Summary
Sequencing technologies are revealing many new non-synonymous single nucleotide variants (nsSNVs) in each personal exome. To assess their functional impacts, comparative genomics is frequently employed to predict if they are benign or not. However, evolutionary analysis alone is insufficient, because it misdiagnoses many disease-associated nsSNVs, such as those at positions involved in protein interfaces, and because evolutionary predictions do not provide mechanistic insights into functional change or loss. Structural analyses can aid in overcoming both of these problems by incorporating conformational dynamics and allostery in nSNV diagnosis. Finally, protein-protein interaction networks using systems-level methodologies shed light onto disease etiology and pathogenesis. Bridging these network approaches with structurally resolved protein interactions and dynamics will advance genomic medicine.
Introduction
Proteins are the remarkable workhorses of life, as they play crucial roles in biological function. They carry out their function through complex, carefully orchestrated protein-protein interactions in a crowded cellular environment. There have been many efforts to understand living systems by identifying protein interactions, including high-throughput methods such as yeast two-hybrid systems [1–3] and high affinity purification followed by mass spectrometry [4]. Moreover, these experimental efforts have been combined with computational approaches, making it possible to generate protein-protein interaction (PPI) networks at different genomic scales, from metabolic pathways to a diversity of species from bacteria to humans [5].
In addition to the tremendous amount of data arising from PPI networks, another front has emerged through genomic sequencing. For the last two decades, scientists have been profiling genomic variations in healthy and diseased individuals. Genome-wide association studies, whole-genome sequencing, and exome sequencing have shown that each personal genome contains millions of variants, thousands of which are non-synonymous single nucleotide variants (nsSNVs). Many of these nSNVs are associated with Mendelian and complex diseases [6]. With the sequencing of each new personal exome, the constellation of nsSNVs is expanding at a fast rate. But the translation of a personal exome variation profile into biomedically relevant information remains a challenge, particularly because a large proportion of novel nsSNVs are rare [7].
In this review, we discuss approaches for diagnosing the potential disease/functional impact of these nSNVs (Figure 1). First, we review methods based on detailed evolutionary and biophysical information, where molecular structures of protein complexes and the corresponding conformational dynamics information are utilized. Then, we review systems-level approaches of PPI networks to identify disease-associated mutations and disease pathology. A unified approach that merges these three major levels of information in diagnosing benign and disease-associated nsSNVs can provide solutions to the current challenges in genomic medicine. [8•,9•].
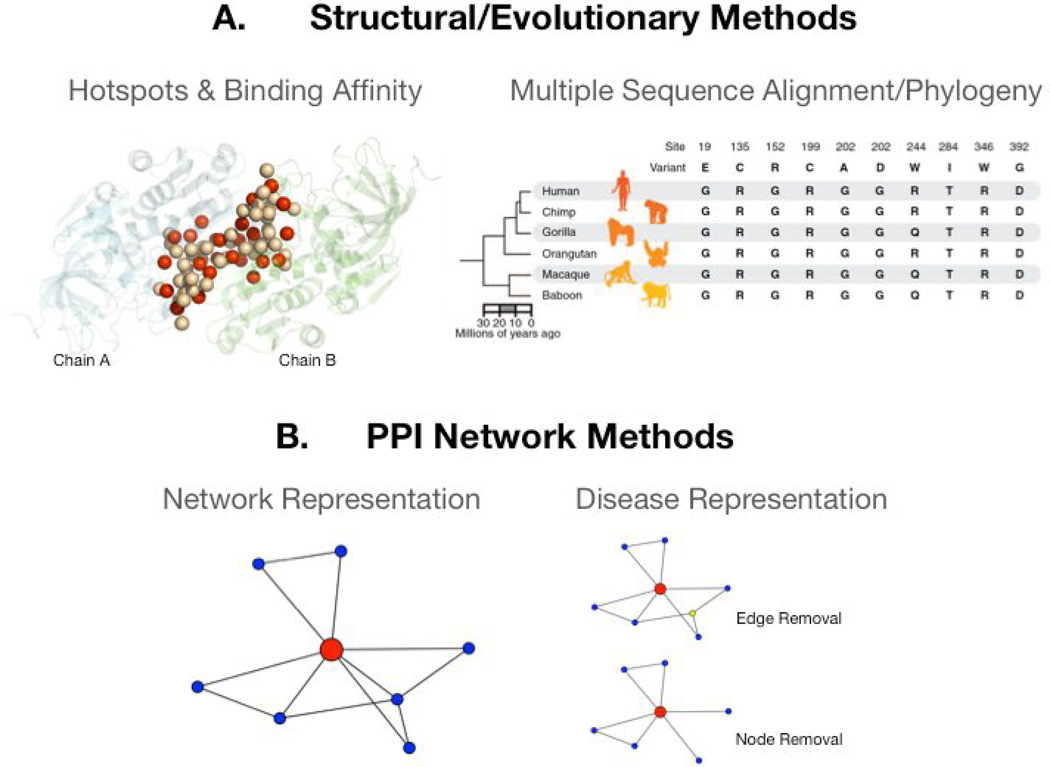
FIGURE 1. Computational tools in genomic medicine.
Two different approaches on different scales are used to determine the impact of nsSNVs on protein-protein interactions. (A) Structural methods use the information provided by hotspots [40,51] and estimate the change in binding affinity upon mutation. Hotspots (red spheres) are functional residues that are critical for binding in protein-protein interactions, while non-hotspot residues (light pink spheres) still participate in interactions at the interface, but are not critical. If a hotspot is mutated, then it will result in a deleterious mutation. Evolutionary metrics leverage positional conservation, which is determined by a multiple sequence alignment of many different species across the tree of life to measure the effect of nsSNVs [11]. Mutations at conserved sites are usually deleterious. The phylogentic tree shown here presents the 10 mutations associated with Miller syndrome, a rare genetic disorder. All mutations occur at slowly evolving or highly conserved sites both in primates and distantly related vertebrates. (B) Protein-protein interaction (PPI) networks represent a systems level approach where each node represents a protein and undirected edges represents their interactions. Disease mutations leading to a loss of an interaction can be shown as a removal of an edge, while knock out mutations can be captured as a node removal. Local and global network metrics can be used to find disease-associated variants.
Evolutionary and Structural Approaches for Prediction of Disease Mutations
A large number of computational tools employ purely evolutionary information to predict the impact of nsSNVs, under the auspices of the neutral theory of molecular evolution [10•,11]. Simply put, evolutionarily permissible substitutions in the amino acid sequence are determined by comparing sequence homologs across the evolution of diverse species. If an nsSNV is not found in the observed variation across the phylogeny, then it may be diagnosed to be putatively disease-associated (i.e., function impacting). To be more precise, probabilistic scoring functions are developed by using amino acid positional conservation and molecular phylegenetics. Current evolution-based diagnosis methods are widely used and are considered to produce good estimates [11–21]. However, they do have blind spots[11] and their accuracies in practical applications is debated because of their need to use training data that may not reflect the distribution of nsSNVs in the application domain [22–24].
Some of the current approaches combine evolutionary considerations with structural information in order to improve the prediction accuracy [21,25–27]. For instance, PolyPhen-2 uses solvent accessibility, secondary structure propensities, and crystallographic B-factors to classify mutational sites [21]. Other approaches consider the change in polarity, volume, and charge of the amino acid. Solvent accessibility has been used in a number of phenotypic prediction studies and has proven to be a useful attribute in disease prediction [26]. Moreover, residue-residue interaction networks of protein structures are used to identify functionally important residues through network topology parameters [27,28] and are utilized in predicting the impacts of observed nsSNVs. While the evolution-based methods are more effective than methods that solely use structural features, their accuracy breaks down at less-conserved positions resulting in true positive rates less than 50% [11,29]. These methods also have great difficulty in diagnosing benign variations at highly conserved positions (<50% rate of correct diagnosis of true negatives) [30]. It has also been shown that in silico tools yield very low accuracy for nsSNVs found to be associated with complex diseases; PolyPhen-2 [29] produced 22% true positives for 757 variants from VARIMED [31]. This low accuracy is due multiple genes having small cooperative effects in complex diseases and disease-associated nsSNVs are often not located at highly conserved positions [32–35].
Beyond evolutionary conservation, there have been many efforts to utilize the structural and network properties to diagnose disease variants. An all-atom structural mapping of observed nsSNVs on human PPI networks revealed that disease-associated nsSNVs are significantly enriched at protein-protein interfaces [34••]. For this reason, some recent methods have focused on modeling interfaces and predicting changes in binding affinities to distinguish the disease-associated nsSNVs from neutral nsSNVs. The proliferation of available experimental structures in the Protein Data Bank [36] and current advancements in homology modeling have facilitated the development of human structural interaction network (HSIN) databases of protein-protein and domain-domain interactions [37]. Mapping neutral and disease-associated nsSNVs on HSIN has shown several important results [32–34••,38,39•]. First, these studies showed that the pleiotropy of disease-associated nsSNVs can be explained by proteins interacting with different proteins at different interfaces [33], where the mutations at these separate interfaces may lead to different diseases and intensities [34••]. Second, nsSNVs at interfaces may disrupt or enhance protein-protein interactions, thus, playing an important role in pathogenesis [38,40]. While the disruption of transient binding interactions can usually affect the protein localization, the loss of obligate interactions due to interface mutations leads to complete loss of function. The mutations that enhance binding interactions may cause aggregation or aberrant recognition, as observed in cancers [39•].
Because interface mutations may alter binding interactions, there have been efforts to predict the effects of these mutations by measuring the difference between the free-energy change upon binding of the wild type and mutant (ΔΔG). Free energy differences upon binding calculated via thermodynamic integration and free energy perturbation approaches combined with molecular dynamics simulations are computationally expensive, particularly for large-scale protein complexes [41]. Therefore, many have developed in silico tools as a fast alternative to estimate ΔΔG using statistical energy functions based on known protein structures [42–44] and/or coupling with machine learning tools using training sets [45–47]. However, these calculations can be rather inaccurate, because local structural changes upon mutations are generally neglected [48,49].
Teng et. al. used an all-atom molecular force field (CHARMM) to investigate the effect of disease and neutral nsSNVs on binding energies for 264 protein-protein complexes with known nsSNVs. They found that disease-associated mutations often destabilize the electrostatic component of the binding energies. Furthermore, the change in physicochemical properties upon mutation, such as large changes in polarity and hydrophobicity, do not significantly alter the binding energy, which makes it challenging to distinguish between disease and benign nsSNVs [50]. Evaluating the importance of a particular interface residue to binding is another approach to predict the impact of nsSNVs.
Experimentally, critical binding sites can be identified by mutating each site to alanine and measuring the change in binding affinity. These positions, called hotspots, are often located at highly conserved positions with large changes in accessible surface area (ASA) upon binding [8•,51]. If a mutation occurs on such a site, it will impact function and, possibly, deleterious. Incorporating biophysical and structural properties of known hotspots into machine learning algorithms have made it possible to distinguish between disease-associated and neutral nsSNVs at protein interfaces [38]. It remains a challenge to predict disease-associated mutations occurring at non-hotspots.
Conformational Dynamics and Allostery in Disease Development
Currently, most machine learning methods that use structural features (e.g., ASA) are based on static 3D structures. This practice neglects protein conformational dynamics. However, protein structure-encoded conformational dynamics, which span a broad timescale of motion from atomic fluctuations and side chain rotations to collective domain movements, underlie a protein’s biological function. Protein evolution studies of several different protein families have shown that changes in conformational dynamics through allosteric regulation lead to new functions(e.g., green fluorescent protein (GFP), beta-lactamase inhibitors, and nuclear receptors [52–54]). Moreover evolutionary rates are strongly correlated with the flexibility of individual positions obtained from conformational dynamics [55–57].
Protein dynamics studies assert that protein function can be explained by analyzing the individual contribution of residues to the conformational dynamics and stability of a protein [55,56,58•]. Therefore, conformational dynamics-based metrics can also be utilized in predicting the impact of nsSNVs on protein function. Gerek et al. used an amino acid site-specific dynamic flexibility index (DFI) metric to evaluate the effect of flexibility of individual positions on biological fitness and function. DFI is a position-specific metric that quantifies the resilience of each residue to a perturbation occurring at another part of the chain, thus identifying the flexible and rigid parts of a protein [55]. Analysis of disease-associated and neutral nsSNVs for more than 100 human proteins revealed that disease-associated nsSNVs occur predominately at low DFI sites (i.e., rigid hinge sites), signifying the importance of hinge sites that control functionally critical motions. In contrast, neutral variants are more abundant at positions with high DFI, suggesting that flexible sites are more robust to mutations [55]. Furthermore, DFI profiles of over a thousand positions harboring mutations revealed that positions at protein interfaces have lower average DFI than those at non-interfaces, suggesting that protein-protein interfaces have less dynamic flexibility[58•]. These results suggest that hinge points at interfaces are critical for binding and mutations at these hinge sites will likely lead to disease.
Allostery is the regulation of cellular functions through the alteration of dynamics and structure upon an action at a distant site, which has been implicated in diseases. There are several disruptions of allosteric regulations that lead to disease development. Mutations can allosterically impair post-translational modification as observed in driver mutations in cancer [59–61]. Disease-associated variances can also change the ON/OFF populations in cell signaling by altering the stability of certain conformations and/or dynamics. Furthermore, they can lead to disease by shifting allosteric pathways, as observed in the mutation that gives rise to hyperekplexia [62]. Finally, mutations farther away from functionally critical sites can allosterically impair hinges (i.e., rigid parts), softening the functionally critical regions and lead to the loss of allosterically regulated conformational dynamics as observed for disease-associated mutations of human ferritin [63•].
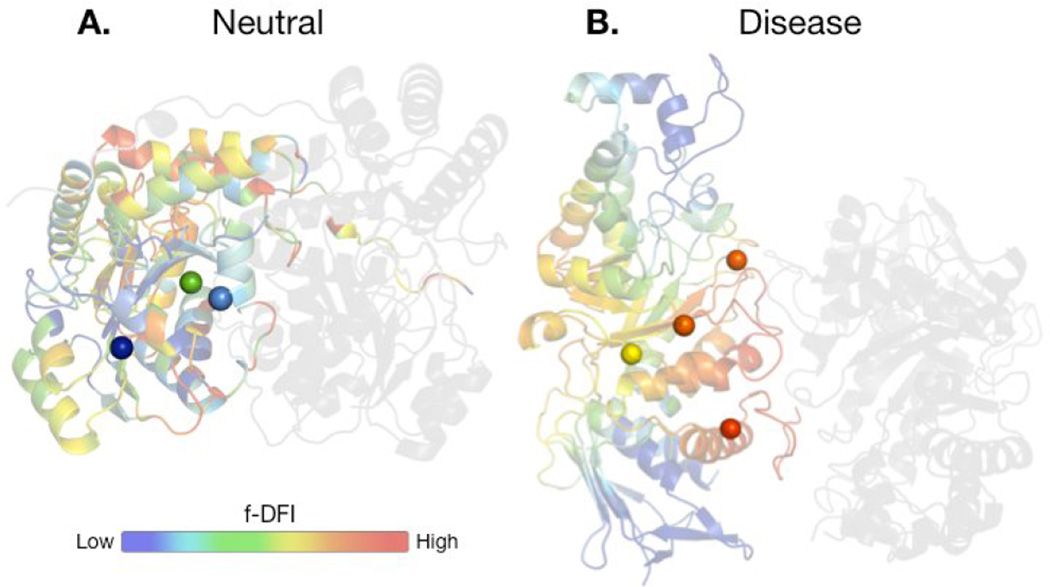
Allostery can elucidate the impact of non-hotspot mutations dynamically linked to hotspots [7]. Hotspots evaluated by the HotPoint server [51] of the protein assemblies in the dataset studied by Butler et al. [58•] indicated that most mutations occurring at hotspots are disease-associated. However, among the 100 disease-associated nsSNVs at interfaces, only half of them were at hotspots. How do non-hotspot sites play a role in disease-association? This can be studied by a new metric called the functional dynamic flexibility index (f-DFI) [63•]. f-DFI quantifies the residue fluctuation response of a position upon the perturbation of a functionally critical distant site. Thus, f-DFI enables the identification of non-hotspots residues that are linked allosterically to hotspots. Interestingly, ~80% of disease-associated mutations at non-hotspots exhibited high f-DFI values (>0.6), indicating they are dynamically coupled to hotspot residues. Figure 2 presents a case study of two protein complexes, alanine:glyoxylate aminotransferase and lysosomal beta-hexosaminidase A. In this case, benign mutations have low f-DFI (< 0.4), despite being in close proximity to hotspots. In contrast, the disease-associated mutations have high f-DFI (> 0.6), indicating they are dynamically linked to hotspots. Two are spatially close to a hotspot, so it is not surprising that they have high f-DFI scores. However, the others are not as close, they are dynamically coupled with hotspots making them critical sites. When non-hotspot sites dynamically coupled to hotspots are mutated, loss of function may occur and result in a potentially detrimental phenotype.
FIGURE 2. Case study using f-DFI for mutations on two protein complexes.
(A) Two protein complexes used in a previous study [58], alanine:glyoxylate aminotransferase (AGT) and (B) lysosomal beta-hexosaminidase A (Hex A), represented by PDB codes 1h0c and 2gk1, respectively. Both are enzymes in the human body that are responsible for functions in the liver and central nervous system. One chain on each dimer is color-coded by its f-DFI score (0 to 1) for each residue, where blue indicates low f-DFI and red high f-DFI [63]. On the left AGT displays 3 neutral mutations, while on the right Hex A shows 4 known disease-associated mutations purported by the Human Gene Mutation Database (HGMD) [87]. None of the mutations are located at hotspots, making them difficult to predict. f-DFI is a metric that can capture the dynamic coupling between mutations at non-hotspots and hotspots despite not being in close proximity.
Network metrics can identify disease-associated proteins in PPI networks
Beyond the joint evolutionary and structural analysis of single proteins, the diagnosis of disease causing nsSNVs for complex diseases would require the analysis of multiple proteins together that are connected in PPI networks. In PPIs, proteins and their interactions are represented as nodes connected by undirected edges [64•–67] without taking into account the details of molecular interactions. PPI networks are described by scale-free networks having hubs with a high degree of connectivity; thus, they have the important property of being resilient to random stochastic effects, a necessary property in biology [68]. Disease can manifest itself in two ways in networks: node removal or edge modification. When a node is removed from a network, it is due to a destabilizing mutation that knocks out a protein. An edge modification is due to removing or adding an edge in the interaction network. It has been experimentally shown that many edgetic mutations are due to mutations on the interface [69]. Edges (interactions) can also be added, leading to gain-of-function mutations [70].
Local and global network metrics combined with known disease-associated proteins can reliably predict unknown disease-associated proteins. The first attempts to identify disease-associated nsSNVs in PPI networks used local metrics such as the Direct Neighbor Counting method (also known as the guilt-by-association method), where it is assumed that candidate proteins that interact with known disease proteins are themselves disease-associated [71,72]. Global metrics can identify disease proteins that do not directly interact with known disease proteins[67]. In the shortest path analysis method, the shortest path between two disease nodes is found. A node in close proximity to multiple disease nodes has a high probability of being disease-associated [73]. It has also been shown that “bottleneck” or “sole-broker” proteins with a high betweeness/centrality (i.e., many shortest paths passing through a node) are also likely to be disease-associated [74,75]. Methods such as diffusion kernel and random walk with restart measure how two non-interacting nodes are related by having random walkers start from a known disease node and diffuse through the network [76]. These global metrics enable the identification of the nodes and edges that are associated to known disease genes by exploiting the full network topology.
Proteins that interact with several disease proteins or proteins that are in proximity to disease proteins will have a higher probability to be encoded by a disease gene [76]. Köhler et al. showed that random walk with restart is superior to local metrics [76]. Although random walk methods produce the most accurate results, they still fail to identify disease variants predicted by local methods. Navlakha and Kingsford were able to create a consensus method using 13 different metrics in tandem in an ensemble of decision trees with a random forest classifier [77]. This method currently has the best accuracy. By incorporating multiple-omics (e.g., genomics, transcriptomics, and proteomics) analysis into network methods, Chen et al. were able to identify biological processes for two viral infections and the development of type 2 diabetes [78••]. The robustness of the PPI network used is critical for higher accuracy of these approaches [79–82]. Guney and Oliva [83] tested several network-based methods with respect to the perturbations of the system using various disease phenotypes from the Online Mendelian Inheritance in Man (OMIM) database. They found that disease proteins are connected via multiples pathways in a PPI network. Even when these networks are significantly perturbed, network-based methods can reveal hidden disease association proteins, particularly in cases of breast cancer and diabetes. In general, the PPI network approaches can identify certain proteins associated with specific disease better than the rest [77,83].
Overall PPI networks represent the simplest networks. They capture whether proteins interact, the architecture of a network, but they do not tell us how or at what rate they interact, and what the parameters of the network are. There have been cases studies were interactions in PPI networks can be parameterized by rate constants [84–86]. Due to difficulties of measuring parameters in a cellular context, parameterization of a proteome wide PPI network in humans has yet to be realized. Thus, the future of network approaches in PPI analysis lies in creating more accurate PPI datasets and integration of different omics
CONCLUSION
Advances in sequencing technologies are providing a myriad of data on human genetic variation. However, distinguishing between neutral variants (with little or no effect on phenotype) from variants conferring disease risk remains elusive. While earlier methods did not consider the role of protein interactions in the identification of disease-associated variants, recent studies about the prevalence of nsSNVs at interfaces provided mechanistic insight about their critical role in interactions. This has led to two different approaches at different length scales: PPI networks at the system level and biophysical methods and evolutionary information at the molecular level. The future in genomic medicine lies in merging these two approaches. By combining how two proteins interact in a PPI network, rather than merely knowing two proteins interact, will provide the next major advancement to undercover disease pathology of Mendelian, particularly complex diseases. As PPI data improves and new nsSNVs are discovered, we get closer to a new phase of genomic medicine.
Highlights.
Protein interfaces are enriched with disease-associated mutations.
Mutations that alter binding interactions lead to disease.
Mutations on evolutionary conserved, hot-spots are usually associated with disease.
Mutations on non-hot spot sites can lead to disease through allosteric regulations.
Disease genes are found at positions that play a critical role in the transmission of information in PPI networks.
Acknowledgments
Support from NIH awards U54GN0945999 and LM011941-01 is gratefully acknowledged by SBO. SK also acknowledges HG002096-12, LM011941-02, and DK098242-03.We thank Dr. Ashini Bolia for a careful review of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
•of special interest
•• of outstanding interest
- 1.Rual J-F, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 2.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 3.Yu H, Tardivo L, Tam S, Weiner E, Gebreab F, Fan C, Svrzikapa N, Hirozane-Kishikawa T, Rietman E, Yang X. Next-generation sequencing to generate interactome datasets. Nat. Methods. 2011;8:478–480. doi: 10.1038/nmeth.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O’Connor L, Li M. Large-scale mapping of human protein–protein interactions by mass spectrometry. Mol. Syst. Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wodak SJ, Vlasblom J, Turinsky AL, Pu S. Protein–protein interaction networks: the puzzling riches. Curr. Opin. Struct. Biol. 2013;23:941–953. doi: 10.1016/j.sbi.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Green ED, Guyer MS, Green ED, Guyer MS, Manolio TA, Peterson JL. Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470:204–213. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- 7.Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, et al. Evolution and Functional Impact of Rare Coding Variation from Deep Sequencing of Human Exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Keskin O, Gursoy A, Ma B, Nussinov R. Principles of Protein–Protein Interactions: What are the Preferred Ways For Proteins To Interact? Chem. Rev. 2008;108:1225–1244. doi: 10.1021/cr040409x. A comprehensive review on protein-protein interaction networks.
- 9. Kuzu G, Keskin O, Gursoy A, Nussinov R. Constructing structural networks of signaling pathways on the proteome scale. Curr. Opin. Struct. Biol. 2012;22:367–377. doi: 10.1016/j.sbi.2012.04.004. The authors detail the use of structural networks that move beyond the PPI network paradigm. In structural networks the types of interactions between proteins are described as well as the nature of how proteins bind with one another. This is in contract to PPI edge and node networks that only describe which protein interact with others.
- 10. Dudley JT, Kim Y, Liu L, Markov GJ, Gerold K, Chen R, Butte AJ, Kumar S. Human genomic disease variants: A neutral evolutionary explanation. Genome Res. 2012;22:1383–1394. doi: 10.1101/gr.133702.111. A perspective on the null hypothesis for evaluating disease-associated variation, which ultimately enables one to use the principles of neutral theory of molecular evolution in diagnosing disease nsSNVs.
- 11.Kumar S, Sanderford M, Gray VE, Ye J, Liu L. Evolutionary diagnosis method for variants in personal exomes. Nat. Methods. 2012;9:855–856. doi: 10.1038/nmeth.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tavtigian SV, Byrnes GB, Goldgar DE, Thomas A. Classification of rare missense substitutions, using risk surfaces, with genetic- and molecular-epidemiology applications. Hum. Mutat. 2008;29:1342–1354. doi: 10.1002/humu.20896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 2011;39:37–43. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi Y, Chan a P. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015 doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katsonis P, Lichtarge O. A formal perturbation equation between genotype and phenotype determines the Evolutionary Action of protein-coding variations on fitness. Genome Res. 2014;24:2050–2058. doi: 10.1101/gr.176214.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B, Krishnan VG, Mort ME, Xin F, Kamati KK, Cooper DN, Mooney SD, Radivojac P. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics. 2009;25:2744–2750. doi: 10.1093/bioinformatics/btp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Capriotti E, Calabrese R, Fariselli P, Martelli P, Altman RB, Casadio R. WS-SNPs&GO: a web server for predicting the deleterious effect of human protein variants using functional annotation. BMC Genomics. 2013;14:S6. doi: 10.1186/1471-2164-14-S3-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas PD, Kejariwal A, Campbell MJ, Mi H, Diemer K, Guo N, Ladunga I, Ulitsky-Lazareva B, Muruganujan A, Rabkin S, et al. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003;31:334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bromberg Y, Yachdav G, Rost B. SNAP predicts effect of mutations on protein function. Bioinformatics. 2008;24:2397–2398. doi: 10.1093/bioinformatics/btn435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adzhubei I, Jordan DM, Sunyaev SR. Predicting Functional Effect of Human Missense Mutations Using PolyPhen-2 [Internet] In: Haines JL, Korf BR, Morton CC, Seidman CE, Seidman JG, Smith DR, editors. Current Protocols in Human Genetics. John Wiley & Sons, Inc.; 2013. pp. 7.20.1–7.20.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum. Mutat. 2011;32:358–368. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- 23.Dorfman R, Nalpathamkalam T, Taylor C, Gonska T, Keenan K, Yuan XW, Corey M, Tsui L-C, Zielenski J, Durie P. Do common in silico tools predict the clinical consequences of amino-acid substitutions in the CFTR gene? Clin. Genet. 2010;77:464–473. doi: 10.1111/j.1399-0004.2009.01351.x. [DOI] [PubMed] [Google Scholar]
- 24.Liu L, Tamura K, Sanderford M, Gray VE, Kumar S. A Molecular Evolutionary Reference for the Human Variome. Mol. Biol. Evol. 2015 doi: 10.1093/molbev/msv198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Espinosa O, Mitsopoulos K, Hakas J, Pearl F, Zvelebil M. Deriving a mutation index of carcinogenicity using protein structure and protein interfaces. PloS One. 2014;9:e84598. doi: 10.1371/journal.pone.0084598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei Q, Xu Q, Dunbrack RL. Prediction of phenotypes of missense mutations in human proteins from biological assemblies. Proteins Struct. Funct. Bioinforma. 2013;81:199–213. doi: 10.1002/prot.24176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye ZQ, Zhao SQ, Gao G, Liu XQ, Langlois RE, Lu H, Wei L. Finding new structural and sequence attributes to predict possible disease association of single amino acid polymorphism (SAP) Bioinformatics. 2007;23:1444–1450. doi: 10.1093/bioinformatics/btm119. [DOI] [PubMed] [Google Scholar]
- 28.Cheng TMK, Lu Y-E, Vendruscolo M, Lio’ P, Blundell TL. Prediction by Graph Theoretic Measures of Structural Effects in Proteins Arising from Non-Synonymous Single Nucleotide Polymorphisms. PLoS Comput. Biol. 2008;4:e1000135. doi: 10.1371/journal.pcbi.1000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar S, Dudley JT, Filipski A, Liu L. Phylomedicine: an evolutionary telescope to explore and diagnose the universe of disease mutations. Trends Genet. 2011;27:377–386. doi: 10.1016/j.tig.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar S, Suleski MP, Markov GJ, Lawrence S, Marco A, Filipski AJ. Positional conservation and amino acids shape the correct diagnosis and population frequencies of benign and damaging personal amino acid mutations. Genome Res. 2009;19:1562–1569. doi: 10.1101/gr.091991.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen R, Davydov EV, Sirota M, Butte AJ. Non-Synonymous and Synonymous Coding SNPs Show Similar Likelihood and Effect Size of Human Disease Association. PLoS ONE. 2010;5:e13574. doi: 10.1371/journal.pone.0013574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishi H, Tyagi M, Teng S, Shoemaker Ba, Hashimoto K, Alexov E, Wuchty S, Panchenko AR. Cancer Missense Mutations Alter Binding Properties of Proteins and Their Interaction Networks. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0066273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kar G, Gursoy A, Keskin O. Human cancer protein-protein interaction network: A structural perspective. PLoS Comput. Biol. 2009;5 doi: 10.1371/journal.pcbi.1000601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang X, Wei X, Thijssen B, Das J, Lipkin SM, Yu H. Three-dimensional reconstruction of protein networks provides insight into human genetic disease. Nat. Biotechnol. 2012;30:159–164. doi: 10.1038/nbt.2106. The work shows that mutations are enriched at the interfaces and their resulting genetic disease can be mapped to the position of the mutation at the interface.
- 35.Safari-Alighiarloo N, Taghizadeh M, Rezaei-Tavirani M, Goliaei B, Peyvandi AA. Protein-protein interaction networks (PPI) and complex diseases. Gastroenterol. Hepatol. Bed Bench. 2014;7:17–31. [PMC free article] [PubMed] [Google Scholar]
- 36.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mosca R, Céol A, Aloy P. Interactome3D: adding structural details to protein networks. Nat. Methods. 2012;10:47–53. doi: 10.1038/nmeth.2289. [DOI] [PubMed] [Google Scholar]
- 38.Schuster-Böckler B, Bateman A. Protein interactions in human genetic diseases. Genome Biol. 2008;9:R9. doi: 10.1186/gb-2008-9-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yates CM, Sternberg MJE. The effects of non-synonymous single nucleotide polymorphisms (nsSNPs) on protein-protein interactions. J. Mol. Biol. 2013;425:3949–3963. doi: 10.1016/j.jmb.2013.07.012. The authors showed how protein-protein interfaces harbor nsSNVs.
- 40.David A, Razali R, Wass MN, Sternberg MJE. Protein-protein interaction sites are hot spots for disease-associated nonsynonymous SNPs. Hum. Mutat. 2012;33:359–363. doi: 10.1002/humu.21656. [DOI] [PubMed] [Google Scholar]
- 41.Klimovich PV, Shirts MR, Mobley DL. Guidelines for the analysis of free energy calculations. J. Comput. Aided Mol. Des. 2015;29:397–411. doi: 10.1007/s10822-015-9840-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dehouck Y, Kwasigroch JM, Rooman M, Gilis D. BeAtMuSiC: Prediction of changes in protein-protein binding affinity on mutations. Nucleic Acids Res. 2013;41:W333–W339. doi: 10.1093/nar/gkt450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li M, Petukh M, Alexov E, Panchenko AR. Predicting the Impact of Missense Mutations on Protein-Protein Binding Affinity. J. Chem. Theory Comput. 2014;10:1770–1780. doi: 10.1021/ct401022c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berliner N, Teyra J, Colak R, Garcia Lopez S, Kim PM. Combining structural modeling with ensemble machine learning to accurately predict protein fold stability and binding affinity effects upon mutation. PloS One. 2014;9:e107353. doi: 10.1371/journal.pone.0107353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moal IH, Fernandez-Recio J. SKEMPI: a Structural Kinetic and Energetic database of Mutant Protein Interactions and its use in empirical models. Bioinformatics. 2012;28:2600–2607. doi: 10.1093/bioinformatics/bts489. [DOI] [PubMed] [Google Scholar]
- 46.Zhao N, Han JG, Shyu C-R, Korkin D. Determining Effects of Non-synonymous SNPs on Protein-Protein Interactions using Supervised and Semi-supervised Learning. PLoS Comput. Biol. 2014;10:e1003592. doi: 10.1371/journal.pcbi.1003592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Z, Wang L, Gao Y, Zhang J, Zhenirovskyy M, Alexov E. Predicting folding free energy changes upon single point mutations. Bioinformatics. 2012;28:664–671. doi: 10.1093/bioinformatics/bts005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Potapov V, Cohen M, Schreiber G. Assessing computational methods for predicting protein stability upon mutation: Good on average but not in the details. Protein Eng. Des. Sel. 2009;22:553–560. doi: 10.1093/protein/gzp030. [DOI] [PubMed] [Google Scholar]
- 49.Khan S, Vihinen M. Performance of protein stability predictors. Hum. Mutat. 2010;31:675–684. doi: 10.1002/humu.21242. [DOI] [PubMed] [Google Scholar]
- 50.Teng S, Madej T, Panchenko A, Alexov E. Modeling Effects of Human Single Nucleotide Polymorphisms on Protein-Protein Interactions. Biophys. J. 2009;96:2178–2188. doi: 10.1016/j.bpj.2008.12.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tuncbag N, Keskin O, Gursoy A. HotPoint: hot spot prediction server for protein interfaces. Nucleic Acids Res. 2010;38:W402–W406. doi: 10.1093/nar/gkq323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Glembo TJ, Farrell DW, Gerek ZN, Thorpe M, Ozkan SB. Collective dynamics differentiates functional divergence in protein evolution. PLoS Comput. Biol. 2012;8:e1002428. doi: 10.1371/journal.pcbi.1002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zou T, Risso VA, Gavira JA, Sanchez-Ruiz JM, Ozkan SB. Evolution of Conformational Dynamics Determines the Conversion of a Promiscuous Generalist into a Specialist Enzyme. Mol. Biol. Evol. 2015;32:132–143. doi: 10.1093/molbev/msu281. [DOI] [PubMed] [Google Scholar]
- 54.Kim H, Zou T, Modi C, Dörner K, Grunkemeyer TJ, Chen L, Fromme R, Matz MV, Ozkan SB, Wachter RM. A Hinge Migration Mechanism Unlocks the Evolution of Green-to-Red Photoconversion in GFP-like Proteins. Structure. 2015;23:34–43. doi: 10.1016/j.str.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nevin Gerek Z, Kumar S, Banu Ozkan S. Structural dynamics flexibility informs function and evolution at a proteome scale. Evol. Appl. 2013;6:423–433. doi: 10.1111/eva.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Y, Bahar I. Sequence Evolution Correlates with Structural Dynamics. Mol. Biol. Evol. 2012;29:2253–2263. doi: 10.1093/molbev/mss097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liberles DA, Teichmann SA, Bahar I, Bastolla U, Bloom J, Bornberg-Bauer E, Colwell LJ, De Koning A, Dokholyan NV, Echave J. The interface of protein structure, protein biophysics, and molecular evolution. Protein Sci. 2012;21:769–785. doi: 10.1002/pro.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Butler BM, Gerek ZN, Kumar S, Ozkan SB. Conformational dynamics of nonsynonymous variants at protein interfaces reveals disease association: The Role of Dynamics in Neutral and Damaging nsSNVs. Proteins Struct. Funct. Bioinforma. 2015;83:428–435. doi: 10.1002/prot.24748. The authors show that conformational dynamics can distinguish disease nsSNVS at interfaces.
- 59.Reimand J, Wagih O, Bader GD. The mutational landscape of phosphorylation signaling in cancer [Internet] Sci. Rep. 2013;3 doi: 10.1038/srep02651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nussinov R, Tsai C-J. “Latent drivers” expand the cancer mutational landscape. Curr. Opin. Struct. Biol. 2015;32:25–32. doi: 10.1016/j.sbi.2015.01.004. This review details the concept of “latent driver” cancer mutations, which are passenger mutations that become driver mutations in the presence of other cancer mutations.
- 61.Nussinov R, Tsai C-J, Ma B. The underappreciated role of allostery in the cellular network. Annu. Rev. Biophys. 2013;42:169–189. doi: 10.1146/annurev-biophys-083012-130257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shan Q, Han L, Lynch JW. Function of hyperekplexia-causing α1R271Q/L glycine receptors is restored by shifting the affected residue out of the allosteric signalling pathway: Restoration of mutant glycine receptor function. Br. J. Pharmacol. 2012;165:2113–2123. doi: 10.1111/j.1476-5381.2011.01701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kumar A, Glembo TJ, Ozkan SB. The Role of Conformational Dynamics and Allostery in the Disease Development of Human Ferritin [Internet] Biophys. J. 2015 doi: 10.1016/j.bpj.2015.06.060. The authors show how dynamics can distinguish between neutral and disease associated mutations in human ferritin protein by allosterically changing the critical functional hinge sites.
- 64.Silverman EK, Loscalzo J. Network medicine approaches to the genetics of complex diseases. Discov. Med. 2012;14:143. [PMC free article] [PubMed] [Google Scholar]
- 65.Barabási A-L, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 66.Furlong LI. Human diseases through the lens of network biology. Trends Genet. 2013;29:150–159. doi: 10.1016/j.tig.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 67. Barabási A-L, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 2011;12:56–68. doi: 10.1038/nrg2918. A comprehensive review of network approaches using networks to predict and understand the mechanics of human disease.
- 68.Seebacher J, Gavin A-C. SnapShot: Protein-Protein Interaction Networks. Cell. 2011;144:1000.e1–1000.e1. doi: 10.1016/j.cell.2011.02.025. [DOI] [PubMed] [Google Scholar]
- 69.Vidal M, Cusick ME, Barabási A-L. Interactome Networks and Human Disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Strachan T, Read AP. Molecular pathology. 1999 [no volume] [Google Scholar]
- 71.Altshuler D, Daly M, Kruglyak L. Guilt by association. Nat. Genet. 2000;26:135–138. doi: 10.1038/79839. [DOI] [PubMed] [Google Scholar]
- 72.Oliver S. Proteomics: guilt-by-association goes global. Nature. 2000;403:601–603. doi: 10.1038/35001165. [DOI] [PubMed] [Google Scholar]
- 73.Krauthammer M, Kaufmann CA, Gilliam TC, Rzhetsky A. Molecular triangulation: Bridging linkage and molecular-network information for identifying candidate genes in Alzheimer’s disease. Proc. Natl. Acad. Sci. 2004;101:15148–15153. doi: 10.1073/pnas.0404315101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cai JJ, Borenstein E, Petrov DA. Broker genes in human disease. Genome Biol. Evol. 2010;2:815–825. doi: 10.1093/gbe/evq064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu H, Kim PM, Sprecher E, Trifonov V, Gerstein M. The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLoS Comput. Biol. 2007;3:e59. doi: 10.1371/journal.pcbi.0030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Köhler S, Bauer S, Horn D, Robinson PN. Walking the Interactome for Prioritization of Candidate Disease Genes. Am. J. Hum. Genet. 2008;82:949–958. doi: 10.1016/j.ajhg.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Navlakha S, Kingsford C. The power of protein interaction networks for associating genes with diseases. Bioinformatics. 2010;26:1057–1063. doi: 10.1093/bioinformatics/btq076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chen R, Mias GI, Li-Pook-Than J, Jiang L, Lam HY, Chen R, Miriami E, Karczewski KJ, Hariharan M, Dewey FE. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–1307. doi: 10.1016/j.cell.2012.02.009. This work shows the power of integrating multiple omics approaches (genomics, transcriptomic, proteomic, and metabolomic) to analyze medical risks, such at type 2 diabetes, and dynamics changes in biological pathways between healthy and diseased individuals.
- 79.Carlson JM, Doyle J. Complexity and robustness. Proc. Natl. Acad. Sci. U.S.A. 2002;99(Suppl 1):2538–2545. doi: 10.1073/pnas.012582499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jeong H, Tombor B, Albert R, Oltvai ZN, Barabási AL. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 81.Huang C-H, Fang J-F, Tsai JJP, Ng K-L. Topological Robustness of the Protein-Protein Interaction Networks [Internet] In: Eskin E, Ideker T, Raphael B, Workman C, editors. Systems Biology and Regulatory Genomics. Springer Berlin Heidelberg; 2006. pp. 166–177. [Google Scholar]
- 82.Whitacre JM. Biological Robustness: Paradigms, Mechanisms, and Systems Principles [Internet] Front. Genet. 2012;3 doi: 10.3389/fgene.2012.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guney E, Oliva B. Analysis of the Robustness of Network-Based Disease-Gene Prioritization Methods Reveals Redundancy in the Human Interactome and Functional Diversity of Disease-Genes. PLoS ONE. 2014;9:e94686. doi: 10.1371/journal.pone.0094686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guido NJ, Wang X, Adalsteinsson D, McMillen D, Hasty J, Cantor CR, Elston TC, Collins JJ. A bottom-up approach to gene regulation. Nature. 2006;439:856–860. doi: 10.1038/nature04473. [DOI] [PubMed] [Google Scholar]
- 85.Kiel C, Serrano L. Cell type-specific importance of ras-c-raf complex association rate constants for MAPK signaling. Sci. Signal. 2009;2:ra38. doi: 10.1126/scisignal.2000397. [DOI] [PubMed] [Google Scholar]
- 86.Ronen M, Rosenberg R, Shraiman BI, Alon U. Assigning numbers to the arrows: parameterizing a gene regulation network by using accurate expression kinetics. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10555–10560. doi: 10.1073/pnas.152046799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cooper DN, Stenson PD, Chuzhanova NA. The Human Gene Mutation Database (HGMD) and Its Exploitation in the Study of Mutational Mechanisms [Internet] In: Baxevanis AD, Petsko GA, Stein LD, Stormo GD, editors. Current Protocols in Bioinformatics. John Wiley & Sons, Inc.; 2006. [DOI] [PubMed] [Google Scholar]