Abstract
We have previously reported that RORγ expression was decreased in ER − ve breast cancer, and increased expression improves clinical outcomes. However, the underlying RORγ dependent mechanisms that repress breast carcinogenesis have not been elucidated. Here we report that RORγ negatively regulates the oncogenic TGF-β/EMT and mammary stem cell (MaSC) pathways, whereas RORγ positively regulates DNA-repair. We demonstrate that RORγ expression is: (i) decreased in basal-like subtype cancers, and (ii) inversely correlated with histological grade and drivers of carcinogenesis in breast cancer cohorts. Furthermore, integration of RNA-seq and ChIP-chip data reveals that RORγ regulates the expression of many genes involved in TGF-β/EMT-signaling, DNA-repair and MaSC pathways (including the non-coding RNA, LINC00511). In accordance, pharmacological studies demonstrate that an RORγ agonist suppresses breast cancer cell viability, migration, the EMT transition (microsphere outgrowth) and mammosphere-growth. In contrast, RNA-seq demonstrates an RORγ inverse agonist induces TGF-β/EMT-signaling. These findings suggest pharmacological modulation of RORγ activity may have utility in breast cancer.
Keywords: Nuclear receptor; RORγ (RORC, RORgamma); Breast cancer; Carcinogenesis; Metastasis; RNA-seq; EMT (epithelial–mesenchymal transition); TGF-β (TGF-beta); Stemness; MaSC (mammary stem cell); DNA repair
Graphical Abstract
Highlights
-
•
RORγ mRNA expression is decreased in the aggressive basal-like subtype and histological grade 3 breast cancers.
-
•
RORγ expression negatively regulates TGF-β/EMT and MaSC pathways, and positively regulates the DNA-repair pathway.
-
•
Agonists of RORγ activity display (in vitro) pharmacological utility against breast cancer.
We have demonstrated that the expression of the druggable and hormone dependent DNA binding protein, the nuclear receptor, RORγ, is decreased in aggressive basal like and histological grade 3 human breast cancers. We then examined the role of RORγ in breast cancer and identified that RORγ: (i) negatively regulates pathways including TGF-β/EMT and mammary stem cell (MaSC)-pathways, that control cell invasion and immortalization, respectively, and (ii) positively regulates pathways necessary for repair of DNA damage. Lastly, we demonstrated that increasing RORγ activity with small molecules (in cells) displays pharmacological utility, suggesting that RORγ could be a target for breast cancer treatment.
1. Introduction
Metastatic breast cancers that acquire therapeutic resistance are associated with high rates of mortality (Gonzalez-Angulo et al., 2007, Singh and Settleman, 2010). The mechanisms underlying the process of metastatic dissemination remain poorly understood owing to the complexity and heterogeneity of breast cancer (Perou et al., 2000). Furthermore, triple negative breast cancers (TNBC) lacking estrogen receptor (ER), progesterone receptor (PR) and HER2 expression (ER −/PR −/HER2 −) are notoriously resistant to hormonal therapy, and associated with poor clinical outcome (Dent et al., 2007). Notably, mesenchymal traits and self-renewal capacity arising from epithelial–mesenchymal transition (EMT) and cancer stem cell (CSC) acquisition are a feature of TNBC (Hennessy et al., 2009). Thus, the identification of new biomarkers underlying mechanisms of tumor dissemination is of paramount importance for TNBC diagnosis and novel therapeutics.
Emerging reports reveal that aberrant expression and function of other nuclear receptors (NRs) are involved in breast cancer (Doan et al., 2014, Muscat et al., 2013). Our previous study identified differential expression of the NR family in ER + ve and ER − ve breast tumors relative to normal breast that underscored the discriminant therapeutic and prognostic value of the NR signature. Importantly, the NR expression profiling study revealed that the expression of RORγ (RORC) was decreased in ER − ve breast cancer and negatively associated with histological grade. Moreover, low RORγ expression correlated with the decreased probability of distant metastasis free survival (DMFS), implying the potential role of RORγ in suppressing advanced breast cancer (Oh et al., 2014). Despite potential evidence for the role of RORγ in breast cancer, the exact molecular mechanisms underlying RORγ-mediated regulation of breast cancer and/or potential pharmacological utility have not been characterized and reported.
Interestingly, RORγ2 (RORγt), a highly related T-cell specific isoform (that has identical DNA- and ligand-binding regions to RORγ1 with minor differences in the N-terminal AB region) plays a critical and hierarchical role in Th17 cell differentiation (Ivanov et al., 2006, Yang et al., 2008). Antagonists of RORγ display utility as anti-inflammatory compounds and for Th17-dependent autoimmune diseases (Solt et al., 2011). However, RORγ agonists are also gaining traction in the therapeutic arena, compounds with increasing activity display pharmacological value in immunotherapeutic approaches to fight cancer by stimulating pro-inflammatory cytokine production and effector T cells (Carter et al., 2015). These characteristics underscore the role of this NR at the nexus of pathways effecting immune suppression (for autoimmune disease) in contrast to activation (for immune-mediated tumor inhibition).
Our study has explored the function of RORγ in negative regulation of metastasis and aggressive tumorigenicity in breast cancer. The study utilizes pharmacological and genetic modulation of RORγ activity and expression (in well-characterized ER + ve and ER − ve cell lines) and demonstrates that (in vitro) induction of RORγ-dependent pathways display anti-cancer efficacy independent of immunotherapeutic pathways. This has revealed that RORγ-dependent programs suppress pathways driving pro-metastatic, EMT and the acquisition of stemness necessary for breast cancer progression and metastasis.
2. Materials and Methods
2.1. Cell Culture and Transfection
T-47D, MCF-7 and MDA-MB-231 cells were initially purchased from ATCC and maintained in RPMI-1640 (for T-47D) or DMEM/F-12 (for MCF-7 and MDA-MB-231) supplemented with 10% FBS. Cell lines were genotyped using STR profiling and tested for mycoplasma contamination. For cells embedded in 3D Matrigel, MCF-7 and MDA-MB-231 cells were grown in DMEM supplemented with 10% FBS and 10 μg/ml of insulin. To knockdown RORγ expression, cells were transfected for 48 h with stealth siRNAs (Invitrogen, HSS109300, HSS109301, HSS109302) that were purchased from Life Technologies. A final concentration of 10 nM of siRNAs was used with RNAiMAX (Invitrogen) following the manufacturer's instruction. A stealth siRNA negative control Med GC (Invitrogen, 12935-300) was used in parallel to normalize the background effect. For over-expression of RORγ, the cDNA vector (Origene, SC121242) was used with Lipofectamin 2000 (Invitrogen), following the manufacturer's guide.
2.2. RNA Isolation and RT-qPCR
RNA was isolated using the Trizol reagent to extract RNA, followed by turbo DNase treatment and RNA clean up with RNeasy RNA column as described previously (Dowhan et al., 2012, Oh et al., 2014). To synthesize complementary DNA, 600 ng of total RNA was used with Taqman reverse transcription from Invitrogen. RT-qPCR was performed using the ViiA7™ RT-qPCR system with primer sets. Primer sequences were presented in Fig. S1. To determine the relative expression compared to control, Ct values were normalized to RPLP0, and relative expression was calculated using the ΔΔCt method. For quantification of LINC00511 expression in breast cancer cell lines, Ct values were normalized to GAPDH, and relative expression was calculated using the ΔCt method.
2.3. Breast Cancer Dataset Analysis
RORγ expression was extracted from the web database of TCGA (TCGA-data-portal) with clinic data. UNC (Harrell et al., 2012) dataset was established as described previously (Oh et al., 2014) and a custom R script was used to retrieve mRNA expression data and phenotype data from UNC dataset. Profiling RORγ expression in the GEO breast cancer collection was performed with GOBO (Ringner et al., 2011) (co.bmc.lu.se/gobo/). Kaplan–Meier survival curves were generated using the KMplot (Györffy et al., 2010) (kmplot.com). RORγ expression in different breast cancer grades was extracted from NKI, UPP, MAINZ and TRANSBIG datasets using a custom R script. The stratified data were analyzed and plotted using the Prism software (version 6). To determine the significance in human cohorts, non-parametric ANOVA with the Kruskal Wallis test was performed in the Prism software. The ChIP-chip data from the Kittler et al. 2013 study including the RORγ (RORC) binding file (Bed format) is available from GSE41995 (GEO).
2.4. Library Preparation and RNA-seq Analysis
Library preparation and sequencing were performed at the IMB Sequencing Facility of the University of Queensland. Total RNA sample libraries were generated using the Illumina TruSeq Stranded mRNA LT sample preparation kit (Illumina, Part no. RS-122-2101 and RS-122-2102), according to the standard manufacturer's protocol (Part no. 15031047 Rev. E October 2013). The mRNA denaturation and elution were performed with 0.1 μg to 0.2 μg of total RNA depending on amount of sample available prior to a heat fragmentation step aimed at producing libraries with an insert size between 120 and 200 bp. cDNA was then synthesized from the enriched and fragmented RNA using SuperScript II Reverse Transcriptase (Invitrogen, Catalog no. 18064014) and random primers. The resulting cDNA was converted into double stranded DNA in the presence of dUTP to prevent subsequent amplification of the second strand and thus maintain the strandedness of the library. Following 3′ adenylation and adaptor ligation, libraries were subjected to 15 cycles of PCR to produce RNA-Seq libraries ready for sequencing. Prior to sequencing, RNA-Seq libraries were qualified via the Agilent Bioanalyzer with the High Sensitivity DNA kit (Integrated Sciences, Part no. 4067–4626). Quantification of libraries for clustering was performed using the KAPA Library Quantification Kit — Illumina/Universal (KAPA Biosystems, Part no. KK4824) in combination with the Life technologies Viia7™ RT-qPCR instrument. Sequencing was performed using the Illumina NextSeq500 (NextSeq control software v1.2/Real Time Analysis v2.1) platform. The library pool was diluted and denatured according to the standard NextSeq500 protocol and sequencing was carried out to generate single-end 76 bp reads using a 75 cycle NextSeq500 High Output reagent Kit (Illumina, Catalog no. FC-404-1005). Reads were mapped against the reference genome (GRCh38.p2) using STAR (Dobin et al., 2013), and read counts for each gene in the Ensembl annotation were generated using htseq-count in the HTSeq python package (Anders et al., 2015) and the GENCODE annotation. Differential expression (n = 3) was detected using the DESeq2 (Love et al., 2014) packages in R. Coding genes and long non-coding genes were separated using the reference of long non-coding genes (GENCODE). It should be noted that there are two isoforms of RORγ, RORγt expression is restricted thymocytes, and RORγ1 is the predominant isoform in other cells and tissues. These isoforms have identical DNA and ligand binding domains, with very minor differences in the N-terminal region. Examination of the RNA-seq data indicates that RORγt expression is absent in the breast cancer cell lines, however, the probes used to interrogate the human breast cancer data sets (which are primarily derived from affymetrix data) cannot discriminate between RORγt and RORγ1 expression.
2.5. Pathway Analysis, Survival (signature) Analysis and Cis-regulatory Analysis
Pathway analysis was conducted utilizing the Ingenuity Pathway Analysis (IPA) and the gene set enrichment analysis (GSEA). For IPA, significantly and differentially expressed genes with fold changes information were loaded into the IPA system (loss of function datasets; FDR < 0.05, Fold changes > 1.3 and gain of function dataset; FDR < 0.05, fold changes > 1.2). IPA computes an activation z-score of each pathways or annotations as previously described (Krämer et al., 2014). We used bias-corrected activation z-score to avoid biased analysis outcome. For GSEA, we downloaded the GSEA tool. All genes (> 50,000) with normalized counts information after DESeq2 was loaded into the GSEA and ran in a default condition with the gene symbol annotation. HALLMARK gene set was used to determine the important pathways (top 5 positively or negatively enriched), using normalized enrichment score (NES). For other gene set analysis, the target gene set was manually downloaded and ran with RNA-seq data in a default condition. To integrate ChIP-chip data to our RNA-seq study, we manually downloaded the RORγ ChIP-chip data that was generated in MCF-7. The annotation of hg18 was transformed into hg19 using the lift-over from the UCSC. RORγ binding was shown using the UCSC web-browser, covisualizing conservation scores (phyloP and phastCon) and histone marks (H3K4Me1, H3K4Me3 and H3K27Ac from 7 cell lines of ENCODE). For survival analysis with RORγ-dependent gene signature, we utilized the genefu package in R as previously described (Oh et al., 2014). For clarification, a low RORγ gene expression signature score indicates that the (RORγ signature) genes significantly decreased in the RORγ knockdown cells are up-regulated relative to the (RORγ signature) genes significantly increased after RORγ knockdown cells. Accordingly, a negative or low RORγ-dependent signature score is suggestive of relatively higher RORγ expression levels and a positive or high RORγ-dependent signature score is indicative of RORγ dysfunction (i.e. lower RORγ expression levels). For cis-regulatory analysis, we utilized the GREAT analysis as previously described (McLean et al., 2010). GREAT computed RORγ binding sites information (hg19) to identify cis-regulatory target genes, RORγ binding location from the TSS and enriched pathways/terms with target genes. To examine the correlation of lncRNA and coding genes, we also utilized the GREAT. Using the location of lncRNA from T-47D, we identified the target coding genes and generated the correlation plot showing both fold changes of lncRNAs and target coding genes.
2.6. PPI Network Analysis
For protein–protein interaction, we utilized the Netgestalt previously described (Shi et al., 2013). We selected the iRef database as a network view and uploaded differentially expressed genes of RNA-seq outcomes with fold changes (log2 scale) in a single binary tract format. The PPI network structure of highlighted pathways from RNA-seq analysis was visualized with fold changes in log2 scale.
2.7. Western Blot Analysis
Cells were lysed using RIPA buffer containing the protease inhibitor (Roche). The concentration of protein samples was measured with the BCA kit (Thermo Scientific). Appropriate amount of protein samples was loaded onto SDS-PAGE and transferred to the PVDF membrane, followed by blocking with 5% skim milk. The membrane was incubated with primary antibody overnight and detected using the ECL reagent, Immobilon Western chemiluminescent horseradish peroxidase substrate (Millipore). The film (Kodak) was exposed with the X-OMAT 2000 film developer (Kodak). Antibodies used for western blotting are: RORγ (Merck Millipore, MABF81), c-MET (Cell Signaling Technology, #8198), SMAD3 (Cell Signaling Technology, #9523), GAPDH (R&D, 2275-PC-100) and HRP anti-rabbit (Cell Signaling Technology, #7074) and HRP anti-mouse (Zymed, 62-6520).
2.8. Immunohistochemistry
Formalin-fixed paraffin-embedded tissue sections of breast cancer (14 ER −, 15 ER +) and normal breast tissues (13 cases) were obtained from the Australian Breast Cancer Tissue Bank (abctb.org.au) and the Victorian Cancer BioBank (viccancerbiobank.org.au). Tissue sections of normal breast tissue biopsies were obtained from the Susan G. Komen for the Cure Tissue Bank at the IU Simon Cancer Center. Normal tissues were from women with no known history of breast disease and were collected following reduction mammoplasty or from volunteers who donated normal breast tissue biopsies. Breast cancer cases were primary invasive ductal carcinomas, with known hormone receptor status, tumor grade and age at diagnosis (Muscat et al., 2013). All tissues were obtained with informed consent from donors, and the use of tissues received approval from the human research ethics committees of the participating institutions. Immunoperoxidase staining was performed using anti-RORγ (NR1F3) rabbit polyclonal antibody (Abcam, Melbourne, Australia, ab78007). Tissues were first heat treated under pressure in citrate buffer, as described previously (Mote et al., 1999), to reveal epitopes. Primary antibody incubations were performed at 4 °C, overnight, in phosphate buffered saline (pH 7.5) containing 0.5% Triton X-100. Primary antibody binding was revealed by subsequent incubation 30 min at room temperature with biotinylated goat anti-rabbit secondary antibody (Dako Australia, Botany, Australia), then one hour, room temperature incubation with streptavidin-horseradish peroxidase conjugate (Dako Australia), followed by color development with diaminobenzidine substrate solution (Dako Australia). Stained sections were scanned using a Hamamatsu Nanozoomer digital slide scanner using a 40 × objective.
2.9. Cell Viability Assay
Cells were plated in quadruplicate with the same confluence. In a following day, cells were treated with RORγ small molecules (10 μM of SR1078/agonist and 5 μM of SR2211/antagonist) over several days without changing the media. Cell viability was measured with the WST-8 (2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt) (Sigma Aldrich, 96992-500TESTS-F), following the manufacturer's instruction.
2.10. Wound Healing Assay
Wound healing assay was performed as previously described (Oh et al., 2014). Cells were transfected with control-siRNA or RORγ-siRNA for 24 h in a 6-well plate. Scratches were created across the monolayers of cell lines using a sterilized yellow tip. Images of wounds were taken at 0-, 24-, and 48-h time points using a 4 × objective of the Nikon Ti–U inverted fluorescence microscope under bright field. Cell migration (gap closure) of scratch wounds was measured using the ImageJ software.
2.11. 3D-Organotypic Growth Assay
MCF-7 and MDA-MB-23 A cells were diluted in complete media supplemented with 5% growth reduced Matrigel (BD Bioscience) and seeded onto solidified Matrigel cushions (150 μl/well) in 29 cm glass bottom dishes (2 × 104 cells/cm2). Cell microsphere outgrowth was observed by Nikon ECLIPSE Ti–U inverted phase contrast microscope. Longitudinal cell growth was normalized to an initial reading taken 24 h after cell plating and addition of SR1078 (10 μM).
2.12. Mammosphere Assay
Mammosphere assay was performed as previously described (Lo et al., 2012). Material and reagent were prepared as followed; a 96-well ultra low attachment plate (Corning) and DMEM/F12 media containing 20 ng/ml epidermal growth factor (Sigma Aldrich), 10 ng/ml basic fibroblast growth factor (Sigma Aldrich), 5 μg/ml insulin (Sigma Aldrich), 50 × B27 supplement (Life technologies) and 0.4% FBS (Gibco). Cells were counted and seeded 200 cells (in 200 μl) per well in the ultra low attachment plate with mixed mammosphere media. For treatment, 10 replicates were seed and incubated with either DMSO or RORγ small molecules for 6 days. Images were taken using the Nikon Ti–U inverted fluorescence microscope under bright field.
2.13. Statistical Analysis
Statistical analysis was performed with Prism software version 6 of GraphPad. All data was presented as mean of S.E.M. For two groups comparison, the two-tailed student's t-test was used. When three or more groups were compared, the one-way analysis of variance (ANOVA) test was used to measure significant effect of one factor. For the statistical analysis in wound healing assay and 3D-organotypic outgrowth assay, the two-way ANOVA was used to determine significant effect of two factors. ANOVA test was followed by Tukey's post hoc test to correct familywise error-rate (FWER). For RORγ expression in human data cohorts, non-parametric ANOVA with the Kruskal Wallis test was performed in the Prism software. Pearson test was used to determine the correlation between RORγ expression and target genes expression. Significance was denoted as NS, *, ** and ***, meaning not significant, P < 0.05, P < 0.01 and P < 0.001, respectively.
3. Results
3.1. RORγ Expression is Decreased in Aggressive Basal-like and Grade 3 Breast Cancers
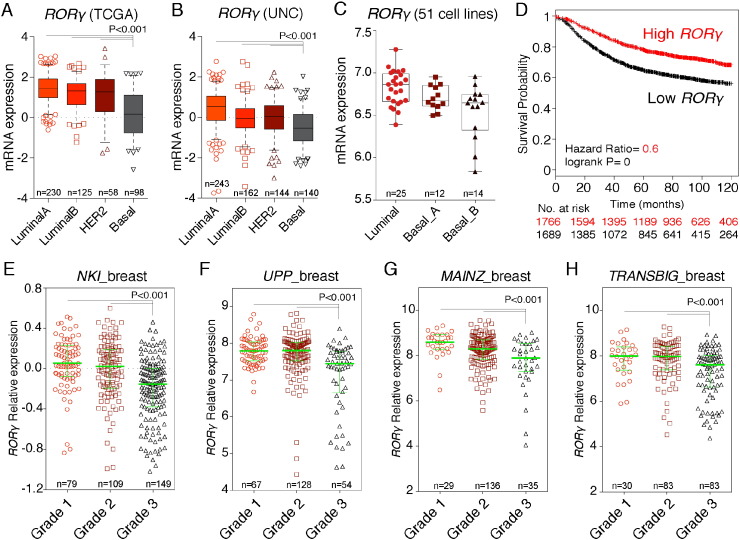
To explore the role of RORγ, we examined the RORγ expression in different human breast cancer subtypes. We observed RORγ mRNA expression is more abundantly expressed in luminal A and B subtypes, and poorly expressed in aggressive basal-like subtype in the TCGA and UNC datasets (P < 0.001) (Fig. 1A and B). Assessment and correlation of RORγ expression levels with intrinsic subtypes was performed using the Hu et al. and PAM50 breast cancer datasets (1881 patients) (Ringner et al., 2011). This analysis demonstrated significantly (P < 0.00001) decreased RORγ expression in basal-like subtypes (Fig. S2A and B). Furthermore, the curated expression data on RORγ in the 51 breast cancer cell lines (Neve et al., 2006) verified that RORγ mRNA expression is poorly expressed in the basal (A and B)-aggressive cell lines relative to expression in the luminal lines (Fig. 1C).
Fig. 1.
Decreased RORγ expression in aggressive basal-like subtype and grade 3 breast cancer.
(A and B) RORγ mRNA expression across molecular subtypes of breast cancer in (A) TCGA and (B) UNC.
(C) Stratified RORγ mRNA expression in molecular subtypes of 51 breast cancer cell lines.(D) Kaplan–Meier survive curve depicting the RFS rate comparison between patient groups with high and low RORγ expression.(E) Decreased RORγ expression in histological grade 3 cancer in four different breast cancer datasets.
Previously, we showed that increased RORγ expression improved the probability of metastasis free survival (Oh et al., 2014). We subsequently explored the significance of RORγ expression on relapse free survival (RFS). Utilizing Kaplan–Meier survival analysis and KMplot (n > 3400), we determined that the patient group with higher RORγ expression displayed increased probability of RFS (Fig. 1D). In this context, we also observed that RORγ expression is negatively associated with histological grade, i.e. RORγ is decreased in grade 3 breast cancer in several human breast cancer cohorts, NKI (van't Veer et al., 2002), UPP (Miller et al., 2005), MAINZ (Schmidt et al., 2008), TRANSBIG (Desmedt et al., 2007) (Fig. 1E–H) and in the combined GEO datasets (Fig. S2C). Furthermore, we confirmed that RORγ expression is significantly decreased in ER − ve cases, compared to ER + ve breast cancer in the combined GEO collection (Fig. S2D), and immunohistochemistry validated RORγ protein expression is decreased in ER − ve breast cancer relative to normal human breast (Fig. S2E). Together, these data underscore decreased expression of RORγ is associated with aggressive basal-like breast cancer and histological grade 3 breast cancer, in accord with the protective effects of increased RORγ expression on clinical outcomes.
3.2. RNA-seq Analysis Revealed that RORγ Negatively Regulates Oncogenic Pathways
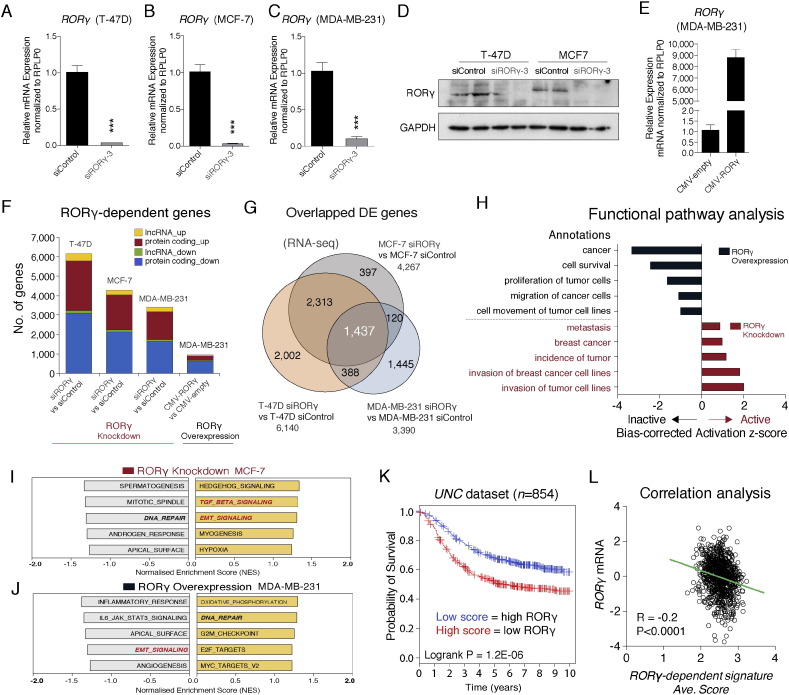
To identify and characterize RORγ-dependent gene expression and signaling pathways relevant to breast carcinogenesis, we performed RORγ siRNA-mediated knockdown and expression profiling in several breast cancer cell lines, including (ER + ve) T-47D, MCF-7 and (ER − ve) MDA-MB-231 cell lines expressing high to low levels of RORγ mRNA. We obtained significant decreases (~ 90%) of RORγ mRNA expression in all three cell lines (Fig. 2A–C). We also validated the siRNA knockdown efficiency on RORγ protein expression by western analysis (Fig. 2D). In parallel, we over-expressed RORγ in MDA-MB-231 (Fig. 2E).
Fig. 2.
Identification of RORγ-dependent genes and pathways by RNA-seq coupled with RORγ gain and loss function.
(A–C) Transfection of RORγ siRNA into (A) T-47D, (B) MCF-7 and (C) MDA-MB-231 significantly decreases RORγ mRNA expression (n = 3).
(D) Western blot analysis confirming depletion of RORγ expression.
(E) Transfection of a RORγ expression vector into MDA-MB-231 significantly increases RORγ mRNA expression (n = 3).
(F) Numbers of significantly differentially expressed transcripts identified after RORγ gain and loss of function in T-47D, MCF-7 and MDA-MB-231 (n = 3).
(G) Overlapping and distinct RORγ-dependent genes (after RORγ siRNA transfection, i.e. loss of function) in T-47D, MCF-7 and MDA-MB-231.
(H) Ingenuity Pathway Analysis (IPA) exhibiting bias-corrected activation z-scores of RORγ-dependent functional pathways after RORγ gain and loss of function in ER − ve MDA-MB-231 cells.
(I and J) GSEA analysis showing top five positive and negative enriched modules derived from HALLMARK gene sets after (I) RORγ depletion and (J) over-expression.
(K) Long-term clinical outcome predicted from RORγ-dependent signature after Kaplan–Meier survival analysis in the UNC cohort depicting the survival rate comparison between high and low RORγ-dependent signatures.
(L) Correlation analysis between RORγ mRNA expression and the RORγ-dependent signature scores in the UNC dataset.
We performed RNA-seq analysis in breast cancer lines (after gain and loss of RORγ function) coupled with STAR (Dobin et al., 2013)-DESeq2 (Love et al., 2014), differential expression (DE) pipeline. We identified that 2000–6000 genes were differentially expressed in a significant and robust manner (FDR < 0.05, Fold change > 1.3) in T-47D, MCF-7 and MDA-μΒ-231 after RORγ knockdown (Fig. 2F). Fig. 2F displays the number of coding and noncoding genes that are differentially (and significantly) up and down regulated after loss of RORγ function. Moreover, the RNA-seq analysis identified RORγ-dependent genes that were differentially expressed and common to both ER + ve and ER − ve breast cancer cell lines (i.e. 1437 genes in T-47D, MCF-7 and MDA-MB-231) (Fig. 2G). In addition, RNA-seq and DE analysis after over-expression of human RORγ in MDA-MB-231 revealed that RORγ over-expression induced the significant differential expression of ~ 800 genes (FDR < 0.05, Fold change > 1.2) (see Fig. 2F).
Importantly, Ingenuity Pathway Analysis (IPA) functional analysis shows the opposing effects of RORγ gain and loss of function. For example, RORγ over-expression in MDA-MB-231 cells suppresses pathways that control cell survival, cell proliferation and migration (Fig. 2H). In contrast, RORγ knockdown activates pathways that induce breast cancer, tumor incidence and metastasis (Fig. 2H). Furthermore, GSEA (Subramanian et al., 2005) confirmed that RORγ knockdown positively enriched transforming growth factor-beta (TGF-β)/EMT signaling modules of hallmark gene sets in MCF-7, T-47D and MDA-MB-231 (Figs. 2I, S3A and B). Conversely, RORγ over-expression suppressed genes of EMT signaling module in MDA-MB-231 cells (Fig. 2J). Notably, GSEA using the genes that were regulated in common in all three cell lines by RORγ depletion (Fig. 2G, n = 1437) still revealed the significantly positive enrichment of TGF-β and EMT modules (Fig. S3C). Hence, these data indicate that RORγ-dependent gene expression in breast cancer cells is associated with genetic programs and pathways that control critical aspects of breast cancer progression and metastasis.
3.3. RORγ-dependent Gene Signature Displays Prognostic Value for Breast Cancer Patient Survival
To extend our understanding of the role of RORγ-dependent genes (i.e. a RORγ-dependent gene signature) on clinical outcome in breast cancer, we utilized the previously applied bioinformatic survival analysis with gene expression signatures (Dowhan et al., 2012, Oh et al., 2014). Using RNA-seq data gene set from T-47D cells, we extracted expression levels of the RORγ-dependent genes from the microarray data of individual patients in the human cohorts (Harrell et al., 2012, Sotiriou et al., 2006). Then we generated the average score to reflect the gene expression changes by RORγ knockdown. With this average score, we separated patients into two groups as high average score and low average score groups and performed Kaplan–Meier survival analysis. Survival analysis shows that the group with higher average score (decreased RORγ expression) has a poorer clinical outcome in the UNC (Harrell et al., 2012) dataset (Fig. 2K). Consistently, we found that in the human cohort, UNT (Sotiriou et al., 2006), there was a similar survival pattern with the RORγ-dependent gene set (Fig. S3D). In keeping with this notion, we observed a significant inverse correlation between the RORγ gene expression levels and RORγ-dependent signature average scores in the UNC (Fig. 2L) and UNT (Fig. S3E) breast cancer data sets. In summary, the RORγ-dependent signature also displays prognostic power in human cohorts.
3.4. RORγ Negatively Modulates TGF-β/SMAD Signaling and EMT Processes: RORγ Directly Targets Critical Genes
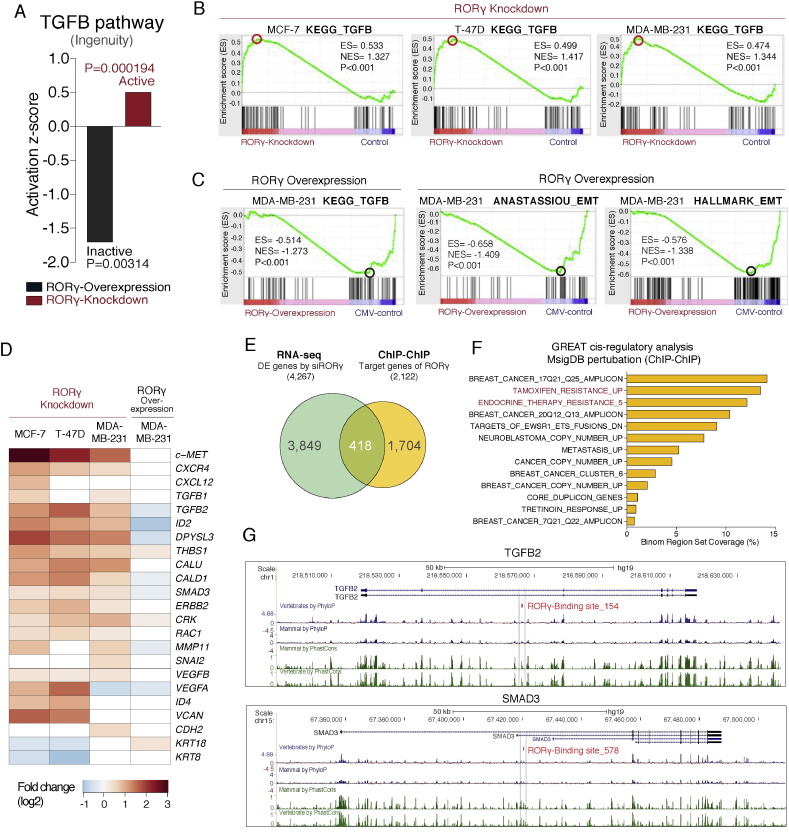
We examined the negative association of RORγ and TGF-β/EMT signaling in more detail. Canonical pathway analysis demonstrated that RORγ over-expression was significantly associated with a negative activation score, indicating suppression of the TGF-β signaling annotation in MDA-MB-231 cells (Fig. 3A). Conversely, RORγ knockdown was associated with an increased activation score, showing induction of the TGF-β signaling annotation (Fig. 3A). Subsequently, we utilized GSEA to mine the RNA-seq data and GSEA clearly demonstrated that the RORγ-dependent gene signature (after depletion of RORγ) increased the enrichment score for the KEGG TGF-β module in MCF-7, T-47D cells, and MDA-MB-231 cells (Fig. 3B). In contrast, GSEA analysis with RORγ over-expression data in MDA-MB-231 identified the inactivation of TGF-β signaling modules (Fig. 3C). This is underscored by the GSEA analysis in MDA-MB-231 cells that identified two well-documented EMT gene signatures (Anastassiou et al., 2011, Liberzon et al., 2011) (Fig. 3C). These observations were further highlighted by the heatmap of increased and decreased expression of EMT-related genes and tumor microenvironment-modifying genes in MDA-MB-231 after RORγ knockdown and over-expression, respectively (Fig. 3D).
Fig. 3.
RORγ expression regulates the TGF-β, EMT and pro-metastatic pathways in breast carcinogenesis.
(A) IPA showing a decreased (negative) and increased (positive) activation score for the TGF-β signaling pathway after RORγ gain and loss of function, respectively, in MDA-MB-231.
(B) GSEA analysis identifies positively enriched TGF-β signaling modules in MCF-7, T-47D and MDA-MB-231 cells after RORγ depletion.
(C) GSEA analysis identifies negatively enriched TGF-β and EMT modules in MDA-MB-231 cells after RORγ over-expression.
(D) Heatmap representation of differentially expressed genes associated with EMT and malignant breast cancer.
(E) Overlap between differentially expressed genes after RORγ knockdown (MCF-7 cells, FDR < 0.05, Fold change > 1.3) and direct RORγ target genes derived from GREAT analysis with RORγ ChIP-chip data from MCF-7.
(F) GREAT analysis of ChIP-chip data significantly highlighted 13 terms of MSigDB perturbation, and identified many breast cancer-related pathways (associated with direct RORγ action).
(G) RORγ binding sites identified in the regulatory regions of the TGFB2 and SMAD3 genome loci. Conservation score in mammal and vertebrate was presented using the UCSC web browser.
We endeavored to gain additional insights into the underlying molecular mechanisms involved in the association between RORγ and TGF-β signaling/EMT transitions. Hence, we utilized RORγ binding information from the previous ChIP-chip study (Kittler et al., 2013) and performed cis-regulatory analysis using the GREAT (McLean et al., 2010). RORγ binding sites (total 3140) from MCF-7 cells were determined with the default region-gene association rule (5 kb upstream and 1 kb downstream, up to 1 mb max extension). Numbers of associated genes (0 to 8) per individual region were identified (Fig. S4A). The total associations between regions and genes were determined and presented by orientation and distance to transcription start sites (TSS), showing RORγ binding sites are primarily located 50–500 kb (Fig. S4B). Furthermore, we identified that total associations hold 2122 genes and the overlap of 418 genes which were differentially expressed after knockdown of RORγ in MCF-7 cells (194 genes were down-regulated and 224 genes were up-regulated) between RNA-seq and GREAT analysis (Fig. 3E). Importantly, the GREAT analysis highlighted 13 significant pathways in the MSigDB perturbation section (FDR < 0.05 in binomial and hypergeometric tests, fold enrichment > 2) (Fig. 3F). Surprisingly, many breast cancer-related terms were predicted as associated with direct RORγ target genes. For example, TAMOXIFEN_RESISTANCE_UP and ENDOCRINE_THERAPY_RESISTANCE_5 modules were enriched (Fig. 3F), and GO analysis of the GREAT data further implicated RORγ in the regulation of mesenchyme development and epithelial–mesenchymal transition (Fig. S4C). Significantly, we found that RORγ binding was associated with several TGF-β-related genes. For example, the ChIP-chip study (from Kevin White and colleagues, Kittler et al. 2013) suggests that RORγ is bound to the conserved regions of TGFB2 and SMAD3 loci (Fig. 3G). This evidence implies that RORγ may potentially be involved in the direct transcriptional regulation of TGFB2 and SMAD3. However, it is well known that there are inconsistencies between ChIP binding data and the significantly decreased amount of primary effects of specific transcription factors on gene regulation/transcript expression (after functional reporter gene analysis). Together, the pathway and integration of RNA-seq and ChIP-chip data analysis suggests that RORγ is involved in the transcriptional regulation of these critical genes in TGF-β signaling.
3.5. RORγ Expression Inversely Regulates Pro-metastatic c-MET Expression
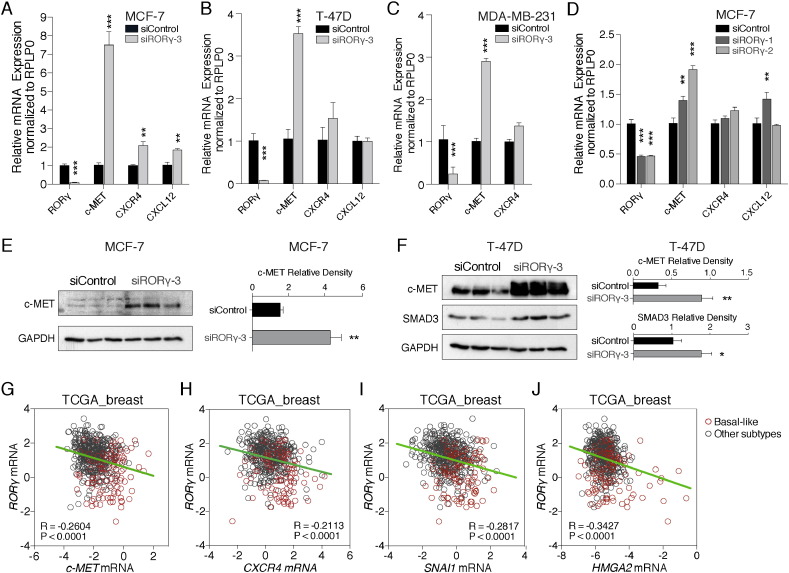
Further analysis of the RNA-seq data revealed RORγ knockdown remarkably increased several other pro-metastatic markers including c-MET (proto-oncogene, receptor tyrosine kinase), and the chemokine and chemokine receptor, CXCL12 and CXCR4 respectively (Fig. 3D). We validated RNA-seq data, and we examined expression of c-MET, CXCR4 and CXCL12 expression by RT-qPCR in the three breast cancer cell lines after RORγ-knockdown (Fig. 4A–C). To demonstrate this is not an off target effect, we also performed RT-qPCR after transfection of two additional RORγ siRNAs, siRORγ-1 and siRORγ-2, into MCF-7 cells (Fig. 4D). This analysis demonstrated that RORγ knockdown by three independent siRNAs increased c-MET expression robustly and significantly in the three breast cancer cell lines. Furthermore, we validated increased c-MET expression after RORγ knockdown in MCF-7 and T-47D breast cancer cells by western analysis (Fig. 4E and F). Significant changes in CXCL12 and CXCR4 were cell line specific.
Fig. 4.
Inverse correlation between the expression of RORγ and the proto-oncogene, c-MET and other pro-metastatic markers.
(A–C) Significant induction of c-MET expression and cell specific changes in chemokine components (CXCR4 and CXCL12) after RORγ depletion in (A) MCF-7, (B) T-47D and (C) MDA-MB-231 (n = 3).
(D) Validation of increased c-MET expression after RORγ depletion with two additional and independent stealth siRNAs targeting RORγ (n = 3).
(E) Western analysis of c-MET protein expression after RORγ knockdown in MCF-7.
(F) Western analysis of c-MET and SMAD3 protein expression after RORγ knockdown in T-47D.
(G) Inverse correlation of RORγ and c-MET in TCGA human cohort depicting lower RORγ expression and higher c-MET expression in basal-like breast cancer subtype (red) compared to other subtypes (gray).
(H–J) Negative association between expression of RORγ and other oncogenes including (H) CXCR4, (I) SNAI1 and (J) HMGA2 in the TCGA breast cancer cohort.
Interestingly, we observed a significant inverse correlation between c-MET and RORγ expression in the TCGA human breast cancer cohort (Fig. 4G). Moreover, we found a significant correlation between increased c-MET and decreased RORγ expression in basal-like aggressive cancers (Fig. 4G). We observed the significant inverse correlation of RORγ and the expression of several other EMT-related genes, including CXCR4, SNAI1 and HMGA2 (Fig. 4H–J), in the TCGA human breast cancer cohort. These genes have been documented to significantly affect clinical survival outcomes (Ponzo et al., 2009, Smith et al., 2004). These suggest that RORγ functions as a negative regulator that controls tumor biology in human breast cancer.
3.6. RORγ Regulates the Genetic Program Controlling DNA-repair
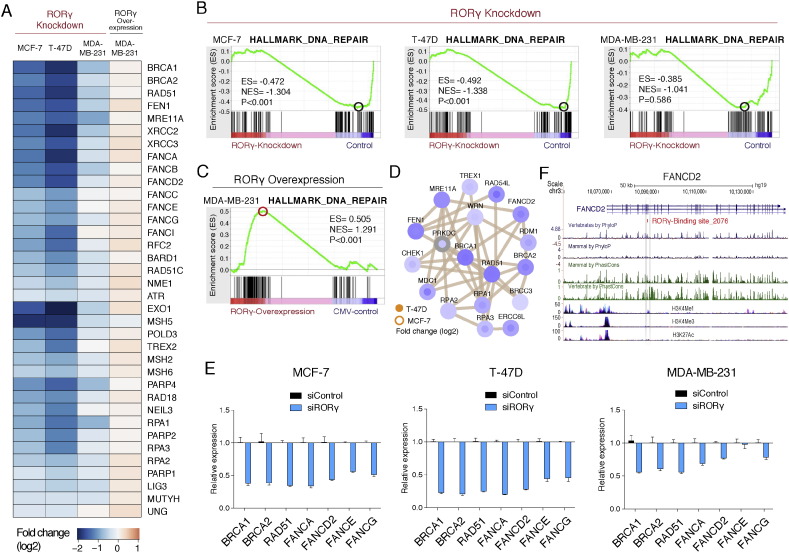
We observed that GSEA also identified enrichment of the gene signature associated with the control of DNA-repair in the DE genes of the RNA-seq datasets (see Fig. 2J). The heatmap (Fig. 5A) clearly demonstrated that RORγ depletion and over-expression in the ER + ve and ER − ve breast cancer cell lines modulated the genes associated with DNA-repair pathways (Fig. 5A). The GSEA analysis demonstrated the negative enrichment of a hallmark DNA-repair signature after RORγ knockdown in three cell lines (Fig. 5B), and the positive enrichment after RORγ over-expression in the aggressive MDA-MB-231 cells (Fig. 5C). The heatmap highlights the differential expression of transcripts involved in DNA-repair including BRCA1, BRCA2, RAD51, several Fanconi anemia genes and other DNA-repair genes involved in the BRCA1 and Fanconi DNA-repair pathway, that was also highlighted by protein–protein interaction (PPI) network analysis (Fig. 5D and Fig. S5A). The importance of these pathways and the effects of RORγ loss of function on the DNA-repair pathway were validated by RT-qPCR (Fig. 5E).
Fig. 5.
RORγ regulates the DNA-repair pathways in ER + ve and ER − ve breast cancer cells.
(A) Heatmap representation of the differentially expressed genes associated with DNA-repair after RORγ knockdown and RORγ over-expression (n = 3) in MCF-7, T-47D and MDA-MB-231 cells.
(B) GSEA analysis demonstrates negative enrichment of the hallmark DNA-repair module in MCF-7, T-47D and MDA-MB-231 after RORγ depletion.
(C) GSEA analysis demonstrates positive enrichment of the hallmark DNA-repair module after RORγ over-expression in MDA-MB-231.
(D) Protein–protein interaction network of the DNA-repair pathway. Netgestalt (iRef database) was used for the PPI network, visualizing fold changes in log2 scale.
(E) Relative mRNA expression of DNA-repair related genes in MCF-7, T-47D and MDA-MB-231 after RORγ depletion (n = 3).
(F) Direct RORγ binding sites associated with the FANCD2 genomic locus.
Subsequently, we utilized RORγ binding information from the previous ChIP-chip (Kittler et al., 2013) study and performed GREAT analysis (McLean et al., 2010). We identified several critical genes in the DNA-repair pathways that were also differentially expressed after RORγ gain and loss of function, and associated with RORγ binding including BRCA2, RAD51C, RAD52, FANCB, FANCC, FANCD2, and FANCI (Fig. S5B). For example, RORγ directly binds conserved regions of FANCD2 (Fig. 5F). In summary, the data clearly implicate RORγ-dependent signaling as a critical regulator of DNA-repair genes.
3.7. RORγ is a Primary Transcriptional Regulator of the Mammary Stem Cell Pathway
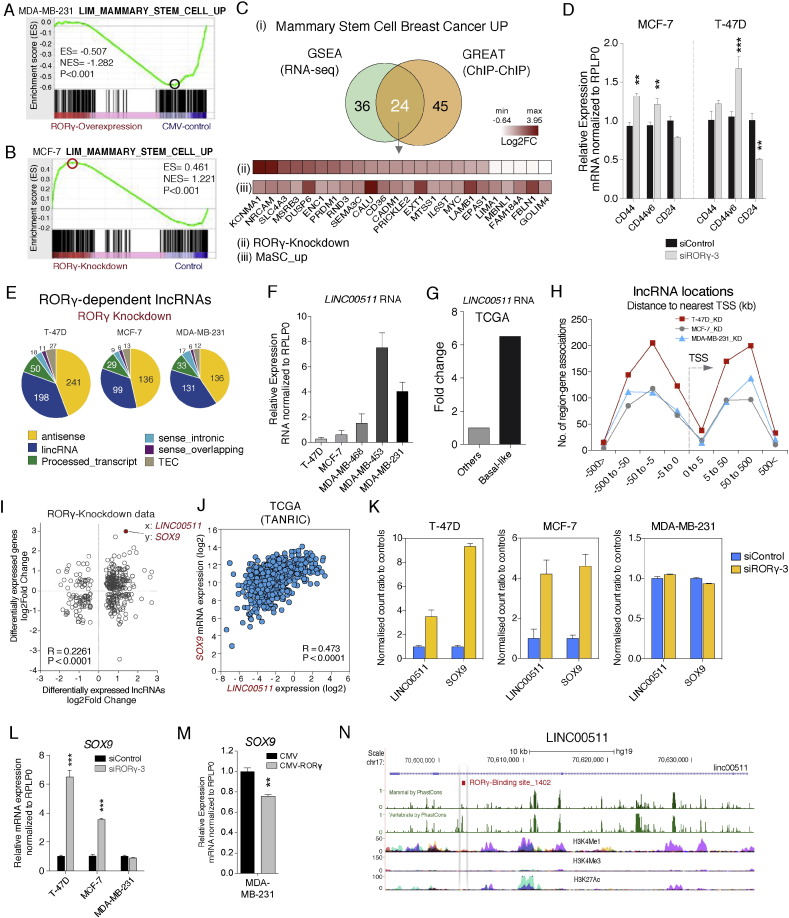
Unexpectedly, we observed that GSEA identified the negative correlation between RORγ expression and the gene signature associated with the acquisition of self-renewal (i.e. mammary stem cell capacity) after RORγ over-expression (i.e. RORγ gain of function) (Fig. 6A). The GSEA clearly showed the significant negative enrichment of genes in the MaSC_up module, suggesting that increased expression of RORγ suppressed the MaSC_up module. In contrast, knockdown of RORγ expression positively enriched the MaSC_up module (Fig. 6B). Notably, we observed that the transcriptome study (Lim et al., supplementary data) comparing MaSC, progenitor and matured luminal cells in human and mouse exhibited loss of RORγ expression in MaSC relative to mature luminal cells (Lim et al., 2010).
Fig. 6.
RORγ regulates the expression of the mammary stem cell gene signature.
(A) GSEA analysis demonstrates negative enrichment of the MaSC up module after RORγ over-expression in MDA-MB-231.
(B) GSEA analysis demonstrates RORγ depletion induces positive enrichment of the MaSC up module in MCF-7.
(C) Overlap between DE genes after RORγ knockdown in the MaSC up module (in MCF-7 cells, FDR < 0.05, Fold change > 1.3) and direct RORγ target genes derived from GREAT analysis of RORγ ChIP-chip data (C-i). Expression of direct targets of RORγ binding in MCF-7 cells after RORγ depletion (C-ii) and in the MaSC_up module (C-iii).
(D) Relative mRNA expression of CD44, CD44v6 and CD24 measured by RT-qPCR after RORγ depletion in MCF-7 and T-47D (n = 3).
(E) Classification of RORγ-dependent long non-coding RNA expression in T-47D, MCF-7 and MDA-MB-231.
(F) Relative expression of LINC00511 RNA expression in five different breast cancer cell lines.
(G) Increased expression of LINC00511 RNA in basal-like breast cancer relative to other subtypes derived from TCGA human cohort (Xu et al. supplementary).
(H) Distance of long non-coding RNAs to nearest TSS (i.e. adjacent genes) defined by GREAT analysis.
(I) Correlation between LINC00511 and SOX9 expression after RORγ depletion identified from scatter plot expression analysis examining correlation between long non-coding RNA (x-axis) and adjacent coding gene (y-axis) in T-47D cells (log2 fold change scale).
(J) A positive correlation between LINC00511 and SOX9 expression in TCGA breast cancer data set. Data were downloaded from the TANRIC lncRNA database (Li et al., 2015).
(K) Expression of LINC00511 and SOX9 in T-47D, MCF-7 and MDA-MB-231 extracted from RNA-seq analysis after RORγ depletion (Normalized count ratio to controls, n = 3).
(L and M) Expression of SOX9 mRNA in T-47D and MCF-7 after RORγ knockdown and RORγ over-expression in breast cancer cells, respectively (n = 3).
(N) Direct RORγ binding sites associated with the LINC00511 genome locus.
We extended this analysis by integrating our RNA-seq datasets to RORγ binding information. GREAT analysis also predicted that > 60 of these genes are enriched in the MaSC gene module. In doing so, we identified the common genes that were enriched in both the RNA-seq/GSEA and ChIP-chip/GREAT analysis (Fig. 6C-i). We identified 24 genes including for example, NRCAM, CALU, and PRICKLE2, etc. in both analyses and the heatmap (Fig. 6C-ii and iii) displays the fold changes (increased) in these RORγ-dependent genes in RORγ-knockdown vs RORγ-control dataset in our study (Fig. 6C-ii), and MaSC vs mature luminal from the Lim et al. dataset (Lim et al., 2010) (Fig. 6C-iii). The comparative data shows similar gene expression patterns between the RORγ-dependent genes and the up-regulated MaSC gene subset, strongly inferring the regulatory role of RORγ signaling in modulating stemness capacity of breast cancer cells.
In this context, we examined the expression of the mRNAs encoding CD44 and CD24 in RORγ-depleted (MCF-7 and T-47D) cells. The phenotype of basal subpopulation displaying increased CD44 and reduced CD24 expression has been linked to poor prognosis and exhibited improved self-renewal capacity (Al-Hajj et al., 2003). Hence, our RT-qPCR showed that loss of RORγ resulted in the increased expression of CD44 and decreased expression of CD24 antigen markers in breast cancer cells (Fig. 6D). We also observed that CD44v6, the stem cell enriched CD44 variant (Todaro et al., 2014), was increased by RORγ knockdown (Fig. 6D).
Furthermore, RNA-seq analysis revealed that RORγ expression controls the expression of the long non-coding RNAs (lncRNA) (see Fig. 2F). We identified that RORγ-dependent lncRNAs are mainly comprised of antisense and long intervening non-coding RNAs (lincRNAs) (Fig. 6E). Moreover, we took the advantage of RNA-seq by comparing control data of three cell lines and defined an MDA-MB-231-specific subset of coding and lncRNA transcripts (by applying a stringent cutoff, FDR < 0.01, Fold change > 2) (Fig. S6). A total of 9743 genes of MDA-MB-231 were differentially expressed against T-47D (controls). Similarly, MDA-MB-231 cells exhibit distinctive expression of 9779 genes compared to MCF-7 (controls), highlighting decreased RORγ expression, but increased (previously described) oncogene expression of c-MET, CXCR4, TGF-β, SMAD3, etc. (Fig. S6). This approach also identified the lncRNA, LINC00511, as highly ranked differentially expressed transcript in ER − ve MDA-MB-231 cells relative to the ER + ve T-47D and MCF-7 cells (Fig. S6). Notably, RNA-seq data from RORγ depleted MCF-7 and T-47D cells revealed that LINC00511 expression was also increased. We further validated this observation in cell lines by RT-qPCR and observed that LINC00511 expression was significantly increased in the metastatic basal breast cancer cell lines (for example, MDA-MB-231 cells), relative to T-47D and MCF-7 luminal cells (Fig. 6F). Underscoring the significance of our in vitro observation, we extracted the LINC00511 data from the TCGA human dataset (Su et al., supplemental data) (Su et al., 2014) and we demonstrated that LINC00511 is highly expressed in the aggressive basal-like breast cancer subtype (Cluster 1 in Su's study) (Fig. 6G).
Recent studies showed that lncRNAs regulate the transcriptional process (Ørom et al., 2010, Yang et al., 2013). Therefore, we identified the adjacent coding genes by utilizing GREAT analysis with locations of RORγ-dependent lncRNAs, and examined the distance between RORγ-dependent lncRNAs and adjacent coding genes (Fig. 6H). Interestingly, scatter plot analysis of correlation between lncRNAs and adjacent coding genes in our RNA-seq dataset identified a significant (and positive) correlation between the increased expression of transcripts encoding LINC00511 and the coding gene, SOX9 among many other correlations between lncRNAs and adjacent coding genes (Fig. 6I, P < 0.0001). Furthermore, we found a strong positive correlation (Pearson correlation score = 0.473, P < 0.0001) between LINC00511 and SOX9 expression in the TCGA breast cancer data (Fig. 6J) after interrogating the TANRIC lncRNA database (Li et al., 2015).
This correlation was further pursued, as SOX9 is involved in the determination of mammary stem cell fate (Guo et al., 2012, Ye et al., 2015) and increased expression implicated in poorer breast cancer survival outcomes (Pomp et al., 2015). In this context, we observed both increased LINC00511 and SOX9 expression in ER + ve, T-47D and MCF-7 cells, but not in ER − ve cells from RORγ knockdown data of RNA-seq (Fig. 6K). Accordingly, we confirmed the increased expression of SOX9 expression by RT-qPCR after RORγ depletion (Fig. 6L), and decreased SOX9 expression after RORγ over-expression in MDA-MB-231 cells (Fig. 6M). Importantly, studies have identified two transcription factors that determine mammary stem cell fate, SOX9 and SNAI2 (Slug), which suffice to induce MaSCs from differentiated luminal cells (Guo et al., 2012). This potentially suggests that RORγ plays a critical role on the negative regulation of SOX9 expression in breast cancer cells (see Fig. 6L and M).
Finally, we further examined whether RORγ is a potential primary (and direct) regulator of LINC00511 or SOX9 transcription by exploiting the ChIP-chip data. To do so, we utilized the GREAT with RORγ binding sites and found the location of RORγ_1402 is in the LINC00511 locus, suggesting RORγ directly binds to cis-regulatory region of SOX9. In addition, we found that RORγ binding site on LINC00511 is highly conserved (Fig. 6N). This observation identifies a probable role of RORγ in the regulation of SOX9 through LINC00511. In summary, the data provide supporting evidence for RORγ-dependent regulation of coding (and long non-coding) RNAs involved in MaSC fate/mammary stem cell capacity, major pathways in the progression of breast carcinogenesis. Significantly, the majority of the coding genes and the lncRNAs associated with the stemness signature and determinants of MaSC capacity have been potentially implicated as targets (and sites) of RORγ binding in the ChIP-chip study. However, as discussed for the TGF-β pathway, definitive evidence for RORγ as a direct transcriptional regulator awaits further functional analysis.
3.8. In Vitro Studies Suggest RORγ Agonists Display Pharmacological Utility in Breast Cancer Cell Lines
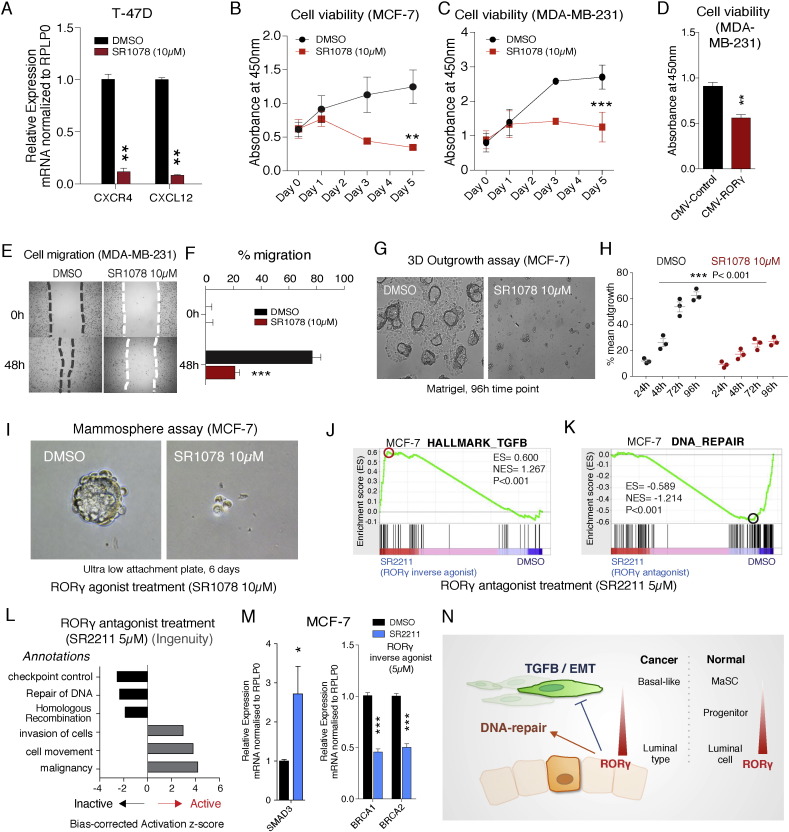
We utilized the well-characterized RORα/γ dual agonist, SR1078, to demonstrate that pharmacological modulation of RORγ activity plays a significant role in modulating breast cancer cell malignancy (Wang et al., 2010). We treated T-47D and MCF-7 cells with the RORγ agonist, SR1078 (10 μM). For example, we examined the effect of the agonist on the expression of the chemokine receptor and ligand, CXCR4 and CXCL12 that significantly influence breast cancer clinical outcomes (Holland et al., 2013). We observed that SR1078 decreased the expression of these components of the chemokine system (Figs. 7A and S7A). We observed that SR1078 significantly decreased cell viability in both MCF-7 and MDA-MB-231 cell lines (Fig. 7B and C). Further confirmation of the selective and specific effect of RORγ expression on cell viability was provided by the demonstration that RORγ over-expression in MDA-MB-231 significantly reduced cell viability (Fig. 7D). These pharmacological and genetic gains of function experiments demonstrated the important role of RORγ in the regulation of cell growth and viability.
Fig. 7.
Anti-cancer activity of RORγ agonist and oncogenic effect of RORγ inverse agonist in breast cancer cells.
(A) Treatment (24 h) of T-47D cells with the RORγ agonist, SR1078 (10 μM) decreased expression of the mRNAs encoding CXCR4 and CXCL12 (n = 3).
(B and C) SR1078 (10 μM) treatment decreased cell viability in (B) MCF-7 and (C) MDA-MB-231 cells (average of absorbance values from 3 independent experiments in triplicate).
(D) RORγ over-expression decreases cell viability of MDA-MB-231 cells (n = 3).
(E and F) SR1078 (10 μM) treatment attenuates cell migration in MDA-MB-231 cells (n = 3).
(G) SR1078 (10 μM) treatment decreases microsphere outgrowth in MCF-7 cells in 3D Matrigel invasion assay (n = 3).
(H) Mean of outgrowth percentage between DMSO and SR1078 treated MCF-7 cells (n = 3).
(I) SR1078 (10 μM) treatment decreases mammosphere growth (n = 3).
(J) GSEA analysis demonstrates that SR2211 (5 μM) treatment (a RORγ selective inverse agonist) of MCF-7 cells positively enriched the TGF-β module after RNA-seq analysis.
(K) GSEA analysis demonstrates that SR2211 (5 μM) treatment negatively enriched the DNA-repair module in MCF-7 cells after RNA-seq analysis.
(L) Ingenuity Pathway Analysis of RNA-seq data derived from SR2211 treatment of MCF-7 cells highlighting positive activation of oncogenic pathways, but showing negative activation of the DNA-repair pathways.
(M) Treatment (24 h) of MCF-7 cells with the selective RORγ inverse agonist, SR2211 (5 μM) increased the mRNA expression of SMAD3 and decreased the expression of mRNAs encoding BRCA1 and BRCA2 (n = 3).
(N) Schematic graph illustrating the probable function of RORγ in breast cancer.
Subsequently, we investigated the effect of modulating RORγ activity and expression on several different aspects of carcinogenesis including cell migration, cell outgrowth and self-renewal. Initially, we performed wound healing assays with SR1078, and observed that SR1078 significantly inhibited cell migration in MDA-MB-231 cells (Fig. 7E and F). In contrast, we demonstrated that depletion of RORγ function by RORγ siRNA increased the rate of cell migration in MDA-MB-231 cells, confirming the specific role of RORγ in this process (Fig. S7B). We then examined the effect of SR1078 on tumor cell invasiveness. For this, we embedded MCF-7 cells in Matrigel and measured their ability to invade from microspheres into the surrounding matrix. Cell outgrowth from microspheres was significantly deceased by 48–96 h treatment with SR1078 treatment compared to DMSO controls (2-way ANOVA, n = 3) (Figs. 7G and H, and S7C). Similarly, SR1078 treatment in MDA-MB-231 cells over 96 h demonstrated that the drug decreased cell number (Fig. S7D). Furthermore, we evaluated the effect of pharmacological modulation of RORγ activity on self-renewal (i.e. the acquisition of mammary stem cell activity) utilizing mammosphere assays on an ultra low attachment plate (Fig. 7I). Increasing RORγ activity in MCF-7 cells has a dramatic inhibitory effect on the self-renewal activity of MCF-7 cells in the mammosphere assay.
Finally, treatment of cells with a selective and specific RORγ inverse agonist, SR2211 (Kumar et al., 2012), followed by RNA-seq and pathway analysis demonstrated that treatment with RORγ inverse agonist resulted in the positive and significant enrichment of the TGF-β signaling pathway (Fig. 7J), but the negative and significant enrichment of DNA-repair (Fig. 7K). Consistently, IPA showed that RORγ inverse agonist led to positive activation scores on oncogenic annotations including invasion of cells, cell movement and malignancy (Fig. 7L). In contrast, the selective inverse agonist significantly repressed activation scores for DNA repair and Homologous recombination annotations. Moreover, RNA-seq analysis after SR2211 treatment identified the specific induction of SMAD3 mRNA expression (a critical mediator of TGF-β signaling) and decreased expression of the mRNAs encoding BRCA1 and 2, that were also validated by RT-qPCR (Fig. 7M).
Together, the effect of (in vitro) pharmacological and genetic modulation of RORγ activity and expression in well-characterized ER + and ER − cell lines on cell migration, the EMT (using microsphere outgrowth assays), and stem cell activity (utilizing mammosphere assays) provides insights into the pharmacological manipulation of RORγ signaling. The role of RORγ in the control of pathways that regulate outgrowth from suppressive and tightly organized epithelial microenvironments and self-renewal activity is fundamental to tumor initiation and metastasis (Fig. 7N), and suggest this NR has therapeutic potential in the treatment of breast cancer.
4. Discussion
In this study, we report on the role of RORγ in the regulation of genetic programs and pathways promoting breast carcinogenesis. The current study evolved from the previous analysis focused on the role of the NR coregulator, PRMT2 (an epigenetic factor) in breast cancer (Oh et al., 2014). The study revealed PRMT2 and RORγ expression were associated with checkpoint control and DNA-repair genes. Increased RORγ expression was associated with the higher probability of metastasis-free survival. Hazard ratio analysis (using Cox regression) revealed in several independent breast cancer datasets a very low hazard (odds) ratio for RORγ, indicating expression is associated with improved clinical outcomes (Oh et al., 2014). Hence, our current investigation was focused on elucidating the protective nature of RORγ signaling in breast cancer. In this context, we observed that RORγ expression is decreased in aggressive basal-like breast cancer and negatively associated with histological grade in several human cohorts that is consistent with our previous observation (Muscat et al., 2013). Furthermore, assessment and correlation of RORγ expression levels with intrinsic breast cancer subtypes assessed using the Hu et al. and PAM50 gene signatures (Ringner et al., 2011) demonstrated (significantly) decreased RORγ expression in basal-like subtypes (ER, PR and HER2 negative). Notably, bioinformatic analysis of all the human breast cancer cell lines demonstrated that the expression of RORγ is decreased in basal-like cell lines, implying that RORγ expression is inversely correlated with breast cancer malignancy.
RNA-seq and bioinformatics analysis of expression data after genetic and pharmacological modulation of RORγ identified roles in EMT and mammary stem cell activity, indicating that RORγ is a fundamental regulator of tumor initiation and metastasis. Specifically, we showed that RORγ attenuates TGF-β/EMT signaling in breast cancer. TGF-β plays a key role in inducing EMT that has been linked to the aggressive breast cancer phenotype (Scheel et al., 2011). Moreover, our study identified that key genes in this pathway, for example, TGF-β and SMAD3, are direct targets of RORγ. To our best knowledge, our study reports for the first time the suppression of TGF-β/EMT signaling by the DNA-binding transcription factor, RORγ, in accord with its effects on survival outcomes. In addition, RORγ depletion in the human breast cancer cell lines resulted in the induction of the CXCR4 chemokine receptor, the chemokine CXCL12 (in selected cell lines) and the induction of c-MET, whereas RORγ agonist treatment significantly decreased CXCR4 and CXCL12 expression in breast cancer cells. Targeting these factors has demonstrated utility in the treatment of basal-like and triple negative breast cancers (Comoglio et al., 2008, Zou et al., 2007). The effect of genetic and pharmacological modulation of RORγ on c-MET and the chemokine system further underscores the significant role of RORγ in regulating the expression of factors controlling metastasis and clinical outcomes.
One unexpected and very exciting aspect of our work was that gain and loss of RORγ function resulted in the negative and positive enrichment of the mammary stem cell signature, respectively, in several breast cancer cell lines. Moreover, the effect of RORγ depletion on the positive enrichment of the stemness signature is underscored by the dramatic effect of increasing RORγ activity (by agonist treatment) on mammosphere growth. These observations are in concordance with the correlations between RORγ expression and survival, and the important links to breast cancer risk and the acquisition of the stem cell-like phenotype. GSEA identified that > 25% of the genes in the MaSC_up signature are enriched by genetic modulation of RORγ. We subsequently integrated the (very significant) expression changes after RNA-seq (~ 60 genes) and the RORγ binding sites, and demonstrated RORγ directly regulates/binds over 20 critical genes in the MaSC module.
The critical role of RORγ signaling in the acquisition of mammary stemness was further underscored by our observation of the dramatic induction of both the non-coding RNA (LINC00511) and its adjacent gene (SOX9) after RORγ depletion. We showed that LINC00511 expression is increased in the aggressive basal-like breast cancer subtype, and the correlation between LINC00511 and SOX9 RNA expression in breast cancer cells and the human TCGA breast cancer cohort (in the TANRIC lncRNA database) (Li et al., 2015). Although the exact mechanism of how LINC00511 controls SOX9 transcription still requires further investigation, our study importantly showed that RORγ potentially binds to the LINC00511 RNA and adjacent of the cis-regulatory region of SOX9. The role of SOX9 has been shown in determining mammary stem cell fate, driving metastasis and its association with poorer survival outcomes (Guo et al., 2012). Our study excitingly provides a link between RORγ expression and SOX9 through the regulation of the non-coding RNA, LINC00511, and provides molecular and cellular evidence highlighting the critical role of RORγ in the regulation of MaSC signature.
DNA-repair is a central system for maintaining genome stability and correcting DNA damage. In our study, both GSEA and IPA analysis (including RT-qPCR validation in three cell lines) significantly highlighted the importance of RORγ signaling in the DNA-repair pathways. Furthermore, RNA-seq coupled with the treatment of the RORγ antagonist SR2211, demonstrated that functional antagonism of RORγ suppressed the majority pathways, and specifically decreased expression of BRCA1&2 mRNA expression (as validated by RT-qPCR), supporting our loss of RORγ function data (i.e. requirement of RORγ for DNA-repair gene expression). We further identified potential RORγ binding to genomic regions of critical genes (including BRCA2), implying the transcriptional (and direct) role of RORγ in DNA-repair. In accord with our previous study (Oh et al., 2014), we identified the inverse relationship between the expression of the histone methyltransferase, PRMT2, and RORγ in breast cancer cells and human cohorts. We demonstrated PRMT2 knockdown increases nucleotide excision repair of UV-induced DNA lesion, and homologous recombination repair of double stranded breaks. This improvement in DNA repair was associated with an ~ 14-fold increase in RORγ mRNA expression (RORγ mRNA induction is one of the top ten changes in differential expression after PRMT2 depletion) (Oh et al., 2014).
In the context of the other RORγ isoform, the role of RORγt in differentiating T 17 cells has been well established and the potential anti-cancer effect of agonists with targeting this mechanism of T 17 cells is emerging as an immunotherapy (Carter et al., 2015, Zou and Restifo, 2010). However, our study focused on the function of RORγ (independent of RORγt) in suppressing breast cancer malignancy. A recent RORγ study (in adenomas) has shown that decreased RORγ expression in patients with somatotroph adenomas is associated with poor clinical recovery after treatment of somatostatin analogs (Lekva et al., 2013) which supports our hypothesis. This correlates with our observation that RORγ negatively regulates EMT. Furthermore, this indicates that the inhibitory regulation of RORγ on TGF-β/EMT can be applicable in other cancer cases. We have explored the role of RORγ in breast cancer biology and identified suppressive effect of RORγ on TGF-β/EMT and mammary stemness and positive regulation on DNA-repair, indicating the critical role of RORγ in breast carcinogenesis.
The pharmacological studies targeting RORγ activity were in accord with the extensive analysis of human datasets, and the human breast cancer cell lines. Future studies in breast cancer cells and mouse models would benefit from (well characterized) RORγ specific agonists rather than the ROR dual agonist to further evaluate the pharmacological utility of this target gene. The cholesterol metabolite, desmosterol has been described as an RORγ agonist (Hu et al., 2015), however, it also activates LXR signaling (Yang et al., 2006) and has not been utilized by other groups. Overall, our study suggests that RORγ agonists have pharmacological utility in vitro, and these compounds may have anti-cancer efficacy against several different critical processes that drive progression of breast cancer.
The following are the supplementary data related to this article.
Primers information used in this study.
Decreased RORγ expression in aggressive basal-like subtype, ER − ve, and grade 3 breast cancer subtypes. (A and B) RORγ mRNA expression in the combined breast cancer datasets based on molecular subtypes defined of Hu et al. (A) and RORγ mRNA expression in the breast cancer dataset based on molecular subtypes defined by PAM50 (B). (C) Decreased RORγ expression of aggressive grade 3 within the combined breast cancer datasets. (D) Comparison of RORγ mRNA expression between ER + ve and ER − ve cases. (E) Immunohistochemistry showing representative (from n = 13-15) RORγ staining in (i and ii) ER-ve cases, (iii and iv) ER + ve breast cancer and (v and vi) normal breast. Insets show the no primary control sections for the matched tissue where the material was available. Scale bars indicate 50 μm for main panels and inset images respectively.
Identification of RORγ-dependent pathways and survival outcomes (A and B) GSEA analysis showing top five positive and negative enriched modules derived from HALLMARK gene sets based on normalized enrichment score after loss of RORγ function in (A) T-47D and (B) MDA-MB-231 breast cancer cells. (C) GSEA analysis identifies positively enriched KEGG_TGF-Β_SIGNALING, EMT_SIGNALING with common overlapping genes among MCF-7, T-47D and MDA-MB-231 cells after loss of RORγ function. (D) The RORγ-dependent signature displays prognostic utility in predicting survival outcomes. A low score, i.e. high RORγ expression predicts a higher probability of survival (in the UNT dataset). (E) Correlation analysis of RORγ1 mRNA expression and the RORγ-dependent signature scores in UNT dataset reveals inverse relationship between RORγ expression and signature scores.
GREAT cis-regulatory analysis of the RORγ ChIP-ChIP data in MCF-7 cells. (A) Percentage of genomic regions based on numbers of associated genes per region. (B) Percentage of region-to-gene association showing many RORγ binding events lie well outside of the proximal promoter. (C) RORγ ChIP-ChIP data highlighted 13 terms of GO biological processes and identified many critical pathways including mesenchyme development, mammary gland development and epithelial to mesenchymal transition. All terms were still enriched after the cutoff (FDR < 0.05 and fold enrichment > 2) in both binomial test and hypergeometric test.
RORγ regulates the DNA-repair pathway in human breast cancer cells. (A) Protein–protein interaction network analysis of the Fanconi anemia pathway. PPI analysis identified the Fanconi anemia pathway network in T-47D and MCF-7 cells. Netgestalt (iRef PPI database) was utilized to visualize the PPI network with fold changes in log2 scale. (B) RORγ binding sites on BRCA2, RAD51C, RAD52, FANCB, FANCC and FANCI.
Identification of differentially expressed genes (coding and long non-coding RNAs) among three breast cancer cell lines, T-47D, MCF-7 and MDA-MB-231. (A) Number of significantly differentially expressed genes in (I) MDA-MB-231 vs T-47D, (ii) MDA-MB-231 vs MCF-7 and (iii) T-47D vs MCF-7. Comparison between an aggressive ER − ve cell line (MDA-MB-231) and two ER + ve luminal types (T-47D or MCF-7) highlighted the high expression of LINC00511, TGFΒ2, SMAD3, c-MET, CXCR4, SOX9 and SNAI2 in basal-like MDA-MB-231 cells. In addition, RNA-seq also identified the decreased expression of RORγ in MDA-MB-231 compared to other cell lines.
Pharmacological and Genetic manipulation of RORγ expression and activity effects breast carcinogenesis in vitro. (A) Treatment (24 h) of ER + ve MCF-7 cells with the RORγ agonist, SR1078 (10 μM) decreased the expression of the mRNAs coding CXCR4 and CXCL12 (n = 3). (B) RORγ depletion increased cell migration in ER − ve MDA-MB-231 cells. Representative images were shown from 3 independent experiments in triplicate. (C and D) The RORγ agonist, SR1078 (10 μM), decreased microsphere outgrowth in ER + ve MCF-7 in 3D Matrigel invasion assay (C) and the RORγ agonist, SR1078 (10 μM) decreased cell growth and outgrowth ER − ve MDA-MB-231 in 3D Matrigel invasion assay (D). (E) UCSC genome-browser showing the RORγ binding site on SMAD3 is overlapped with one of ENCODE regulator binding regions.
Conflicts of Interests
The authors declare there are no conflicts of interest and/or competing interests to disclose.
Author Contributions
T.G.O., S-C.M.W., B.R.A., J.M.G. and J.D.G. performed experiments. T.G.O. conducted bioinformatic analysis, and was involved in the preparation and writing of the manuscript. J.D.G. and C.L.C. performed immunohistochemistry experiments and contributed to the manuscript. B.R.A. and A.S.Y. performed 3D-organotypic outgrowth assay and contributed to the manuscript. G.E.O.M. conceived and supervised the project, was involved in experimental analysis and interpretation and wrote the manuscript.
Funding Acknowledgement
This research was supported by Core institutional support from the Institute of Molecular Bioscience (IMB), The University of Queensland, The Cancer Council Queensland's Thomas and Dorothy Nicholson Research Grant (GEOM/App#10711301, 2014-15), The Cancer Council Queensland's Project Grant (GEOM/App#1100250, 2016-17) and Human Frontiers Science Program (ASY/RGP 0023/2014). TGO is a University of Queensland PhD candidate. GEOM (APP#1059341), CLC (APP#1081334) and ASY (APP#1044041) are National Health and Medical Research Council (of Australia) Principal Research Fellows.
Accession Numbers
The RNA-sequencing data sets have been deposited in the Gene Expression Omnibus (GEO) database under accession GSE74981.
References
- Al-Hajj M., Wicha M.S., Benito-Hernandez A., Morrison S.J., Clarke M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastassiou D., Rumjantseva V., Cheng W., Huang J., Canoll P.D., Yamashiro D.J., Kandel J.J. Human cancer cells express slug-based epithelial–mesenchymal transition gene expression signature obtained in vivo. BMC Cancer. 2011;11:529. doi: 10.1186/1471-2407-11-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. HTSeq–a python framework to work with high-throughput sequencing data. Bioinformatics (Oxf., Engl.) 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter L., Moisan J., Majchrzak K., Hu X., Morgan R., Liu X., Demock K., Wang Y., Lesch C., Sanchez B. Novel RORgamma agonists enhance anti-tumor activity of adoptive T cell therapy (TUM2P. 1010) J. Immunol. 2015;194:69.67. [Google Scholar]
- Comoglio P.M., Giordano S., Trusolino L. Drug development of MET inhibitors: targeting oncogene addiction and expedience. Nat. Rev. Drug Discov. 2008;7:504–516. doi: 10.1038/nrd2530. [DOI] [PubMed] [Google Scholar]
- Dent R., Trudeau M., Pritchard K.I., Hanna W.M., Kahn H.K., Sawka C.A., Lickley L.A., Rawlinson E., Sun P., Narod S.A. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin. Cancer Res. 2007;13:4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- Desmedt C., Piette F., Loi S., Wang Y., Lallemand F., Haibe-Kains B., Viale G., Delorenzi M., Zhang Y., d'Assignies M.S. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin. Cancer Res. 2007;13:3207–3214. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- Doan T.B., Eriksson N.A., Graham D., Funder J.W., Simpson E.R., Kuczek E.S., Clyne C., Leedman P.J., Tilley W.D., Fuller P.J., et al. Breast cancer prognosis predicted by nuclear receptor-coregulator networks. Mol. Oncol. 2014 doi: 10.1016/j.molonc.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxf., Engl.) 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowhan D.H., Harrison M.J., Eriksson N.A., Bailey P., Pearen M.A., Fuller P.J., Funder J.W., Simpson E.R., Leedman P.J., Tilley W.D., et al. Protein arginine methyltransferase 6-dependent gene expression and splicing: association with breast cancer outcomes. Endocrinol. Relat. Cancer. 2012;19:509–526. doi: 10.1530/ERC-12-0100. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Angulo A.M., Morales-Vasquez F., Hortobagyi G.N. Overview of resistance to systemic therapy in patients with breast cancer. Adv. Exp. Med. Biol. 2007;608:1–22. doi: 10.1007/978-0-387-74039-3_1. [DOI] [PubMed] [Google Scholar]
- Guo W., Keckesova Z., Donaher J.L., Shibue T., Tischler V., Reinhardt F., Itzkovitz S., Noske A., Zurrer-Hardi U., Bell G., et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Györffy B., Lanczky A., Eklund A.C., Denkert C., Budczies J., Li Q., Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- Harrell J.C., Prat A., Parker J.S., Fan C., He X., Carey L., Anders C., Ewend M., Perou C.M. Genomic analysis identifies unique signatures predictive of brain, lung, and liver relapse. Breast Cancer Res. Treat. 2012;132:523–535. doi: 10.1007/s10549-011-1619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy B.T., Gonzalez-Angulo A.M., Stemke-Hale K., Gilcrease M.Z., Krishnamurthy S., Lee J.S., Fridlyand J., Sahin A., Agarwal R., Joy C., et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69:4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland J.D., Gyorffy B., Vogel R., Eckert K., Valenti G., Fang L., Lohneis P., Elezkurtaj S., Ziebold U., Birchmeier W. Combined wnt/beta-catenin, met, and CXCL12/CXCR4 signals characterize basal breast cancer and predict disease outcome. Cell Rep. 2013;5:1214–1227. doi: 10.1016/j.celrep.2013.11.001. [DOI] [PubMed] [Google Scholar]
- Hu X., Wang Y., Hao L.Y., Liu X., Lesch C.A., Sanchez B.M., Wendling J.M., Morgan R.W., Aicher T.D., Carter L.L., et al. Sterol metabolism controls T(H)17 differentiation by generating endogenous RORgamma agonists. Nat. Chem. Biol. 2015;11:141–147. doi: 10.1038/nchembio.1714. [DOI] [PubMed] [Google Scholar]
- Ivanov I.I., McKenzie B.S., Zhou L., Tadokoro C.E., Lepelley A., Lafaille J.J., Cua D.J., Littman D.R. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17 + T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Kittler R., Zhou J., Hua S., Ma L., Liu Y., Pendleton E., Cheng C., Gerstein M., White K.P. A comprehensive nuclear receptor network for breast cancer cells. Cell Rep. 2013;3:538–551. doi: 10.1016/j.celrep.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Krämer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics (Oxf., Engl.) 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N., Lyda B., Chang M.R., Lauer J.L., Solt L.A., Burris T.P., Kamenecka T.M., Griffin P.R. Identification of SR2211: a potent synthetic RORgamma-selective modulator. ACS Chem. Biol. 2012;7:672–677. doi: 10.1021/cb200496y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekva T., Berg J.P., Heck A., Fougner S.L., Olstad O.K., Ringstad G., Bollerslev J., Ueland T. Attenuated RORC expression in the presence of EMT progression in somatotroph adenomas following treatment with somatostatin analogs is associated with poor clinical recovery. PLoS One. 2013;8 doi: 10.1371/journal.pone.0066927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Han L., Roebuck P., Diao L., Liu L., Yuan Y., Weinstein J.N., Liang H. TANRIC: an interactive open platform to explore the function of lncRNAs in Cancer. Cancer Res. 2015;75:3728–3737. doi: 10.1158/0008-5472.CAN-15-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A., Subramanian A., Pinchback R., Thorvaldsdottir H., Tamayo P., Mesirov J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics (Oxf., Engl.) 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E., Wu D., Pal B., Bouras T., Asselin-Labat M.L., Vaillant F., Yagita H., Lindeman G.J., Smyth G.K., Visvader J.E. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res. 2010;12:R21. doi: 10.1186/bcr2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo P.K., Kanojia D., Liu X., Singh U.P., Berger F.G., Wang Q., Chen H. CD49f and CD61 identify Her2/neu-induced mammary tumor-initiating cells that are potentially derived from luminal progenitors and maintained by the integrin-TGFbeta signaling. Oncogene. 2012;31:2614–2626. doi: 10.1038/onc.2011.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L.D., Smeds J., George J., Vega V.B., Vergara L., Ploner A., Pawitan Y., Hall P., Klaar S., Liu E.T. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc. Natl. Acad. Sci. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mote P.A., Balleine R.L., McGowan E.M., Clarke C.L. Colocalization of progesterone receptors A and B by dual immunofluorescent histochemistry in human endometrium during the menstrual cycle. J. Clin. Endocrinol. Metab. 1999;84:2963–2971. doi: 10.1210/jcem.84.8.5928. [DOI] [PubMed] [Google Scholar]
- Muscat G.E., Eriksson N.A., Byth K., Loi S., Graham D., Jindal S., Davis M.J., Clyne C., Funder J.W., Simpson E.R., et al. Research resource: nuclear receptors as transcriptome: discriminant and prognostic value in breast cancer. Mol. Endocrinol. 2013;27:350–365. doi: 10.1210/me.2012-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve R.M., Chin K., Fridlyand J., Yeh J., Baehner F.L., Fevr T., Clark L., Bayani N., Coppe J.P., Tong F., et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh T.G., Bailey P., Dray E., Smith A.G., Goode J., Eriksson N., Funder J.W., Fuller P.J., Simpson E.R., Tilley W.D., et al. PRMT2 and RORgamma expression are associated with breast cancer survival outcomes. Mol. Endocrinol. 2014;28:1166–1185. doi: 10.1210/me.2013-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ørom U.A., Derrien T., Beringer M., Gumireddy K., Gardini A., Bussotti G., Lai F., Zytnicki M., Notredame C., Huang Q., et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou C.M., Sorlie T., Eisen M.B., van de Rijn M., Jeffrey S.S., Rees C.A., Pollack J.R., Ross D.T., Johnsen H., Akslen L.A., et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Pomp V., Leo C., Mauracher A., Korol D., Guo W., Varga Z. Differential expression of epithelial–mesenchymal transition and stem cell markers in intrinsic subtypes of breast cancer. Breast Cancer Res. Treat. 2015 doi: 10.1007/s10549-015-3598-6. [DOI] [PubMed] [Google Scholar]
- Ponzo M.G., Lesurf R., Petkiewicz S., O'Malley F.P., Pinnaduwage D., Andrulis I.L., Bull S.B., Chughtai N., Zuo D., Souleimanova M., et al. Met induces mammary tumors with diverse histologies and is associated with poor outcome and human basal breast cancer. Proc. Natl. Acad. Sci. 2009;106:12903–12908. doi: 10.1073/pnas.0810402106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringner M., Fredlund E., Hakkinen J., Borg A., Staaf J. GOBO: gene expression-based outcome for breast cancer online. PLoS One. 2011;6:e17911. doi: 10.1371/journal.pone.0017911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel C., Eaton E.N., Li S.H., Chaffer C.L., Reinhardt F., Kah K.J., Bell G., Guo W., Rubin J., Richardson A.L., et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145:926–940. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M., Böhm D., von Törne C., Steiner E., Puhl A., Pilch H., Lehr H.-A., Hengstler J.G., Kölbl H., Gehrmann M. The humoral immune system has a key prognostic impact in node-negative breast Cancer. Cancer Res. 2008;68:5405–5413. doi: 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- Shi Z., Wang J., Zhang B. NetGestalt: integrating multidimensional omics data over biological networks. Nat. Methods. 2013;10:597–598. doi: 10.1038/nmeth.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.C., Luker K.E., Garbow J.R., Prior J.L., Jackson E., Piwnica-Worms D., Luker G.D. CXCR4 regulates growth of both primary and metastatic breast Cancer. Cancer Res. 2004;64:8604–8612. doi: 10.1158/0008-5472.CAN-04-1844. [DOI] [PubMed] [Google Scholar]
- Solt L.A., Kumar N., Nuhant P., Wang Y., Lauer J.L., Liu J., Istrate M.A., Kamenecka T.M., Roush W.R., Vidovic D., et al. Suppression of TH17 differentiation and autoimmunity by a synthetic ROR ligand. Nature. 2011;472:491–494. doi: 10.1038/nature10075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou C., Wirapati P., Loi S., Harris A., Fox S., Smeds J., Nordgren H., Farmer P., Praz V., Haibe-Kains B. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J. Natl. Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- Su X., Malouf G.G., Chen Y., Zhang J., Yao H., Valero V., Weinstein J.N., Spano J.P., Meric-Bernstam F., Khayat D., et al. Comprehensive analysis of long non-coding RNAs in human breast cancer clinical subtypes. Oncotarget. 2014;5:9864–9876. doi: 10.18632/oncotarget.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaro M., Gaggianesi M., Catalano V., Benfante A., Iovino F., Biffoni M., Apuzzo T., Sperduti I., Volpe S., Cocorullo G., et al. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell. 2014;14:342–356. doi: 10.1016/j.stem.2014.01.009. [DOI] [PubMed] [Google Scholar]
- van't Veer L.J., Dai H., Van De Vijver M.J., He Y.D., Hart A.A., Mao M., Peterse H.L., van der Kooy K., Marton M.J., Witteveen A.T. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Wang Y., Kumar N., Nuhant P., Cameron M.D., Istrate M.A., Roush W.R., Griffin P.R., Burris T.P. Identification of SR1078, a synthetic agonist for the orphan nuclear receptors RORalpha and RORgamma. ACS Chem. Biol. 2010;5:1029–1034. doi: 10.1021/cb100223d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., McDonald J.G., Patel A., Zhang Y., Umetani M., Xu F., Westover E.J., Covey D.F., Mangelsdorf D.J., Cohen J.C., et al. Sterol intermediates from cholesterol biosynthetic pathway as liver X receptor ligands. J. Biol. Chem. 2006;281:27816–27826. doi: 10.1074/jbc.M603781200. [DOI] [PubMed] [Google Scholar]
- Yang X.O., Pappu B.P., Nurieva R., Akimzhanov A., Kang H.S., Chung Y., Ma L., Shah B., Panopoulos A.D., Schluns K.S., et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Lin C., Jin C., Yang J.C., Tanasa B., Li W., Merkurjev D., Ohgi K.A., Meng D., Zhang J., et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500:598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Tam W.L., Shibue T., Kaygusuz Y., Reinhardt F., Ng Eaton E., Weinberg R.A. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature. 2015;525:256–260. doi: 10.1038/nature14897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W., Restifo N.P. TH17 cells in tumour immunity and immunotherapy. Nat. Rev. Immunol. 2010;10:248–256. doi: 10.1038/nri2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H.Y., Li Q., Lee J.H., Arango M.E., McDonnell S.R., Yamazaki S., Koudriakova T.B., Alton G., Cui J.J., Kung P.P., et al. An orally available small-molecule inhibitor of c-met, PF-2341066, exhibits cytoreductive antitumor efficacy through antiproliferative and antiangiogenic mechanisms. Cancer Res. 2007;67:4408–4417. doi: 10.1158/0008-5472.CAN-06-4443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers information used in this study.
Decreased RORγ expression in aggressive basal-like subtype, ER − ve, and grade 3 breast cancer subtypes. (A and B) RORγ mRNA expression in the combined breast cancer datasets based on molecular subtypes defined of Hu et al. (A) and RORγ mRNA expression in the breast cancer dataset based on molecular subtypes defined by PAM50 (B). (C) Decreased RORγ expression of aggressive grade 3 within the combined breast cancer datasets. (D) Comparison of RORγ mRNA expression between ER + ve and ER − ve cases. (E) Immunohistochemistry showing representative (from n = 13-15) RORγ staining in (i and ii) ER-ve cases, (iii and iv) ER + ve breast cancer and (v and vi) normal breast. Insets show the no primary control sections for the matched tissue where the material was available. Scale bars indicate 50 μm for main panels and inset images respectively.
Identification of RORγ-dependent pathways and survival outcomes (A and B) GSEA analysis showing top five positive and negative enriched modules derived from HALLMARK gene sets based on normalized enrichment score after loss of RORγ function in (A) T-47D and (B) MDA-MB-231 breast cancer cells. (C) GSEA analysis identifies positively enriched KEGG_TGF-Β_SIGNALING, EMT_SIGNALING with common overlapping genes among MCF-7, T-47D and MDA-MB-231 cells after loss of RORγ function. (D) The RORγ-dependent signature displays prognostic utility in predicting survival outcomes. A low score, i.e. high RORγ expression predicts a higher probability of survival (in the UNT dataset). (E) Correlation analysis of RORγ1 mRNA expression and the RORγ-dependent signature scores in UNT dataset reveals inverse relationship between RORγ expression and signature scores.
GREAT cis-regulatory analysis of the RORγ ChIP-ChIP data in MCF-7 cells. (A) Percentage of genomic regions based on numbers of associated genes per region. (B) Percentage of region-to-gene association showing many RORγ binding events lie well outside of the proximal promoter. (C) RORγ ChIP-ChIP data highlighted 13 terms of GO biological processes and identified many critical pathways including mesenchyme development, mammary gland development and epithelial to mesenchymal transition. All terms were still enriched after the cutoff (FDR < 0.05 and fold enrichment > 2) in both binomial test and hypergeometric test.
RORγ regulates the DNA-repair pathway in human breast cancer cells. (A) Protein–protein interaction network analysis of the Fanconi anemia pathway. PPI analysis identified the Fanconi anemia pathway network in T-47D and MCF-7 cells. Netgestalt (iRef PPI database) was utilized to visualize the PPI network with fold changes in log2 scale. (B) RORγ binding sites on BRCA2, RAD51C, RAD52, FANCB, FANCC and FANCI.
Identification of differentially expressed genes (coding and long non-coding RNAs) among three breast cancer cell lines, T-47D, MCF-7 and MDA-MB-231. (A) Number of significantly differentially expressed genes in (I) MDA-MB-231 vs T-47D, (ii) MDA-MB-231 vs MCF-7 and (iii) T-47D vs MCF-7. Comparison between an aggressive ER − ve cell line (MDA-MB-231) and two ER + ve luminal types (T-47D or MCF-7) highlighted the high expression of LINC00511, TGFΒ2, SMAD3, c-MET, CXCR4, SOX9 and SNAI2 in basal-like MDA-MB-231 cells. In addition, RNA-seq also identified the decreased expression of RORγ in MDA-MB-231 compared to other cell lines.
Pharmacological and Genetic manipulation of RORγ expression and activity effects breast carcinogenesis in vitro. (A) Treatment (24 h) of ER + ve MCF-7 cells with the RORγ agonist, SR1078 (10 μM) decreased the expression of the mRNAs coding CXCR4 and CXCL12 (n = 3). (B) RORγ depletion increased cell migration in ER − ve MDA-MB-231 cells. Representative images were shown from 3 independent experiments in triplicate. (C and D) The RORγ agonist, SR1078 (10 μM), decreased microsphere outgrowth in ER + ve MCF-7 in 3D Matrigel invasion assay (C) and the RORγ agonist, SR1078 (10 μM) decreased cell growth and outgrowth ER − ve MDA-MB-231 in 3D Matrigel invasion assay (D). (E) UCSC genome-browser showing the RORγ binding site on SMAD3 is overlapped with one of ENCODE regulator binding regions.