Abstract Abstract
Lonchophylla Thomas, 1903 is a Neotropical bat genus that comprises 12 species, with little cytogenetic information available. Here we present the description of the karyotype of three species collected in Southeastern Brazil. Lonchophylla bokermanni Sazima, Vizotto & Taddei, 1978, Lonchophylla dekeyseri Taddei, Vizotto & Sazima, 1983, and Lonchophylla peracchii Dias, Moratelli & Esberard, 2013 showed the same diploid number 2n = 28 and the same autosomal fundamental number FNa = 50, in both Lonchophylla bokermanni and Lonchophylla peracchii. We observed that the karyotypes were also cytogenetically similar when we compared the studied species with other species within the same genus. It is therefore not possible to differentiate the species using only karyotypes with conventional staining. However, this information increases the knowledge of the genus and can be one more important character for a better phylogenetic comprehension of this taxon.
Keywords: Karyology, chromosomes, Bokermann’s Nectar Bat, Atlantic Forest, Cerrado, Endangered species, Lonchophyllinae, range extension
Introduction
In recent years, new species and a genus of the subfamily Lonchophyllinae were described: Lonchophylla peracchii Dias, Moratelli & Esberard, 2013, Lonchophylla inexpectata Moratelli & Dias, 2015, and Hsunycteris Parlos, Timm, Swier, Zeballos & Baker, 2014 (Dias et al. 2013, Parlos et al. 2014, Moratelli and Dias 2015). For the description of bat species, morphological and morphometric characteristics are usually employed, but the use of other tools such as cytogenetic analysis can provide essential information for evolutionary relationships of bats (Varella-Garcia and Taddei 1989, Garcia and Pêssoa 2010), as already seen for rodents, for example (Romanenko and Volobouev 2012). Although there are few cytogenetic data for Lonchophyllinae, they were nevertheless informative for systematic rearrangements of this taxon (see Parlos et al. 2014).
In Brazil, there are records for five species of this genus: Lonchophylla bokermanni Sazima, Vizotto & Taddei, 1978, Lonchophylla dekeyseri Taddei, Vizotto & Sazima, 1983, Lonchophylla inexpectata, Lonchophylla mordax Thomas, 1903 and the new species, Lonchophylla peracchii mentioned above. There are karyotype data available until now for the two congeneric taxa from outside the country, Lonchophylla robusta Miller, 1912 and Lonchophylla concava Goldman, 1914 (Parlos et al. 2014), but no cytogenetic data were available for Brazilian species. Therefore, this study is the first to describe the karyotype of Lonchophylla bokermanni, Lonchophylla dekeyseri and Lonchophylla peracchii.
Material and methods
Five individuals of Lonchophylla were collected and four were karyotyped: one adult female (MN79997) and one adult male of Lonchophylla bokermanni (MN81467), one adult female of Lonchophylla dekeyseri (MN80002) and one adult male of Lonchophylla peracchii (MN81468).
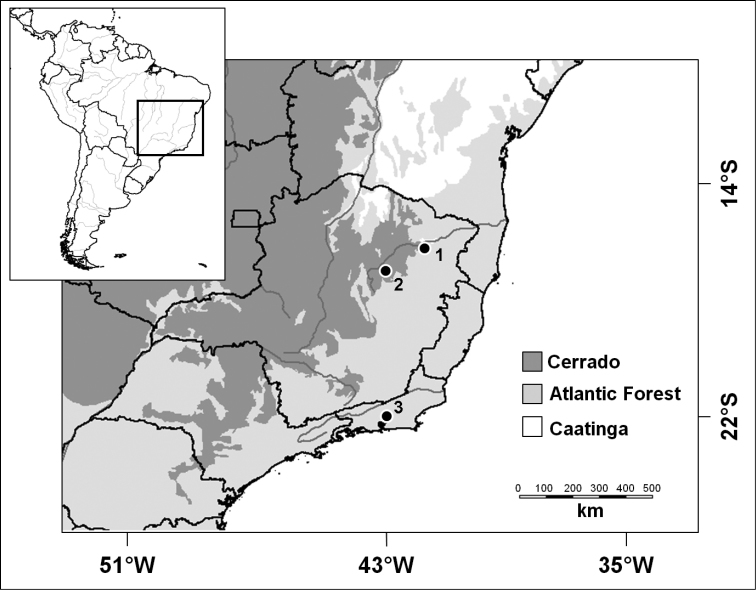
Lonchophylla bokermanni was captured in Fazenda Santa Cruz, Diamantina municipality (18°16'11"S; 43°23'04"W, 1.129 m a.s.l), in the Vale do Jequitinhonha, Minas Gerais State (Figure 1). The locality has a Cerrado vegetation classified as arboreal savanna with enclaves of deciduous forest (IBGE 2012). Sampling occurred in March 2011 using 13 to 15 mist-net (9 × 3 m, 35 mm mesh), which remained open in the first six hours after the sunset for six consecutive nights.
Figure 1.
Localities of Lonchophylla species records: 1 Itinga, Minas Gerais 2 Dimantina, Minas Gerais 3 Magé, Rio de Janeiro.
Lonchophylla dekeyseri was captured in Fazenda Ilha, Itinga municipality (16°38'05"S; 41°50'54"W, 240 m a.s.l), Vale do Jequitinhonha, Minas Gerais State (Figure 1). The locality is in the Cerrado, with vegetation classified as open savanna in transition with Dry Forest (IBGE 2012). The sampling procedures were performed in March 2012, using 12 to 16 mist-nets (9 × 3 m, 35 mm mesh) that remained open in the first six hours after the sunset for seven consecutive nights.
Lonchophylla peracchii was captured in Reserva Particular do Patrimônio Natural El Nagual, Magé municipality (22°32'55"S; 43°03'20"W, 197 m a.s.l), Rio de Janeiro State (Figure 1). The locality is in the Atlantic Forest, with vegetation classified as Ombrophilous Dense Forest (IBGE 2012). Sampling occurred in August 2012 using two mist-nets (12 × 3 m, 30 mm mesh) that remained open throughout night period (± 12 hours) for two consecutive nights.
Chromosomes in metaphases were obtained through in vitro bone marrow culture grown in Dulbecco´s MEM with 10% fetal bovine serum and colchicine for 2 hours, following by an incubation in KCl 0.075M solution at 37 °C by 30 minutes, centrifuged, fixed in Carnoy solution (methanol: acetic acid, 3:1). The fixation step was repeated three times. Preparation was done by dropping one drop by distance onto clean microscope slides and air-dried. Conventional staining with Giemsa 5% was used to observe (2n) and (FNa) and chromosome morphology variation. This analysis was carried out using an optic photomicroscope (Nikon Eclipse 50i), in a 1,000 increase – lenses of 100 plus 10 ocular lenses.
Captures were authorized by IBAMA (1785/89-IBAMA) and SISBIO (4156/95-46 in the Vale of Jequitinhonha and 3893-1/28717 in Magé).
Results and discussion
All three species showed the same diploid number 2n = 28 and an autosomal fundamental number FNa = 50 was observed (Figure 2). The autosomal complement of males Lonchophylla bokermanni and Lonchophylla peracchii consists of 12 pairs of meta/submetacentrics varying from large to small, and a pair of small acrocentric chromosomes (FNa = 50). Two size classes of autosomal chromosomes can be observed – the eight first are all large chromosomes, and in the second row (Figure 2A–C), with smaller ones, including four metacentric and the smallest chromosome of the karyotype, the only acrocentric ones. The X chromosome is a medium sized metacentric and the Y is a minute acrocentric, smaller than the last pair of autosomal complement. Similarly, the karyotype of Lonchophylla dekeyseri can be characterized but the identification of the sex chromosome pair was impossible in the sole collected specimen which was a female.
Figure 2.
Giemsa-stained karyotypes of A Lonchophylla bokermanni 2n = 28, FNa = 50 (male, MN81467) B Lonchophylla peracchii 2n = 28, FNa = 50 (male, MN81468) and C Lonchophylla dekeyseri 2n = 28 (female, MN80002).
Karyotype comparison is considered as an important tool to establish phylogenetic relationships and as a taxonomic tool to confirm some species identities (Baker 1970, Silva et al. 2006, Urdampilleta et al. 2013). However, the resolution power of the cytogenetic method is not the same for all groups. Sometimes, it is necessary to analyze as many as possible the species’ karyotypes (Garcia and Pessoa 2010). In bats, the few available published karyotype data from South America (Moratelli and Morielle-Versute 2007) make it difficult to propose, using such kind of information, new or different taxonomic arrangements and a better comprehension of the systematics of Neotropical bats (Garcia and Pessoa 2010).
Three new karyotypes here described for Lonchophylla bokermanni, Lonchophylla dekeyseri and Lonchophylla peracchii are similar to those known for Lonchophylla robusta (Baker 1973, Baker 1979) and Lonchophylla concava (Parlos et al. 2014). A species currently allocated to the genus Hsunycteris and previously described as Lonchophylla thomasi, presents different karyotype compositions: 2n = 30, FNa = 34; 2n = 32, FNa = 34, 38 and 40; 2n = 36, FNa = 48. Additionally, this species also presented an increased number of acrocentric chromosomes, whereas in other Lonchophylla species, only a pair of small acrocentric is observed (Pair 13 in Figure 2) (Parlos et al. 2014).
The karyotype conservatism in Microchiroptera has been observed in other studies (Varella-Garcia et al. 1989, Sousa and Araújo 1990) which well corroborate with our results. Even if species distinction is not evident for representatives of the Lonchophylla genus through the conventional chromosome characteristics, the generic separation of Lonchophylla – Hsunycteris is supported by their different karyotypes.
Acknowledgments
Julia L Luz, Maíra SM Godoy, Luciana M Costa, Luciana G Pereira, Edvandro A Ribeiro and André C Siqueira helped in the fieldwork; Luana Azamor helped in laboratory work; to FAPERJ for IC Grant toBrunna Almeida and for scholarship for Renan F. Souza; and CNPq for financial support. Lena Geise has CNPq and Uerj/Prociencia fellowship.
Citation
Almeida B, Novaes RLM, Aguieiras M, Souza RF, Esbérard CEL, Geise L (2016) Karyotype of three Lonchophylla species (Chiroptera, Phyllostomidae) from Southeastern Brazil. Comparative Cytogenetics 10(1): 109–115. doi: 10.3897/CompCytogen.v10i1.6646
References
- Baker RJ. (1970) Karyotypic trends in bats. In: Wimsatt WA. (Eds) Biology of bats. Academic Press Inc, New York, 65–97. doi: 10.1016/b978-0-12-758001-2.50007-1 [Google Scholar]
- Baker RJ. (1973) Comparative cytogenetics of the New World leaf-nosed bats (Phyllostomatidae). Periodicum Biologorum 75: 37−45. [Google Scholar]
- Baker RJ. (1979) Karyology. In: Baker RJ, Jones K, Carter DC. (Eds) Biology of bats of the New World family Phyllostomatidae, Part III Special Publications of the Museum of Texas Tech University, 107−155.
- Dias D, Peracchi AL. (2008) Quirópteros da Reserva Biológica do Tinguá, estado do Rio de Janeiro, sudeste do Brasil (Mammalia: Chiroptera). Revista Brasileira de Zoologia 25(2): 333−369. doi: 10.1590/s0101-81752008000200023 [Google Scholar]
- Dias D, Esbérard CEL, Moratelli R. (2013) A new species of Lonchophylla (Chiroptera, Phyllostomidae) from the Atlantic Forest of southeastern Brazil, with comments on L. bokermanni. Zootaxa 3722(3): 347−360. doi: 10.11646/zootaxa.3722.3.4 [DOI] [PubMed] [Google Scholar]
- Garcia JP, Pessôa LM. (2010) Karyotypic composition of bats from the Brazilian nuclear power plants, state of Rio de Janeiro. Chiroptera Neotropical 16(1): 617–628. [Google Scholar]
- Geise L. (2014) Procedimentos genéticos iniciais na captura e preparação de mamíferos. In: Reis NR, Peracchi AL, Rossaneis BK, Fregonezi MN. (Eds) Técnicas de estudos aplicadas aos mamíferos silvestres brasileiros. Technical Books Editora, Rio de Janeiro, 221−235. [Google Scholar]
- Griffiths TA, Gardner AL. (2008) Subfamily Lonchophyllinae. In: Gardner AL. (Ed.) Mammals of South America, volume 1: marsupials, xenarthrans, shrews, and bats. University of Chicago Press, Chicago and London, 244–255. [Google Scholar]
- IBGE (2012) Manual técnico da vegetação brasileira. Second Edition Instituto Brasileiro de Geografia e Estatística, Rio de Janeiro, 274 pp. [Google Scholar]
- Moratelli R, Dias D. (2015) A new species of nectar-feeding bat, genus Lonchophylla, from the Caatinga of Brazil (Chiroptera, Phyllostomidae). ZooKeys 514: 73–91. doi: 10.3897/zookeys.514.10013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moratelli R, Morielle-Versute E. (2007) Métodos e aplicações da citogenética na taxonomia de morcegos brasileiros. In: Reis NR, Peracchi AL, Pedro WA, Lima IP. (Eds) Morcegos do Brasil. Editora da UEL, Londrina, 197–228. [Google Scholar]
- Parlos JA, Timm RM, Swier VJ, Zeballos H, Baker RJ. (2014) Evaluation of paraphyletic assemblages within Lonchophyllinae, with description of a new tribe and genus. Ocassional Papers of the Museum of Texas Tech University 320: 1–23. [Google Scholar]
- Ribeiro NAB, Nagamachi CY, Pieczarka JC, Rissino JD, Nevez ACB, Gonçalves ACO, Marques-Aguiar S, Assis MFL, Barros RMS. (2003) Cytogenetic analysis in species of the subfamily Glossophaginae (Phyllostomidae, Chiroptera) supports a polyphyletic origin. Caryologia 56(1): 85–96. doi: 10.1080/00087114.2003.10589311 [Google Scholar]
- Romanenko SA, Volobouev V. (2012) Non-Sciuromorph rodent karyotypes in evolution. Cytogenentic and Genome Research 137: 233–245. doi: 10.1159/000339294 [DOI] [PubMed] [Google Scholar]
- Sikes RS, Gannon WL, The Animal Care and Use Committee of the American Society of Mammalogists (2011) Guidelines of the American Society of Mammalogists for the use of wild mammals in research. Journal of Mammalogy 92(1): 235–253. doi: 10.1644/10-MAMM-F-355.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MJ, Patton JL, Yassuda-Yonenaga Y. (2006) Phylogenetic relationships and karyotype evolution in the sigmodontine rodent Akodon (2n = 10 and 2n = 16) from Brazil. Genetics and Molecular Biology 29(3): 469–474. doi: 10.1590/S1415-47572006000300012 [Google Scholar]
- Sousa MJ, Araújo MCP. (1990) Conservative pattern of the G-bands and diversity of C-banding patterns and NORs in Stenodermatinae bats (Chiroptera-Phyllostomatidae). Revista Brasileira de Genética 13(2): 255–268. [Google Scholar]
- Urdampilleta JD, Coulleri JP, Ferrucci MS, Forni-Martins ER. (2013) Karyotype evolution and phylogenetic analyses in the genus Cardiospermum L. (Paullinieae, Sapindaceae). Plant Biology 15(5): 868–881. doi: 10.1111/j.1438-8677.2012.00679.x [DOI] [PubMed] [Google Scholar]
- Varella-Garcia M, Morielle-Versute E, Taddei VA. (1989) A survey of cytogenetic data on Brazilian bats. Revista Brasileira de Genética 12: 761–793. [Google Scholar]