Abstract
Fbxo30 is an orphan member of the F-box protein family with no known substrate or function. Here, we report that while Fbxo30−/− mice exhibit normal development, growth, life span, and fertility, the females fail to nurture their offspring due to defective mammopoiesis. Mass spectrometry analysis of Fbxo30-associated proteins revealed that Fbxo30 specifically interacts with the bipolar spindle kinesin EG5 (encoded by Kif11). As a result, Fbxo30 targets Eg5 for ubiquitinylation and controls its oscillation during the cell cycle. Correlated with EG5 dysregulation, Fbxo30−/− mammary epithelial cells exhibit multiple defects in centrosome homeostasis, mitotic spindle formation, and proliferation. Effects on proliferation, centrosome homeostasis, and mammopoiesis in the Fbxo30−/− mice were rescued through normalization of Eg5 activity using shRNA and/or an EG5 inhibitor. Our data reveal the Fbxo30-Eg5 interaction as a critical checkpoint in mammopoiesis and a critical role for ubiquitinylation-regulated Eg5 oscillation in the cell cycle.
Keywords: Mammopoiesis, Bipolar Mitotic Kinesin Eg5, F-Box proteins
Introduction
F-Box proteins (FBX) are defined by the shared protein-protein interaction domain called F-Box. Based on the existence of additional structural motifs, FBX are divided into three subfamilies, including FBXW (for WD40 domain), FBXL (for leucine-rich motif) and FBXO (for other motifs) (Jin et al., 2004). FBX often interact with a specific motif on the substrate called degron. Since the interactions between the best characterized FBX and their substrates are regulated by phosphorylation of the degron (Koivomagi et al., 2011; Liu et al., 2002; Nash et al., 2001; Wei et al., 2005), the term phosphodegron was coined to epitomize regulation between FBXs and their substrates (Skaar et al., 2013). However, it is increasingly clear that many FBX-substrate interactions are either based on recognition of unmodified degron sequence (D’Angiolella et al., 2012; D’Angiolella et al., 2010) or recognition of degron with other covalent modification such as glycosylation (Yoshida et al., 2002). More recently, phosphorylation of degron has been shown to inhibit FBXL12 binding to p85β subunit of PI3K (Kuchay et al., 2013).
The biological function of FBX is informed by their specific substrates. By conferring substrate specificity to the SCF ubiquitin E3 ligases, FBXs regulate a variety of cellular functions, including cell cycle (Carrano et al., 1999; Frescas and Pagano, 2008), signal transduction (Koivomagi et al., 2011; Kuchay et al., 2013; Liu et al., 2002; Nash et al., 2001; Wei et al., 2005), DNA repair (D’Angiolella et al., 2012) and genomic instability (Puklowski et al., 2011) among others (Skaar et al., 2013). Despite the extensive effort, the substrates for the majority of FBX remain to be identified. This gap prevents us from fully understanding the function and mechanism of action by FBX. Identification of the substrates for the orphan FBX is a major focus in cell biology as these efforts not only clarify the function of the orphan FBXs, but also elucidate novel cellular regulatory pathways.
FBXO30 is an orphan FBX with no known substrates. Here we take a reverse genetic approach to evaluate the function of Fbxo30 in mice. Our data revealed an unexpected function of the Fbxo30 gene in mamopoiesis, including branch morphogenesis, and defective production of mature luminal epithelial cells. Mass spectrometry analysis of Fbxo30-associated proteins revealed motor protein Eg5 as Fbxo30 substrate. By regulating the Eg5 abundance, Fbxo30 regulates mitosis and mammopoiesis. Our data not only revealed an unexpected function of Fbxo30 in organogenesis, but also identified it as a crtical regulator for mitosis.
With identification of mammary stem cells (Shackleton et al., 2006; Stingl et al., 2006), the mouse has emerged as a major tool to study mammopoiesis. Accumulating data lead to a developmental pathway for differentiation of mammary stem cells into progenitors for basal myoepithelial and luminal epithelial cells, respectively (Shackleton et al., 2006; Stingl et al., 2006; Visvader, 2009; Visvader and Stingl, 2014). Genetic studies indicate critical roles for Slug and Sox9 as key determinants of mammary stem cell function (Guo et al., 2012). Commitment to luminal vs basal fates is influenced by the expression of p63 and Notch 1 and 3, respectively (Bouras et al., 2008; Raouf et al., 2008; Yalcin-Ozuysal et al., 2010). Elf5 has emerged as a key in specifying luminal fate (Chakrabarti et al., 2012; Oakes et al., 2008), while GATA3 directs differentiation to luminal epithelial cells (Asselin-Labat et al., 2007). Our data presented herein revealed a critical role for Fbxo30-EG5 interaction mammary gland morphogenesis, and thus suggesting a critical checkpoint in mammopoiesis.
Results
Mice with targeted mutation of Fbxo30 revealed a critical role for Fbxo30 in mouse mammopoiesis
With the exception of a small stretch of 68 amino acids encoded by exon 3, the majority of the 746 amino acids of Fbxo30are encoded by exon 2. We replaced the 3.5 kb of genomic DNA encompassing the entire region of exon 2 (2.1 kb) and part of intron 2 (1.4 kb) with an in-frame GFP cDNA followed by a Neomycin cassette for selection (supplemental Fig. S1). The heterozygous mutant embryonic stem cells (ES cells) of C57BL/6 origin were used to generate chimeric mice. The null allele was germline-transmitted at a Mendelian ratio, suggesting that the Fbxo30 gene was not essential for mouse development. Moreover, Fbxo30−/− and Fbxo30+/+ littermates had normal fertility and grew at a similar rate with indistinguishable life-span. Surprisingly, regardless of the paternal genotypes, mutation of the Fbxo30 in the nursing female reduced the survival of the offspring, as the survival rates of offspring from Fbxo30−/−, Fbxo30+/−, and Fbxo30+/+ females were 0-12%, 55-69%, and 82-84% respectively (Table S1). Only a small number of offspring were successfully nurtured by Fbxo30−/− mice, usually after multiple pregnancies.
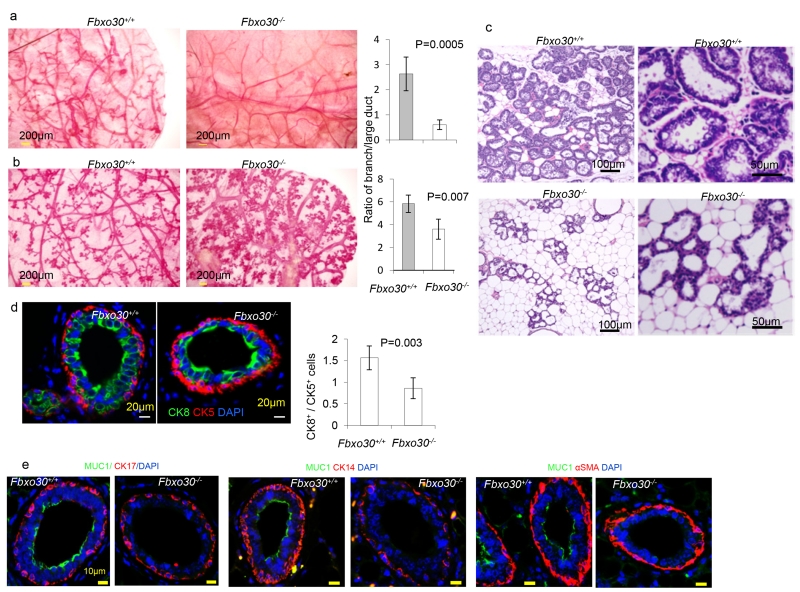
Since the offspring of Fbxo30−/− females developed normally with foster mothers and since necropsy revealed that the pups died of starvation (data not shown), we examined the mammary glands by histology and flow cytometry. In mammary gland whole mounts, virgin Fbxo30−/− mice had atrophic mammary glands, characterized by both reduced branching and atrophic large ducts (images in Fig. 1a). The ratio between lateral branches and large ducts was decreased by Fbxo30 deletion (Fig. 1a, right panel). Pregnancy of WT mice is associated with increased branching and formation of alveolar structures in the branches. However, the Fbxo30−/− mammary glands showed diminished branching with the alveoli forming adjacent to the large ducts (Fig. 1b). Histological analysis revealed that while wild-type mammary glands had proliferated massively at day 2 postpartum, those in Fbxo3−/− mice remained largely atrophic (Fig. 4c). Furthermore, while most of the glandules in Fbxo30+/+ mice were filled with milk droplets, very few glandule in the Fbxo30−/− mice contained milk droplets. These findings explained why the Fbxo30−/− female failed to nurture the offspring.
Fig. 1.
Defective mammopoiesis in Fbxo30−/− mice. a. Morphological defects of virgin Fbxo30−/− mammary glands by whole-mount Carmine alum staining shows reduced branching and atrophic large ducts. The images are representative from three independent experiments, each of which involves three mice per group. Bar graphs shown depict ratios of length of lateral branches to that of the large ducts. Length was measured by Neurolucida Tracing software (MBF Bioscience, Williston, VT 05495). Data shown are means and SEM (P = 0.0005). Data presented are summary data from one experiment involving three mice per group. One fat pad per mouse was analyzed. The data have been reproduced in another independent experiment. b. Morphological defects of mammary glands in 12-day pregnant Fbxo30−/− mice and WT littermates were visualized in whole mount sections. Reduction in mammary gland branching in pregnant Fbxo30−/−female mice were determined as in a, except that the mammary glands were isolated from day 12 of the first pregnancy (P = 0.007). Data presented are summary data from one experiment involving three mice per group. One fat pad per mouse was analyzed. The data have been reproduced in another independent experiment. c. Histology analysis of postpartum mammary gland reveals atrophy and lack of milk droplets in the Fbxo30−/− female mammary glands. Sample collected at day 2 postpartum were fixed in formalin and stained with hematoxylin and eosin. Similar results were obtained in 8 mice analyzed, including 3 WT and 5 Fbxo30−/− mice. d. Reduction of luminary epithelial cells in the Fbxo30−/− mammary gland. Formalin-fixed mammary gland tissue sections were stained with antibodies specific for either cytokeratin 5 (CK5, basal myoepithelial cells) or cytokeratin 8 (CK8, luminary epithelial cells). The images in the upper panels are representative of 4 independent experiments involving one mouse per group (Scale bar, 20μM). Data shown in the right panel are the ratios of K8+/K5+ cells. The error bar depicts the standard errors of means (n=10 glands from one fatpad in each group). Data have been reproduced three times. e. Defective production of musin by luminal epithelial cells in Fbxo−/−mammary glands. As in d, except that the mammary sections were stained with antibodies against CK14, SMA and CK17, respectively in conjunction with Muc1.
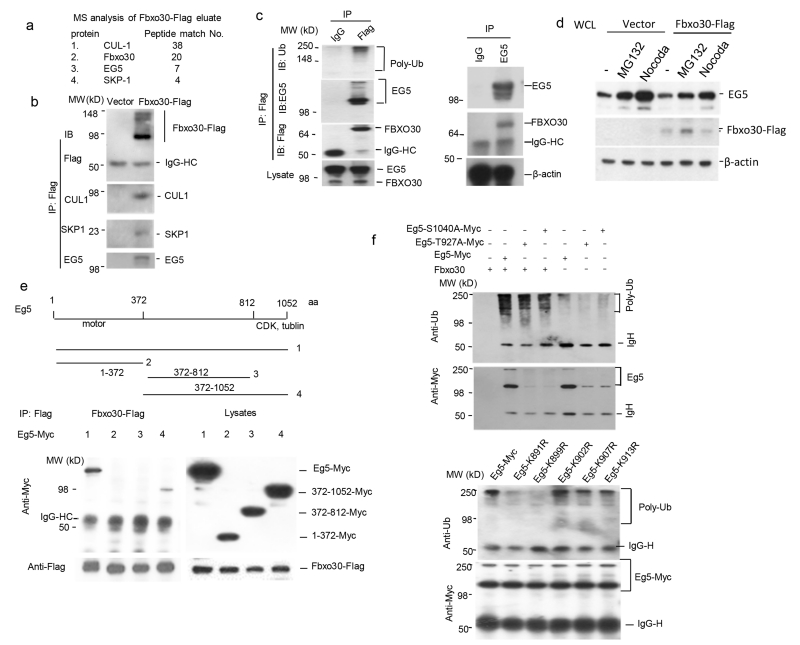
Fig. 4.
Fbxo30 is an E3 ligase for EG5. a. Identification of EG5 and other SCF components in a complex by mass spectrometry analysis. 293T cells were transfected with cDNA encoding Flag-tagged Fbxo30 or an unrelated protein (Laforin). Flag-tagged proteins were purified by immunoprecipitating with anti-Flag. Immunoprecipitated proteins were eluted with Flag peptide. The Fbxo30-associated proteins were concentrated by SDS-PAGE. The gel slices of approximately 1cm were submitted to Taplin Biological Mass Spectrometry Facility at the Harvard Medical School (https://taplin.med.harvard.edu) using the microcapillary LC/MS/MS techniques. The numbers of unique peptides associated with Fbxo30 are listed, while peptides shared with Laforin are excluded. b. Confirmation of EG5-SCF complex by co-IP. cDNA encoding Flag-tagged Fbxo30 was transfected into 293T cells. After IP with anti-Flag, the precipitates were probed for EG5, SKP1, and CUL1 by their specific antibodies. c. Association between endogenous EG5 and Fbxo30. The endogenous FBXO30 was tagged with a 3× Flag epitope by homologous knock-in (see supplemental Fig. S2). The left panel shows that anti-Flag-Fbxo30 co-precipitated endogenous EG5; the right panel shows co-precipitation of endogenous FBXO30 by anti-EG5. d. Impact of proteasome inhibitor MG132 and M-phase arrest on endogenous EG5 levels: effect of ectopically expressed Fbxo30. After 24 hours transfected with empty vector or Fbxo30-Flag expressing vector, the 293T cells were treated with 2.5μM MG132 or 50 nM nocodazole for an additional 6 hours before being lysed for Western-blot. Note that in vector control, EG5 levels is increased after MG132 treatment and that EG5 over-expression decrease EG5 at M-phase. e. Eg5 C-terminal region interacts with Fbxo30. As in b, except either full-length or indicated truncation mutants were used and that Eg5 were detected by anti-Myc antibody. The left panel shows the IP-western blot data, while the right panel depicts the expression levels in the lysates. f. In vitro ubiquitinylation of Eg5 by Fbxo30. Data in upper panels show phosphorylation-independent ubiquitylation, while those in the lower panels show selectivity of ubiquitylation. Fbxo30-Flag protein was purified from HEK293 cells transfected with Flag-tagged Fbxo30 and incubated with bead-bound EG5 isolated from EG5-Myc transfected cells and with a ubiquitinylation system containing E1, E2, and proteasomal components. After removing the unbound components by re-immunoprecipitating Eg5 with a c-Myc antibody, the extent of Eg5 ubiquitinylation was determined by immunoblot with an anti-poly-ubiquitin antibody (top), while the amount of EG5 was determined with an anti-Myc antibody (bottom). WT Eg5 were used in the upper panel, while the K>R mutants were compared with WT Eg5 in lower panel. Data in b-e are representative images and have been repeated 2 times.
To identify the relative number of luminary epithelial vs myoepithelial cells, we stained mammary sections with antibodies specific for cytokeratin (CK) 8 (luminary epithelial cells) or CK5 (basal myoepithelial cells). A reduction in the ratio of CK8+/CK5+cells was observed in the Fbxo30−/− mammary glands (Fig. 1d). The normal development of myoepithelial cells is confirmed by using a panel of different markers, including CK14, CK17 and αSMA (Fig. 1e). Remarkably, production of mucin by the luminal epithelial cells is largely abrogated by Fbxo30 deletion (Fig. 1e).
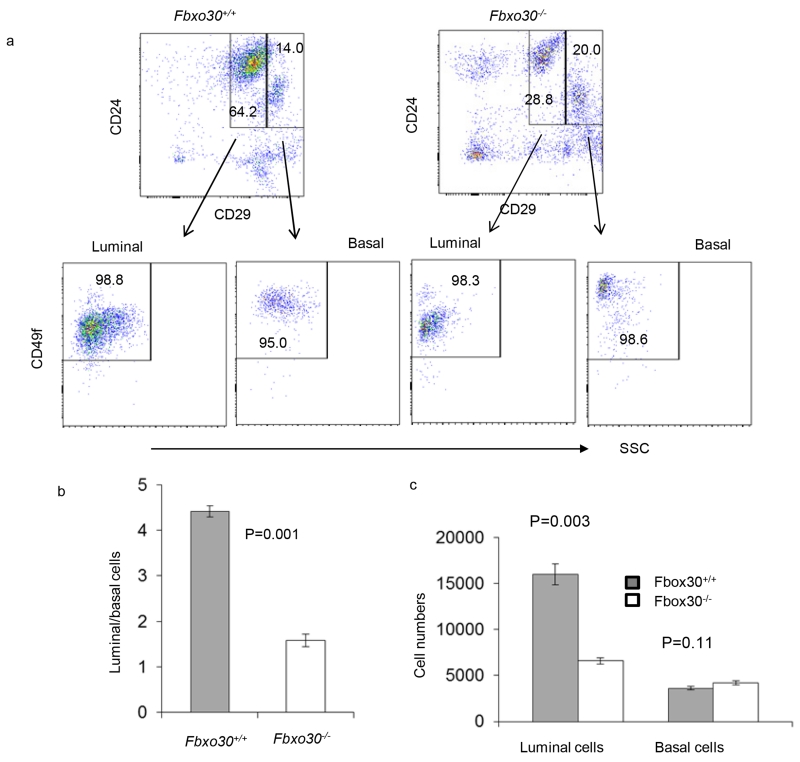
We analyzed the cellular composition of the mammary glands using CD29, CD49f, CD61, and CD24 markers (Visvader, 2009). CD45 and CD31 were used to mark hematopoietic cells and vascular endothelial cells respectively for exclusion. We used flow cytometry to characterize the frequency of major cell types in the mammary gland. Representative FACS profiles are shown in Fig. 2a, while summary data of the major mammary gland cell types are shown in Fig. 2b and Fig. 2c. Among the CD45− CD31− cells, we observed a reduction in the proportion of CD24+CD29lo cells (Fig. 2a, upper panels). Since essentially all of the CD24+CD29lo cells express high levels of CD49f (Fig. 2a, lower panels), they belong to luminal epithelial cells. Conversely, the percent of CD24+CD29hiCD49+myoepithelial cells was increased (Fig. 2a). In combination, the increase of myoepithelial and decrease of luminal epithelial cells result in a nearly 2-fold decrease in the relative abundance of luminal lineages (Fig. 2b). Correspondingly, the absolute numbers of luminal epithelial cells recovered from the Fbxo30−/− is more severely reduced (Fig. 2c).
Fig. 2.
Fbxo30 deficiency affects luminal vs basal lineage choice. a. Representative FACS profiles depicting selective reduction in luminal epithelial cells in the mammary gland of the Fbxo30−/− mice. The markers and gating strategy used to define specific mammary gland cell types are indicated. b. Reduction in luminal/basal ratio among Fbxo30−/− mammary epithelial cells. c. Number of recoverable luminal and basal epithelial cells in mammary gland per mice. Data shown are number of epithelial cells with either luminal or basal epithelial cell markers based on FACS staining. Data shown in this figure are representative data from an experiment involving 3 mice in each group and have been reproduced three times.
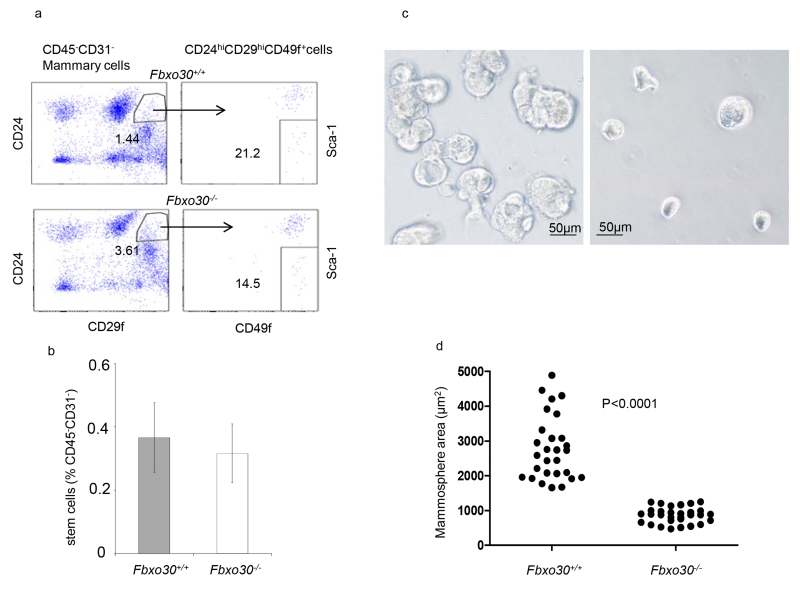
We used the accepted CD31−CD45−CD24hiCD29hiSca1− markers to assess the amounts of mammary stem cells in WT and Fbxo30−/− mice. Among CD45−CD31− cells from mammary glands. A reduction of the CD24hiCD29fhi cells was observed in the Fbxo30−/− mammary gland, as demonstrated in Fig. 3a left panels. However, the CD49fhiSca-1− populations were reduced in the Fbxo30−/− mammary gland. As a result, the percent of CD31−CD45−CD24hiCD29hiSca1− mammary stem cells were unaffected. Therefore, Fbxo30 mutation does not affect the number of mammary stem cells (Fig. 3b). Despite their normal numbers, the size of the Fbxo30−/− mammosphere was markedly reduced (Fig. 3c and d).
Fig. 3.
Impact of Fbxo30 deficiency on number and differentiation of mammary stem cells. a. Representative FACS profiles depicting a WT and an Fbxo30−/− mammary epithelial cells. The stem cells were defined as CD29hiCD49fhiCD24hi Sca-1−. b. Summary data on % of mammary stem cells in WT and Fbxo30−/− mammary gland. Data shown are means and SEM, n=5 mice per group, summarized from three independent experiments. c. Fbxo30−/− stem cells have diminished production of progeny in mammosphere culture. Representative images depicting the reduction in mammosphere size (Scale bar, 20μM). d. Areas of the representative mammosphere. Each dot represents the area of an individual sphere, as calculated using Scion software. These data have been reproduced three times.
Taken together, the data in Fig. 1-3 reveal that while Fbxo30 is not required for mammary stem cell maintenance, it is required for differentiation and/or expansion of luminary progenitors into ductal and/or alveolar cells.
Fbxo30 is the E3 ligase of EG5
Fbxo30 is an orphan F-Box protein with no known substrate. To fill in this gap, we isolated the Flag-tagged Fbxo30-associated proteins by coimmunoprecipitation with anti-Flag antibodies. The proteins eluted with free Flag peptides were digested with trypsin and analyzed by mass spectrometry. In addition to those from Fbxo30, peptides from Skp1 and Cullin were also prominently represented, as expected. Interestingly, 7 unique peptides from the bipolar kinesin motor protein EG5 were identified (Fig. 4a). To confirm this interaction, we probed anti-Flag immunoprecipitates of either vector or Fbxo30-Flag-transfected 293T cells. As shown in Fig. 4b, anti-Flag specifically co-precipitated EG5, Skp1, and Cul1. Since there are no commercial antibodies that satisfactorily immunoprecipitate FBXO30, we knocked-in a 3× Flag tag immediately before the stop codon in the FBXO30 open reading frame (Zhang et al., 2008) (Supplemental Fig. S2) and used an anti-Flag mAb to determine whether endogenous FBXO30 associates with EG5. As shown in Fig. 4c left panel, anti-Flag precipitated FBXO30 and EG5 as well as polyubiquitinated proteins. Reciprocally, anti-EG5 specifically co-precipitated FBXO30 (Fig. 4c, right panel). These results confirmed the endogenous association between EG5 and FBXO30. Moreover, inhibition of proteasome increased EG5 in whole cell lysate (Fig.4d). Using truncation mutant of Eg5, we observed that Fbxo30 interacts with the C-terminal region rather than the N- and central part of Eg5. The binding sites likely reside between AA812-1052 (Fig. 4e).
To confirm that Fbxo30 is the E3 ligase for EG5, we isolated the Fbxo30 proteins from Fbxo30-Flag transfectants and incubated them with Myc-tagged EG5 in the ubiquitinylation reaction mixture containing an E1 ubiquitin-activating enzyme and an E2 ubiquitin-conjugating enzyme. The extent of Eg5 ubiquitinylation was determined by immunoblot with an anti-poly-ubiquitin antibody. As shown in Fig. 4f, upper panel, incubation of EG5 with Fbxo30 in the presence of E1 and E2 resulted in robust EG5 ubiquitinylation. After normalizing the amount of input protein, it is clear that EG5 ubiquitinylation did not require phosphorylation at either T927 or S1040, two phosphorylation sites critical for regulation of EG5 activity by CDK1 and Plk1-NEK9-NEK6/7, respectively (Bertran et al., 2011; Blangy et al., 1995; Rapley et al., 2008), as the relative amounts of polyubiquitinated EG5 increased even though the amounts of EG5 was reduced. The mechanism of reduced accumulation of mutant EG5 remains to be explained.
To identify potential ubiquitylation sites, we have identified a patch of Lysine (K) between AA891-913. As shown in Fig. 4f lower panel, while mutations of K891R and K899R substantially reduce polyubiqutylation of EG5, other mutations have minimal effect. These data further support ubiquitylation of EG5 by Fbxo30, although they do not necessarily rule out additional ubiquitylation sites on EG5.
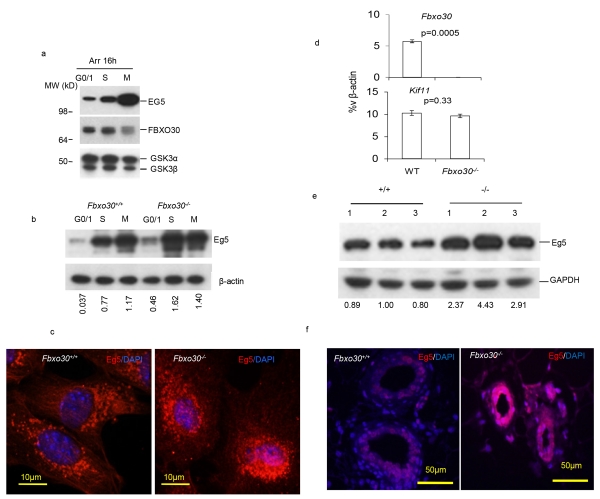
Eg5 activities are tightly regulated during cell cycle by phosphorylation (Bertran et al., 2011; Blangy et al., 1995; Rapley et al., 2008). Since it is unclear whether Eg5 oscillates during cell cycle, we arrested, at G0/1, S and M phases, the HCT116 cells expressing a Flag tagged Fbxo30 under control of its endogenous promoter. As shown in Fig. 5a, EG5 levels increased progressively from G0/1, S to M phases but reduced after the cells are released from the M phase arrest. The peak EG5 level at M phase correlated with the lowest FBXO30 level. To test whether Fbxo30 is responsible for Eg5 accumulation, we arrested WT and Fbxo30−/− MEFs at G1/G0, S, and M phase and compared the Eg5 levels. As shown in Fig. 5b, Eg5 levels increase from G1 to S phase but peaked at the M-phase in the WT MEF. Targeted mutation of Fbxo30 accelerated the increase of Eg5, as it peaked at S rather than M phase. Since Fbxo30 is expressed at lower levels at M phase, it is expected that deletion of this gene does not significantly affect EG5 levels (Fig. 5b). The mechanism for fluctuations of Fbxo30 during cell cycles is currently unknown.
Fig. 5.
FBXO30 regulate EG5 oscillation during cell cycle. a. Increased EG5 protein levels at mitosis correlates with reduced Fbxo30. HCT116 cells with Fbxo30-3×-Flag knock-in alleles were arrested at G0/1 phase with serum-free medium, early S phase with 1 μg/ml aphidicolin, or M phase with 50 ng/ml nocodazole. All treatments were performed for 16 hours. The lysates were analyzed for the endogenous levels of EG5 and Fbxo30 by Western blot. GSK3 levels were used as a loading control. Data shown have been reproduced 3 times. b. Fbxo30 deletion greatly increases Eg5 levels at S and M phase of the Fbxo30−/− MEFs. WT and Fbxo30−/− MEFs were arrested for 16 hours in serum-free media, 1 μg/ml aphidicolin, or 50 ng/ml nocodazole. The lysates were analyzed for the endogenous levels of EG5 by Western blot. β-actin levels were used as a loading control. The relative amounts of EG5 protein are provided underneath each lane. The numbers indicate the ratio of signals from anti-EG5 over anti-β–actin. Data shown have been reproduced 3 times. c. Elevated Eg5 levels in Fbxo30−/− MGEs. WT and Fbxo30−/− MGEs were arrested at S phase with 1 μg/ml aphidicolin for 16 hours. Images shown are representative of those obtained from three independent experiments. d. Normal expression of the Kif11(Eg5-encoding) gene in Fbxo30−/− mammary glands. RNA isolated from single cell suspension of WT and Fbxo30−/− mammary glands were analyzed for expression of Fbxo30 and Kif11. While deletion of Fbxo30 abolished expression of Fbxo30, it had no effect on the Kif11 transcript, as determined by real-time PCR. e. Western blot data using lysates MGE (n=3). The images of the Western blot were shown in the top panel. The relative amounts of Eg5 protein are provided underneath each lane. The numbers indicate the ratio of signals from anti-Eg5 over anti-Gapdh. These data have been reproduced twice. f. Eg5 staining in WT and Fbxo30−/− mammary as determined by immunofluorescence. The nuclei are visualized by the DNA dye DAPI. Data shown are representative images from 3 experiments, each involving 1 fatpad from 1 mouse per group.
To determine whether the Fbxo30 regulate Eg5 in mammary gland epithelial (MGE) cells, we arrested WT and Fbxo30−/− MGE at S phase, and determined Eg5 accumulation using immunofluorescence in conjunction with DAPI (Fig. 5c). S-phase synchronized Fbxo30−/− MGE exhibited markedly increased overall Eg5 levels, as reflected by huge increases of Eg5+ spots. Despite comparable mRNA levels of Kif11 gene which encodes EG5 (Fig. 5d), freshly isolated MGE have substantially elevated Eg5 protein as revealed by Western blot (Fig. 5e). To determine whether Fbxo30 regulates Eg5 in vivo, we compared the cellular levels of Eg5 in mammary glands from WT and Fbxo30−/− mice by immunofluorescence. This analysis revealed a marked increase in the cellular concentration of EG5 in Fbxo30−/− mammary gland (Fig. 5f).
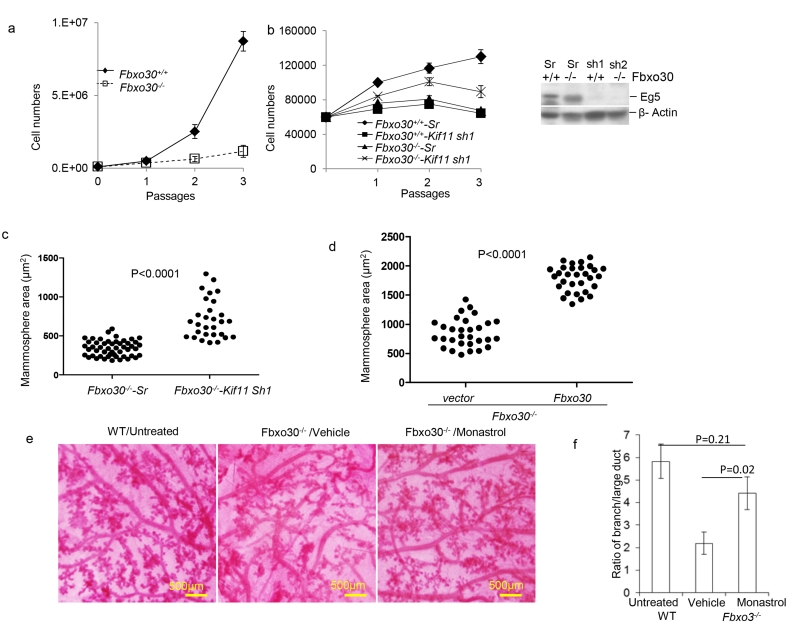
As an ATPase motor protein, Eg5 is targeted to the mitotic spindle and centrosome and plays a critical role in mitosis (Mardin and Schiebel, 2012; Nigg and Stearns, 2011). Consistent with the abnormal levels of Eg5 in Fbxo30−/− MGE (Fig. 5), proliferation rate of Fbxo30−/− MGE was also markedly reduced over 3 passages (Fig. 6a). To determine whether the defective proliferation in MGE cells is attributable to increased EG5 activity, we used shRNA to silence Kif11 gene in the Fbxo30−/−MGE cells (Fig. 6b, right panel). Consistent with a critical role for Eg5 in cell proliferation (Bertran et al., 2011; Blangy et al., 1995; Rapley et al., 2008), growth of WT MGE was inhibited by shRNA specific for the Kif11 gene. In contrast, shRNA silencing of Kif11 increased the proliferation of Fbxo30−/− MGE (Fig. 6b). Furthermore, Kif11 shRNA significantly increased the size of mammospheres (Fig. 6c).
Fig. 6.
Fbxo30 regulates cell proliferation and mammopoiesis through EG5. a. Fbxo30 is required for propagation of mammary epithelial cells in vitro. Data shown are the number of WT and Fbxo30−/− MGE over 3 passages. b. ShRNA silencing of Kif11 reduced proliferation of WT MGEs, but modestly enhanced proliferation of Fbxo30−/− MGEs. Freshly isolated WT or Fbxo30−/− MGE were transduced with lentiviral vectors with either scrambled or Kif11 shRNA for 24 hours. Equal number of transduced cells were re-plated and counted over 3 passages. The efficiency of Kif11 shRNA silencing is shown in the right panel. c. ShRNA silencing of Kif11 in Fbxo30−/− MGEs increases the size of mammospheres. Transduced primary MGEs were cultured for mammosphere assay. Data shown are the surface area of individual spheres from the two groups. P value was calculated by student t-test. Data shown in a-c are representative of 3 independent cultures. d. Ectopic expression of Fbxo30 partially restored sizes of Fbxo30−/− mammospheres. Fbxo30−/− epithelial cells were transduced with lentiviral vector alone or those with Fbxo30 cDNA prior to culture. These data have been reproduced three times. e and f. Restoration of mammopoiesis of pregnant Fbxo30−/− mice by monastrol, including increased secondary branching and reduced alveoli associated with the large ducts. Day 6 pregnant Fbxo30−/− mice received 3 injections of either vehicle control or monastrol (5mg/kg, every other day, intraperitoneal injection) over a 6 day period. Mammary glands were harvested on day 12 of pregnancy and used for whole mount sections. e. Representative images of whole mount sections from one of 3 mice in each group, 1 fatpad is analyzed per mouse. This experiment has been repeated 2 times. A whole mount section of WT mammary gland is included for comparison. f. Summary data of the ratio of the length of lateral branches to that of the large ducts, as measured by Neurolucida Tracing software. Data shown are from one of two independent experiments. n=3 mice per group, one fat pad per mice is analyzed. Error bars are SEM.
Reintroduction of WT Fbxo30 to the mammary epithelial cells through ectopic expression partially restored the mammosphere formation (Fig. 6d). While these data confirmed the function of Fbxo30, it also suggests that ectopic expression of Fbxo30 does not fully recapitulated the function of endogenous alleles.
To determine whether increased EG5 is responsible for the defective mammopoiesis during pregnancy, we treated day 6 pregnant Fbxo30−/− mice with the Eg5 inhibitor monastrol (Mayer et al., 1999) and analyzed the mammary gland morphology at day 12. As shown in Fig. 6e, f, a short-term treatment of the Fbxo30−/− mice with monastrol substantially rescued mammopoiesis, as revealed by the increased branching and distance between the alveoli and the large ducts. Therefore, elevated Eg5 activity is responsible for the defective mammopoiesis of Fbxo30−/−MGE in both virgin and pregnant mice.
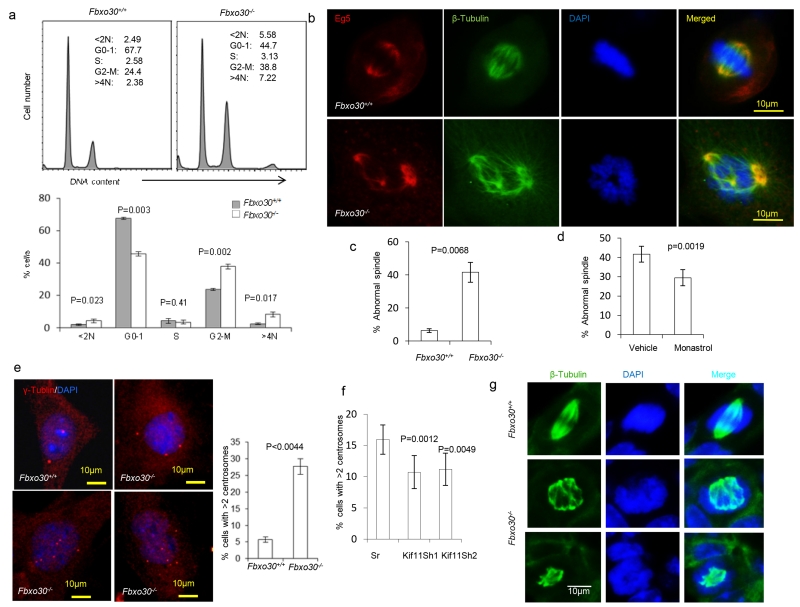
Given the role of Eg5 in centrosome separation (Mardin et al., 2011; Mardin and Schiebel, 2012), we evaluated the impact of Fbxo30 mutation on mitosis. Based on DNA content, it was clear that deletion of Fbxo30 caused G2/M arrest and polyploidy (Fig. 7a). To understand how Fbxo30 defects affect genome instability and G2/M arrest, we analyzed Eg5 distribution and mitotic spindles in mitotic WT MGE cells. We found that Eg5 was targeted to bipolar mitotic spindles, with a higher concentration at the two poles of the MGE cells. However, in the Fbxo30−/− MGE, Eg5 was targeted to multipolar spindles or bipolar multiple spindles, resulting in abnormal separation of chromosomes (Fig. 7b, c). This was due to over-active Eg5 as the number of cells with abnormal spindles was normalized by Eg5 inhibitor (Fig. 7d). Consistent with increased multipolar spindles, targeted mutation caused centrosome amplification, as indicated by the percent of MGE with more than two centrosomes (Fig. 7e). This is mediated by increased Eg5, as reduction of Eg5 through shRNA silencing of Kif11 attenuated the defects (Fig. 7f).
Fig. 7.
Fbxo30 deletion causes chromosomal instability and spindle defects in vivo and in vitro. a. DNA content analysis based on propidium iodide staining of ethanol-fixed MGE cells after 2 passages in vitro. Representative FACS profiles are shown on top and the summary data involving 3 mouse samples per group shown in the bottom. Note a major increase in cells with > 4N in DNA content. b. Deletion of Fbxo30 causes formation of multipolar mitotic spindles. WT (upper panels) and Fbxo30−/− (lower panels) MGE were stained for EG5, tubulin, and DNA. c. Summary data for three different cultures (n=3). d. Monastrol treatment reduces % of MGE cells with abnormal spindles. As in c, except that the MGE cells prepared from Fbxo30−/− mice were treated with either vehicle control or monastrol (30 nM) for 48 hours prior to staining. e. Fbxo30 deletion causes abnormal centrosome amplification. The images on the left depict γ-tubulin distribution in a WT and 3 Fbxo30−/− MGE cells, while the right panel shows the mean and SEM of percent cells with greater than 2 centrosomes (n=15) and has been repeated 3 times. f. ShRNA silencing of the Kif11 gene restores centrosome homeostasis in Fbxo30−/−MGE cells. Fbxo30−/− MGE cells were cultured in the presence of either vehicle or a low dose of monastrol (30 nM) for 48 hours. The centrosome number was determined by γ-tubulin staining. Data shown are means and SEM of percent cells with >2 centrosomes (n=10) and have been repeated 3 times). g. Accumulation of multi-polar spindle mitotic cells in the Fbxo30−/− mammary glands, as revealed by staining with anti-β-tubulin mAb and DAPI staining of frozen mammary gland sections. The top panels show a cell with a normal bipoplar spindle in WT mammary gland; the lower panels show a cell in Fbxo30−/− mammary gland with multipolar mitotic spindle and abnormal distribution of chromatin. Cells with mitotic spindle are difficult to find in vivo. Only 1 such cell was found in five slides prepared from 5 virgin mice, while more than 50 cells with mitotic spindle were found in 2 of the 5 slides from the 5 virgin Fbx30−/− mice. None of the mitotic spindles have normal morphology. Seven such cells are shown in supplemental Fig. S3.
A critical issue is whether defective spindle formation can be found to increase in the Fbxo30−/− mammary glands. To address this issue, we stained mammary gland with anti-β-tubulin antibodies and DAPI and searched for cells with mitotic spindle in mammary glands. Mitotic cells were readily observed in the mammary glands from the Fbxo30−/− mice but rarely in those from wild-type mice, which is consistent with a G2/M arrest of Fbxo30−/− MGE in vivo. Remarkably, while the WT MGE has normal bipolar mitotic spindle, essentially all of the mitotic cells in the Fbxo30−/− mammary glands contained abnormal spindles, including those with mono- or multi-polar spindles with abnormal donut-type chromatin (Fig. 7g, Fig. S3). The increase of cells with either mono- and multi-polar spindles in mammary glands phenocopied what was observed in MEF cultured from transgenic mice that over-express Eg5 (Castillo et al., 2007).
Discussion
Taken together, we have demonstrated that Fbxo30 is an E3 ligase that specifically ubiquitinylates and regulates Eg5 levels in mammary epithelial cells and mouse embryonic fibroblasts. The Fbxo30-Eg5 pathway controls centrosome homeostasis and genome stability. Since cells with abnormal mitotic spindles undergo apoptosis (Ganem et al., 2009), the defective cell growth and genome instability observed in the Fbxo30−/− cells are intrinsically connected.
Previous studies have demonstrated that EG5 targeting to the spindle pole is regulated by phosphorylation at sites T927 and S1040 by Cdk1 and Plk1-Ned6, respectively (Bertran et al., 2011; Blangy et al., 1995; Rapley et al., 2008) and their phosphorylation is critical for correct formation of mitotic spindle. Our data demonstrate that phosphorylation of neither site is required for EG5 ubiquitinylation. Thus, Eg5 is subject to at least two layers of regulation: Cdk1 and Plk1-Nek9-Nek6/7 mediated phosphorylation, which regulates localization but not Eg5 levels, and the Fbxo30-mediated ubiquitinylation, which controls Eg5 levels. Since phospho-peptide analysis revealed that endogenous Eg5 is phosphorylated only at the T927 and S1040 sites in vivo (Rapley et al., 2008), and since mutation of neither site abolish Eg5 ubiquitinylation, it is likely that Fbxo30-mediated Eg5 degradation occurs by a phosphorylation-independent pathway, which would be similar to the Fbxo1-CP110 and Fbx011-BCL6 interactions (D’Angiolella et al., 2010; Duan et al., 2012). Two recent studies demonstrated the APC/C-CDH1 complex also cause ubiquitinylation of EG5 (Drosopoulos et al., 2014; Eguren et al., 2014). Given the specific activation of the APC/C-CDH1 during mitosis, and given the relatively low expression and small effect of Fbxo30 deficiency on EG5 levels at M phase, we suggest that Fbxo30 and APC/C-CDH1 may function at different phases of cell cycle in regulating EG5 levels, although the possibility that these different E3 may function in different cell types cannot be ruled out.
The mitotic spindle defects observed in the Fbxo30−/− mammary epithelial cells is a direct consequence of Eg5 over-expression as it can be normalized by shRNA silencing of EG5. Moreover, since normalization of Eg5 activity by monastrol and/or Kif11 shRNA rescued defects in centrosome homeostasis, cellular proliferation, mammosphere formation and mammopoiesis, the defective regulation of EG5 levels is likely the root cause of genome instability and defective mammopoiesis in the Fbxo30−/− mice.
Apart from a critical function of Fbxo30 in mammopoiesis described herein, a recent report suggests that Fbxo30 is negatively regulated by the BMP signaling pathway and contributes to loss of muscle mass caused by defective BMP signaling (Sartori et al., 2013). However, the Fbxo30 substrate involved in regulation of muscle mass was not identified. Paradoxically, mice lacking Fbxo30 have normal development, fertility and life span. Since the developmental pathway for mammary gland is similar to that of T-cell differentiation (Watson and Khaled, 2008), we investigated the possible role for EG5 in development and accumulation of T cells in the thymus and spleen. We found that cellular compositions of spleen and thymus were largely unaffected by Fbxo30 deletion (supplemental Fig. S4&5). As a potential explanation for lack of function of Fbxo30 in spleen cells, we found that expression of Fbxo30 among spleen cells is considerably lower than in the mammary epithelial cells (supplemental Fig. S6a). Correspondingly, the levels of EG5 is largely unaffected by deletion of the Fbxo30 gene (Fig. S6b). With recent identification of APC/C-CDH as an E3 ligase for EG5 (Drosopoulos et al., 2014; Eguren et al., 2014), it is possible that this or other E3 may serve to regulate EG5 depending on the cell types studied.
Accumulating data have demonstrated that some of the most critical regulators of centrosome homeostasis, including Plk4, SAS6, and Cp110 are regulated by SCF-type E3 ligase(Silverman et al., 2012). Our data demonstrate that Eg5 abundance, as well as its oscillation during cell cycle, are also regulated by an E3 ligase. It is of note that Eg5 levels are frequently elevated in human cancer and that such elevation correlates with poor prognosis (Ding et al., 2011; Saijo et al., 2006). Since transgenic expression of Eg5 leads to abnormal centrosome homeostasis, genetic instability, and cancer development (Castillo et al., 2007), it would be of interest to test whether Fbxo30 defects may contribute to Eg5 elevation in human cancer.
Experimental Procedures
Generation of Fbxo30−/− mice
The mice were generated by inGenious Targeting Laboratories through a research contract. The constructs generated are diagrammed in Fig. S1. Briefly, a 13.2 kb region used to construct the targeting vector was first subcloned from a positively identified B6 BAC clone using a homologous recombination-based technique. The region was designed such that the short homology arm (SA) extended 1.9 kb to the 5′ Neo cassette. The long homology arm (LA) started at the 5′ end of the GFP cassette and was 7.8 kb. The GFP/Neo cassette was inserted 3 bp before the ATG of exon 1 and replaced ~3.5 kb of the gene sequence. The targeting vector was confirmed by restriction analysis after each modification step and by sequencing using primers designed to read from the LacZ/Neo cassette into the 5′ end of the SA (N1) and the 3′ end of the LA (GFP1) or from primers that anneal to the vector sequence, P6 and T7, and read into the 5′ and 3′ ends of the BAC subclone. The BAC was subcloned into a ~2.4kb backbone vector containing an ampicillin selection cassette for retransformation of the construct prior to electroporation. The total size of the targeting construct (including vector backbone and GFP/Neo cassette) was 14.7kb. Ten micrograms of the targeting vector was linearized by AscI and then transfected by electroporation of iTL C57 BL/6 embryonic stem cells. After selection in G418, surviving clones were expanded for PCR analysis to identify recombinant ES clones. Primers, A1 was designed downstream (3′) to the short homology arm (SA) outside the region used to generate the targeting construct. ES cells with a targeted locus were injected into albino C57BL/6 blastocysts to generate chimeric mice. Chimera were identified by coat color and used to generate mice with germline transmission of the targeted Fbxo30−/− locus.
Generation of HCT116 cell line with Flag-tagged endogenous Fbxo30 gene
The vector construction and knock-in by homologous recombination were based on a report (Zhang et al., 2008). Briefly, the targeting vector was constructed by polymerase chain reaction (PCR) using genomic DNA as the template for the homologous left arm. Homologous recombination was screened by PCR with primers derived from the neomycin resistance gene and the upstream region of the left homologous arm. Two clones were identified from two 96-well-plates of G418 resistant clones. The neomycin resistance gene was excised by Cre recombinase. The primers used were: 3flag KI FBXO30 Left arm F, GGGAAAG/ideoxyU/atttgtaaagatgaggggcttg;
3flag KI FBXO30 Left arm R,
GGAGACA/ideoxyU/GcAAGTACAGGTTTTAAAACTGAGCG
3flag KI FBXO30 right arm F, GGTCCCA/ideoxyU/AAGTTGTAATATTACTAGCACATA;
3flag KI FBXO30 right arm R, GGCATAG/ideoxyU/ACTTTCTTGGTGGTGAAGCCTA;
3flag KI FBXO30 Left Screening F; tctaggtctgtcctgctctgc;
3flag KI FBXO30 Right screening R, tttttaagcagaccacatcagg
3flag KI FBXO30 cre F, GAAGGACGTTCACTACGCTCA;
3flag KI FBXO30 Cre R, TTCCAAAAATTTCACACATTTCA
Immunofluorescence
After deparaffinization and rehydration of slides with Xylene and ethanol, the mammary gland sections from Fbxo30 knockout and WT mice were treated with 10 mM sodium citrate buffer, pH 6.0. The sections were permeabilized with 0.3% Triton X-100 in 10 mM Tris-HCl buffer for 30 min. After blocking with 2% bovine serum albumin (BSA) for 60 minutes, sections were incubated with primary antibody diluted in 10 mM Tris-HCl buffer containing 2% BSA at 4°C, overnight, with subsequent staining with secondary antibody in BSA-Tris-HCl buffer at room temperature for 2-4 hours. The nuclei were stained with DAPI. Slides were mounted with Prolong Antifade mounting buffer (Invitrogen, Carlsbad, CA 92008). Antibodies for EG5 (NB500-181, Novus), β-tubulin (F2043, Sigma), γ-tubulin (T5326, Sigma), Keratin 5 (AF138, Covance), Keratin 8 (TROMA-I, DSHB, Iowa city, IA52242), Keratin K14 (ab7800, Abcam), Keratin K17 (ab53707, Abcam), and αSMA (ab5694, Abcam), were used for immunofluorescence.
Transplantation
Three-week-old female mice were anesthetized with Ketamine and Xylazine. An inverted Y incision was made from the midline point between the fourth set of nipples and ending at a point between the fourth and fifth sets. The fourth and fifth fat pads on each side were exposed using forceps to separate the skin from the body cavity. Half of No.4 mammary fad pads from the fourth nipple to the lymph node was removed using scissors to clear the mammary epithelium and 1×106 mammary epithelial cells suspended in 30 μl of Hanks balanced solution plus 2% FBS were injected into the mammary epithelium cleared fat pads. After cell transplantation, the incision was sutured with 9–mm autoclips and the mice were placed in a clean cage and were monitored for a short period of time. Staples were removed after 2 weeks of surgery.
Isolation and culture of mammary gland epithelia
Mammary glands were minced with razor blades on a sterile surface and then transferred to a tube containing dissociation solution (EpiCult-B medium with collagenase and Hyaluronidase, Stemcell Tech, Vancouver, Canada) and incubated for 7 hours at 37°C with occasional pipetting and vortexing. After dissociation, the cells were collected by centrifugation at 350 × g. 1-5 mL of pre-warmed Trypsin-EDTA were added to the partially-dissociated tissue and mixed by pipetting for 1 to 3 minutes. 10 mL of cold Hanks’ Balanced Salt Solution supplemented with 2% FBS was added to stop the digestion and the cells were again collected by centrifugation at 350 × g for 5 minutes. To dissociate the clumps, 2 mL of pre-warmed 5 mg/mL Dispase (Catalog #07913 Stemcell Tech) and 200 μL of 1 mg/mL DNase I (Catalog #07900 Stemcell Tech) was added to the pellet and was then pipetted for 1 minute with a P1000 micropipetor to further dissociate cell clumps. The cell suspension was then diluted with an additional 10 mL of cold Hanks’ Balanced Salt Solution supplemented with 2% FBS and filtered through a 40 μm cell strainer into a new 50 mL centrifuge tube. Cells were collected after centrifugation and red blood cells were lysed as needed.
Flow cytometry
The isolated cells from the mammary glands of WT and Fbxo30−/− mice were stained with fluorochrome-conjugated anti-CD31, CD45.2, CD61, CD24, CD49f, CD29, Sca-1, and CD133 monoclonal antibodies (eBioscience, San Diego, CA 92121). The stained cells were analyzed by a fluorescence activated cell analyzer (LSR II).
Mammosphere assay
The isolated cells from the mammary glands of either WT or Fbxo30−/− mice were suspended in the EpiCult-B (Stemcell Tech, Vancouver, Canada) and seeded in ultra-low attachment 6-well plates (Corning, NY14831). The cells were cultured for 7 days and the formed mammospheres were photographed. The surface area of the mammospheres were measured based on analysis of the images using Scion image software (Scion Corporation, Frederick, Maryland, USA).
Whole-mount staining
Whole-mount preparations of mammary glands were performed according to the protocol in the NIH Biology of Mammary Gland website (http://mammary.nih.gov/histological/histology/index.html). Briefly, inguinal #4 mammary glands were dissected, spread on glass slides, and fixed overnight in Carnoy’s fixative. The next day, the tissues were hydrated and then stained in carmine-aluminum stain overnight. The tissue were dehydrated, cleared in xylene, and mounted between two glass slides using Permount mounting media.
shRNA silencing
The lentiviral vector pLenti6/V5-TOPO, obtained from Invitrogen, was modified by replacing the CMV promoter with a U6 promoter to drive a shRNA-expressing cassette and inserting a pGK-driven EGFP cassette and a blasticidin-resistant cassette. The sequence used to silence Kif11 was 5′-TGCAGGTCAGATTTACACT-3′ (sh1) and TATTGTCTTCAGGTCTTCA (Sh2). The mammary gland epithelial cell cultures were infected with either scrambled shRNA control lentivirus or lentivirus encoding Kif11-shRNA for 24 hours. After recovery for an additional 48 hours, the cultures were used for assays.
In vitro Ubiquitinylation assay
All the reagents used for the in vitro ubiquitinylation assay were purchased from Boston Biochem (Cambridge, MA 02139, catalog no. K930). Ubiquitinylation was performed according to standard protocols provided by the supplier, with modifications. In brief, HEK293 cells were transfected with Eg5-Myc or its phosphorylation site mutants. Anti-Myc immunoprecipitate beads were mixed with components of the ubiquitinylation system in the presence or absence of Fbxo30 proteins, which were eluted using Flag peptide from anti-Flag immunoprecipitates of transfected HEK293 cells using Flag peptide. After 2 hours of incubation at 37°C, the beads were washed with a 1% Triton-×100 Tris-HCl buffer, resuspended with a 2% SDS Tris-HCl buffer and re-immunoprecipitated with an anti-Myc antibody after a 10 fold dilution with a 1% Triton-×100 Tris-HCl buffer. The final immunoprecipitates were resuspended in SDS loading buffer, heated for 3 minutes at 90°C, and resolved by 10% SDS-PAGE.
Immunoprecipitation and Western blot
Cells or tissues were lysed with a 1% Triton X-100 buffer containing 20 mM Tris-HCl (pH7.4), 150 mM NaCl, 40 mM NaF, 2 mM DTT, and a protease and phosphatase inhibitor cocktail (Sigma). The cleared lysates were immunoprecipitated with protein G beads and antibody at 4°C with overnight shaking. After washing with the lysis buffer for three times, the beads were treated with SDS loading buffer and the eluted proteins were analyzed by Western blot. The EG5 antibodies used for Western blot and immunoprecipitation were a rabbit antibody (ab61199, Abcam) and a mouse antibody (NB100-78467, Novus), respectively.
Measurement of duct and branch lengths
The ratio of branch over duct length was measured using a method developed for measuring neurite and axon lengths (Kim et al., 2013; Sterne et al., 2015). The Neurolucida software was used to trace and measure the length of duct and that of the branches in a double blind fashion. Total lengths of either ducts or branches in all area captured in low power images (see Fig. 1a, b and Fig. 6d as examples) were integrated to calculate the branch over duct ratios.
Statistics
Student t-test was used to determine statistical significance between two groups of samples.
Supplementary Material
Acknowledgement
We thank Dr. Jane Visvader and Bing Ye for valuable advice, Drs. Junlin Guan, Yuan Zhu, Kaoru Sakebe for critical reading of the manuscript, and Ms Morgan Daley for editing the manuscript. This study is supported by grants from National Institute of Health, USA and those from the US Department of Defense.
Footnotes
Author contributions
YanL and YW performed the experiments. YangL, PZ and YW supervised the study. YanL, YW, PaZheng and YangL prepared the manuscript. ZD and XY generated the FOXP3-Flag tag knockin cell line.
References
- Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, Hartley L, Robb L, Grosveld FG, van der Wees J, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- Bertran MT, Sdelci S, Regue L, Avruch J, Caelles C, Roig J. Nek9 is a Plk1-activated kinase that controls early centrosome separation through Nek6/7 and Eg5. The EMBO journal. 2011;30:2634–2647. doi: 10.1038/emboj.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangy A, Lane HA, d’Herin P, Harper M, Kress M, Nigg EA. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83:1159–1169. doi: 10.1016/0092-8674(95)90142-6. [DOI] [PubMed] [Google Scholar]
- Bouras T, Pal B, Vaillant F, Harburg G, Asselin-Labat ML, Oakes SR, Lindeman GJ, Visvader JE. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008;3:429–441. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol. 1999;1:193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- Castillo A, Morse HC, 3rd, Godfrey VL, Naeem R, Justice MJ. Overexpression of Eg5 causes genomic instability and tumor formation in mice. Cancer research. 2007;67:10138–10147. doi: 10.1158/0008-5472.CAN-07-0326. [DOI] [PubMed] [Google Scholar]
- Chakrabarti R, Wei Y, Romano RA, DeCoste C, Kang Y, Sinha S. Elf5 regulates mammary gland stem/progenitor cell fate by influencing notch signaling. Stem Cells. 2012;30:1496–1508. doi: 10.1002/stem.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angiolella V, Donato V, Forrester FM, Jeong YT, Pellacani C, Kudo Y, Saraf A, Florens L, Washburn MP, Pagano M. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell. 2012;149:1023–1034. doi: 10.1016/j.cell.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angiolella V, Donato V, Vijayakumar S, Saraf A, Florens L, Washburn MP, Dynlacht B, Pagano M. SCF(Cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature. 2010;466:138–142. doi: 10.1038/nature09140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S, Xing N, Lu J, Zhang H, Nishizawa K, Liu S, Yuan X, Qin Y, Liu Y, Ogawa O, et al. Overexpression of Eg5 predicts unfavorable prognosis in non-muscle invasive bladder urothelial carcinoma. International journal of urology: official journal of the Japanese Urological Association. 2011;18:432–438. doi: 10.1111/j.1442-2042.2011.02751.x. [DOI] [PubMed] [Google Scholar]
- Drosopoulos K, Tang C, Chao WC, Linardopoulos S. APC/C is an essential regulator of centrosome clustering. Nature communications. 2014;5:3686. doi: 10.1038/ncomms4686. [DOI] [PubMed] [Google Scholar]
- Duan S, Cermak L, Pagan JK, Rossi M, Martinengo C, di Celle PF, Chapuy B, Shipp M, Chiarle R, Pagano M. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature. 2012;481:90–93. doi: 10.1038/nature10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguren M, Alvarez-Fernandez M, Garcia F, Lopez-Contreras AJ, Fujimitsu K, Yaguchi H, Luque-Garcia JL, Fernandez-Capetillo O, Munoz J, Yamano H, et al. A synthetic lethal interaction between APC/C and topoisomerase poisons uncovered by proteomic screens. Cell reports. 2014;6:670–683. doi: 10.1016/j.celrep.2014.01.017. [DOI] [PubMed] [Google Scholar]
- Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zurrer-Hardi U, Bell G, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes and Development. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Wang X, Coolon R, Ye B. Dscam expression levels determine presynaptic arbor sizes in Drosophila sensory neurons. Neuron. 2013;78:827–838. doi: 10.1016/j.neuron.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivomagi M, Valk E, Venta R, Iofik A, Lepiku M, Balog ER, Rubin SM, Morgan DO, Loog M. Cascades of multisite phosphorylation control Sic1 destruction at the onset of S phase. Nature. 2011;480:128–131. doi: 10.1038/nature10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchay S, Duan S, Schenkein E, Peschiaroli A, Saraf A, Florens L, Washburn MP, Pagano M. FBXL2- and PTPL1-mediated degradation of p110-free p85beta regulatory subunit controls the PI(3)K signalling cascade. Nat Cell Biol. 2013;15:472–480. doi: 10.1038/ncb2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, Zhang Z, Lin X, He X. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- Mardin BR, Agircan FG, Lange C, Schiebel E. Plk1 controls the Nek2A-PP1gamma antagonism in centrosome disjunction. Current biology: CB. 2011;21:1145–1151. doi: 10.1016/j.cub.2011.05.047. [DOI] [PubMed] [Google Scholar]
- Mardin BR, Schiebel E. Breaking the ties that bind: new advances in centrosome biology. The Journal of cell biology. 2012;197:11–18. doi: 10.1083/jcb.201108006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Stearns T. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol. 2011;13:1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes SR, Naylor MJ, Asselin-Labat ML, Blazek KD, Gardiner-Garden M, Hilton HN, Kazlauskas M, Pritchard MA, Chodosh LA, Pfeffer PL, et al. The Ets transcription factor Elf5 specifies mammary alveolar cell fate. Genes and Development. 2008;22:581–586. doi: 10.1101/gad.1614608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puklowski A, Homsi Y, Keller D, May M, Chauhan S, Kossatz U, Grunwald V, Kubicka S, Pich A, Manns MP, et al. The SCF-FBXW5 E3-ubiquitin ligase is regulated by PLK4 and targets HsSAS-6 to control centrosome duplication. Nat Cell Biol. 2011;13:1004–1009. doi: 10.1038/ncb2282. [DOI] [PubMed] [Google Scholar]
- Raouf A, Zhao Y, To K, Stingl J, Delaney A, Barbara M, Iscove N, Jones S, McKinney S, Emerman J, et al. Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell Stem Cell. 2008;3:109–118. doi: 10.1016/j.stem.2008.05.018. [DOI] [PubMed] [Google Scholar]
- Rapley J, Nicolas M, Groen A, Regue L, Bertran MT, Caelles C, Avruch J, Roig J. The NIMA-family kinase Nek6 phosphorylates the kinesin Eg5 at a novel site necessary for mitotic spindle formation. Journal of cell science. 2008;121:3912–3921. doi: 10.1242/jcs.035360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo T, Ishii G, Ochiai A, Yoh K, Goto K, Nagai K, Kato H, Nishiwaki Y, Saijo N. Eg5 expression is closely correlated with the response of advanced non-small cell lung cancer to antimitotic agents combined with platinum chemotherapy. Lung Cancer. 2006;54:217–225. doi: 10.1016/j.lungcan.2006.06.018. [DOI] [PubMed] [Google Scholar]
- Sartori R, Schirwis E, Blaauw B, Bortolanza S, Zhao J, Enzo E, Stantzou A, Mouisel E, Toniolo L, Ferry A, et al. BMP signaling controls muscle mass. Nature genetics. 2013;45:1309–1318. doi: 10.1038/ng.2772. [DOI] [PubMed] [Google Scholar]
- Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Silverman JS, Skaar JR, Pagano M. SCF ubiquitin ligases in the maintenance of genome stability. Trends in biochemical sciences. 2012;37:66–73. doi: 10.1016/j.tibs.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nature reviews Molecular cell biology. 2013;14:369–381. doi: 10.1038/nrm3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterne GR, Kim JH, Ye B. Dysregulated Dscam levels act through Abelson tyrosine kinase to enlarge presynaptic arbors. eLife. 2015;4 doi: 10.7554/eLife.05196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- Visvader JE. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes and Development. 2009;23:2563–2577. doi: 10.1101/gad.1849509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE, Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes and Development. 2014;28:1143–1158. doi: 10.1101/gad.242511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson CJ, Khaled WT. Mammary development in the embryo and adult: a journey of morphogenesis and commitment. Development. 2008;135:995–1003. doi: 10.1242/dev.005439. [DOI] [PubMed] [Google Scholar]
- Wei W, Jin J, Schlisio S, Harper JW, Kaelin WG., Jr. The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 2005;8:25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Yalcin-Ozuysal O, Fiche M, Guitierrez M, Wagner KU, Raffoul W, Brisken C. Antagonistic roles of Notch and p63 in controlling mammary epithelial cell fates. Cell death and differentiation. 2010;17:1600–1612. doi: 10.1038/cdd.2010.37. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Chiba T, Tokunaga F, Kawasaki H, Iwai K, Suzuki T, Ito Y, Matsuoka K, Yoshida M, Tanaka K, et al. E3 ubiquitin ligase that recognizes sugar chains. Nature. 2002;418:438–442. doi: 10.1038/nature00890. [DOI] [PubMed] [Google Scholar]
- Zhang X, Guo C, Chen Y, Shulha HP, Schnetz MP, LaFramboise T, Bartels CF, Markowitz S, Weng Z, Scacheri PC, et al. Epitope tagging of endogenous proteins for genome-wide ChIP-chip studies. Nat Methods. 2008;5:163–165. doi: 10.1038/nmeth1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.