from the Quorum Signal Autoinducer-2 Promotes Helicobacter pylori Biofilm Dispersal. mBio [Internet] 2015; 6:e00379; PMID:26152582; http://dx.doi.org/ 10.1128/mBio.00379-15 Available from: http://mbio.asm.org/content/6/4/e00379-15.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zargar A, Quan DN, Carter KK, Guo M, Sintim HO, Payne GF, Bentley WE. Bacterial Secretions of Nonpathogenic Escherichia coli Elicit Inflammatory Pathways: a Closer Investigation of Interkingdom Signaling. mBio [Internet] 2015; 6:e00025; PMID:25759496; http://dx.doi.org/ 10.1128/mBio.00025-15 Available from: http://mbio.asm.org/content/6/2/e00025-15.abstract; [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Christiaen SEA, O'Connell Motherway M, Bottacini F, Lanigan N, Casey PG, Huys G, Nelis HJ, van Sinderen D, Coenye T. Autoinducer-2 plays a crucial role in gut colonization and probiotic functionality of Bifidobacterium breve UCC2003. PloS One 2014; 9:e98111; PMID:24871429; http://dx.doi.org/ 10.1371/journal.pone.0098111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Miller MB, Skorupski K, Lenz DH, Taylor RK, Bassler BL. Parallel Quorum Sensing Systems Converge to Regulate Virulence in Vibrio cholerae. Cell 2002; 110:303-14; PMID:12176318; http://dx.doi.org/ 10.1016/S0092-8674(02)00829-2 [DOI] [PubMed] [Google Scholar]
- [45].Rossmann FS, Racek T, Wobser D, Puchalka J, Rabener EM, Reiger M, Hendrickx APA, Diederich A-K, Jung K, Klein C, et al.. Phage-mediated Dispersal of Biofilm and Distribution of Bacterial Virulence Genes Is Induced by Quorum Sensing. PLoS Pathog 2015; 11:e1004653; PMID:25706310; http://dx.doi.org/ 10.1371/journal.ppat.1004653 [DOI] [PMC free article] [PubMed] [Google Scholar]


