Abstract
Treatment of disseminated tuberculosis in children ≤ 6 years has not been optimized. The pyrazinamide-containing combination regimen used to treat disseminated tuberculosis in babies and toddlers was extrapolated from adult pulmonary tuberculosis. Due to hepatotoxicity worries, there are no dose–response studies in children. We designed a hollow fiber system model of disseminated intracellular tuberculosis with co-perfused three-dimensional organotypic liver modules to simultaneously test for efficacy and toxicity. We utilized pediatric pharmacokinetics of pyrazinamide and acetaminophen to determine dose-dependent pyrazinamide efficacy and hepatotoxicity. Acetaminophen concentrations that cause hepatotoxicity in children led to elevated liver function tests, while 100 mg/kg pyrazinamide did not. Surprisingly, pyrazinamide did not kill intracellular Mycobacterium tuberculosis up to fourfold the standard dose as monotherapy or as combination therapy, despite achieving high intracellular concentrations. Host-pathogen RNA-sequencing revealed lack of a pyrazinamide exposure transcript signature in intracellular bacteria or of phagolysosome acidification on pH imaging. Artificial intelligence algorithms confirmed that pyrazinamide was not predictive of good clinical outcomes in children ≤ 6 years who had extrapulmonary tuberculosis. Thus, adding a drug that works inside macrophages could benefit children with disseminated tuberculosis. Our in vitro model can be used to identify such new regimens that could accelerate cure while minimizing toxicity.
Keywords: Disseminated tuberculosis, Pediatric, Hepatotoxicity, 3D liver hollow fiber model
Highlights
-
•
We designed a pre-clinical of disseminated for simultaneous identification of toxicity and efficacy in children.
-
•
The system is a co-culture of infected monocytes and 3 dimensional organotypic liver recapitulating children pharmacokinetics.
-
•
Pyrazinamide, central drug in treatment regimen, had no effect as monotherapy or contribute to the combination therapy.
Due to fear of toxicity children are often not involved in clinical trials, and as a result the optimal treatment regimens are often lacking. As an example, toddlers and babies develop disseminated tuberculosis but are treated with regimens designed for adults with lung cavity disease. We designed a “glass-mouse” model of disseminated tuberculosis that simultaneously tests for the efficacy and toxicity of the anti-tuberculosis drugs for children with disseminated disease. We found that while not causing dose-dependent liver toxicity, one of the central drugs used to treat this children is likely not efficacious.
1. Introduction
Recent estimates are that one million children developed tuberculosis (TB) in 2010 alone (Jenkins et al., 2014). In babies and toddlers, TB manifests as disseminated disease, attributed to poor immune containment (Newton et al., 2008). In disseminated disease, Mycobacterium tuberculosis (Mtb) is predominantly intracellular as opposed to the predominantly extracellular bacteria in cavitary pneumonia in adults. However, therapeutic regimens used to treat pediatric TB were copied from those for adults with pulmonary TB. For multi-drug resistant TB (MDR-TB) and extensively drug-resistant TB in children, no regimens have been developed as of yet and in some parts of the world such children are left untreated, and have been deemed “invisible” (Becerra and Swaminathan, 2014, Dheda et al., 2014). There has been reluctance to perform dose–response studies and first in human studies in children because of concerns for toxicity such as drug induced liver injury (DILI). Indeed, the incidence of DILI among children treated for TB ranges between 1.7–8%, making this a legitimate concern (Devrim et al., 2010, Roy et al., 2006) Recently, the World Health Organization and STOP-TB have advocated for the design of new and shorter treatment regimens for children, leading to zero TB deaths in children. Thus the problem is pressing. Recently Momper et al. (2015) summarized reasons for failed clinical trials in all pediatric drug development as mainly due to lack of efficacy, poor dosing, or toxicity; they proposed use of pre-clinical models to tackle these failures so that results could be directly translated to the clinic. No pre-clinical model of intracellular TB has been designed as of yet. On the other hand, preclinical toxicity requires multiple animal model species testing. Regardless, rodent models which are commonly used as one of the animals'models have predictive rates of less than 40% for human DILI and resulted in second highest discontinuation rates for several drugs (Olson et al., 2000). In “Preventing drug-induced liver injury: how useful are animal models?” Ballet argues that no animal models are useful for predicting the most common forms of human DILI at all and that humans themselves are best models for DILI (Ballet, 2015). Indeed, no good pre-clinical model of DILI in which human concentration-time profiles of pharmaceuticals are examined exists as of yet. Here, we designed a model of intracellular TB, fitted a 3-dimensional human organoid liver (3D-liver), and dialed human pediatric-like pharmacokinetics, for performance of pediatric dose–response studies to estimate both safety and DILI at the same time.
One of the pivotal drugs in the regimen used to treat children with both drug-susceptible and MDR-TB, based on equivalent use in adults, is pyrazinamide (Donald et al., 2012). The doses for use in children are as yet unclear; based on computer aided clinical trial simulations we and others have proposed that the doses be doubled for children with pulmonary disease (Gumbo et al., 2009a, Thee et al., 2011). However, this could lead to more DILI. On the other hand, there has been a debate as to whether pyrazinamide kills intracellular Mtb at all even in adults (Crowle et al., 1991, Gumbo et al., 2009a). Contrariwise, it has been argued that pyrazinamide could work as an immunomodulatory agent in the context of interferon-gamma (IFN-γ) secretion in granulomas (Ahmad et al., 2011). If the former assertion is correct, pyrazinamide as used currently in children with disseminated disease would not contribute to efficacy of the regimen. However, if the immunomodulatory effect hypothesis is correct, then pyrazinamide could be a vital component of treatment for disseminated TB. Here, we examined the efficacy and toxicity of pyrazinamide in our novel model for disseminated pediatric disease, and found that the drug does not kill intracellular Mtb even when IFN-γ and tumor necrosis factor α (TNF-α) are supplemented. Moreover, novel host-pathogen RNA sequencing (RNA-Seq) revealed no transcriptomic or immunological signature of pyrazinamide exposure.
2. Materials and Methods
2.1. Bacterial Strain and Growth Conditions
Prior to each experiment stock Mtb culture (H37Ra, ATCC# 25177; H37Rv ATCC# 27294) was thawed and grown in Middlebrook 7H9 broth supplemented with 10% oleic acid-dextrose-catalase (OADC) at 37 °C under 5% CO2 and shaking conditions. For the pH 5.8 studies the log-phase growth bacteria were further cultured for 4 more days in Middlebrook 7H9 broth acidified and buffered to a pH of 5.8, as described in the past (Gumbo et al., 2009a).
2.2. Tissue Culture
Human derived THP-1 cell line (ATCC TIB-202) and murine macrophage cell line (J774A.1, ATCC TIB-67) were grown in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) at 37 °C under 5% CO2.
2.3. Reagents and Supplies
Hollow fiber system (HFS) cartridges were purchased from FiberCell (Frederick, MD). Pyrazinamide, isoniazid, rifampin, and acetaminophen (APAP) were purchased from SIGMA (USA).
Liver enzymes such as aspartate transaminase (AST), alanine transaminase (ALT), and lactate dehydrogenase (LDH) were measured using Vitros 250 system. Change in the Cyp3A4 was measured using RT-PCR while Bioplex Multiplex System (Bio RAD, USA) was used for measurement of interleukin 8 (IL-8), monocyte chemotactic protein 1 (MCP-1), vascular endothelial growth factor (VEGF), and platelet-derived growth factor (PDGF).
2.3.1. Three Dimensional Organotypic Liver Hollow Fiber Model of Mtb
The HFS-TB model consists of (a) a peripheral or pharmacodynamics compartment for the Mtb inoculum and (b) a central drug containing or pharmacokinetic compartment (Srivastava and Gumbo, 2011). In the past, we developed a HFS models of extracellular Mtb and intracellular Mycobacterium avium complex (MAC) in which concentration-time profiles of antibiotics encountered in adult patients were recapitulated (Deshpande et al., 2010a, Deshpande et al., 2010b, Gumbo et al., 2004, Gumbo et al., 2007a, Gumbo et al., 2007b, Gumbo et al., 2009a, Schmalstieg et al., 2012, Srivastava et al., 2010, Srivastava et al., 2011a). For pediatric disseminated TB, we infected THP-1 macrophages with Mtb. We further improved the intracellular HFS-TB model for assessment of hepatotoxicity by retrofitting 3D liver organoid cultures. The 3D-liver cultures were made of 106 HEpG2 embedded in alginate beads (total volume of 400 μL), which were cultured in 3D perfusion in the 3DKUBE™ (KIYATEC, Greenville, SC). The 3D KUBES were fitted in to the flow path of the HFS between the central reservoir, where the drug is delivered, and peripheral compartment. The drug was infused into the central compartment via a syringe pump at a predefined rate to achieve the peak concentrations and systemic clearances encountered in humans treated with the equivalent doses.
Several preparatory experiments demonstrated that monocytes would only live for a few days if the virulent Mtb H37Rv was used to infect the macrophages; however the isogenic strain H37Ra produced a stable infection for over 4 weeks. For infection, prior to each intracellular Mtb study, THP-1 monocyte cells were grown in RPMI 1640 medium supplemented with 10% FBS to a density of 1.5 ∗ 106 CFU/mL. Next, log phase Mtb culture was at a bacterial density of 1.5 ∗ 107 CFU/mL was co-incubated with the THP-1 cells for 6 h at 37 °C under 5% CO2, giving a multiplicity of infection of 1:10. Infected THP-1 cells were then washed twice with warm RPMI 1640 by centrifugation at 100 × g for 5 min and examined in a hemocytometer for cell counts and viability after staining with trypan blue. The THP-1 cell counts before and after infection with Mtb were also verified with an automated cell counter (Sceptor, EMD Milipore). We then inoculated the peripheral compartment of each HFS with 20 mL of infected THP-1 cells (3.0 ∗ 107 cells/HFS) into the peripheral compartment of the HFS.
2.4. Intracellular HFS-TB Studies Using Pediatric Pharmacokinetics of Pyrazinamide for Efficacy and DILI
The THP-1 cells were infected with Mtb as described above and inoculated into the HFS retrofitted with 3D KUBES. The systems were then treated with one of the five pyrazinamide doses (0, 1/4, 1/2, 1, 2, and 4 times standard dose) administered daily for 28 days via computer-controlled syringe pumps, mimicking a 6 h half-life as encountered in babies (Donald et al., 2012). As a positive control for DILI, we utilized daily APAP (t1/2 = 3 h) with a goal of 4 h post dose levels of > 150 mg/L, known to cause DILI in children based on the Rumack-Matthews monogram (Rumack and Matthew, 1975, Rumack et al., 1981). RPMI 1640 supplemented with 10% FBS was infused using computer controlled peristaltic pump at a predefined rate to maintain the THP-1 monocyte and HepG2 cells in the 3D KUBES and to achieve the half-life of each drug in each HFS. Samples from the peripheral compartment were collected on day 0, 7, 14, 21, and 28, washed twice with normal saline to remove any carry over drug, and cultured on Middlebrook 7H10 agar supplemented with 10% OADC to enumerate the total bacillary burden. To confirm the intended pharmacokinetic profile of the drug achieved in each system, the central and peripheral compartments were sampled seven times over 24 h to measure the drug concentration. Samples from the central compartment and the 3D KUBES were also subjected to measure the level of AST, ALT, LDH, Cyp3A4, IL-8, MCP-1, VEGF, and PDGF to analyze the DILI on days 0, 2, 5, 21 and 26.
2.5. Role of Pyrazinamide in the Combination Therapy Against Intracellular Mtb
We performed HFS studies to determine the extent of pyrazinamide contribute to the standard three drug (isoniazid, rifampin, and pyrazinamide) regimen in disseminated TB. The HFSs were treated with either dual regimen (isoniazid–rifampin) or triple regimen (isoniazid–rifampin–pyrazinamide) over 28 days, in triplicate. The systems were sampled at predetermined time point for enumeration of the total bacterial burden as described above. In addition the concentration-time profile of the drugs was confirmed by sampling the central and peripheral compartments seven times over 24 h and concentrations of the isoniazid, rifampin, and pyrazinamide were measured using the previously validated method (Gumbo et al., 2009a, Srivastava et al., 2011a, Srivastava et al., 2011b).
2.6. Pyrazinamide Activity Against Extracellulat Mtb and Effect of Cytokines on Pyrazinamide Efficacy Against Intracellular Mtb
We examined the effect of several concentrations of pyrazinamide (1/8 to 8 × of the standard dose) against extracellular Mtb in Middlebrook broth that had been acidified to pH 5.8 (Gumbo et al., 2009b) over a period of 14 days. On day 14 the cultures were washed twice to remove any carry over drug, serially diluted and cultured on Middlebrook agar, and incubated at 37 °C with 5% CO2 for at least 21 days before the colonies were counted.
It has been argued that pyrazinamide works against intracellular Mtb in humans in concert with secreted IFN-γ by granulomas; indeed in the BALB/c mouse model of TB in which Mtb is predominantly intracellular pyrazinamide shows dramatic efficacy (Ahmad et al., 2011). To test this hypothesis, we first activated the THP-1 macrophages using phorbol myristate ester (PMA) and then infected the adherent cells with Mtb as described above, and performed the dose–response study, with the same dose ranges as used for the extracellular experiments, in the presence of IFN-γ at a concentration of 5 μg/mL and the IFN-γ antagonist dexamethasone at 1 μM concentration. The study was performed twice. Next, we repeated the experiment but this time with TNF-α at a concentration of 5 ng/mL, and a TNF-α antagonist (rabbit anti-human TNF-α) at 10 μg/mL. The experiment was performed twice. Finally, the same study was repeated with pyrazinamide (but no cytokines), but instead of adherent THP-1 cells we infected adherent mouse macrophage cells, the J774A.1 cell line, originally isolated from BALB/c mice.
2.7. Host-pathogen RNA-Seq
Mtb cultures grown in Middelbrook broth on day 4 of logarithmic phase growth and from semidormant bacteria on day 4 of incubation in broth at pH 5.8 (Gumbo et al., 2009a), were treated with pyrazinamide concentration of 294 mg/L (> EC80) for four days, and RNA extracted. Next, during the combination therapy hollow fiber studies above, samples for enumeration of Mtb cultures described above were used to extract RNA on the various sampling days described above.
Following extraction, the RNA samples were depleted of ribosomal RNA, and subsequently used for sequencing library preparation for whole transcriptome analysis using TruSeq RNA sample preparation v2 kit (Illumina Inc. San Diego, CA). The size selected libraries were quantified by PicoGreen assay (Life Technologies, Carlsbad, CA) and visualized with an Agilent Bioanalyzer using a DNA 1000 kit (Agilent Technologies, Waldbronn, Germany). This was followed by cluster generation step in an Illumina cBOT TM instrument following the manufacturer's protocol using a TruSeqTM SR Cluster Kits v3 (Illumina Inc.; San Diego, CA). Finally 6 indexed libraries were loaded into each flow cell lane and sequencing (75 bp single end) was performed on an Illumina HiSeqH2000 instrument (Illumina, Inc.) according to the manufacturer's protocol. Multiplexed single-read runs were carried out with a total of 57 cycles per run (including 7 cycles for the index sequences). Illumina HCS 2.0.12/RTA 1.17.21.3/SAV 1.8.20 Software was used for real-time image processing and base calling.
The data was analyzed for the quality control of the reads, and alignment of reads was made to Mtb reference genome NC_000962, and HumanRefSeqBuilt37 (GRCh37p13). CLC Genomic workbench (v 8) was used for alignment of sequencing reads. The data was normalized and statistical test was performed to find the significantly differentially expressed genes (DEG) with statistically significant p value (< 0.05) with Bonferroni post-test correction. We defined DEG as ≥ 2.0-fold change with a significant p-value < 0.05. Analyses were performed using CLC Genomics workbench 8.0.
2.8. Imaging for Changes in the Intracellular pH
THP-1 and J774A.1 cells were infected with Mtb, as described earlier. At 24, 48, 72 and 168 h post infection, cells were washed and stained for 30 min with pHrodo Green as per the manufacturer's protocol (Life Technologies, USA). The stained cells were fixed with 4% formalin for 15 min and subjected to fluorescent microscopy (Cytation 3 Cell Imaging Multi-mode Reader, BioTek Instruments Inc., USA) to determine the changes in the intracellular pH as relative increase or decrease in the fluorescence intensity. We used the FITC filter sets with excitation and emission at 509/533. Images were taken at 10-times magnification and Gene5 data analysis software was used for image acquisition and analysis (BioTek Instruments Inc., USA). In addition to the qualitative analysis of the images for the changes in the green fluorescence, we also recorded the number of cells, size and mean fluorescence values for each sample to compare the differences between the uninfected and Mtb infected cells. The experiment was performed in triplicate on three different occasions. In order to make sure that findings were not simply due to the fact that Mtb H37Ra was avirulent, we repeated the studies with Mtb H3Rv, with the same mammalian cell lines, with up to 72 h post-infection observation.
2.9. Statistical Analysis for the Laboratory Experiments
Two-way analysis of variance (ANOVA) was used to compare bacterial burden at each time point in GraphPad Prism version 6.0 (GraphPad Software, CA). Linear regression was used to compare the kill slopes of different treatment regimens and for dose-response studies we used the inhibitory sigmoid Emax model.
2.10. Recruitment of Children in India and Artificial Intelligence Analyses
Children were enrolled for pharmacokinetic analysis and clinical outcome ascertainment in two prospective studies in India (Ramachandran et al., 2013, Ramachandran et al., 2015). Recruitment details and ethical considerations for the children were as published before (Ramachandran et al., 2013, Ramachandran et al., 2015). Briefly, parents or guardians gave informed written consent for all children ≤ 6 years-old, while all children aged > 7 years gave assent. The study protocols were approved by the Institutional Ethics Committees of each study site. Children were recruited at multiple hospitals and sites in India, as specified before (Ramachandran et al., 2013, Ramachandran et al., 2015). Tuberculosis case definitions and clinical care were according to India's Revised National TB Control Program (RNTCP), as were definitions of outcome such as treatment completion, failure, and non-adherence. Individual drugs were made available for the entire treatment duration. The treatment doses were as follows: pyrazinamide 30–35 mg/kg, rifampin 10 mg/kg, isoniazid 10 mg/kg, ethambutol 30 mg/kg, and streptomycin 15 mg/kg, three times a week by parent/guardian. Children were admitted for blood draw of samples for pharmacokinetic analyses; treatment was administered under supervision of a study nurse, who watched the children swallow the medicines, after which blood was drawn at 0 (pre-dose), 2, 4, and 8 h. HIV-infected children were treated with antiretroviral therapy as recommended by the National AIDS Control Organization (NACO) guidelines. Drug concentration assays were as described before (Ramachandran et al., 2013, Ramachandran et al., 2015). Patients were followed for at least 6 months.
We utilize several artificial intelligence (AI) approaches and ensembles to identify predictors of outcome in TB, as described in full elsewhere (Gumbo et al., 2014b, Gumbo et al., 2014a, Pasipanodya and Gumbo, 2013). We defined good clinical outcomes in children as completion of therapy that required no further treatment. We excluded children who were non-adherent from the definition of poor outcomes since we have found elsewhere that non-adherence is a poor surrogate of poor microbial or clinical outcomes (Pasipanodya and Gumbo, 2011, Pasipanodya and Gumbo, 2013, Srivastava et al., 2011a). We specified 16 potential predictors, including all clinical and demographic factors, measures of nutrition, HIV-coinfection, and isoniazid, rifampicin, and pyrazinamide concentrations. The classification and regression tree (CART) algorithms search through all these potential predictors and perform binary recursive partitioning in order to classify patients into two groups of maximum homogeneity at a node, the first being the primary node. The process continues to create daughter nodes and branches, hence trees. A variable importance score was assigned to each subsequent daughter node, which was a percentage of how much the daughter nodes improved when added to the primary node. We employed Gini criterion function for splitting nodes and attaining the minimum cost tree. Finally, the deep trees created were pruned to improve the predictive value of the models by using the receiver operator curve (ROC) score. CART employs a cross validation procedure in which new data sets are created by randomly splitting the main dataset into learning and test databases, and calculating the test ROC based on the best tree chosen. However, CART is notorious for over fitting and a bias for multi-way splits. Bootstrap aggregating (bagging) is an ensemble of algorithms that reduce variance and overfitting, reduce bias, and create stable trees (Breiman, 1996). Random Forest performs this function by averaging out the CART decision training trees (i.e., by trees “voting”), to give a modal output (Breiman, 2001). We validated the Random Forest model by setting aside a third of the data, then used the area under the ROC to compare models after running 300 to 500 classification trees. We utilized Salford Predictive Miner System software (San Diego, CA) for the AI analyses.
3. Result
3.1. An Intracellular Mtb and 3D Organotypic Liver Hollow Fiber Co-culture System
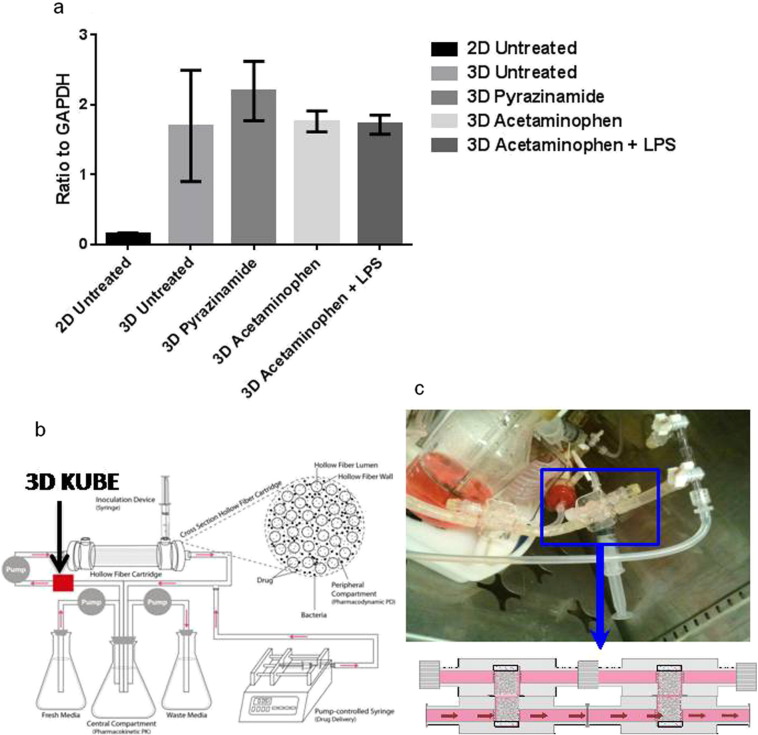
First, we compared the effect of static concentrations of pyrazinamide and APAP (positive control), at the same 24 h area under the concentration-time curve (AUC) concentrations encountered in patients, on 3D-liver cultures compared to HEpG2 monolayers, in triplicate. CYP3A4 was chosen for study because it is responsible for metabolizing more than 50% of all of all pharmaceutical compounds in humans, including APAP (Sevrioukova and Poulos, 2015). Fig. 1a shows quantitative PCR results for the Cytochrome P450 3 A4 (CYP3A4). HepG2 cells had a four-fold increase in CYP3A4 expression in 3D co-culture as compared with the monolayer (Fig. 1a). The intent of the experiment was not to examine the cytochrome P450-mediated metabolism of pyrazinamide, but instead this was a surrogate readout of the 3-D model's overall metabolic capacity as a better approximation of in vivo events, which was higher than in monolayers. These 3DKUBEs were then utilized as modules to examine toxicity in the HFS model. We retrofitted pairs of engineered 3DKUBEs into the media flow path of the central compartment of each HFS, are shown in Fig. 1b–c. The infected human-derived THP-1 cells and the hepatocytes were viable for up to 4 weeks.
Fig. 1.
The intracellular Mtb hollow fiber model of tuberculosis with 3 dimensional organotypic liver modules. (a) 3D KUBES versus a monolayer of HepG2 cells (2-D) were treated with either pyrazinamide 60 μg/mL, or APAP150 μM, or LPS 1 μg/mL; RNA was extracted and subjected to quantitative PCR for CYP3A4. There was higher expression of CYP34A in 3D KUBES compared to 2D cell model (p = 0.0001). Error bars are standard deviation. The experiments were done in triplicate and performed twice to confirm the findings. (b) Schematic diagram of the HFS showing retrofitted 3D KUBEs in the media flow path. (c) An image of 3D KUBEs attached to the flow path of the hollow fiber model system.
3.1.1. Pyrazinamide Dose-effect Studies for Efficacy and DILI Using Pediatric Pharmacokinetics
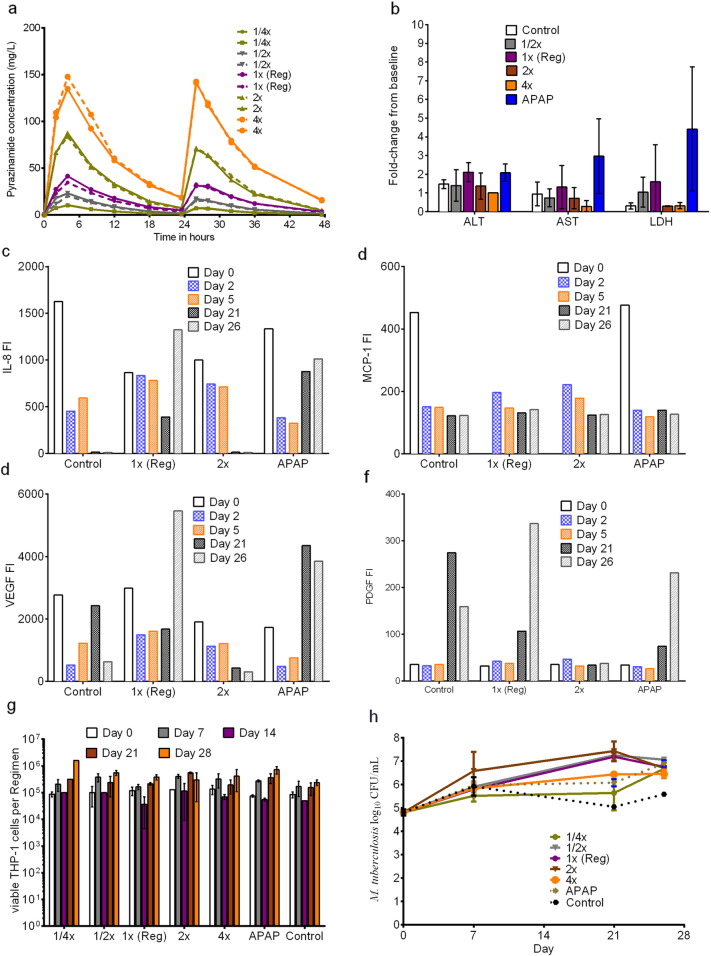
Next, we examined the effect of five pyrazinamide doses and one APAP dose. Each HFS had four 3DKUBEs in the flow path, which were exposed to pyrazinamide concentration-time profiles shown in Fig.2a. The figure shows good recapitulation of serum concentration-time profiles of pyrazinamide in children in each HFS replicate. The pyrazinamide elimination rate constant was 0.117 ± 0.011 h− 1, which is identical to that encountered in children < 6 years old and translates to a half-life of 5.92 ± 0.58 h. Each HFS was sampled once every 7 days for AST, ALT, and LDH concentrations, with results shown in Fig.2b. APAP, the DILI positive control based on the Rumack-Matthews nomogram target (Rumack and Matthew, 1975), was associated with at least 3-fold change for LDH (p < 0.05) and increase in both AST and ALT. On the other hand, there was actually a slight decrease in AST (r2 = 0.21) and ALT (r2 = 0.32) with increased pyrazinamide dose. This was verified not be due to decreased numbers of liver cells from toxicity. Cytokine analysis on the cell culture supernatant from the 3D-liver modules was performed at different time points; results for selected cytokines such as IL8, MCP-1, VEGF, and PDGF, are shown in Fig. 2c–f. The figures show that the pattern of IL-8 and VEGP for APAP treated HFS differ from non-treated controls. The results mean that for pyrazinamide doses up to 100 mg/kg human equivalent (4 times standard), there was no evidence of increased DILI or cytokine release consistent with liver damage, while the positive control APAP demonstrated DILI by these criteria.
Fig. 2.
Assessment of pyrazinamide DILI and efficacy in the HFS. (a) Concentration time profile of each of the five pyrazinamide doses as achieved in the HFS. Shown are the actual observed concentration-time profiles for each dose, demonstrating faithful recapitulation of pyrazinamide pharmacokinetics in pediatric circulatory system in each HFS replicate. (b)When each HFS was sampled every seven days to measure the changes in LFTs, there was virtually no difference between different pyrazinamide doses and untreated controls in enzyme concentrations. However, the hepatotoxicity by APAP was evident based on the significantly higher AST, ALT, and LDH than untreated controls. (c–f) Levels of IL-8, MCP-1, VEGF, and PDGF measured on different days as indicators of hepatotoxicity. Statistically significant increases in levels of IL-8 and VEGF (p < 0.05) were encountered with APAP, known to cause hepatotoxicity, but not with any pyrazinamide dose. (g) Numbers of the viable THP-1 cells in the HFS were not significantly different between control, pyrazinamide and APAP treatment (p > 0.05). (h) Comparison of the kill curves of pyrazinamide dose up to four times of the standard human equivalent dose shows that all doses failed to control the growth of intracellular Mtb.
The number of viable macrophages in the peripheral compartments of the HFS replicates at each sampling time point are shown in Fig. 2g, which demonstrates continued viability up to 4 weeks. The Mtb burden is shown in Fig.2h, which demonstrates that for both the standard and high dose pyrazinamide, the day-to-day Mtb burden was no better than sham treatment. Thus, despite high doses, whose concentrations were validated by direct measurement, pyrazinamide daily doses did not kill intracellular Mtb in the HFS over 28 days of experiment. The minimum inhibitory concentration (MIC) of pyrazinamide against the Mtb isolate, examined at pH 5.8, was 25 mg/L on several occasions, so that failure is not due to intrinsic pyrazinamide resistance of the laboratory strain we used.
3.2. Interferon-γ and TNF-α Do Not Enhance Efficacy or Potency of Pyrazinamide
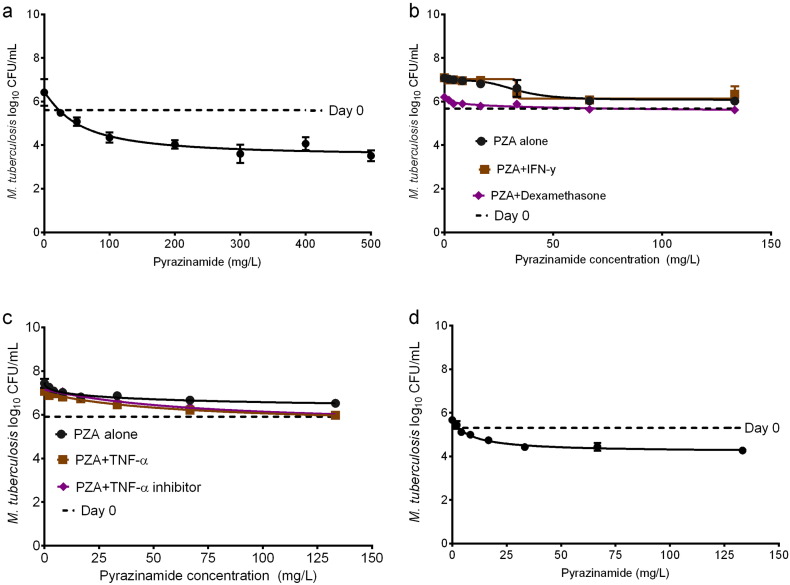
In order to determine if IFN-γ or TNF-α improved pyrazinamide effectiveness, we first examined the effect of several concentrations of pyrazinamide against extracellular Mtb in Middlebrook broth that had been acidified to pH 5.8, with results shown in Fig.3a. The figure shows concentration-dependent killing of Mtb by pyrazinamide with 14 days incubation; the pyrazinamide markedly killed Mtb by about 2-log below day 0 burden (stasis). Based on inhibitory sigmoid Emax model the mean (± standard error) concentration mediating 50% of maximal kill (potency) was a 51.95 ± 15.79 mg/L and maximal kill (efficacy) was 2.93 ± 0.46 log10 CFU/mL on day 14.
Fig. 3.
Effect of IFN-γ on intracellular activity of pyrazinamide. In each experiment, cultures were treated with pyrazinamide concentrations shown for 14 days. There were 3 replicates, and each study was performed. Weighting was by 1/Y2. (a) Pyrazinamide killed extracellular Mtb at pH 5.8, as shown by maximal kill below the day 0 bacterial burden (stasis). (b) In THP-1 human-derived cells treated with either pyrazinamide alone, or with IFN-γ or its inhibitor, the Mtb grew above stasis. (c) Similar results were encountered with exogenous TNF-α or TNF-α antagonist. (d) Pyrazinamide killed the intracellular Mtb in J774A.1 mouse macrophage cell line below stasis.
Next, we activated THP-1 macrophages and performed the same dose–response study with and without IFN-γ or the IFN-γ inhibitor, with day 14 results shown in Fig.3b. There figure shows that there was no net kill of Mtb, with maximal effect occurring at bacterial burdens higher than on day 0, which means bacteria treated with pyrazinamide actually grew. Comparison of inhibitory sigmoid Emax regression fits revealed that the three conditions (pyrazinamide alone versus with IFN-γ versus with IFN-γ inhibitor) shared the EC50 of 30.24 ± 2.72 mg/L and the slope (Hill factor) of 3.03 ± 0.79, while the Emax was actually higher without IFN-γ (1.05 ± 0.13 log10 CFU/mL) compared to with IFN-γ (0.80 ± 0.08 log10 CFU/mL). Similarly, supplementation with either TNF-α or a TNF-α inhibitor resulted in no decrease in microbial burden below stasis (Fig. 3c). All three treatment conditions shared the 3 parameters of EC50, Hill factor, and Emax. Thus, pyrazinamide had no antimicrobial effect in activated and infected macrophages on 6 occasions (each experiment was performed twice), and neither did IFN-γ nor TNF-α improve that. On the other hand, since pyrazinamide kills Mtb in BALB/c mice, we performed the same 14 day pyrazinamide dose-effect study, substituting murine-derived J774A.1 cells for human derived THP-1 cells. Fig.3d shows that pyrazinamide killed Mtb in the murine-derived macrophage cell-line below stasis. The EC50 was 8.65 ± 2.02 mg/L, which is several fold-lower that of pyrazinamide against extracellular Mtb at pH 5.8, while Emax was lower at 1.47 ± 0.14 log10 CFU/mL, but the Hill factor of 1.18 ± 0.44 was similar to results in Fig.3a. Thus pyrazinamide is effective in murine macrophages and extracellularly at pH 5.8, but not in activated human-derived cells.
3.3. Lack of Pyrazinamide-effect in Combination Therapy
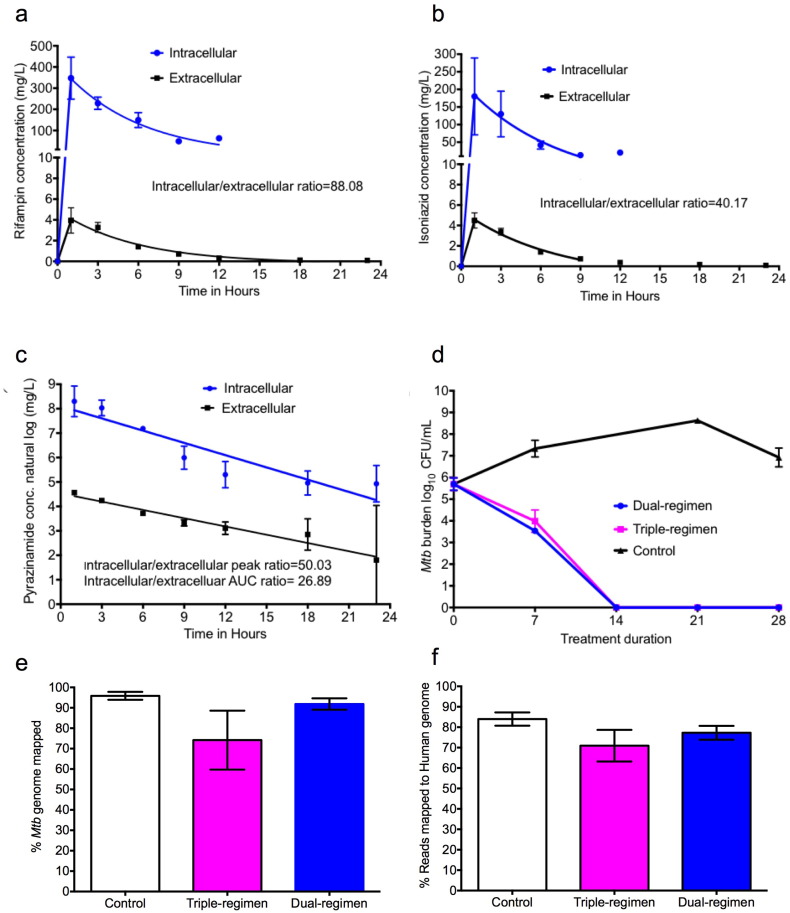
Pyrazinamide could still kill intracellular Mtb when in combination regimens while demonstrating no efficacy as monotherapy due to higher order non-linear interactions between pyrazinamide and rifampin seen in adults (Chigutsa et al., 2015, Pasipanodya and Gumbo, 2013). Therefore, we examined the effect of the three-drug regimen of isoniazid, rifampin, and pyrazinamide (triple-regimen) and compared it to the two-drug regimen of isoniazid and rifampin (dual) in the HFS model. Pyrazinamide was administered at double the standard dose. The concentration-time profiles achieved are shown in Fig. 4a–c, which demonstrate good pharmacokinetic recapitulation of each drug. The figures also demonstrate the capability to produce different half-lifes of drugs in the same HFS, an aspect vital to studying the effects of combination therapy regimens. Importantly, we also simultaneously measured the intracellular concentration-time profile of each of the three antibiotics in the macrophages in the HFS. Fig. 4a–c demonstrates that in fact, at steady state, after 4 weeks of daily dosing, the intracellular concentration-time profiles of rifampin, isoniazid and the pyrazinamide were dramatically higher inside macrophages. For rifampin, the elimination rate constant (and hence half-life) was similar between the intracellular and extracellular pharmacokinetics (0.17 versus 0.19 h− 1), as was the case with isoniazid (0.19 versus 0.21 h− 1) [p = 0.99]. On the other hand, pyrazinamide elimination constant was 0.253 ± 0.112 versus 0.136 ± 0.005 h− 1and volume was 8.379 ± 3.310 × 10− 2 versus 38.8 ± 0.40 × 10− 2 L for intracellular versus extracellular (central compartments), respectively. Fig.4c shows the range of pyrazinamide concentrations on the natural logarithm scale, which reveals a difference in the slopes, indicative of differences in half-life of 3.20 versus 5.14 h. This suggests that an active process eliminates or metabolizes pyrazinamide from mammalian cells faster than it is washed out of the HFS; and that intracellular milieu creates a different pharmacokinetic system within the extracellular pharmacokinetic system. Fig.4d demonstrates the change in bacterial burden with time for the dual versus triple regimens. The Fig.4d shows that there was no difference in kill-rates between the triple versus dual regimens. Thus, pyrazinamide did not contribute to microbial kill of the intracellular Mtb by the combination regimen during the 28-day experiment, despite achieving higher intracellular peak concentrations and 0-24 h AUCs. The failure of therapy is therefore not due to achieving poor drug concentration, but suggests that some intracellular process may be limiting the effectiveness of pyrazinamide.
Fig. 4.
Pharmacokinetic profiles and transcriptome of combination therapy. This experiment was performed twice. Concentration-time profile of (a) isoniazid and (b) rifampin. (c) Given the range of pyrazinamide concentrations from 15 to about 5000 mg/L, we used the natural log transformed concentrations for pyrazinamide in a linear regression. The pyrazinamide concentrations demonstrate 2 different slopes, hence half-life, for pyrazinamide. (d) The dual drug regimen (without pyrazinamide) and the triple drug regimen (with pyrazinamide) had the same kill rates and slopes in the HFS model of intracellular Mtb. Thus, pyrazinamide does not contribute to the microbial kill in this regimen, nor does it lead to faster microbial kill rates.(e) HFS contents were subjected to RNA-Seq and sequences re-aligned against the Mtb genome. The figure shows good overall coverage of the Mtb genome in the different HFS cultures. (f) The percentage of the reads that mapped to the human genome in contents from the dual control, dual-, and triple-drug regimen is shown; the mean was ~ 77%.
3.4. Lack of Pyrazinamide Transcriptomic Signature in Intracellular Mtb
In order to identify the RNA gene-signature specific to pyrazinamide exposure, we exposed extracellular Mtb to pyrazinamide for 4 days, extracted RNA and performed RNA-Seq. There were 106 DEGs based on a non-adjusted p-value: 61genes were up-regulated and 45 down-regulated. The full list of genes and their expression is shown in Supplementary Data Set 1. We compared this to the transcriptome of cultures extracted from HFS treated with either the dual or triple regimen. Fig.4e shows that we were able to re-align the Mtb reads > 90%of the bacterium's genome in HFS contents from infected non-treated, dual, and triple drug regimens. Fig.4f shows the reads that mapped to percentage of the human genome. We then sought to identify the 106-gene pyrazinamide signature in the triple treatment regimen compared to non-treated HFS contents. No single DEG from the 106-transcript pyrazinamide exposure signature was identified in the triple therapy regimen, even with a non corrected p-value. We also compared the Mtb transcriptome of the dual therapy HFS contents to the triple therapy regimen to identify any DEGs between the dual versus triple drug regimens. The transciptomes were virtually identical, with no single DEG to differentiate the two regimens on p-value post-test correction. Thus, we found no transcript evidence of effective pyrazinamide exposure in intracellular Mtb.
As regards to human genes, there were no DEGs identified between dual and triple regimens, with post-test correction. When no post-test correction was performed, which leads to a high false positive discovery rate, there were 103 down-regulated and 118 up-regulated DEGs. Pathway analysis of these DEGs using REACTOME mapped to olfactory transduction (15 genes), metabolic pathways (9 genes) and neuroactive ligands (4 genes). We found no human gene signature indicative of induction of pro-inflammatory pathways, nor a single DEG that mapped to these pathways, in the three-drug regimen. Thus, we found no transcript evidence of induction of pro-inflammatory pathways in human macrophage derived cell line, despite high intracellular pyrazinamide concentrations.
3.5. Mtb Neutralizes the Phagosome Acid pH in Human But Not Mouse Macrophages
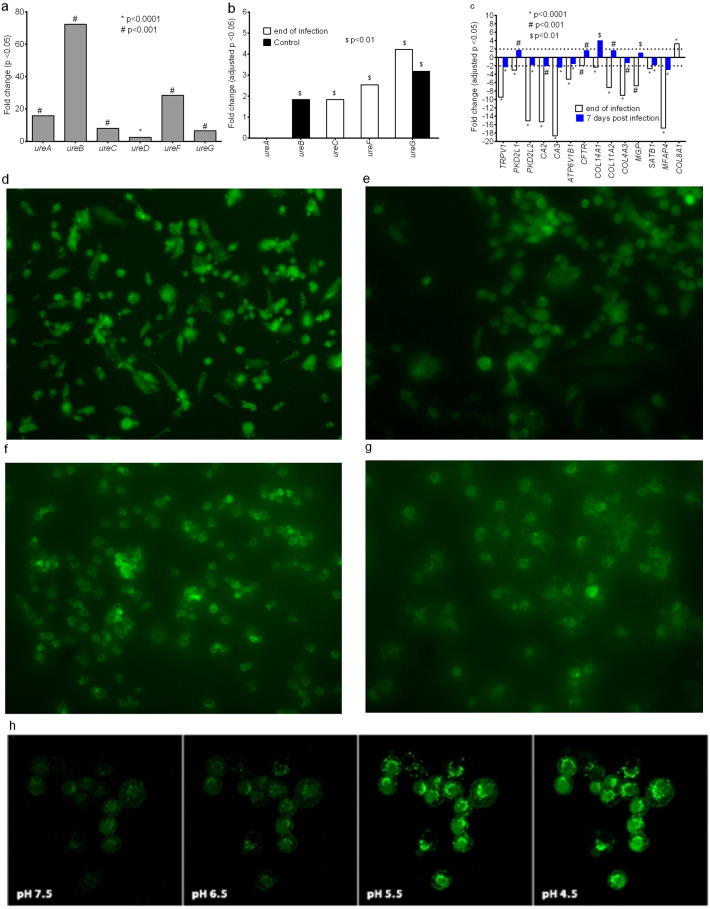
In order to kill Mtb, pyrazinamide is first activated by acidic conditions; the relationship between pyrazinamide effect and pH is described by the Henderson-Hasselbach equation, with better Mtb kill at low pH (McDermott and Tompsett, 1954, Zhang et al., 1999). We hypothesized that one reason for lack of pyrazinamide microbial kill could be that the pH in infected macrophage phagolysosomes is neutralized by Mtb overtime, as proposed by Zhang et al. for cultures in test tubes (Zhang et al., 2002). We exposed Mtb to Middlebrook broth at pH 5.8 buffered using acetic acid, as well as to the same broth at neutral pH with no buffer, and extracted RNA for RNA-Seq after 4 days. Fig.5a demonstrates up-regulation of Mtb genes which encode for urease enzymes that neutralize acidic pH. We sought for the expression pattern of the same urease genes in the Mtb infected PMA-activated THP-1 cells at the beginning of infection (i.e., at the end of bacterial-macrophage co-incubation), and at subsequent time points in the HFS model of intracellular infection. There was up-regulation of some of these same Mtb urease genes at the beginning of infection as was encountered in the extracellular Mtb exposed to acidic pH, suggesting that at transcript level, that the Mtb pH neutralization machinery was up-regulated (Fig. 5b). The genes continued being up-regulated up to 7 days, as shown in Fig.5b. Since this was host-pathogen RNA-seq, we were able to simultaneously examine the human macrophage gene expression. Fig.5c shows the expression of human genes that encode cellular pH sensing proteins in Mtb infected macrophages compared to non-infected macrophages. Fig.5c shows the differential expression of genes encoding intracellular pH proteins that are known to be down-regulated under acidic conditions: TRPV1, PKD2L1, PKD2L2, CA2, CA3,and ATPV1B1(Ishimaru and Matsunami, 2009, Karet, 2002, Nakanishi et al., 2010, Sherry et al., 1994). These genes were down-regulated at the beginning of infection, consistent with low pH conditions, but the direction changed and there was reversal by day 7 (Fig. 5c). On the other hand, COL8A1 is up-regulated under acidic conditions (Bumke et al., 2003), as was the case at the beginning of infection, but by day 7 was no longer significantly up-regulated (Fig. 5c).
Fig. 5.
Intracellular pH of Mtb-infected macrophage. (a) Mtb urease genes were up-regulated under acidic condition. (b) Intracellular Mtb in HFS cultures shows up-regulation of the same Mtb urease genes after 6 h co-culture of the Mtb and THP-1 macrophages, i.e., at the end of macrophage infection and on day 14, demonstrating ongoing attempt to neutralize phagolysosome environment.(c) Macrophage genes encoding pH sensing proteins were down-regulated at the end of the 6 h infection, consistent with an acidic intracellular pH, which reversed on day 7 in the HFS. Similarly COL8A1, whose expression is in the opposite direction, suggests change in the intracellular pH from acidic pH to neutral. (d–e) Neutralization of intracellular pH in Mtb-infected THP-1 cells was confirmed using the pHrodo Green staining. Representative images at 72 h post-infection indicate that the intracellular pH of the non-infected THP-1 cells was still acidic whereas that of the Mtb infected THP-1 macrophages was alkaline. (f–g) In Mtb infected J774A.1 cells there was no change in the intracellular pH at 72 h post-infection, and even on day 7. (h) Fluorescence intensity versus the intracellular pH scale [*, p < 0.05; **, p < 0.01; ***, p < 0.001].
Adopted from life Technologies product manual, cat#P35373.
In order to confirm the RNA expression changes at the level of phenotype, we infected THP-1 cells as well as the mouse JA774A.1 cell line with Mtb and directly measured intracellular pH changes using the pHrodo Green dye. At 24 h post-infection, there were no differences in the intracellular pH of infected and non-infected THP-1 cells. However, Fig. 5d–g show that starting 72 h post infection there was neutralization of acidic pH in the human derived macrophage cell-line, but not in the murine derived cell-line (Fig. 5f–g). The mean green fluorescent intensity in the non-infected THP-1 cells was 24,311 ± 1666 compared to the 9976 ± 587 in the Mtb infected THP-1 cells at 72 h (p < 0.01, r2 = 0.97). Reduction in green fluorescence intensity occurs with increase in pH. On the other hand, the mean fluorescence intensity in the non-infected J774A.1 cells was 24,425 ± 1122 versus 21,070 ± 28.50 in Mtb-infected J774A.1 cells (p = 0.10, r2 = 0.82). Using the virulent Mtb H37Rv, with observations up to 72 h revealed similar results (shown in Supplementary Fig. 1). The fluorescence intensity at 72 h was 25,925 ± 3708 in J774A.1 cells alone compared to 21,069 ± 40 with Mtb-infection (p > 0.05), while in THP-1 cells alone it was 21,917 ± 1492 with Mtb infection (p < 0.005). Taken together, these data mean that there is neutralization of the acid pH in infected human macrophages with time, and since pyrazinamide is activated by acid pH, this could be one reason for lack of efficacy.
3.6. Translation of HFS Results to Children in the Clinic
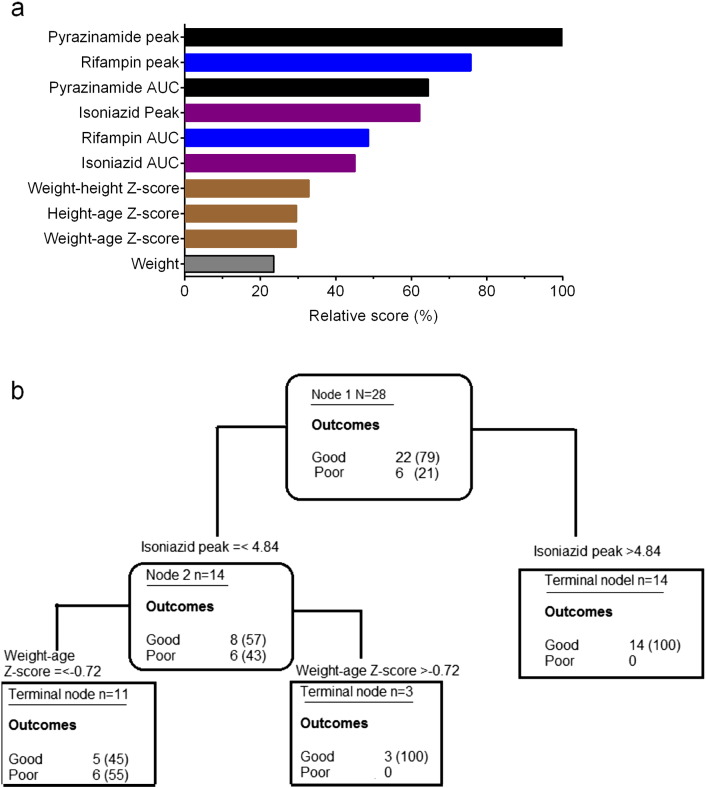
We wanted to confirm if results from our HFS model translate to actual children with extrapulmonary TB treated with pyrazinamide-containing regimens, based on prospective study from India (Ramachandran et al., 2013, Ramachandran et al., 2015). The clinical and demographic characteristics of the 81 children with extrapulmonary TB are shown in Table 1. When children of all ages were examined using the AI method of Random Forests, the most important predictors of good clinical outcome (out of the 16 the model clinical, demographic, and drug concentration factors) were as shown in Fig.6a. The highest r2 for the covariance of concentrations such as AUC and peak concentration within each drug was 0.49, which was encountered with rifampin peak and AUC. Pyrazinamide, isoniazid, and rifampin peak and AUC concentrations, in that order, were the most highly ranked predictors of outcome (Fig. 6a). The findings are virtually similar to our findings in adults with pulmonary TB (Pasipanodya et al., 2013), which means the machine learning algorithms performed adequately.
Table 1.
Comparison of children's clinical and demographic characteristics by outcomes.
| Total N = 81 | Favorable n = 66 | Unfavorable n = 15 | p-Value | ||
|---|---|---|---|---|---|
| Gender | Female n (%) | 39 (48) | 31 (47) | 8 (53) | 0.656 |
| Male n (%) | 42 (52) | 35 (53) | 7 (47) | ||
| HIV test results | Negative n (%) | 55 (68) | 45 (68) | 10 (67) | 0.910 |
| Positive n (%) | 26 (32) | 21 (32) | 5 (33) | ||
| Disease site | Extra-pulmonary | 79 (98) | 64 (97) | 15 (100) | 0.792 |
| Pulmonary and extra-pulmonary | 2 (2) | 2 (3) | 0 | ||
| Treatment category | New | 54 (67) | 44 (67) | 10 (67) | 1.00 |
| Previously treated | 27 (33) | 22 (33) | 5 (33) | ||
| Age | Median (range) | 8 (1.5, 14) | 8 (1.5, 14) | 7 (2, 14) | 0.946 |
Fig. 6.
Pharmacokinetic analysis and clinical outcome as predictors of treatment success. (a) The best predictors of treatment outcome in all 81 children with tuberculosis using the clinical data and machine learning algorithm were the peak and AUC of pyrazinamide, followed by isoniazid, followed by rifampin. Also identified were various measures of nutritional status, based on Z-scores. (b) When only children < 6 years old were analyzed using similar methods, the role of pyrazinamide as a predictor vanished, and only isoniazid concentrations and Z-scores were still significant.
However, when the same analyses were confined to children < 6 years old who had extrapulmonary TB (i.e., babies and toddlers), the findings were as shown in Fig.6b. Only isoniazid peak concentrations were the main drivers of good clinical outcome, followed by weight-for-height Z score (a measure of malnutrition) which was the terminal node. The r2 on cross-validation was 64% compared to 89% on the learn sample, which is reassuring. Based on frequentist statistical inference, 8/14 (57%) children of these children who had an isoniazid peak concentration less than 4.8 mg/L had good outcomes, compared to 14/14 (100%) with concentrations > 4.8 mg/L, an odds ratio of 37.92 (95% confidence interval: 1.89–761.1; p = 0.002). Specifically, pyrazinamide concentrations were no longer drivers of good clinical outcome in toddlers and infants with extrapulmonary TB, consistent with them playing a minimal to no role in predicting outcome.
4. Discussion
The HFS for extracellular Mtb in lung cavities of adult patients, which has been qualified by the European Medicine Agency and received editorial endorsement from the Food and Drug Administration (FDA), has a forecasting accuracy of within 94% of the quantitative clinical values such as optimal exposures (Gumbo et al., 2015a, Gumbo et al., 2015b, Pasipanodya et al., 2015). Since the Mtb in babies and toddlers is intracellular as a result of poor immune containment, our model mimics the same physiological condition of Mtb and the relative lack of an immune system in children. This co-perfused model permits repetitive sampling to enumerate intracellular bacteria, macrophage viability, and serial liver function tests, during exposure to dynamic concentration-time profiles of the drugs specific to young children. This allows for a direct pharmacometric-based translation of efficacy and toxicity from the laboratory to children (Pasipanodya and Gumbo, 2011). Since most of the drugs approved by FDA were tested for DILI in adults, and the hepatotoxicity in children is identified essentially as post-licensing surveillance, our model allows pre-clinical dose testing for hepatotoxicity for the anti-TB compounds. Indeed, toxicity can be examined not just for antibiotics, but for any pharmaceutical compound intended for use in children.
We found a surprising lack of contribution to efficacy from pyrazinamide in the treatment of disseminated TB. Specifically, we demonstrated the lack of pyrazinamide activity inside the human macrophage cell line. The HFS allowed us to delineate two pyrazinamide pharmacokinetic systems, interrelated, systemic versus intracellular, or “wheels within wheels.” Thus, failure was not due to lack of good pyrazinamide penetration into human macrophages, but likely due to a non-acidic pH inside the Mtb-infected macrophages several days post-infection. We found no enhancement of the pyrazinamide effect after adding IFN-γ; nor induction of genes mapping to pro-inflammatory immune pathways on pyrazinamide exposure (Manca et al., 2013). Regardless, the reason why babies and toddlers get disseminated TB is lack of immune containment (i.e., they are effectively immunosuppressed), and thus we would expect little to no help for pyrazinamide from the children's immune systems. This means that pyrazinamide has to exert a direct antibiotic effect in order to be effective. When all 81 children in our study were examined, in a population dominated by older children (> 65%) who often have a pulmonary component of TB, pyrazinamide peak and AUC were the most important predictors of outcome, as was the case in adults in the past (Chigutsa et al., 2015, Pasipanodya et al., 2013). Which one of the two, AUC or peak, is more important would require a separate experimental design. In the past we have found that in South African children, there were low correlations between the peak concentration and AUC for rifampicin and isoniazid, similar what we found here; the exception being pyrazinamide correlation between AUC and peak which was moderate (Hiruy et al., 2015). The important finding, however, is that when children < 6 years old were examined, who are most likely to present with disseminated TB, pyrazinamide was no longer a driver, and none of the pyrazinamide concentrations had any variable importance score. This, lack of pyrazinamide effect based on an agnostic AI algorithm, is consistent with HFS findings for disseminated pediatric tuberculosis. These findings demonstrate the importance of considering the differences in pathological lesions and microbial physiology, between adults and babies and young children. A regimen that specifically caters for these differences could have a better possibility at shortening therapy duration in disseminated tuberculosis.
Finally, we advance some molecular techniques such as host-pathogen RNA-seq, especially given the high GC content of Mtb, and the relative amounts of mammalian RNA to bacterial RNA: proverbial needles in a haystack. Host-pathogen RNA-seq has hitherto been discussed mainly as a theoretical possibility. We were able to map RNA back to ~ 95% of the Mtb genome. This technique, together with the repetitive sampling provided by the HFS, allows use of this “digital” read count information as a pharmacodynamic measure for intracellular bacterial infections, in the context of dynamic drug concentrations. The changes in drug concentration with time and the simultaneous changes in RNA digital reads can then be utilized to build system equations that would allow us to scale from the level of RNA dynamics to bacterial populations to the whole child using the same type of inhomogeneous differential equations we introduced 12 years ago as well as those recently introduced by others (Gumbo et al., 2004, Magombedze and Mulder, 2012, Magombedze and Mulder, 2013).
The following are the supplementary data related to this article.
Intracellular pH changes upon infection with M. tuberculosis H37Rv. Studies were done in replicates of three, and experiments were performed twice. A shows uninfected THP-1 human macrophage cell 72 h post-infection and shows that the intracellular pH was still acidic, while B shows Mtb H37Rv infected macrophages changing towards basic pH at the same time point. C shows uninfected mouse-derived JA774A.1 macrophages at 72 h of incubation while D shows the same cell line that had been Mtb H37Rv infected at the same time point.
Supplementary material
Competing Interests
T.G founded Jacaranda Biomed and serves as a consultant for LuminaCare Solutions. H.E.C and S.Sh are employees of KIYATEC.
Acknowledgments
Funding: This work was supported by the National Institutes of Health (NIH) via DP2 OD001886 and R01AI079497. Author contributions: S.S and T.G. designed the study; S.S., D.D., C.M.S., and K.N.C. performed the experiments; S.S. and T.G. performed RNA extraction, sequencing, and bioinformatic analysis; H.E.C and S.Sh. prepared the 3D KUBES; S.Sw. and G.R. performed the clinical studies; and T.G., J.G.P., S.Sw., and G.R. compiled the data and interpretation. All authors wrote the paper.
References
- Ahmad Z., Fraig M.M., Bisson G.P., Nuermberger E.L., Grosset J.H., Karakousis P.C. Dose-dependent activity of pyrazinamide in animal models of intracellular and extracellular tuberculosis infections. Antimicrob. Agents Chemother. 2011;55:1527–1532. doi: 10.1128/AAC.01524-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballet F. Preventing drug-induced liver injury: how useful are animal models? Dig. Dis. 2015;33:477–485. doi: 10.1159/000374093. [DOI] [PubMed] [Google Scholar]
- Becerra M.C., Swaminathan S. Commentary: a targets framework: dismantling the invisibility trap for children with drug-resistant tuberculosis. J. Public Health Policy. 2014;35:425–454. doi: 10.1057/jphp.2014.35. [DOI] [PubMed] [Google Scholar]
- Breiman L. Bagging predictors. Mach. Learn. 1996;24:123–140. [Google Scholar]
- Breiman L. Random forests. Mach. Learn. 2001;45:5–32. [Google Scholar]
- Bumke M.A., Neri D., Elia G. Modulation of gene expression by extracellular pH variations in human fibroblasts: a transcriptomic and proteomic study. Proteomics. 2003;3:675–688. doi: 10.1002/pmic.200300395. [DOI] [PubMed] [Google Scholar]
- Chigutsa E., Pasipanodya J.G., Visser M.E., van Helden P.D., Smith P.J., Sirgel F.A., Gumbo T., McIlleron H. Impact of nonlinear interactions of pharmacokinetics and MICs on sputum bacillary kill rates as a marker of sterilizing effect in tuberculosis. Antimicrob. Agents Chemother. 2015;59:38–45. doi: 10.1128/AAC.03931-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowle A.J., Dahl R., Ross E., May M.H. Evidence that vesicles containing living, virulent Mycobacterium tuberculosis or Mycobacterium avium in cultured human macrophages are not acidic. Infect. Immun. 1991;59:1823–1831. doi: 10.1128/iai.59.5.1823-1831.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande D., Srivastava S., Meek C., Leff R., Gumbo T. Ethambutol optimal clinical dose and susceptibility breakpoint identification by use of a novel pharmacokinetic–pharmacodynamic model of disseminated intracellular Mycobacterium avium. Antimicrob. Agents Chemother. 2010;54:1728–1733. doi: 10.1128/AAC.01355-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande D., Srivastava S., Meek C., Leff R., Hall G.S., Gumbo T. Moxifloxacin pharmacokinetics/pharmacodynamics and optimal dose and susceptibility breakpoint identification for treatment of disseminated Mycobacterium avium infection. Antimicrob. Agents Chemother. 2010;54:2534–2539. doi: 10.1128/AAC.01761-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devrim I., Olukman O., Can D., Dizdarer C. Risk factors for isoniazid hepatotoxicity in children with latent TB and TB: difference from adults. Chest. 2010;137:737–738. doi: 10.1378/chest.09-2120. [DOI] [PubMed] [Google Scholar]
- Dheda K., Gumbo T., Gandhi N.R., Murray M., Theron G., Udwadia Z., Migliori G.B., Warren R. Global control of tuberculosis: from extensively drug-resistant to untreatable tuberculosis. Lancet Respir. Med. 2014;2:321–338. doi: 10.1016/S2213-2600(14)70031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald P.R., Maritz J.S., Diacon A.H. Pyrazinamide pharmacokinetics and efficacy in adults and children. Tuberculosis. 2012;92:1–8. doi: 10.1016/j.tube.2011.05.006. (Edinb.) [DOI] [PubMed] [Google Scholar]
- Gumbo T., Louie A., Deziel M.R., Parsons L.M., Salfinger M., Drusano G.L. Selection of a moxifloxacin dose that suppresses drug resistance in Mycobacterium tuberculosis, by use of an in vitro pharmacodynamic infection model and mathematical modeling. J. Infect. Dis. 2004;190:1642–1651. doi: 10.1086/424849. [DOI] [PubMed] [Google Scholar]
- Gumbo T., Louie A., Deziel M.R., Liu W., Parsons L.M., Salfinger M., Drusano G.L. Concentration-dependent Mycobacterium tuberculosis killing and prevention of resistance by rifampin. Antimicrob. Agents Chemother. 2007;51:3781–3788. doi: 10.1128/AAC.01533-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbo T., Louie A., Liu W., Brown D., Ambrose P.G., Bhavnani S.M., Drusano G.L. Isoniazid bactericidal activity and resistance emergence: integrating pharmacodynamics and pharmacogenomics to predict efficacy in different ethnic populations. Antimicrob. Agents Chemother. 2007;51:2329–2336. doi: 10.1128/AAC.00185-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbo T., Dona C.S., Meek C., Leff R. Pharmacokinetics-pharmacodynamics of pyrazinamide in a novel in vitro model of tuberculosis for sterilizing effect: a paradigm for faster assessment of new antituberculosis drugs. Antimicrob. Agents Chemother. 2009;53:3197–3204. doi: 10.1128/AAC.01681-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbo T., Siyambalapitiyage Dona C.S., Meek C., Leff R. Pharmacokinetics–pharmacodynamics of pyrazinamide in a novel in vitro model of tuberculosis for sterilizing effect: a paradigm for faster assessment of new antituberculosis drugs. Antimicrob. Agents Chemother. 2009;53:3197–3204. doi: 10.1128/AAC.01681-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbo T., Chigutsa E., Pasipanodya J., Visser M., van Helden P.D., Sirgel F.A., McIlleron H. The pyrazinamide susceptibility breakpoint above which combination therapy fails. J. Antimicrob. Chemother. 2014;69:2420–2425. doi: 10.1093/jac/dku136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbo T., Pasipanodya J.G., Wash P., Burger A., McIlleron H. Redefining multidrug-resistant tuberculosis based on clinical response to combination therapy. Antimicrob. Agents Chemother. 2014;58:6111–6115. doi: 10.1128/AAC.03549-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbo T., Pasipanodya J.G., Nuermberger E., Romero K., Hanna D. Correlations between the hollow fiber model of tuberculosis and therapeutic events in tuberculosis patients: learn and confirm. Clin. Infect. Dis. 2015;61:S18–S24. doi: 10.1093/cid/civ426. [DOI] [PubMed] [Google Scholar]
- Gumbo T., Pasipanodya J.G., Romero K., Hanna D., Nuermberger E. Forecasting accuracy of the hollow fiber model of tuberculosis for clinical therapeutic outcomes. Clin. Infect. Dis. 2015;61:S25–S31. doi: 10.1093/cid/civ427. [DOI] [PubMed] [Google Scholar]
- Hiruy H., Rogers Z., Mbowane C., Adamson J., Ngotho L., Karim F., Gumbo T., Bishai W., Jeena P. Subtherapeutic concentrations of first-line anti-TB drugs in South African children treated according to current guidelines: the PHATISA study. J. Antimicrob. Chemother. 2015;70:1115–1123. doi: 10.1093/jac/dku478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru Y., Matsunami H. Transient receptor potential (TRP) channels and taste sensation. J. Dent. Res. 2009;88:212–218. doi: 10.1177/0022034508330212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins H.E., Tolman A.W., Yuen C.M., Parr J.B., Keshavjee S., Perez-Velez C.M., Pagano M., Becerra M.C., Cohen T. Incidence of multidrug-resistant tuberculosis disease in children: systematic review and global estimates. Lancet. 2014;383:1572–1579. doi: 10.1016/S0140-6736(14)60195-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karet F.E. Inherited distal renal tubular acidosis. J. Am. Soc. Nephrol. 2002;13:2178–2184. doi: 10.1097/01.asn.0000023433.08833.88. [DOI] [PubMed] [Google Scholar]
- Magombedze G., Mulder N. A mathematical representation of the development of Mycobacterium tuberculosis active, latent and dormant stages. J. Theor. Biol. 2012;292:44–59. doi: 10.1016/j.jtbi.2011.09.025. [DOI] [PubMed] [Google Scholar]
- Magombedze G., Mulder N. Understanding TB latency using computational and dynamic modelling procedures. Infect. Genet. Evol. 2013;13:267–283. doi: 10.1016/j.meegid.2012.09.017. [DOI] [PubMed] [Google Scholar]
- Manca C., Koo M.S., Peixoto B., Fallows D., Kaplan G., Subbian S. Host targeted activity of pyrazinamide in Mycobacterium tuberculosis infection. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0074082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott W., Tompsett R. Activation of pyrazinamide and nicotinamide in acidic environments in vitro. Am. Rev. Tuberc. 1954;70:748–754. doi: 10.1164/art.1954.70.4.748. [DOI] [PubMed] [Google Scholar]
- Momper J.D., Mulugeta Y., Burckart G.J. Failed pediatric drug development trials. Clin. Pharmacol. Ther. 2015;98:245–251. doi: 10.1002/cpt.142. [DOI] [PubMed] [Google Scholar]
- Nakanishi M., Hata K., Nagayama T., Sakurai T., Nishisho T., Wakabayashi H., Hiraga T., Ebisu S., Yoneda T. Acid activation of Trpv1 leads to an up-regulation of calcitonin gene-related peptide expression in dorsal root ganglion neurons via the CaMK-CREB cascade: a potential mechanism of inflammatory pain. Mol. Biol. Cell. 2010;21:2568–2577. doi: 10.1091/mbc.E10-01-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton S.M., Brent A.J., Anderson S., Whittaker E., Kampmann B. Paediatric tuberculosis. Lancet Infect. Dis. 2008;8:498–510. doi: 10.1016/S1473-3099(08)70182-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson H., Betton G., Robinson D., Thomas K., Monro A., Kolaja G., Lilly P., Sanders J., Sipes G., Bracken W., Dorato M., Van D.K., Smith P., Berger B., Heller A. Concordance of the toxicity of pharmaceuticals in humans and in animals. Regul. Toxicol. Pharmacol. 2000;32:56–67. doi: 10.1006/rtph.2000.1399. [DOI] [PubMed] [Google Scholar]
- Pasipanodya J., Gumbo T. An oracle: antituberculosis pharmacokinetics–pharmacodynamics, clinical correlation, and clinical trial simulations to predict the future. Antimicrob. Agents Chemother. 2011;55:24–34. doi: 10.1128/AAC.00749-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasipanodya J.G., Gumbo T. A meta-analysis of self-administered vs directly observed therapy effect on microbiologic failure, relapse, and acquired drug resistance in tuberculosis patients. Clin. Infect. Dis. 2013;57:21–31. doi: 10.1093/cid/cit167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasipanodya J.G., McIlleron H., Burger A., Wash P.A., Smith P., Gumbo T. Serum drug concentrations predictive of pulmonary tuberculosis outcomes. J. Infect. Dis. 2013;208:1464–1473. doi: 10.1093/infdis/jit352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasipanodya J.G., Nuermberger E., Romero K., Hanna D., Gumbo T. Systematic analysis of hollow fiber model of tuberculosis experiments. Clin. Infect. Dis. 2015;61:S10–S17. doi: 10.1093/cid/civ425. [DOI] [PubMed] [Google Scholar]
- Ramachandran G., Hemanth Kumar A.K., Bhavani P.K., Poorana G.N., Sekar L., Vijayasekaran D., Banu Rekha V.V., Ramesh K.S., Ravichandran N., Mathevan G., Swaminathan S. Age, nutritional status and INH acetylator status affect pharmacokinetics of anti-tuberculosis drugs in children. Int. J. Tuberc. Lung Dis. 2013;17:800–806. doi: 10.5588/ijtld.12.0628. [DOI] [PubMed] [Google Scholar]
- Ramachandran G., Kumar A.K., Bhavani P.K., Kannan T., Kumar S.R., Gangadevi N.P., Banurekha V.V., Sekar L., Ravichandran N., Mathevan G., Sanjeeva G.N., Dayal R., Swaminathan S. Pharmacokinetics of first-line antituberculosis drugs in HIV-infected children with tuberculosis treated with intermittent regimens in India. Antimicrob. Agents Chemother. 2015;59:1162–1167. doi: 10.1128/AAC.04338-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B., Ghosh S.K., Sutradhar D., Sikdar N., Mazumder S., Barman S. Predisposition of antituberculosis drug induced hepatotoxicity by cytochrome P450 2E1 genotype and haplotype in pediatric patients. J. Gastroenterol. Hepatol. 2006;21:784–786. doi: 10.1111/j.1440-1746.2006.04197.x. [DOI] [PubMed] [Google Scholar]
- Rumack B.H., Matthew H. Acetaminophen poisoning and toxicity. Pediatrics. 1975;55:871–876. [PubMed] [Google Scholar]
- Rumack B.H., Peterson R.C., Koch G.G., Amara I.A. Acetaminophen overdose. 662 cases with evaluation of oral acetylcysteine treatment. Arch. Intern. Med. 1981;141:380–385. doi: 10.1001/archinte.141.3.380. [DOI] [PubMed] [Google Scholar]
- Schmalstieg A.M., Srivastava S., Belkaya S., Deshpande D., Meek C., Leff R., van Oers N.C., Gumbo T. The antibiotic-resistance arrow of time: efflux pump induction is a general first step in the evolution of mycobacterial drug-resistance. Antimicrob. Agents Chemother. 2012;56:4806–4815. doi: 10.1128/AAC.05546-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevrioukova I.F., Poulos T.L. Current approaches for investigating and predicting cytochrome P450 3A4–ligand interactions. Adv. Exp. Med. Biol. 2015;851:83–105. doi: 10.1007/978-3-319-16009-2_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry A.M., Cuppoletti J., Malinowska D.H. Differential acidic pH sensitivity of delta F508 CFTR Cl-channel activity in lipid bilayers. Am. J. Physiol. 1994;266:C870–C875. doi: 10.1152/ajpcell.1994.266.3.C870. [DOI] [PubMed] [Google Scholar]
- Srivastava S., Gumbo T. In vitro and in vivo modeling of tuberculosis drugs and its impact on optimization of doses and regimens. Curr. Pharm. Des. 2011;17:2881–2888. doi: 10.2174/138161211797470192. [DOI] [PubMed] [Google Scholar]
- Srivastava S., Musuka S., Sherman C., Meek C., Leff R., Gumbo T. Efflux-pump-derived multiple drug resistance to ethambutol monotherapy in Mycobacterium tuberculosis and the pharmacokinetics and pharmacodynamics of ethambutol. J. Infect. Dis. 2010;201:1225–1231. doi: 10.1086/651377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S., Pasipanodya J.G., Meek C., Leff R., Gumbo T. Multidrug-resistant tuberculosis not due to noncompliance but to between-patient pharmacokinetic variability. J Infect. Dis. 2011;204:1951–1959. doi: 10.1093/infdis/jir658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S., Sherman C., Meek C., Leff R., Gumbo T. Pharmacokinetic mismatch does not lead to emergence of isoniazid- or rifampin-resistant Mycobacterium tuberculosis but to better antimicrobial effect: a new paradigm for antituberculosis drug scheduling. Antimicrob. Agents Chemother. 2011;55:5085–5089. doi: 10.1128/AAC.00269-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thee S., Seddon J.A., Donald P.R., Seifart H.I., Werely C.J., Hesseling A.C., Rosenkranz B., Roll S., Magdorf K., Schaaf H.S. Pharmacokinetics of isoniazid, rifampin, and pyrazinamide in children younger than two years of age with tuberculosis: evidence for implementation of revised World Health Organization recommendations. Antimicrob. Agents Chemother. 2011;55:5560–5567. doi: 10.1128/AAC.05429-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Scorpio A., Nikaido H., Sun Z. Role of acid pH and deficient efflux of pyrazinoic acid in unique susceptibility of Mycobacterium tuberculosis to pyrazinamide. J. Bacteriol. 1999;181:2044–2049. doi: 10.1128/jb.181.7.2044-2049.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Permar S., Sun Z. Conditions that may affect the results of susceptibility testing of Mycobacterium tuberculosis to pyrazinamide. J. Med. Microbiol. 2002;51:42–49. doi: 10.1099/0022-1317-51-1-42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Intracellular pH changes upon infection with M. tuberculosis H37Rv. Studies were done in replicates of three, and experiments were performed twice. A shows uninfected THP-1 human macrophage cell 72 h post-infection and shows that the intracellular pH was still acidic, while B shows Mtb H37Rv infected macrophages changing towards basic pH at the same time point. C shows uninfected mouse-derived JA774A.1 macrophages at 72 h of incubation while D shows the same cell line that had been Mtb H37Rv infected at the same time point.
Supplementary material