Graphical abstract
Keywords: Bacillus subtilis, Characterization, Phytase, pH stability, Thermostability, Catalytic activity
Abstract
In this study, an extracellular alkali-thermostable phytase producing bacteria, Bacillus subtilis B.S.46, were isolated and molecularly identified using 16S rRNA sequencing. Response surface methodology was applied to study the interaction effects of assay conditions to obtain optimum value for maximizing phytase activity. The optimization resulted in 137% (4.627 U/mL) increase in phytase activity under optimum condition (56.5 °C, pH 7.30 and 2.05 mM sodium phytate). The enzyme also showed 60–73% of maximum activity at wide ranges of temperature (47–68 °C), pH (6.3–8.0) and phytate concentration (1.40–2.50 mM). The partially purified phytase demonstrated high stability over a wide range of pH (6.0–10.0) after 24 h, retaining 85% of its initial activity at pH 6 and even interestingly, the phytase activity enhanced at pH 8.0–10.0. It also exhibited thermostability, retaining about 60% of its original activity after 2 h at 60 °C. Cations such as Ca2+ and Li+ enhanced the phytase activity by 10–46% at 1 mM concentration. The phytase activity was completely inhibited by Cu2+, Mg2+, Fe2+, Zn2+, Hg2+ and Mn2+ and the inhibition was in a dose dependent manner. B. subtilis B.S.46 phytase had interesting characteristics to be considered as animal feed additive, dephytinization of food ingredients, and bioremediation of phosphorous pollution in the environment.
Introduction
Phytic acid (myo-inositol 1,2,3,4,5,6-hexakisphosphate) or its salt, phytate is the major storage form of phosphorus in plants and represents 1–1.5% of weight and 60–80% of total phosphorus in cereals, legumes, and oil seeds [1]. Phytate is considered an anti-nutritional factor because of its high negatively charged structure and strong ability to chelate and bind minerals such as calcium, magnesium, zinc and iron [2]. It is also known to form complexes with proteins under both acidic and alkaline pH conditions affecting the proteins’ structure, thus decreasing the enzymatic activity, protein solubility and digestibility [3]. Phytate phosphorus is poorly utilized by non-ruminant animals such as pigs, poultry, human, and fish because of insufficient or lack of natural phytase activity in their gastrointestinal tract [4]. Animal feedstuffs are mainly of plant origin and therefore have a lot of phytate, but phytate phosphorous is not available for them and consequently, its excretion causes several environmental problems such as water pollution and eutrophication especially in areas of intensive livestock production [5], [6].
Phytases (myo-inositol 1,2,3,4,5,6-hexakisphosphate phosphohydrolases: EC 3.1.3.8 and EC 3.1.3.26) are a group of enzymes, which catalyze the stepwise removal of phosphates from phytic acid to less phosphorylated myo-inositol intermediates and inorganic phosphate. The presence of phytases has been reported in plants, animal tissues, and microorganisms [7]. Numerous researchers have shown that microbial phytases are more promising for the commercial production of phytase [7], [8], [9]. Although several strains of bacteria [10], yeasts [11], and fungi [9] have been isolated and studied for phytase production, currently commercial scale feed phytases are mainly derived from Aspergillus niger (3-phytase), Peniophora lycii and Escherichia coli (6-phytase) [7], [12]. However, according to strict substrate specificity, higher heat stability, wide pH profile, and resistant to proteolysis, Bacillus phytases are potential alternatives to fungal ones [8], [13], [14]. Several Bacillus phytases isolated from different sources have been characterized [15], [16], [17]. There is no single phytase as an ideal phytase and therefore, there has been a continuous effort to isolate new bacterial strains producing novel and efficient phytases. Phytases are also of great interest for other applications including processing and reduction of phytate in food industry, production of individual myo-inositol phosphate derivatives for human health and medicine, environmental protection, soil nutrient enhancement and aquaculture [18], [19], [20].
To our knowledge, no study has been published on the application of response surface methodology (RSM) for optimizing the catalytic activity of phytase. In the present study, phytase activity of Bacillus subtilis B.S.46, isolated from the phyllosphere of rice plant, was optimized by RSM. Furthermore, characterization of partially purified phytase was also investigated.
Material and methods
Chemicals
All of the chemicals and reagents used in this study were purchased from Merck (Darmstadt, Germany) and Sigma Chemical Co. (St. Louis, MO, USA).
Bacterial strain, inoculum preparation and phytase production
Submerged fermentation was used to evaluate the phytase activity of 70 microbial isolates obtained from the rhizosphere and phyllosphere of different fields and orchards in Iran (Agricultural Biotechnology Research Institute of Iran, Karaj, Iran). The isolates were first cultured on agar plates (g/L: nutrient broth (NB) 8, yeast extract 1, K2HPO4 1, KH2PO4 0.25, glucose 0.4, MgSO4 0.12, and agar 18) and incubated at 30 °C for 24 h. Inoculum was prepared by transferring a loop of fresh culture from the agar plate into a 50-mL tube containing 10 mL of sterile NB and incubated in a shaker incubator at 170 rpm and 30 °C for 18 h [21]. Next, each of the isolates was inoculated at the concentration of 2% into a 100-mL Erlenmeyer flask containing 25 mL of phytase production medium (g/L: sodium phytate 10, dextrin 12, yeast extract 4, meat extract 3, MgSO4 0.3). Pre-sterilized CaCl2 solution was added at a final concentration of 0.01% before inoculation. The initial pH of the culture was adjusted to 7.5 before autoclaving at 121 °C for 15 min. After inoculation, the flasks were incubated at 30 °C for 48 h and 170 rpm using a shaker incubator.
Enzyme extraction and phytase assay
At the end of fermentation, the cultures were harvested by centrifugation at 10,000×g (Suprema 25, TOMY, Japan) for 20 min at 4 °C, and the clear cell-free supernatants were used for phytase assay. Phytase activity was determined by measuring the amount of phosphate released from sodium phytate during enzymatic reaction using the ammonium molybdate method [22]. Briefly, a reaction mixture of 400 μL of 1.5 mM sodium phytate in 100 mM Tris–HCl buffer (pH 7.0) and 100 μL of crude enzyme was incubated at 55 °C for 30 min. The reaction was stopped by adding 400 μL of color reagent solution (1.5:1.5:1 ratio of 0.24% ammonium vanadate, 10% ammonium molybdate, 65% nitric acid) and the samples were centrifuged at 15,000×g (Biofuge pico, Kendro, Germany) for 10 min at room temperature. The yellow color developed due to phytase activity was determined spectrophotometrically at 415 nm (Microplate reader, infinite M200 Pro, Tecan, Switzerland) using the standard curve prepared from KH2PO4. One unit of phytase activity is defined as the amount of enzyme liberating 1 μmol of inorganic phosphorus per minute under assay conditions.
Molecular identification using 16S rRNA
The selected strain B.S.46 was cultured in 10 mL Luria Broth medium at 28 °C for 18 h. About 1.5 mL of culture (at the final optical density of 1 at 600 nm) was concentrated by centrifugation at 13,000×g for 10 min. Total DNA was extracted from the microbial pellet using Dneasy Blood and Tissue Kit (QIAGEN. Cat. No. 69504). The optical density of the extracted DNA was measured by nanodrop (OD 260 = 43 ng/μL) and then, it was stored at −20 °C.
Amplification of 16S rDNA gene was performed using bacterial universal primers PAF (3′-AGAGTTTGATCCTGGCTCAG-5′) and PAR (3′-AAGGAGGTGATCCAGCCGCA-5′) [23]. The temperature profile for PCR consisted of a first denaturation step of 5 min at 94 °C, followed by 35 cycles of 40 S/94 °C for denaturation, 40 S/58 °C for annealing, 40 S/72 °C for extension and a final extension step of 10 min at 72 °C. The PCR product was purified using High pure PCR Product purification Kit (Roche. Cat. No. 11.732.668.001) and used for DNA sequencing. DNA sequencing was performed using ABI 3730XL DNA Analyzers by BioNeer Company (Bioneer Co, South Korea). The 16S rRNA sequence was aligned with the reference sequences in the GenBank database using the BLAST search facility at the National Center for Biotechnology Information (NCBI). The 16S rRNA gene sequence alignment was done using the CLUSTAL W and Phylogenetic tree was created using neighbor-joining method [24] applying the Kimura-2-parameter model [25] as implemented in MEGA4 [26] with 1000 replicates [27].
Optimization and modeling of B. subtilis B.S.46 phytase activity by RSM
A central composite design (CCD) consisting of 20 experimental runs with 6 replications at center point to determine the effects of the three independent variables in 5 levels was used to optimize the crude enzyme activity (Table 1). The independent variables were temperature (X1, °C), pH (X2), and phytate concentration (X3, mM) and the response was crude phytase activity (Y, U/mL). The experimental design and results of CCD are listed in Table 2. The experimental data were fitted in accordance with Eq. (1) as a second-order polynomial equation including linear and interaction effects of each variable:
| (1) |
where Y is the predicted response, Xi and Xj are independent variables, β0 is the offset term, βi is the ith linear coefficient, βii is the ith quadratic coefficient, and βij is the ijth interaction coefficient. The statistical software package, Design Expert Version 7.1 (Stat-Ease Inc., Minneapolis, MN, USA), was used for the experimental design, the analysis of variance (ANOVA), estimated coefficients and standard errors, and generation of surface plots. The goodness of fit of the regression model obtained was given by coefficient of determination (R2). Validation of the experimental model including the optimum value of three independent variables for maximum response was done using the numerical optimization package of the software.
Table 1.
Independent variables and their levels used for CCD.
| Independent variables | Unit | Symbol | Coded levels |
||||
|---|---|---|---|---|---|---|---|
| −α | −1 | 0 | +1 | +α | |||
| Temperature | °C | X1 | 40.00 | 47.00 | 57.50 | 68.00 | 75.00 |
| pH | – | X2 | 5.50 | 6.30 | 7.50 | 8.70 | 9.50 |
| Phytate concentration | mM | X3 | 0.50 | 1.00 | 1.75 | 2.50 | 3.00 |
Table 2.
The CCD plan and actual phytase activity results by B. subtilis B.S.46.
| Run no. | Independent variables |
Phytase activity (U/mL) | ||
|---|---|---|---|---|
| X1 | X2 | X3 | ||
| 1 | 0 | 0 | 0 | 4.368 |
| 2 | +1 | +1 | −1 | 0.409 |
| 3 | −1 | +1 | +1 | 0.606 |
| 4 | 0 | +α | 0 | 0.351 |
| 5 | +α | 0 | 0 | 0.820 |
| 6 | 0 | 0 | 0 | 4.223 |
| 7 | +1 | −1 | +1 | 0.962 |
| 8 | +1 | +1 | +1 | 0.658 |
| 9 | 0 | −α | 0 | 1.955 |
| 10 | −1 | +1 | −1 | 1.106 |
| 11 | 0 | 0 | 0 | 4.332 |
| 12 | +1 | −1 | −1 | 2.627 |
| 13 | 0 | 0 | −α | 1.654 |
| 14 | 0 | 0 | +α | 3.479 |
| 15 | 0 | 0 | 0 | 4.207 |
| 16 | 0 | 0 | 0 | 4.370 |
| 17 | −1 | −1 | +1 | 2.632 |
| 18 | 0 | 0 | 0 | 4.591 |
| 19 | −1 | −1 | −1 | 2.560 |
| 20 | -α | 0 | 0 | 1.603 |
Partial purification and characterization of phytase
Partial purification of the enzyme was done by solid ammonium sulfate precipitation and dialysis in the cold room (4 °C). First, 3 mL of 1 M Tris–HCl buffer (pH 7.2) containing 1 mM CaCl2 was added to 25 mL of crude enzyme and 10.11 g of solid ammonium sulfate was used to reach the final saturation of 60%. This amount was slowly added during 1 h and the resulting solution was allowed to mix for 1 h with constant stirring. Then, the content was centrifuged at 16,000×g and 4 °C for 30 min and the supernatant was collected for the second step. Subsequently, 5.54 g of solid ammonium sulfate was added to obtain the final saturation of 85% following the exact procedure as for the first step. After centrifugation, the pellet was suspended in 1 mL of 20 mM Tris–HCl buffer (pH 7.5) and dialyzed (Dialysis cellulose tubing, MWCO12.4 kDa, Sigma–Aldrich) against 20 mM Tris–HCl buffer (pH 7.5) for 24 h (the buffer was replaced every 8 h). Finally, the active fraction was distributed in the vials and stored at −20 °C for characterization experiments. Protein content was determined according to Bradford’s approach using BSA as the standard [28].
The pH stability was determined by pre-incubation of the partially purified enzyme with various pH buffers ranging from 3.0 to 10.0 at 4 °C for different time periods (30 min to 24 h). The temperature stability was determined with pre-incubation of the partially purified phytase at different temperatures of 40, 50, and 60 °C from 10 min to 7 days. The effects of different metal ions at 1, 2, 5, and 10 mM concentrations on phytase activity using sodium phytate as the substrate were studied. The residual and relative activities for both experiments were then measured at the following conditions (56.5 °C, pH 7.3, 2.05 mM sodium phytate).
Results and discussion
Selection and molecular identification of the best isolate
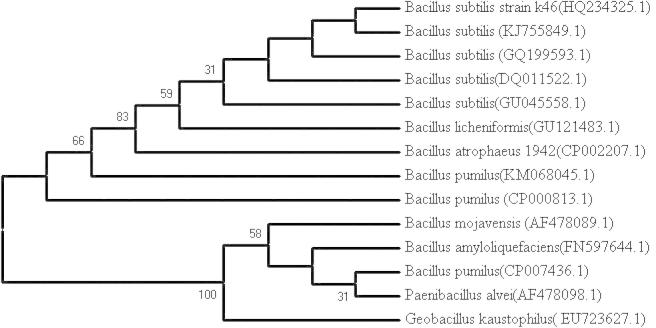
Several microbial isolates, which were isolated from different fields and orchards in Iran, were screened for phytase production on broth medium supplemented with phytic acid. The results showed that the isolate B.S.46 was the most efficient phytase producing isolate (1.952 U/mL) (data not shown). The amount of phytase production obtained by the isolate B.S.46 was higher than the amounts of 0.64, 0.40, and 0.2 U/mL reported for B. subtilis US417, B. subtilis VTTE 68013, and Bacillus sp. KHU-10, respectively [16], [29], [30], but lower than the values of about 3 U/mL stated for B. laevolacticus and B. amyloliquefaciens US573 [17], [31]. Therefore, the isolate B.S.46 was molecularly identified using 16S rRNA. The 16S rRNA gene was amplified by PCR using 16S rDNA. Alignment of the partial 16S rDNA sequences with those available in NCBI GenBank exhibited that the isolate B.S.46 matched the closest with B. subtilis PY79 (GenBank: CP006881.1), B. subtilis strain ET (GenBank: HQ266669.1), B. subtilis subsp. subtilis strain BAB-1 (GenBank: CP004405.1), B. subtilis strain shu-3 (GenBank: HM470251.1), and B. subtilis strain Em7 (GenBank: GU258545.1) with 95% similarity (accessed on 08-NOV-2010). The sequencing result was deposited in the GenBank with accession No. HQ234325.1. The phylogenetic analysis on the basis of 16S rDNA gene revealed that the isolate B.S.46 was closely related to other B. subtilis retrieved from NCBI GenBank (Fig. 1).
Fig. 1.
Phylogenetic relationships of B. subtilis strain B.S.46 and the 13 reference sequences retrieved from NCBI GenBank. Phylogenetic tree was constructed using the neighbor joining method (MEGA 4.0). The confidence of branching was assessed by computing 1000 bootstrap. The reference sequences are marked with GenBank accession numbers in parenthesis.
Optimization and modeling of B. subtilis B.S.46 phytase activity by RSM
Preliminary tests showed that phytase activity significantly increased at 65 °C, while considerably decreased at 40 °C. Also, neutral to alkaline pH values had a similar positive effect, but acidic pH levels showed a negative impact (data not shown). Table 2 shows design matrix and corresponding results for RSM experiments. The data from CCD were fitted to Eq. (1) and the following reduced cubic Eq. (2) was obtained from the coded data:
| (2) |
where Y is the phytase activity (U/mL); X1, temperature (°C); X2, pH and X3, phytate concentration (mM).
The highest phytase activity (4.591 U/mL) was recorded at run 18 at 57.5 °C, pH 7.5 and 1.75 mM of sodium phytate, while the minimum phytase activity (0.351 U/mL) was obtained at run 4 at 57.5 °C, pH 9.5 and 1.75 mM of sodium phytate. It also showed approximately 40% of maximal activity at run 9 indicating the enzyme can hydrolysis phytate at acidic pH value (5.50). In addition, the enzyme displayed good activity (60–73% of maximum value) at wide ranges of temperature (47–68 °C), pH (6.30–8.00) and phytate concentration (1.40–2.50 mM). The results indicated the sensitivity of phytase activity to experimental conditions and the importance of finding optimum conditions. Accordingly, B. subtilis B.S.46 phytase appears most promising for hydrolyzing phytates in the small intestine [32], [33].
The results for ANOVA analysis are summarized in Table 3. It shows that the fitted model is significant at 99% of confidence level (P < 0.0001). The coefficient of determination (R2), which is an estimate of the fraction of overall variation in the data accounted by the model, was calculated as 0.9979; thus the model is capable of explaining 99.79% of the variation in response. It ensures a satisfactory adjustment of the reduced cubic model to the experimental data. The ‘adjusted R2’ was 0.9932 indicating that the model is highly significant. The predicted R2 of 0.9788 was in complete agreement with the adjusted R2 showing an excellent correlation between the experimental and predicted values. Furthermore, the high value of adequate precession (37.346) that represents signal (response) to noise (deviation) ratio indicates an adequate signal suggesting that the model can be used to navigate the design space. The statistical significance of the model equation and its terms was supported by the high F-value of 214.45 (Table 3).
Table 3.
ANOVA results of the developed model for B. subtilis B.S.46 phytase activity.
| Source | Sum of squares | DOF | Mean square | F-value | P-value |
|---|---|---|---|---|---|
| Model | 46.11 | 13 | 3.55 | 214.45 | <0.0001 |
| Residual | 0.099 | 6 | 0.017 | ||
| Lack of fit | 3.819E−003 | 1 | 3.819E−003 | 0.20 | 0.6733 |
| Pure error | 0.095 | 5 | 0.019 | ||
| Cor total | 46.21 | 19 |
R2 = 0.9979, Adj R2 = 0.9932, Pred R2 = 0.9788, Adeq Precision = 37.346.
The estimated coefficients of regression model (Eq. (2)), standard errors and corresponding P-value are given in Table 4. The significance of each coefficient was determined by F-value and P-value. The smaller the magnitude of the P-value, the more significant is the corresponding coefficient. The negative coefficients for and and the positive coefficient for indicated negative and positive linear effects on phytase activity, respectively. The negative coefficients for , and demonstrated negative quadratic effects. While the interaction influence of temperature and phytate concentration was negative, and terms showed positive effects on the response. In addition, the positive cubic coefficient for and the negative cubic coefficients for and contributed positively and negatively on phytase activity, respectively.
Table 4.
The estimated coefficients for B. subtilis B.S.46 phytase activity.
| Source | Coefficient | Standard error | F-value | P-value |
|---|---|---|---|---|
| Intercept | 4.35 | 0.052 | ||
| X1 – Temp. | −0.23 | 0.054 | 18.54 | 0.0051 |
| – pH | −0.48 | 0.054 | 77.78 | 0.0001 |
| – Conc. | 0.54 | 0.054 | 100.69 | <0.0001 |
| 0.12 | 0.045 | 6.94 | 0.0389 | |
| −0.12 | 0.045 | 7.38 | 0.0348 | |
| 0.17 | 0.045 | 13.61 | 0.0102 | |
| −1.12 | 0.034 | 1086.15 | <0.0001 | |
| −1.14 | 0.034 | 1126.77 | <0.0001 | |
| −0.64 | 0.034 | 354.01 | <0.0001 | |
| 0.31 | 0.045 | 46.71 | 0.0005 | |
| −0.27 | 0.071 | 14.97 | 0.0083 | |
| −0.77 | 0.071 | 119.75 | <0.0001 |
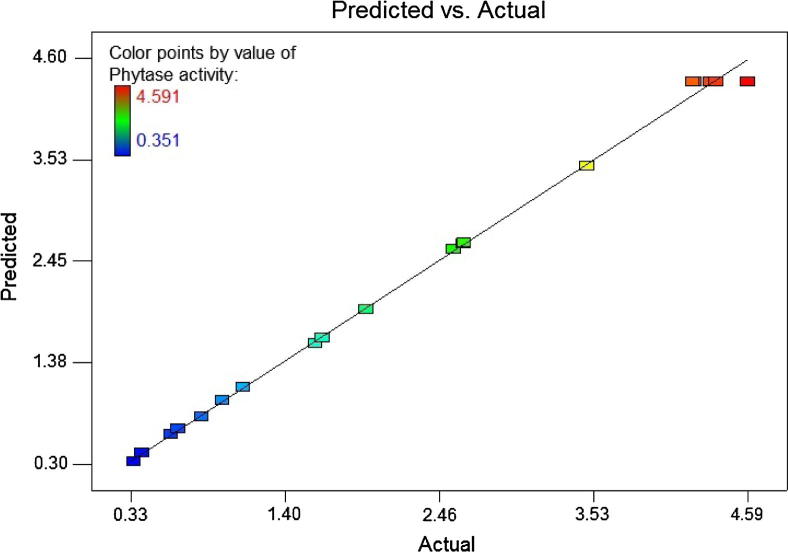
The parity plot for phytase activity indicated an excellent correlation between the actual and predicted values under different assay conditions (Fig. 2). The graph showed a good fit to the model and displayed an acceptable variation between the experimental and predicted values in the range of the selected independent variables according to the scattering pattern of points around the sloping line.
Fig. 2.
The actual versus predicted phytase activities by B. subtilis B.S.46.
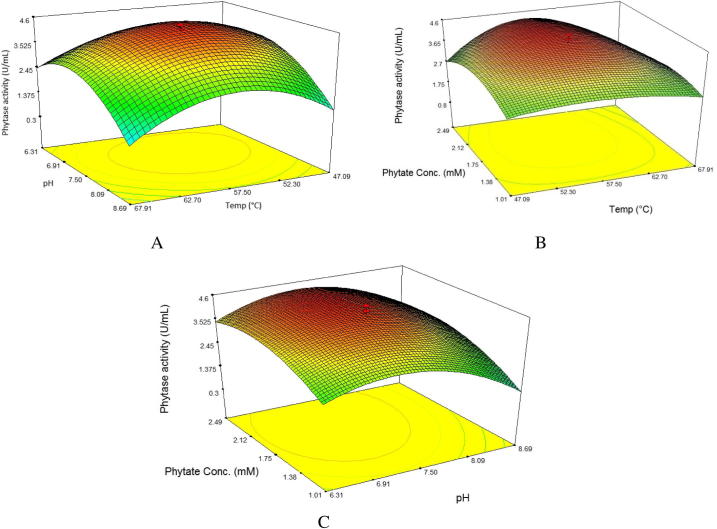
To show the interaction effects of independent variables, the predicted values were plotted as surface plots, in which their shapes indicated no positive interaction between each two factors. Maximum phytase production was recorded approximately in the middle levels of each independent variables while further increase in the levels resulted in a gradual decrease in phytase activity (Fig. 3). As can be seen from Fig. 3A, intermediate levels of temperature and pH resulted in an increase in phytase activity and the highest activity was obtained at 55–57.5 °C and pH 7.0–7.5. The surface plot in Fig. 3B shows that the moderate levels of temperature and phytate concentration increased the enzyme activity, whereas their low or high levels had negative effects. Temperature and phytate concentration ranges of 55–57.5 °C and 1.75–2.12 mM gave the maximum phytase activity (Fig. 3B). Fig. 3C shows the surface plot of the interaction effect of pH and phytate concentration on phytase activity. Neutral pH and average concentration of phytate led to the optimum phytase activity. The inhibitory effects of high or low levels of these two factors can also be seen in Fig. 3C. Maximum phytase activity was obtained at pH (7.0–7.5) and phytate concentration (1.75–2.12 mM). In addition, it can be seen from Fig. 3 that the enzyme had good catalytic activity with broad temperature (47–68 °C), pH (6.3–8.7), and phytate concentration (1.00–2.50 mM) optima retaining about 50% of its activity compared with the maximum activity. The model was validated under several conditions including the optimum point predicted by the numerical optimization tool in the software. As can be seen in Table 5, the actual phytase activities were in the range and close to the predicted phytase activities indicating that proposed model was greatly powerful to navigate and predict design space. The optimal conditions for maximal phytase activity suggested by the model were 56.5 °C, pH 7.3, and 2.05 mM sodium phytate; the results showed a strong agreement between the predicted (4.521 U/mL) and experimental (4.315–4.627 U/mL) responses confirming the adequacy of model. Therefore, optimization by RSM led to 137% enhancement in phytase activity compared with the non-optimized assay conditions (1.952 U/mL) demonstrating the significance of assay condition optimization.
Fig. 3.
Surface plots showing the interaction effects of (A) temperature and pH, (B) temperature and phytate concentration, (C) pH and phytate concentration on phytase activity by B. subtilis B.S.46.
Table 5.
Validation experiments including the optimum point with the corresponding predicted and actual phytase activities.
| Run no. | Temperature (°C) | pH | Phytate concentration (mM) | Predicted phytase activity (U/mL) | Actual phytase activity (U/mL) |
|---|---|---|---|---|---|
| 1 | 50.0 | 6.5 | 1.50 | 3.586 | 3.337 |
| 2 | 60.0 | 7.0 | 1.00 | 3.203 | 3.631 |
| 3 | 45.0 | 6.0 | 2.00 | 2.454 | 2.522 |
| 4a | 56.5 | 7.3 | 2.05 | 4.661 | 4.627 |
Optimum point.
It is very important to know the characteristics of phytases especially for their industrial applications because phytases from various sources display different properties [7]. The results of our study are in agreement with those obtained for B. subtilis phytase [29], Bacillus sp. KHU-10 phytase (10 mM CaCl2) [30], and recombinant phytase (rePhyCm) [34]. However, Gulati et al. [17], El-Toukhy et al. [35], Borgi et al. [36], and Nuñal et al. [37] reported different findings for B. laevolacticus, B. subtilis MJA, B. licheniformis ATCC 14580 (PhyL), and different Bacillus strains, respectively. B. amyloliquefaciens US573 had a similar optimum pH of 7.5, but higher optimum temperature (70 °C) than B. subtilis B.S.46 [31]. The results showed that increasing phytate concentration up to about 2.1 mM had a positive effect, while higher levels inhibited the phytase activity as previously reported in Shigella sp. CD2 and Schizophyllum commune [38], [39].
Partial purification and characterization of B. subtilis B.S.46 phytase
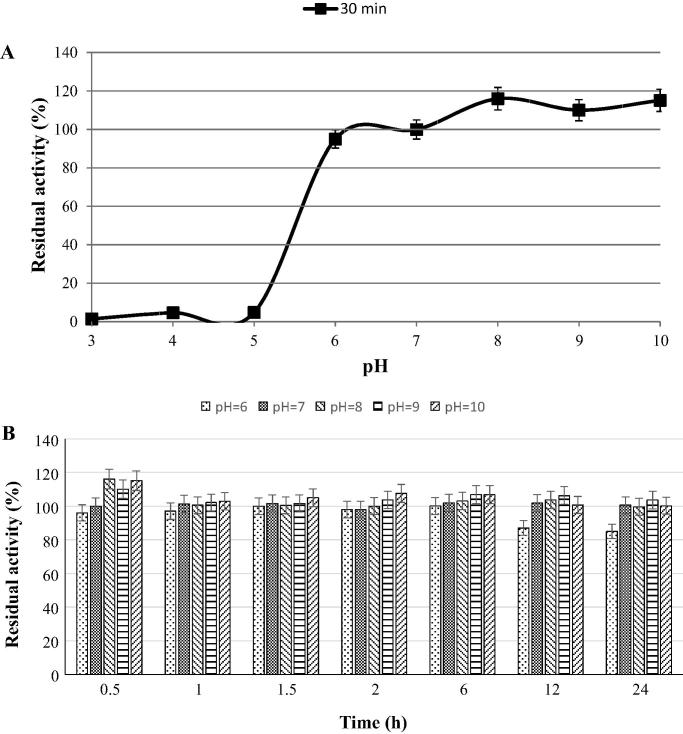
The phytase from B. subtilis B.S.46 was partially purified using solid ammonium sulfate precipitation and dialysis. The enzyme exhibited a specific activity of 5.45 U/mg proteins (data not shown). The results of pH stability of phytase are shown in Fig. 4. After 30 min, the enzyme completely inactivated at pH values of 3.0–5.0, while 95–100% of its initial activity was retained at pH 6.0–7.0 and interestingly, even the activation of enzyme up to 1.16-fold occurred at pH 8.0–10.0 (Fig. 4A). The enzyme was highly stable at slightly acidic to alkaline pH ranging from 6.0 to 10.0 and maintained 85–100% of its initial activity after 24 h (Fig. 4B), which is in consistent with the reported pH ranges for recombinant phytase (rePhyCm) (5.0–9.0) and B. laevolacticus phytase (7.0–10.0) [17], [34]. This interesting characteristic as feed additive can be potentially useful for hydrolyzing phytic acid in the intestine of animals and some fish having slightly neutral to alkaline pH in their digestive tract [33]. In contrast, B. amyloliquefaciens US573 exhibited good stability at pH value ranging from 3 to 9 after 1 h at 37 °C [31]. Borgi et al. [36] showed that after 4 h, the B. licheniformis phytase in the presence of 0.6 mM Ca2+ retained 80% of its activity at pH 7.0 and 7.5, while considerably suppressed at pH 6.0. Nuñal et al. [37] reported that after pre-incubation at 25 °C for 1 h, the Bacillus phytases showed the maximal stability at pH 6 and decreasing and/or increasing pH (3.0–11) gradually reduced their activities. The B. subtilis MJA phytase retained more than 80% of its initial activity over a wide pH range (2.0–8.0) after 4 h, but exposing it to pH values of 2 or 8 for 24 h resulted in the 50% inhibition of enzyme activity [35]. Choi et al. [30] showed that Bacillus sp. KHU-10 phytase retained 80% of it activity at pH 6.5–10 after 30 min with 10 mM CaCl2.
Fig. 4.
Effect of pH on the stability of B. subtilis B.S.46 phytase (A) during 30 min and (B) 24 h.
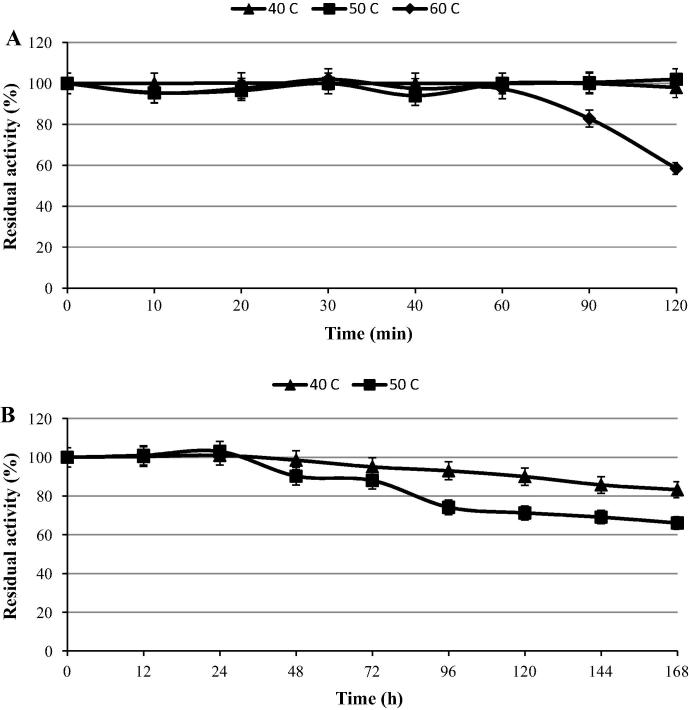
Thermostability is particularly an important trait since feed pelleting is commonly performed at temperatures between 65 and 95 °C and therefore the enzyme should withstand inactivation due to high temperatures [33]. The results of thermostability showed that the enzyme retained 83% and 60% of its initial activity at 60 °C after 90 and 120 min (Fig. 5A). The temperature stability of phytase from the present study was higher than the ones reported for Bacillus phytases [37], rePhyCm [34], shiitake mushroom phytase [40], B. subtilis MJA phytase [35] and Bacillus sp. KHU-10 [30], but lower than B. amyloliquefaciens US573 [31], B. licheniformis ATCC 14580 (PhyL) [36] and B. laevolacticus phytases [17]. However, the enzyme was also highly stable at 40 °C and 50 °C after 168 h (7 days); the relative activities reached 83% and 66%, respectively (Fig. 5B).
Fig. 5.
Effect of temperature on the stability of B. subtilis B.S.46 phytase (A) during 120 min and (B) 168 h.
The influence of various metal ions on phytase activity is presented in Table 6. The phytase activity was significantly increased by Ca2+ at 1 and 2 mM (about 50% and 30% respectively), while higher concentrations had an inhibitory impact. Li+ at different concentrations also increased the phytase activity by 10–20%. Various concentrations of Hg2+ completely inhibited the phytase activity. Cu2+ and Mg2+ at 1 mM showed little effect on phytase activity, while higher concentrations considerably inhibited its activity. Fe2+, Zn2+, and Mn2+ at 1 mM inhibited the enzyme by 70%, 37%, and 18%, respectively and increasing the concentration greatly inhibited the enzyme activity. The activity of phytase was slightly changed at different concentrations of K+ and Na+. Similar results were previously reported indicating the activating effect of Ca2+ and the inhibitory role of Fe2+, Cu2+, Zn2+, Mn2+, and Mg2+ on phytase activity [31], [34], [35], [36]. In agreement with our study, Choi et al. [30], Gulati et al. [17], and Salmon et al. [39] showed that K+ and Na+ had insignificant effects on Bacillus sp. KHU-10, B. laevolacticus and S. commune phytase, respectively. Several studies have been done on the metal dependency of Bacillus phytases indicating that the loss of enzymatic activity is most likely due to a conformational change, as the circular dichroism spectra of the holoenzyme and metal-depleted enzyme were significantly different [41], [42], [43].
Table 6.
Effect of metal ions on B. subtilis B.S.46 phytase activity.
| Reagents | Relative activity (%) |
|||
|---|---|---|---|---|
| 1 mM | 2 mM | 5 mM | 10 mM | |
| None | 100 | 100 | 100 | 100 |
| CaCl2 | 146 | 127 | 46 | 10 |
| LiCl | 110 | 120 | 119 | 111 |
| NaCl | 95 | 93 | 96 | 92 |
| KCl | 95 | 90 | 88 | 91 |
| CuSO4 | 92 | 39 | 8 | 5 |
| MgCl2 | 94 | 16 | 10 | 4 |
| FeSO4 | 70 | 0 | 0 | 0 |
| ZnSO4 | 37 | 14 | 12 | 9 |
| MnCl2 | 18 | 15 | 0 | 0 |
| HgCl2 | 0 | 0 | 0 | 0 |
Conclusions
A thermo-stable alkaline phytase was isolated from the phyllosphere of rice plant and identified using 16S rRNA sequencing as B. subtilis B.S.46. The RSM optimization of catalytic activity of B. subtilis B.S.46 phytase resulted in a 137% increase (4.627 U/mL) with optimal temperature, pH and phytate concentration of 56.5 °C, 7.3, and 2.05 mM, respectively. It displayed broad pH stability at pH 6.0–10.0 retaining about 85% of the initial activity after 7 days at pH 6.0. Temperature stability showed that B. subtilis B.S.46 phytase was highly stable at 40 and 50 °C for 7 days and it retained 60% of its initial activity after 120 min at 60 °C. The results demonstrated that calcium and lithium ions had stimulating effects on phytase activity (10–46%), while heavy metal ions especially at high concentration (10 mM) completely inhibited the enzyme activity. The B. subtilis B.S.46 phytase demonstrated interesting properties to be considered for potential industrial applications. Further work is underway for the optimization of culture conditions for increasing phytase production as well as studies on the ability of B. subtilis B.S.46 phytase to release inorganic phosphorous from food and feed ingredients.
Conflict of Interests
The authors declare that they have no conflict of interest.
Compliance with Ethics Requirements
This article doesnot contain any studies with human or animal subjects.
Acknowledgments
The authors would like to thank Ms. Hoseini, Ms. Bazrafshan, and Ms. Moteshaffi for their assistance during the course of this research. The authors are grateful to the Department of Microbial Biotechnology and Biosafety, Agricultural Biotechnology Research Institute of Iran (ABRII, Karaj, Iran) for financial support of this project.
Footnotes
Peer review under responsibility of Cairo University.
Contributor Information
Maryam Hashemi, Email: hashemim@abrii.ac.ir.
Mohammad Safari, Email: msafari@ut.ac.ir.
References
- 1.Lei X.G., Weaver J.D., Mullaney E., Ullah A.H., Azain M.J. Phytase, a new life for an “old” enzyme. Annu Rev Anim Biosci. 2013;1(1):283–309. doi: 10.1146/annurev-animal-031412-103717. [DOI] [PubMed] [Google Scholar]
- 2.Roohani N., Hurrell R., Wegmueller R., Schulin R. Zinc and phytic acid in major foods consumed by a rural and a suburban population in central Iran. J Food Compost Anal. 2012;28(1):8–15. [Google Scholar]
- 3.Pallauf J., Rimbach G. Nutritional significance of phytic acid and phytase. Arch Anim Nutr. 1997;50(4):301–319. doi: 10.1080/17450399709386141. [DOI] [PubMed] [Google Scholar]
- 4.Suhairin A., Manap A., Yazid M., Hussin M., Shobirin A., Mustafa S. Phytase: application in food industry. Int Food Res J. 2010;17:13–21. [Google Scholar]
- 5.Bohn L., Meyer A.S., Rasmussen S.K. Phytate: impact on environment and human nutrition. A challenge for molecular breeding. J Zhejiang Univ Sci B. 2008;9(3):165–191. doi: 10.1631/jzus.B0710640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vats P., Bhattacharyya M.S., Banerjee U.C. Use of phytases (myo-inositolhexakisphosphate phosphohydrolases) for combatting environmental pollution: a biological approach. Critical Rev Environ Sci Technol. 2005;35:469–486. [Google Scholar]
- 7.Yao M.Z., Zhang Y.H., Lu W.L., Hu M.Q., Wang W., Liang A.H. Phytases: crystal structures, protein engineering and potential biotechnological applications. J Appl Microbiol. 2012;112(1):1–14. doi: 10.1111/j.1365-2672.2011.05181.x. [DOI] [PubMed] [Google Scholar]
- 8.Fu S., Sun J., Qian L., Li Z. Bacillus phytases: present scenario and future perspectives. Appl Biochem Biotechnol. 2008;151(1):1–8. doi: 10.1007/s12010-008-8158-7. [DOI] [PubMed] [Google Scholar]
- 9.Yin Q., Zheng Q., Kang X. Biochemical characteristics of phytases from fungi and the transformed microorganism. Anim Feed Sci Technol. 2007;132(3):341–350. [Google Scholar]
- 10.Jorquera M., Martínez O., Maruyama F., Marschner P., de la Luz Mora M. Current and future biotechnological applications of bacterial phytases and phytase-producing bacteria. Microbes Environ. 2007;23(3):182–191. doi: 10.1264/jsme2.23.182. [DOI] [PubMed] [Google Scholar]
- 11.Kaur P., Satyanarayana T. Yeast acid phosphatases and phytases: production, characterization and commercial prospects. In: Satyanarayana T., Kunze G., editors. Yeast biotechnology: diversity and applications. Springer; Netherlands: 2009. pp. 693–714. [Google Scholar]
- 12.Singh B., Satyanarayana T. Fungal phytases: characteristics and amelioration of nutritional quality and growth of non-ruminants. J Anim Physiol Anim Nut. 2014:1–15. doi: 10.1111/jpn.12236. [DOI] [PubMed] [Google Scholar]
- 13.Cheng C., Lim B. Beta-propeller phytases in the aquatic environment. Arch Microbiol. 2006;185(1):1–13. doi: 10.1007/s00203-005-0080-6. [DOI] [PubMed] [Google Scholar]
- 14.Konietzny U., Greiner R. Bacterial phytase: potential application, in vivo function and regulation of its synthesis. Braz J Microbiol. 2004;35(1–2):12–18. [Google Scholar]
- 15.Roy T., Banerjee G., Dan S.K., Ray A.K. vol. 661. Springer; 2013. Optimization of fermentation conditions for phytase production by two strains of Bacillus licheniformis (LF1 and LH1) isolated from the intestine of Rohu, Labeo rohita (Hamilton) pp. 27–35. (Proceedings of the zoological society). [Google Scholar]
- 16.Farhat A., Chouayekh H., Farhat M.B., Bouchaala K., Bejar S. Gene cloning and characterization of a thermostable phytase from Bacillus subtilis US417 and assessment of its potential as a feed additive in comparison with a commercial enzyme. Mol Biotechnol. 2008;40(2):127–135. doi: 10.1007/s12033-008-9068-1. [DOI] [PubMed] [Google Scholar]
- 17.Gulati H., Chadha B., Saini H. Production and characterization of thermostable alkaline phytase from Bacillus laevolacticus isolated from rhizosphere soil. J Ind Microbiol Biotechnol. 2007;34(1):91–98. doi: 10.1007/s10295-006-0171-7. [DOI] [PubMed] [Google Scholar]
- 18.Kumar V., Sinha A.K., Makkar H.P., Becker K. Dietary roles of phytate and phytase in human nutrition: a review. Food Chem. 2010;120(4):945–959. [Google Scholar]
- 19.Hussin A.S.M., Farouk A.E., Sulleb H.M. Phytate-degrading enzyme and its potential biotechnological application: a review. J Agrobiotechnol. 2010;1:1–16. [Google Scholar]
- 20.Caipang C., Dechavez R.B., Amar M.J.A. Potential application of microbial phytase in aquaculture. Extreme Life Biospeol Astrobiol. 2011;3(1):55–66. [Google Scholar]
- 21.Hashemi M., Razavi S.H., Shojaosadati S.A., Mousavi S.M., Khajeh K., Safari M. Development of a solid-state fermentation process for production of an alpha amylase with potentially interesting properties. J Biosci Bioeng. 2010;110(3):333–337. doi: 10.1016/j.jbiosc.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 22.Engelen A.J., Heeft F.C., Randsdorp P.H., Somers W.A., Schaefer J., van der Vat B.J. Determination of phytase activity in feed by a colorimetric enzymatic method: collaborative interlaboratory study. J AOAC Int. 2001;84(3):629–633. [PubMed] [Google Scholar]
- 23.Zakaria M.R., Tabatabaei M., Ghazali F.M., Abd-Aziz S., Shirai Y., Hassan M.A. Polyhydroxyalkanoate production from anaerobically treated palm oil mill effluent by new bacterial strain Comamonas sp. EB172. World J Microbiol Biotechnol. 2010;26(5):767–774. [Google Scholar]
- 24.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 25.Kimura K. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 26.Tamura K., Dudley J., Nei M., Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 27.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 28.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 29.Kerovuo J., Lauraeus M., Nurminen P., Kalkkinen N., Apajalahti J. Isolation, characterization, molecular gene cloning, and sequencing of a novel phytase from Bacillus subtilis. Appl Environ Microbiol. 1998;64(6):2079–2085. doi: 10.1128/aem.64.6.2079-2085.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi Y.M., Suh H.J., Kim J.M. Purification and properties of extracellular phytase from Bacillus sp. KHU-10. J Protein Chem. 2001;20(4):287–292. doi: 10.1023/a:1010945416862. [DOI] [PubMed] [Google Scholar]
- 31.Boukhris I., Farhat-Khemakhem A., Blibech M., Bouchaala K., Chouayekh H. Characterization of an extremely salt-tolerant and thermostable phytase from Bacillus amyloliquefaciens US573. Int J Biol Macromol. 2015;80:581–587. doi: 10.1016/j.ijbiomac.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 32.Simon O., Igbasan F. In vitro properties of phytases from various microbial origins. Int J Food Sci Technol. 2002;37:813–822. [Google Scholar]
- 33.Igbasan F.A., Männer K., Miksch G., Borriss R., Farouk A., Simon O. Comparative studies on the in vitro properties of phytases from various microbial origins. Arch Anim Nutr. 2000;53(4):353–373. doi: 10.1080/17450390009381958. [DOI] [PubMed] [Google Scholar]
- 34.Wang Q., Fu S.J., Sun J.Y., Weng X.Y. Characterization of a thermostable alkaline phytase from Bacillus licheniformis ZJ-6 in Pichia pastoris. World J Microbiol Biotechnol. 2011;27(5):1247–1253. [Google Scholar]
- 35.El-Toukhy N.M., Youssef A.S., Mikhail M.G. Isolation, purification and characterization of phytase from Bacillus subtilis MJA. Afr J Biotechnol. 2013;12(20):2957–2967. [Google Scholar]
- 36.Borgi M.A., Boudebbouze S., Aghajari N., Szukala F., Pons N., Maguin E. The attractive recombinant phytase from Bacillus licheniformis: biochemical and molecular characterization. Appl Microbiol Biotechnol. 2014;98(13):5937–5947. doi: 10.1007/s00253-013-5421-9. [DOI] [PubMed] [Google Scholar]
- 37.Nuñal S.N., Serrano A.E., Jr., Seraspe E.B., Maeda H. Biochemical properties of extracellular phytases from Bacillus spp. Mem Fac Fish. Kagoshima Univ. 2008;Special Issue:49–55. [Google Scholar]
- 38.Roy M.P., Poddar M., Singh K.K., Ghosh S. Purification, characterization and properties of phytase from Shigella sp. CD2. Indian J Biochem Biophys. 2012;49:266–271. [PubMed] [Google Scholar]
- 39.Salmon D.N.X., Piva L.C., Binati R.L., Rodrigues C., de Souza Vandenberghe L.P., Soccol C.R. A bioprocess for the production of phytase from Schizophyllum commune: studies of its optimization, profile of fermentation parameters, characterization and stability. Bioprocess Biosyst Eng. 2012;35:1067–1079. doi: 10.1007/s00449-012-0692-6. [DOI] [PubMed] [Google Scholar]
- 40.Zhang G.Q., Wu Y.Y., Ng T.B., Chen Q.J., Wang H.X. A phytase characterized by relatively high pH tolerance and thermostability from the shiitake mushroom lentinus edodes. BioMed Res Int. 2013:1–7. doi: 10.1155/2013/540239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao M.Z., Lu W.L., Chen T.G., Wang W., Fu Y.J., Yang B.S. Effect of metals ions on thermostable alkaline phytase from Bacillus subtilis YCJS isolated from soybean rhizosphere soil. Ann Microbiol. 2013;64(3):1123–1131. [Google Scholar]
- 42.Oh B.C., Chang B.S., Park K.H., Ha N.C., Kim H.K., Oh B.H. Calcium-dependent catalytic activity of a novel phytase from Bacillus amyloliquefaciens DS11. Biochemistry. 2001;40(32):9669–9676. doi: 10.1021/bi010589u. [DOI] [PubMed] [Google Scholar]
- 43.Kerovuo J., Lappalainen I., Reinikainen T. The metal dependence of Bacillus subtilis phytase. Biochem Biophys Res Commun. 2000;268:365–369. doi: 10.1006/bbrc.2000.2131. [DOI] [PubMed] [Google Scholar]