Abstract
The Stegomyia aegypti mosquito (=Aedes aegypti; Diptera: Culicidae) is the primary vector of viruses that cause Yellow fever, Dengue and Chikungunya fever. In the absence of effective vaccines, reduction of these diseases relies on vector control strategies. The success of these strategies is tightly linked to the population dynamics of the target populations. In the present study, fourteen collections from St. aegypti populations separated by periods of 1 to 13 years were analysed to determine their temporal genetic stability. Although temporal structure is discernible in most populations, the degree of temporal differentiation is dependent on the population and does not obscure the geographic structure between populations. The results suggest that performing detailed studies in the years prior to and after population reduction or modification-based control interventions at each target field site may be useful in assessing the probability of success.
Keywords: Aedes (Stegomyia) aegypti, Yellow fever, Dengue, Chikungunya, temporal dynamics, population stability
Stegomyia aegypti, the yellow fever mosquito, is the primary vector of dengue virus and also transmits Chikungunya virus, both agents of emergent diseases for which there are no effective available vaccines (World Health Organization, 2014). Vector control is thus the most effective way of reducing disease transmission. Elimination of mosquito populations historically has involved the use of insecticides, supplemented by public education campaigns to promote behavioural changes and the elimination of mosquito breeding sites. Excessive and/or inconsistent use of insecticides has led to an increase in resistance against these compounds (Pietrantonio et al., 2000; Ponce-García G. et al., 2009; Alvarez et al., 2013). Non-conventional genetic control strategies that include vector population suppression and replacement have been proposed as alternatives to overcome this problem and are currently in trial phases (for a detailed review see Gabrieli et al., 2014). Population suppression may employ the release of males rendered sterile via chemicals or irradiation [sterile insect technique; SIT] or the release of genetically engineered males that are either sterile, or able to introduce lethal genes into natural populations. Population replacement strategies also rely on the release of transgenic mosquitoes, but aim to spread genes that will render natural populations refractory to pathogens. Regardless of the control strategy to be implemented, its success depends on an understanding of the target population dynamics.
Parameters such as connectivity between neighbouring locations, population size, and temporal genetic stability, directly impact the results of mosquito control campaigns. For example, the initial failure of SIT for vector control in El Salvador (Anopheles albimanus) and India (Culex quinquefasciatus) has been attributed to the migration of mated females into the target area, which as a consequence would have rescued the target population (Editorial, 1975; Benedict and Robinson, 2003). By studying the temporal genetic dynamics of a population its degree of genotype turnover can be inferred, either as a consequence of immigration from neighbouring populations or due to long-distance human mediated movement. These data will help inform the type of intervention that will be most effective at that specific location.
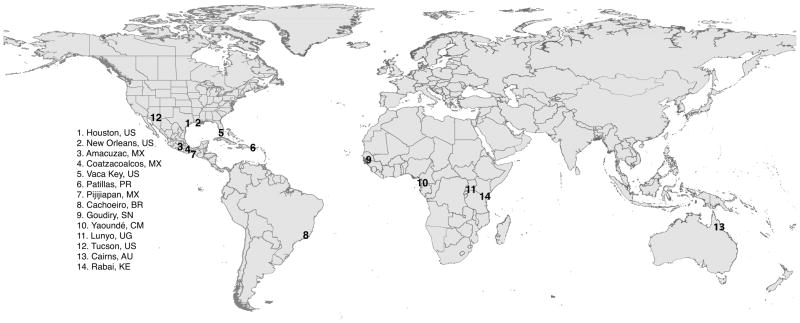
The present study addresses the temporal population dynamics of 14 populations of St. aegypti which were sampled 2 or 3 times at intervals ranging from 1 to 13 years, between 1998 and 2014 (Table 1, Figure 1). Mosquitoes came directly from the field and from multiple oviposition traps or larval breeding sites to avoid sampling siblings, with the exception of the two samples from Pijijiapan (Mexico), which each passed through one generation in the laboratory prior to genetic analyses but which are expected to be highly representative of the respective field populations since they were established from hundreds of individuals from the field.
Table 1.
Collection information and population genetics parameters.
| Location | Year | N | Ho | He | AR* | p(SMM) | p(TPM) |
|---|---|---|---|---|---|---|---|
| Rabai, KE | 2006 | 31 | 0.549 | 0.721 | 5.87 | 0.14746 | 0.00244 |
| Rabai, KE | 2009 | 37 | 0.650 | 0.705 | 5.81 | 0.2334 | 0.00806 |
| Rabai, KE | 2012 | 16 | 0.627 | 0.574 | 3.76 | 0.00488 | 0.00098 |
| Goudiry, SN | 2007 | 46 | 0.600 | 0.505 | 2.69 | 0.00098 | 0.00098 |
| Goudiry, SN | 2012 | 54 | 0.498 | 0.585 | 3.53 | 0.00195 | 0.00098 |
| Lunyo, UG | 2012 | 54 | 0.656 | 0.602 | 4.34 | 0.00049 | 0.00049 |
| Lunyo, UG | 2013 | 53 | 0.577 | 0.669 | 4.75 | 0.01221 | 0.00342 |
| Cachoeiro, BR | 2008 | 23 | 0.399 | 0.404 | 3.06 | 0.32031 | 0.14746 |
| Cachoeiro, BR | 2010 | 47 | 0.510 | 0.535 | 3.16 | 0.15137 | 0.06396 |
| Cachoeiro, BR | 2012 | 47 | 0.463 | 0.495 | 3.23 | 0.09229 | 0.04248 |
| Cairns, AU | 2009 | 48 | 0.631 | 0.549 | 3.24 | 0.0061 | 0.00122 |
| Cairns, AU | 2013 | 51 | 0.581 | 0.593 | 3.53 | 0.05225 | 0.04248 |
| Yaoundé, CM | 2005 | 10 | 0.450 | 0.603 | 4.08 | 0.00098 | 0.00098 |
| Yaoundé, CM | 2009 | 47 | 0.632 | 0.639 | 4.88 | 0.20605 | 0.01611 |
| Yaoundé, CM | 2014 | 54 | 0.548 | 0.629 | 4.81 | 0.20361 | 0.06396 |
| Pijijiapan, CHP, MX | 2006 | 48 | 0.493 | 0.464 | 2.68 | 0.00049 | 0.00049 |
| Pijijiapan, CHP, MX | 2008 | 47 | 0.391 | 0.458 | 2.94 | 0.03418 | 0.0061 |
| Coatzacoalcos, VER, MX | 2003 | 35 | 0.483 | 0.464 | 3.07 | 0.00928 | 0.00928 |
| Coatzacoalcos, VER, MX | 2008 | 50 | 0.418 | 0.341 | 2.4 | 0.00684 | 0.00488 |
| Houston, TX, US | 1998 | 47 | 0.408 | 0.370 | 2.2 | 0.00049 | 0.00049 |
| Houston, TX, US | 2009 | 29 | 0.546 | 0.451 | 2.84 | 0.01221 | 0.00684 |
| Houston, TX, US | 2011 | 19 | 0.439 | 0.416 | 2.82 | 0.00684 | 0.00488 |
| New Orleans, NO, US | 2011 | 46 | 0.570 | 0.617 | 4.12 | 0.10986 | 0.0105 |
| New Orleans, NO, US | 2012 | 63 | 0.591 | 0.603 | 4.24 | 0.20361 | 0.06396 |
| Vaca Key, FL, US | 2006 | 42 | 0.589 | 0.620 | 3.96 | 0.12939 | 0.01343 |
| Vaca Key, FL, US | 2009 | 43 | 0.512 | 0.589 | 3.93 | 0.17627 | 0.00806 |
| Tucson, AZ, US | 2012 | 53 | 0.545 | 0.589 | 3.7 | 0.38037 | 0.03418 |
| Tucson, AZ, US | 2013 | 54 | 0.554 | 0.557 | 3.63 | 0.15137 | 0.10986 |
| Patillas, PR | 2012 | 54 | 0.600 | 0.594 | 4.06 | 0.0105 | 0.00464 |
| Patillas, PR | 2014 | 54 | 0.574 | 0.536 | 3.78 | 0.00488 | 0.00342 |
| Amacuzac, MOR, MX | 2012 | 54 | 0.540 | 0.546 | 3.53 | 0.17627 | 0.01343 |
| Amacuzac, MOR, MX | 2013 | 54 | 0.569 | 0.530 | 3.3 | 0.03418 | 0.0061 |
| Amacuzac, MOR, MX | 2014 | 53 | 0.560 | 0.558 | 3.51 | 0.33936 | 0.26611 |
| Mean | 0.538 | 0.549 | 3.680 |
Ho = observed heterozygosity; He = expected heterozygosity; AR = Allelic Richness estimated by rarefaction (N=20 genes), p(SMM) and p(TPM)= probability of bottleneck under the Stepwise Mutation Model or under the Two Phase Model with variance =0.36 [38]. Values in bold indicate significance after Bonferroni correction.
Figure 1.
Sampling locations of Stegomyia aegypti collections used in this study. Samples were collected at each location in two or three occasions within the period of 1998 – 2014. Time intervals between the temporal collections per location ranged from 1 to 13 years.
DNA was extracted using the DNeasy Blood and Tissue kit (Qiagen). The samples from Coatzacoalcos, Houston (1998), and Goudiry (2007), were kindly provided as DNA extractions by Dr. W. C. Black at the Colorado State University. Individual mosquitoes were genotyped for microsatellites as described in Brown et al. (2011). The resulting products were processed for fragment analysis at the DNA Analysis Facility at Science Hill at Yale University, using GS 500 Rox internal size standard (Applied Biosystems). Microsatellite alleles were scored using GeneMapper v4.0 (Applied Biosystems).
A total of 43 out of 384 (11.2%) within-population deviations from Hardy-Weinberg equilibrium (HWE) were identified after sequential Bonferroni correction. The two Lunyo (Uganda) collections had the highest percentage of loci out of HWE with 41.7% and 58.3% respectively, followed by Patillas 2014 (Puerto Rico) with 45.5%, and Goudiry 2007 (Senegal) with 40%. The 12 microsatellite markers used in this study have been extensively validated by previous studies (Brown et al., 2011, Gloria-Soria et al., 2014) and deviations from HWE are attributed to the presence of null alleles at frequencies that do not affect the assessment of population structure.
Genetic distance, measured by pairwise FST values, was lower between temporal collections than between geographic localities (mean FST = 0.1010 and mean FST = 0.2092 respectively; File S1), with the exception of Houston and Coatzacoalcos, which had FST values exceeding the mean values across localities (File S1). A significant correlation was found between genetic distance and time between collections (R2= 0.4192, p=0.0123). However, this trend was driven by the Houston 1998 collection, the oldest collection in our dataset. When Houston 1998 was removed, or substituted for the Houston 2011 collection, the correlation disappeared (R2=0.1423, p=0.2038; and R2= 0.1166, p=0.2322). In agreement with the FST results, Houston and Coatzacoalcos were the most differentiated populations at the temporal scale as estimated by DEST (Jost, 2008) (DEST> 0.3, File S1).
Temporal continuity was considered equivalent to the connectivity among neighbour populations and measured by the amount of gene flow between temporal collections, that is, the number of migrants (Nm) across temporal samples. Nm values were estimated using the private allele method from Genepop v. 4.2 (File S1), which uses rare alleles to estimate gene flow and assumes that they have reached a quasi-equilibrium in the population (Barton & Slatkin, 1986). These results were consistent with the FST and DEST results (File S1), indicating that Goudiry, Coatzacoalcos, and Houston are the most differentiated populations through time (Nm < 0.2) and also pointing to New Orleans as the most stable population (Nm = 14.68; File S1).
Analysis of Molecular Variance (AMOVA) on all temporal collections to estimate the contribution of the temporal and geographic components to the variability observed in the data indicates that 79.24% of the overall genetic variation is explained by individual variation within the populations, 7.99% is explained by temporal variation, and 12.77% can be attributed to geographic variation (p < 0.001). However, temporal variation has a larger effect (>10%) in the diversity observed in Coatzacoalcos, Goudiry, Cachoeiro, and Houston, relative to the rest of the populations when the AMOVA is applied to each geographic location individually (data not shown).
Analyses to detect recent population size reductions using BOTTLENECK v. 1.2.02 (File S1), a software that tests each population for an excess in heterozygosity relative to the allelic diversity observed, found evidence of bottlenecks in Lunyo (2012), Pijijiapan (2006), and Houston (1998). The results were independent of the mutation model considered (infinite alleles model: IAM, a two-phase model: TPM, and the Stepwise Mutation model: SMM) and remained significant after multiple test correction (Table 1).
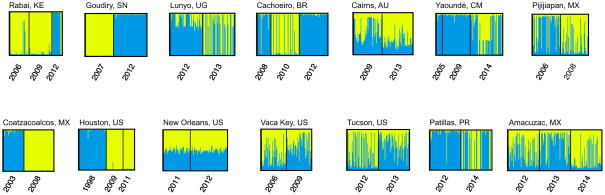
Bayesian clustering of individual localities was implemented in STRUCTURE v. 2.3 (File S1), a program that identifies genetic clusters and assigns individuals to these clusters with no a priori information of sample location. Based on 20 independent runs and 500,000 iterations, this method supports K=2 as the optimal number of clusters for most populations, consistent with temporal differentiation. Exceptions were New Orleans and Cairns, which show no genetic structure (K=1); as shown in Figure 2. Principal component analysis of allele frequencies described a similar pattern, with clear temporal differentiation observed in Goudiry, Coatzacoalcos, and Houston and different degrees of temporal overlap in the rest of the locations (Figure S1).
Figure 2.
Genetic structure of Stegomyia aegypti collections. STRUCTURE bar plots indicating relatedness of St. aegypti collections based on 12 microsatellite loci. Each vertical bar represents an individual. The height of each bar represents the probability of assignment to each of K clusters (indicated by different colours). Analyses of individual sites identified an optimal number of clusters of K = 2 for all populations except Cairns, AU and New Orleans, US (K=1).
This analysis clearly shows that Houston, Coatzacoalcos, and Goudiry, have experienced genotype turnover across the years sampled. In 1998 elevated levels of resistance to malathion were detected in Culex mosquitoes of Houston (Pietrantonio et al., 2000), forcing vector control programs to switch to pyrethroids in subsequent years. Although malathion remained effective against St. aegypti in most regions (NMTCB, 2011; Alvarez et al., 2013), it is possible that mosquitoes may have exhibited low levels of insecticide resistance and that the introduction of a new insecticide initially had a major impact on the population, with the effect of subsequent treatments tapering over time as resistance to pyrethroids developed. In contrast, the temporal change of Coatzacoalcos does not appear to involve insecticide use. Pyrethroid use in Mexico began in 2000 and although the frequency of the pyrethroid resistant allele (Ile1016) in Coatzacoalcos went from zero in 2003 to 27% in 2006 (Ponce-García et al., 2009), we found no signature of a bottleneck. Information regarding insecticide use in Goudiry was not available at the time this paper was written.
Immigration of novel genotypes by human transportation could also explain the high genotype turnover observed in these locations. Houston and Coatzacoalcos are major port cities and Goudiry sits on the Dakar-Niger railroad line, the main east-west commerce route that connects the major port of Senegal (Dakar), with Mali and Niger. Nevertheless, we cannot discard the possibility that the apparent temporal structure in Goudiry is in fact the result of a pre-existing population structure, as the temporal samples were collected from different areas of the city. The 2007 samples were collected from the east side of Goudiry, while the 2012 collections were from the central Market because the original breeding sites had disappeared by that time (W. C. Black, personal communication). Goudiry is an isolated site with a hot and dry climate, so high genetic structure as a consequence of low gene flow would not be surprising.
Allele frequencies at all other sites were fairly constant over several years. Though differentiation did occur between collections (Figure 2 and File S1), populations at each site remained in the same genetic grouping over time (Figure S1). The temporal stability of New Orleans had previously been detected with isozymes between 1976 and 1979 (Tabachnick, 1982). This pattern suggests that vector control strategies have a minimal impact on the New Orleans St. aegypti population, that the population is large and local elimination of mosquitoes is rapidly followed by re-colonization by mosquitoes from neighbouring areas that are genetically similar, or that migration of novel genotypes does not occur or has reached an equilibrium and no longer affects the genetic composition of the recipient population. In New Orleans, dengue is not a major public health concern and vector control efforts have focused on control of Culex spp. to prevent transmission of West Nile Virus, and not of St. aegypti (NMTCB, 2011 and 2012). Similar arguments can be made to explain the stability of the Amacuzac population, although in this example, the local vector control specifically targets St. aegypti to control the spread of dengue. The weather conditions in Amacuzac are suitable for sustaining a large mosquito population all year long and insecticide use in this locality is infrequent (approximately every 4 months) unless a possible disease outbreak is reported. It is important to note that the ability of this study to detect migrants is limited to the detection of incoming individuals that are genetically distinct from the population being analysed. Migrants that are not genetically differentiated will not be identified by this method.
The switch in insecticide use can explain the bottleneck signature observed in Houston, and it is likely that the bottleneck and temporal differentiation observed in Pijijiapan are consequences of the laboratory passage despite efforts to minimize such effects. The causes of the bottleneck signature in Lunyo are still unclear.
In conclusion, these results suggest that St. aegypti populations vary markedly in their degree of temporal stability in different geographic locations. Some populations remain substantially unchanged over time, others display temporal differentiation while maintaining their geographic fingerprints, and yet others, display dramatic shifts in gene frequencies over time.
Temporal genetic changes in St. aegypti populations most likely occur due to fluctuations in population size followed by recolonization events or genetic drift. Such fluctuations may be natural, due to dry-wet seasons or temperature extremes (e.g. Huber et al., 2002), or human-driven, for example, the result of vector elimination campaigns. Genetic changes may also be affected by immigration of mosquitoes via human trade and transport.
The observation of heterogeneity in genetic temporal stability among populations suggests that a detailed study at each field site in the years prior to and after the release of any modified mosquitoes for either population elimination or modification-based control strategies will be desirable to increase their long-term chances of success. Vector control efforts are likely to fail without accurate ecological information, including knowledge of temporal dynamics of mosquito populations. Sites with temporally stable St. aegypti populations are potential optimal locations for genetically based population replacement programs as they are likely to facilitate the effective spread of genetic modifications and allow such changes in the population to be maintained over time. In contrast, temporally unstable locations have a higher probability for reinvasion of mosquito populations, even if initial efforts at eliminating populations are successful.
Supplementary Material
A - Scatter plot of the first two principal components of 14 geographic locations based on 12 microsatellite loci. Each site was sampled at two or three different time points separated by periods of 1 to 13 years. Each temporal collection is plotted with a different colour. Scores are displayed for each individual plot.
Supplementary tables: Allele numbers and frequency table, Locus-by-locus Chi-square test on allelic frequencies between time points, FST statistics, Population differentiation parameters (Jost DEST values and Nm), Bottleneck analysis results, references of the analysis software.
Acknowledgments
This study was possible thanks to various colleagues that supplied the collections used in this study. Collections not previously reported were kindly provided by S. Ritchie (Cairns), and R. Barrera (Puerto Rico). Samples from Coatzacoalcos, Houston (1998), and Goudiry (2007) came from W. C. Black’s collection at the Colorado State University. This work was funded by the US NIH RO1 AI101112 (JRP) and US NIH 3R01AI091646-04S1 (AG-S).
References
- Alvarez LC, Ponce G, Oviedo M, Lopez B, Flores AE. Resistance to malathion and deltamethrin in Aedes aegypti (Diptera: Culicidae) from western Venezuela. Journal of Medical Entomology. 2013;50(5):1031–1039. doi: 10.1603/me12254. [DOI] [PubMed] [Google Scholar]
- Barton NH, Slatkin M. A quasi-equilibrium theory of the distribution of rare alleles in a subdivided population. Heredity. 1986;56:409–415. doi: 10.1038/hdy.1986.63. [DOI] [PubMed] [Google Scholar]
- Benedict MQ, Robinson AS. The first releases of transgenic mosquitoes: an argument for the sterile insect technique. Trends in Parasitology. 2003;19(8):349–355. doi: 10.1016/s1471-4922(03)00144-2. [DOI] [PubMed] [Google Scholar]
- Brown JE, McBride CS, Johnson P, Ritchie S, Paupy C, Bossin H, Lutomiah J, Fernandez-Salas I, Ponlawat A, Cornel AJ, Black WC, IV, Gorrochotegui-Escalante N, Urdaneta-Marquez L, Sylla M, Slotman M, Murray KO, Walker C, Powell JR. Worldwide patterns of genetic differentiation imply multiple ‘domestications’ of Aedes aegypti, a major vector of human diseases. Proceedings of the Royal Society of London B. 2011;278:2446–2454. doi: 10.1098/rspb.2010.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrieli P, Smidler A, Catteruccia F. Engineering the control of mosquito-borne infectious diseases. Genome Biology. 2014;15(535) doi: 10.1186/s13059-014-0535-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloria-Soria A, Brown JE, Kramer V, Hardstone Yoshimizu M, Powell JR. Origin of the Dengue Fever Mosquito, Aedes aegypti, in California. PLoS Neglected Tropical Diseases. 2014;8(7):e3029. doi: 10.1371/journal.pntd.0003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber K, Le Loan L, Huu Hoang T, Khanh Tien T, Rodhain F, Failloux A-B. Temporal genetic variation in Aedes aegypti populations in Ho Chi Minh City (Vietnam) Heredity. 2002;89(1):7–14. doi: 10.1038/sj.hdy.6800086. [DOI] [PubMed] [Google Scholar]
- Jost L. GST and its relatives do not measure differentiation. Molecular Ecology. 2008;17:4015–4026. doi: 10.1111/j.1365-294x.2008.03887.x. [DOI] [PubMed] [Google Scholar]
- Monteiro FA, Schama R, Martins AJ, Gloria-Soria A, Brown JE, Powell JR. Genetic Diversity of Brazilian Aedes aegypti : Patterns following an Eradication Program. PLoS Neglected Tropical Diseases. 2014;8(9):e3167. doi: 10.1371/journal.pntd.0003167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NMTCB. New Orleans Mosquito & Termite Control Board Annual Report. 2011 and 2012. [Google Scholar]
- Oh, New Delhi; oh, Geneva. Nature. 1975;256:355–357. Editorial. [Google Scholar]
- Pietrantonio PV, Gibson G, Nawrocki S, Carrier F, Knight WP. Insecticide resistance status, esterase activity, and electromorphs from mosquito populations of Culex quinquefasciatus Say (Diptera: Culicidae), in Houston (Harris County), Texas. Journal of Vector Ecology. 2000;25(1):74–89. [PubMed] [Google Scholar]
- Ponce-García G, Flores AE, Fernández-Salas I, Saavedra-Rodríguez K, Reyes-Solis G, Lozano-Fuentes S, Guillermo Bond J, Casas-Martínez M, Ramsey JM, García-Rejón J, Domínguez-Galera M, Ranson H, Hemingway J, Eisen L, Black IVWC. Recent Rapid Rise of a Permethrin Knock Down Resistance Allele in Aedes aegypti in México. PLoS Neglected Tropical Diseases. 2009;3(10):e531. doi: 10.1371/journal.pntd.0000531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabachnick WJ. Geographic and temporal patterns of genetic variation of Aedes aegypti in New Orleans. American Journal of Tropical Medicine and Hygiene. 1982;31(4):849–853. doi: 10.4269/ajtmh.1982.31.849. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A - Scatter plot of the first two principal components of 14 geographic locations based on 12 microsatellite loci. Each site was sampled at two or three different time points separated by periods of 1 to 13 years. Each temporal collection is plotted with a different colour. Scores are displayed for each individual plot.
Supplementary tables: Allele numbers and frequency table, Locus-by-locus Chi-square test on allelic frequencies between time points, FST statistics, Population differentiation parameters (Jost DEST values and Nm), Bottleneck analysis results, references of the analysis software.