Intellectual disability (ID) is defined as limitations in both mental capacity and adaptive behaviour, and affects 2–3% of the population worldwide. A subset of ID, thought to be 5–10% of ID in males, is caused by mutations on the X chromosome (X-linked intellectual disability; XLID). Improvements in sequencing technologies have enabled the association of numerous genetic variants with ID, but the underlying etiology remains poorly understood, and diagnosis and treatment remain limited. Mutations in KDM5C (also known as JARID1C and SMCX) were first found in XLID patients in 2005 (Jensen et al., 2005). Additional KDM5C mutations were subsequently identified and they are now thought to account for 1–4% of X-linked ID. This is a substantial proportion; in comparison, Fragile-X syndrome is the most common cause of XLID and accounts for less than 10%. KDM5C-linked ID patients (OMIM 300534) often display short stature (~ 60% patients) and aggressive behaviour (~ 30% patients) in addition to mild to severe ID.
KDM5C encodes a histone demethylase specific for histone 3 lysine 4 di- and tri-methylation (H3K4me2/3; Iwase et al., 2007). Histone proteins organize the genome, with DNA wrapping around histone octamers to form the basic unit of chromatin. Post-translational modification of histones influences chromosome compaction and provides binding sites for specific proteins, thereby regulating nuclear processes. Distinct histone modifications correlate with different transcriptional states; the KDM5C target H3K4me3 marks active promoters. Patient KDM5C mutations lower protein levels through nonsense-mediated decay or diminished protein stability, and missense mutations reduce demethylase activity (Brookes et al., 2015). This suggests a loss-of-function disease mechanism, but the cellular and molecular changes are not fully understood.
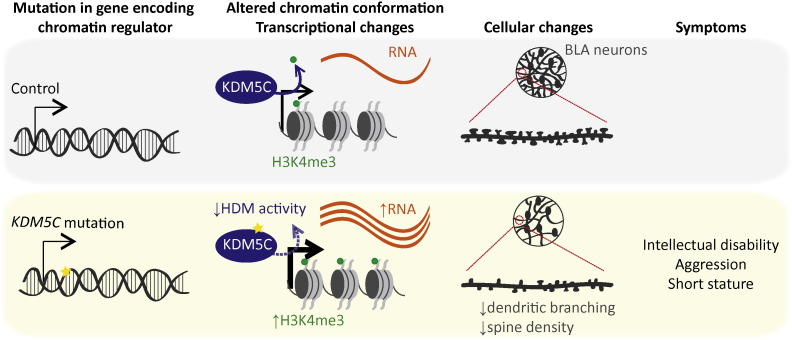
In a recent study, Yang Shi and colleagues illuminated KDM5C-linked ID through generating and characterising a Kdm5c-knockout mouse (Iwase et al., 2016). Importantly, behavioural tests confirm that the mouse is a good model for the disorder, with Kdm5c-knockout mice showing heightened aggression, reduced anxiety, social aversion, and defects in learning and memory. The gross architecture of the brain was unaffected in Kdm5c-knockout mice, but abnormal neuronal morphology was noted in specific regions related to aggression (the basolateral amygala; BLA) and learning (BLA and cortex). Dendritic spine density was affected in both of these regions, whilst dendritic arborisation was reduced in the BLA (Fig. 1). Defects in dendritic branching and spine morphology have been identified in human post-mortem samples of different neurological disorders (Kulkarni and Firestein, 2012), suggesting that these deficits may be a morphological correlate of KDM5C-linked ID.
Fig. 1.
Model for how mutations in chromatin regulator genes cause disease, exemplified by KDM5C mutations. Mutations in chromatin regulator genes affect protein level or activity, such as reduced histone demethylase (HDM) activity of mutant KDM5C. This results in alterations in chromatin conformation at target genes, such as increased H3K4me3 at KDM5C target promoters. Resultant changes in transcription induce cellular phenotypes, to which neurons seem particularly susceptible, leading to disease symptoms.
To understand how loss of a histone demethylase results in the cellular defects observed, the authors examined genome-wide changes in gene expression in Kdm5c-knockout amygdala and cortex (Iwase et al., 2016). Multiple pathways were misregulated, including pathways important for neurological development and function. Exploration of the contribution of each altered gene/pathway to the behavioural phenotypes should provide insight into potential therapeutic targets for patients. Interestingly, in addition to influencing transcription, KDM5C has been shown to regulate DNA replication (Rondinelli et al., 2015). Although neurons are post-mitotic, this could have important implications during development, when ID is thought to arise.
To explore the mechanisms of Kdm5c transcriptional regulation, Iwase et al. used primary neuronal cultures from wildtype and Kdm5c-KO littermates (Iwase et al., 2016). Kdm5c binding sites across the genome were enriched for CpG islands and high levels of H3K4me3. This finding is consistent with a recent study showing that Drosophila Lid, the Kdm5 family ortholog, recognises H3K4me2/3 through its PHD (plant homeodomain) finger, and co-localises with the mark genome-wide (Liu and Secombe, 2015). Intriguingly, only the Kdm5c-bound genes with the lowest expression levels show significant expression changes in Kdm5c-knockout neurons (Iwase et al., 2016). Chromatin modifiers are often postulated to ‘fine-tune’ expression, and this may explain why robustly transcribed genes show little alteration following its removal. Genes with altered expression levels in Kdm5c-knockout neurons predominantly show up-regulation and enhanced H3K4me3, in keeping with Kdm5c acting as a repressor through removal of an active chromatin modification (Fig. 1). A previous study of Kdm5c-knockout embryonic stem cells similarly found that Kdm5c acts at promoters to repress genes, but also suggested that Kdm5c is able to activate genes through the same demethylase activity at enhancers (Outchkourov et al., 2013). It remains to be seen whether enhancer regulation contributes to KDM5C patient symptoms; down-regulated genes were identified in Kdm5c-knockout brain and cultured neurons (Iwase et al., 2016) but the mechanisms were not examined.
One intriguing facet of KDM5C-linked XLID is that whilst the mutations are germline and therefore present in all cells, the most prominent symptoms are neurological. Changes in gene expression (Jensen et al., 2010) have also been were identified in blood samples from patients with KDM5C mutations (Jensen et al., 2010). This suggests that changes in chromatin modification may occur in many tissues without eliciting a phenotype, whereas the highly complex and more recently evolved central nervous system is susceptible. This observation is by no means unique to KDM5C mutations; germline mutations in many chromatin modifiers give rise to disorders with a neurological component. Of note, chromatin changes in more easily accessible tissues may be useful as molecular markers for diagnosis and treatment. This is exemplified in a recent study of Sotos Syndrome, a syndromic ID disorder caused by mutations in the histone methyltransferase NSD1, which determined a signature of DNA methylation in peripheral blood that could be used to diagnose sufferers (Choufani et al., 2015).
Mutations in genes encoding chromatin regulators are increasingly being recognized as an important contributor to neurological disorders including ID, autism spectrum disorders and schizophrenia. Furthermore, changes to chromatin can also be instigated by the environment and may contribute to disease susceptibility and prognosis. Thus, chromatin modifications should be considered as promising biomarkers and potential therapeutic targets. Good mouse models, such as the Kdm5c-knockout mice described here (Iwase et al., 2016), provide valuable tools to advance clinical practice.
References
- Brookes E., Laurent B., Ounap K., Carroll R., Moeschler J.B., Field M., Schwartz C.E., Gecz J., Shi Y. Mutations in the intellectual disability gene KDM5C reduce protein stability and demethylase activity. Hum. Mol. Genet. 2015;24:2861–2872. doi: 10.1093/hmg/ddv046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choufani S., Cytrynbaum C., Chung B.H., Turinsky A.L., Grafodatskaya D., Chen Y.A., Cohen A.S., Dupuis L., Butcher D.T., Siu M.T., et al. NSD1 mutations generate a genome-wide DNA methylation signature. Nat. Commun. 2015;6:10207. doi: 10.1038/ncomms10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S., Lan F., Bayliss P., de la Torre-Ubieta L., Huarte M., Qi H.H., Whetstine J.R., Bonni A., Roberts T.M., Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Iwase S., Brookes E., Agarwal S., Badeaux A.I., Ito H., Vallianatos C.N., Tomassy G.S., Kasza T., Lin G., Thompson A., et al. A mouse model of X-linked intellectual disability associated with impaired removal of histone methylation. Cell Rep. 2016;14:1000–1009. doi: 10.1016/j.celrep.2015.12.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen L.R., Amende M., Gurok U., Moser B., Gimmel V., Tzschach A., Janecke A.R., Tariverdian G., Chelly J., Fryns J.P., et al. Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am. J. Hum. Genet. 2005;76:227–236. doi: 10.1086/427563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen L.R., Bartenschlager H., Rujirabanjerd S., Tzschach A., Numann A., Janecke A.R., Sporle R., Stricker S., Raynaud M., Nelson J., et al. A distinctive gene expression fingerprint in mentally retarded male patients reflects disease-causing defects in the histone demethylase KDM5C. PathoGenetics. 2010;3:2. doi: 10.1186/1755-8417-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni V.A., Firestein B.L. The dendritic tree and brain disorders. Mol. Cell. Neurosci. 2012;50:10–20. doi: 10.1016/j.mcn.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Liu X., Secombe J. The histone demethylase KDM5 activates gene expression by recognizing chromatin context through its PHD reader motif. Cell Rep. 2015;13:2219–2231. doi: 10.1016/j.celrep.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outchkourov N.S., Muino J.M., Kaufmann K., van Ijcken W.F., Groot Koerkamp M.J., van Leenen D., de Graaf P., Holstege F.C., Grosveld F.G., Timmers H.T. Balancing of histone H3K4 methylation states by the Kdm5c/SMCX histone demethylase modulates promoter and enhancer function. Cell Rep. 2013;3:1071–1079. doi: 10.1016/j.celrep.2013.02.030. [DOI] [PubMed] [Google Scholar]
- Rondinelli B., Schwerer H., Antonini E., Gaviraghi M., Lupi A., Frenquelli M., Cittaro D., Segalla S., Lemaitre J.M., Tonon G. H3K4me3 demethylation by the histone demethylase KDM5C/JARID1C promotes DNA replication origin firing. Nucleic Acids Res. 2015;43:2560–2574. doi: 10.1093/nar/gkv090. [DOI] [PMC free article] [PubMed] [Google Scholar]