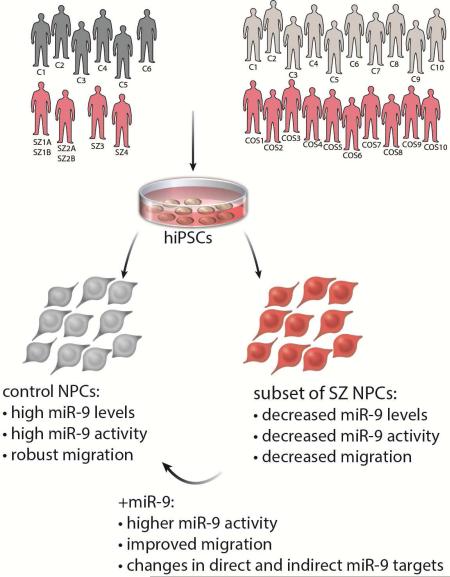
Abstract
Converging evidence indicates that microRNAs (miRNAs) may contribute to disease risk for schizophrenia (SZ). We show that miR-9 was abundantly expressed in control NPCs, but also significantly down-regulated in a subset of SZ NPCs. We observed a strong correlation between miR-9 expression and miR-9 regulatory activity in NPCs as well as between miR-9 levels/activity, neural migration and diagnosis. Overexpression of miR-9 was sufficient to ameliorate a previously reported neural migration deficit in SZ NPCs, whereas knockdown partially phenocopied aberrant migration in control NPCs. Unexpectedly, proteomic- and RNAseq-based analysis revealed that these effects were mediated primarily by small changes in expression of indirect miR-9 targets, rather than large changes in direct miR-9 targets; these indirect targets are enriched for migration-associated genes. Together these data indicate that aberrant levels and activity of miR-9 may be one of the many factors that contribute to SZ risk, at least in a subset of patients.
Keywords: microRNA-9, human induced pluripotent stem cell, neural progenitor cells, schizophrenia
Graphical Abstract
INTRODUCTION
SZ is hypothesized to be a neurodevelopmental condition (Weinberger, 1987) arising from dysregulated development of neural circuitry (Jarskog et al., 2007). Moreover, SZ has a highly heritable component (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014), and recent studies have demonstrated that transcripts expressed in the developing prefrontal cortex are enriched for those harboring SZ risk alleles (Gulsuner et al., 2013; Lin et al., 2015), suggesting that disruptions in fetal cortical development may underlie SZ. Multiple lines of strong genetic evidence suggest that miRNAs, particularly miR-137, are involved in the etiology of SZ (Ripke et al., 2013; Ripke et al., 2011; Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). Additionally, the coding region for a component of miRNA biogenesis, DiGeorge syndrome critical region gene 8 (DGCR8), lies within 22q11.2, the most common and penetrant of all recurrently observed SZ associated copy number variants (CNVs) (reviewed (Karayiorgou et al., 2010)).
As patient hiPSC-derived NPCs and neurons most resemble fetal brain tissue (Brennand et al., 2015), they are an indispensable tool in the study of possible molecular aspects of SZ predisposition. Using hiPSCs, we previously reported aberrant migration in SZ NPCs (Brennand et al., 2015; Lee et al., 2015) and diminished neuronal connectivity and synaptic activity in SZ neurons (Brennand et al., 2011; Yu et al., 2014), mirroring postmortem pathological findings (Wong and Van Tol, 2003).
Herein we show that miR-9 was not only highly expressed in control NPCs, but also significantly down-regulated in a subset of SZ NPCs. We find a correlation between miR-9 expression, miR-9 regulatory activity and neural migration. Retroviral expression of miR-9 was sufficient to rescue a neural migration deficit in SZ NPCs, whereas knockdown phenocopied aberrant migration in control NPCs. Finally, decreased miR-9 levels were corroborated in a subset of cases from a second and larger cohort comprised of ten cases and ten controls, indicating the robustness of this observation. We posit that dysregulation of miR-9 may contribute to risk for SZ, at least in a subset of patients.
RESULTS
Expression profile of 800 annotated miRNAs in SZ NPCs
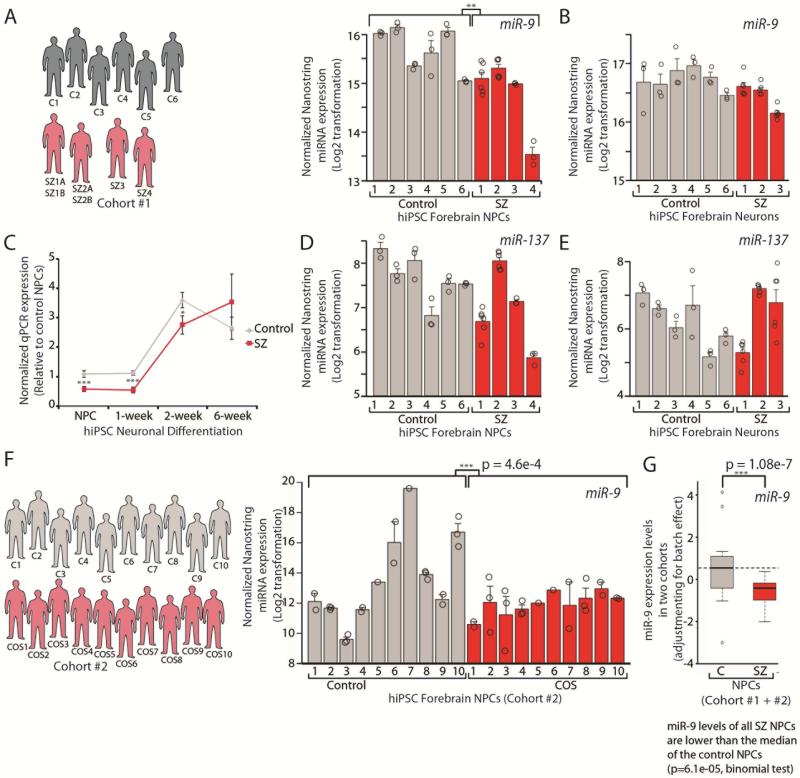
Unbiased comparison of 800 annotated miRNAs between our previously validated NPCs derived from four SZ patients and six controls (see SI Experimental Procedures for available clinical information and cell line descriptions) using the Nanostring nCounter expression platform identified hsamiR-9-5p (miR-9), a regulator of neurogenesis in neural stem cells (Zhao et al., 2009), as the most abundant and the most down-regulated miRNA (Bonferroni corrected student's t-test p < 0.02; p = 1.08e−07, nested ANOVA) (Table 1, SI Table 1, Fig. 1A), qPCR validated (Fig. 1C, SI Fig. 1). Values from independent replicates of the same NPC line for all individuals are indicated by circles (as well as from two independent NPC lines from SZ1 and SZ2), showing that miR-9 levels were consistent between biological samples (Fig. 1A). In contrast, miR-137 was not significantly perturbed in SZ NPCs and neurons (Tables 1-2, SI Tables 1, Fig. 1D-E).
Table 1.
miRNAs with highly significant (Bonferroni-corrected) perturbations in expression in SZ hiPSC forebrain NPCs.
| Abundance | Fold-Change (SZ/Control) | Student's t-test | Bonferroni corrected p-value | Function | Target genes perturbed by microarray | Predicted activity (Z-score) | |
|---|---|---|---|---|---|---|---|
| hsa-miR-98 | low | −1.94 | 6.0E-05 | 0.0479 | cancer progression; immune modulation; Alzheimer's disease | yes | 1.648 |
| hsa-miR-9-5p | high | −1.70 | 2.4E-05 | 0.0194 | neural differentiation; neural migration; EMT | yes | 3.958 |
| hsa-miR-27b-3p | high | −1.69 | 5.2E-05 | 0.0419 | myogenesis; angiogenesis; adipogenesis | yes | 0.984 |
| hsa-miR-532-5p | low | −1.66 | 4.5E-05 | 0.0362 | neuronal growth cones; myogenesis | yes | --- |
| hsa-miR-130a-3p | moderate | 1.30 | 3.5E-05 | 0.0279 | EMT, hypoxia response; autophagy-lysosomal pathway | yes | 1.167 |
Fig. 1. Decreased miR-9 levels occur in SZ NPCs but not SZ hiPSC forebrain neurons.
A,B,D,E. Nanostring nCounter analysis of normalized miR-9 (A,B) and miR-137 (D,E) expression levels in SZ NPCs (A,D) and neurons (B,E). Values from independent replicates of the same NPC line for all individuals are indicated by circles (as well as from two independent NPC lines from SZ1,2 (A,D) and two independent NPC lines from SZ1,2,3 (B,E)). C. qPCR validation of normalized miR-9 expression during the differentiation of SZ NPCs into 1-, 2- and 6-week-old neurons. F. Nanostring nCounter analysis of normalized miR-9 expression levels in NPCs from ten COS patients and ten unrelated controls. Values from biological replicates of NPC lines differentiated from independent hiPSC clones are indicated by circles (C1,2,3,4,8,9,10; COS1,2,3,5,6,7,8,10) as well as independent replicates from the same NPC lines (C2,3,9; COS2,3,4,8). G. Samples from two SZ hiPSC cohorts were combined after adjusting for the batch effect with linear regression, where average miR-9 expression level was calculated for each sample. Error bars are s.e., *P < 0.05, **P < 0.01, ***P < 0.001.
Table 2.
miRNAs with highly significant (Bonferroni-corrected) perturbations in expression in SZ hiPSC forebrain neurons.
| Abundance | Fold-Change (SZ/Control) | Student's t-test | Bonferroni corrected p-value | Function | Target genes perturbed by microarray | Predicted activity (Z-score) | |
|---|---|---|---|---|---|---|---|
| hsa-miR-28-5p | moderate | −2.76 | 4.2E-05 | 0.0337 | cellular proliferation | yes | --- |
| hsa-miR-191-5p | high | −2.26 | 1.0E-05 | 0.0081 | spine remodelling in LTP; regulator of BDNF, cancer | no | 0.452 |
| hsa-miR-148b-3p | high | −2.22 | 1.1E-05 | 0.0092 | tumor progression | yes | 0.664 |
| hsa-miR-1180 | high | −2.13 | 2.9E-06 | 0.0023 | unknown | no | --- |
| hsa-miR-450a-5p | moderate | −1.72 | 1.5E-05 | 0.0122 | tumor suppressor | no | --- |
| hsa-let-7d-5p | moderate | −1.70 | 6.4E-06 | 0.0051 | neurogenesis; apoptosis | yes | 1.619 |
| hsa-miR-374a-5p | high | −1.52 | 1.6E-05 | 0.0129 | neurogenesis; retinal ganglion cell differentiation | yes | --- |
| hsa-miR-374b-5p | moderate | 1.63 | 1.5E-05 | 0.0118 | neurogenesis; retinal ganglion cell differentiation | yes | --- |
| hsa-miR-92a-3p | moderate | 1.70 | 4.9E-06 | 0.0039 | neuronal homeostatic scaling; apoptosis; proliferation | yes | 1.518 |
| hsa-miR-34a-5p | high | 1.81 | 5.5E-05 | 0.0438 | neurite outgrowth; neural migration; memory consolidation | yes | 4.157 |
| hsa-miR-30b-5p | moderate | 1.85 | 5.0E-05 | 0.0401 | tumor suppressor, stress reponse | yes | 2.527 |
This finding was corroborated by a replication hiPSC cohort derived from ten childhood-onset-SZ (COS) patients and ten unrelated controls, all of whom are part of an ongoing longitudinal study conducted at the NIH (see SI Experimental Procedures for available clinical information). COS patients are thought to represent a subset of adult onset SZ patients defined by onset and severity; at adulthood, there are no genetic or clinical differences between COS patients and those with chronic poor outcome adult onset SZ (reviewed (Rapoport et al., 2012)). Cohort 2 hiPSCs showed normal karyotypes, robust self-renewal and expression of pluripotency markers (SI Fig. 2A-C); 2-3 validated hiPSC lines were generated per person (see SI Experimental Procedures for cell line descriptions). Cohort 2 NPCs expressed high levels of NESTIN and SOX2 (SI Fig. 2D). Using Nanostring nCounter analysis, we again observed decreased miR-9 levels in COS NPCs (0.38-fold, p=4.6e−04, nested ANOVA) (Fig. 1F). Values from biological replicates of NPC lines differentiated from independent hiPSC clones are indicated by circles (C1,2,3,4,8,9,10; COS1,2,3,5,6,7,8,10) as well as independent replicates from the same NPC lines (C2,3,9; COS2,3,4,8) (Fig. 1F). Moreover, because the reduced miR-9 level in a subset of SZ NPCs is beyond the inter-individual and intra-individual variations (Fig. 1F,G), we posit that perturbations in miR-9 may be more broadly relevant to SZ and not a result of variability reflected in culture (SI Table 2).
Although significantly decreased miR-9 levels was observed on average between SZ and control NPCs across both hiPSC cohorts, miR-9 expression differences were not correlated with diagnosis. Samples from the two cohorts were combined after adjusting for batch effect with linear regression (see Methods). As shown in Fig. 1G, miR-9 levels measured by Nanostring nCounter, were significantly lower in SZ NPCs than control NPCs (p = 1.08e-07, nested ANOVA). Moreover, we found that miR-9 levels in NPCs derived from all 14 SZ patients were lower than the median in control NPCs (p = 6.1e-05, binomial test), and 7 out of 14 SZ NPCs are lower than the 25% quantile of control NPCs (p = 0.038, binomial test). These observations indicate that lower miR-9 levels in SZ NPCs relative to control NPCs are largely driven by a significant subset of SZs, which is not unexpected given the heterogeneity of a complex disorder like SZ.
In order to leverage existing datasets, our subsequent mechanistic experiments in this report focused only on our first cohort of NPCs.
miR-9 targets enriched for differentially expressed genes in SZ NPCs
In an effort to elucidate miR-9 targets enriched in our previous NPC (GEO GSE40102) (Brennand et al., 2015) and neuron (GEO GSE25673) (Brennand et al., 2011) microarray gene expression datasets we used Partek Genomics Suite, accepting the caveat that the output generated would be a very large list of significant miRNAs (SI Table 3), likely confounded by the large number of overlaps among putative target genes of different miRNAs (Friedman et al., 2009). Known miR-9 targets were significantly enriched in the differential expression (DE) list (in total 84 miR-9 targets (9.3% of predicted targets) were perturbed (p<1e−45), 56% up-regulated and 44% down-regulated) (SI Table 3).
Global integrative modeling suggests miR-9 is a regulator in SZ NPCs
To adjust for overlaps among putative target genes of different miRNAs, and to integrate both transcriptional and post-transcriptional regulation of gene expression, we performed global modeling of putative miRNA and transcription factor (TF) targets (Balwierz et al., 2014) with an RNAseq dataset generated from control and SZ NPCs (GEO GSE63738) (Topol et al., 2015b) and neurons (GEO GSE63734) (SI Table 4). The linear regression-based model was first proposed to predict TF regulatory activities and motifs from yeast gene expression data (Conlon et al., 2003) and further extended to infer the miRNA regulatory activities (Setty et al., 2012). The method regresses the fold-change of a gene to its (multiple) putative regulatory miRNAs and TFs (Fig. 2A). The coefficient (z-score) of a miRNA or a TF, estimated using genome-wide fold changes and predicted targets of all miRNAs and TFs, represents the regulatory activity change of the miRNA/TF between SZ and control NPCs. Owing to the inherent properties of a linear regression model, the activity change reflects the non-redundant effect of the miRNA/TF with respect to all the other miRNAs and TFs (Fig. 2B). It is worth noting that although putative miRNA/TF targets likely contain false positive predictions and do not capture cell-type specific miRNA/TF gene binding, regulatory activities estimated from the regression model are very reliable in general (Balwierz et al., 2014). (Refer to the Methods for further details of the model.)
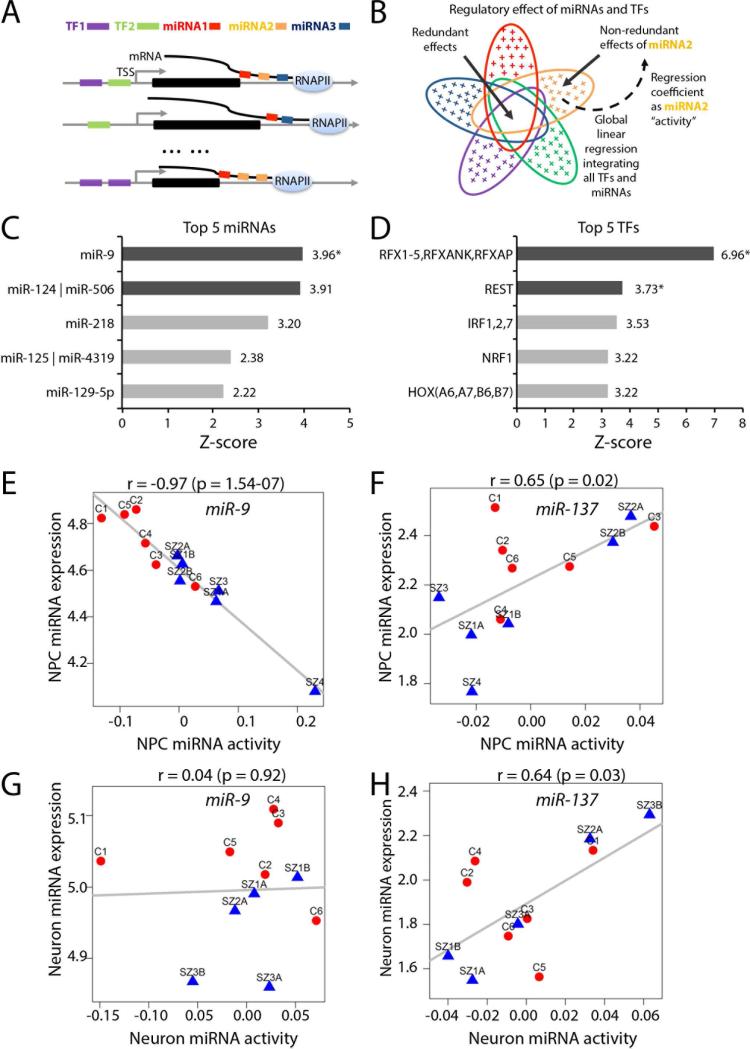
Fig. 2. A global integrative model identified miRNA and transcription factor (TF) candidates for regulating gene expression changes between SZ and control NPCs.
A. An illustrative example of regulatory relationships between five regulators (two TFs and three miRNAs, color coded) and three genes. The expression of each gene can be regulated by one or multiple miRNAs and TFs. B. Linear regression is used to model global regulatory relationships between all TFs and miRNAs and genome-wide mRNA expression and to predict change of activity (importance) of each miRNA and TF, conditional on all the other regulators to remove redundant effects, as illustrated for miRNA2. The color codes for miRNAs and TFs are consistent with (A). An ellipse denotes the downstream regulatory effect of a miRNA or a TF. The blank overlapping region denotes the redundant effect shared among miRNAs and TFs; the colored crosses in each ellipse represent the non-redundant regulatory effect for the corresponding miRNA or TF. Each regression coefficient reflects the importance (change of activity) of an miRNA or a TF between SZ and controls NPCs. C-D. Top five miRNAs (C) and top five TFs (D) with change of activities predicted by the model. The dark gray bars mark miRNAs/TFs with Bonferroni corrected p-value <0.05; the stars mark the miRNAs/TFs whose change of activities are also significantly correlated with their expression. E-F. Scatter plots showing relationship between expression and predicted change of activities for miRNA-9 (E) and miR-137 (F) in control and SZ NPCs. G-H. Scatter plots show no relationship between expression and predicted change of activities for miRNA-9 (G) and miR-137 (H) in control and SZ hiPSC neurons.
Several miRNAs and TFs with significant activity changes were discovered (Fig. 2C,D, SI Tables 5-6). miR-9 had the greatest activity z-score (3.96; Bonferroni corrected p=0.0032) (Fig. 2C, Table 1). For each of the top five miRNAs (Fig. 2C; SI Fig. 3A-F) and top five TFs identified (Fig. 2D; SI Fig. 3G-K), we correlated RNAseq-derived sample-specific activity (Methods) to expression levels of miRNAs (from Nanostring nCounter) and TFs (from RNAseq) in matched samples. Overall, the most significant correlation between miRNA activity and miRNA expression (r=−0.97, p=1.54e−07) was observed for miR-9 (Fig. 2E). In contrast, miR-137 did not show significant activity change (z=1.38; p=0.08, before Bonferroni correction) and showed only a modest correlation between miRNA expression and activity (r=0.65, p=0.02) (Fig. 2F). REST, a target of miR-9, was observed to have significant activity change amongst TFs (SI Fig. 3H).
Collectively, multiple analyses suggest that miR-9 may contribute to differential gene expression between SZ and control NPCs. Importantly, our SZ cohort was not selected on the basis of genetic variants at the miR-9 locus; moreover, we identified no broadly shared haplotype in any of the miR-9 loci that would explain these consistent results (see SI Experimental Procedures for subject rs181900 genotypes). Previously reported CNV genotyping did not detect any specific genetic changes at the miR-9 locus that might be predicted to contribute to SZ risk (Brennand et al., 2011).
Decreased miR-9 activity is specific to SZ NPCs and not detected in SZ hiPSC neurons
When our global integrative analysis was extended to neurons, we observed no substantial miR-9 activity (miR-9 z=2.40, Bonferroni corrected p=0.70; miR-137 z=1.40, Bonferroni corrected p=1) or correlation between activity and expression (miR-9 r=0.036, p=0.92; miR-137 r=0.64, p=0.03) in control and SZ hiPSC neurons (Fig. 2G,H; SI Tables 5-6). Nanostring (Fig. 1B, Table 2, SI Table 1) and qPCR (Fig. 1C) analysis confirmed that aberrant miR-9 levels were restricted to SZ NPCs as no significant differences in miR-9 levels were observed between SZ and control hiPSC 6-week-old neurons. Temporal expression profiling of miR-9 expression throughout neuronal differentiation across all four patients and six controls demonstrated significantly decreased miR-9 levels in SZ NPCs and 1-and 2-week-old neurons, which normalized as miR-9 expression increased to control levels over extended neuronal differentiation (Fig. 1C; SI Fig. 1), suggesting that the significantly lower expression level of miR-9 in SZ NPCs may not persist in the adult human brain.
Decreased miR-9 regulatory activity correlates to reduced migration in SZ NPCs
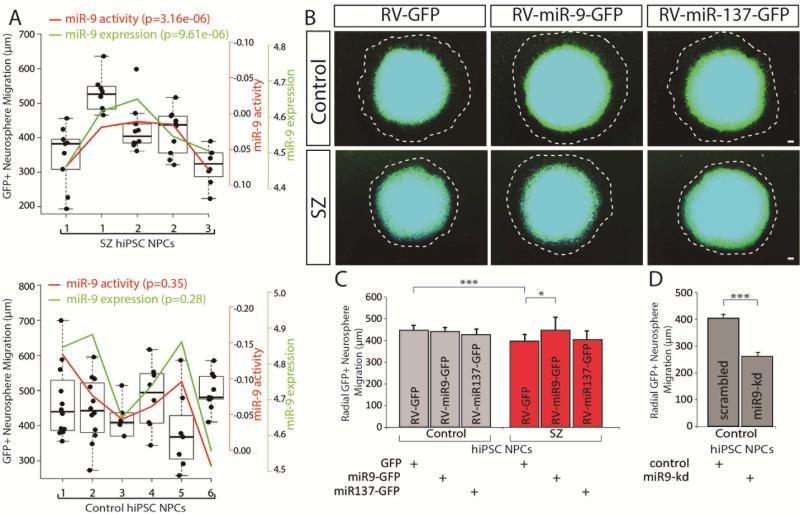
As stated above, miR-9 regulates neurogenesis and neuronal maturation in vivo (Shibata et al., 2011; Zhao et al., 2009) and plays a role in neural migration in vitro as well (Delaloy et al., 2010). Using neurosphere outgrowth as an assay for neural migration, we have previously shown migration deficits in SZ NPCs (Brennand et al., 2015; Lee et al., 2015) and postulated that decreased miR-9 levels may contribute to this phenotype. We observed a significant positive correlation between radial migration and both endogenous miR-9 activity (p=3.16e−06, nested ANOVA) and endogenous miR-9 expression (p=9.61e−06, nested ANOVA) in SZ NPC neurospheres (Fig. 3A).
Fig. 3. Aberrant migration in SZ NPCs rescued by restoration of miR-9 levels.
A. Correlation between miR-9 level (both endogenous expression and regulatory activity inferred from RNAseq data) and radial neurosphere migration in control and SZ NPCs. B. Representative florescent images of hiPSC forebrain NPC neurosphere outgrowth assay, following stable transduction with RV-GFP, RV-miR-9-GFP or RV-miR137-GFP. The average distance between the radius of the inner neurosphere (dense aggregate of nuclei) and outer circumference of cells (white dashed line) was calculated. DAPI-stained nuclei (blue). Scale bar 100 m. C. Radial neurosphere migration by control and SZ NPCs, following stable transduction with RV-GFP, RV-miR-9-GFP or RV-miR137-GFP. D. Radial neurosphere migration by control and SZ NPCs, following transient reduction of miR-9 levels. Error bars are s.e., *P < 0.05, **P < 0.01, ***P < 0.001.
Modulating miR-9 levels affects neural migration
Using a previously characterized retrovirus, RV-MDH-GFP-miR-9 (De Bosscher et al., 2010) hereafter referred to as RV-miR-9-GFP, we increased levels of active miR-9 in control and SZ NPCs (SI Fig. 4A). Comparison of 1,539 neurospheres (375 RV-GFP control, 287 RV-miR-9-GFP control, 416 RV-GFP SZ and 328 RV-miR-9-GFP SZ) revealed that overexpression of miR-9 in SZ NPCs, but not control NPCs, significantly increased total radial migration (p<0.0399) (Fig. 3B,C; SI Fig. 5; see SI Experimental Procedures for quantification of the total number of neurospheres analyzed in each assay). In addition to miR-9, we manipulated a second miRNA, miR-137. The miR-137 locus has a well-established and significant genome wide signal associated with SZ (Ripke et al., 2013; Ripke et al., 2011; Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014) and is brain-enriched with the ability to modulate neural stem cell proliferation and neural differentiation (Smrt et al., 2010; Sun et al., 2011). miR-137 was not significantly differentially expressed between control and SZ NPCs (Fig. 1D) or neurons (Fig. 1E). Moreover, overexpression of miR-137 using an engineered retrovirus MDH-GFP-miR-137 vector (RV-miR-137-GFP) (SI Fig. 4B) had no apparent effect on the migration of neurospheres derived from SZ patients (p=0.72) (Fig. 3B-C) (132 RV-GFP control, 133 RV-miR-137-GFP control, 151 RV-GFP SZ and 151 RV-miR-137-GFP SZ). Both miR-9 and miR-137 also regulate the replication of NPCs (reviewed in (Meza-Sosa et al., 2014)); however, we observed only a small insignificant increase in proliferation with RV-miR-9-GFP or RV-miR-137-GFP transduction in either control or SZ hiPSC NPCs using two independent assays (SI Fig. 6A-C). Additionally, transient knockdown of miR-9 using locked nucleic acid (LNA) probes in control NPCs significantly decreased total radial migration (166 scrambled control and 154 miR-9-knockdown control) (p<0.0009) (Fig. 3D; SI Fig. 5; see SI Experimental Procedures for quantification of the total number of neurospheres analyzed in each assay).
Effect of manipulating miR-9 levels on global gene expression
When we first attempted to quantify changes in ten empirically validated miR-9 target genes (see SI Experimental Procedures) in control and SZ NPCs stably and efficiently transduced with RV-GFP or RV-miR-9-GFP, we observed that persistent overexpression of miR-9 did not lead to significantly decreased expression of any of these well-characterized candidate miR-9 target genes (SI Fig. 6E, SI Tables 7-8). However, we did detect ameliorated expression of NRXN1 (p=0.0092), NRXN2 (p=0.0200) and NCAM1 (p=0.0541) (SI Fig. 6D, SI Tables 7-8).
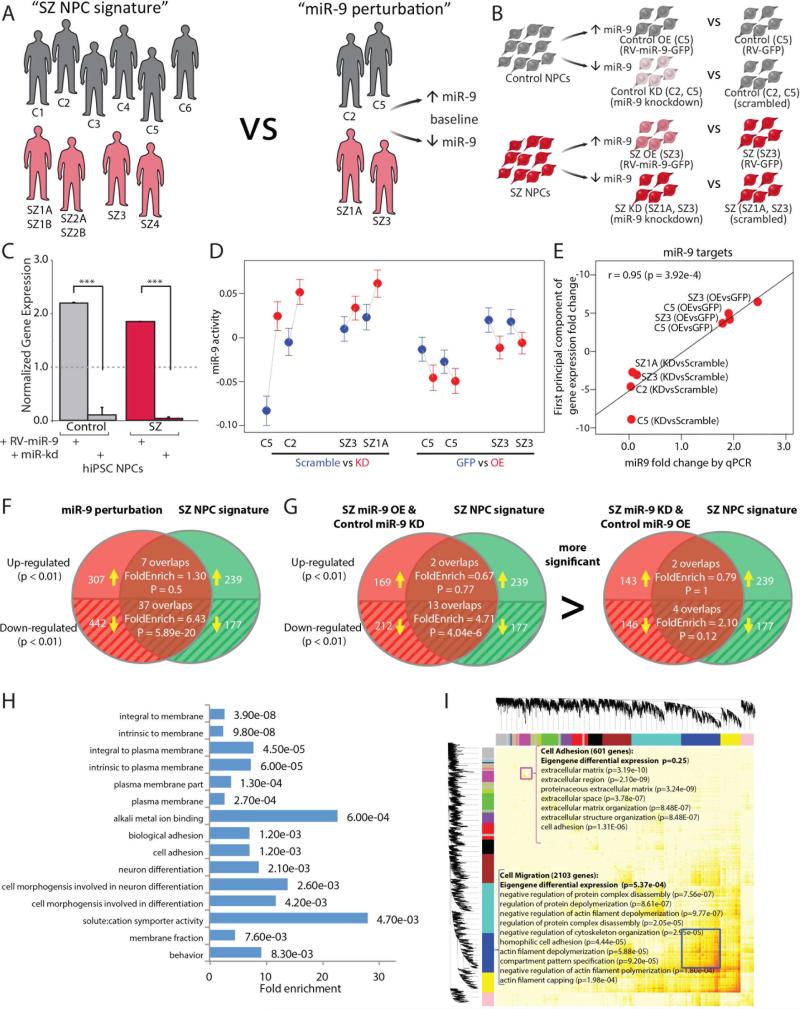
To more broadly investigate the effect of manipulating miR-9 levels on miR-9 regulatory activity, we performed transcriptomic profiling to examine global gene expression changes following RV-overexpression of miR-9 in control and SZ NPCs (Fig. 4A,B). Biological duplicates of passage-matched NPCs from 1 control (female) and 1 SZ patient (female) were transduced with either RV-GFP or RV-miR-9-GFP; GFP-positive NPCs were purified by fluorescent activated cell sorting (FACS) (SI Fig. 4C) and expanded for two passages. In parallel, passage-matched NPCs from 2 controls (1 male, 1 female) and 2 SZ patients (1 male, 1 female) were transiently transfected with either scrambled or miR-9 LNA probes. In both instances, miR-9 perturbation was confirmed by qPCR (Fig. 4C). Because RV-overexpression was stable and persistent, analyzed after FACS purification and expansion of stable lines, this experiment most likely captured indirect miR-9 targets. Conversely, as miR-9 knockdown was transient and samples were collected within 48 hours, this experiment most likely better reflected direct miR-9 targets. Although RNAseq analysis revealed large inter-individual heterogeneity, we were able to resolve several functional consistencies in the effects of our miR-9 perturbations: i) the change in miR-9 activity was consistent with the inhibitory role of miR-9 (Fig. 4D), ii) the gene expression fold-change of miR-9 target genes (between each perturbation and its corresponding control, summarized by the first principal component; see Methods) was correlated (r=0.95, p=3.92e−04) with miR-9 fold change (Fig. 4E) and iii) the differentially expressed (DE; p <0.01) gene list resulting from miR-9 perturbation (paired t-test; see Methods) was enriched for miR-9 targets (1.53-fold, p=1.2e−5).
Fig. 4. Effect of manipulating miR-9 levels on global gene expression.
A. Schematic demonstrating the integration of RNAseq datasets for “SZ NPC signature” (6 controls; 4 SZ patients) with “miR-9 perturbation” (2 controls, 2 SZ patients). B. Experimental design for comparing the effects of stable miR-9 overexpression and transient miR-9 knockdown in control and SZ NPCs. C. qPCR validation of RV-miR-9 overexpression and transient miR-9 knockdown in NPCs. D. miR-9 activity for each sample in the perturbation dataset, inferred from RNAseq data. E. Correlation between miR-9 perturbation fold-change (validated by qPCR) and miR-9 target gene fold-change (first principal component; see Methods). F. Overlap between DE genes in the “miR-9 perturbation” dataset and the “SZ NPC signature” RNAseq datasets. G. Overlap between DE genes in two subsets of “miR-9 perturbation” dataset and the “SZ NPC signature” RNAseq datasets. H. DAVID Gene Ontology analysis for the genes significantly differentially expressed in the similar direction between “miR-9 perturbation” and “SZ NPC signature” datasets. I. WGCNA for the “miR-9 perturbation” RNAseq dataset identified 17 modules. Error bars are s.e., *P < 0.05, **P < 0.01, ***P < 0.001.
We integrated the miR-9 perturbation RNAseq data with our existing RNAseq datasets contrasting control and SZ hiPSC NPC expression from our cohort 1 (six controls, four patients), to ask whether there was any relationship between the “SZ NPC signature” and “miR-9 perturbation” datasets (Fig. 4A; SI Experimental Procedures). Indeed, we observed that the DE (p-value <0.01) in “SZ NPC signature” is enriched for DE (fdr<0.01) in “miR-9 perturbation” (the overall enrichment is 2.31-fold (p=9.39e−09)) (Fig. 4F); there is significant correlation between DE fold-change in these two datasets (overall genes r=0.188; p<10e−50). The significance observed in the above correlation and enrichment analysis between our integrated datasets suggests that perturbation of miR-9 partially contributes to the difference between SZ and control NPCs. Among the genes that showed significant changes in the upward direction in both “miR-9 perturbation” and “SZ NPC signature” DE, only three (MTHFD2, MYO1C, SDC1) are putative miR-9 target genes supported by both TargetScan 6.0 (Friedman et al., 2009) and a published starBase CLIP-Seq dataset (Yang et al., 2011), suggesting that few established miR-9 targets are directly being up-regulated in low miR-9 conditions (SI Table 9).
To gain further mechanistic insight into the migration phenotype under “miR-9 perturbation” we next considered indirect targets. First, we observed that the enrichment of DE genes between the “SZ NPC signature” and “miR-9 perturbation” datasets is focused in the downward direction in low miR-9 conditions (SZ/control and miR-9-knockdown/miR-9-overexpression; 6.43-fold enrichment, p=5.89e−20), indicating that these are not direct miR-9 targets, which would be expected to be up-regulated by miR-9 knockdown (Fig. 4F). Second, we found that this enrichment was driven by a subset of the “miR-9 perturbation” dataset: SZ versus SZ-miR-9-overexpression and control-miR-9-knockdown versus control (Fig. 4G); this is consistent with the observed changes in migration when manipulating miR-9 levels (Fig. 3C,D). Third, we identified 37 common genes within the DE of both “miR-9 perturbation” and “SZ NPC signature” (SI Table 10), putative indirect targets of miR-9. DAVID GO enrichment analysis of this overlapping gene list showed enrichment for plausible migration-associated categories, such as plasma membrane (p-value 4.5e−05), cell morphogenesis (p-value 2.6e−03) and biological adhesion (p-value 1.2e−03) pathways (Fig. 4H). Last but not least, functional modular analysis by weighted gene coexpression network analysis (WGCNA) (Zhang and Horvath, 2005) identified 17 modules among which two were enriched for cell adhesion, extracellular matrix and membrane part GO terms (Fig. 4I; SI Fig. 7). Collectively, these analyses reinforce the functional relevance of miR-9 to our observed amelioration of SZ NPC migration and suggest that this cellular phenotype is mediated by a large network of genes that are regulated by miR-9. These data also suggest that miR-9 overexpression in SZ NPCs shifts towards a more control-like NPC signature and miR-9 knockdown in control NPCs shifts towards a more SZ-like NPC signature, at least with respect to migration-related genes and modules.
Because over-expression of miR-9 in SZ NPCs restored only a subset of SZ DEGs, predominantly annotated as migration and adhesion related genes, it is important to stress that manipulating miR-9 is not sufficient to achieve a full rescue of the molecular phenotype. Rather, we posit that miR-9 may be one (of the many) post-transcriptional regulator(s) of genes that are perturbed in SZ NPCs.
Effect of stable miR-9 overexpression on the proteome
Though we acknowledge that many miRNAs may act through mRNA-destabilization rather than translational-repression (Guo et al., 2010), we nevertheless conducted an unbiased global proteomic comparison following RV-miR-9 overexpression in control and SZ NPCs, in order to examine whether migration-related DE changes identified from the RNAseq data could be detected at the protein level. We used label-free mass spectrometry (LC MS/MS) for a quantitative comparison of protein levels across 90 samples, identifying 2562 proteins across control (one NPC line each derived from 6 controls) and SZ NPCs (two NPC lines each derived from 4 SZ patients) stably transduced with either RV-GFP or RV-miR-9-GFP (GFP-positive NPCs were not FACS-purified). 51 proteins were significantly (p<0.05) altered in control miR-9 overexpression NPCs (SI Table 11) and 45 in SZ miR-9 overexpression NPCs (SI Table 11), though none remained significant following FDR correction. In both cases, the most perturbed DAVID (http://david.abcc.ncifcrf.gov) pathways included actin cytoskeleton, protein localization and RNA processing (SI Tables 12-14).
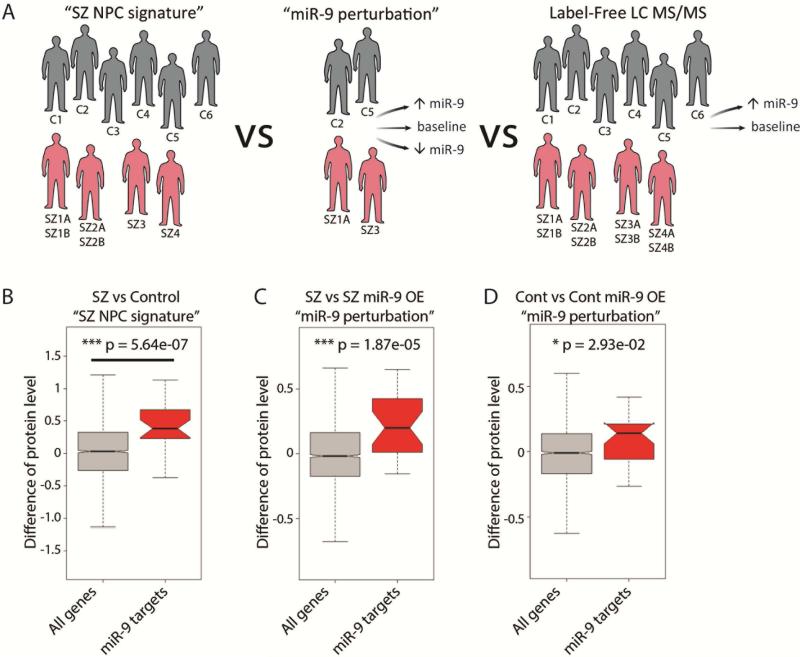
We integrated both the “SZ NPC signature” and “miR-9 perturbation” datasets with our LC MS/MS proteomic data (Fig. 5A) to further explore the effects of miR-9 overexpression on indirect miR-9 target genes. First, we observed that proteomic changes were enriched for putative miR-9 targets in each of the “SZ NPC signature” (p=5.64e−07) (Fig. 5B), SZ miR-9 overexpression (p=1.87e−05) (Fig. 5C) and control miR-9 overexpression (p=2.93e−02) (Fig. 5D) RNAseq datasets (SI Table 9). Here, the putative miR-9 targets are referred to as the predicted miR-9 targets (based on sequence information from TargetScan (Friedman et al., 2009), and further filtered by the correlation (negative; p<0.01) between miR-9 and gene expression from our RNAseq data (SZ vs. control hiPSC NPCs). Second, we explored whether the 37 indirect miR-9 target genes identified in the “miR-9 perturbation” dataset also had altered protein levels following miR-9 overexpression; only 9 of these 37 genes were detected, of which 5 were also changed at the protein level, confirming their potential relevance (SI Table 10).
Fig. 5. Effect of manipulating miR-9 levels on proteome.
A. Schematic demonstrating integration of “SZ NPC signature” (6 controls; 4 SZ patients) with “miR-9 perturbation” (2 controls, 2 SZ patients) and “Label-free LC MS/MS” (6 controls; 4 SZ patients) B-D. Putative miR-9 targets enriched for proteomic changes in “SZ NPC signature.” The direction of changes were also consistent with the RNAseq data: significant in both comparisons of SZ/Control (B) and SZ+RV-GFP/SZ+RV-miR-9 (C), and modest significant in comparison of control+RV-GFP/control+RV-miR-9 (D).
Overall, our global RNAseq and proteomic analyses of miR-9 perturbations suggest that small changes in direct and indirect miR-9 targets may occur as a result of reduced miR-9 levels in a subset of SZ NPCs.
Genetic association between miR-9 and SZ
Pre-miR-9 can be transcribed from chromosomes 1 (pre-miR-9-1), 5 (pre-miR-9-2), and 15 (pre-miR-9-3); the mature miRNA sequence generated from all three loci is identical. Although our expression analyses did not distinguish between these immature forms, only pre-miR-9-2 is expressed in neural stem cells differentiated from hiPSCs (Delaloy et al., 2010) and neither miR-9-1 or miR-9-3 show robust expression in the developing human brain; miR-9-2 expression peaks by 16 weeks post conception (Miller et al., 2014).
Based on a recent GWAS (Hauberg et al., 2016; Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014), miR-9-2 is near rs181900 (within the r2=0.6 linkage disequilibrium region), a SNP approaching genome-wide significance (p=7.1×10−8) (Hauberg et al., 2016). Moreover, a gene-set enrichment analysis using the summary statistics from PGC2 found an enrichment of predicted miR-9 targets amongst SZ-associated genes (Hauberg et al., 2016). This suggests that genetic variants in both miR-9 and its targets are associated with increased risk of SZ. Leveraging our “miR-9 perturbation” dataset, which reflects both direct and indirect miR-9 targets, we performed an integrative competitive gene-set enrichment analysis (Hauberg et al., 2016) (see SI Experimental Procedures) with the PGC2 GWAS summary statistics (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). We observed that the “miR-9 perturbation” DE list was enriched for genes associated with SZ (p=7.46×10−4). Furthermore, focusing specifically on indirect miR-9 targets (those genes in the “miR-9 perturbation” DE list that were not in the TargetScan list of predicted miR-9 targets) also revealed enrichment amongst genes associated with SZ (p=0.0020). Together, both our NPC-derived data and the integrative GWAS analysis support the link between miR-9 and SZ.
DISCUSSION
Using unbiased miRNA expression analysis we report decreased miR-9 levels in a subset of SZ NPCs from two independent SZ patient cohorts (totalling 14 SZ patients and 16 controls) reprogrammed and differentiated independently and using different methodologies. We observed a strong correlation between miR-9 expression and miR-9 regulatory activity in NPCs as well as between miR-9 levels/activity, neural migration and diagnosis; while in comparison to GWAS the sample sizes achievable in hiPSC studies remain small, our findings suggest that aberrant levels and activity of miR-9 may be one of the many factors that contribute to SZ risk, at least in a subset of patients. All SZ NPCs (16 individuals) had reduced miR-9 levels relative to the median value of control NPCs. We further defined a subset (50%) of the SZ NPCs (using the 25% quantile of the control NPCs) that seem to be driving the signal of decreased miR-9 levels. Of course, owing to the sample sizes involved, these findings must necessarily be considered preliminary until they can be confirmed in much larger hiPSC and/or post-mortem cohorts.
Because rs181900 genotype does not explain differences in miR-9 levels and activity in our SZ NPCs (see SI Experimental Procedures for subject rs181900 genotypes), we posit that there may be both miR-9 genotype- and non-genotype-dependent changes in the miR-9 controlled regulatory network that affect SZ risk in a subset of patients; one or more SZ risk factors may act either upstream or downstream of miR-9. If the impact of these genetic variations is indeed during early cortical development, this would be consistent with our inability to detect perturbed miR-9 levels in post-mortem DLPFC obtained from ten SZ patients and ten controls (SI Fig. 1E), as well as array-based miRNA studies of the Stanley Brain Bank, which found no significant difference in miR-9 expression levels in post-mortem cortical tissue from 38 controls and 37 SZ patients (Miller et al., 2012; Perkins et al., 2007).
Interestingly, we found that manipulating miR-9 levels resulted in small changes in the transcript and protein levels of many genes, many of which are not predicted to be direct miR-9 targets. This is consistent with a recent RNAseq dataset generated by over-expressing miR-137 in human neural stem cells (Collins et al., 2014) and other reports that perturbations in miRNAs tend to yield only subtle changes, but in many genes (Guo et al., 2010). The stronger correlation of miR-9 expression, activity and migration in SZ NPCs was unexpected and may imply that the genetic (or epigenetic) background of SZ patients has epistasis with miR-9 activity, reflecting genetic polymorphisms or perturbations in expression networks prevalent in SZ. These findings support a model of an extremely complex genetic architecture underlying SZ risk, one in which miR-9 is just one of many factors contributing to SZ predisposition in a subset of patients. In line with this hypothesis, our gene-set enrichment analyses using the PGC2 GWAS showed that both direct and indirect targets of miR-9 are enriched for SZ risk loci. Given our findings of reduced miR-9 levels in a subset of SZ patients, it is important to consider whether genetic and/or clinical characteristics (such as various research domain criteria (RDoC)) correlate to miR-9 status. More broadly, a future question for investigation will concern the extent to which miR-9 dysregulation occurs in larger hiPSC cohorts generated from SZ, bipolar disorder and autism spectrum disorder patients.
Although hiPSC-based RNAseq studies have small sample sizes, the computational methods used in our study demonstrate that integrative analysis leveraging the rich and increasing collection of miRNA targets and transcription factor targets have great power and specificity to predict regulators. We expect the general computational and integrative methodology presented in this study to have broader impact in studies of other diseases using hiPSCs.
METHODS
Description of SZ patients, hiPSC reprogramming and NPC differentiation
(see SI Experimental Procedures for available clinical information and cell line descriptions)
For cohort #1, patient and control NPCs were differentiated from hiPSCs reprogrammed from fibroblasts obtained from Coriell or ATCC. In total, NPCs from four patients and six controls were compared, as described previously (Brennand et al., 2015; Brennand et al., 2011). A summary of Cohort 1 NPC lines used in each assay can be found in SI Experimental Procedures.
For cohort #2, fibroblast biopsies were obtained from patients with childhood-onset-SZ (COS) and unrelated controls that were recruited as part of a longitudinal study Dr. Judith Rapoport (NIMH). hiPSCs were derived via sendal viral reprogramming methods as described previously (Lee et al., 2015).Cohort 2 COS NPCs were generated in a similar method to Cohort 1 (Brennand et al., 2015; Brennand et al., 2011), although differentiated with dual-SMAD inhibition (Chambers et al., 2009) to improve yield, as described (Topol et al., 2015a); five controls from Cohort 2 were published previously (Lee et al., 2015). NPCs were maintained at high density, grown on Matrigel in NPC media (DMEM/F12, 1x N2, 1x B27-RA (Invitrogen), 1 μg/ml Laminin (Invitrogen) and 20 ng/ml FGF2 (Invitrogen) and split approximately 1:3-1:4 every week with Accutase (Millipore) (Brennand et al., 2011). Detailed description can be found in SI Experimental Procedures.
Gene and miRNA expression analysis
Gene expression analysis was performed on passage-matched NPCs cultured on Matrigel (see SI Experimental Procedures for cell line and passage information). Cells were lysed in RNA BEE (Tel-test, Inc). RNA was chloroform extracted, pelleted with isopropanol, washed with 70% ethanol and resuspended in water. RNA was treated with RQ1 RNase-free DNase (Promega) for 30 minutes at 37°C and then the reaction was inactivated by incubation with EGTA Stop buffer at 65°C for 10 minutes. Total RNA for RNAseq was purified using the miRNeasy Mini Kit (Qiagen).
For RNAseq, samples were prepared and run by the Genomics Core at Icahn School of Medicine at Mount Sinai. The Illumina HiSeq 2500 RNA kit was used for 100nt/single end reads, four samples were run per lane. Raw cDNA reads were aligned to the hg19 reference with the spliced gap aligner STAR, with count-based quantitation carried out via the Subread package featureCounts at both the gene and exon levels for UCSC and ensemble annotation builds. The count data were normalized and modeled as over-dispersed Poisson data using a negative binomial model in the Bioconductor package edgeR (Robinson et al., 2010). Fold changes, p-values and false discovery rates (FDRs) are obtained from the same package for integrative analysis.
For Nanostring miRNA studies, we used nCounter Human v2 miRNA (NS_H_MIR_V2.1) (based on miRBase v18). The public R package ‘NanoStringNorm’ was used to select the best normalization method for Nanostring miRNA expression based on the consistency between technical replicates (Waggott et al., 2012). When combining the two SZ cohorts, we regressed away the batch effect with a simple regression model, and used residuals as the miR9 expression.
Global integrative modeling of miRNAs, TFs and differential gene expression
The regression model is defined as: where fg is the fold change of gene g between two conditions; Mig is the number of binding sites of miRNA i on the 3’ UTR of the gene g; Njg is the number of binding sites of TF j on the promoter of gene g; A, B and C (a constant) can be inferred based on the values of f, M and N for all the genes in the RNAseq data. The z-scores of coefficients Ai and Bj represent the activity changes of miRNA i and TF j, respectively. Global miRNA binding sites represented by 86 miRNA seed families are based on TargetScan 6.0 (Friedman et al., 2009). Global TF binding sites represented by 190 position weighted matrices (PWMs) covering 350 mammalian TFs were based on the union of JASPAR (Sandelin et al., 2004), TRANSFAC (Matys et al., 2003), and additional motifs from ChIP-chip and ChIP-seq data collected by (Balwierz et al., 2014). The initial regression analysis was done using ISMARA (Balwierz et al., 2014) before further integrative analysis with miRNA expression data; sample-specific miRNA/TF activity was estimated by the same regression model where the fold changes were calculated between a single sample and all the samples combined together. These sample-specific miRNA/TF activities were correlated with miRNA/TF expressions for the matched sample as presented in the main text.
Overexpression of miR-9
The human miR-9-3 gene was previously amplified by PCR from normal genomic DNA and cloned into the MDH1–PGK–GFP 2.0 retroviral vector (http://www.addgene.org/25036) (De Bosscher et al., 2010). We similarly amplified and cloned human miR-137 into the MDH1–PGK–GFP 2.0 retroviral vector. High-titer retroviral supernatant was generated by co-transfection of miRNA expression vector together with PUMVC (http://www.addgene.org/8449/) and CMV-VSVG to package retrovirus in HEK-293T cells.
Knockdown of miR-9
hiPSC-derived NPCs were transiently transfected with miRCURY LNA knockdown probes (Exiqon) with Lipofectamine RNAiMAX (Life Technology) in Optimem (ThermoFisher Scientific) for 24 hours. A scrambled miRNA, which bears no homology to any known miRNA or mRNA sequences in human, mouse, and rat, was used as negative control, and hsa-anti-miR-9 (410014-08) (100 nM) was used to specifically inhibit miR-9. 24 hours after transfection, NPCs were dissociated with accutase in order to generate neurospheres. Neurospheres were additionally cultured in miRCURY LNA knockdown probes (Exiqon) with Lipofectamine RNAiMAX (Life Technology) in NPC media.
Neurosphere migration assay
48-hour radial neurosphere migration was assayed as previously described (Brennand et al., 2015; Lee et al., 2015). A quantification of the total number of neurospheres analyzed in each assay can be found in SI Experimental Procedures . Detailed description can be found in SI Experimental Procedures.
LC MS/MS quantitative mass spectrometry
To compare global protein levels in control and SZ NPCs (stably transduced with RV-GFP or RV-miR-9-GFP), we used quantitative label free LC-MS/MS analysis. We injected 2μg of protein from each control (one NPC line each derived from six controls) and SZ (two NPC lines each derived from four SZ patients) hiPSC forebrain NPC line, in triplicate, on a Thermo Q-Exactive mass spectrometer equipped with a Dionex Ultimate 3000 (RSLCnano) chromatography system. Detailed description can be found in SI Experimental Procedures.
Statistical analysis
For phenotypic and qPCR analysis, statistical analysis was performed using JMP (Carey, NC). Box-Cox transformation of raw data was performed to correct non-normal distribution of the data and residuals. Improvements were assessed by Shapiro-Wilk W test of the transformed data and residuals. Means were compared within diagnosis by Oneway analysis using both Student's T test and Tukey Kramer HSD. Finally, a nested analysis of values for individual patients was performed using standard least squares analysis comparing means for all pairs using Student's T test for specific pairs and Tukey Kramer HSD for multiple comparisons. Student's T tests were used to test statistical differences between control and SZ Nanostring and LC MS/MS analyses, with subsequent correction for multiple hypotheses done by Bonferroni and/or Benjamini Hochberg methods, as noted. In all figures, data presented are from a single representative experimental replicate where pooled data represent biological replicate samples within a given experiment: error bars represent standard error. *P < 0.05, **P < 0.01, ***P < 0.001.
Supplementary Material
HIGHLIGHTS.
miR-9 is highly expressed in NPCs and down-regulated in a subset of SZ NPCs.
miR-9 expression level is strongly correlated with miR-9 regulatory activity.
Manipulation of miR-9 impacts neural migration.
miR-9 effects seem to be mediated by small changes in indirect miR-9 targets.
ACKNOWLEDGMENTS
Kristen Brennand is a New York Stem Cell Foundation - Robertson Investigator. The Brennand Laboratory is supported by the Brain and Behavior Research Foundation, NIH grant R01 MH101454 and the New York Stem Cell Foundation. The Fang Laboratory is supported by NIH grants R01 MH097276 and R01 GM114472. The Cotter Laboratory is supported by Health Research Board Clinical Scientist Award. FACS purification was conducted at the ISMMS Flow Cytometry Center Of Research Excellence and RNAseq was conducted at the ISMMS Genomics Core Facility. This work was supported in part through the computational resources and staff expertise provided by the Department of Scientific Computing at the Icahn School of Medicine at Mount Sinai.
ABBREVIATIONS
- SZ
schizophrenia
- hiPSC
human induced pluripotent stem cell
- NPC
neural progenitor cell
- miRNA
microRNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
A.T. contributed to experimental design and completed the NPC experiments. S.Z. analyzed all RNAseq data, Nanostring nCounter data, proteomic data and completed miRNA functional activity modeling; Y-C.W. and H.S. performed RNAseq read alignment. B.J.H differentiated and validated COS and control NPCs, P.A.G. and J.R. contributed COS fibroblasts; C.R. and N.T. assisted with analysis of the replication and neurosphere migration experiments. J.E., G.C. and D.C. completed the mass spectrometry experiments. M.E.H. and M.M. integrated findings into the PGC2 GWAS. A.S. identified the first differences in miR-9 levels in SZ NPCs with K.J.B and F.H.G. Y.H. contributed to qPCR. D.R., J.J. and P.S. genotyped the SZ patients included in this study. B.R. and J.D. designed and performed the drug enrichment analysis. G.F. and K.J.B. designed the experiments, supervised data analysis and wrote the manuscript.
AUTHOR INFORMATION
As per our agreement with Coriell Cell Repository, some hiPSC lines generated from control and SZ fibroblasts will be available from Coriell. Additionally, all control, SZ and COS hiPSCs are currently being deposited with the NIMH Center For Collaborative Studies Of Mental Disorders At RUCDR.
The authors have declared that no competing interests exist.
REFERENCES
- Balwierz PJ, Pachkov M, Arnold P, Gruber AJ, Zavolan M, van Nimwegen E. ISMARA: automated modeling of genomic signals as a democracy of regulatory motifs. Genome research. 2014;24:869–884. doi: 10.1101/gr.169508.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand K, Savas JN, Kim Y, Tran N, Simone A, Hashimoto-Torii K, Beaumont KG, Kim HJ, Topol A, Ladran I, et al. Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol Psychiatry. 2015;20:361–368. doi: 10.1038/mp.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, Li Y, Mu Y, Chen G, Yu D, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011 doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins AL, Kim Y, Bloom RJ, Kelada SN, Sethupathy P, Sullivan PF. Transcriptional targets of the schizophrenia risk gene MIR137. Translational psychiatry. 2014;4:e404. doi: 10.1038/tp.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon EM, Liu XS, Lieb JD, Liu JS. Integrating regulatory motif discovery and genome-wide expression analysis. Proceedings of the National Academy of Sciences. 2003;100:3339–3344. doi: 10.1073/pnas.0630591100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bosscher K, Haegeman G, Elewaut D. Targeting inflammation using selective glucocorticoid receptor modulators. Current opinion in pharmacology. 2010;10:497–504. doi: 10.1016/j.coph.2010.04.007. [DOI] [PubMed] [Google Scholar]
- Delaloy C, Liu L, Lee JA, Su H, Shen F, Yang GY, Young WL, Ivey KN, Gao FB. MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell. 2010;6:323–335. doi: 10.1016/j.stem.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome research. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulsuner S, Walsh T, Watts AC, Lee MK, Thornton AM, Casadei S, Rippey C, Shahin H, Nimgaonkar VL, Go RC, et al. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–529. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauberg ME, Roussos P, Grove J, Borglum AD, Mattheisen M, Schizophrenia Working Group of the Psychiatric Genomics, C. Analyzing the Role of MicroRNAs in Schizophrenia in the Context of Common Genetic Risk Variants. JAMA Psychiatry. 2016 doi: 10.1001/jamapsychiatry.2015.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarskog LF, Miyamoto S, Lieberman JA. Schizophrenia: new pathological insights and therapies. Annu Rev Med. 2007;58:49–61. doi: 10.1146/annurev.med.58.060904.084114. [DOI] [PubMed] [Google Scholar]
- Karayiorgou M, Simon TJ, Gogos JA. 22q11.2 microdeletions: linking DNA structural variation to brain dysfunction and schizophrenia. Nat Rev Neurosci. 2010;11:402–416. doi: 10.1038/nrn2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IS, Carvalho CMB, Douvaras P, Ho SM, Hartley BJ, Zuccherato LW, Ladran IG, Siegel AJ, McCarthy S, Malhotra D, et al. Characterization of molecular and cellular phenotypes associated with a heterozygous CNTNAP2 deletion using patient-derived hiPSC neural cells. NPJ Schizophrenia. 2015;1:15019. doi: 10.1038/npjschz.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin GN, Corominas R, Lemmens I, Yang X, Tavernier J, Hill DE, Vidal M, Sebat J, Iakoucheva LM. Spatiotemporal 16p11.2 protein network implicates cortical late mid-fetal brain development and KCTD13-Cul3-RhoA pathway in psychiatric diseases. Neuron. 2015;85:742–754. doi: 10.1016/j.neuron.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gößling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV. TRANSFAC®: transcriptional regulation, from patterns to profiles. Nucleic acids research. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meza-Sosa KF, Pedraza-Alva G, Perez-Martinez L. microRNAs: key triggers of neuronal cell fate. Frontiers in cellular neuroscience. 2014;8:175. doi: 10.3389/fncel.2014.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BH, Zeier Z, Xi L, Lanz TA, Deng S, Strathmann J, Willoughby D, Kenny PJ, Elsworth JD, Lawrence MS, et al. MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proc Natl Acad Sci U S A. 2012;109:3125–3130. doi: 10.1073/pnas.1113793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, Parker JS, Jin J, Hammond SM. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome biology. 2007;8:R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport JL, Giedd JN, Gogtay N. Neurodevelopmental model of schizophrenia: update 2012. Mol Psychiatry. 2012;17:1228–1238. doi: 10.1038/mp.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, O'Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S, Bergen SE, Collins AL, Crowley JJ, Fromer M, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, Lin DY, Duan J, Ophoff RA, Andreassen OA, et al. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engström P, Wasserman WW, Lenhard B. JASPAR: an open - access database for eukaryotic transcription factor binding profiles. Nucleic acids research. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty M, Helmy K, Khan AA, Silber J, Arvey A, Neezen F, Agius P, Huse JT, Holland EC, Leslie CS. Inferring transcriptional and microRNA-mediated regulatory programs in glioblastoma. Mol Syst Biol. 2012;8:605. doi: 10.1038/msb.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Nakao H, Kiyonari H, Abe T, Aizawa S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J Neurosci. 2011;31:3407–3422. doi: 10.1523/JNEUROSCI.5085-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrt RD, Szulwach KE, Pfeiffer RL, Li X, Guo W, Pathania M, Teng ZQ, Luo Y, Peng J, Bordey A, et al. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells. 2010;28:1060–1070. doi: 10.1002/stem.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Ye P, Murai K, Lang MF, Li S, Zhang H, Li W, Fu C, Yin J, Wang A, et al. miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat Commun. 2011;2:529. doi: 10.1038/ncomms1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topol A, Tran NN, Brennand KJ. A guide to generating and using hiPSC derived NPCs for the study of neurological diseases. Journal of visualized experiments : JoVE. 2015a:e52495. doi: 10.3791/52495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topol A, Zhu S, Tran N, Simone A, Fang G, Brennand KJ. Altered WNT Signaling in Human Induced Pluripotent Stem Cell Neural Progenitor Cells Derived from Four Schizophrenia Patients. Biol Psychiatry. 2015b doi: 10.1016/j.biopsych.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggott D, Chu K, Yin S, Wouters BG, Liu FF, Boutros PC. NanoStringNorm: an extensible R package for the pre-processing of NanoString mRNA and miRNA data. Bioinformatics. 2012;28:1546–1548. doi: 10.1093/bioinformatics/bts188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger DR. Implications of normal brain development for the pathogenesis of schizophrenia. Arch Gen Psychiatry. 1987;44:660–669. doi: 10.1001/archpsyc.1987.01800190080012. [DOI] [PubMed] [Google Scholar]
- Wong AH, Van Tol HH. Schizophrenia: from phenomenology to neurobiology. Neurosci Biobehav Rev. 2003;27:269–306. doi: 10.1016/s0149-7634(03)00035-6. [DOI] [PubMed] [Google Scholar]
- Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39:D202–209. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu DX, Di Giorgio FP, Yao J, Marchetto MC, Brennand K, Wright R, Mei A, McHenry L, Lisuk D, Grasmick JM, et al. Modeling hippocampal neurogenesis using human pluripotent stem cells. Stem Cell Reports. 2014;2:295–310. doi: 10.1016/j.stemcr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol. 2009;16:365–371. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.