Abstract
Cancer cells frequently up-regulate DNA replication and repair proteins such as the multifunctional DNA2 nuclease/helicase, counteracting DNA damage due to replication stress and promoting survival. Therefore, we hypothesized that blocking both DNA replication and repair by inhibiting the bifunctional DNA2 could be a potent strategy to sensitize cancer cells to stresses from radiation or chemotherapeutic agents. We show that homozygous deletion of DNA2 sensitizes cells to ionizing radiation and camptothecin (CPT). Using a virtual high throughput screen, we identify 4-hydroxy-8-nitroquinoline-3-carboxylic acid (C5) as an effective and selective inhibitor of DNA2. Mutagenesis and biochemical analysis define the C5 binding pocket at a DNA-binding motif that is shared by the nuclease and helicase activities, consistent with structural studies that suggest that DNA binding to the helicase domain is necessary for nuclease activity. C5 targets the known functions of DNA2 in vivo: C5 inhibits resection at stalled forks as well as reducing recombination. C5 is an even more potent inhibitor of restart of stalled DNA replication forks and over-resection of nascent DNA in cells defective in replication fork protection, including BRCA2 and BOD1L. C5 sensitizes cells to CPT and synergizes with PARP inhibitors.
Abbreviations: BLM, Bloom Syndrome; CPT, camptothecin; DNA2, DNA2 nuclease/helicase; DSB, double-strand break; EMSA, electrophoretic mobility shift assay; EXO1, exonuclesae 1; FA, Fanconi anemia; FEN1, flap endonuclease 1; HDR, homology direct repair; HTS, high throughput screening; MES, mouse embryonic stem; MSFC, Multiple Stage Full Coverage; NCI DTP, National Cancer Institute Developmental Therapeutics Program; RMSD, root mean square deviation; RT, radiotherapy; SSA, single-strand annealing; SSB, single-strand break; WRN, Werner Syndrome
Keywords: DNA2 inhibitor, Nuclease, Helicase, DNA binding, DNA end resection, DNA replicatoin fork protection, Chemotherapy, Sensitizer, Camptothecin, PARP inhibitor, Cancer
Highlights
-
•
C5 binds to the helicase domain of DNA2 and inhibits its nuclease, ATPase, and helicase activities.
-
•
C5 inhibits DNA end resection for DNA double strand break repair and restart of stalled replication forks.
-
•
C5 suppresses over-resection of nascent DNA in cells defective in fork protection.
-
•
C5 sensitizes cancer cells to chemotherapeutic agents.
Most chemotherapeutics introduce DNA lesions that block DNA replication to kill cancer cells. Upregulation of DNA repair proteins in cancer cells is a major reason for human cancer to become resistant to chemotherapeutics. DNA repair proteins such as DNA2 nuclease/helicase have long been proposed as targets for sensitization of cancer cells to chemotherapy. We identify a selective DNA2 inhibitor (C5) and demonstrate that DNA2 inhibition by C5 suppresses cancer cells to rescue stalled replication forks. Consequently, it sensitizes cancer cells to replication fork stalling agents such as camptothecin. Thus, C5 is a promising lead compound for developing new anticancer drugs.
1. Introduction
DNA replication is the central process of all actively dividing cells. Blocking this process can result in cell cycle arrest, senescence, and apoptosis. Therefore, DNA replication forks are the targets of most cancer chemotherapeutics, including agents that induce DNA lesions, such as camptothecin (CPT) and cisplatin, plus those that stall forks, such as gemcitabine and 5-fluorouracil (Le Tourneau et al., 2010). In addition, radiotherapy (RT), which is used to treat ~ 50% of all cancers, induces DNA damage. If not repaired, this DNA damage may block or collapse DNA replication forks and kill cancer cells (Begg et al., 2011). However, a drawback of these therapies is that the cancer cell may become resistant to the radiation or chemotherapy. Reasons for resistance include increased tolerance for DNA lesions, and enhanced capacity for DNA damage response and repair. Many cancer cells that are resistant to RT or chemotherapeutic drugs have abnormally high DNA repair capacity, and inhibition of DNA repair has successfully sensitized the cancer cells to cytotoxicity from chemotherapeutic drugs.
One major conserved DNA repair enzyme is the DNA2 helicase/nuclease (DNA2). Complete inactivation of either the helicase or nuclease activity of DNA2 in cells from a wide range of organisms, including yeast and humans, induces cell cycle arrest and cell death (Budd and Campbell, 1995, Kang et al., 2000, Kim et al., 2005, Duxin et al., 2009, Duxin et al., 2012, Wawrousek et al., 2010, Karanja et al., 2012, Lin et al., 2013). Disruption of DNA2 has been associated with human disease. A splice-site mutation that causes decreased levels of human DNA2 gives rise to Seckel syndrome, a primordial dwarfism syndrome. Other mutations are linked to breast cancers (Strauss et al., 2014). Interestingly, the DNA2-deficient Seckel cells show markers of senescence, where cells are viable but cease to proliferate (Shaheen et al., 2014).
DNA2 plays three key roles that allow the cancer cells to resist the intrinsic and extrinsic DNA replication stresses induced by chemotherapy or RT (radiotherapy): flap removal during DNA replication, double-strand break (DSB) resection during repair, and stabilization and restart of reversed replication forks (Wanrooij and Burgers, 2015). During replication, DNA2 removes the long 5′ RNA/DNA “flaps” that arise during Okazaki fragment processing in difficult-to-replicate genomic regions (Bae et al., 2001, Budd and Campbell, 1997, Kang et al., 2010, Kao et al., 2004a, Kao et al., 2004b, Masuda-Sasa et al., 2008, Stewart et al., 2008, Stewart et al., 2010). In yeast, ScDNA2 is probably the major nuclease for RNA primer removal during Okazaki fragment maturation, in collaboration with flap endonuclease 1 (FEN1 or Rad27) (Bae et al., 2001, Budd and Campbell, 1997). For DSB repair, DNA2 acts in one of the two major DSB resection pathways. DNA2 acts with the Bloom Syndrome (BLM) helicase or Werner Syndrome (WRN) helicase in DSB end resection at a critical early step after licensing by limited cleavage by the MRN/CtIP complex (Budd et al., 2005, Imamura and Campbell, 2003, Zhu et al., 2008, Budd and Campbell, 2009, Niu et al., 2009, Cejka et al., 2010, Nimonkar et al., 2011, Symington and Gautier, 2011, Sturzenegger et al., 2014, Liao et al., 2008, Liao et al., 2011). BLM (or WRN), moving on the 3′ terminated strand, unwinds the duplex end to create a “fork”; DNA2 acts as a nuclease on the complementary strand and degrades the 5′ end to produce 3′ ssDNA tails for strand invasion during homology-directed repair (HDR) and S phase checkpoint activation. This resection activity functions in parallel to and independently of resection by Exonuclease 1 (EXO1), downstream of MRE11 (Shibata et al., 2014). At stalled replication forks, DNA2 acts to stabilize, repair and restart forks to allow completion of replication (Hu et al., 2012, Karanja et al., 2014, Thangavel et al., 2015, Weitao et al., 2003a, b). DNA2 also acts in signaling, as both an activator and a target of checkpoint kinases. For instance, DNA2 is required to directly activate the yeast master signaling kinase ATR (Kumar and Burgers, 2013). Furthermore, DNA2 is a target of checkpoint effector kinase Rad53/Chk1/2, and is required to regulate potentially deleterious fork reversal and template switching during replication fork stalling in yeast and humans (Hu et al., 2012, Lai and Foiani, 2012, Thangavel et al., 2015). DNA2 can also play a negative role when the RAD51, BRCA1, BRCA2 and the FA/BRCA (Fanconi anemia/Breast cancer) pathway is impaired (Hashimoto et al., 2010, Schlacher et al., 2011, Schlacher et al., 2012, Petermann et al., 2010). Like MRE11, which functions upstream of DNA2, DNA2 is involved in the excessive resection seen in cells deficient in fork protection (Karanja et al., 2014, Wang et al., 2015, Higgs et al., 2015). Thus, DNA2 must be highly regulated to protect genome stability. In this work, we describe a potent inhibitor of these DNA2 DNA replication and repair activities that sensitizes cancer cells to chemotherapies.
Mechanistically, DNA2 is well studied, but nevertheless poorly defined. Biochemical and genetic experiments have demonstrated an intricate interaction between the nuclease and helicase (Bae et al., 2001, Budd and Campbell, 2000, Budd and Campbell, 2009, Kao et al., 2004a, Kao et al., 2004b, Levikova et al., 2013). Furthermore, biochemical studies indicate that there is a major DNA binding site interacting at the junction of the flap and downstream duplex DNA that is required for both the nuclease and helicase activities (Stewart et al., 2010). Which motifs in the DNA2 protein govern this major binding site have been elusive, but the inhibitor work we report here clarifies these relationships and both provides functional support and is in turn supported by the recently published X-ray crystal structure of murine DNA2 (Zhou et al., 2015). The conclusions of our mechanistic studies using the inhibitor correlate with the multi-domain interaction of DNA2 with DNA revealed in the X-ray crystal structure of murine DNA2 (Zhou et al., 2015), and our mutational studies of inhibitor susceptibility specifically provide functional support for the model proposed for the role of helicase domain DNA binding contacts in nuclease activation in that study (Zhou et al., 2015).
2. Materials & Methods
2.1. 3-D Modeling of Human DNA2 and High Throughput Virtual Screening of the Inhibitors
The methodology for 3-D modeling of human DNA2 and high throughput virtual screening of the inhibitors has been described in the Results section.
2.2. Protein Purification and Nuclease Activity Assay
All WT and mutant DNA2 proteins in this study were expressed as 3X Flag-tagged recombinant proteins in 293T cells and were purified using affinity chromatography as previously described (Ronchi et al., 2013). The nuclease assay was conducted as previously described (Zheng et al., 2008).
2.3. ATPase Assays and EMSA Assays
The ATPase assay was conducted as previously described (Masuda-Sasa et al., 2006). WT DNA2 nuclease cleaves the DNA substrate, which makes it technically difficult to display the helicase and ATPase activities. We therefore chose to use the D294A mutant, which is defective in nuclease activity but has similar ATPase activity to WT DNA2, to test the inhibitory effects of C5 (Masuda-Sasa et al., 2006). The EMSA was performed as described previously (Hellman and Fried, 2007).
2.4. Inhibition Mode and Nonlinear Regression to Determine the Inhibitor Ki
See detailed methods in the on-line supplemental experimental procedures.
2.5. Cell Culture, Measurement of IC50, Clonogenic Assay, and Cell Proliferation Assay
See detailed methods in the on-line supplemental experimental procedures.
2.6. Immunofluorescence Staining
Cells (on cover-slips) with or without various drug treatments for 24 h were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X100, blocked with the Image iT FX signal enhancer (Invitrogen), and incubated (1.5 h, room temperature) with the indicated primary antibodies. The antibodies against γ-H2AX and phosphorylated RPA2 were from Sigma Aldrich. The cells were then washed with PBS buffer and incubated (1 h, room temperature) with the corresponding secondary antibodies (1:200, Invitrogen). The slides were then washed with PBS buffer, counter stained with DAPI and analyzed with a fluorescence microscope (Olympus AX70).
2.7. DSB Repair Reporter Assays
HDR-GFP and SSA-GFP were integrated in U2OS cells (Gunn and Stark, 2012, Howard et al., 2015). The U2OS cells were transfected with the I-SceI expression vector, or the empty vector and GFP vector as negative and positive controls, respectively. At 3 h after transfection, we changed the culture medium to fresh medium with or without compound C5, and cultured the cells for 3 more days. The cells were harvested and the GFP + frequencies (repair frequencies) were determined by flow cytometry using a CyAn ADP Analyzer (Beckman Coulter, Inc.).
2.8. DNA Fiber Assay
See detailed methods in the on-line supplemental experimental procedures.
2.9. Knockdown of Gene Expression in MCF7, A549, and U2OS Cells
Cells were transfected with scrambled siRNA oligos (siControl) or siRNA oligos against human DNA2, BOD1L, and BRCA2 (Sigma). The residual protein levels were detected by western blot.
3. Results
3.1. Elimination of the DNA2 Gene Sensitizes Cells to Radio- and Chemo-Therapeutic Agents
We were interested in DNA2 as a target of inhibition because yeast and human cells that are depleted of DNA2 are sensitive to agents that cause replication stress, such as CPT and cisplatin (Budd and Campbell, 2000, Karanja et al., 2012, Peng et al., 2012). DNA2 is recruited to ionizing-radiation (IR)-induced subnuclear foci in chicken and human cell lines (Hoa et al., 2015). To further investigate the biological roles of DNA2 in mammals, we established DNA2 knockout (dna2−/−) mouse ES (MES) cells. The dna2−/− MES cells were viable, presumably due to backup repair pathways, perhaps involving EXO1, as in yeast; although their proliferation rate was approximately 50% of the WT MES cells. To investigate if DNA2 knockout caused the cells to be more sensitive to DNA damaging agents, we treated WT and dna2−/− MES cells with γ-irradiation (IR) and CPT and observed that dna2−/− MES cells were significantly more sensitive than WT cells to both IR and CPT (Figure S1a and S1b). Taken together with previous work, these findings suggested that DNA2 is a useful candidate for sensitizing cancer cells to DNA damage-inducing therapeutic agents.
3.2. Virtual High Throughput Screening and Experimental Validation for Inhibitors of DNA2
We employed a three-dimensional structural model for virtual screening of small molecules that bind to DNA2. The homology model for the DNA2 helicase domain was based on the crystal structures of the Upf1–RNA U15 complex (PDB 2XZL (Chakrabarti et al., 2011)) and the human Upf1–ADP complex (PDB 2GK6 (Cheng et al., 2007)), which have high sequence identity with DNA2. Although the sequence identity (30%) between DNA2 and Upf1 was at the lower limit for homology modeling, we were able to successfully build the Upf1-based DNA2 model structure (Fig. 1a). (See also Ref. Ronchi et al. (2013) for an earlier version of the model.) After aligning the DNA2 helicase domain with UPF1, the homology model was built with the SWISS-MODEL tool and refined with the Schrodinger Protein Preparation Wizard to fix missing residues and hydrogen positions (Fig. 1a). We predicted druggable sites on the DNA2 model using an in-house-developed Druggable Site Prediction by FDA-approved drugs (DSP) methodology, which uses a diverse subset of 100 FDA-approved drug molecules to dock around the protein surface and predict best binding sites on the protein surface. The definition of the “best binding site” is based on the numbers of the tested drugs bound to the protein pockets. We identified three docking pockets for screening, designated as Sites 1, 2, and 3 with 53%, 24%, and 12% of the tested drugs bound to them, respectively (Fig. 1a). Sites 1 and 3 are predicted to make close contact with DNA and Site 2 is close to the ATP binding and hydrolysis motifs of the helicase domain. The proposed Site 3 is composed of the limited residues conserved between DNA2 and UPF1 in the N-terminal model sequence and interacts with the DNA and Site 1 (Fig. 1a). More importantly, molecular dynamics simulation results further indicated the DNA2 model was stable and reasonable, making it a good choice for in silico screening of DNA2 small molecule inhibitors. Refinement of the structure by molecular dynamics simulation showed that the site centers are stable and that the N-terminal domain stays as shown in Fig. 1b. The average root-mean-square deviation (RMSD) of site centers was only 2–3 Å during simulation, although the RMSD of some flexible loop regions could be more than 15 Å, which indicates that the predicted sites are stabilized to allow ligand binding. Based on an alignment of helicase family members (Cheng et al., 2007), there are seven highly conserved helicase motifs including I, Ia, II, III, IV, V, and VI in DNA2. The I, II, and IV motifs are for ATP binding and hydrolysis, whereas the Ia, III, and V motifs are the DNA binding domains in other helicases. Domains IV and VI coordinate the DNA unwinding and hydrolysis. One predicted site of drug binding, Site 1, contains amino acids in the Ia motif. Site 2 contains residues in motif I, as well as in motifs V and VI (Fig. 1c). This information allowed us to focus our search for candidate DNA2 inhibitors on the most likely small molecule binding sites. It also guided us to a pocket that might further illuminate how DNA2 interacts with its DNA substrate.
Fig 1.
Three dimensional human DNA2 model and potential pockets for screening small molecule DNA2 inhibitors (see also Figures S1 and Table S1).
a. A homology model for human DNA2 in complex with single-stranded DNA (ssDNA). Orange: ssDNA; Cyan: nuclease domain; Green: ATP binding domain; Pink: helicase domain. Three potential drug binding pockets are specified as Sites 1–3.
b. Refinement of the DNA2 model structure by molecular dynamics simulation (50 ns). Root mean square deviation (RMSD) values during simulation at three potential drug binding sites are shown as fluctuating bars. The secondary structures of the sites are represented by color, which is specified at the bottom of the graph.
c. The linear domain and motif structures and the drug binding sites of human DNA2. Upper panel: the DNA2 functional domain structure; Middle panel: the three putative drug binding sites; and Bottom panel: the secondary structure motifs.
d. Inhibition of DNA2 nuclease activity by chemical compounds that were selected from the virtual screen. Recombinant flag-tagged DNA2 (10 nM) was mixed with 32P-labeled flap DNA substrates (500 fmol) in the absence or presence of the potential DNA2 binding chemical compound (250 μM each). The image shows a representative biochemical reaction (37 °C, 15 min) that was resolved using 15% denaturing polyacrylamide gel electrophoresis (PAGE). The locations of substrates and products on the gel are indicated.
e. The chemical name and structure of C5.
We then conducted a virtual high throughput screening (vHTS) for molecules binding to Site 1 because Site 1 gave the best FDA drug screen score and is predicted to affect DNA binding, making it a favorable target site. Our model also suggested that Site 1, which interacts with Site 3, might be a shared component of both the helicase and nuclease activity and that inhibitors bound to Site 1 might clarify the coordination of the nuclease and helicase activities of DNA2. We used an in-house-developed Multiple-Stage Full-Coverage (MSFC-VS) algorithm to screen an in silico a library of 260,071 compounds from the National Cancer Institute Developmental Therapeutics Program (NCI DTP) library for binding to Site 1. This generated a list of 40 compounds (Table S1). Although Site 1 is located in the putative helicase domain, we chose to search for inhibitors that might affect both the nuclease and helicase activities, because previous studies suggested that they may compete for the same DNA binding site. We therefore screened these compounds biochemically for their inhibition of DNA2 nuclease activity using purified recombinant hDNA2 and a well-defined flap substrate. Among the 40 identified compounds, 4 molecules inhibited the DNA2 nuclease activity (Fig. 1d and Table S1). Among these compounds, 4-hydroxy-8-nitroquinoline-3-carboxylic acid, designated as compound C5, had the top Glide XP Docking Score (− 8.3 kcal/mol), and displayed the highest cytotoxicity to human cancer cells (Fig. 1e and Table S1). To determine if C5 specifically targets DNA2, we tested if C5 will inhibit the enzyme activities of two similar structure specific nucleases, FEN1 and EXO1. We found that C5 poorly inhibited the activities of the other two nucleases, in contrast to DNA2 (Figure S1c–f). Taken together, C5 is a specific inhibitor of DNA2 nuclease activity in vitro.
3.3. Kinetic Analysis of C5 in Inhibiting DNA2 Activity
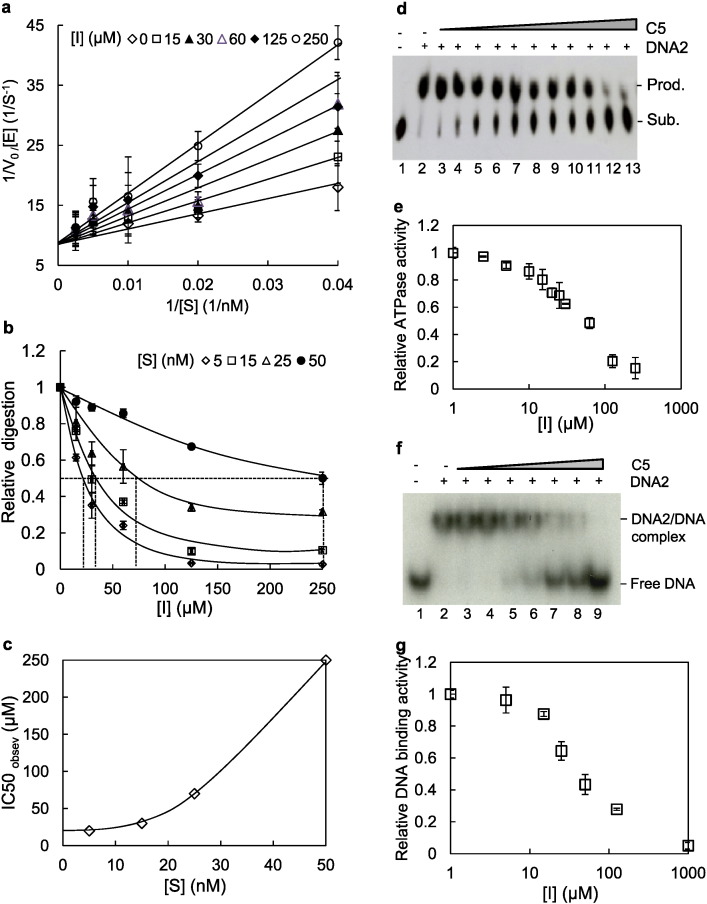
In order to determine the IC50 of C5 for inhibition of the nuclease activity of DNA2, we conducted kinetic analyses. Using a flap DNA substrate, we first studied the time course of the nuclease activity in order to determine the proper time interval for kinetic analysis at various substrate concentrations (Figure S2a). We found that the enzyme activity was linear at 1–10 min, and we conducted the assays in this range. To evaluate the mechanism of inhibition of nuclease activity by C5, we measured the nuclease activity of DNA2 at various concentrations of inhibitor and substrate (Fig. 2a). Using a Lineweaver-Burk plot, we evaluated the competition patterns using competitive, noncompetitive, and uncompetitive models. The competitive inhibition model fits best to the inhibition data with C5. To extract the intrinsic inhibition constant of C5 for DNA2, we obtained apparent inhibition constant, or IC50, values, for C5 at a series of substrate concentrations (Fig. 2b). Extrapolation of the observed IC50 values to limiting substrate concentrations, as described in the Materials & Methods section, gave an inhibition constant of 20 μM (Fig. 2c).
Fig. 2.
Inhibitory kinetics of DNA2 nuclease activity, and C5 inhibitory effects to ATPase activity and DNA substrate binding capacity (see also Figure S2).
a–c. The nuclease activity of DNA2 was analyzed in the presence of varying concentrations of DNA2 enzyme (0.5–5 nM), flap DNA substrate (5–50 nM), and DNA2 inhibitors C5 (0–250 μM).
a. Lineweaver-Burk plot of DNA2 nuclease activity in the presence of various concentrations of flap DNA substrate (x axis) and C5 (μM, designated as [I]).
b. DNA2 nuclease activity in the presence of various concentrations of compound C5 (x axis) and flap DNA substrate (nM, designated as [S]). The C5 concentration (IC50observed) that inhibits 50% of the DNA2 nuclease activity at a given concentration of DNA substrates is indicated with dotted lines. The inhibited nuclease activities were normalized to the DMSO control, set as 1.
c. Plot of IC50observed versus [S] for determination of IC50. The values of IC50observed obtained from panel B and corresponding DNA substrate concentrations were plotted. When [S] is zero, the derived corresponding IC50observed value is the theoretical IC50 of C5. In panels a–c, the values are means ± s.d of three independent experiments.
d. The representative TLC image showing C5 inhibition of the ATPase activity of DNA2. The DNA2 enzyme concentration used was 10 nM; ATP substrate concentration used was 200 μM; DNA concentration used was 200 nM; the inhibitor C5 concentrations used was in a range of 0 to 250 μM.
e. Quantification of inhibition of DNA2 in the ATPase activity, the relative ATPase activities normalized to DMSO control. The values shown are the means ± s.d. of three independent assays.
f. The representative EMSA image showing C5 inhibition of DNA2 binding to the DNA substrate. The DNA2 enzyme concentration used was 50 nM; the 32P labeled DNA concentration used was 1 nM; the compound C5 concentrations used ranged from 0 to 1000 μM.
g. Quantification of inhibition of DNA2 substrate binding, the relative binding activities normalized to DMSO control. The values shown are the means ± s.d. of three independent assays.
The kinetic studies suggested that C5 acts as a competitive inhibitor of DNA2 nuclease activity. A competitive model predicts C5 binding to DNA2 should block binding of DNA2 to the DNA substrate. Consistent with the model, we found that C5 inhibited the ATPase activity of DNA2, which is dependent on DNA binding (Fig. 2d and e). To further test this model, we evaluated DNA2 substrate binding in the presence of various C5 concentrations directly, by electrophoretic mobility shift assay (EMSA). We found that DNA2 effectively bound flap DNA substrates, leading to reduced electrophoretic mobility; but the addition of C5 reduced the formation of the DNA2–substrate complex (Fig. 2f and g). The inhibitor concentration needed to reduce the DNA2–substrate complex formation to 50% is 30 μM, which is comparable to the IC50 value for inhibition of the DNA2 nuclease activity. Finally, we tested the ability of C5 to inhibit DNA2 helicase activity. Using the DNA2 helicase substrate in which an M13 phage DNA is hybridized to a 5′ tailed oligonucleotide (Masuda-Sasa et al., 2006), we found, as expected by our finding of inhibition of DNA-dependent ATPase activity, that C5 inhibits the 5′ to 3′, end–dependent DNA helicase activity of DNA2 (Figure S2b, compare lanes 3 and 6, 4 and 7, 5 and 8) (Masuda-Sasa et al., 2006, Balakrishnan et al., 2010). Based on our results, we suggest that although our homology structure does not show the nuclease domain, that contacts in both ATPase and helicase domains are required for nuclease activity, consistent with the recent X-ray crystal structure of murine DNA2 (80% identical to human DNA2) (Zhou et al., 2015).
3.4. Validation of Site 1 on DNA2 as the Binding Pocket for C5 and Rationale for its Effect on DNA Binding
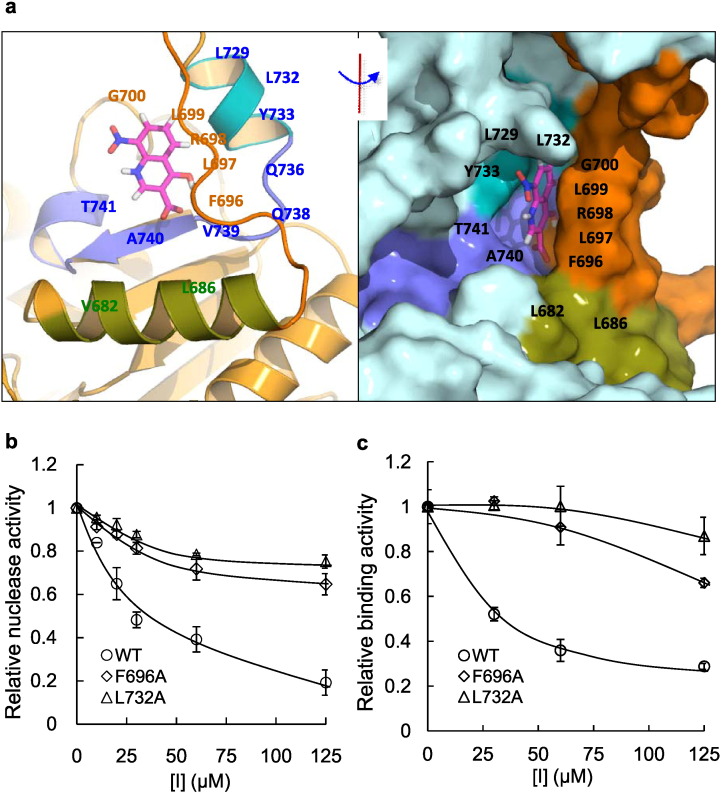
C5 was identified by virtual screening for small molecules that bind to a pocket (Site 1) near the DNA binding site in our computational DNA2 model, which superimposes on the DNA binding site in the X-ray crystal structure. Based on our 3-D model, we searched the residues within 6 Å spheres around the predicted C5 binding site. We identified multiple residues, including V682, L686, F696, L697, R698, L699, G700, L729, L732, Y733, Q736, Q738, V739, and T741, that may play a key role in coordinating C5 binding (Fig. 3a). Superposition of our model for human DNA2 on the mouse DNA2 structure, shows that these putative C-5 interacting amino acids could directly affect those involved in DNA binding (Zhou et al., 2015). To test if C5 indeed binds to/affects the putative pocket at Site 1, we substituted residues predicted to be near C5 with alanine. The mutant and WT DNA2 proteins were purified and the nuclease activities were assayed. Although Site 1 lies in the helicase domain, many of the mutations altered the nuclease activity in the absence of inhibitor, presumably because the Site 1 pocket is close to the putative DNA substrate binding pocket. These are the first mutations within the helicase domain that have been demonstrated to concomitantly affect the nuclease activity, strengthening our results for dual DNA2 helicase and nuclease inhibition by C5. Other mutants, however, maintained intact nuclease activity (Figure S3a and S3b). We chose F696A and L732A with enzyme activity similar to the WT to test their sensitivity to compound C5. Both the nuclease activity and DNA binding activity of the two mutants were less sensitive to C5 than wild type (Fig. 3b and c and Figure S3c). The compound C5 IC50s for nuclease activities and DNA substrate binding were greater than 250 μM for F696A and L732A, compared to 30 μM for the WT. This suggests that the mutations impair the interaction of C5 with the designated binding site (Site 1) of DNA2 and that this site does affect substrate DNA binding in the helicase portion of DNA2 that is critical for nuclease activity.
Fig. 3.
DNA2 mutations at Site 1 impair C5 inhibition of DNA2 nuclease activity (see also Figure S3).
a. Three dimensional structure of the Site 1 small molecule binding pocket of DNA2. The left panel shows a cartoon view and the right panel shows a surface view of Site 1. The C5 inhibitor is shown as a pink stick. The residues 680–691 are green, 692–710 are orange, 729–735 are cyan and 736–745 are light-blue.
b. The 14 residues within 6 Å spheres around compound C5 that form Site 1 pocket were identified. Among them, F696A and L732A, which did not affect the nuclease activity of DNA2 (Figure S3), reduced C5 inhibition of DNA2 nuclease activity. The nuclease activity of WT, F696A, and L732A (1 nM) was assayed in the presence of various concentrations of C5 (indicated as [I] in a range from 0 to 250 μM) and quantified, the DNA substrate concentration was 15 nM. We added DMSO without the inhibitor C5 as a control where the relative nuclease activity was set as 1. The values shown are the means ± s.d. of three independent experiments.
c. The DNA binding activity of F696A and L732A is resistant to C5 inhibitor. We added DMSO without the inhibitor C5 as a control where the relative binding activity was set as 1. The DNA2 enzyme concentration used was 50 nM. The DNA concentration used was 1 nM. The inhibitor C5 concentrations ranged from 0 to 125 μM. The values shown are the means ± s.d. of three independent experiments.
3.5. C5 Displays On-Target Effects on DNA2
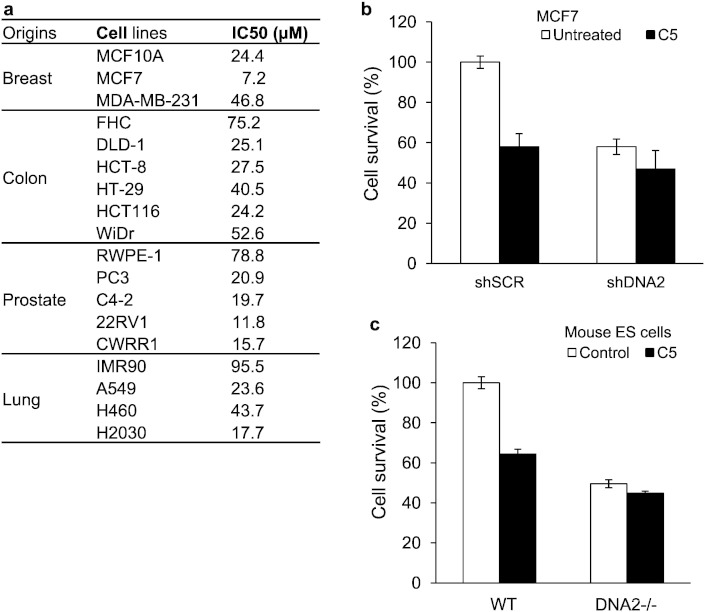
Next, we determined the cytotoxicity of DNA2 inhibitor C5 and evaluated whether the compound had on-target effects on DNA2 in cultured cells. We have measured IC50 values of C5 with a panel of 18 cell lines of 4 major types of cancers by a cell proliferation assay (Chou, 2010). The IC50 values among the different cell lines varied from 7 μM to 70 μM, which is comparable to the estimated enzymatic IC50 value of 20 μM (Fig. 4a). Furthermore, we considered that if the toxic effects of C5 were due to targeting DNA2, then cells lacking DNA2 should be resistant to the effects of compound C5. As anticipated, we found a reduced spontaneous survival rate in human and mouse cells with either shRNA-mediated knockdown (Fig. 4b) or knockout (Fig. 4c) of DNA2. We found that WT MES cells and human MCF7 cancer cells treated with C5 showed a 60% survival rate, compared to the untreated WT control (Fig. 4b and c), a result similar to the effects of DNA2 knockdown and knockout. Importantly, we found that treatment of dna2−/− MES or DNA2 knockdown human cancer cells with C5 did not further reduce the survival rate (Fig. 4b and c). On the other hand, C5 and Mre11 knockdown displayed strong potentiation in cell killing (Figure S4b). These findings suggest that the cytotoxic effects of compound C5 were primarily due to specific effects on DNA2 at the given concentration. This suggests that due to residual viability, those normal cells may be less sensitive to inhibitors than cancer cells, with repair and checkpoint defects. This also suggests that other enzymes, such as FEN1, EXO1, or Mre11 cannot completely compensate for the loss of DNA2, indicating that DNA2 inhibitors are likely to have significant physiological effects in cancer cells. The results suggest that C5 is a potent and specific inhibitor of DNA2 and that virtual docking is a valid method to help identify DNA2-specific drugs.
Fig. 4.
IC50 and on-target cytotoxic effects of C5 in human cancer cells and mouse embryonic stem (MES) cells (see also Figure S4).
a. IC50 values of C5 with a panel of 18 cell lines from 4 major types of cancers. Human non-cancerous or cancer cells were seeded on a 96-well plate and incubated in culture medium containing 0 to 80 μM C5 for 7 days. The IC50 was calculated using the CompuSyn software(Chou, 2010). Values are the average of two independent assays.
b. Control (shSCR) or DNA2 knockdown MCF7cells were cultured in medium containing 0 or 1 μM C5 for 4 days. The live cells were counted. The cell survival was calculated by normalizing the number of live cells from each culture to that of the control MCF7 cells (shSCR), which was arbitrarily set as 100.
c. The same experiment as in A was performed on MES cells from WT and DNA2 knockout mice, which were cultured in medium containing 0 or 1 μM C5 for 4 days. The values shown are the means ± s.d. of three experiments.
3.6. C5 Suppresses Key DNA2 DNA Replication and Repair Functions
To further validate C5 as a DNA2 inhibitor, we tested its effects on DNA2 functions known to be affected by knockdown or deletion of DNA2 in previous studies (Peng et al., 2012, Howard et al., 2015, Karanja et al., 2012, Karanja et al., 2014). We first determined the effect of C5 on two recombination pathways, SSA (single-strand annealing) and homologous recombination (HR), using I-SceI/GFP-based reporter assays (Gunn and Stark, 2012, Howard et al., 2015, Bryant et al., 2009, Pierce et al., 1999). Since these pathways are most active in S/G2 (Figure S5a), we determined GFP positive cells in the G2 population. We found that both SSA and HR were reduced by C5 in a dose-dependent manner (Fig. 5a). For comparison, at 60 μM C5, SSA and HR were reduced to the same level as in an siRNA DNA2 knockdown carried out in parallel (Figure S5b).
Fig. 5.
Inhibitor C5 suppresses resection-related homology directed repair (HDR) and single-stand annealing (SSA) and causes accumulation of phosphorylated RPA foci (see also Figure S5).
a. C5 inhibits HDR and SSA frequency. The U2OS cells carrying the GFP reporter gene for HDR or SSA assay were transfected with I-Sce I expression vector. The cells were then incubated in medium containing 0, 10, 20, 40, and 60 μM C5. After 72 h, the cells were harvested and the GFP positive cells were analyzed by flow cytometry. In the DNA2 knockdown experiment, the U2OS cells were transfected with 10 nM of scrambled or DNA2 siRNA oligos for 24 h. The cells were then transfected with the I-SceI expression vector. After 48 h, the cells were harvested and the GFP positive cells were analyzed by flow cytometry. Knockdown of DNA2 in the engineered U2OS cells was confirmed by western blot (Figure S5b). Values are mean ± s.d. of three independent experiments.
b. DNA2 inhibition by siRNA or C5 impairs replication fork-related DNA end resection in MCF7 cells at similar levels. MCF7 cells were untreated or treated with 10 μM C5 for 24 h (left panels) or treated with scrambled siRNA (siControl) or siRNA against DNA2 (siDNA2) for 72 h (right panels). The knockdown efficiency of DNA2 was checked by western blotting. The cells were then treated with 1 μM CPT for 4 h. The levels of γ-H2AX and phosphorylated RPA (S33) were analyzed by western blot using antibodies against γH2AX (Millipore) and phosphorylated RPA (S33) (Abcam). Total level of RPA and β-actin were used as controls, which were detected using antibodies against RPA32 (Abcam) and β-actin (GeneTex).
c–h. DNA2 inhibition by siRNA or C5 impairs replication fork-related DNA end resection in A549 cells at similar levels. A549 cells were untreated or treated with 10 μM C5 for 24 h (c-e) or treated with scrambled siRNA (siControl) or siRNA against DNA2 (siDNA2) for 48 h (f–h panels). The knockdown efficiency of DNA2 was checked by western blotting (Figure S5). The cells were then treated with 1 μM CPT for 4 h. Panels c and f: Representative images. Panels d, e, g, and h: Quantifications: the levels of γ-H2AX and phosphorylated RPA (S33) were quantified by ImagePro Premier, and the relative P-RPA or γH2AX per nucleus was calculated. Values are means ± s.d. of three independent assays.
We next wanted to verify if the defects in recombination assays were due to inhibition of end resection. During the early steps of recombination, DNA2 in complex with BLM or WRN protein resects DSB ends, producing ssDNA 3′ overhangs (Karanja et al., 2012, Nimonkar et al., 2011, Sturzenegger et al., 2014). The ssDNA overhangs are coated with RPA, which is then phosphorylated by ATR (Zou and Elledge, 2003). To measure resection, we determined the level of phosphorylated RPA2 (S33 or S4/8) in cells treated with C5 in the presence or absence of CPT. CPT stabilizes cleavable complex intermediates in topoisomerase I reactions which collapse into DSBs when encountered by a replication fork (Hsiang et al., 1989, Patel et al., 2012). CPT increased phosphorylated RPA2 (P-RPA), as measured on western blots and by immunofluorescence of P-RPA2 foci (Fig. 5b–e). C5 significantly reduced the CPT-induced P-RPA level in γH2AX-positive cells (Fig. 5b–e). The level of C5 used here reduced RPA phoshporylation to the same extent as we observed in parallel studies using siRNA DNA2 knockdown (compare Fig. 5c–e and f–h) consistent with previous reports (Duxin et al., 2009, Duxin et al., 2012, Karanja et al., 2012, Peng et al., 2012). Interestingly, we also noted that C5 alone caused background increase in γH2AX (Fig. 5b–h), presumably because the DNA2 inhibitor itself causes replication stress, similarly as shown previously for shDNA2 knockdown (Duxin et al., 2009, Duxin et al., 2012, Karanja et al., 2012).
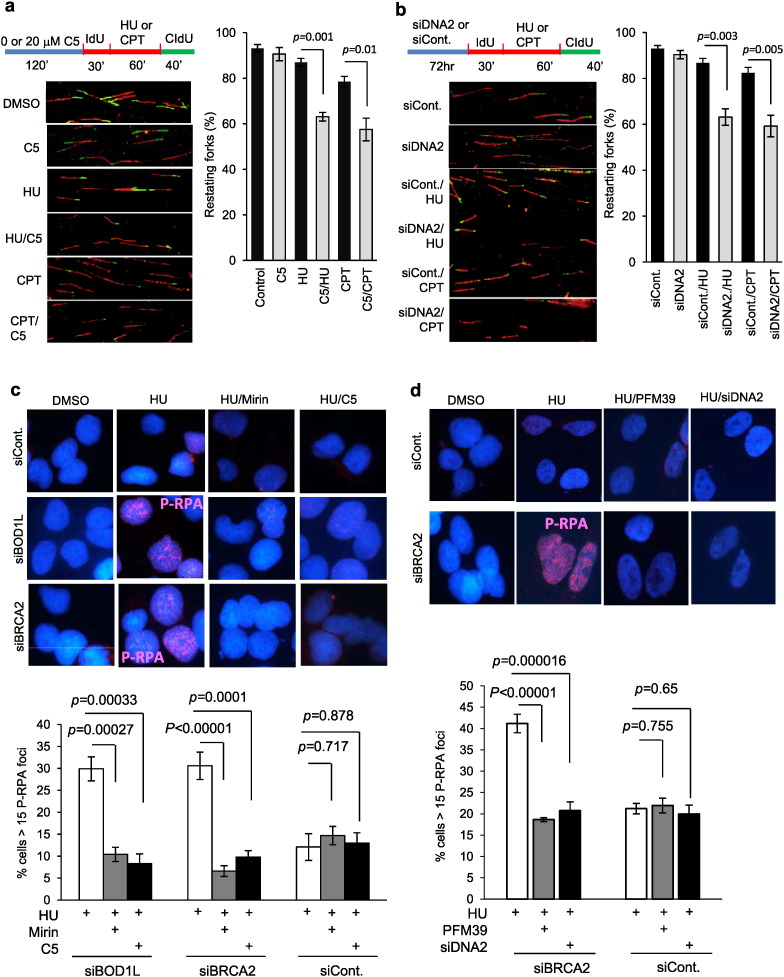
When DNA replication forks are stalled in S-phase, DNA2 plays an important role in stabilizing the stalled forks, in preventing DSB formation, and in resection of the nascent strand to prepare for the restart of replication (Karanja et al., 2012, Thangavel et al., 2015, Weitao et al., 2003a, Weitao et al., 2003b, Hu et al., 2012). A recent study using DNA fiber assays indicated that knockdown of DNA2 by siRNAs inhibits replication fork restart in HU-treated cells or cells treated with low levels of CPT (Thangavel et al., 2015). We have conducted DNA fiber assays to evaluate the effect of C5 on restart of forks treated with low levels of CPT, which in contrast to high CPT levels does not cause DSBs but results in increased positive supercoiling and fork slowing and stalling (Ray Chaudhuri et al., 2012), or with HU, which inhibits production of nucleotide precursors. In absence of C5, 80% or 75% of replication forks could restart (red–green tracts) in HU-treated or CPT-treated cells, respectively (Fig. 6a). However, 20 μM C5 reduced the percentage of restarting forks to 60% and 50% in the HU-treated or CPT-treated cells, respectively (Fig. 6a). This level of inhibition at 20 μM C5 was equivalent to knockdown of DNA2 using siRNA (Fig. 6b). These results extend previous studies on DNA2 functions during replication restart (Ray Chaudhuri et al., 2012, Thangavel et al., 2015, Teicher, 2008) and show that DNA2 is more sensitive to inhibition at stalled replication forks than at a single DSB (Fig. 5). C5 also inhibited restart in cells treated with high levels of CPT, resulting in replication associated DSBs (Figure S6a).
Fig. 6.
C5 suppresses restart of stalled DNA replication forks and over-resection of nascent DNA in cells defective in fork protection (see also Figure S6).
a. and b. C5 inhibits DNA replication fork restart at similar levels as DNA2 knockdown. a. A549 cells were mock or pretreated with C5 (20 μM) for 2 h, labeled with IdU (red) for 30 min, and co-cultured with IdU and the indicated drugs (20 μM C5, 150 nM CPT or 2 mM HU) or drug combinations (20 μM C5 combined with 150 nM CPT or 2 mM HU) for 1 h, and then washed and labeled with CIdU (green) for 40 min. Percentage of restarting forks (red-green tracks) was calculated by dividing the red-green tracks by the sum of the red-green and red only tracks. At least 150 tracks counted for each sample as shown in left panels were calculated. Values are means ± s.d. from three independent experiments (right panel). The p value was calculated by the Student's t-test. b. For the control experiment in which knockdown of DNA2 was employed, A549 cells were transfected with scrambled or DNA2 siRNAs for 72 h, and the ability of the cells to restart replication after CPT or HU fork stalling was determined as in panel a. Western blotting confirmed an efficient knockdown of DNA2 at 72 h post siRNA transfections (Figure S5). In both panels a and b, red tracks represent synthesis before addition of HU or CPT. Red/green tracks represent molecules that recovered from fork stalling. Green only tracks represent initiations after removal of HU or CPT.
c–d. C5 prevents single-stranded DNA accumulation in BOD1L-depleted or BRCA2-depleted U2OS cells upon replication stalling with HU at similar level as DNA2 knockdown. c. Cells with more than 15 P-RPA foci indicative of single-stranded DNA were scored in U2OS cells transfected with siRNA against BRCA2 or BOD1L or scrambled siRNA, as indicated, followed by treatment with 4 mM hydroxyurea (HU) for 5 h. Cells were pretreated with MRE11 inhibitor mirin (50 μM) or DNA2 inhibitor C5 (20 μM). In the absence of HU, no cells with greater than 15 foci were observed. d. Cells with more than 15 P-RPA foci indicative of single-stranded DNA were scored in U2OS cells transfected with siRNA against BRCA2 or scrambled siRNA, as indicated, followed by treatment with 4 mM HU for 5 h. Cells were pretreated with MRE11 inhibitor, PFM39 or siRNA against DNA2. In both c and d, top panels show the representative immunofluorescence images, and the bottom panel shows the quantifications. Error bars represent the SEM. p-Values were calculated with the Student's t-test. Western blots of knockdowns are shown in Figure S6.
Fork protection is a DNA break repair-independent pathway suppressing genomic instability (Schlacher et al., 2011, Schlacher et al., 2012). The fork protection pathway is mediated by BRCA2, BRCA1, RAD51, members of the Fanconi anemia pathway, and BOD1L. In the absence of any of these factors, excessive nascent DNA degradation occurs at stalled replication forks. This degradation is prevented if the end-resection protein MRE11 is inhibited either by siRNA knockdown or by inhibition of MRE11 nuclease by the small molecule mirin (Schlacher et al., 2011, Schlacher et al., 2012). MRE11 functions upstream of DNA2 in resection of double-strand breaks, and DNA2, in addition to MRE11 has been implicated in over-resection at stalled replication forks in BOD1L and in RAD51 impaired cells, since over-resection is suppressed by knockdown of DNA2 in the absence of BOD1L or in a RAD51 mutant cell line (Higgs et al., 2015, Wang et al., 2015). To determine if C5, like DNA2 knockdown, suppressed nascent DNA degradation and resulting accumulation of ssDNA at stalled forks, we monitored phospho-RPA (P-RPA) foci in BRCA2- or BOD1L-depleted or mock transfected U2OS cells with and without HU treatment (Fig. 6c and d). In the absence of HU, in mock-depleted cells we saw few cells with greater than 15 P-RPA foci/per cell, and neither mirin nor C5 significantly decreased the number of P-RPA positive cells, consistent with the fork protection pathway being intact (Fig. 6c). After treatment of the BRCA2- or BOD1L-depleted U2OS cells with HU, which stalls forks but is not expected to produce DSBs (Schlacher et al., 2011, Schlacher et al., 2012, Petermann et al., 2010), and which are fork protection defective, we observed a dramatic increase in P-RPA positive cells indicative of nascent DNA degradation upon treatment with HU (Fig. 6c and Figure S6b). This degradation was suppressed by C5. The potency of C5 was estimated by comparing the effect of mirin, the MRE11 inhibitor, in the same experiment. Remarkably, 20 μM C5 has a comparable potency to 50 μM mirin in reducing this degradation (Fig. 6c, d). To exclude off-target effects of mirin in our experiments, we showed that another potent MRE11 inhibitor, PFM39 (Shibata et al., 2014), reduced P-RPA foci to the same extent as mirin in the BRCA2-deficient cells (Fig. 6d). Finally, we showed in a parallel experiment that the level of reduction in P-RPA foci caused by C5 in BRCA2-deficient cells is also equivalent to that observed in a DNA2 knockdown (Fig. 6d and Figure S6b). Our results are in keeping with previous work showing that knockdown of DNA2 counteracts excessive nascent strand degradation in both BOD1L and in RAD51 impaired cells (Higgs et al., 2015, Wang et al., 2015). Importantly, extending previous reports, we show that MRE11 and DNA2 both are responsible for degrading stalled replication forks in fork protection defective cells, including BOD1L and BRCA2. We conclude that C5 suppresses the fork protection defect of the BRCA2- or BOD1L-deficient cells by inhibition of DNA2. Taken together, the data shows that C5 is a specific inhibitor of DNA2 activities at stalled replication forks in vivo.
3.7. C5 Sensitizes Cells to PARP Inhibitors
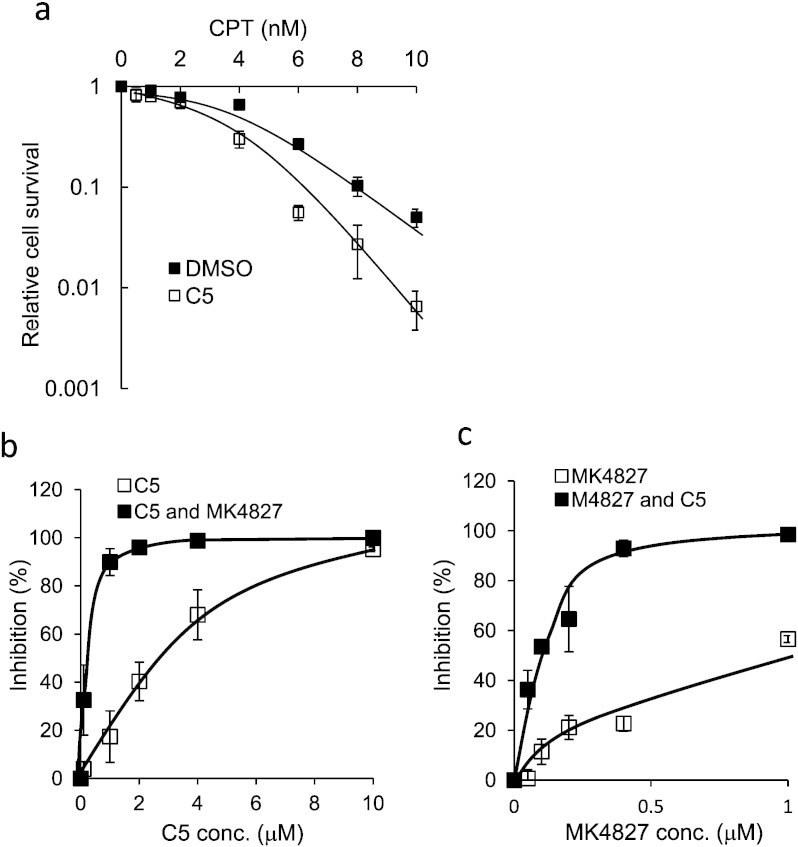
To test our hypothesis that inhibition of DNA2 synergizes with other chemotherapeutic agents, we treated cells with CPT and C5. We found the two agents to be synergistic for cellular lethality (Fig. 7a). These data uncover the potential for C5 in increasing killing efficiencies of DNA damaging chemotherapeutics.
Fig. 7.
DNA2 inhibitor C5 synergistically kills breast cancer cells MCF7 with CPT and PARP inhibitor MK4827.
a. C5 sensitizes MCF cells to CPT. Clonogenic assays were conducted to evaluate the survival rate of MCF cells treated with different concentrations of CPT in the absence or presence of C5 (1 μM). The survival rate of the cells treated with various concentrations of CPT was calculated by normalizing the number of colonies to that of the cells without CPT treatment. The survival rate of the cells without CPT treatment was arbitrarily set as 1. The values shown are the means ± s. d. of three independent experiments.
b–c. The synergy between the DNA2 inhibitor C5 and the PARP inhibitor MK4827 was assayed by clonogenic assay. The values are means ± s.d. of three independent clonogenic assays. The IC50 and combination index (CI) was calculated using the Compusyn program.
b. Representative inhibition curve of varying concentrations of C5 from 0 to 10 μM in combination with MK4827 (0 or 1 μM).
c. Representative inhibition curve of varying concentrations of MK4827 from 0 to 1 μM in combination with C5 (0 or 2 μM).
PARPs have been shown to play an important role in DNA single-strand break (SSB) repair and at stalled replication forks (Bryant et al., 2009, Yang et al., 2004, Hu et al., 2014, Ying et al., 2012), and inhibition of PARPs resulted in accumulation of SSBs and consequent DSBs in the cells (Fisher et al., 2007, Okano et al., 2003). More recently, PARPs have also been implicated in loading DNA damage response proteins to DSB sites (Hu et al., 2014, Li and Yu, 2013). Because DNA2 appears to function in many of the same pathways as the BRCA breast cancer genes, including DSB repair and replication fork protection, and PARP inhibition shows synthetic lethality with BRCA1 or BRCA2 mutants (Fathers et al., 2012, Fong et al., 2008), we considered that PARP inhibition might increase the effectiveness of the DNA2 inhibitor. To test this hypothesis, we determined the survival of MCF7 breast cancer cells cultured in the medium containing compound C5 and/or MK4827, a PARP inhibitor (Jones et al., 2009). We found that MK4827 and C5 had a strong synergistic effect in inhibiting the survival of MCF7 cells (Fig. 7b and c). The IC50 for MK4827 and C5 was 0.8 μM and 1.9 μM. The combination index for MK4827 (1 μM) and C5 (2 μM) was 0.13, indicating a very strong synergy between two drugs. These data support a model where DNA2 and PARP collaboratively participate or have complementary roles in DNA damage response, DSB repair, BER repair pathways, and replication fork protection, which will be an important issue to dissect in future studies.
4. Discussion
We have used a partial DNA2 protein structure based on the homology between the helicase domain of DNA2 and yeast Upf1–RNA U15 complex and human Upf1–ADP complex (Chakrabarti et al., 2011, Cheng et al., 2007) to identify 3 well defined pockets (Sites 1, 2, and 3), where drug like molecules can preferentially dock. We then used a virtual screen consisting of docking of 260,721 NCI deposited small molecules to these sites to identify DNA2 inhibitors. This screen is similar to a previous virtual screen used to identify inhibitors of ribonucleotide reductase (Chen et al., 2015, Zhou et al., 2013). We characterized one inhibitor, C5, which we demonstrate biochemically inhibits nuclease, DNA dependent ATPase, helicase, and DNA binding activities of DNA2. Through a series of functional analyses, we have pinpointed the specific functions of DNA2 that C5 targets to explain its cellular toxicity. C5 suppresses replication-coupled DSB end resection and restart of either HU- or CPT-stalled DNA replication forks. C5 also inhibits over-resection of nascent DNA in cells defective in replication fork protection, such as BRCA2. All these data support our conclusion that virtual screening can be efficient, and that C5 is a promising lead compound to develop sensitizers for cancer chemotherapeutics that cause replication stress.
It is interesting to note that C5, which our model and data suggest binds to the helicase domain of DNA2, can suppress the nuclease activity. We propose that this occurs because C5 can reduce DNA substrate binding to a site in the helicase domain necessary to activate the nuclease. The dramatic effect of the inhibitor on the nuclease activity, its predicted binding site, and more importantly, the effect of mutations we identified in helicase domain 1A on the nuclease activity and DNA binding reveal that the DNA binding site in helicase domain 1A (counterintuitively) is indeed critical for nuclease activity. This in turn suggests that DNA contacts in the nuclease site are not sufficiently strong to promote nuclease activity; multiple domains of DNA2 have to interact with DNA to elicit nuclease activity. These major new insights into the structure/function mechanism of this important class of enzyme, fused helicase/nuclease (including AddAB and RecB), extend recent reports on the X-ray crystal structure of murine DNA2 (80% identity to human DNA2 in the helicase domain)(Zhou et al., 2015). The structure shows DNA threading through a tunnel in the enzyme, first contacting the nuclease. The helicase domain follows and binding occurs as domain 1A and then 2A contact the DNA in the tunnel (Zhou et al., 2015). This explains why both nuclease and helicase utilize threading mechanisms (Bae et al., 2002, Balakrishnan et al., 2010, Kao et al., 2004a). Our work in turn provides direct evidence for requirement for both these domains for nuclease and helicase activity. Taken together with previous biochemical studies (Kao et al., 2004a, Bae et al., 2002, Stewart et al., 2010, Balakrishnan et al., 2010), a “thread, bind, and cleave” model now best explains how DNA2 nuclease works. C5 could either block threading, so that domain 1A does not come into contact with DNA or C5 could block binding to 1A or 2A. While we know about the active sites, the inhibitor will be useful in studying the role of the helicase, which is still conjectural. Co-crystals of DNA2 with C5 should be very informative with respect to the mechanism we propose for C5 function based on the putative binding to Site 1 in our homology structure. The putative common DNA binding site, predicted by our results and shown in the crystal structure, explains how the nuclease and helicase compete for the same substrate, as proposed by Levikova et al. (Levikova et al., 2013) and that the nuclease catalytic site contacts must be disrupted for helicase to be active when duplex DNA is encountered. Therefore, in addition to possible therapeutic uses, the C5 inhibitor and well-designed derivatives will be valuable in future studies of how the helicase and nuclease activities are co-regulated and integrated.
Our study of C5 adds cogent support to more circumstantial evidence that has accumulated that a major, and not minor, function of DNA2 both in yeast and human is to participate in the protection, remodeling and restart of stalled replication forks. We first reported this when we showed, using 2D gels, that yeast dna2-1 mutants led to replication fork collapse or remodeling into intermediates thought to be chi-structured (recombination) intermediates or reversed forks and DSBs, when replication forks stalled at the endogenous FOB1 protein-mediated replication fork barrier in the rDNA (Weitao et al., 2003a, b). In Schizosaccharomyces pombe, furthermore, it was shown that DNA2 was acted on by the checkpoint to prevent fork collapse upon stalling at a similar barrier (Hu et al., 2012). More recently, human DNA2 has been shown in elegant DNA spreading experiments using knockdowns to be crucial for restart of forks stalled by CPT and specifically to promote limited resection necessary for restart (Thangavel et al., 2015). This activity must be tightly controlled, however, because the restoration of fork protection by inhibition of DNA2 in both BRCA2- and BOD1L depleted cells, at very low levels of the DNA2 inhibitor C5 (20 μM) and to the same extent as knockdown of DNA2 (Fig. 6), confirms previous reports that, if not properly regulated, over-resection by DNA2 can lead to excessive fork degradation and genome instability (Wang et al., 2015, Higgs et al., 2015). This coincides with the demonstration that depletion of DNA2 can suppress the cisplatin sensitivity of FANCD2−/− cells, which we proposed was due to over-resection at stalled forks (Karanja et al., 2014). By contrast to these replication functions, we found that higher levels of C5 were required to achieve the same level of inhibition as by knockdown of DNA2 (Fig. 5) when measuring SSA and HR in the GFP reporter assays, which are not thought to depend on on-going replication,
Until this study, there were no known small molecule inhibitors of DNA2. Discovering and testing additional DNA2 inhibitors will not only be invaluable for characterizing the integrated DNA2 enzymatic activities but will also enhance the preparation of advanced inhibitors for anticancer regimens, either alone or in combination with other chemotherapeutics. Importantly, chemical inhibition of DNA2 displayed cytotoxicity to DNA2-proficient mouse and human cells but not toward dna2−/− MES DNA2-deficient cells of human cells after DNA2 knockdown, supporting the assertion that we have found a DNA2-specific inhibitor. The use of inhibitors plus mutations to elucidate the mechanism of DNA2 nuclease/helicase activation as a basis for understanding its regulation in vivo is critical to design new therapeutic regimens. Inhibitors allow one to monitor the acute response of cells to the absence of DNA2, which is an important distinction from genetic studies, whose interpretation is always made difficult when studying essential genes such as DNA2. In the current study, we tested if targeting DNA2 may exploit a specific vulnerability in the cancer cells, assuming that normal cells are better protected by intact checkpoints and redundant repair processes.
We also tested the possible potentiation of PARP inhibitors by DNA2 inhibitor C5, because previous work suggested that DNA2 and PARP might participate in overlapping repair and replication stress response pathways (Bryant et al., 2009, Wanrooij and Burgers, 2015, Yang et al., 2004, Ying et al., 2012). PARP inhibitors have proved promising in treating BRCA-deficient tumors but fall short in that many such tumors are resistant. We tested if DNA2 might be partially responsible by performing a complementary function using the C5 DNA2 inhibitor. Indeed, C5 potentiated PARPi and vice versa. This “synthetic lethality” suggests novel approaches to applications of PARPi therapy. It also increases confidence that DNA2 and PARP function in response to replication stress that might occur in response to oncogene activation or treatment with DNA damaging agents such as CPT. Our demonstration of the stimulation of PARP inhibition by C5 may suggest that DNA2 plays a complementary role with PARP at stalled replication forks, where PARP has been shown to mediate replication fork restart in conjunction with MRE11, a nuclease that acts upstream of DNA2 in resection functions (Shibata et al., 2014, Bryant et al., 2009). In sum, the data presented here show that DNA2 inhibitors sensitize cancer cells to DNA damaging agents and additional agents used in current therapy and therefore may be feasible anti-cancer agents.
Funding Sources
The work was supported by an NIH grant RO1 CA085344, the Margaret E. Early Medical Research Trust grant ADF-1481M to B.H.S., Margaret Early Medical Research Trust grant and ARO W81XWH-09-1-0041 to J.L.C., a Caltech-City of Hope Biomedical Research Initiative to B.H.S. and J.L.C., and K22 grant 5K22CA175262–04 to K.S. Wenpeng Liu is supported by the China Scholarship Council (CSC) during his visit to California Institute of Technology and City of Hope, and K.S. is a CPRIT scholar in cancer biology.
Conflict of Interest Statement
We declare no financial or other relationships that may lead to a conflict of interest in this study.
Author Contributions
L.Z., J.L.C. and B.S. designed the study and supervised the entire project. L.Z., J.L.C., B.S. and K.S. wrote the manuscript. H.L. established the 3-D model and virtually screened the chemical inhibitors. W.L., P.P., J.L.C., Q.W., C.L, and K.K.J. purified the proteins and performed all of the biochemical experiments in the current study. K.S. and V.P. designed and executed the fork protection experiments. Z.L. performed the DNA fiber experiments. Correspondence concerning fork protection experiments may be addressed to K.S. and V.P. M.Z., Z.L., H.D., K.K.J., L.C., performed the cellular functional (SSA, HR, P-RPA resection, replication fork restart) and toxicological assays. S.S. supervised the enzyme kinetics experiments.
Acknowledgments
We thank the City of Hope Microscopy and Bioinformatics Core Facilities, National Cancer Institute (NCI) designated cancer center support grant P30 CA033572, and Nancy Linford, Ph.D. for her critical reading and editing of the manuscript and S. Howard and J. Stark for their technical support.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2016.02.043.
Contributor Information
Li Zheng, Email: lzheng@coh.org.
Judith L. Campbell, Email: jcampbel@caltech.edu.
Binghui Shen, Email: bshen@coh.org.
Appendix A. Supplementary data
Supplementary experimental procedures and figures.
References
- Bae S.H., Bae K.-H., Kim J.A., Seo Y.S. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature. 2001;412:456–461. doi: 10.1038/35086609. [DOI] [PubMed] [Google Scholar]
- Bae S.-H., Kim D.W., Kim J., Kim J.-H., Kim D.-H., Kim H.-D., Kang H.-Y., Seo Y.-S. Coupling of DNA helicase and endonuclease activities of yeast Dna2 facilitates Okazaki fragment processing. J. Biol. Chem. 2002;277:26632–26641. doi: 10.1074/jbc.M111026200. [DOI] [PubMed] [Google Scholar]
- Balakrishnan L., Polaczek P., Pokharel S., Campbell J.L., Bambara R.A. Dna2 exhibits a unique strand end-dependent helicase function. J. Biol. Chem. 2010;285:38861–38868. doi: 10.1074/jbc.M110.165191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg A.C., Stewart F.A., Vens C. Strategies to improve radiotherapy with targeted drugs. Nat. Rev. Cancer. 2011;11:239–253. doi: 10.1038/nrc3007. [DOI] [PubMed] [Google Scholar]
- Bryant H.E., Petermann E., Schultz N., Jemth A.S., Loseva O., Issaeva N., Johansson F., Fernandez S., Mcglynn P., Helleday T. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009;28:2601–2615. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd M.E., Campbell J.L. A new yeast gene required for DNA replication encodes a protein with homology to DNA helicases. Proc. Natl. Acad. Sci. U. S. A. 1995;92:7642–7646. doi: 10.1073/pnas.92.17.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd M.E., Campbell J.L. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol. Cell. Biol. 1997;17:2136–2142. doi: 10.1128/mcb.17.4.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd M.E., Campbell J.L. The pattern of sensitivity of yeast dna2 mutants to DNA damaging agents suggests a role in DSB and postreplication repair pathways. Mutat. Res. 2000;459:173–186. doi: 10.1016/s0921-8777(99)00072-5. [DOI] [PubMed] [Google Scholar]
- Budd M.E., Campbell J.L. Interplay of Mre11 nuclease with Dna2 plus Sgs1 in Rad51-dependent recombinational repair. PLoS One. 2009;4:e4267. doi: 10.1371/journal.pone.0004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd M.E., Tong A.H., Polaczek P., Peng X., Boone C., Campbell J.L. A network of multi-tasking proteins at the DNA replication fork preserves genome stability. PLoS Genet. 2005;1:634–650. doi: 10.1371/journal.pgen.0010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P., Cannavo E., Polaczek P., Masuda-Sasa T., Pokharel S., Campbell J.L., Kowalczykowski S.C. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010;467:112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S., Jayachandran U., Bonneau F., Fiorini F., Basquin C., Domcke S., Le Hir H., Conti E. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol. Cell. 2011;41:693–703. doi: 10.1016/j.molcel.2011.02.010. [DOI] [PubMed] [Google Scholar]
- Chen M.C., Zhou B., Zhang K., Yuan Y.C., Un F., Hu S., Chou C.M., Chen C.H., Wu J., Wang Y., Liu X., Smith D.L., Li H., Liu Z., Warden C.D., Su L., Malkas L.H., Chung Y.M., Hu M.C., Yen Y. The novel ribonucleotide reductase inhibitor COH29 inhibits DNA repair in vitro. Mol. Pharmacol. 2015;87:996–1005. doi: 10.1124/mol.114.094987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Muhlrad D., Lim M.K., Parker R., Song H. Structural and functional insights into the human Upf1 helicase core. EMBO J. 2007;26:253–264. doi: 10.1038/sj.emboj.7601464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T.C. Drug combination studies and their synergy quantification using the Chou–Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- Duxin J.P., Dao B., Martinsson P., Rajala N., Guittat L., Campbell J.L., Spelbrink J.N., Stewart S.A. Human Dna2 is a nuclear and mitochondrial DNA maintenance protein. Mol. Cell. Biol. 2009;29:4274–4282. doi: 10.1128/MCB.01834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxin J.P., Moore H.R., Sidorova J., Karanja K., Honaker Y., Dao B., Piwnica-Worms H., Campbell J.L., Monnat R.J., Stewart S.A. Okazaki fragment processing-independent role for human Dna2 during DNA replication. J. Biol. Chem. 2012:21980–21991. doi: 10.1074/jbc.M112.359018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fathers C., Drayton R.M., Solovieva S., Bryant H.E. Inhibition of poly(ADP-ribose) glycohydrolase (PARG) specifically kills BRCA2-deficient tumor cells. Cell Cycle. 2012;11:990–997. doi: 10.4161/cc.11.5.19482. [DOI] [PubMed] [Google Scholar]
- Fisher A.E., Hochegger H., Takeda S., Caldecott K.W. Poly(ADP-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol. Cell. Biol. 2007;27:5597–5605. doi: 10.1128/MCB.02248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong Y., Chou S.J., Hung K.F., Wu H.T., Kao S.Y. An investigation of the differential expression of Her2/neu gene expression in normal oral mucosa, epithelial dysplasia, and oral squamous cell carcinoma in Taiwan. J. Chin. Med. Assoc. 2008;71:123–127. doi: 10.1016/S1726-4901(08)70003-0. [DOI] [PubMed] [Google Scholar]
- Gunn A., Stark J.M. I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods Mol. Biol. 2012;920:379–391. doi: 10.1007/978-1-61779-998-3_27. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y., Chaudhuri A.R., Lopes M., Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat. Struct. Mol. Biol. 2010;17:1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman L.M., Fried M.G. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2007;2:1849–1861. doi: 10.1038/nprot.2007.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs M.R., Reynolds J.J., Winczura A., Blackford A.N., Borel V., Miller E.S., Zlatanou A., Nieminuszczy J., Ryan E.L., Davies N.J., Stankovic T., Boulton S.J., Niedzwiedz W., Stewart G.S. BOD1L Is required to suppress deleterious resection of stressed replication forks. Mol. Cell. 2015;59:462–477. doi: 10.1016/j.molcel.2015.06.007. [DOI] [PubMed] [Google Scholar]
- Hoa N.N., Kobayashi J., Omura M., Hirakawa M., Yang S.H., Komatsu K., Paull T.T., Takeda S., Sasanuma H. BRCA1 and CtIP are both required to recruit Dna2 at double-strand breaks in homologous recombination. PLoS One. 2015;10:e0124495. doi: 10.1371/journal.pone.0124495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard S.M., Yanez D.A., Stark J.M. DNA damage response factors from diverse pathways, including DNA crosslink repair, mediate alternative end joining. PLoS Genet. 2015;11:e1004943. doi: 10.1371/journal.pgen.1004943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiang Y.H., Lihou M.G., Liu L.F. Arrest of replication forks by drug-stabilized topoisomerase I-DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res. 1989;49:5077–5082. [PubMed] [Google Scholar]
- Hu Y., Petit S.A., Ficarro S.B., Toomire K.J., Xie A., Lim E., Cao S.A., Park E., Eck M.J., Scully R., Brown M., Marto J.A., Livingston D.M. PARP1-driven poly-ADP-ribosylation regulates BRCA1 function in homologous recombination-mediated DNA repair. Cancer Discov. 2014;4:1430–1447. doi: 10.1158/2159-8290.CD-13-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Sun L., Shen F., Chen Y., Hua Y., Liu Y., Zhang M., Hu Y., Wang Q., Xu W., Sun F., Ji J., Murray J.M., Carr A.M., Kong D. The intra-s phase checkpoint targets dna2 to prevent stalled replication forks from reversing. Cell. 2012;149:1221–1232. doi: 10.1016/j.cell.2012.04.030. [DOI] [PubMed] [Google Scholar]
- Imamura O., Campbell J.L. The human Bloom syndrome gene suppresses the DNA replication and repair defects of yeast dna2 mutants. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8193–8198. doi: 10.1073/pnas.1431624100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P., Altamura S., Boueres J., Ferrigno F., Fonsi M., Giomini C., Lamartina S., Monteagudo E., Ontoria J.M., Orsale M.V., Palumbi M.C., Pesci S., Roscilli G., Scarpelli R., Schultz-Fademrecht C., Toniatti C., Rowley M. Discovery of 2-{4-[(3S)-piperidin-3-yl]phenyl}-2H-indazole-7-carboxamide (MK-4827): a novel oral poly(ADP-ribose)polymerase (PARP) inhibitor efficacious in BRCA-1 and -2 mutant tumors. J. Med. Chem. 2009;52:7170–7185. doi: 10.1021/jm901188v. [DOI] [PubMed] [Google Scholar]
- Kang J.-Y., Choi E., Bae S.-H., Lee K.-H., Gim B.-S., Kim H.-D., Park C., Macneill S.A., Seo Y.-S. Genetic analyses of Schizosaccharomyces pombe dna2+ reveal that Dna2 plays an essential role in Okazaki fragment metabolism. Genetics. 2000;155:1055–1067. doi: 10.1093/genetics/155.3.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.H., Lee C.H., Seo Y.S. Dna2 on the road to Okazaki fragment processing and genome stability in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 2010;45:71–96. doi: 10.3109/10409230903578593. [DOI] [PubMed] [Google Scholar]
- Kao H.-I., Campbell J.L., Bambara R.A. Dna2p helicase/nuclease is a tracking protein, like FEN1, for flap cleavage during Okazaki fragment maturation. J. Biol. Chem. 2004;279:50840–50849. doi: 10.1074/jbc.M409231200. [DOI] [PubMed] [Google Scholar]
- Kao H.I., Veeraraghavan J., Polaczek P., Campbell J.L., Bambara R.A. On the roles of Saccharomyces cerevisiae Dna2p and FEN1 in Okazaki fragment processing. J. Biol. Chem. 2004;279:15014–15024. doi: 10.1074/jbc.M313216200. [DOI] [PubMed] [Google Scholar]
- Karanja K.K., C S.W., Duxin J.P., Stewart S.A., Campbell J.L. DNA2 and EXO1 in replication-coupled homology directed repair and in the interplay between HDR and the FA/BRCA network. Cell Cycle. 2012;11:3983–3996. doi: 10.4161/cc.22215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanja K.K., Lee E.H., Hendrickson E.A., Campbell J.L. Preventing over-resection by DNA2 helicase/nuclease suppresses repair defects in Fanconi anemia cells. Cell Cycle. 2014;13:1540–1550. doi: 10.4161/cc.28476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.-H., Lee K.-H., Kim J.-H., Ryu G.-H., Bae S.-H., Lee B.-C., Moon K.-Y., Byun S.-M., Koo H.-S., Seo Y.-S. Enzymatic properties of the Caenorhabditis elegans Dna2 endonuclease/helicase and a species-specific interaction between RPA and Dna2. Nucleic Acids Res. 2005;33:1372–1383. doi: 10.1093/nar/gki255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Burgers P.M. Lagging strand maturation factor Dna2 is a component of the replication checkpoint initiation machinery. Genes Dev. 2013;27:313–321. doi: 10.1101/gad.204750.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.S., Foiani M. Dna2 offers support for stalled forks. Cell. 2012;149:1181–1183. doi: 10.1016/j.cell.2012.05.021. [DOI] [PubMed] [Google Scholar]
- Le Tourneau C., Stathis A., Vidal L., Moore M.J., Siu L.L. Choice of starting dose for molecularly targeted agents evaluated in first-in-human phase I cancer clinical trials. J. Clin. Oncol. 2010;28:1401–1407. doi: 10.1200/JCO.2009.25.9606. [DOI] [PubMed] [Google Scholar]
- Levikova M., Klaue D., Seidel R., Cejka P. Nuclease activity of Saccharomyces cerevisiae Dna2 inhibits its potent DNA helicase activity. Proc. Natl. Acad. Sci. U. S. A. 2013;E1992-2001 doi: 10.1073/pnas.1300390110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Yu X. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell. 2013;23:693–704. doi: 10.1016/j.ccr.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S., Toczylowski T., Yan H. Identification of the Xenopus DNA2 protein as a major nuclease for the 5′–> 3′ strand-specific processing of DNA ends. Nucleic Acids Res. 2008;36:6091–6100. doi: 10.1093/nar/gkn616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S., Toczylowski T., Yan H. Mechanistic analysis of Xenopus EXO1's function in 5′-strand resection at DNA double-strand breaks. Nucleic Acids Res. 2011;39:5967–5977. doi: 10.1093/nar/gkr216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Sampathi S., Dai H., Liu C., Zhou M., Hu J., Huang Q., Campbell J., Shin-Ya K., Zheng L., Chai W., Shen B. Mammalian DNA2 helicase/nuclease cleaves G-quadruplex DNA and is required for telomere integrity. EMBO J. 2013;32:1425–1439. doi: 10.1038/emboj.2013.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda-Sasa T., Imamura O., Campbell J.L. Biochemical analysis of human Dna2. Nucleic Acids Res. 2006;34:1865–1875. doi: 10.1093/nar/gkl070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda-Sasa T., Polaczek P., Peng X.P., Chen L., Campbell J.L. Processing of G4 DNA by Dna2 helicase/nuclease and RPA provides insights into the mechanism of Dna2/RPA substrate recognition. J. Biol. Chem. 2008;283:24359–24373. doi: 10.1074/jbc.M802244200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar A.V., Genschel J., Kinoshita E., Polaczek P., Campbell J.L., Wyman C., Modrich P., Kowalczykowski S.C. BLM-DNA2-RPA-MRN- and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H., Raynard S., Sung P. Multiplicity of DNA end resection machineries in chromosome break repair. Genes Dev. 2009;23:1481–1486. doi: 10.1101/gad.1824209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano S., Lan L., Caldecott K.W., Mori T., Yasui A. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell. Biol. 2003;23:3974–3981. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A.G., Flatten K.S., Schneider P.A., Dai N.T., Mcdonald J.S., Poirier G.G., Kaufmann S.H. Enhanced killing of cancer cells by poly(ADP-ribose) polymerase inhibitors and topoisomerase I inhibitors reflects poisoning of both enzymes. J. Biol. Chem. 2012;287:4198–4210. doi: 10.1074/jbc.M111.296475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G., Dai H., Zhang W., Hsieh H.J., Pan M.R., Park Y.Y., Tsai R.Y., Bedrosian I., Lee J.S., Ira G., Lin S.Y. Human nuclease/helicase DNA2 alleviates replication stress by promoting DNA End resection. Cancer Res. 2012;72:2802–2813. doi: 10.1158/0008-5472.CAN-11-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E., Orta M.L., Issaeva N., Schultz N., Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce A.J., Johnson R.D., Thompson L.H., Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri A., Hashimoto Y., Herrador R., Neelsen K.J., Fachinetti D., Bermejo R., Cocito A., Costanzo V., Lopes M. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat. Struct. Mol. Biol. 2012;19:417–423. doi: 10.1038/nsmb.2258. [DOI] [PubMed] [Google Scholar]
- Ronchi D., Di Fonzo A., Lin W., Bordoni A., Liu C., Fassone E., Pagliarani S., Rizzuti M., Zheng L., Filosto M., Ferro M.T., Ranieri M., Magri F., Peverelli L., Li H., Yuan Y.C., Corti S., Sciacco M., Moggio M., Bresolin N., Shen B., Comi G.P. Mutations in DNA2 link progressive myopathy to mitochondrial DNA instability. Am. J. Hum. Genet. 2013;92:293–300. doi: 10.1016/j.ajhg.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K., Christ N., Siaud N., Egashira A., Wu H., Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K., Wu H., Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen R., Faqiih E., Ansari S., Abdel-Salam G., Al-Hassnan Z.N., Al-Shidi T., Alomar R., Sogaty S., Alkuraya F.S. Genomic analysis of primorial dwarfism reveals novel disease genes. Genome Res. 2014;24:291–299. doi: 10.1101/gr.160572.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata A., Moiani D., Arvai A.S., Perry J., Harding S.M., Genols M.-M., Maity R., Rossum-Fikkert S.V., Kertokallo A., Romoli F., Ismail A., Ismalaj E., Petricci E., Neale M.J., Bristorw R.G., Masson J.-Y., Wyman C., Jeggo P.A., Tainer J.A. DNA Double-Strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol. Cell. 2014;53:7–18. doi: 10.1016/j.molcel.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart J.A., Campbell J.L., Bambara R.A. Dna2 is a structure-specific nuclease, with affinity for 5′-flap intermediates. Nucleic Acids Res. 2010;38:920–930. doi: 10.1093/nar/gkp1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart J.A., Miller A.S., Campbell J.L., Bambara R.A. Dynamic removal of replication protein A by Dna2 facilitates primer cleavage during Okazaki fragment processing in Saccharomyces cerevisiae. J. Biol. Chem. 2008;283:31356–31365. doi: 10.1074/jbc.M805965200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss C., Kornowski M., Benvenisty A., Shahar A., Masury H., Ben-Porath I., Ravid T., Arbel-Eden A., Goldberg M. The DNA2 nuclease/helicase is an estrogen-dependent gene mutated in breast and ovarian cancers. Oncotarget. 2014;5:9396–9409. doi: 10.18632/oncotarget.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturzenegger A., Burdova K., Kanagaraj R., Levikova M., Pinto C., Cejka P., Janscak P. DNA2 cooperates with the WRN and BLM RecQ helicases to mediate long-range DNA end resection in human cells. J. Biol. Chem. 2014;289:27314–27326. doi: 10.1074/jbc.M114.578823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L.S., Gautier J. Double-Strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- Teicher B.A. Next generation topoisomerase I inhibitors: rationale and biomarker strategies. Biochem. Pharmacol. 2008;75:1262–1271. doi: 10.1016/j.bcp.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Thangavel S., Berti M., Levikova M., Pinto C., Gomathinayagam S., Vujanovic M., Zellweger R., Moore H., Lee E.H., Hendrickson E.A., Cejka P., Stewart S., Lopes M., Vindigni A. DNA2 drives processing and restart of reversed replication forks in human cells. J. Cell Biol. 2015;208:545–562. doi: 10.1083/jcb.201406100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A.T., Kim T., Wagner J.E., Conti B.A., Lach F.P., Huang A.L., Molina H., Sanborn E.M., Zierhut H., Cornes B.K., Abhyankar A., Sougnez C., Gabriel S.B., Auerbach A.D., Kowalczykowski S.C., Smogorzewska A. A dominant mutation in human RAD51 reveals its function in DNA Interstrand crosslink repair independent of homologous recombination. Mol. Cell. 2015;59:478–490. doi: 10.1016/j.molcel.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanrooij P.H., Burgers P.M. Yet another job for Dna2: checkpoint activation. DNA Repair (Amst) 2015;32:17–23. doi: 10.1016/j.dnarep.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrousek K.E., Fortini B.K., Polaczek P., Chen L., Liu Q., Dunphy W.G., Campbell J.L. Xenopus DNA2 is a helicase/nuclease that is found in complexes with replication proteins and-1/Ctf4 and Mcm10 and DSB response proteins Nbs1 and ATM. Cell Cycle. 2010;9:1156–1166. doi: 10.4161/cc.9.6.11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitao T., Budd M., Campbell J.L. Evidence that yeast SGS1, DNA2, SRS2, and FOB1 interact to maintain rDNA stability. Mutat. Res. 2003;532:157–172. doi: 10.1016/j.mrfmmm.2003.08.015. [DOI] [PubMed] [Google Scholar]
- Weitao T., Budd M., Hoopes L.L., Campbell J.L. Dna2 helicase/nuclease causes replicative fork stalling and double-strand breaks in the ribosomal DNA of Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:22513–22522. doi: 10.1074/jbc.M301610200. [DOI] [PubMed] [Google Scholar]
- Yang Y.G., Cortes U., Patnaik S., Jasin M., Wang Z.Q. Ablation of PARP-1 does not interfere with the repair of DNA double-strand breaks, but compromises the reactivation of stalled replication forks. Oncogene. 2004;23:3872–3882. doi: 10.1038/sj.onc.1207491. [DOI] [PubMed] [Google Scholar]
- Ying S., Hamdy F.C., Helleday T. Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer Res. 2012;72:2814–2821. doi: 10.1158/0008-5472.CAN-11-3417. [DOI] [PubMed] [Google Scholar]
- Zheng L., Zhou M., Guo Z., Lu H., Qian L., Dai H., Qiu J., Yakubovskaya E., Bogenhagen D.F., Demple B., Shen B. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol. Cell. 2008;32:325–336. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Pourmal S., Pavletich N.P. Dna2 nuclease-helicase structure, mechanism and regulation by RPA. Elife. 2015;4 doi: 10.7554/eLife.09832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Su L., Hu S., Hu W., Yip M.L., Wu J., Gaur S., Smith D.L., Yuan Y.C., Synold T.W., Horne D., Yen Y. A small-molecule blocking ribonucleotide reductase holoenzyme formation inhibits cancer cell growth and overcomes drug resistance. Cancer Res. 2013;73:6484–6493. doi: 10.1158/0008-5472.CAN-13-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Chung W.H., Shim E.Y., Lee S.E., Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Elledge S.J. Sensing DNA damage through ATRIP recognition of RPA–ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary experimental procedures and figures.