Abstract
The human alphacoronaviruses HCoV-NL63 and HCoV-229E are commonly associated with upper respiratory tract infections (URTI). Information on their molecular epidemiology and evolutionary dynamics in the tropical region of southeast Asia however is limited. Here, we analyzed the phylogenetic, temporal distribution, population history, and clinical manifestations among patients infected with HCoV-NL63 and HCoV-229E. Nasopharyngeal swabs were collected from 2,060 consenting adults presented with acute URTI symptoms in Kuala Lumpur, Malaysia, between 2012 and 2013. The presence of HCoV-NL63 and HCoV-229E was detected using multiplex polymerase chain reaction (PCR). The spike glycoprotein, nucleocapsid, and 1a genes were sequenced for phylogenetic reconstruction and Bayesian coalescent inference. A total of 68/2,060 (3.3%) subjects were positive for human alphacoronavirus; HCoV-NL63 and HCoV-229E were detected in 45 (2.2%) and 23 (1.1%) patients, respectively. A peak in the number of HCoV-NL63 infections was recorded between June and October 2012. Phylogenetic inference revealed that 62.8% of HCoV-NL63 infections belonged to genotype B, 37.2% was genotype C, while all HCoV-229E sequences were clustered within group 4. Molecular dating analysis indicated that the origin of HCoV-NL63 was dated to 1921, before it diverged into genotype A (1975), genotype B (1996), and genotype C (2003). The root of the HCoV-229E tree was dated to 1955, before it diverged into groups 1–4 between the 1970s and 1990s. The study described the seasonality, molecular diversity, and evolutionary dynamics of human alphacoronavirus infections in a tropical region.
Introduction
Human coronaviruses were first reported in the mid-1960s and are known to be associated with acute upper respiratory tract infections (URTI) or the common cold.1–3 According to the International Committee for Taxonomy of Viruses, human coronavirus NL63 (HCoV-NL63) and 229E (HCoV-229E) belong to the alphacoronavirus genus, a member of the Coronaviridae family. Coronaviruses are positive-strand RNA viruses with the largest genome of approximately 27–31 kb in size.4 In previous studies, analysis of the spike (S) glycoprotein, nucleocapsid (N), and 1a genes of HCoV-NL63 and HCoV-229E revealed evidence of genetic recombination, genetic drift, and positive selection events as part of the evolution of the virus.5,6 Phylogenetically, HCoV-NL63 and HCoV-229E are more closely related to each other than to any other human coronavirus.7
HCoV-NL63 and HCoV-229E account for about 5% of all acute URTI,7–9 and in some cases, a small proportion of infections are associated with hospital admission.10,11 URTI symptoms such as cough and sore throat are often observed in patients infected with either HCoV-NL63 or HCoV-229E.12,13 The prevalence of HCoV-NL63 varies from one study to another; however, in most temperate and tropical countries, it appears to peak around September–April, whereas HCoV-229E is usually detected at low rates throughout the year.14–16 In spite of the clinical importance of HCoV infections,17 the prevalence, seasonality, clinical, and phylogenetic characteristics of HCoVs remain mostly unreported from the tropical region of southeast Asia. On the basis of the S, N, and 1a genes of the HCoV-NL63 and HCoV-229E sequences from Malaysia and also worldwide, we describe the genetic history and phylodynamic profiles of both human alphacoronaviruses using a set of phylogenetic tools.
Materials and Methods
Ethics statement.
The study was approved by the University of Malaya Medical Ethics Committee (MEC890.1). Standard, multilingual consent forms permitted by the Medical Ethics Committee were used. Written consent was obtained from all study participants.
Clinical specimens.
A total of 2,060 consenting outpatients who presented with acute URTI symptoms were recruited at the Primary Care Clinic of University Malaya Medical Center in Kuala Lumpur, Malaysia, between March 2012 and February 2013. Demographic data such as age, gender, and ethnicity were acquired before the collection of nasopharyngeal swabs. The severity of the URTI symptoms (sneezing, nasal discharge, nasal congestion, headache, sore throat, voice hoarseness, muscle ache, and cough) was graded according to criteria described earlier.18–21 The nasopharyngeal swabs were transferred to the laboratory in universal transport media and stored at −80°C.
Molecular detection of HCoV-NL63 and HCoV-229E.
Extraction of total nucleic acids from the nasopharyngeal swabs was carried out using the magnetic bead–based protocols applied in the NucliSENS easyMAG automated nucleic acid extraction system (BioMérieux, Marcy I'Etoile, France).22,23 The presence of respiratory viruses in specimens was examined using the xTAG Respiratory Virus Panel FAST multiplex reverse transcriptase polymerase chain reaction (RT-PCR) assay (Luminex Molecular Diagnostics, Toronto, Canada), which can identify HCoV-NL63, HCoV-229E, HCoV-OC43, HCoV-HKU1, and other respiratory viruses and subtypes.24
Genetic analysis of HCoV-NL63 and HCoV-229E.
Gene fragment sequencing of the S (S1 domain), complete N, and partial 1a (nsp3) genes was performed for HCoV-NL63 and HCoV-229E specimens. The S1 is a highly variable receptor-binding domain, whereas the N and nsp3 are conserved regions within the coronavirus genome, and these three regions are therefore efficiently used for genotyping.5,6 Viral RNA was reverse transcribed into complementary DNA (cDNA) using the SuperScript III kit (Invitrogen, Carlsbad, CA) with random hexamers (Applied Biosystems, Carlsbad, CA). The partial S gene (S1 domain) (HCoV-NL63: 1,383 nt [20,413–21,796] and HCoV-229E: 855 nt [20,819–21,674]), complete N gene (HCoV-NL63: 1,133 nt [26,133–27,266] and HCoV-229E: 1,330 nt [25,673–27,003]), and partial 1a (nsp3) gene (HCoV-NL63: 781 nt [5,811–6,592] and HCoV-229E: 766 nt [5,898–6,664]) were amplified through PCR using 10 μM of newly designed or previously published primers listed in Table 1. The PCR mixture (25 μL) contained cDNA, PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 3 mM MgCl, and 0.01% gelatin), 100 μM (each) deoxynucleoside triphosphates, Hi-Spec additive and 4 U/μL BIO-X-ACT Short DNA polymerase (BioLine, London, United Kingdom). The cycling conditions were as follows: initial denaturation at 95°C for 5 minutes followed by 40 cycles of 94°C for 1 minute, 54.5°C for 1 minute, 72°C for 1 minute, and a final extension at 72°C for 10 minutes. PCR reactions were performed in a C1000 Touch automated thermal cycler (Bio-Rad, Hercules, CA). Nested/semi-nested PCR was performed if necessary, under the same cycling conditions at 30 cycles. Purified PCR products were sequenced using the ABI PRISM 3730XL DNA Analyzer (Applied Biosystems). The nucleotide sequences were codon aligned with relevant complete and partial HCoV-NL63 and HCoV-229E reference sequences retrieved from the GenBank.5,6,28–31
Table 1.
Polymerase chain reaction primers for HCoV-NL63 and HCoV-229E
| Target gene | HCoV | Primer | Location* | Sequence (5′–3′) | Reference |
|---|---|---|---|---|---|
| Spike (S) | NL63 | SP1F | 20,390–20,412 | Forward: TGAGTTTGATTAAGAGTGGTAGG | 25 |
| SP2F | 20,397–20,418 | Forward (nested): GATTAAGAGTGGTAGGTTGTTG | 25 | ||
| SP1R | 21,809–21,828 | Reverse: CAAACTGCAAGTGCTCACAC | 25 | ||
| SP2R | 21,797–21,816 | Reverse (nested): GCTCACACTGCAACTTTTCA | 25 | ||
| 229E | LPS1 | 20,732–20,751 | Forward: AATAATTGGTTCCTTCTAAC | 26 | |
| JH1 | 20,797–20,818 | Forward (nested): TTTGTTGCTTAATTGCTTATGG | 26 | ||
| LPR | 21,710–21,728 | Reverse: AACATACACTGCCAAATTT | This study | ||
| JH2 | 21,675–21,694 | Reverse (nested): TTTGCCAAAAGAAAAAGGGC | 26 | ||
| Nucleocapsid (N) | NL63 and 229E | αN-F | 26,102–26,127 (NL63) | Forward: ARRTTGCTTCATTTWWTCTAA | This study |
| 25,652–25,672 (229E) | |||||
| αN-Fn | 26,112–26,132 | Forward (nested): ATTTWWTCTAAACTAAACRAA | This study | ||
| NL63 | NL-NR | 27,278–27,299 | Reverse: ATAATAAACAKTCAACTGGAAT | This study | |
| NL-NRn | 27,267–27,287 | Reverse (nested): CAACTGGAATTACAAAACAAT | This study | ||
| 229E | E-NR | 27,046–27,063 | Reverse: GATCCTTGTCAAGCCAAA | This study | |
| E-NRn | 26,882–26,900 | Reverse (nested): AAAATTCCAACTAAAGCCT | This study | ||
| 1a | NL63 | SS5852-5Pf | 5,778–5,798 | Forward: CTTTTGATAACGGTCACTATG | 27 |
| P3E2-5Pf | 5,789–5,810 | Forward (semi-nested): GGTCACTATGTAGTTTATGATG | 27 | ||
| NL-1aR | 6,593–6,616 | Reverse: CTCATTACATAAAACATCRAACGG | This study | ||
| 229E | E-1aF | 5,865–5,585 | Forward: CTGTTGAYAAAGGTCATTATA | This study | |
| E-1aFn | 5,876–5,897 | Forward (semi-nested): GGTCATTATACTGTTTATGAYA | This study | ||
| E-1aR | 6,665–6,688 | Reverse: TTCATCACAAATAACATCAAATGG | This study |
Maximum clade credibility (MCC) trees for the partial S (S1 domain), complete N, and partial 1a (nsp3) genes were reconstructed in BEAST (version 1.7).32 MCC trees were produced using a relaxed molecular clock, assuming uncorrelated lognormal distribution under the general time-reversible nucleotide substitution model with a proportion of invariant sites (GTR+I) and a constant coalescent/exponential tree model. The Markov chain Monte Carlo run was set at 6 × 106 steps long sampled every 10,000 state. The trees were annotated using Tree Annotator program included in the BEAST package, after a 10% burn-in, and visualized in FigTree (version 1.3.1).33 The evolutionary history and divergence time (in calendar year) for the HCoV-NL63 and HCoV-229E genotypes were also assessed. The mean divergence time and the 95% highest posterior density regions were evaluated. The best-fitting model was determined by the Bayes factor using marginal likelihood analysis implemented in Tracer (version 1.5).32 The substitution rate of 3.3 × 10−4 substitutions/site/year for the S gene of human alphacoronavirus estimated previously was used for the divergence time inference.5
Maximum likelihood (ML) phylogenetic trees were also reconstructed for the three regions in the phylogenetic analysis using parsimony (PAUP 4.0) software,34 with a Hasegawa–Kishino–Yano nucleotide substitution model plus discrete gamma categories. The statistical robustness and reliability of the branching orders were evaluated by a bootstrap analysis of 1,000 replicates. To investigate the genetic relatedness among the HCoV-NL63 and HCoV-229E genotypes, inter-genotype pairwise nucleotide distances were estimated for the S gene using MEGA 5.1.35 Such analysis was not implemented for the N and 1a genes due to their high genetic invariability across HCoV-NL63 and HCoV-229E genotypes.5,6
Statistical analysis.
All categorical variables were analyzed using the two-tailed Fisher's exact test/χ2 test by the Statistical Package for the Social Sciences (release 16.0; IBM Corp., Chicago, IL). P values < 0.05 were considered significant.
Nucleotide sequences.
HCoV-NL63 and HCoV-229E nucleotide sequences produced in the study have been deposited in GenBank under the accession nos. KT359730-KT359913.
Results and Discussion
Detection of HCoV-NL63 and HCoV-229E in nasopharyngeal swabs.
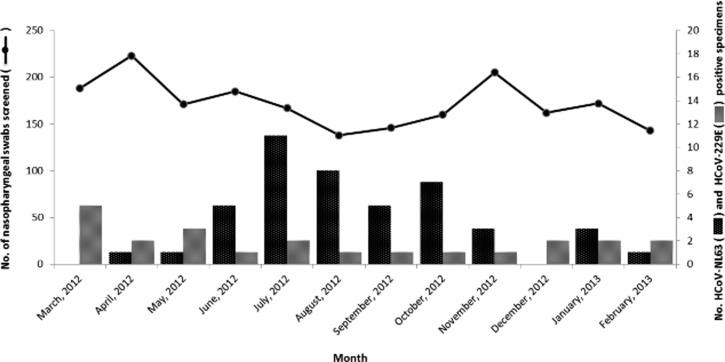
In the current cross-sectional study, a total of 2,060 nasopharyngeal swab specimens collected from Kuala Lumpur, Malaysia, throughout a 12-month study period (March 2012 to February 2013), were screened for the presence of HCoV-NL63 and HCoV-229E using the multiplex RT-PCR method, as an alternative approach to other detection methods such as cell culture.36 Human alphacoronavirus was identified in 68 (3.3%) subjects; HCoV-NL63 and HCoV-229E were detected in 45/2,060 (2.2%) and 23/2,060 (1.1%) patients, respectively. These findings are consistent with the global average prevalence of human alphacoronavirus, which ranges between 1% and 10%, with HCoV-229E generally detected at lower rates than HCoV-NL63.8–10,27,37–40 In contrast to an earlier study,41 no coinfection of alpha- and betacoronavirus (HCoV-OC43 and HCoV-HKU1) was observed within an individual. Age, gender, and ethnicity of the patients were summarized in Table 2. A peak in the number of HCoV-NL63 infections was recorded for the period between June and October 2012, although the number of patients with URTI symptoms screened during those months was relatively low (Figure 1 ). This pattern of virus prevalence corroborates with that observed in neighboring country Thailand, in which a peak of HCoV-NL63 incidence was recorded in September.14 In contrast, studies from temperate regions commonly reported a higher prevalence of HCoV-NL63 during winter seasons.7–9,42 However, the number of HCoV-229E infections detected in Malaysia was low, with no significant peak observed throughout the year, similar to other studies reported worldwide.14,38,43 It is important to note that the study was performed in a relatively short duration, therefore limiting the epidemiological and disease trend comparison with reports from other countries.
Table 2.
Demographic data of 68 adult outpatients infected with human alphacoronavirus in Kuala Lumpur, Malaysia, 2012–2013
| Factor | HCoV-NL63 (N = 45) | HCoV-229E (N = 23) | P value |
|---|---|---|---|
| Gender | 0.80 | ||
| Male | 25 (55.6%) | 12 (52.2%) | |
| Female | 20 (44.4%) | 11 (47.8%) | |
| Age | 0.45 | ||
| < 40 | 13 (28.9%) | 7 (30.4%) | |
| 40–60 | 10 (22.2%) | 8 (34.8%) | |
| > 60 | 22 (48.9%) | 8 (34.8%) | |
| Symptoms | 0.99 | ||
| Sneezing | 42 (93.3%) | 20 (87.0%) | |
| Nasal discharge | 38 (84.4%) | 19 (82.6%) | |
| Nasal congestion | 29 (64.4%) | 15 (65.2%) | |
| Headache | 23 (51.1%) | 13 (56.5%) | |
| Sore throat | 32 (68.9%) | 14 (60.9%) | |
| Hoarseness of voice | 35 (77.8%) | 15 (65.2%) | |
| Muscle ache | 27 (60.0%) | 16 (69.6%) | |
| Cough | 43 (95.6%) | 20 (87.0%) | |
| Ethnicity | 0.08 | ||
| Malay | 11 (24.5%) | 5 (21.8%) | |
| Chinese | 24 (53.3%) | 7 (30.4%) | |
| Indian | 10 (22.2%) | 11 (47.8%) | |
Figure 1.
Annual distribution of HCoV-NL63 and HCoV-229E among adults with acute upper respiratory tract infections in Kuala Lumpur, Malaysia. The total number of nasopharyngeal swabs screened and the monthly distribution of HCoV-NL63 and HCoV-229E between March 2012 and February 2013 were presented.
Phylogenetic analysis of the S, N, and 1a genes.
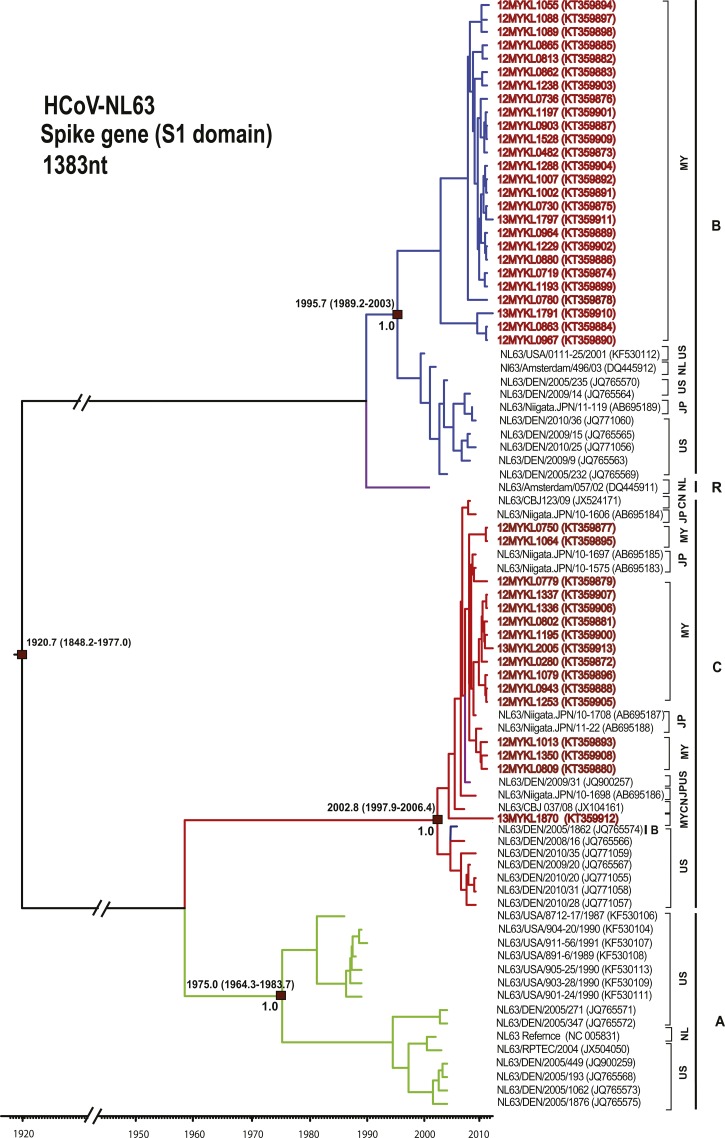
A total of 42/45 (93.3%) partial S (S1 domain) and 43/45 (95.6%) of each complete N and partial 1a (nsp3) genes were successfully sequenced from HCoV-NL63 specimens. Amplification of these genes was difficult for two xTAG-positive HCoV-NL63 specimens, possibly due to their low viral copy number. Phylogenetic analysis of HCoV-NL63 (Figure 2 and Supplemental Figure 1) showed that 27 subjects (27/43, 62.8%) in the study belonged to genotype B (supported by a posterior probability of 1.0 and bootstrap value of 100% at the internal nodes of the MCC and ML trees of the S gene, respectively, with an intra-group pairwise genetic distance of 0.6% ± 0.1%) together with previously reported sequences from the United States, Europe, and Asia.5,25,28,29 Another 16 subjects (16/43, 37.2%) were found to be grouped under genotype C (supported by a posterior probability of 1.0 and bootstrap value of 67% at the internal nodes of the MCC and ML trees of the S gene, respectively, with an intra-group pairwise genetic distance of 0.2% ± 0.1%) with recently described global sequences.25,28,29 Discordance in phylogenetic clustering among the S, N, and 1a genes of the HCoV-NL63 Malaysian sequences had been observed (Supplemental Figure 1). On the basis of the S (S1domain) gene analysis, 26 Malaysian strains (26/42; 61.9%) belong to genotype B while another 16 Malaysian strains (16/42; 38.1%) were classified within genotype C. In contrast, sequences of the three HCoV-NL63 genotypes (A, B, and C) appear to be intermingled in the N and 1a phylogenetic trees. Such discordance was similarly reported in earlier studies where it was confirmed that such phylogenetic pattern was resulted from multiple recombination events along the HCoV-NL63 genome, in addition to the fact that the S1 region sequenced in this study is considered the most variable along the genome, while the N and 1a (nsp3) genes are too conserved.5 To estimate the genetic diversity between HCoV-NL63 genotypes A, B, and C, inter-genotype pairwise genetic distance was assessed for the S gene (Table 3). Genetic distances between genotypes A versus B and B versus C were high (more than 5.0%), compared with that between genotypes A versus C, which was at 2.1%. This is consistent with the phylogenetic tree topology in which genotypes A and C were more closely related and probably shared a common ancestor.
Figure 2.
Maximum clade credibility tree of HCoV-NL63. Spike gene (S1 domain) sequences (1,383 nucleotides) were analyzed under the relaxed molecular clock with a GTR+I substitution model and a constant size coalescent model implemented in BEAST. Posterior probability values and the estimation of the time of the most recent common ancestors with 95% highest posterior density were indicated on major nodes. The HCoV-NL63 sequences obtained in this study were color coded and HCoV-NL63 genotypes A–C were indicated; green = genotype A, blue = genotype B, and red = genotype C. The recombinant genotype is indicated by purple color. The sampling site for each sequence was indicated by codes for the representation of countries. Country codes are as follows; MY = Malaysia; US = United States; JP = Japan; NL = Netherlands; CN = China.
Table 3.
The genetic diversity among alphacoronavirus genotypes in the spike gene
| HCoV | Genetic distance | ||||
|---|---|---|---|---|---|
| NL63 | genotype A | genotype B | genotype C | ||
| genotype A | - | 0.8† | 0.5 | ||
| genotype B | 7.6* | - | 0.6 | ||
| genotype C | 2.1 | 6.7 | - | ||
| 229E | group 1 | group 2 | group 3 | group 4 | |
| group 1 | - | 0.4 | 0.6 | 0.7 | |
| group 2 | 1.5 | - | 0.3 | 0.4 | |
| group 3 | 2.5 | 1.2 | - | 0.3 | |
| group 4 | 3.5 | 2.6 | 1.5 | - | |
Pairwise genetic distances are expressed in percentage (%) of nucleotide difference.
Standard error estimates of the mean genetic distances are shown in the upper diagonal.
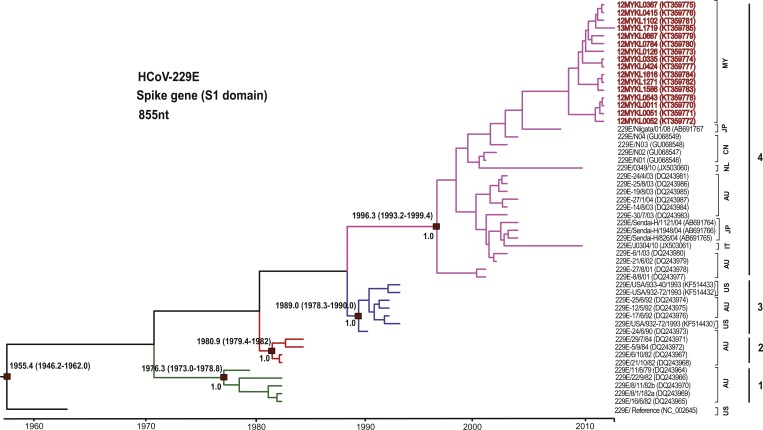
At least one gene (S, N, and/or 1a) was successfully sequenced from 23 positively tested HCoV-229E specimens (16, 18, and 22 of HCoV-229E S, N, and 1a genes, respectively). Phylogenetic analysis revealed that all of the HCoV-229E sequences obtained in this study were classified with group 4, which includes isolates that have been globally circulating since 2001 (Figure 3 and Supplemental Figure 2).6,30,31 The group was supported by a posterior probability of 1.0 and bootstrap value of 100% at the internal nodes of the MCC and ML trees of the S gene, respectively, with an intra-group pairwise genetic distance of 0.3% ± 0.1%. Such phylogenetic data were comparable to those obtained from the N tree, resulted from the hot substitution spots in the S1 and N regions of the HCoV-229E genome.30 The four HCoV-229E groups could not be clearly defined within the 1a gene tree because of the limited number of reference sequences available in the public database (Supplemental Figure 2). Inter-genotype pairwise genetic distance was generally low (below 5.0%) in the S gene among groups 1–4 (Table 3).
Figure 3.
Maximum clade credibility tree of HCoV-229E. Spike gene (S1 domain) sequences (855 nucleotides) were analyzed under the relaxed molecular clock with a GTR+I substitution model and a constant size coalescent model implemented in BEAST. Posterior probability values and the estimation of the time to the most recent common ancestors with 95% highest posterior density were indicated on the major nodes. The HCoV-229E sequences obtained in this study were color coded and the HCoV-229E groups 1–4 were indicated green = genotype 1, red = genotype 2, blue = genotype 3, and purple = genotype 4. The sampling site for each sequence was indicated by codes for the representation of countries. Country codes are as follows; MY = Malaysia; US = United States; JP = Japan; NL = Netherlands; CN = China; AU = Australia; IT = Italy.
Estimation of divergence times.
The molecular clock analysis of HCoV-NL63 and HCoV-229E was performed using the coalescent-based Bayesian relaxed molecular clock under the constant and exponential tree models (Figures 2 and 3). The mean evolutionary rates for the S gene of HCoV-NL63 and HCoV-229E were newly estimated based on the constant tree model at 4.3 × 10−4 (2.3–6.7 × 10−4) and 3.9 × 10−4 (1.3–6.4 × 10−4) substitutions/site/year, respectively. These results were similar to the previously reported substitution rate of the alphacoronavirus S gene (3.3 × 10−4 substitutions/site/year).5 The evolutionary analysis indicated that the time of the most recent common ancestor (tMRCA) of HCoV-NL63 was dated back to the 1920s, while the estimated divergence time of genotype A was dated to 1975, followed by genotype B around 1996 and genotype C in 2003 (Figure 2). Furthermore, the divergence time of HCoV-229E (Figure 3) was estimated around 1955 while the tMRCA of group 1 diverged in 1976, followed by that of group 2 in 1981, group 3 in 1989, and group 4 in 1996. The appearance of groups 1–4 in a timely ordered manner would give strength to the earlier reported hypothesis that positive selection and genetic drift play a major role in the evolution of HCoV-229E.6,30 To the best of our knowledge, this is the first study that reported the divergence times of human alphacoronavirus genotypes. In addition, the most recently reported HCoV-229E strains (between 2001 and 2013) from major parts of the world belong to group 4. In accordance with earlier studies, genotype replacement is evident within HCoV-229E, although sampling bias may also influence the results.6,30 Bayes factor analysis showed insignificant differences (Bayes factor less than 3.0) between the constant and exponential coalescent models of demographic analysis, in which the divergence times estimated using the constant coalescent tree model were similar to those calculated using the exponential model (Supplemental Table 1).
Clinical symptoms assessment.
Clinical findings of the URTI symptoms (sneezing, nasal discharge, nasal congestion, headache, sore throat, hoarseness of voice, muscle ache, and cough) and their severity levels (none, moderate, and severe) were analyzed using the two-tailed Fisher's exact test. The association between symptom severity and HCoV-NL63/HCoV-229E infection was insignificant (P values > 0.05) (Supplemental Table 2). In line with previous clinical studies,10,44,45 the majority of patients infected with HCoV-NL63 and HCoV-229E presented with at least one respiratory symptom that was moderately severe.
In summary, this study provides insight into the phylogeny and evolution of the HCoV-NL63 and HCoV-2293E genotypes. Genetic characterization of human alphacoronavirus isolates currently circulating in Malaysia indicates the circulation of globally prevalent genotypes in the tropical region of southeast Asia. This study has detailed the genetic history of HCoV-NL63 and HCoV-229E genotypes. Since alphacoronavirus evolve through recombination, positive selection, and genetic drift events, continuous molecular surveillance of human alphacoronavirus is warranted to keep track on the evolution of the virus in southeast Asia.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Nyoke Pin Wong, Nur Ezreen Syafina, Farhat A. Avin, Chor Yau Ooi, Sujarita Ramanujam, Nirmala K. Sambandam, Nagammai Thiagarajan, and See Wie Teoh for assistance and support.
Disclaimer: The outcomes and conclusions that this study has reported were drawn by the authors and do not necessarily represent the views or policies of the institutions of affiliation.
Footnotes
Financial support: This work was supported by grants from the Ministry of Education, Malaysia: High Impact Research UM.C/625/1/HIR/MOE/CHAN/02/02 to Kok Keng Tee.
Authors' addresses: Maryam Nabiel Al-Khannaq, Kim Tien Ng, Xiang Yong Oong, Yong Kek Pang, and Adeeba Kamarulzaman, Department of Medicine, Faculty of Medicine, University of Malaya, Kuala Lumpur, Malaysia, E-mails: maryasala@gmail.com, kim_honcho@yahoo.com, oxy123@siswa.um.edu.my, ykpang@ummc.edu.my, and adeeba@ummc.edu.my. Yutaka Takebe, Department of Medicine, Faculty of Medicine, University of Malaya, Kuala Lumpur, Malaysia, AIDS Research Center, National Institute of Infectious Diseases, Toyama, Shinjuku-ku, Tokyo, Japan, and School of Medicine, Yokohama City University, Yokohama, Kanagawa, Japan, E-mail: takebe@niid.go.jp. Jack Bee Chook, Department of Medicine, Faculty of Medicine, University of Malaya, Kuala Lumpur, Malaysia, and Department of Health Sciences, Faculty of Health and Life Sciences, Management and Science University, Selangor, Malaysia, E-mail: jackbee2002@um.edu.my. Nik Sherina Hanafi, Department of Primary Care Medicine, Faculty of Medicine, University of Malaya, Kuala Lumpur, Malaysia, E-mail: sherina@ummc.edu.my. Kok Keng Tee, Department of Medical Microbiology, Faculty of Medicine, University of Malaya, Kuala Lumpur, Malaysia, E-mail: k2tee@um.edu.my.
References
- 1.Tyrrell DAJ, Bynoe ML. Cultivation of a novel type of common-cold virus in organ cultures. Br Med J. 1965;5448:1467–1470. doi: 10.1136/bmj.1.5448.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McIntosh K, Becker WB, Chanock RM. Growth in suckling-mouse brain of “IBV-like” viruses from patients with upper respiratory tract disease. Proc Natl Acad Sci USA. 1967;58:2268–2273. doi: 10.1073/pnas.58.6.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hendley J, Fishburne H, Gwaltney J., Jr Coronavirus infections in working adults. Eight-year study with 229 E and OC 43. Am Rev Respir Dis. 1972;105:805–811. doi: 10.1164/arrd.1972.105.5.805. [DOI] [PubMed] [Google Scholar]
- 4.Lai M, Perlman S, Anderson J. Coronaviridae. In: Knipe D, Howley PM, editors. Fields Virology. 5th edition. Philadelphia, PA: Lippincott Williams and Wilkins; 2006. pp. 1305–1335. [Google Scholar]
- 5.Pyrc K, Dijkman R, Deng L, Jebbink MF, Ross HA, Berkhout B, van der Hoek L. Mosaic structure of human coronavirus NL63, one thousand years of evolution. J Mol Biol. 2006;364:964–973. doi: 10.1016/j.jmb.2006.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chibo D, Birch C. Analysis of human coronavirus 229E spike and nucleoprotein genes demonstrates genetic drift between chronologically distinct strains. J Gen Virol. 2006;87:1203–1208. doi: 10.1099/vir.0.81662-0. [DOI] [PubMed] [Google Scholar]
- 7.Dijkman R, van der Hoek L. Human coronaviruses 229E and NL63: close yet still so far. J Formos Med Assoc. 2009;108:270–279. doi: 10.1016/S0929-6646(09)60066-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerna G, Percivalle E, Sarasini A, Campanini G, Piralla A, Rovida F, Genini E, Marchi A, Baldanti F. Human respiratory coronavirus HKU1 versus other coronavirus infections in Italian hospitalised patients. J Clin Virol. 2007;38:244–250. doi: 10.1016/j.jcv.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vabret A, Dina J, Gouarin S, Petitjean J, Tripey V, Brouard J, Freymuth F. Human (non-severe acute respiratory syndrome) coronavirus infections in hospitalised children in France. J Paediatr Child Health. 2008;44:176–181. doi: 10.1111/j.1440-1754.2007.01246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bastien N, Anderson K, Hart L, Van Caeseele P, Brandt K, Milley D, Hatchette T, Weiss EC, Li Y. Human coronavirus NL63 infection in Canada. J Infect Dis. 2005;191:503–506. doi: 10.1086/426869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Elden LJ, van Loon AM, van Alphen F, Hendriksen KA, Hoepelman AI, van Kraaij MG, Oosterheert JJ, Schipper P, Schuurman R, Nijhuis M. Frequent detection of human coronaviruses in clinical specimens from patients with respiratory tract infection by use of a novel real-time reverse-transcriptasepolymerase chain reaction. J Infect Dis. 2004;189:652–657. doi: 10.1086/381207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapikian AZ, James HD, Jr, Kelly SJ, Dees JH, Turner HC, McIntosh K, Kim HW, Parrott RH, Vincent MM, Chanock RM. Isolation from man of “avian infectious bronchitis virus-like” viruses (coronaviruses) similar to 229E virus, with some epidemiological observations. J Infect Dis. 1969;119:282–290. doi: 10.1093/infdis/119.3.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Hoek L. Human coronaviruses: what do they cause? Antivir Ther. 2007;12:651–658. [PubMed] [Google Scholar]
- 14.Dare RK, Fry AM, Chittaganpitch M, Sawanpanyalert P, Olsen SJ, Erdman DD. Human coronavirus infections in rural Thailand: a comprehensive study using real-time reverse-transcription polymerase chain reaction assays. J Infect Dis. 2007;196:1321–1328. doi: 10.1086/521308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaunt ER, Hardie A, Claas E, Simmonds P, Templeton KE. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J Clin Microbiol. 2010;48:2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leung TF, Li CY, Lam WY, Wong GW, Cheuk E, Ip M, Ng PC, Chan PK. Epidemiology and clinical presentations of human coronavirus NL63 infections in Hong Kong children. J Clin Microbiol. 2009;47:3486–3492. doi: 10.1128/JCM.00832-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garbino J, Crespo S, Aubert JD, Rochat T, Ninet B, Deffernez C, Wunderli W, Pache JC, Soccal PM, Kaiser L. A prospective hospital-based study of the clinical impact of non-severe acute respiratory syndrome (non-SARS)-related human coronavirus infection. Clin Infect Dis. 2006;43:1009–1015. doi: 10.1086/507898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson GG, Dowling HF, Spiesman IG, Boand AV. Transmission of the common cold to volunteers under controlled conditions. I. The common cold as a clinical entity. AMA Arch Intern Med. 1958;101:267–278. doi: 10.1001/archinte.1958.00260140099015. [DOI] [PubMed] [Google Scholar]
- 19.Turner RB, Wecker MT, Pohl G, Witek TJ, McNally E, St George R, Winther B, Hayden FG. Efficacy of tremacamra, a soluble intercellular adhesion molecule 1, for experimental rhinovirus infection: a randomized clinical trial. JAMA. 1999;281:1797–1804. doi: 10.1001/jama.281.19.1797. [DOI] [PubMed] [Google Scholar]
- 20.Yale SH, Liu K. Echinacea purpurea therapy for the treatment of the common cold: a randomized, double-blind, placebo-controlled clinical trial. Arch Intern Med. 2004;164:1237–1241. doi: 10.1001/archinte.164.11.1237. [DOI] [PubMed] [Google Scholar]
- 21.Zitter JN, Mazonson PD, Miller DP, Hulley SB, Balmes JR. Aircraft cabin air recirculation and symptoms of the common cold. JAMA. 2002;288:483–486. doi: 10.1001/jama.288.4.483. [DOI] [PubMed] [Google Scholar]
- 22.Boom R, Sol C, Salimans M, Jansen C, Wertheim-van Dillen P, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan KH, Yam WC, Pang CM, Chan KM, Lam SY, Lo KF, Poon LL, Peiris JM. Comparison of the NucliSens easyMAG and Qiagen BioRobot 9604 nucleic acid extraction systems for detection of RNA and DNA respiratory viruses in nasopharyngeal aspirate samples. J Clin Microbiol. 2008;46:2195–2199. doi: 10.1128/JCM.00315-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jokela P, Piiparinen H, Mannonen L, Auvinen E, Lappalainen M. Performance of the Luminex xTAG Respiratory Viral Panel Fast in a clinical laboratory setting. J Virol Methods. 2012;182:82–86. doi: 10.1016/j.jviromet.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kon M, Watanabe K, Tazawa T, Watanabe K, Tamura T, Tsukagoshi H, Noda M, Kimura H, Mizuta K. Detection of human coronavirus NL63 and OC43 in children with acute respiratory infections in Niigata, Japan, between 2010 and 2011. Jpn J Infect Dis. 2012;65:270–272. doi: 10.7883/yoken.65.270. [DOI] [PubMed] [Google Scholar]
- 26.Hays JP, Myint SH. PCR sequencing of the spike genes of geographically and chronologically distinct human coronaviruses 229E. J Virol Methods. 1998;75:179–193. doi: 10.1016/S0166-0934(98)00116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJ, Wolthers KC, Wertheim-van Dillen PM, Kaandorp J, Spaargaren J, Berkhout B. Identification of a new human coronavirus. Nat Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dominguez SR, Sims GE, Wentworth DE, Halpin RA, Robinson CC, Town CD, Holmes KV. Genomic analysis of 16 Colorado human NL63 coronaviruses identifies a new genotype, high sequence diversity in the N-terminal domain of the spike gene and evidence of recombination. J Gen Virol. 2012;93:2387–2398. doi: 10.1099/vir.0.044628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geng H, Cui L, Xie Z, Lu R, Zhao L, Tan W. Characterization and complete genome sequence of human coronavirus NL63 isolated in China. J Virol. 2012;86:9546–9547. doi: 10.1128/JVI.01457-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farsani SM, Dijkman R, Jebbink MF, Goossens H, Ieven M, Deijs M, Molenkamp R, van der Hoek L. The first complete genome sequences of clinical isolates of human coronavirus 229E. Virus Genes. 2012;45:433–439. doi: 10.1007/s11262-012-0807-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shirato K, Kawase M, Watanabe O, Hirokawa C, Matsuyama S, Nishimura H, Taguchi F. Differences in neutralizing antigenicity between laboratory and clinical isolates of HCoV-229E isolated in Japan in 2004–2008 depend on the S1 region sequence of the spike protein. J Gen Virol. 2012;93:1908–1917. doi: 10.1099/vir.0.043117-0. [DOI] [PubMed] [Google Scholar]
- 32.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214–219. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rambaut A. FigTree, A Graphical Viewer of Phylogenetic Trees. 2007. http://tree.bio.ed.ac.uk/software/figtree Available at. Accessed July 1, 2014.
- 34.Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (* And Other Methods). Version 4. Sunderland, United Kingdom: MA Sinauer Associates; 2003. [Google Scholar]
- 35.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schildgen O, Jebbink MF, de Vries M, Pyrc K, Dijkman R, Simon A, Müller A, Kupfer B, van der Hoek L. Identification of cell lines permissive for human coronavirus NL63. J Virol Methods. 2006;138:207–210. doi: 10.1016/j.jviromet.2006.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cabeca TK, Bellei N. Human coronavirus NL-63 infection in a Brazilian patient suspected of H1N1 2009 influenza infection: description of a fatal case. J Clin Virol. 2012;53:82–84. doi: 10.1016/j.jcv.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaunt ER, Hardie A, Claas EC, Simmonds P, Templeton KE. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J Clin Microbiol. 2010;48:2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moes E, Vijgen L, Keyaerts E, Zlateva K, Li S, Maes P, Pyrc K, Berkhout B, van der Hoek L, Van Ranst M. A novel pancoronavirus RT-PCR assay: frequent detection of human coronavirus NL63 in children hospitalized with respiratory tract infections in Belgium. BMC Infect Dis. 2005;5:6. doi: 10.1186/1471-2334-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matoba Y, Abiko C, Ikeda T, Aoki Y, Suzuki Y, Yahagi K, Matsuzaki Y, Itagaki T, Katsushima F, Katsushima Y, Mizuta K. Detection of the human coronavirus 229E, HKU1, NL63, and OC43 between 2010 and 2013 in Yamagata, Japan. Jpn J Infect Dis. 2014;68:138–141. doi: 10.7883/yoken.JJID.2014.266. [DOI] [PubMed] [Google Scholar]
- 41.Vabret A, Mourez T, Dina J, van der Hoek L, Gouarin S, Petitjean J, Brouard J, Freymuth F. Human coronavirus NL63, France. Emerg Infect Dis. 2005;11:1225–1229. doi: 10.3201/eid1108.050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Razuri H, Malecki M, Tinoco Y, Ortiz E, Guezala MC, Romero C, Estela A, Breña P, Morales ML, Reaves EJ, Gomez J, Uyeki TM, Widdowson MA, Azziz-Baumgartner E, Bausch DG, Schildgen V, Schildgen O, Montgomery JM. Human coronavirus-associated influenza-like illness in the community setting in Peru. Am J Trop Med Hyg. 2015;93:1038–1040. doi: 10.4269/ajtmh.15-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lau SK, Woo PC, Yip CC, Tse H, Tsoi HW, Cheng VC, Lee P, Tang BS, Cheung CH, Lee RA, So LY, Lau YL, Chan KH, Yuen KY. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J Clin Microbiol. 2006;44:2063–2071. doi: 10.1128/JCM.02614-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abdul-Rasool S, Fielding BC. Understanding human coronavirus HCoV-NL63. Open Virol J. 2010;4:76–84. doi: 10.2174/1874357901004010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu R, Zhang L, Tan W, Zhou W, Wang Z, Peng K, Ruan L. Characterization of human coronavirus 229E infection among patients with respiratory symptom in Beijing, Oct–Dec, 2007. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2009;23:367–370. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.