Abstract
Dengue is currently regarded as a major public health problem worldwide. In a hyperendemic region during an outbreak, we detected the co-circulation of all Dengue virus (DENV) serotypes including two different genotypes of DENV-3 and DENV-4, and concurrent infections with up to three serotypes were identified in symptomatic patients. A total of 49 acute phase plasma samples from patients clinically suspected of dengue were collected during the 4 weeks of May 2013. DENV-1-4 was detected by reverse transcriptase semi-nested polymerase chain reaction in 33 samples (67.3%), of which 26 DNA fragments were sequenced. Twenty samples (76.9%) were identified with a single DENV serotype and six (23.1%) with more than one serotype. DENV-3 was the predominant serotype of the outbreak. On the basis of phylogenetic analyses, DENV-1 isolates belong to genotype V, DENV-2 to American–Asian genotype, DENV-3 to genotypes I and III, and DENV-4 to genotypes I and II.
Dengue is currently regarded globally as the most important mosquito-borne viral disease as it is a major public health problem worldwide.1 The disease is caused by four antigenically and genetically distinct viruses designated dengue virus 1–4 (DENV-1-4), which are subdivided into distinct genotypes.2,3 DENVs are transmitted by mosquito vectors, primarily Aedes aegypti.4 DENV infection is often unapparent but can lead to a wide range of clinical manifestations from mild disease to severe dengue cases with bleeding, plasma leakage, and shock.5
In dengue-endemic countries, the co-circulation of multiple DENV serotypes in the same area has been described with concurrent infections.1,6 The first reported case of concurrent infection with two serotypes (DENV-1 and DENV-4) occurred in Puerto Rico in 1982.4 Since this time, dual infections have been reported in locals in New Caledonia, Taiwan, China, Singapore, and India. In 1999, an infection with three serotypes (DENV-1/DENV-3/DENV-4) was detected in two patients from Indonesia and one from Mexico.4,6
In this study, we report, for the first time in Brazil, the co-circulation and coinfection of DENV-1-4 during a severe outbreak in an endemic area of southeast Brazil. Our data raise new questions about biological and public health aspects related to dengue occurrence in Brazil.
The study was performed in Contagem, Minas Gerais, which borders the city of Belo Horizonte (state capital), and it has an area of 195,268 km2 with approximately 603,442 inhabitants. In 2013, Contagem underwent a dengue epidemic with the highest number of notified cases that has been ever recorded (23,436) and three deaths. In May 2013, a total of 49 acute phase plasma samples were collected during 4 weeks from clinically suspected dengue patients admitted to Geraldo Pinto Vieira Hospital in Contagem. Until this month, 22,808 (97.3%) cases had been reported. This research was approved by the Ethical Committee of UFMG. The residential addresses of DENV-positive patients are plotted on the map of Contagem shown in Figure 1 .
Figure 1.
Spatial distribution of Dengue virus positive infections. QUANTUM GIS version 2.0.1 software was used, and the database of Contagem was provided by Prefeitura Municipal de Contagem. For addresses outside this city, Google Earth was used.
Viral RNA was extracted from plasma samples using the QIAmp® Viral RNA Extraction Kit (QIAGEN Inc., Valencia, CA) and used to synthesize complementary DNA (cDNA) with random primer (Invitrogen, Carlsbad, CA). The semi-nested polymerase chain reaction (PCR) assay targeting the C-prM region of DENV genome used in this study distinguishes the four serotypes by the size of the products, as described by Lanciotti and others, with modifications.7 In the first step, a highly conserved primer pair D1 (forward) and D2 (reverse) is used to amplify a 511-bp external product. A second round PCR reaction is performed using the external product as template and the D1 primer in combination with each of the four serotype-specific reverse primers TS1, TS2, TS3, and TS4 separately. The expected sizes of the PCR products are 482 bp (D1 and TS1 for DENV-1), 119 bp (D1 and TS2 for DENV-2), 290 bp (D1 and TS3 for DENV-3), and 392 bp (D1 and TS4 for DENV-4). To obtain larger DNA fragments for sequencing, the cDNA of some DENV-2 and DENV-3 positive samples were used as template in an alternative semi-nested PCR with the forward primer D1.1 (5′-CGA GAA ACC GCG TGT CAA C-3′) in a second round reaction, designed to amplify a 490-bp product.8
The DNA fragments obtained from semi-nested PCR were purified (MinElute PCR Purification Kit; QIAGEN Inc.) and sequenced in both directions (Applied Biosystems 3730 DNA sequencer; Life Technologies, Carlsbad, CA). A basic local alignment search tool (BLAST) search was carried out to confirm the identity of the viruses. Nucleotide sequences were aligned using the software MUSCLE (European Molecular Biology Laboratory, European Bioinformatics Institute, Hinxton, Cambridge, United Kingdom). We performed an analysis based on the maximum likelihood (ML) method. Selection of the best-fit nucleotide substitution model was performed using jModelTest (Free Software Foundation, Inc., Boston, MA).9 Phylogenetic tree reconstruction using ML methods was performed in PhyML v.3.1 (French National Institute of Bioinformatics, Gif-sur-Yvette, France) with 100 bootstrap replicates with the use of the Akaike information criterion.10
From 49 tested samples, 33 (67.3%) were positive for DENV RNA by reverse transcriptase semi-nested PCR, of which 26 DNA fragments were sequenced. Twenty samples (76.9%) were identified with a single DENV serotype: eight (40%) as DENV-3, five (25%) as DENV-4, four (20%) as DENV-2, and three (15%) as DENV-1. Six samples were identified with more than one serotype resulting in an overall rate of concurrent infections of 23.1% with the following distribution: four (15.4%) with two DENV serotypes (50% with DENV-2 and DENV-3, 25% with DENV-3 and DENV-4, and 25% with DENV-2 and DENV-4); and two cases (7.7%) with three DENV serotypes (DENV-2, DENV-3, and DENV-4). DENV-3 was detected in 13 (50%) samples, including those with single or concurrent infections, and was the predominant serotype of the outbreak. These sequences have been deposited in GenBank.
Considering a number of 22 identified patients (Table 1) with equal gender rate, only three samples were from adolescents, while the others were from adults. The onset of symptoms of all patients was between epidemiological weeks 19 and 22 of 2013. Samples from 21 patients were subjected to serological tests, of which 12 were IgM positive. The two patients with DENV-1 detection had the lowest platelet counts. All patients with a clinical diagnosis in the notification form were considered to have dengue without warning signs, even the patients with coinfections.
Table 1.
Clinical and epidemiological data from DENV positive patients
| Patient ID | DENV serotype | DENV genotype | Age (years)/sex | Onset of symptoms | EW/2013 | Blood collection for IgM | IgM | Platelet count/mm3 |
|---|---|---|---|---|---|---|---|---|
| MG2/2013 | 1 | V | 16/M | May 11, 2013 | 19 | May 17, 2013 | R | 54,000 |
| MG13/2013 | 1 | V | 37/F | May 14, 2013 | 20 | May 20, 2013 | R | 89,000 |
| MG16/2013 | 1 | V | NA | NA | NA | NA | NA | NA |
| 34C | 2 | A/A | 40/F | May 22, 2013 | 21 | May 27, 2013 | NR | 210,000 |
| 28C | 2 | A/A | 24/F | May 11, 2013 | 19 | May 16, 2013 | R | 153,000 |
| 27C | 2 | A/A | NA | NA | NA | NA | NA | NA |
| 29C | 2 | A/A | 45/F | May 12, 2013 | 20 | May 17, 2013 | R | 140,000 |
| 14C | 3 | I | 47/M | May 13, 2013 | 20 | May 17, 2013 | R | 159,000 |
| MG10/2013 | 3 | I | 52/M | May 20, 2013 | 21 | May 26, 2013 | NR | 249,000 |
| MG11/2013 | 3 | III | 40/F | May 27, 2013 | 22 | May 27, 2013 | R | 226,000 |
| MG18/2013 | 3 | III | 53/M | May 10, 2013 | 19 | May 16, 2013 | R | NA |
| MG19/2013 | 3 | III | NA | NA | NA | NA | NA | NA |
| MG23/2013 | 3 | III | 20/F | May 10, 2013 | 19 | May 16, 2013 | NR | NA |
| MG50/2013 | 3 | III | NA | NA | NA | NA | NA | NA |
| 33BH | 3 | III | 39/M | May 13, 2013 | 20 | May 18, 2013 | R | 197,000 |
| MG49/2013 | 4 | II | 50/M | May 26, 2013 | 22 | May 27, 2013 | NR | 186,000 |
| 7C | 4 | II | 44/F | May 23, 2013 | 21 | May 31, 2013 | R | 190,000 |
| 21BH | 4 | I | 52/F | May 11, 2013 | 19 | May 16, 2013 | NR | NA |
| MG22/2013 | 4 | I | 21/M | May 10, 2013 | 19 | May 16, 2013 | NR | NA |
| 31C | 4 | II | 26/F | May 10, 2013 | 19 | May 16, 2013 | R | 292,000 |
| MG38/2013 | 2, 3 | A/A, I | 43/F | May 11, 2013 | 19 | May 17, 2013 | R | 144,000 |
| MG36/2013 | 2, 3 | A/A, I | 38/M | May 17, 2013 | 20 | May 23, 2013 | NR | 223,000 |
| 40BH | 2, 4 | A/A, I | 17/M | May 24, 2013 | 21 | June 3, 2013 | NR | 161,000 |
| MG25/2013 | 3,4 | III, II | 15/M | May 10, 2013 | 19 | May 15, 2013 | NR | NA |
| MG39/2013 | 2, 3, 4 | A/A, I, II | 23/M | May 12, 2013 | 20 | NA | TND | 178,000 |
| MG41/2013 | 2, 3, 4 | A/A, I, I | 66/F | May 11, 2013 | 19 | May 17, 2013 | R | 209,000 |
A/A = American/Asian; DENV = dengue virus; EW = epidemiological week of onset of symptoms; ID = identification; NA = not available; NR = not reactive; R = reactive; TND = test not done.
The positive dengue cases were concentrated in the Eldorado and Industrial health districts of Contagem (Figure 1), which are the most populous and urbanized areas. Patients from other cities were also admitted in the hospital because they were working in Contagem.
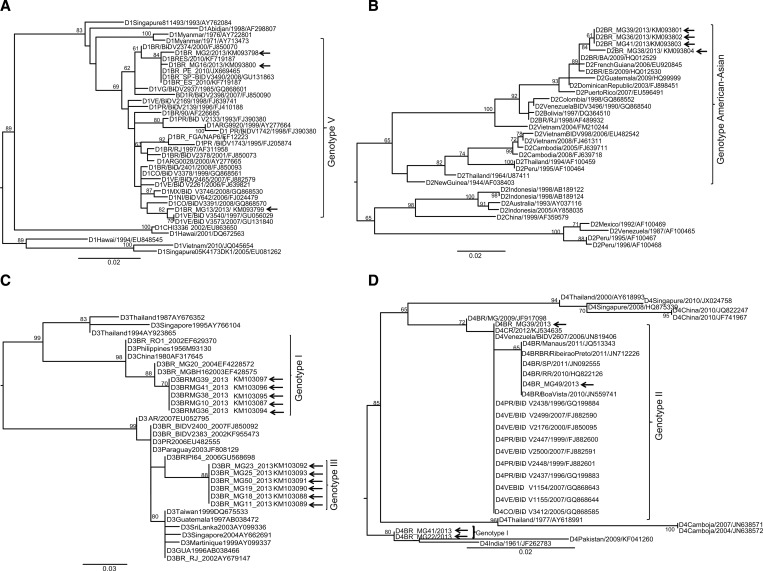
On the basis of phylogenetic analyses, the DENV-1 isolates belong to genotype V (Figure 2A ). The DENV-2 sequences grouped with the American–Asian genotype (Figure 2B), the DENV-3 samples to genotypes I and III (Figure 2C), and the DENV-4 samples to genotype I and II (Figure 2D). All these DENV genotypes are circulating in Brazil.
Figure 2.
Phylogenetic trees of dengue virus (DENV) 1, 2, 3, and 4 based on the partial sequence of the C-prM DENV gene. Phylogenetic tree construction using maximum likelihood (ML) methods was performed in PhyML. Bootstrap supporting values greater than 50 are shown at the nodes. Roman numerals denote the different genotypes of DENV-1-4. The phylogenetic trees were based on the partial nucleotide sequence of the C-prM gene. The best-fit model was based on Akaike information criterion. Phylogenetic trees: (A) DENV-1: TIM1 with gamma correction (TIM1 + G). (B) DENV-2: TIM1 with estimated invariable site (TIM3 + I). (C) DENV-3: TIM1 with estimated invariable site (TIM2 + I). (D) DENV-4: TPM2 + G with TIM1 with gamma correction (TIM3 + I). Samples from Contagem are indicated by an arrow.
In areas where multiple DENV serotypes are transmitted concurrently, clinical cases caused by more than one serotype of DENV may be more common than previously thought. The high attack rates that occur during epidemics also provide opportunities for mosquitoes to become infected with two or more DENV serotypes.6
To our knowledge this is the first report that clearly demonstrates the presence of the four DENV serotypes and more than one genotype for DENV-3 and DENV-4, in addition to the occurrence of concurrent infections with two or three serotypes during a short period in a hyperendemic region.
In Brazil, a coinfection with DENV-1 and DENV-2 was reported in 2001, DENV-2 and DENV-3 in 2005, and DENV-3 and DENV-4 among 674 patients with acute undifferentiated fever from the Tropical Medicine Reference Center of Manaus, Amazonas, between 2005 and 2010. Hence, coinfection with distinct DENV serotypes during outbreaks may be expected.11
It has been postulated that concurrent infections by multiple DENV serotypes may influence the clinical course of the disease. This is considered as a single major factor for the emergence of severe dengue, but larger studies are needed to prove this association.12
In a hyperendemic scenario, besides coinfections, the number of secondary infections also increases. Secondary infections with a different DENV serotype are major risk factors for severe diseases because of antibody-dependent enhancement mechanism.2
The co-circulation of genetically distinct DENV, even at the genotype level, detected in Contagem in 2013 and the occurrence of concurrent infections could impact recombination events and lead to the emergence of more virulent isolates.13 The introduction of a new DENV serotype and a distinct genotype into a new area highlights the potential for epidemics. Indeed the presence of a serotype against which the population is not immunized indicates a risk for a sharp increase in the number of DENV infections, including severe cases.14 DENV-3 genotype I, detected in six patients in this study, has the potential to cause a more severe disease and has a neurotrophic potential in mice model, nonetheless these patients were selected based on classical dengue symptoms.15
The study revealed the co-circulation of DENV-1-4 in a hyperendemic region, and DENV-3 was dominant during this 4-week survey. Concurrent infections occurred at a rate of 23.1% in dengue cases in which the patients recovered without complications. Six different genotypes were found co-circulating during the outbreak. DENV-3 genotypes I and III and DENV-4 genotypes I and II were detected along with genotypes V and American–Asian for DENV-1 and DENV-2, respectively.
These results highlight the genetic variability of circulating DENV strains and contribute to better comprehend the dengue epidemiology. Furthermore, these data strengthen the need for improvements on vector surveillance and control and represent a challenge for efficient multivalent vaccine development as well.
ACKNOWLEDGMENTS
We thank technician of Secretaria Municipal de Saúde da Prefeitura de Contagem, especially, Marco Túlio de Oliveira and Izabela Vasconcelos Vieira for support with the blood collection, patient data, and address database.
Footnotes
Financial support: This work was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq)–Pronex and INCT em Dengue, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG), and Ministério da Saúde, DECIT.
Disclosure: Erna G. Kroon and Paulo C. P. Ferreira are researchers from CNPq.
Authors' addresses: Elisa H. P. Andrade, Ana P. P. Vilela, Júlio C. C. Rosa, Daniela P. J. Miranda, Paulo C. P. Ferreira, and Erna G. Kroon, Departamento de Microbiologia, Universidade Federal de Minas Gerais, Belo Horizonte, Brazil, E-mails: elisahpandrade@yahoo.com.br, pessoavilela58@gmail.com, jcbhrama@yahoo.com.br, dmiranda@superig.com.br, peregrinopc48@gmail.com, and kroone@icb.ufmg.br. Leandra B. Figueiredo and Jonatas S. Abrahão, Laboratório de Vírus, Departamento de Microbiologia, Universidade Federal de Minas Gerais, Gerais, Minas, Brazil, E-mails: leandrabio@yahoo.com.br and jonatas.abrahao@gmail.com. Jaquelline G. Oliveira, Laboratório de Imunologia Celular e Molecular, Centro de Pesquisas René Rachou, Fundação Oswaldo Cruz (FIOCRUZ), Belo Horizonte, Brazil, E-mail: jaque@cpqrr.fiocruz.br. Elisa H. P. Andrade, Hassan M. Zibaoui, and Valdelaine E. M. Araújo, Secretaria de Saúde, Prefeitura Municipal de Contagem, Contagem, Brazil, E-mails: elisahpandrade@yahoo.com, hassan.zibaoui@contagem.mg.gov.br, and valdelainearaujo@yahoo.com.br.
References
- 1.Jiang T, Yu X-D, Hong W-X, Zhou W-Z, Yu M, Deng Y-Q, Zhu S-Y, Qin E-D, Wang J, Qin C-F, Zhang F-C. Co-circulation of two genotypes of dengue virus serotype 3 in Guangzhou, China. Virol J. 2009;9:1–8. doi: 10.1186/1743-422X-9-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Das S, Pingle MR, Muñoz-Jordán J, Rundell MS, Rondini S, Granger K, Chang GJ, Kelly E, Spier EG, Larone D, Spitzer E, Barany F, Golightly LM. Detection and serotyping of dengue virus in serum samples by multiplex reverse transcriptase PCR–ligase detection reaction assay. J Clin Microbiol. 2008;46:3276–3284. doi: 10.1128/JCM.00163-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weaver SC, Vasilakis N. Molecular evolution of dengue viruses: contributions of phylogenetics to understanding the history and epidemiology of the preeminent arboviral disease. Infect Genet Evol. 2009;9:523–540. doi: 10.1016/j.meegid.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chinnawirotpisan P, Mammen MP, Jr, Nisalak A, Thaisomboonsuk B, Narupiti S, Thirawuth V, Putnak R, Zhang C. Detection of concurrent infection with multiple dengue virus serotypes in Thai children by ELISA and nested RT-PCR assay. Arch Virol. 2008;153:2225–2232. doi: 10.1007/s00705-008-0249-9. [DOI] [PubMed] [Google Scholar]
- 5.World Health Organization Dengue and Severe Dengue. 2013. http://www.who.int/mediacentre/factsheets/fs117/en/ Available at. Accessed January 13, 2014.
- 6.Loroño-Pino MA, Cropp CB, Farfán JA, Vorndam AV, Rodríguez-Angulo EM, Rosado-Paredes EP, Flores-Flores LF, Beaty BJ, Gubler DJ. Common occurrence of concurrent infections by multiple dengue virus serotypes. Am J Trop Med Hyg. 1999;61:725–730. doi: 10.4269/ajtmh.1999.61.725. [DOI] [PubMed] [Google Scholar]
- 7.Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase polymerase chain reaction. J Clin Microbiol. 1992;30:545–551. doi: 10.1128/jcm.30.3.545-551.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vilela APP, Figueiredo LB, Santos JR, Eiras AE, Bonjardim CA, Ferreira PCP, Kroon EG. Dengue virus 3 genotype I in Aedes aegypti mosquitoes and eggs, Brazil, 2005–2006. Emerg Infect Dis. 2010;16:989–992. doi: 10.3201/eid1606.091000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Posada D. jModelTest: Phylogenetic Model Averaging. 2011. https://code.google.com/p/jmodeletest2/ Available at. Accessed June 11, 2014. [DOI] [PubMed]
- 10.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 11.Figueiredo RMP, Naveca FG, Oliveira CM, Bastos MS, Mourão MPG, Viana SS, Melo MN, Itapirema EF, Saatkamp CJ, Farias IP. Co-infection of dengue virus by serotypes 3 and 4 in patients from Amazonas, Brazil. Rev Inst Med Trop Sao Paulo. 2011;53:321–323. doi: 10.1590/s0036-46652011000600004. [DOI] [PubMed] [Google Scholar]
- 12.Bharaj P, Chahar HS, Pandey A, Diddi K, Dar L, Guleria R, Kabra SK, Broor S. Concurrent infections by all four dengue virus serotypes during an outbreak of dengue in 2006 in Delhi, India. Virol J. 2008;12:542–549. doi: 10.1186/1743-422X-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vinodkumar CS, Kalapannavar NK, Basavarajappa KG, Sanjay D, Gowli C, Nadig NG, Prasad BS. Episode of coexisting infections with multiple dengue virus serotypes in central Karnataka, India. J Infect Public Health. 2013;6:302–306. doi: 10.1016/j.jiph.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Nunes MRT, Faria NR, Vasconcelos HB, Medeiros DBA, Lima CPS, Carvalho VL, Silva EVP, Cardoso JF, Sousa EC, Nunes KNB, Rodrigues SG, Abecasis AB, Suchard MA, Lemey P, Vasconcelos PFC. Phylogeography of dengue virus serotype 4, Brazil, 2010–2011. Emerg Infect Dis. 2012;18:1858–1864. doi: 10.3201/eid1811.120217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Souza KPR, Silva EG, Rocha ESO, Figueiredo LB, Almeida-Leite CM, Arantes RM, Assis Silva Gomes J, Ferreira GP, de Oliveira JG, Kroon EG, Campos MA. Nitric oxide synthase expression correlates with death in an experimental mouse model of dengue with CNS involvement. Virol J. 2013;10:1–10. doi: 10.1186/1743-422X-10-267. [DOI] [PMC free article] [PubMed] [Google Scholar]