Abstract
Ubiquitination is a central process affecting all facets of cellular signaling and function1. A critical step in ubiquitination is the transfer of ubiquitin from an E2 ubiquitin-conjugating enzyme to a substrate or a growing ubiquitin chain, which is mediated by E3 ubiquitin ligases. RING-type E3 ligases typically facilitate the transfer of ubiquitin from the E2 directly to the substrate2,3. The RBR family of RING-type E3 ligases, however, breaks this paradigm by forming a covalent intermediate with ubiquitin similarly to HECT-type E3 ligases4–6. The RBR family includes Parkin4 and HOIP, the central catalytic factor of the linear ubiquitin chain assembly complex (LUBAC)7. While structural insights into the RBR E3 ligases Parkin and HHARI in their overall autoinhibited forms are available8–13, no structures exist of intact fully active RBR E3 ligases or any of their complexes. Thus, the RBR mechanism of action has remained largely enigmatic. Here we present the first structure of the fully active HOIP-RBR in its transfer complex with an E2~ubiquitin conjugate, which elucidates the intricate nature of RBR E3 ligases. The active HOIP-RBR adopts a conformation markedly different from that of autoinhibited RBRs. HOIP-RBR binds the E2~ubiquitin conjugate in an elongated fashion, with the E2 and E3 catalytic centers ideally aligned for ubiquitin transfer, which structurally both requires and enables a HECT-like mechanism. In addition, surprisingly, three distinct helix–IBR-fold motifs inherent to RBRs form ubiquitin-binding regions that engage the activated ubiquitin of the E2~Ub conjugate as well as an additional regulatory ubiquitin molecule. The features uncovered reveal critical states of the HOIP-RBR E3 ligase cycle, and comparison with Parkin and HHARI suggests a general mechanism for RBR E3 ligases.
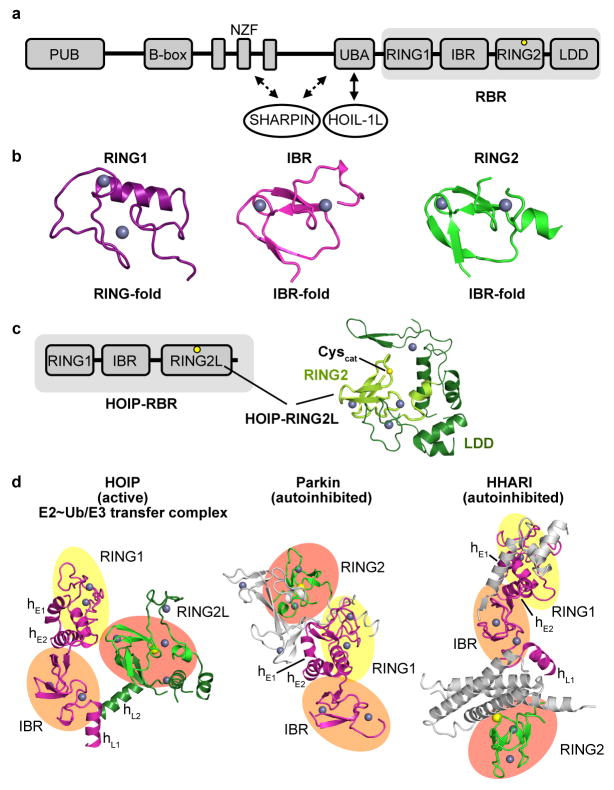
RBR E3 ligases are characterized by an extended RING domain (RING1) followed by an ‘in-between RING’ (IBR) domain and the catalytic domain, which is structurally an IBR domain but is commonly designated RING2 (Extended Data Fig. 1a,b)8–11,14. HOIP, one of the most studied RBRs, is the key E3 ligase of the LUBAC. It is a prototypical RBR yet contains an extended RING2 domain that includes the linear ubiquitin chain determining domain (LDD), enabling the selective formation of linear ubiquitin linkages (Extended Data Fig. 1c)5–7,14, and is thus denoted RING2L. The HOIP-RBR is kept in an autoinhibited state by the HOIP-UBA domain, whose sequestration by the LUBAC constituent HOIL-1L activates HOIP to trigger, together with SHARPIN, NF-κB signaling and other cellular processes5–7,15–20. To obtain the first insight into an active RBR in a key catalytic complex, we generated a stable E2~ubiquitin conjugate (UbcH5B C85K~ubiquitin)21 and isolated its complex with HOIP-RBR. The subsequent addition of free ubiquitin proved necessary for crystal formation, allowing us to solve the HOIP-RBR/UbcH5B~ubiquitin transfer complex structure at 3.5 Å resolution (Fig. 1a; Extended Data Figs. 2, 3).
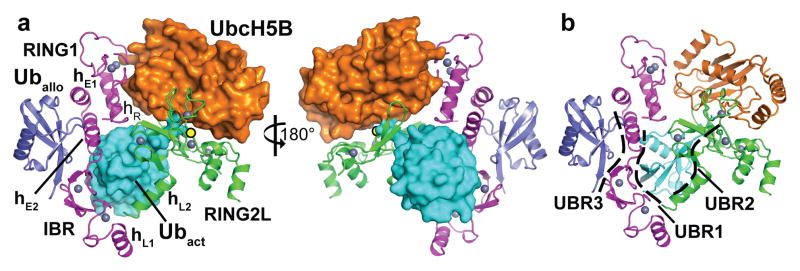
Figure 1. Structure of the HOIP-RBR/UbcH5B~ubiquitin transfer complex.
a, Structure with key elements annotated. The RBR-RING1 and IBR together with two RING1 extension helices (hE1, hE2) form an arm-like unit (magenta; for domain annotations see Extended Data Fig. 1). The RING1–IBR and the catalytic RING2L (green) are connected by two linker helices (hL1, hL2) and together engage the E2~ubiquitin conjugate (UbcH5B~Ubact; orange~cyan) positioning it for ubiquitin transfer onto the RBR catalytic cysteine (yellow circle). An allosteric ubiquitin molecule (Uballo, blue) binds to the RING1–IBR across from the activated ubiquitin. Zinc ions are shown as grey spheres; hR denotes the RING1 helix. b, Location of the three ubiquitin-binding regions (UBRs, dashed lines) in the complex.
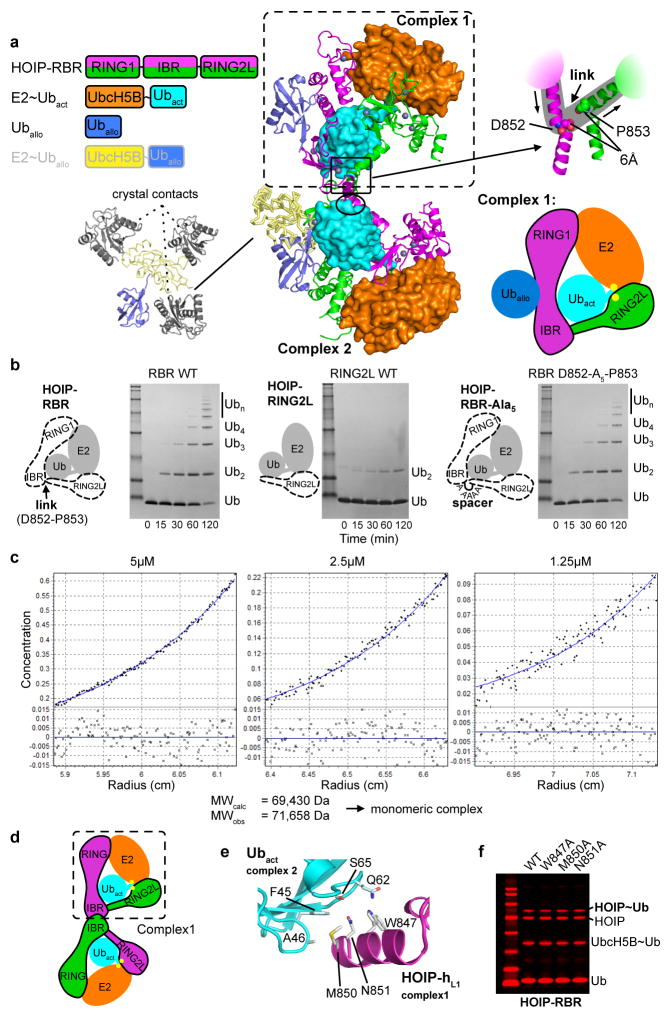
The asymmetric unit contains two HOIP-RBR molecules interacting with two UbcH5B~ubiquitin conjugates and an additional ubiquitin or E2~ubiquitin conjugate, arranged in a swapped dimer configuration (Extended Data Fig. 3a). While this arrangement could have functional relevance, analysis of interfaces and biophysical examination (Extended Data Fig. 3b–f) indicate a monomeric assembly of the HOIP/E2~ubiquitin loading complex (Fig. 1), represented in the crystal structure by the RING1–IBR module (residues 699–852) from one HOIP molecule and the RING2L (residues 853–1072) from the second HOIP molecule in the asymmetric unit. In this assembly, the RING1–IBR module forms an elongated arm-like unit (Fig. 2a) that together with the RING2L embraces the E2~ubiquitin conjugate in a clamp-like manner (Fig. 1a). This active HOIP-RBR conformation is markedly different from previous structures of autoinhibited RBRs (Extended Data Fig. 1d) and enables an astounding array of features inherent to the active RBR. Most notably, three distinct helix–IBR-fold motifs function as essential discrete ubiquitin-binding regions (UBR) (Fig. 1b).
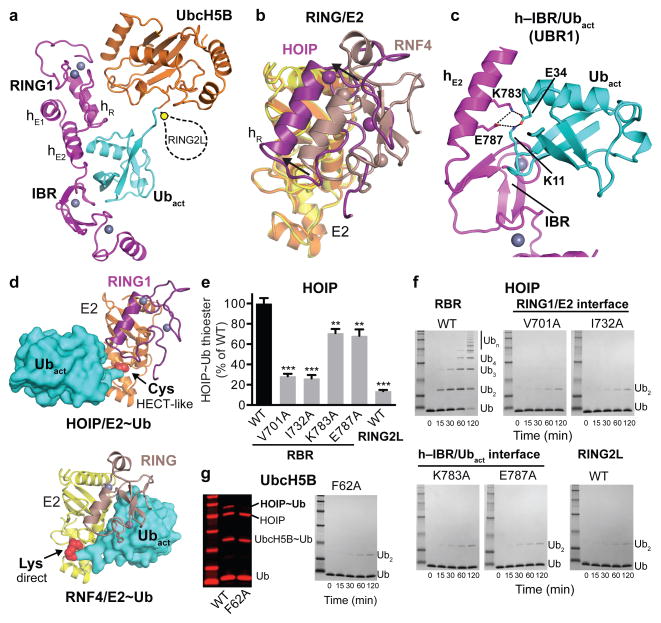
Figure 2. The HOIP-RING1–IBR coordinates the UbcH5B~ubiquitin conjugate in a bipartite manner tailored to a HECT-like mechanism.
a, Coordination of the UbcH5B~ubiquitin conjugate (orange and cyan) by the RING1–IBR (magenta). HOIP-RING2L is indicated schematically. b, The HOIP-RING1 coordinates the E2 in a shifted position compared to classic RINGs. Overlay of HOIP-RING1/UbcH5B (magenta/yellow) and RNF4-RING/UbcH5A (brown/orange, PDBid: 4AP421). c, RBR-hE2 and IBR form UBR1 binding the activated ubiquitin. A central salt bridge system connects hE2 and ubiquitin. d, Comparison of E2~ubiquitin binding by HOIP-RING1 (top) and a classic RING (bottom, RNF4 PDBid: 4AP421) highlights the differences in E2~ubiquitin thioester positioning (red spheres). The directionality of the thioester attacking residue (active site cysteine of RBR in the HECT-like transfer and lysine of the substrate in RING-mediated transfer) is indicated. e/f, Quantitative thioester-transfer (e) (mean activity ± s.e.m. (n=3); one-way ANOVA followed by Tukey’s post-test, **: P<0.01, ***: P<0.001; Supplementary Fig. 1) and linear polyubiquitination (f) assays of HOIP-RBR WT and interface mutants, and HOIP catalytic domain (RING2L). g. Thioester transfer and linear ubiquitin assays for UbcH5B WT and RING1 interaction mutant F62A (Coomassie-stained bands in red).
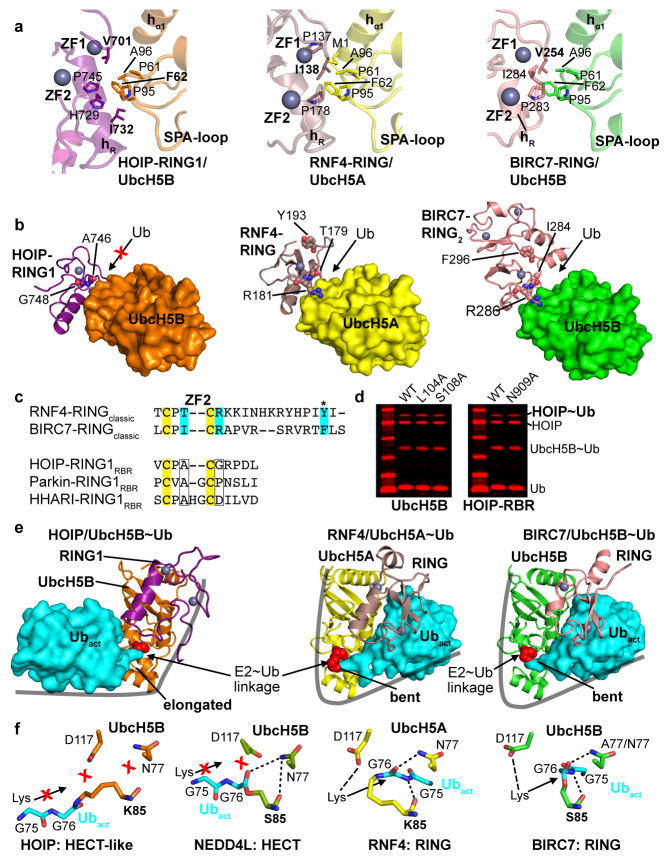
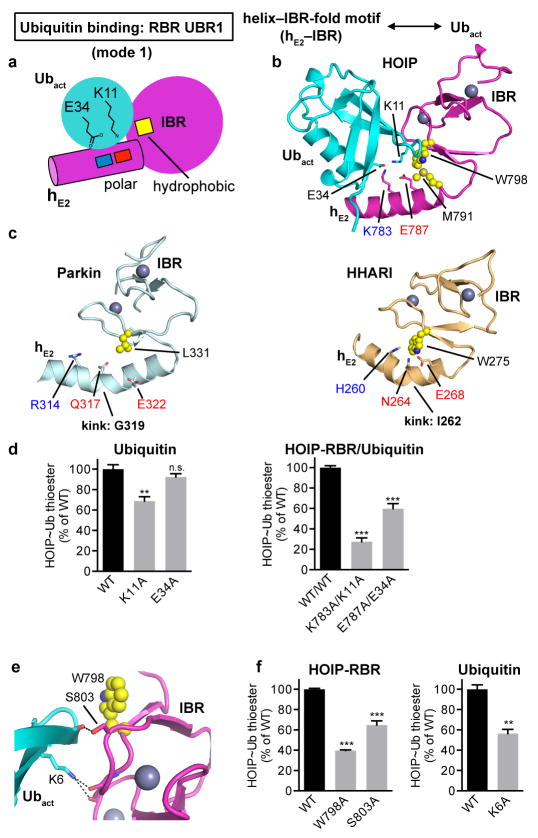
The HOIP-RING1/E2 interaction is tailored towards a HECT-like mechanism, setting it apart from classic RING E3 ligases. While RING/E2 interactions of both classic RING and RBR E3 ligases utilize similar surfaces (Extended Data Fig. 4a)21–26, the position of the HOIP-RING1 domain relative to the E2 is shifted compared to classic RING/E2 complexes (Fig. 2b). Therefore, the RBR-RING1 and the E2 do not form a composite surface to bind the E2-conjugated activated ubiquitin (Ubact, Extended Data Fig. 4b,c,e), which is key to the mechanism of classic RING E3 ligases21,24,27. Instead, two extension helices (hE1, hE2) link the RBR-RING1 to the IBR domain (Figs. 1, 2a)8–11, and helix hE2 with the IBR forms an UBR (UBR1) that engages the activated ubiquitin (Fig. 2c; Extended Data Fig. 5a,b). UBR1 binds ubiquitin in a distinctive mode (mode 1) that utilizes a salt bridge system involving HOIP-hE2 residues K783 and E787 and ubiquitin residues K11 and E34, with further support from the HOIP-IBR (Fig. 2c; Extended Data Fig. 5b). Thus, in HOIP the entire RING1–IBR arm mediates bipartate binding of the E2~ubiquitin conjugate, with RING1 binding E2 and the hE2–IBR module binding activated ubiquitin (Fig. 2a). Sequence and structural comparisons with Parkin and HHARI suggest conservation of this mechanism among RBR E3 ligases (Extended Data Figs. 4c, 5c). Importantly, the bipartite binding mode results in an elongated conformation of the E2~ubiquitin conjugate with its thioester linkage not suited for direct attack by the amine function of a substrate (Fig. 2d; Extended Data Fig. 4e–f). The consequence is an entirely different catalytic arrangement compared to classic RING-supported catalysis, as emphasized by the observed lack of effect of mutations in UbcH5B L104 and S108, two residues crucial for classic RING/E2 catalysis21,25,26 (Extended Data Fig. 4d). Instead, the E2~ubiquitin thioester is ideally positioned for transfer of the activated ubiquitin onto HOIP-RING2L, thus both enabling and requiring a HECT-like mechanism. The significance of each interaction site is demonstrated in thioester transfer and polyubiquitination assays, where mutation of key RING1 and UbcH5B residues drastically impairs RBR activity (Fig. 2e–g). Single mutations in the hE2 salt bridge moderately diminish activity in thioester assays but dramatically impair polyubiquitination activity (Fig. 2e,f), indicating a cumulative effect due to a potential role of UBR1 in coordinating Ubact in steps subsequent to its initial transfer to HOIP. However, removal of the salt-bridge in HOIP/ubiquitin double mutants and mutation of the HOIP-IBR/Ubact interface cause the expected drastic reduction in thioester transfer activity (Extended Data Fig. 5d–f).
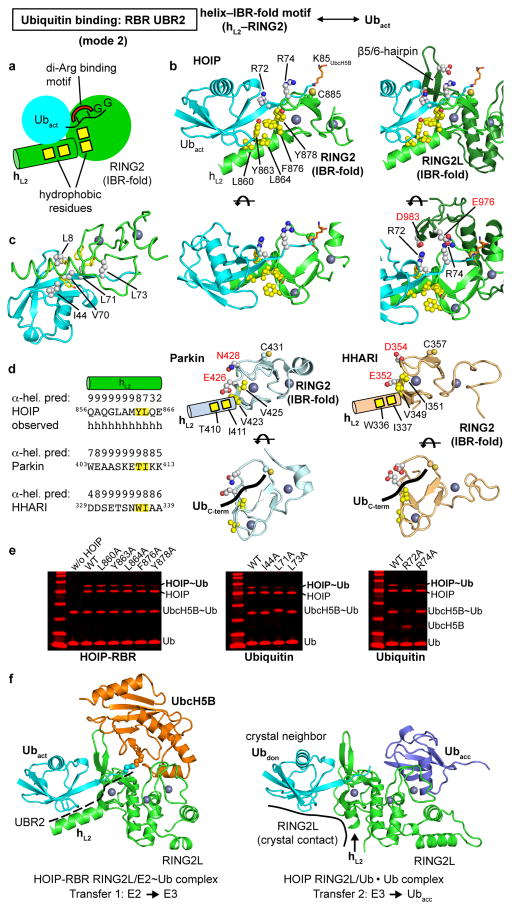
The other portion of the RBR/E2~ubiquitin embrace is centered around the catalytic HOIP-RING2L (Figs.1a; 3a). Here a helix–IBR-fold motif consisting of helix hL2 from the IBR–RING2L linker and RING2L form a second UBR (UBR2) binding the activated ubiquitin (Fig. 3a; Extended Data Fig. 6a–c). UBR2, which is conserved in Parkin and HHARI (Extended Data Fig. 6d), uses a hydrophobic pattern in helix hL2 and RING2 to interact with the canonical and a second hydrophobic patch28 of ubiquitin (Extended Data Fig. 6b–e). These interactions support the engagement of the ubiquitin R72/R74 di-Arg motif by polar residues, which is the key characteristic of UBR binding mode 2. This ultimately places the ubiquitin C-terminus onto RING2 (Extended Data Fig. 6b,d) and thus the ubiquitin/E2-thioester linkage onto the RBR active site. A previous structure14 of the isolated HOIP-RING2L with two ubiquitin molecules bound in a linear non-covalent arrangement mimics the final HOIP-RING2L donor/acceptor ubiquitin transfer complex (Fig. 3b; Extended Data Fig. 6f). Remarkably, in this structure the donor ubiquitin adopts a position identical to the activated ubiquitin in HOIP-UBR2 despite lacking the hL2 interaction (Extended Data Fig. 6f). This indicates that the binding mode of the activated ubiquitin observed in the HOIP/E2~ubiquitin complex persists from the E2~ubiquitin/E3 HOIP transfer complex to the HOIP~ubiquitin/acceptor ubiquitin transfer complex. The E2 portion of the E2~ubiquitin conjugate instead interacts with a region of RING2L that overlaps with that observed for the acceptor ubiquitin in the second transfer reaction (Fig. 3b; Extended Data Fig. 6f). Thus, the binding of E2~ubiquitin and of acceptor ubiquitin/substrate are mutually exclusive, requiring formation of a covalent HECT-like RBR~ubiquitin intermediate in the RBR E3 ligase cycle.
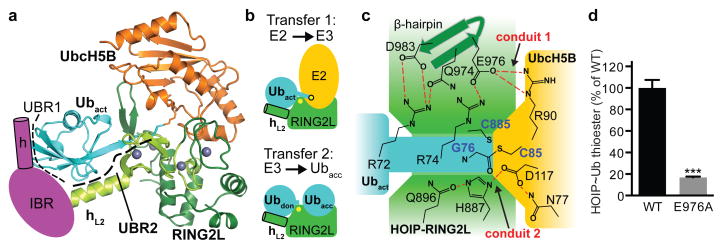
Figure 3. Mechanism of E2~ubiquitin/HOIP-RBR ubiquitin transfer.
a, UbcH5B~ubiquitin conjugate bound to HOIP-RING2L (RING2, light green and LDD extension, dark green). UBR1 (schematic of helix–IBR, magenta) co-operates with UBR2 (comprising hL2 and IBR-fold of RING2L) to bind the activated ubiquitin. UbcH5B interacts with RING2L in a region designated for the acceptor ubiquitin14 (displayed in b). b, Positioning of E2~Ubact or donor(don)/acceptor(acc) ubiquitin onto RING2L. c, Ternary HOIP-RBR/E2~ubiquitin catalytic transfer complex. Two main contact conduits (red dashes) position the RBR catalytic cysteine (C885) near the E2 C85-ubiquitin thioester linkage. d, HOIP~ubiquitin thioester formation assay with WT/E976A RBR, supports the HOIP/UbcH5B/Ub link in conduit 1 (mean activity ± s.e.m. (n=3); two-tailed unpaired Student’s t-test, ***: P<0.001; Supplementary Fig. 1).
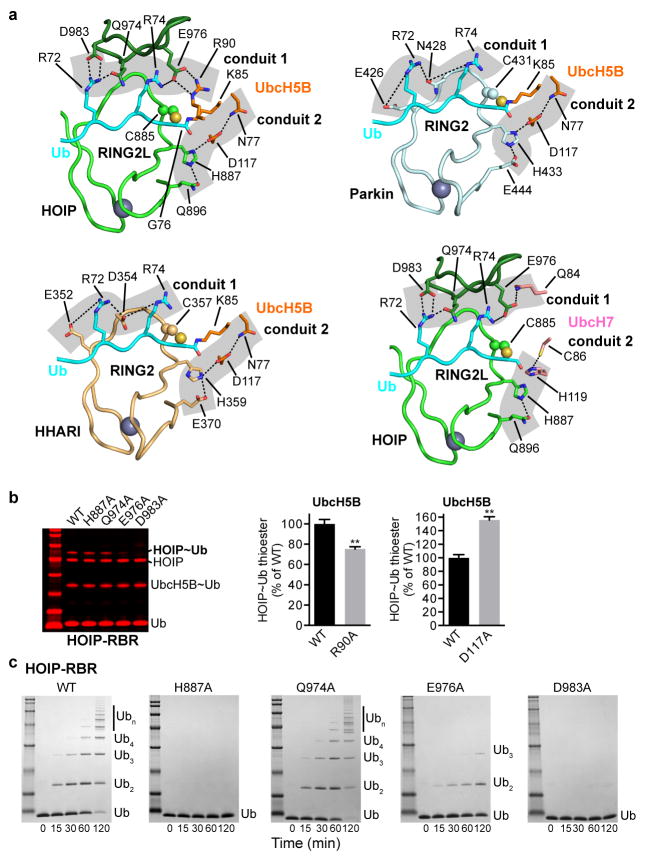
Importantly, the HOIP-RBR/E2~ubiquitin complex structure lacks the spatial gap between the E2 and E3 catalytic centers that is frequently observed in HECT/E2 complex structures and that was also predicted for RBR/E2~ubiquitin transfer complexes8–11,28,29. Thus, except for the ~3.5 Å spacer due to the C85 to lysine substitution in the E2~ubiquitin conjugate, the HOIP-RBR/E2~ubiquitin structure accurately depicts the immediate transfer complex. Here the catalytic centers of HOIP-RING2L and E2 come in close proximity via two contact conduits involving all three proteins (Fig. 3c; Extended Data Fig. 7a). The first conduit consists of ubiquitin R72, which interacts with D983 and Q974 in the β5/6-hairpin of HOIP-RING2L. Additionally, E976 in this hairpin mediates a salt bridge with ubiquitin R74 and UbcH5B R90, thus facilitating interactions among all three proteins.
The second conduit consists of catalytic residues of UbcH5B (N77, D117) and HOIP (H887, Q896)5,6,10,14,21–24,26. These residues appear permissive to close proximity between the reaction centers, yet not crucial for transesterification because, for example, a H887A mutation does not affect the thioester transfer reaction10,14 (Extended Data Fig. 7b). Surprisingly mutation to alanine of UbcH5B D117, a critical residue for classic RING-supported catalysis21,24, enhances transesterification (Extended Data Fig. 7b) further underlining the vastly different catalytic mechanism of RBR E3 ligases. This finding also points to a trade-off in the E2 active site to support both classic RING and HECT-type RBR E3 ligases. Interestingly UbcH7, which is specialized for HECT-like E3 catalysis4, features a histidine instead of D117 (Extended Data Fig. 7a). Mutational analysis demonstrates a crucial role for conduit 1 and also indicates that the close proximity between the ubiquitin thioester (and thus C85 of UbcH5B) and HOIP catalytic cysteine C885 is the driving factor for E2/RBR E3 ubiquitin transfer (Fig. 3c,d; Extended Data Fig. 7). Analysis of Parkin and HHARI shows conservation of the conduits (Extended Data Fig. 7a). However, while Parkin and HHARI lack the β5/6-hairpin indigenous to HOIP-RING2L, they instead possess a pair of conserved polar residues in the RING2 active site loop that are capable of binding the di-Arg motif in conduit 1.
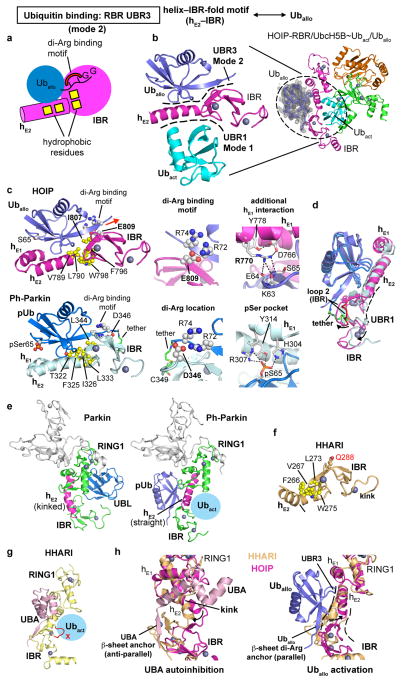
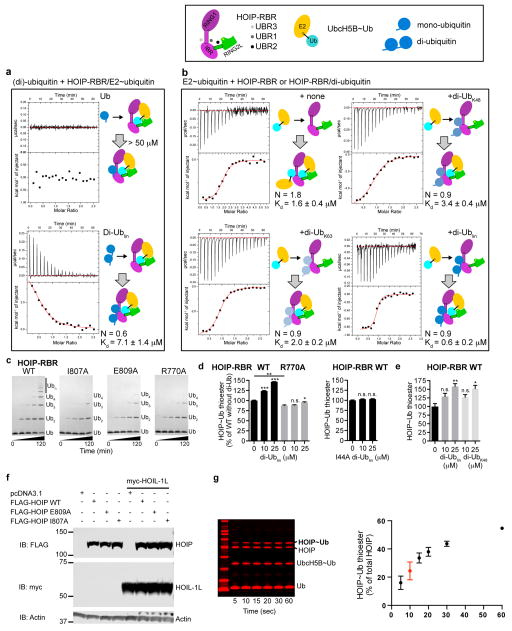
Surprisingly, our structure reveals that an additional allosteric ubiquitin molecule (Uballo) interacts with a third HOIP-UBR. UBR3 is located in the RING1–IBR arm immediately across UBR1 and Ubact (Figs.1b,4a; Extended Data Fig. 8a,b). Uballo uses a binding mode similar to that of Ubact with UBR2 (mode 2), characterized by hydrophobic interactions and a di-arginine binding clamp (Extended Data Fig. 8a). Uballo interacts with helix hE2 of the extended RING1 and with the IBR, and makes additional interactions with helix hE1 (Fig. 4a, Extended Data Fig. 8c). Through this binding, Uballo induces a “straight” conformation of helix hE2 locking RING1 and IBR in their relative position, forming UBR1 to accommodate the activated ubiquitin (Fig. 4a; Extended Data Fig. 8b–d). Notably, UBR3 in the HOIP-RBR/UbcH5B~ubiquitin complex binds linear di-ubiquitin (Kd=7μM) better than mono-ubiquitin (Kd>50 μM) (Extended Data Fig. 9a). Importantly, pre-incubation of HOIP-RBR with linear di-ubiquitin leads to improved binding of UbcH5B~ubiquitin (Extended Data Fig. 9b), emphasizing the allosteric function of UBR3. Accordingly, HOIP-UBR3 I807A and E809A mutants show moderately decreased activity in thioester transfer assays but more pronounced effects in polyubiquitination assays, where linear di-ubiquitin/polyubiquitin are intrinsically produced (Fig. 4b; Extended Data Fig. 9c). Importantly, N- and C-terminally capped linear di-ubiquitin increases HOIP-RBR thioester transfer activity in a dose-dependent manner, but cannot activate HOIP-UBR3 mutants (Fig. 4b; Extended Data Fig. 9d). Moreover, the linear di-ubiquitin I44A mutant also fails to activate HOIP-RBR (Extended Data Fig. 9d).
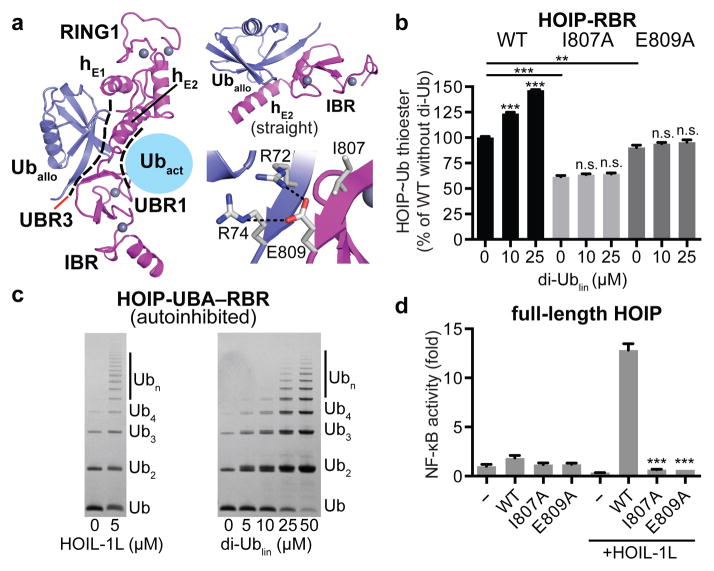
Figure 4. An allosteric ubiquitin interacts with UBR3 in the RING1–IBR arm and is crucial for HOIP activity.
a, An allosteric ubiquitin (Uballo, blue) binds to UBR3 across the activated ubiquitin. Left: Overview depicting UBR3/Uballo. Top right: UBR3 hE2–IBR/Uballo interaction. Bottom right: Close-up on the HOIP ubiquitin di-Arg binding motif (E809) anchoring a parallel ubiquitin/IBR β-sheet. b, Linear di-ubiquitin increases HOIP-RBR activity. Thioester transfer assays of HOIP WT and UBR3 mutants (mean activity ± s.e.m. (n=3); one-way ANOVA followed by Tukey’s post-test, **: P<0.01, ***: P<0.001, n.s.: not significant; Supplementary Fig. 1). c, Polyubiquitination assays showing release of HOIP-UBA–RBR autoinhibition by HOIL-1L or linear di-ubiquitin. d, Effect of HOIP WT or UBR3 mutants in NF-κB reporter assays using HEK293T cells expressing full-length HOIP with/without HOIL-1L (mean activity ± s.e.m. of three biological replicates each with three technical replicates; one-way ANOVA followed by Tukey’s post-test, ***: P<0.001; Extended Data Fig. 9f).
Excitingly, the interaction of Uballo with UBR3 is structurally similar to that recently reported for phospho-ubiquitin in a tethered complex with ΔUBL-Pediculus humanus Parkin (Ph-Parkin)13 (Extended Data Fig. 8c–e). Binding of phospho-ubiquitin leads to a straight conformation of Parkin helix hE2 and an accompanying reorientation of RING1 and IBR, indicating a general role of UBR3 and ubiquitin in allosteric regulation of RBR proteins. Functionally, binding of phospho-ubiquitin activates Parking by counteracting the autoinhibitory function of the Parkin UBL domain (Extended Data Fig. 8c–e)13. In HOIP, the UBA domain exerts intramolecular autoinhibition5,6. While the structure of autoinhibited HOIP is unknown, the structure of autoinhibited HHARI shows binding of its UBA domain to a region analogous to UBR3 (Extended Data Fig. 8f–h)11. To determine if linear di-ubiquitin can overcome HOIP autoinhibition, we examined its effect on HOIP-UBA–RBR. As expected, HOIP-UBA–RBR alone exhibits low E3 activity but is activated by HOIL-1L (Fig. 4c). Strikingly, linear di-ubiquitin can also remove HOIP-UBA autoinhibition and at high concentrations allows the processive formation of polyubiquitin chains by HOIP-UBA–RBR (Fig. 4c). Importantly, in HEK293T cells expressing full-length HOIP, the HOIP-UBR3 I807A and E809A mutants fail to activate NF-κB regardless of HOIL-1L expression, demonstrating an essential physiological role of UBR3 (Fig. 4d). Thus, UBR3 likely serves as a critical sensor of ubiquitin chains that regulates LUBAC function. Whether this role is tailored to linear ubiquitin chains or ubiquitin chains in general (Extended Data Fig. 9b,e) needs further investigation in the context of other LUBAC constituents and binding partners.
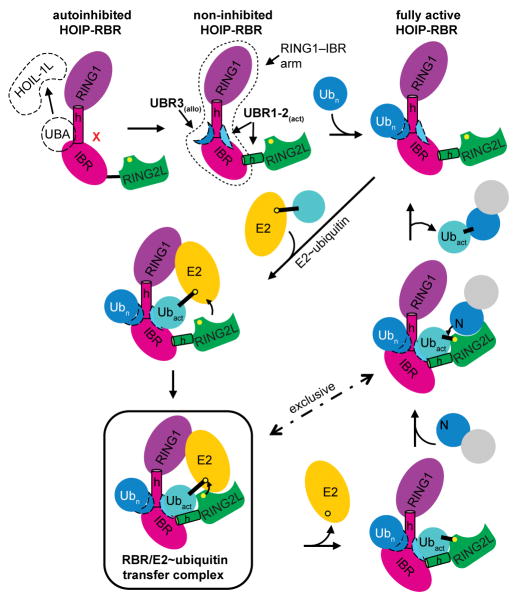
The features revealed by the HOIP-RBR/E2~Ubact/Uballo complex structure provide the missing links in our understanding of these enigmatic multidomain E3 ligases8–11,14 and yield a mechanistic model for the RBR E3 ubiquitin ligase cycle, as summarized in Extended Data Fig. 10. Furthermore, the conservation of key mechanistic features in HOIP, HHARI, Parkin and other RBRs (Supplementary Data 2) underlines the general nature of the catalytic RBR cycle unraveled in this study.
Methods
No statistical methods were used to predetermine sample size.
Constructs
Human HOIP and HOIL-1L cDNA were purchased from Open Biosystems (cloneIDs 4653017 and 3877587, respectively). HOIP-RBR (residues 696–1072), HOIP-RING2L (residues 853–1072) and full-length HOIL-1L were cloned into the pET-NKI-6xHis-3C-LIC vector30 coding for an N-terminal 6x-His-tag with a 3C protease cleavage site. HOIP-UBA-RBR (residues 475–1072) was cloned into a pET-NKI-6xHis-eGFP-3C-LIC vector that codes for a 3C-cleavable His-tagged enhanced green-fluorescent protein (eGFP) followed by the HOIP sequence. Human UbcH5B and Cdc34 DNA were a gift of M. Petroski. Coding sequences for UbcH13 and Uev1a were extracted out of a human cDNA library (Agilent Megaman). For crystallization, UbcH5B (residues 2–147) with the mutations S22R (to prevent backside ubiquitin binding31) and C85K (to enable covalent ubiquitin linkage21) was cloned into the pET-NKI-6xHis-3C-LIC vector. UbcH5B without S22R and C85K mutations (used for enzymatic assays), Cdc34, UbcH13 and Uev1a were cloned into the same vector. Untagged mono-ubiquitin with native N- and C-termini, used for crystallization and linear ubiquitination assays, was cloned into the pET29 vector (Novagen) using NdeI/XhoI restriction sites. N-terminally blocked mono-ubiquitin used for thioester assays was cloned in the pET-NKI-6xHis-3C-LIC vector. Untagged linear di-ubiquitin was cloned with overlap extension PCR and ligated into the pET29 vector (Novagen) using NdeI/XhoI restriction sites. N- and C-terminally blocked di-ubiquitin with a N-terminal His-tag and a C-terminal Ala-Ser sequence was cloned into the pET-NKI-6xHis-3C-LIC vector. Human ubiquitin activating enzyme E1 (Ube1) was cloned into a pET28 vector resulting in an N-terminal His-tag. For NFκB assays full-length HOIP with an N-terminal FLAG-tag and HOIL-1L with an N-terminal myc-tag were cloned into pcDNA3.1(+) (Invitrogen) using EcoRI/NotI restriction sites. Mutations in UbcH5B, ubiquitin and HOIP were introduced using standard site-directed mutagenesis techniques.
Protein expression and purification
All proteins were expressed in BL21(DE3) E. coli after induction with 0.5 mM IPTG overnight at 20°C. For expression of HOIP and HOIL-1L constructs, 0.5 mM ZnCl2 was added to the cultures before induction. Bacteria were harvested by centrifugation, lysed by addition of lysozyme and sonication in the presence of protease inhibitors (PMSF and leupeptin) and DNase. Lysates were cleared by centrifugation and His-tagged proteins were initially purified using Ni-NTA agarose (Qiagen). For HOIP RBR used for crystallization, and UbcH5B, Cdc34, UbcH13, Uev1a, wild-type ubiquitin to generate K48-linked di-ubiquitin and HOIL-1L His tags were removed by addition of 3C protease overnight at 4 °C. HOIP-RBR and HOIL-1L were further purified using Superdex 200 10/300 GL or HiLoad 16/600 Superdex 200 pg size exclusion chromatography columns (GE Healthcare) equilibrated in protein buffer (10 mM HEPES pH 7.9, 100 mM NaCl). UbcH5B used for biochemical assays was further purified on a Superdex 75 10/300 GL size exclusion chromatography column (GE Healthcare) equilibrated in protein buffer. HOIP mutants for activity assays, and Cdc34, UbcH13 and Uev1a were desalted into protein buffer directly after Ni-NTA purification using PD MidiTrap G-25 desalting columns (GE Healthcare). Ube1 for biochemical assays was further purified using ion exchange chromatography (Source Q) in 10 mM HEPES pH 7.9, 10 mM NaCl and eluted with a gradient from 10 – 500 mM NaCl. N-terminally His-tagged (di)-ubiquitin was purified using Ni-NTA as described above followed by size exclusion chromatography using a Superdex 75 10/300 GL column (GE Healthcare) equilibrated in protein buffer or buffer exchange into protein buffer using PD MidiTrap G-25 desalting columns. To purify untagged mono- or di-ubiquitin, 0.5 mM EDTA and 100 mM sodium acetate pH 4.5 were added to the bacterial lysates and lysates were cleared by centrifugation, diluted 7-fold with 50 mM sodium acetate pH 4.5 and applied to a Source S 10/100 ion exchange column (GE Healthcare) equilibrated in 50 mM sodium acetate pH 4.5. Ubiquitin was eluted with a 0 – 500 mM NaCl gradient and further purified by size exclusion chromatography on a Superdex 75 10/300 GL column (GE Healthcare) equilibrated in protein buffer. His-eGFP-HOIP was purified using size exclusion chromatography as described for HOIP-RBR, followed by 3C cleavage and removal of His-eGFP via a second round of size exclusion chromatography. All proteins were generally flash frozen in liquid nitrogen in small aliquots and stored at −80°C.
UbcH5B~ubiquitin linkage
UbcH5B~ubiquitin linkage was performed based on published methods21. Briefly, Ube1, UbcH5B(S22R/C85K) and ubiquitin were mixed and buffer exchanged into 50 mM Tris pH 10, 150 mM NaCl using PD-10 desalting columns (GE Healthcare). 10 mM MgCl2, 5 mM ATP and 1 mM TCEP were added and the protein was incubated at 37°C for 16 h. The completeness of the reaction was monitored using SDS-PAGE and covalently linked UbcH5B~ubiquitin was purified from unreacted proteins and Ube1 using a Superdex 75 10/300 GL size exclusion chromatography column (GE Healthcare) equilibrated in protein buffer.
HOIP-RBR/UbcH5B~ubiquitin complex formation
HOIP-RBR was mixed with a 1.3-fold molar excess of UbcH5B~ubiquitin and applied to a Superdex 200 10/300 GL size exclusion chromatography column equilibrated in protein buffer. Complex formation and purity was confirmed using SDS-PAGE, and complex containing fractions were pooled and concentrated to ~12 mg/ml for crystallization.
HOIP/UbcH5B~ubiquitin/ubiquitin crystallization
Crystallization was performed using the vapour diffusion technique in sitting drop MRC 96-well plates (Molecular Dimensions). Initial crystals were obtained mixing HOIP/UbcH5B~ubiquitin complex solution with an equimolar amount of free ubiquitin in the Morpheus Screen (Molecular Dimensions). Subsequently, 2 μl of the protein complex were mixed with 0.6 μl reservoir solution (0.1 M Morpheus Buffer 3 pH 8.5 (Tris/Bicine), 0.12 M Morpheus Alcohols Mix (0.02 M each of 1,6-Hexanediol; 1-Butanol; 1,2-Propanediol (racemic); 2-Propanol; 1,4-Butanediol; 1,3-Propanediol), 30% Morpheus P550MME_P20K mix (20% PEG550MME, 10% PEG20K) and 8% glycerol) in MRC 48-well plates (Molecular Dimensions). Crystals appeared after about one week at 12°C and were cryo-cooled, and evaluated on a rotating anode X-ray generator (Rigaku FR-E superbright). Seeding and dehydration of the crystals was performed to improve crystal diffraction. For successful dehydration, reservoir was slowly added to the protein drop (3x 0.5 μl within ~2 h) and subsequently equilibrated overnight at 12°C against a reservoir solution with increased P550MME_P20K concentration by adding 11 μl 60% Morpheus P550MME_P20K stock solution to 50 μl reservoir solution. The new reservoir solution was then slowly added to the protein drop (3x 0.5 μl, followed by 2x 1 μl with removal of 1 μl each in the last steps). After further overnight equilibration, crystals were harvested from the drop and directly cryo-cooled in a cryogenic nitrogen stream at 100 K. Crystals diffracted in house to 4–6 Å. Complete diffraction data were measured at 100 K at beamline 23ID-D of the General Medical Sciences and Cancer Institutes Structural Biology Facility at the Advanced Photon Source (GM/CA @ APS), Argonne National Laboratory. Despite their size (common dimensions of ~200 × 140 × 100 μm3) crystals exhibited substantial inhomogeneity resulting in split and smeared diffraction spots. Using raster scans32, a suitable region for data collection could be identified at the edge of the crystal. Utilizing a small (20 μm diameter) beam, split spots could be separated to allow reliable indexing and integration. Utilization of a small beam necessitated higher flux to retain reliable diffraction. To mitigate the radiation damage, the total dose was distributed over a 100-μm stretch of the crystal by using the “helical” mode of “vector” data collection as implemented in JBluIce33. Data were measured at 1.282 Å wavelength with a Pilatus3 6m pixel array detector with a 1 mm-thick sensor (Dectris).
Data processing and structure solution
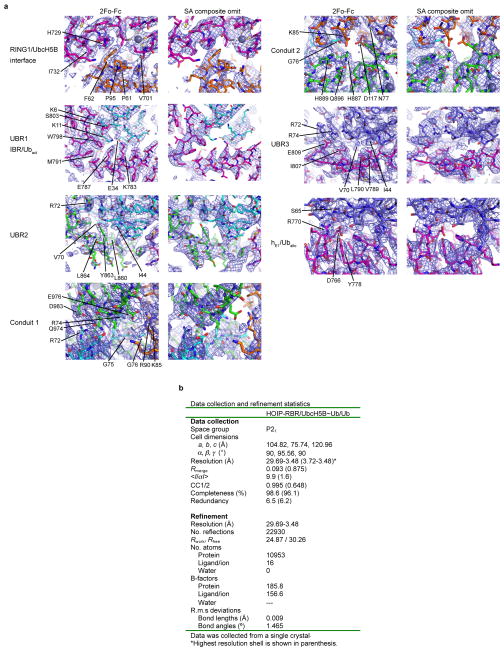
Data were collected from a single crystal and indexed, integrated and scaled in XDS/XSCALE34. Data were further processed using AIMLESS35 from the CCP4 suite36 with a resolution cut-off of 3.48 Å, resulting in an <I/σI> = 1.6 and CC1/2 = 0.648 in the highest resolution shell. Phasing was done in Phaser37 using the MR-SAD protocol as implemented in PHENIX38. For this, independent molecular replacement searches were initially performed for the RING2L domain of HOIP (from PDBid: 4LJP14), UbcH5B (from PDBid: 3A3339), and ubiquitin (from PDBid: 4LJP14) with the four C-terminal residues deleted. Various ambiguous solutions were identified that could not be separated, and Zn2+ sites could not be identified using MR-SAD due to incompleteness of resultant models. However, manual inspection revealed that some MR solutions contained ubiquitin oriented near identically to the symmetry-related donor ubiquitin observed in the HOIP-RING2L/ubiquitin-ubiquitin transfer complex (PDBid: 4LJP14). Based on this observation, a trimmed search model was created that contained a complex of the core of HOIP-RING2L (with residues 906–924 and 949–999 removed) and C-terminally truncated ubiquitin. An MR search using this model found a single solution for two copies of the complex. After successful iterative searches for two UbcH5B molecules and two further ubiquitin molecules, MR-SAD using Phaser identified 15 distinct Zn2+ sites including the known Zn2+ sites in the RING2L domain of HOIP. Further molecular replacement in Phaser using a single unit of the initial HOIP-RING2L/UbcH5B~ubiquitin complex (without the additional second ubiquitin), and the NMR structure of HOIP-IBR (zinc atoms removed, deposited in Protein Data Bank40 under PDBid: 2CT7, unpublished) correctly placed a single HOIP-IBR domain, which was then manually copied to the other NCS-related HOIP in the asymmetric unit. For molecular replacement of the HOIP-RING1, Sculptor41 was used to generate a search model based on the structure of the RING1 domain of HHARI (PDBid: 4KBL11). However, Phaser was not able to correctly place this domain, probably due to the low sequence conservation of only 27% identity. However, since mutational analysis of HOIP suggested that the RING/E2 interaction is preserved between RING-type E3 ligases and RBR-type E3 ligases5, we overlaid the E2 of the published RNF4-RING/UbcH5A~ubiquitin structure (PDBid: 4AP421) with the E2 in our structure and then used this overlay to add the RING1 model generated by Sculptor. This overlay placed the HOIP-RING1 Zn2+-coordinating residues near the last remaining free Zn2+-ions found earlier by Phaser MR-SAD, indicating correct placement of the RING1 domain. In the final round of molecular replacement, the two additional ubiquitin (Uballo) molecules were reinstated at the RING1-IBR interface. At this stage, Refmac42 was used for refinement using settings optimized for low-resolution refinement43 including “jelly body refinement” and Babinet scaling. ProSMART44 was used to generate external restraints against high-resolution structures (PDBid: 4LJO14 for HOIP-RING2L and ubiquitin, and PDBid: 2ESK45 for UbcH5B). After this, clear extra electron density became visible for the unmodelled helical linker regions of the RING1-IBR and IBR-RING2L transitions and for other regions omitted in the initial search models. Further model building and refinement was manually performed in Coot46 and Refmac. During refinement additional clear positive difference map electron density became visible and Phaser was used to place one additional UbcH5B molecule (UbcH5Badd) into this density. TLS restraints were generated using the TLSMD server47 and NCS restraints were used throughout refinement. One overall B-factor was refined in Refmac. In later rounds of refinement the PDB_REDO server48 was used for refinement optimization and MolProbity49 was used for structure validation. Data processing and refinement statistics are summarized in Extended Data Fig. 2b. Ramachandran statistics were calculated using MolProbity and 94.8% of all residues are in favoured regions, 4.9% in allowed regions and 0.3% are outliers. The final structure has a MolProbity score of 1.75 (100th percentile). In the final structure the two HOIP-RBR molecules (see also Extended Data Fig. 3) are defined by electron density from residues 699 to 707, 711 to 948, 969 to 991, and 996 to 1011 (chain A) and 699 to 754, 760 to 957, 967 to 1015, 1019 to 1035 and 1054 to 1066 (chain B). The catalytic UbcH5B~ubiquitin conjugates are defined from UbcH5B residues 3 to 147 and ubiquitin residues 1 to 76 (chains C and E), and UbcH5B residues 2 to 147 and ubiquitin residues 1 to 76 (chains D and F). The allosteric ubiquitin chains (chains G and H) are defined from residues 1 to 76 and the additional UbcH5B (chain I) is defined from residues 2 to 146. Phenix was used to calculate simulated annealing (SA) composite omit maps and feature enhanced maps (FEM). All molecular figures were prepared in PyMOL (Schroedinger, LLC).
K48-linked and K63-linked ubiquitin chain formation
K48-linked and K63-linked ubiquitin chains were formed through a linkage specific enzymatic reaction using Cdc34 and UbcH13/Uev1a E2 ubiquitin-conjugating enzymes, respectively, as described in the literature50. Ubiquitin chains were separated using ion exchange chromatography as described above for purification of mono-ubiquitin. Purified K48-linked di-ubiquitin was directly desalted into protein buffer using PD-10 desalting columns, whereas K63-linked di-ubiquitin was further purified on a Superdex 75 10/300 GL size exclusion chromatography column equilibrated in protein buffer. Native ubiquitin without additional residues was used to generate di-ubiquitin chains for ITC experiments, whereas N-terminally blocked ubiquitin was used to form K48-linked di-ubiquitin for testing allosteric activation of HOIP RBR.
Linear polyubiquitination assay
Linear ubiquitin formation assays were performed in 50mM HEPES pH7.9, 100 mM NaCl, 10 mM MgCl2 and 0.6 mM DTT using 200 nM E1, 1 μM UbcH5B, 1 μM HOIP-RBR or HOIP-RING2L and 40 μM untagged ubiquitin. Reactions were started by addition of 10 mM ATP and were incubated at 30°C for 2 h. Samples were taken at the indicated time points and treated with 50 mM sodium acetate pH4.5 as described previously6, mixed with SDS sample buffer and analysed by SDS-PAGE using 12% Bolt Bis-Tris gels (Life technologies). Proteins were visualized with Coomassie Brilliant blue dye. To test the activating effect of linear di-ubiquitin on autoinhibited HOIP-UBA-RBR, 5μM HOIP-UBA-RBR was pre-incubated with N- and C-terminally blocked linear di-ubiquitin or HOIL-1L at the indicated concentrations before addition of the remaining assay components. Samples were taken after 60 min and subsequently treated as described above.
UbcH5B~ubiquitin to HOIP-RBR ubiquitin transfer assay
To monitor HOIP~ubiquitin thioester ubiquitin transfer from UbcH5B to HOIP, Ube1 (100 nM), UbcH5B (4 μM) and N-terminally blocked ubiquitin (32 μM) were mixed in 50 mM HEPES pH 7.9, 100 mM NaCl, 10 mM MgCl2 and 5 mM ATP and incubated at 25°C for 5 min when 2 μM HOIP-RBR was added. Samples were taken 10 s after HOIP addition, quenched by addition of pre-heated SDS protein loading buffer without DTT, and run on a 12% SDS-PAGE gel (Life Technologies). The 10-second time point used was empirically determined with a time-course experiment (Extended Data Fig. 9g). Gels were stained with Coomassie Brilliant blue dye and scanned on a Li-COR Odyssey scanner using the 700 nm (red) channel. For the thioester transfer assay shown in Fig. 3d, 200 nM Ube1, 2 μM UbcH5B, 8 μM HOIP-RBR, 8 μM N-terminally blocked ubiquitin and 10 mM ATP were used and samples taken after 30 sec. Proteins were transferred to a PVDF membrane and ubiquitin was visualized on a LI-COR Odyssey scanner at 800 nm using an anti-ubiquitin antibody (P4D1, Santa Cruz, 1:200 dilution in TBST [50 mM Tris pH 7.4, 150 mM NaCl, 0.05% Tween-20]) followed by an IRDye 800CW secondary antibody (LI-COR, 1:10,000 dilution in TBST). All quantitative experiments shown in graphs were performed in triplicates and band intensities were quantified using the ImageStudio software (LI-COR). HOIP thioester transfer activity was calculated as the fraction of HOIP~Ub to total HOIP for each mutant and normalized against thioester transfer activity of WT HOIP. Data were analysed in GraphPad Prism using two-tailed unpaired Student’s t-test or one-way ANOVA followed by Tukey’s post-test.
Allosteric activation of HOIP-RBR by di-ubiquitin
To test the allosteric activation of HOIP-RBR by linear di-ubiquitin, a modified ubiquitin transfer assay was performed. HOIP-RBR was pre-incubated with N- and C-terminally blocked linear di-ubiquitin at the indicated final concentrations for 5 min at 25°C. At the same time, Ube1, UbcH5B, ubiquitin and ATP were premixed and incubated for 5 min at 25°C, resulting in fully loaded UbcH5B~Ub. Both mixtures were subsequently mixed together, resulting in final concentrations of 100 nM Ube1, 2 μM UbcH5B, 8 μM N-terminally blocked ubiquitin and 2 μM HOIP-RBR in the final buffer described for the standard ubiquitin transfer assay. Samples were taken after 30 s and further treated as described for the standard transfer assay. A 30 sec time point was determined to give the best results in this assay, in which lower E2 and mono-ubiquitin concentrations were used resulting in an overall slower reaction rate. The experiments comparing the effects of linear versus K48-linked di-ubiquitin (Extended Data Fig. 9e) were performed similarly, with the difference that all samples were incubated with apyrase (Sigma) for 5 min to deplete ATP before addition of HOIP/di-ubiquitin and prevent E2-loading of K48-linked di-ubiquitin, which features a free C-terminus on one of the ubiquitin units.
Analytical ultracentrifugation (AUC)
Sedimentation equilibrium experiments were performed in a ProteomeLab XL-I (Beckman Coulter) analytical ultracentrifuge. HOIP-RBR/UbcH5B~ubiquitin as used for crystallization was loaded into a 6-channel equilibrium cell at 5.0, 2.5 and 1.25 μM concentration and centrifuged at 10,000 rpm, 20°C in an An-50 Ti 8-place rotor until equilibrium was achieved. Data were analysed using HeteroAnalysis software (J.L. Cole and J.W. Lary, University of Connecticut; http://www.biotech.uconn.edu/auf/).
Isothermal titration calorimetry (ITC)
ITC experiments were performed on an ITC200 calorimeter (Microcal). Aliquots (2 μl each) of 500–650 μM UbcH5B~ubiquitin or di-ubiquitin solution were injected into the cell containing 40–50 μM HOIP-RBR or HOIP-RBR/di-ubiquitin complexes. The experiments were performed at 23°C in buffer containing 10 mM HEPES pH 7.9, 100 mM NaCl. For titrations of UbcH5B~ubiquitin into HOIP-RBR/di-ubiquitin complexes, HOIP-RBR was pre-incubated with an equimolar amount of di-ubiquitin before the ITC experiments. Data were analysed using the Origin software (Microcal).
NF-κB luciferase assay
Human embryonic kidney (HEK) 293T cells (ATCC) were co-transfected with NF-κB-luc reporter plasmid that contains an NF-κB response element upstream of the promoter driving the luciferase reporter gene, pGL4.74[hRluc/TK] control vector (Promega) and epitope tagged FLAG-HOIP or myc-HOIL-1L pcDNA3.1(+) plasmids in 6-well plates in triplicates using Lipofectamine 2000 transfection reagent. Since this assay could be carried out in a variety of cellular contexts, HEK293T cells were used because they are easy to transfect and suitable for the assay. The cells tested negative for mycoplasma contamination. Empty pcDNA3.1(+) vector was used as control. After 36 h, cells were lysed and 20 μl cell lysates were used to measure firefly luciferase and Renilla luciferase (transfection control) signals using the dual luciferase reporter assay system according to the manufacturer’s protocol (Promega). Data were analysed in GraphPad Prism and one-way ANOVA followed by Tukey post-tests were used for statistical analysis. Immunoblotting was performed with anti-FLAG (clone M2, Sigma-Aldrich) and anti-myc (clone 9E10, Sigma-Aldrich) antibodies, to confirm equivalent WT and mutant protein expression levels.
Extended Data
Extended Data Figure 1. HOIP domain organization and nomenclature.
a, Domain organization of HOIP as commonly outlined in the literature. HOIP consists of a PNGase/ubiquitin-associated (PUB) domain followed by a B-box zinc-finger (B-box) domain51, NPL4 zinc-fingers (NZF), the autoinhibitory UBA domain and the RING-between-RING module (RBR, grey background). The HOIP-RBR module contains the typical RING1, in-between RING (IBR) and RING2 domains, and a HOIP-specific additional linear ubiquitin chain determining domain (LDD). A yellow circle indicates the RBR catalytic cysteine (C885) forming the HECT-like thioester intermediate with ubiquitin. The binding sites of the other LUBAC constituents HOIL-1L and SHARPIN are also indicated. b, The RBR-RING2 domain has the topology of an IBR domain. The individual HOIP RING1, IBR and RING2 domains from the HOIP-RBR/E2~Ub/Ub structure are shown to enable direct comparison of their folds. This illustrates that the zinc finger domain designated RING2 in fact adopts the topology of an IBR, as multiple groups have reported for various RBR E3 ligases previously8–11,14,52,53. The terms RBR and RING2 however are used in this study for consistency with the widely accepted nomenclature. c, The HOIP-RING2–LDD region. HOIP features an extension of its catalytic RING2 domain termed LDD, which adds two zinc fingers and a helical arrangement to the RING2. The LDD is usually denoted as a domain following RING2. Rittinger and colleagues14 however showed that the LDD is intertwined with the HOIP-RING2 to form a single extended domain that contains a central canonical RBR-RING2 with the additional features of the LDD ensuring the linear ubiquitin chain formation characteristic of HOIP. This domain will thus be designated RING2L (for RING2–LDD). The RING2L from the current HOIP structure is displayed with RING2 in light green and LDD in dark green. d, The structural arrangement of active HOIP-RBR in the HOIP/E2~ubiquitin complex is markedly different from that of autoinhibited RBRs. Left: Active HOIP-RBR from the HOIP/E2~ubiquitin complex. The RING1–IBR region and the RING2L are colored magenta and green respectively. The individual RBR domains are also highlighted: RING1, yellow circle; IBR, orange circle; RING2 red circle. The RING1 extension helices (hE1, hE2) and IBR–RING2 linker helices (hL1 and hL2) are labeled. Middle and right: Analogous representations of autoinhibited Parkin and HHARI (PDBid: 4I1H8 and PDBid 4KBL11). Additional domains and regions besides the RBR of Parkin and HHARI are colored grey.
Extended Data Figure 2. Quality of crystallographic data and electron density maps.
a, Final 2Fo-Fc (left) and simulated annealing (SA) composite omit (right) electron density maps of select interfaces of the HOIP/UbcH5B~Ub/Ub complex contoured at 1σ. Proteins are shown in sticks and colored according to Fig. 1. b, Data collection and refinement statistics.
Extended Data Figure 3. Complexity of the crystallographic asymmetric unit and structure of the HOIP/UbcH5B~ubiquitin/ubiquitin E2-E3 transfer complex.
a, The asymmetric unit contains two transfer complexes. Left: Colour schematic of the proteins present in the asymmetric unit. Middle: Structure of the asymmetric unit. The asymmetric unit contains two HOIP-RBR/UbcH5B~ubiquitin complex arrangements (complex 1, 2). The two UbcH5B~ubiquitin conjugates are coloured orange~cyan and are bound to two HOIP-RBR molecules (magenta and green), which cross over between the complexes. Additional allosteric ubiquitin (Uballo, blue) and UbcH5B~ubiquitin (UbcH5B~Uballo, yellow~blue) molecules are bound to the HOIP-RBRs in complex 1 and complex 2, respectively. Since Uballo makes all contacts with the RBRs and the UbcH5B of the UbcH5B~Uballo conjugate solely mediates crystal contacts (bottom left), only Uballo of complex 1 is displayed and discussed in the text and figures in terms of the additional ubiquitin binding. The black oval indicates an additional HOIP-hL1/Ubact inter-complex interaction discussed in panels e/f. Right: Close-up of the region where the two RBRs of HOIP cross over between the complexes. The close-up shows that residues D852 and P853 of the respective RBRs come in 6Å proximity suggesting a continuity in the biological complex in which the two residues from the respective RBRs are linked (as indicated by the grey background), resulting in the monomeric complex schematically illustrated underneath and discussed in panels c – f. b, The RING1–IBR and RING2L form two distinct entities to bind E2~ubiquitin. The monomeric complex as displayed for complex 1 assumes a flexible linkage between the autonomous units of the RING1–IBR arm and the RING2L from the two different RBR molecules in the asymmetric unit. This linkage is formed by residues D852 and P853 connecting IBR and RING2L (schematically illustrated in the cartoon). To test the structural integrity of the assumed link and the autonomy of the RING1–IBR arm on one side and the RING2L on the other side, we introduced a spacer comprising five alanine residues between D852 and P853 in HOIP-RBR (see cartoon) and measured the activity of the RBR D852-Ala5-P853 insertion mutant in polyubiquitination assays. The assays show that the mutant (right) retains an activity similar to the wild-type RBR (left) indicating that indeed RING1-IBR and RING2L act as autonomous units. The dramatically reduced activity of HOIP-RING2L alone (residues P853 to end) is also shown for reference (middle). c, The HOIP-RBR/UbcH5B~ubiquitin complex is monomeric at concentrations of 1.25 – 5 μM. To determine if the HOIP-RBR/UbcH5B~ubiquitin complex is indeed monomeric in solution, we analyzed the isolated HOIP-RBR/UbcH5B~ubiquitin complex protein material that was used for crystallization by sedimentation equilibrium analytical ultracentrifugation (SE-AUC). SE-AUC provides an absolute, shape-independent measurement of molecular weight, thus allowing accurate determination of the oligomeric state. The three SE-AUC experiments performed on the HOIP-RBR/UbcH5B~ubiquitin complex yielded an absolute molecular weight (MW) of 71,658 Da, indicating a monomeric complex. At an order of magnitude higher concentrations (12.5 – 50 μM), SE-AUC results indicate the formation of a dimer with MW ~144kDa, although curve fitting residuals also show significant presence of aggregates (data not shown). These results indicate that the biological complex in solution is monomeric at physiological low μM concentrations such as those used for the thioester transfer assays and polyubiquitination assays. However, the dimeric arrangement observed in the crystal structure might be relevant in a high concentration setting such as within the LUBAC complex. Here, a high local concentration of HOIP-RBR could favor binding of the E2~ubiquitin between the RING1–IBR and RING2L of two neighboring molecules. Importantly, all mechanisms depicted in this article hold true for both the monomeric and dimeric states (as illustrated in panel d). This means that the deduced mechanism is in principle applicable to different RBR E3 ligases of which some might function as dimers in local high concentration assemblies (such as within the LUBAC), whereas others might be active in a monomeric setting. d, Schematic illustration of the dimeric arrangement as observed in the asymmetric unit. The schematic shows that all features deduced (Figs. 1 – 4 and Extended Data Figs. 4 – 10) are also valid for the dimeric case (binding of Uballo is omitted for clarity). e, Asymmetric unit dimer-related interactions between HOIP-hL1 and Ubact. The dimeric arrangement contains no additional protein-protein interfaces compared to the monomeric assemblies with the exception of hL1 residues W847, M850 and N851, which in the asymmetric unit contact the activated ubiquitin of the other complex (indicated by an oval in panel a). f, Mutational analysis of HOIP-hL1/Ubact interactions. Mutations of HOIP-hL1 residues that interact with Ubact have no effect on thioester transfer activity (Coomassie-stained bands in red), indicating that this “trans” complex interaction is not critical for the RBR mechanism, in line with the model of a monomeric arrangement.
Extended Data Figure 4. The HOIP-RING1–IBR applies an altered binding mode compared to classic RING E3s necessitating a HECT-like mechanism.
a, The HOIP RBR-RING1 uses an E2 interaction pattern similar to classic RINGs, which however results in a shifted binding. Shown are the details of the RING/E2 interaction in the HOIP-RING1RBR/UbcH5B~ubiquitin complex (left), the RNF4-RINGclassic/UbcH5A~ubiquitin complex (middle; PDBid: 4AP421), and the BIRC7-RINGclassic/UbcH5B~ubiquitin complex (right; PDBid: 4AUQ24). The HOIP-RBR-type RING1 uses a pattern of hydrophobic residues as the core of the interaction with E2 that is similar to that in classic RING E3 ligases. Subtle differences however support a shifted binding mode (see also Fig. 2b). The main features of the RING and E2 as well as HOIP residues mutated in Fig. 2e – g are displayed in bold. Zinc finger (ZF) 1 and 2 of the RING domains and the SPA-loop of the E2 containing a conserved Ser-Pro-Ala motif are annotated. For the following panels the same structures and color codes as in panel a are used. b, The shift in binding and altered surface residues in HOIP-RING1 do not support the composite RING/E2 binding site for activated ubiquitin utilized by classic RING/E2 complexes. UbcH5A/B E2s are rendered as surface representation and the RING domains in ribbon representation. Residues crucial for classic RING E3s to recruit the activated ubiquitin in the composite RING/E2 ubiquitin-binding surface21,24 are depicted (middle, right). In HOIP-RING1 (left) equivalent residues are not conserved (displayed), indicating that HOIP cannot accommodate the activated ubiquitin in its RING1/UbcH5B complex. For illustration purposes, only one monomer of the dimeric RNF4 is shown (although Y193 from the other RING molecule is still displayed)21; for BIRC7 the RING dimer (RING2) is displayed24. c, Alignment of HOIP, Parkin and HHARI RING1 domains with classic RING domains centered around residues displayed in panel b. Residues crucial for ubiquitin binding in classic RINGs are highlighted in cyan and their structural equivalents in the RING1 domains of the RBR E3 ligases HOIP, Parkin and HHARI are indicated by boxes, attesting to the absence of a composite RING1/E2 ubiquitin binding site in RBR ligases (* indicates ubiquitin interaction residues from the other RING molecule in the dimers formed by the classic RINGs RNF4 and BIRC7). The T/I-C-R sequence observed in the dimeric RINGs of RNF4 and BIRC7 represents the highly conserved Φ-x-R/K motif, where Φ is a hydrophobic residue and x is either a Cys in RING E3 ligases or a polar residue in U-box ligases26. This motif is not only critical for E3-mediated catalysis by dimeric RING ligases (such as RNF4 or BIRC7) but is also necessary for E2-mediated catalysis by simpler monomeric RING and U-Box E3 ligases26. The fact that this motif is not conserved in HOIP, HHARI and Parkin further confirms the mechanistic differences between RBRs and classic RING domains. d. Thioester transfer assays show that E2 residues critical for classic RING-supported catalysis are not important for HOIP catalysis. Left: HOIP-RBR thioester transfer assays show similar activity of UbcH5B wild-type and L104A and S108A mutants. This is in stark contrast to the reported effects of these mutations on classic RING-supported catalysis21,24, underlying the fundamentally different mechanism of the HECT-like catalysis by HOIP-RBR. Right: Mutation to Ala of HOIP-RBR N909, which would be in the vicinity of the activated ubiquitin if the E2~ubiquitin conjugate were bound in a bent manner (see panel e), also shows no effect on HOIP thioester formation (Coomassie-stained bands in red). e, The altered E2~ubiquitin binding mode of RBR-RING1 results in the requirement for a HECT-like mechanism. Displayed are the entire RING/UbcH5~ubiquitin complexes with RINGs and E2s depicted in ribbon representation and the activated ubiquitin in surface representation. The bipartite binding mode used by the HOIP-RING1–hE2–IBR arm (see also Fig. 2a) results in an elongated E2~ubiquitin conformation (left, only the RING1 domain of HOIP is depicted) while formation of a composite RING/E2 binding surface in the case of classic RING E3 ligases (middle, right) results in binding of the activated ubiquitin in a compact manner with a bent E2~ubiquitin conformation. Importantly, this bent conformation places the thioester-link in a specific position relative to the catalytic machinery of the E2, allowing direct attack by the lysine/amine function of a substrate or growing ubiquitin chain. The Lys85/Ser85 residues mediating the E2~ubiquitin linkage and mimicking UbcH5A/B catalytic cysteine C85 are displayed as red spheres. In the elongated E2~ubiquitin conformation propagated by the HOIP-RBR, this attack is not possible. The linkage is however ideally positioned for the attack by the RBR catalytic cysteine in a HECT–like mechanism (see also Fig. 3 and Extended Data Fig. 7a). f, Close-up of the catalytic centers in E2~ubiquitin linkages. Details of the catalytic centers resulting from the E2~ubiquitin conjugate conformations outlined in panel e and, for comparison, the HECT-type E3 NEDD4L/UbcH5B~Ub structure (PDBid: 3JW028), with the directionality of an attacking amine indicated as previously proposed21. In the HECT-like RBR arrangement, the UbcH5B~ubiquitin linkage is not aligned correctly relative to the E2 catalytic machinery for a direct attack by an amine function. This is similar in the HECT-type arrangement in the NEDD4L complex but completely different from the arrangement in classic RING-supported E2 catalysis. Additionally, the ubiquitin C-terminal residues G75-G76 reside in a position that would overlap with the attacking amine. The available structure of the BIRC7/UbcH5B~Ub complex (PDBid: 4AUQ24) features an UbcH5B N77A mutant and the remainder of the Asn side-chain has been manually added based on wild-type UbcH5B from PDBid: 2ESK45 (right).
Extended Data Figure 5. Helix–IBR-fold motifs constitute new ubiquitin-binding regions (UBR) in active RBR proteins: Binding of the activated ubiquitin by UBR1 using binding mode 1.
a, Schematic illustrating binding mode 1 utilized by hE2–IBR to bind the activated ubiquitin in the RING1–IBR arm. The general principle of this binding mode is that the RING1 extension helix 2 (hE2) preceding the IBR presents a pattern of charged/polar residues (indicated by blue and red squares, which symbolize K, R, H and E, Q, N residues respectively) that interact with ubiquitin E34 and K11. These interactions are supported by the IBR surface, with a particular contribution of hydrophobic residues (yellow square) flanking the salt-bridge system. b, Coordination of the activated ubiquitin by HOIP-hE2–IBR in mode 1. HOIP-hE2 residues K783 and E787 bind ubiquitin residues E34 and K11 and are flanked by hydrophobic residues M791 and W798 from HOIP-IBR. c, Structurally equivalent residues in Parkin and HHARI. Displayed are the hE2–IBR modules from autoinhibited Parkin (PDBid: 5C1Z12) and HHARI (PDBid: 4KBL11) with residues equivalent to HOIP residues in panel b depicted, illustrating the general conservation of UBR1. It should be noted that these structures feature autoinhibited forms of the RBR proteins, which exhibit a kink in hE2 of UBR1. This kink would sterically hinder ubiquitin binding to UBR1 and likely participates in the RBR autoinhibition mechanism (see also Extended Data Fig. 8). d, Thioester-transfer assays of ubiquitin and UBR1 salt bridge mutants. In agreement with the observed four-residue salt bridge system in panel b, the single K11A or E34A ubiquitin mutations show only a slight to moderate effect since the remaining charged residue can still coordinate the two oppositely charged residues of HOIP. In contrast, elimination of both similarly charged residues in the complex by combining the HOIP K783A and ubiquitin K11A or HOIP E787A and ubiquitin E34A mutations results in a more dramatic loss of activity (mean activity ± s.e.m (n=3), one-way ANOVA followed by Tukey’s post-test, **P<0.01, ***: P<0.001, n.s.: not significant; representative gels shown in Supplementary Fig. 1). e, f, Role of the IBR in UBR1. Close-up of the additional IBR/Ubact interactions in stick representation (e) shows that HOIP S803 and ubiquitin K6 coordinate the backbone carbonyl functions of ubiquitin T12 and HOIP A800/K829 respectively. W798, which is involved in hydrophobic interactions, is also displayed in sphere representation. Quantitative thioester transfer assays (f) show that alanine mutants of residues outlined in panel e cause a significant loss of activity (mean activity ± s.e.m (n=3), left: one-way ANOVA followed by Tukey’s post-test, right: two-tailed unpaired Student’s t-test, **P<0.01, ***: P<0.001; representative gels shown in Supplementary Fig. 1).
Extended Data Figure 6. Binding of the activated ubiquitin by UBR2 using binding mode 2 and exclusive binding of E2 and acceptor ubiquitin.
a, Schematic illustrating binding mode 2 utilized by a helix–IBR-fold motif (hL2–RING2) to bind the activated ubiquitin and position the thioester linkage for the transfer reaction. The second helix (hL2) of the linker between the IBR domain and the catalytic RING2 domain uses a pattern of two or three hydrophobic residues (yellow squares) to interact with the ubiquitin canonical hydrophobic patch surrounding I4428 (not shown). Hydrophobic residues of the RING2 IBR-fold complete the hydrophobic interaction network by coordinating residues L71 and L73 in the second hydrophobic patch28 of ubiquitin (not shown). The central hallmark of this binding mode is the coordination of the characteristic di-Arg (R72, R74) motif in the ubiquitin C-terminus, resulting in a firm placement of the C-terminus on ZF1 of RING2. b, Structure of the interaction of the helix–IBR-fold in HOIP-hL2–RING2 (UBR2) with the activated ubiquitin. Left: hL2 residues L860, Y863 and L864 (yellow spheres) interact with ubiquitin residues L8, I44 and V70 (not shown). Additionally, hydrophobic residues F876 and Y878 (yellow spheres) from the IBR-fold of the minimal catalytic RING2 (light green, see also Extended Data Fig. 1c) coordinate ubiquitin residues L8, L71 and L73 (not shown). Right: Display of the full HOIP-RING2 including the LDD insertion (RING2L). The coordination of the ubiquitin di-Arg motif is achieved by HOIP residues D983 and E976 from the LDD insertion that is part of the catalytic HOIP-RING2L (dark green). This results in the placement of the E2~Ub thioester linkage (K85 replacing UbcH5B C85 is shown as orange sticks) in the vicinity of the catalytic HOIP C885. Bottom: alternatively oriented views of the interaction. c, The two hydrophobic patches of ubiquitin engaged by UBR2. The hydrophobic residues of HOIP-RING2 interacting with ubiquitin as highlighted in panel b are shown as yellow sticks. The interaction residues on the canonical hydrophobic patch of ubiquitin (L8, I44, V70) and the second hydrophobic patch (L71, L73)28 are displayed as grey spheres. d, The RING2 domains of Parkin and HHARI also contain a helix–IBR-fold (hL2–RING2) module with patterns of residues consistent with the formation of a UBR2. Left: Helical predictions for the region preceding the RING2 domains of HOIP, Parkin and HHARI. The structures of Parkin and HHARI in their autoinhibited forms do not display a helix equivalent to hL2 because this region is either not defined (in the crystal structures of HHARI and most Parkin structures) or adopts an extended conformation (in two other Parkin structures)8–11. However, a helical prediction reliability score (with 1 lowest to 9 highest score) calculated using JPred454 shows a strong helical probability for the segment of Parkin and HHARI preceding the RING2 domain. In fact, the score is similar to that of HOIP, which is displayed with the observed helical secondary structure, pointing to the presence of an equivalent of hL2 in active forms of Parkin and HHARI. These RBR E3 ligases also contain residues capable of interacting with the hydrophobic patch in ubiquitin in positions equivalent to HOIP Y863 and L864 (highlighted in yellow). Right: Structures of the RING2 domains of PARKIN (PDBid: 4I1H8) and HHARI (PDBid: 4KBL11) showing hydrophobic residues (yellow sphere representation) in structurally equivalent positions to HOIP F876 and Y878 and residues (labeled red) capable of interacting with the di-Arg motif in their catalytic RING2. Helix hL2 with the conserved hydrophobic residues not present in the crystal structures as discussed above is indicated schematically. Bottom: Different orientations with the putative placement of the ubiquitin C-terminus indicated schematically. e, The effect of UBR2 alanine mutations in thioester transfer assays increases with their proximity to the di-Arg motif. Left: Mutation of HOIP-UBR2 hydrophobic residues to alanine. The hL2 L860A and L864A mutations show little effect on activity, while the Y863A mutation and particularly the RING2L F876 and Y878 mutations, which reside proximal to the di-Arg binding motif formed by D983 and E976 (see also Extended Data Figure 7) show a marked reduction in activity. Middle, Right: Mutation of complementary ubiquitin residues involved in UBR2 binding. Similarly to the HOIP mutations, the ubiquitin I44A, L71A and L73A mutations show increasing effects with a closer location to the di-Arg motif. Furthermore, the ubiquitin R74A mutation shows a strong effect on activity, emphasizing the importance of its interaction with HOIP and of the resulting placement of the ubiquitin C-terminus linked to the E2. The ubiquitin R72A mutant failed to form an UbcH5B~ubiquitin conjugate, thus preventing analysis. Coomassie-stained bands are in red. f, Overlap of the UbcH5B binding site on HOIP-RING2L with the binding site of the acceptor ubiquitin. Left: UbcH5B~ubiquitinact (orange/cyan) interaction with RING2L (green) from the HOIP-RBR/E2~ubiquitin complex. Right: RING2L interaction with two ubiquitin molecules arranged in linear fashion, mimicking the HOIP-RING2L~ubiquitindonor to ubiquitinacceptor (Ubdon,Ubacc) transfer complex (PDBid: 4LJP14). Despite the fact that the placement of the donor ubiquitin in the 4LJP structure results from a crystal contact, this ubiquitin exhibits a position identical to that of the activated ubiquitin bound to UBR2 in the HOIP-RBR/UbcH5B~Ub complex. It should be noted that the UBR2 interaction with hL2 is missing because the RING2L from the crystal neighbor presenting the donor ubiquitin pushes hL2 into a different conformation. Importantly, in the HOIP-RBR/UbcH5B~ubiquitin complex (left) the E2 binds RING2L in a region that overlaps with the binding site for the acceptor ubiquitin (Ubacc, dark blue) in the RING2L~ubiquitindonor to ubiquitinacceptor transfer complex (right). This highlights how the E2~ubiquitin conjugate and the acceptor ubiquitin (which is the substrate of the E3 reaction) cannot bind the RBR at the same time, thus making a HECT-like transfer a requirement in the E3 ligase mechanism of RBR proteins.
Extended Data Figure 7. Catalytic center of the E2~ubiquitin/HOIP-RBR E3 transfer complex.
a, Close-up view of the catalytic center of the transfer complex shows conservation of the contact conduits. Top left: Close-up view of the catalytic center in the HOIP/UbcH5B~ubiquitin transfer complex. Contact conduits 1 and 2 are highlighted by grey background. HOIP catalytic cysteine C885 is depicted in sphere representation. K85 replacing the catalytic cysteine (C85) in UbcH5B and ubiquitin G76 are displayed in stick representation, featuring the UbcH5B~ubiquitin linkage. Top right: Model of the conduits in a Parkin/UbcH5B~ubiquitin complex. The structure of Parkin-RING2 (from autoinhibited Parkin, PDBid: 4I1H8) was overlaid on that of HOIP-RING2 indicating equivalent contact conduits. Bottom left: Analogous model for HHARI using RING2 from autoinhibited HHARI (PDBid: 4KBL11). Bottom Right: Model of the conduits in a HOIP/UbcH7~ubiquitin complex. The model was generated from PDB entry 4Q5E55 with UbcH7 (with the free catalytic cysteine C86 displayed) overlaid on UbcH5B of the HOIP/UbcH5B~ubiquitin transfer complex. The structure of the HOIP/UbcH5B~ubiquitin transfer complex and the other models depicted indicate a conservation of the contact conduits. Mechanistically, the conduits allow for the RBR catalytic cysteine and the E2 catalytic cysteine~ubiquitin linkage to be in close proximity, which serves as main driving force of the transesterification reaction. A reaction driven mainly by proximity is also in agreement with the chemical nature of the catalytic cysteine, which has a pKa of ~8 for the free amino acid. This allows the cysteine to naturally deprotonate, without an absolute need for HOIP H88710,14, prior to attack of the ubiquitin G76 carbonyl function. In addition, the thioester linkage is far more labile than for example an amide bond, thus further facilitating a proximity-mediated reaction56. However, in light of the geometric arrangement observed, additional subtle catalytic contributions of H887 in supporting the transition state of the reaction and/or re-protonation of the E2 catalytic cysteine are in principle possible. This prospect is particularly intriguing because UbcH7 exhibits a potential break in conduit 2 (between H887 and H119), yet provides its “own” histidine (H119) to the catalytic center. b, Thioester transfer assays for HOIP contact conduit mutants. Left: Thioester transfer assays show that the D983A and E976A mutations strongly affect activity. This is consistent with D983 forming the di-Arg binding motif in UBR2 (see Extended Data Fig. 6) and E976 bridging the three proteins in the complex. In contrast, the H887A mutation does not have a marked effect, in agreement with published results10,14. The Q974A mutation also does not have a strong effect, pointing to a weak auxiliary function of this residue in support of the critical D983 (Coomassie-stained bands in red). Right: The UbcH5B R90A mutation shows a moderate yet significant effect, in line with the structure, which suggests a more pronounced effect for HOIP E976A than for UbcH5B R90A. Surprisingly, mutation of the catalytic D117 in UbcH5B, which is essential for classic RING-supported catalysis, shows a positive effect on the HECT-like thioester transfer, further emphasizing a separate mechanism for RBR HECT-like catalysis (as outlined in Fig. 2 and Extended Data Fig. 4). The gain of function of the D117A mutation also points to a trade-off for this E2 residue to participate in the classic RING-supported versus RBR HECT-like E2/E3 mechanisms (mean activity ± s.e.m (n=3), two-tailed unpaired Student’s t-test, **P<0.01; representative gels shown in Supplementary Fig. 1) c. Polyubiquitination assays for HOIP contact conduit alanine mutants. These assays show similar activity profiles as the thioester transfer assays except for the H887A mutation, which is essential for amide bond formation in the second transfer reaction10,14.
Extended Data Figure 8. The hE2–IBR module contains an additional UBR (UBR3) that binds an allosteric ubiquitin using binding mode 2.
a, UBR3 binds the allosteric ubiquitin (Uballo) using binding mode 2. Cartoon depicting the overall features of binding mode 2 in UBR3. The binding of Uballo is largely analogous to the binding of the activated ubiquitin by UBR2 in the helix–IBR-fold of hL2–RING2L (Extended Data Fig. 6a). Yellow squares indicate hydrophobic patches. b, Location of the UBR3/Uballo interface in the overall complex and its relation to the UBR1/Ubact interface. The additional ubiquitin (Uballo) binds to UBR3 in the hE2–IBR module immediately across UBR1 and the activated ubiquitin. The electron density map (feature-enhanced map57, grey, contour level σ=1) for the additional ubiquitin is also depicted on the right. c, Details of the UBR3/Uballo interaction and its similarity to the interaction of phospho-ubiquitin (pUb) with Pediculus humanus Parkin (Ph-Parkin). Top left: Close-up of HOIP-UBR3. Uballo binds to the hE2–IBR module with additional contacts to hE1. Depicted are hydrophobic residues of HOIP (V789, L790, F796, W798 and I807) interacting with the ubiquitin canonical hydrophobic patch (L8, I44 and V70/not shown) and a second hydrophobic patch (L71 and L73/not shown), with the critical HOIP I807 emphasized. The di-Arg binding motif is also depicted, with E809 coordinating ubiquitin R72 and R74 and aligning the ubiquitin C-terminus in a parallel manner with sheet β2 of the IBR. A red arrow indicates that UBR3 sterically allows the binding of di-ubiquitin/polyubiquitin chains on the C-terminal side of the bound ubiquitin (see also Extended Data Fig. 9). Ubiquitin Ser65 is indicated for comparison with the pUb/Ph-Parkin structure (bottom). Top middle: Close-up on the di-Arg binding motif. Top right: Close-up on the additional contacts between hE1 and Uballo. HOIP R770 interacts with D766, which makes contacts to Y778, and the backbone carbonyl functions of ubiquitin K63 and E64. Bottom: The recent structure of phospho-ubiquitin bound to Ph-Parkin (PDBid: 5CAW13) reveals a similar mechanism. Left: Close-up with the residues corresponding to those in HOIP depicted. Middle: Close-up of the di-Arg motif. Ph-Parkin D346 coordinates R72 similar to HOIP-UBR3/Uballo. The chemical tether introduced in the pUb/Ph-Parkin structure between the phospho-ubiquitin C-terminus and a non-conserved Cys in Ph-Parkin (C349) shifts ubiquitin R74 away from Ph-Parkin D346 indicating that the di-Arg binding motifs of HOIP and Parkin undergo a similar interaction with ubiquitin or phospho-ubiquitin respectively. Right: The Ph-Parkin interaction equivalent to the HOIP hE1 interaction involves a ubiquitin phospho-serine 65 (pSer65) binding pocket in Ph-Parkin. Phospho-serine 65 is directly coordinated by R307 and Y314 and also H304, which is positioned similarly to HOIP D766. d. The binding of phosphoUb propagates the formation of UBR1 in Parkin. Overlay of Uballo (slate) bound to HOIP (magenta) and phosphoUb (light blue) bound to Ph-Parkin (grey blue). Ubiquitin binding propagates a straight conformation of hE2 and an opening of the IBR relative to the extended RING1 (only hE1 is shown). The tether introduced in the phosphoUb/Ph-Parkin interaction appears to exert a strain on IBR loop 2 and thus the IBR. This suggests that in the absence of the artificial tether phosphoUb can propagate the formation of a fully functional UBR1 (binding Ubact) in Parkin concomitantly to relieving the UBL autoinhibition, as elegantly demonstrated by Komander and colleagues13. e, The interaction of phospho-ubiquitin with a site analogous to UBR3 in Ph-Parkin causes a straight conformation of hE2 and reorientation of the IBR relative to RING1 as prerequisite to accommodate the activated ubiquitin. Left: Full-length autoinhibited Parkin (PBDid: 5C1Z12). Right: Tethered phospho-ubiquitin in complex with ΔUBL-Ph-Parkin (PDBid: 5CAW13). f, Conservation of UBR3 in HHARI. HHARI features a helix–IBR module similar to that of HOIP, with conserved hydrophobic patches and a polar residue (Q288) in the position of HOIP E809 that is in principle capable of binding the ubiquitin di-Arg motif. The hE2–IBR region from autoinhibited HHARI (PDBid: 4KBL11) is displayed. As for autoinhibited Parkin, a key difference from active HOIP is the kink in helix hE2 of autoinhibited HHARI (see also panel g and Extended Data Fig. 5c). Taken together, these features indicate an overall similarity and the existence of a UBR3 that is allosterically linked to UBR1 in HOIP, Parkin and HHARI and potentially also other RBR proteins (see alignment in Supplementary Data 2). g, HHARI-UBA domain (pink) binds HHARI-RBR (yellow) in an equivalent position as Uballo binding to HOIP, but promotes an inhibitory conformation of RING1–IBR that cannot bind the activated ubiquitin (PDBid: 4KBL11). h, UBR3 is a regulatory hotspot and UBR3/UBR1 crosstalk. Parkin autoinhibition is facilitated by an UBL domain and is inherently linked to the equivalent of UBR3 and counteraction by phospho-ubiquitin binding. Different to Parkin, no structure of autoinhibited HOIP-RBR is available and thus the conformation of autoinhibited HOIP is not known. However HHARI is, like HOIP, autoinhibited by its UBA domain and the autoinhibited structure of HHARI has been solved previously (PDBid: 4KBL11). This structure reveals that unlike the UBL of Parkin, the UBA of HHARI directly utilizes the region of UBR3 for binding and autoinhibition, which includes the kink in hE2 and a relative RING1–IBR positioning that is incompatible with binding of Ubact as observed in this study, further underlying a regulatory “hotspot” function of the RBR UBR3/UBR1. Left: the UBA domain (pink) of HHARI utilizes an anti-parallel β-sheet anchor with strand β2 of the HHARI-IBR (light orange) positioning the UBA to induce a kink in helix hE2 compared to its conformation in active HOIP (magenta) and counteracting the formation of a productive UBR3 and UBR1. Right: Binding of the Uballo to UBR3 in mode 2 utilizes a parallel β-sheet anchor (centered around the di-Arg binding interaction) with strand β2 of the IBR in active HOIP (magenta) inducing a straight conformation of hE2 and a conformation of UBR1 suited to bind the activated ubiquitin of the E2~Ubact conjugate. Putative shifts for an analogous UBR1 formed in HHARI are indicated. Of note, the structure of autoinhibited HOIP is not known and therefore the placement of the UBA in the schematic illustration of Extended Data Fig. 10 is deduced based on the similar autoinhibition of HOIP and HHARI by their UBA domains, which still needs to be demonstrated.
Extended Data Figure 9. UBR3 interacts with di-ubiquitin and allosterically promotes E2~ubiquitin binding and HOIP-RBR activation.
a, ITC experiments analyzing the binding of mono-ubiquitin or linear di-ubiquitin to an isolated HOIP-RBR/E2~ubiquitin complex. While the binding of mono-ubiquitin is below the sensitivity of the experimental setting (Kd > 50μM), the binding of linear di-ubiquitin exhibits a Kd of 7.1 μM. b, ITC experiments analyzing the binding of UbcH5B~ubiquitin to HOIP-RBR in the absence and presence of different di-ubiquitin chains. Top left: The binding stoichiometry of UbcH5B~ubiquitin and WT HOIP-RBR is n = 1.8, indicating that two UbcH5B~ubiquitin molecules interact with one RBR through UBR1/2 (catalytic binding) and UBR3 (binding of the ubiquitin moiety of the E2~ubiquitin conjugate) with a combined overall Kd of 1.6 μM. The graph shows a single titration step, indicating “crosstalk” between UBR3 and UBR1. Top right: The presence of K48-linked di-ubiquitin leads to 1:1 binding (n = 0.9) of UbcH5B~ubiquitin to HOIP-RBR, indicating that di-ubiquitin occupies UBR3 and limits UbcH5B~ubiquitin binding to only its bona fide catalytic binding site (with ubiquitin binding to UBR1/2 and UbcH5B binding to RING1/RING2L). However, the presence of K48-linked di-ubiquitin results in the lowest affinity (Kd = 3.4 μM) for the binding of the conjugate to the RBR, indicating a negative effect of this linkage compared to the other di-ubiquitin entities tested. Bottom left: K63-linked di-ubiquitin has a more favorable effect on UbcH5B~ubiquitin binding (Kd = 2.0 μM), Bottom right: The strongest allosteric effect is observed in the presence of linear di-ubiquitin, which enables sub-micromolar binding (Kd = 600 nM) of UbcH5B~ubiquitin. These results show that linear di-ubiquitin functions as a potent activator of HOIP-RBR by binding to UBR3 (see also below and Fig. 4). While the structure depicts the interactions of one ubiquitin unit with UBR3, a second ubiquitin C-terminal to the UBR3-interacting ubiquitin may undergo further interactions with the IBR (as indicated by the arrow in Extended Data Fig. 8c). The cartoon representations summarize the configuration of each ITC experiment. c, Polyubiquitination assays of UBR3 mutants. The HOIP-RBR I807A, E809A and R770A mutants exhibit a marked reduction in activity, supporting the importance of UBR3 in HOIP function. d, Activation of HOIP-RBR by di-ubiquitin. While wild-type HOIP-RBR is activated by the presence of increasing concentrations of WT linear di-ubiquitin, WT linear di-ubiquitin only has a weak effect on activation of the UBR3 R770A mutant (similar to the I807A and E809A mutants in Fig. 4b). Additionally the di-ubiquitin mutant I44A, which is mutated at a critical UBR3-interacting residue in both ubiquitin units, does not have an activating effect on WT HOIP-RBR thioester activity (mean activity ± s.e.m (n=3), one-way ANOVA followed by Tukey’s post-test, *: P<0.05, **P<0.01, ***: P<0.001, n.s.: not significant; representative gels shown in Supplementary Fig. 1). e, Effect of linear versus K48-linked di-ubiquitin on HOIP-RBR thioester transfer activity. In contrast to the ITC binding studies in panel b, linear and K48-linked di-ubiquitin are both able to increase the thioester transfer activity of HOIP-RBR (although the experimental setup necessary to investigate the K48-linkage resulted in larger error; see Methods; mean activity ± s.e.m (n=3), one-way ANOVA followed by Tukey’s post-test, *: P<0.05, **P<0.01, n.s.: not significant; representative gels shown in Supplementary Fig. 1). These results show an UBR3-dependent activating effect of di-ubiquitin, and thus potentially of polyubiquitin chains. However, whether HOIP-UBR3 acts as a universal ubiquitin sensor or has a preference for linear ubiquitin over other types of linkage needs to be further examined through careful investigations also including full-length proteins of the LUBAC in cellular settings. Additionally, although there is a substantial gap between UBR3 and the position of the acceptor ubiquitin, longer acceptor ubiquitin chains might be able to bridge this gap and mediate a cooperative effect between the two sites. This would be consistent with a recent publication showing that the presence of K63-linked ubiquitin chains is frequently necessary for the formation of linear polyubiquitin chains58. f, Protein expression levels for the NF-κB reporter assays in cells shown in Fig. 4d. Shown are anti-FLAG immunoblots of HOIP WT and mutants and anti-myc immunoblots of HOIL-1L, demonstrating similar protein expression levels in different cell lysates. Lysates were also probed by immunoblotting for actin as a loading control. Uncropped blots are shown in Supplementary Fig. 1. g, Time-course of HOIP~ubiquitin thioester transfer assay. (Left) SDS-PAGE showing time-course of HOIP~ubiquitin thioester transfer assay. Coomassie-stained bands in red visualized using LI-COR Odyssey at 700 nm. (Right) Plot of quantified HOIP~ubiquitin thioester transfer assay time-course (mean ± s.e.m., n = 2). The 10-second time point used in the end-point assays throughout the study is highlighted in red.
Extended Data Figure 10. Schematic of RBR mechanism: HOIP-RBR activation and E3 ligase cycle.
HOIP-RBR is initially autoinhibited by its UBA domain. Sequestration of the autoinhibitory HOIP-UBA domain by HOIL-1L5–7,18 releases the conformational restraint exerted by the UBA, allowing formation of UBR1 and UBR3. Binding of a ubiquitin entity such as a linear ubiquitin chain to UBR3 stabilizes the active conformation of UBR1 and the RING1–IBR arm, facilitating binding of the E2~ubiquitin conjugate. In the subsequent HOIP/E2~ubiquitin transfer complex, the E2~ubiquitin conjugate is engaged in a clamp-like manner bringing the RBR active cysteine and the E2~ubiquitin thioester in close proximity, ultimately leading to the transfer of the ubiquitin to the RBR cysteine. The E2 then vacates the complex, freeing the site for binding of Ubact, whose N-terminal amine attacks the RBR thioester7,14. Once the ubiquitin chain linkage is formed, the ubiquitinated substrate/growing ubiquitin chain must exit RING2L to enable binding of a new E2~ubiquitin conjugate for the next loading of the RBR in the HECT-like E3 ligase cycle. The growing ubiquitin chain could be retained near the RBR by the HOIP NZF domains, HOIL-1L or SHARPIN15–17,59, directly linking the HECT-like mechanism to co-operative processes within the LUBAC.
Supplementary Material
Acknowledgments
The authors thank A. Bobkov (SBP Protein Analysis Facility) for performing ITC and AUC experiments, M. Petroski (SBP) for providing UbcH5B and Cdc34 constructs, J. Badger (DeltaG technologies) for assistance in model evaluation and E. Pasquale (SBP) for help with manuscript writing. This work was supported by NIH grant R01AA017238 and institutional funding (S.J.R.), an EMBO Long-term Postdoctoral Fellowship (B.C.L.), a Rutherford Discovery Fellowship from the New Zealand government administered by the Royal Society of New Zealand (P.D.M.) and NCI Cancer Center Support Grant P30CA030199 (SBP Protein Analysis Core Facility). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. GM/CA@APS has been funded in whole or in part with Federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006).
Footnotes
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
Supplementary Information is available in the online version of the paper.
Author Contributions B.C.L. designed and carried out all experiments (except for the cell-based experiments), including crystallization, structure solution and refinement, and wrote the manuscript. M.K.D. expressed proteins and performed initial purification. P.D.M. participated in early stages of the study, structure solution and writing of the manuscript. R.S. collected and processed diffraction data. A.R. performed the HEK293T cell experiments under the supervision of C.F.W. S.J.R. oversaw and actively participated in all steps of the study and wrote the manuscript.
Coordinates and structure factors have been deposited in the Protein Data Bank under accession code 5EDV.
The authors declare no competing financial interests.
References
- 1.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 2.Metzger MB, Pruneda JN, Klevit RE, Weissman AM. RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim Biophys Acta. 2014;1843:47–60. doi: 10.1016/j.bbamcr.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berndsen CE, Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol. 2014;21:301–307. doi: 10.1038/nsmb.2780. [DOI] [PubMed] [Google Scholar]
- 4.Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474:105–108. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smit JJ, Monteferrario D, Noordermeer SM, van Dijk WJ, van der Reijden BA, Sixma TK. The E3 ligase HOIP specifies linear ubiquitin chain assembly through its RING-IBR-RING domain and the unique LDD extension. EMBO J. 2012;31:3833–3844. doi: 10.1038/emboj.2012.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stieglitz B, Morris-Davies AC, Koliopoulos MG, Christodoulou E, Rittinger K. LUBAC synthesizes linear ubiquitin chains via a thioester intermediate. EMBO Rep. 2012;13:840–846. doi: 10.1038/embor.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 2006;25:4877–4887. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riley BE, Lougheed JC, Callaway K, Velasquez M, Brecht E, Nguyen L, Shaler T, Walker D, Yang Y, Regnstrom K, Diep L, Zhang Z, Chiou S, Bova M, Artis DR, Yao N, Baker J, Yednock T, Johnston JA. Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat Commun. 2013;4:1982. doi: 10.1038/ncomms2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trempe JF, Sauve V, Grenier K, Seirafi M, Tang MY, Menade M, Al-Abdul-Wahid S, Krett J, Wong K, Kozlov G, Nagar B, Fon EA, Gehring K. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science. 2013;340:1451–1455. doi: 10.1126/science.1237908. [DOI] [PubMed] [Google Scholar]
- 10.Wauer T, Komander D. Structure of the human Parkin ligase domain in an autoinhibited state. EMBO J. 2013;32:2099–2112. doi: 10.1038/emboj.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duda DM, Olszewski JL, Schuermann JP, Kurinov I, Miller DJ, Nourse A, Alpi AF, Schulman BA. Structure of HHARI, a RING-IBR-RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism. Structure. 2013;21:1030–1041. doi: 10.1016/j.str.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar A, Aguirre JD, Condos TE, Martinez-Torres RJ, Chaugule VK, Toth R, Sundaramoorthy R, Mercier P, Knebel A, Spratt DE, Barber KR, Shaw GS, Walden H. Disruption of the autoinhibited state primes the E3 ligase parkin for activation and catalysis. EMBO J. 2015 doi: 10.15252/embj.201592337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wauer T, Simicek M, Schubert A, Komander D. Mechanism of phospho-ubiquitin-induced PARKIN activation. Nature. 2015;524:370–374. doi: 10.1038/nature14879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stieglitz B, Rana RR, Koliopoulos MG, Morris-Davies AC, Schaeffer V, Christodoulou E, Howell S, Brown NR, Dikic I, Rittinger K. Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP. Nature. 2013;503:422–426. doi: 10.1038/nature12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerlach B, Cordier SM, Schmukle AC, Emmerich CH, Rieser E, Haas TL, Webb AI, Rickard JA, Anderton H, Wong WW, Nachbur U, Gangoda L, Warnken U, Purcell AW, Silke J, Walczak H. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471:591–596. doi: 10.1038/nature09816. [DOI] [PubMed] [Google Scholar]
- 16.Ikeda F, Deribe YL, Skanland SS, Stieglitz B, Grabbe C, Franz-Wachtel M, van Wijk SJ, Goswami P, Nagy V, Terzic J, Tokunaga F, Androulidaki A, Nakagawa T, Pasparakis M, Iwai K, Sundberg JP, Schaefer L, Rittinger K, Macek B, Dikic I. SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature. 2011;471:637–641. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tokunaga F, Nakagawa T, Nakahara M, Saeki Y, Taniguchi M, Sakata S, Tanaka K, Nakano H, Iwai K. SHARPIN is a component of the NF-kappaB-activating linear ubiquitin chain assembly complex. Nature. 2011;471:633–636. doi: 10.1038/nature09815. [DOI] [PubMed] [Google Scholar]
- 18.Yagi H, Ishimoto K, Hiromoto T, Fujita H, Mizushima T, Uekusa Y, Yagi-Utsumi M, Kurimoto E, Noda M, Uchiyama S, Tokunaga F, Iwai K, Kato K. A non-canonical UBA-UBL interaction forms the linear-ubiquitin-chain assembly complex. EMBO Rep. 2012;13:462–468. doi: 10.1038/embor.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sasaki K, Iwai K. Roles of linear ubiquitinylation, a crucial regulator of NF-kappaB and cell death, in the immune system. Immunol Rev. 2015;266:175–189. doi: 10.1111/imr.12308. [DOI] [PubMed] [Google Scholar]
- 20.Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, Nakagawa T, Kato M, Murata S, Yamaoka S, Yamamoto M, Akira S, Takao T, Tanaka K, Iwai K. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol. 2009;11:123–132. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- 21.Plechanovova A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mace PD, Linke K, Feltham R, Schumacher FR, Smith CA, Vaux DL, Silke J, Day CL. Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment. J Biol Chem. 2008;283:31633–31640. doi: 10.1074/jbc.M804753200. [DOI] [PubMed] [Google Scholar]
- 23.Plechanovova A, Jaffray EG, McMahon SA, Johnson KA, Navratilova I, Naismith JH, Hay RT. Mechanism of ubiquitylation by dimeric RING ligase RNF4. Nat Struct Mol Biol. 2011;18:1052–1059. doi: 10.1038/nsmb.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat Struct Mol Biol. 2012;19:876–883. doi: 10.1038/nsmb.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 26.Pruneda JN, Littlefield PJ, Soss SE, Nordquist KA, Chazin WJ, Brzovic PS, Klevit RE. Structure of an E3:E2~Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol Cell. 2012;47:933–942. doi: 10.1016/j.molcel.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Branigan E, Plechanovova A, Jaffray EG, Naismith JH, Hay RT. Structural basis for the RING-catalyzed synthesis of K63-linked ubiquitin chains. Nat Struct Mol Biol. 2015 doi: 10.1038/nsmb.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamadurai HB, Souphron J, Scott DC, Duda DM, Miller DJ, Stringer D, Piper RC, Schulman BA. Insights into ubiquitin transfer cascades from a structure of a UbcH5B~ubiquitin-HECT(NEDD4L) complex. Mol Cell. 2009;36:1095–1102. doi: 10.1016/j.molcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang L, Kinnucan E, Wang G, Beaudenon S, Howley PM, Huibregtse JM, Pavletich NP. Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade. Science. 1999;286:1321–1326. doi: 10.1126/science.286.5443.1321. [DOI] [PubMed] [Google Scholar]
- 30.Luna-Vargas MP, Christodoulou E, Alfieri A, van Dijk WJ, Stadnik M, Hibbert RG, Sahtoe DD, Clerici M, Marco VD, Littler D, Celie PH, Sixma TK, Perrakis A. Enabling high-throughput ligation-independent cloning and protein expression for the family of ubiquitin specific proteases. J Struct Biol. 2011;175:113–119. doi: 10.1016/j.jsb.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 31.Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell. 2006;21:873–880. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 32.Hilgart MC, Sanishvili R, Ogata CM, Becker M, Venugopalan N, Stepanov S, Makarov O, Smith JL, Fischetti RF. Automated sample-scanning methods for radiation damage mitigation and diffraction-based centering of macromolecular crystals. Journal of synchrotron radiation. 2011;18:717–722. doi: 10.1107/S0909049511029918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stepanov S, Makarov O, Hilgart M, Pothineni SB, Urakhchin A, Devarapalli S, Yoder D, Becker M, Ogata C, Sanishvili R, Venugopalan N, Smith JL, Fischetti RF. JBluIce-EPICS control system for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2011;67:176–188. doi: 10.1107/S0907444910053916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakata E, Satoh T, Yamamoto S, Yamaguchi Y, Yagi-Utsumi M, Kurimoto E, Tanaka K, Wakatsuki S, Kato K. Crystal structure of UbcH5b~ubiquitin intermediate: insight into the formation of the self-assembled E2~Ub conjugates. Structure. 2010;18:138–147. doi: 10.1016/j.str.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 40.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bunkoczi G, Read RJ. Improvement of molecular-replacement models with Sculptor. Acta Crystallogr D Biol Crystallogr. 2011;67:303–312. doi: 10.1107/S0907444910051218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nicholls RA, Long F, Murshudov GN. Low-resolution refinement tools in REFMAC5. Acta Crystallogr D Biol Crystallogr. 2012;68:404–417. doi: 10.1107/S090744491105606X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nicholls RA, Fischer M, McNicholas S, Murshudov GN. Conformation-independent structural comparison of macromolecules with ProSMART. Acta Crystallogr D Biol Crystallogr. 2014;70:2487–2499. doi: 10.1107/S1399004714016241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ozkan E, Yu H, Deisenhofer J. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc Natl Acad Sci U S A. 2005;102:18890–18895. doi: 10.1073/pnas.0509418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 48.Joosten RP, Long F, Murshudov GN, Perrakis A. The PDB_REDO server for macromolecular structure model optimization. IUCrJ. 2014;1:213–220. doi: 10.1107/S2052252514009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dong KC, Helgason E, Yu C, Phu L, Arnott DP, Bosanac I, Compaan DM, Huang OW, Fedorova AV, Kirkpatrick DS, Hymowitz SG, Dueber EC. Preparation of distinct ubiquitin chain reagents of high purity and yield. Structure. 2011;19:1053–1063. doi: 10.1016/j.str.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 51.Elliott PR, Nielsen SV, Marco-Casanova P, Fiil BK, Keusekotten K, Mailand N, Freund SM, Gyrd-Hansen M, Komander D. Molecular basis and regulation of OTULIN-LUBAC interaction. Mol Cell. 2014;54:335–348. doi: 10.1016/j.molcel.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spratt DE, Martinez-Torres RJ, Noh YJ, Mercier P, Manczyk N, Barber KR, Aguirre JD, Burchell L, Purkiss A, Walden H, Shaw GS. A molecular explanation for the recessive nature of parkin-linked Parkinson’s disease. Nat Commun. 2013;4:1983. doi: 10.1038/ncomms2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spratt DE, Mercier P, Shaw GS. Structure of the HHARI catalytic domain shows glimpses of a HECT E3 ligase. PLoS ONE. 2013;8:e74047. doi: 10.1371/journal.pone.0074047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Drozdetskiy A, Cole C, Procter J, Barton GJ. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grishin AM, Condos TE, Barber KR, Campbell-Valois FX, Parsot C, Shaw GS, Cygler M. Structural basis for the inhibition of host protein ubiquitination by Shigella effector kinase OspG. Structure. 2014;22:878–888. doi: 10.1016/j.str.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 56.Lehninger AL, Nelson DL, Cox MM. Principles of Biochemistry. 2. Worth Publishers; 1993. [Google Scholar]
- 57.Afonine PV, Moriarty NW, Mustyakimov M, Sobolev OV, Terwilliger TC, Turk D, Urzhumtsev A, Adams PD. FEM: feature-enhanced map. Acta Crystallogr D Biol Crystallogr. 2015;71:646–666. doi: 10.1107/S1399004714028132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emmerich CH, Ordureau A, Strickson S, Arthur JS, Pedrioli PG, Komander D, Cohen P. Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc Natl Acad Sci U S A. 2013;110:15247–15252. doi: 10.1073/pnas.1314715110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sato Y, Fujita H, Yoshikawa A, Yamashita M, Yamagata A, Kaiser SE, Iwai K, Fukai S. Specific recognition of linear ubiquitin chains by the Npl4 zinc finger (NZF) domain of the HOIL-1L subunit of the linear ubiquitin chain assembly complex. Proc Natl Acad Sci U S A. 2011;108:20520–20525. doi: 10.1073/pnas.1109088108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.