Abstract
Ets1 is a sequence-specific transcription factor that plays an important role during hematopoiesis, and is essential for the transition of CD4−/CD8− double negative (DN) to CD4+/CD8+ double positive (DP) thymocytes. Using genome-wide and functional approaches, we investigated the binding properties, transcriptional role and chromatin environment of Ets1 during this transition. We found that while Ets1 binding at distal sites was associated with active genes at both DN and DP stages, its enhancer activity was attained at the DP stage, as reflected by levels of the core transcriptional hallmarks H3K4me1/3, RNA Polymerase II and eRNA. This dual, stage-specific ability reflected a switch from non-T hematopoietic toward T-cell specific gene expression programs during the DN-to-DP transition, as indicated by transcriptome analyses of Ets1−/− thymic cells. Coincidentally, Ets1 associates more specifically with Runx1 in DN and with TCF1 in DP cells. We also provide evidence that Ets1 predominantly binds distal nucleosome-occupied regions in DN and nucleosome-depleted regions in DP. Finally and importantly, we demonstrate that Ets1 induces chromatin remodeling by displacing H3K4me1-marked nucleosomes. Our results thus provide an original model whereby the ability of a transcription factor to bind nucleosomal DNA changes during differentiation with consequences on its cognate enhancer activity.
INTRODUCTION
Erythroblast Transformation-Specific 1 (Ets1) is a sequence-specific transcription factor (TF) whose timely expression plays an important role in the development of hematopoietic cells, particularly in T- and natural killer (NK) lineages (1–3). In T-cells, loss of Ets1 results in impaired β-selection, deficient T-cell activation, altered Th1 immune response as well as aberrant Th17 differentiation (4–7). Besides these developmental properties, Ets1 is a characterized oncogene as a result of its transposition in acute leukemias, angiogenic effects and overexpression in luminal breast cancers (8–10).
Recent studies have pointed toward Ets1 being one of the potentially important upregulated TFs during the CD4−/CD8− double negative (DN) to CD4+/CD8+ double positive transition (DP) (11). The DN to DP transition involves major changes in gene expression that are correlated with the production of a functional T-cell receptor (TCR) β chain and thus pre-TCR at the DN3 stage, which triggers internal signaling upon activation (12,13). The main milestones of this transition include irrevocable commitment towards the αβ T-cell lineage, by downregulation of alternative lineage-potential genes. Repression of hallmark DN genes, such as Il2ra, Cd44, Il7r and the pre-TCR, Ptcra, also marks this step (14–16). Conversely, αβ T-cell lineage specific genes are upregulated at this point, via de-repression or upregulation mechanisms such as that of respectively Cd4 and Cd8 (17,18). These subsequently play a key role at the DP stage, where productive recombination and activation of a Tcra locus is linked with positive selection, a process which favors multiplication of cells that can recognize MHC I/II molecules via interaction with CD8 and CD4, respectively. With the exception of key loci such as the Tcra and Tcrb loci where Ets1 binds to their respective enhancers (19,20), direct evidence of Ets1 involvement with transactivation of many other critical T-cell genes has however remained elusive.
Large-scale analyses studying binding and sequence-specific properties of Ets1 have been performed using chromatin immunoprecipitation (ChIP)-chip and ChIP-seq in cell lines, as well as in vitro studies using Selex (21–24). Overall, these studies yielded two dominant motifs, CGGAAG (canonical Ets1) and AGGAAG (Pu.1-like) for Ets1. Importantly, another study determined that both motifs correspond to binding associated with distinct ontologies for Ets family members (25). However, while previous studies encompassed the role of Ets1 in primary murine cells and cell lines (26–30), to date, no high-throughput analyses hinging on Ets1 have been carried out in primary cells in a stage-specific manner in order to describe both endogenous Ets1 binding and motif dynamics. Further, while single-stage associations of TFs with Ets1 as co-factor have been described (22,26,29), the dynamics of these have not been investigated with Ets1 as the centerpiece. It has also recently been shown that certain Ets family members such as Pu.1 can lead to chromatin remodeling genome-wide (31,32). Finally, previous reports indicated that Ets1 can bind both nucleosomal DNA and nucleosome-depleted regions (NDRs) (33,34), although genome-wide meta-analyses in human cell lines suggested an overall association with NDRs (35), leaving the question open of whether this TF contacts nucleosomes in vivo.
To elucidate the role of Ets1 during DN to DP in vivo, and to investigate its role in gene regulation during this critical transition, we performed multiple genome-wide analyses, including ChIP-seq, transcriptome analyses and MNase-seq, using Rag2−/− (mainly DN3) and wild-type DP murine models (36), as well as Ets1−/− DP cells. Our results indicate that Ets1, essentially as an activator, is associated with a shift from a general hematopoietic to a thymic T-cell -specific gene expression program. Although many distal Ets1 binding sites remain occupied during this transition, their properties and co-factor association change dramatically. Furthermore, whereas Runx1 generally co-associates with DN-bound sites, TCF1 preferentially joins and correlates with Ets1 sites in DP cells. Finally, Ets1 happens to bind both nucleosome and NDRs at enhancer locations at both differentiation stages. Interestingly however, Ets1 bound a majority of nucleosome-occupied regions (NORs) in DN while it bound more NDRs in DP at distal sites, reflecting a shift of binding property for this TF during thymic differentiation. Knock-down experiments in DP model cells further show local H3K4me1 remodeling at Ets1 target sites but not at other regulatory regions.
MATERIALS AND METHODS
Cell preparation and sorting
Thymuses were extracted from 5–6-week-old C57BL/6 Rag2−/− knockout and wild-type as previously described (37,38). Ets1−/− DP thymocytes were generated as previously described (4). P5424 cells were grown in RPMI-1640 with 100 units/ml penicillin, 100 mg/ml streptomycin and 2 mM L-glutamine at 37°C and with 5% CO2. Isolation of CD4+ CD8+ DP cells was performed via AutoMACS. Cell purities were obtained via FACS for Rag2−/- and DP thymocytes (Supplementary Figure S1A). See Supplementary Materials and Methods for further details.
ChIP-Seq and MNase-Seq
ChIP and Micrococcal Nuclease (MNase) digestion of permeabilzed cells were essentially performed as previously described (37,38). All ChIP conditions and antibodies are described in Supplementary Table S1. Sizes of immunoprecipitated fragments were checked using a Bioanalyzer (Agilent, Supplementary Figure S1B). Samples were sequenced using Illumina GA II, HiSeq 2000 and SOLiD 4, 500XL sequencers according to manufacturer's instructions. Further details are described in Supplementary Materials and Methods.
Ets1 knockdown in P5424 cells
P5424 cells were transfected with vector expressing shRNA against Ets1 (for knockdown experiments) or a scrambled sequence (for control experiments). For each transfection reaction, 25 μg vector DNA was electroporated into 5 million cells with a Neon® Transfection System (ThermoFisher Scientific) by using a pulse at 1600V for 20 ms in a volume of 100 μl. Forty-eight hours after electroporation, cells were either cross-linked for ChIP experiments or processed for other assays (WB etc.).
Western blot
Cells were washed once with ice cold phosphate buffered saline. To detect Ets1 by western blotting, protein equivalent of 100 000 cells were resuspended in 50 μl 1× Laemmli buffer (0.25 M Tris–HCL pH 6.8, 2.2% sodium dodecyl sulphate, 10% Glycerol, 2% 2-mercaptoethanol and bromophenol blue) and boiled for 5 min followed by electrophoresis using sodium dodecyl sulphate-polyacrylamide gel electrophoresis and immunoblotted with anti-Ets1 antibody (Santa Cruz sc-350×).
Short RNA-seq
Short RNA-seq preparation was conducted as previously described (38). Briefly, total RNA was extracted using TRIzol (Invitrogen, USA) according to manufacturer's instructions with some modifications to ensure higher recovery rates of small RNAs, which were subsequently isolated using a denaturating urea gel (see Supplementary Materials and Methods).
Gene expression microarray assays and analysis
Gene expression microarray assays were carried out in triplicate as per manufacturer's instructions and as previously described (39) for the Affymetrix Mouse Gene 1.0 and 2.1 ST platforms. Expression data were processed in R via the affy package. Further details are available in Supplementary Materials and Methods.
High-throughput sequencing data processing and peak detection
Raw data pre-processing was performed according to manufacturer's instructions for Illumina GA II, HiSeq 2000, SOLiD 4/5500XL. Briefly, data was aligned to the mm9 genome using CASAVA Eland, bowtie and bfast for these platforms, respectively. Aligned reads were processed using a previously described in-house ChIP-seq analysis pipeline (37,38). Peak detection was carried out using CoCAS (40) for ChIP-Seq datasets. For short RNA-Seq, data was processed using the MIRO pipeline and GEM aligner. Full details of used parameters are available in Supplementary Materials and Methods.
Ets1-fold change ranking and corresponding heatmaps and average profiles
The union of DN + DP datasets was essentially obtained as previously described for DNaseI hypersensitive sites (DHSs) (41,42). In brief, summits of Ets1 DN and DP peaks were merged, annotated as proximal or distal sites to the nearest gene and sorted by Ets1 DP/DN fold change after retrieving counts in both datasets at summits. Corresponding heatmaps and average profiles of other high-throughput sequencing data were retrieved using this order via a previously described R pipeline (38). Tag count and expression fold change heatmaps were generated using Java TreeView (43). Further details of used parameters are available in Supplementary Materials and Methods.
Correlations between gene expression and Ets1-fold change
Direct correlations between ChIP and expression fold change were carried out as previously described for DHS and expression fold changes (42) using 100 classes. Venn diagram overlaps were plotted using pybedtools; associated hypergeometric P-values were computed in R via ChIPpeakAnno (44). Further details are available in Supplementary Materials and Methods.
Motif discovery and co-occurrence clustering
Motif discovery in each population was carried out using Homer (45) ±200 bp around the Ets1 summit. Identified motif matrices corresponded to the closest Homer built-in matrix match (collated from public databases). For motif co-occurrence clustering, motif positions were extended by 200 bp, intersected and enrichment of co-occurrences was calculated against a random background in transcriptionally active sites. Full details of used parameters are available in Supplementary Materials and Methods.
Nucleosome depleted region determination and ranking
Peak detection in inverted MNase data was carried out to identify NDRs using CoCAS. For classes of NDR sites, regions were ranked by increasing DN and DP MNase signal ±100 bp around Ets1 DN + DP summits, intersecting with Ets1 or not. For increasing H3K4me1 nucleosome NDRs, signal ±500 bp around Ets1 was used. Further details of used parameters are available in Supplementary Materials and Methods.
Clustering and PCA analyses
All clustering analyses were conducted using cluster 3.0 (46). Heatmaps were generated using Java TreeView. Principal component analysis (PCA) were performed in R via the prcomp function. For full details on individual analyses, see Supplementary Materials and Methods.
Gene ontology and tissue specificity analyses
Gene ontology analyses were performed using DAVID (47,48) and Homer. Tissue specificity analyses were conducted using DAVID. For full details for individual analyses, see Supplementary Materials and Methods.
Downloaded datasets
High-throughput sequencing data from our previously published studies were retrieved from GEO series GSE29362, GSE38577, GSE45014, GSE60029 and used in addition to the data generated in this study (37,38,49,50). Other ChIP-Seq datasets for Ets1 in other cell types were retrieved from GEO series GSE20898, GSE31331, GSE40684 and GSE36030 (26–30). Other retrieved datasets included TCF1 in EML and whole thymus (51–53), E47 in DN cells (54), Pu.1 in DN2 and DP (11), ENCODE thymus DNAse-Seq (30), in vitro reconstituted MNase-Seq (32), as well as Immunological Genome Project (55), other WT DN3 and DP (56), Th1 and Th17 (57), and Treg expression microarray data (58,59) from GEO series GSE31221, GSE46662, GSE30518, GSE31233, GSE37074, GSE50762, GSE37301, GSE15907, GSE46090, GSE48657, GSE49166 and GSE66332, respectively. Datasets were processed as high-throughput sequencing and expression microarray data presented in this study.
RESULTS
Ets1 participates in T-cell lineage commitment by differentially binding cis-regulatory regions of hallmark, stage-specific genes
To characterize the genome-wide binding properties of Ets1 and its role in transcription during the DN to DP transition, we carried out ChIP-seq for Ets1, RNA Polymerase II (Pol II), as well as for histone tail modifications (H3K4me1, H3K4me3 and H3K27me3) in Rag2−/− DN thymocytes (essentially blocked in DN3, see Supplementary Table S1–S3 and Figure S1A and B for experimental controls, conditions and Ets1 ChIP-seq peak output). Note that Ets1 is slightly induced both at the RNA and protein levels in DP cells (Supplementary Figure S1C and D). We subsequently compared these results to our previously described DP datasets for these marks (37,38). We also performed gene expression microarrays in DP thymocytes, identifying DP/DN up- and downregulated genes via comparison to our previously published DN microarrays (39). Interestingly, we found more Ets1 ChIP-seq peaks in DN than in DP (5893 versus 3401). The majority of these elements were located in distal regions (located at least 5 Kbp away from the transcription start site, TSS) in both cases (Supplementary Figure S1E and F). Overall, 2028 elements were common in DN and DP populations, for which distinguishing promoter/distal status yielded comparable fractions of shared elements (Supplementary Figure S1G). We examined ChIP-seq profiles for Pol II and histone modification marks in areas surrounding Ets1 binding in DN and DP, and found that for highly up- or downregulated, stage-specific loci such as Cd8 and Tcrg, gains or losses of Ets1 were mostly correlated with corresponding gains and losses of H3K4me1/3 and Pol II (Figure 1A). In the case of Ets1 recruitment (Cd8), gain of Ets1 mostly correlated with gain of histone marks and Pol II, as well as with loss of the repressive mark H3K27me3, Cd8 being a previously described Polycomb target (60). Most importantly, we noted that positive and negative changes in Ets1 signal in turn corresponded to gains or losses of gene expression for these loci (Figure 1A, bottom track). Interestingly, these findings also held true genome-wide, whereby gene expression of genes closest to Ets1 peaks generally showed higher gene expression than the average of all genes, suggesting an overall association of Ets1 with transactivation (Supplementary Figure S1H and I). At both stages, the majority of genes adjacent to Ets1 peaks were expressed.
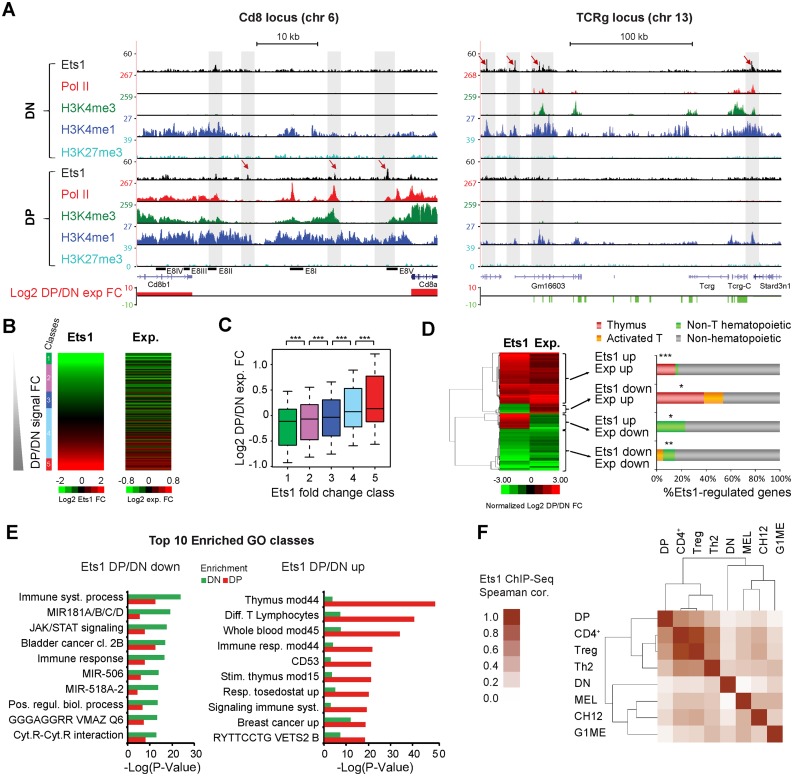
Figure 1.
Ets1 is dynamically associated with positive regulation of stage-specific genes. (A) Examples of Ets1 gain (Cd8) or loss (Tcrg) at critical T-cell genes during DN to DP transition. Genome browser screenshots of the Cd8 and Tcrg loci (left, right) showing Ets1, Pol II, H3K4me1/3, H3K27me3 ChIP-seq coverages and gene expression fold changes in DN and DP. (B) Correlation between Ets1 and gene expression fold changes. Heatmaps of increasing DP/DN Ets1 (left) and corresponding gene expression fold changes (right). (C) Significance of gene expression differences by increasing Ets1-fold change. Expression fold changes for all classes, shown as a boxplot. Significance levels for each comparison are indicated above, with 1 star denoting P < 0.05, 2 stars denoting P < 0.01 and 3 stars denoting P < 0.005. (D) Genes highly differentially regulated by Ets1 correspond to tissue-specific genes. Left: hierarchical clustering of significantly differentially regulated and Ets1 bound genes showing 4 distinct gene clusters. Right: tissue specificity of genes expressed as percentages of each cluster. Significance levels for enrichments are indicated as in (C). (E) Genes differentially regulated by Ets1 correspond to stage-specific genes. Gene ontology analysis of groups of bottom and top 1000 DP/DN enriched genes bound by Ets1 (left, right). For each group, the top 10 gene ontology classes are shown with their corresponding enrichment in the other group. (F) DN and DP Ets1 datasets correlate with Non T-cell hematopoietic and T-cell lineages, respectively. Hierarchical clustering of pair-wise Spearman correlation coefficients of Ets1 ChIP-seq tag counts from DN, DP, non T-cell hematopoietic and T-cell lineages in distal Ets1 DN + DP peaks.
We next focused essentially on distal Ets1 sites, since they represented the majority of peaks, and since previous studies by others and us established a more specific role for distal Ets1 sites during T-cell differentiation (22,37). This was also in light of previous reports having linked in vivo T-cell differentiation with distal rather than proximal regulation (26). We computed and sorted the union of distal peaks by increasing Ets1 DP/DN fold change (DN + DP peaks, 3900 sites, see Supplementary Table S4 for a complete list) and subdivided them into five distinct classes (also corresponding to unique densities of Ets1 DN or DP signal, see Supplementary Figure S2A and B). Known hallmark genes included Il2ra, for which Ets1 binding and gene expression were strongly reduced in DP (DN-specific class 1), and Il7r (DN-specific class 2), moderately reduced, Cd3g, stable for both (shared class 4), and Satb1, for which both Ets1 and gene expression strongly increased (DP-specific class 5). To gain further insights as to the role of Ets1 in transactivation of gene expression, we computed and plotted expression fold changes of genes nearest to distal peaks and observed a noticeable parallel between Ets1 and gene expression fold changes (Figure 1B), whereby each class of increasing Ets1-fold change showed significantly higher expression fold change compared to its neighbor (Figure 1C, P = 1.9 × 10−3, P = 4 × 10−3, P = 2.6 × 10−4 and P = 7.5 × 10−9, respectively). Not only did this prove to be a significantly positive correlation, but also interestingly, gene expression fold change likewise seemed to be positively correlated with changes in the number of Ets1 peaks nearest the gene (Supplementary Figure S2C and D).
To ascertain that Rag2−/− thymocytes were a valid model for DN3 thymocytes, we retrieved mRNA expression datasets from the Immunologial Genome project (61). We performed hierarchical correlation clustering in differentially regulated genes and noted that Rag2−/− thymocytes were most closely related to DN3 thymocytes (Supplementary Figure S3A). Additionally, we ranked DN3 and DP expression data from another study (56) by increasing Ets1 DP/DN fold change and also observed a direct correlation with expression fold change (Supplementary Figure S3B), albeit slightly lower.
We next aimed to identify genes regulated differentially by Ets1 between DN and DP, as well as potentially diverse ontologies reflected by trends of changes in gene expression. To this end, we selected 203 genes varying significantly in both Ets1 binding and gene expression (≥2σ significance). Hierarchical clustering allowed us to identify four groups of genes corresponding to different combinations of Ets1 and gene expression fold changes. Positive and negative changes of gene expression mostly corresponded to positive and negative changes to Ets1 binding, respectively, with a minority of negatively correlated Ets1 and gene expression fold change clusters (Figure 1D). When examining tissue specificities of genes encompassed by these clusters, we noticed that DP/DN upregulated genes were highly enriched in thymus-specific genes (P = 8.6 × 10−5), while downregulated genes were enriched in non T-cell hematopoietic genes (P = 2 × 10−3). Minor patterns of peaks associated with genes showing discordant changes in Ets1 and expression levels were also observed. Upregulated genes losing Ets1 were enriched in thymus genes (P = 0.019) while downregulated genes acquiring Ets1 were enriched in non T-cell hematopoietic genes (P = 0.033). Finally, we proceeded to identify potentially enriched ontology classes in Ets1 DP/DN down- and upregulated genes using the DP/DN top- and bottom-most enriched distal sites. Importantly, our results showed that Ets1 DN-enriched sites were preferentially associated with genes involved in non T-cell hematopoietic processes whereas DP-enriched sites were associated with thymus and T-cell specific genes. (Figure 1E). Analysis of Ets1 ChIP-seq datasets from non T-cell hematopoietic lineages (G1ME, CH12, Mel, (27,28,30)) as well as fully differentiated T-cell lineages (resting CD4+, Treg, Th2, (26,29)) revealed high similarity between mature T-cell lineages and DP but not DN stages, which, along with non T-cell hematopoietic lineages, were more distantly related (Figure 1F and Supplementary Figure S3C). The heterogeneous group encompassing DN thymocytes and other lineages exhibited Ets1 binding to general hematopoietic genes such as Cbfa2t3 (which encodes for Eto-2), downregulated in DP and mature T-cells, and conversely DP/mature T-cell genes such as Tox were bound by Ets1 at these stages but not in DN and other lineages (Supplementary Figure S3D).
Overall, we show that distal Ets1 sites are associated with active genes in both DN and DP, and could mediate a switch from non- T-cell hematopoietic towards T-cell specific gene expression programs during the DN to DP transition.
Loss of Ets1 impairs activation of thymus gene expression programs and reduces commitment to the T-cell lineage
To better ascertain the functional role of Ets1 and its involvement in the switch of expression programs in the thymus, we generated Ets1−/− DP and control DP gene expression microarray datasets. For this, we used DP cells sorted from Rag2−/− thymuses reconstituted with Ets1−/− fetal liver cells (4). Strikingly, and in line with our observations described above, we observed that DP Ets1−/− versus WT upregulated genes corresponded to general hematopoietic genes, while the ones lost correspond to thymus and T-cell specific genes (Figure 2A). We confirmed these findings by retrieving gene expression data from the Immunological Genome project (55) for all DN, DP, T-cell stages and for other relevant hematopoietic lineages: common lymphoid progenitors (CLP), CD19+ B-cells, dendritic cells (DC), granulocyte macrophage progenitors and NK cells, as well as other T-cell datasets not present at the Immunological Genome Project: Th1, Th2, Th17 and Treg (57–59). Importantly, we performed PCA on merged datasets using expression data from all DP Ets1−/− up- and downregulated genes and observed that DP Ets1−/− gene expression values correlated with values from earliest thymic progenitors (ETP) as well as from other hematopoietic lineages (Figure 2B). As expected, the DP WT dataset clustered with other DP datasets. Upregulated genes In DP Ets1−/- included genes from the B-cell lineage, such as Cd19, Ms4a1, Cd79b, IgH, IgK and SpiB, as well genes from myeloid lineages, such as Fcgr3, Itgam, Cd14, Tlr9 and DN genes, such as Ptcra, Ctla4, Cd44, Il2ra and Il7r (Supplementary Table S5). Conversely, downregulated genes included DP and post β-selection genes, such as Tcra, Tcrb, Ets2, Ltb, Plxnd1, Cd5 and Tnfsf9. Of note, Cd8a, Cd8b1, Cd4 and Cd28 expression was not affected, which suggests a compensatory mechanism in the absence of Ets1. These effects were also visible when using changes in gene expression against DP (Supplementary Figure S4A). These trends were confirmed by gene expression levels in DP Ets1−/− which were high and matched those in other lineages such as DC, CLP and CD19+ B-cells in DP Ets1−/− upregulated genes, and were lower than those of DP-related stages in DP Ets1−/− downregulated genes (Figure 2C). This indicates that (i) genes transactivated by Ets1 at the DP stage are DP-specific genes (ii) in the absence of Ets1, DP cells are still primed toward other hematopoietic lineages, suggesting that the loss of Ets1 reduces commitment to the T-cell lineage altogether.
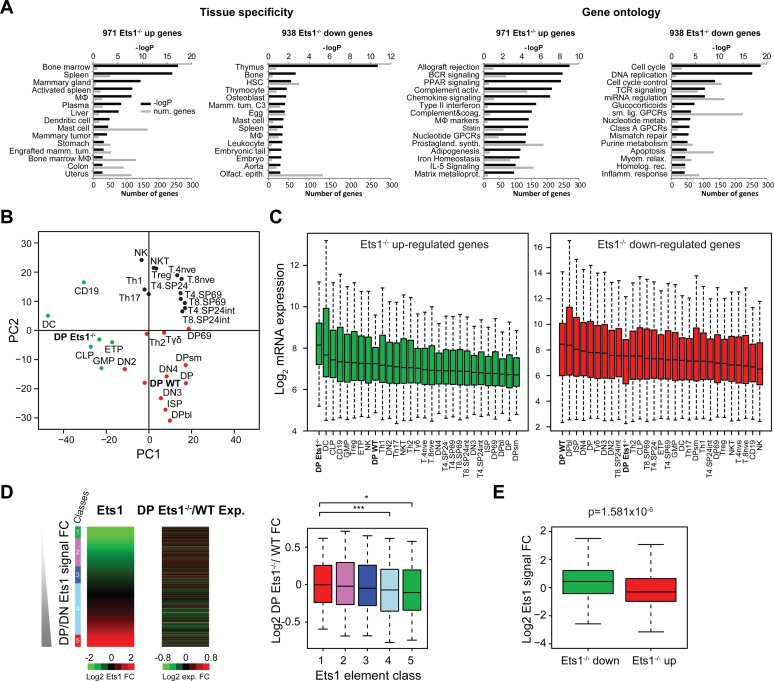
Figure 2.
Loss of Ets1 impairs the thymus and T-cell gene expression program switch. (A) Tissue-specificity and gene ontology (Wikipathways) of Ets1−/− up- and downregulated genes. –Log P-values are indicated in black; numbers of genes for each term are indicated in gray. MΦ: macrophage. (B) PCA of microarray gene expression values in DP Ets1−/− and WT datasets, as well relevant Immunological Genome datasets in T-cells and in other hematopoietic lineages with the addition of Th1, Th2, Th17 and Treg datasets from other studies in both Ets1−/− up- and downregulated genes. DP Ets1−/− and WT cells are indicated in bold. (C) Gene expression boxplots for datasets described in (B) in Ets1−/− -up (green) and -downregulated genes (red). Datasets are ranked by decreasing median expression value. DP Ets1−/− and WT datasets are indicated in bold. (D and E) Anticorrelation of WT DP/DN Ets1-fold change (D, left heatmap) and Ets1−/−/WT DP gene expression change (D, right heatmap). Ets1−/− / WT gene expression fold change values are shown as boxplots for each class of Ets1 DP/DN fold change (D, right). Significance levels for enrichments are indicated as in Figure 1C. (E) Boxplot showing Ets1 DP/DN fold change for Ets1−/− up- (green) and downregulated genes (red).
We next wanted to relate these changes with those observed in Ets1 binding between the DN and DP stages. Crucially, by retrieving DP Ets1−/−/WT fold changes for Ets1 DN + DP peaks, we noted an inverse correlation between the two, indicating that the loss of Ets1 results in a block of gene expression programs, whereby the thymus-specific program genes bound by Ets1 at DP-specific (class 5) were not activated and the DN program (classes 1–3) was not repressed (Figure 2D and E). Significant decreases in gene expression could be seen for genes associated with shared, class 4 sites and DP-specific compared to DN-specific class 1 sites (P = 5.1 × 10−4 and P = 0.037, respectively). Genes already bound in DN and lost with the deletion of Ets1 included thymus genes likely corresponding to DN/DP shared genes, as well as other general hematopoietic genes (Supplementary Figure S4B and C). This was also the case for such genes bound in DP. However, genes upregulated by the loss of Ets1 corresponded to genes for other lineages and not T-cells. A number of genes bound in DP and up- or downregulated in DP Ets1−/− cells were also bound in DN, however the overlap with Ets1 bound genes and genes up-regulated by the loss of Ets1 was not significant, likely indicating the up-regulation of those as an off-target effect rather than a direct lift of repression by Ets1 (Supplementary Figure S4C).
Overall, our findings confirm, at the genome-wide scale, previous findings that the loss of Ets1 impairs the DN to DP transition (4) by also reducing commitment to the T-cell lineage. They imply that Ets1 plays a critical role in the transactivation of T-cell and thymus genes, with its loss corresponding to loss of gene expression. These results also suggest that the transactivating properties of Ets1 are more prominent at the DP stage where its deletion directly causes loss of expression of bound genes, while genes bound by Ets1 at the DN are expressed at this stage but can also be expressed in the absence of Ets1 at the DP stage.
Dynamic of core transcriptional and epigenetic features at Ets1 binding sites
We previously described distal T-cell specific elements, which were characterized by the presence of Pol II and both epigenetic marks H3K4me1 and me3, as well as being enriched for Ets1 in DP cells (37). Since we observed differential transactivation of Ets1-bound genes during the DN to DP transition, we next wished to determine whether these transcriptional and epigenetic hallmarks, reflecting active and tissue-specific enhancers, also followed this differential pattern. Since H3K4me1 and me3 levels also reflect enhancer activity, we also sought to determine whether differences in those levels could be seen in DN-specific, DN-DP shared and DP-specific sites in light of the differential effect of the loss of Ets1 pointing to altered transcriptional properties for Ets1 between those stages. We focused on Ets1 binding sites and plotted distal H3K4me1, H3K4me3 as well as Pol II profiles sorted by Ets1 DP/DN fold change as before. Signal variations for these marks broadly mirrored those of Ets1 essentially in DP, and to a lesser extent in DN (Figure 3A). This did not seem to be the case at promoter regions, for which signal for these marks stayed overall unchanged as ranked by Ets1 DP/DN signal or as total signal in DN and DP, as opposed to distal regions (Supplementary Figure S5A and B). When further examining signals of histone modification marks at distal elements for both stages, we noted that while elements from class 1–3 displayed high levels of H3K4me1, H3K4me3 levels were comparatively lower in DN cells (Figure 3A). Since H3K4me3 has been associated with active enhancers (39), this could indicate a less active state of chromatin, perhaps poised for repression of the nearest gene at the next stage of differentiation. Further, we noted that distal DP-specific class 5 sites bore substantial levels of H3K4me1 and H3K4me3 in DN, and that conversely DN-specific class 1 sites showed residual H3K4me1 but much lower H3K4me3 (Figure 3A and B). This suggests H3K4me1/3 priming in class 5 at the DN stage and de-methylation of H3K4me3 for class 1 at the DP stage. As exemplified at the E8II enhancer of the Cd8 locus, and at the Tcra and Tcrb enhancers, Ets1 in fact seemed to recruit Pol II as well as promoting higher H3K4me3 in some cases (Figure 1A, Supplementary Figure S5C). While overall, changes in Ets1 correlated with those of active transcriptional hallmarks (Figure 5D), correlation plots between signals for Ets1 signal and these marks showed overall higher correlation at the DP stage (Supplementary Figure S5E). Thus, at the DP stage, Ets1 binding correlates with transcriptional and epigenetic marks whereas Ets1 variations at the DN stage are weak in class 1–4 and show lower correlation with these marks.
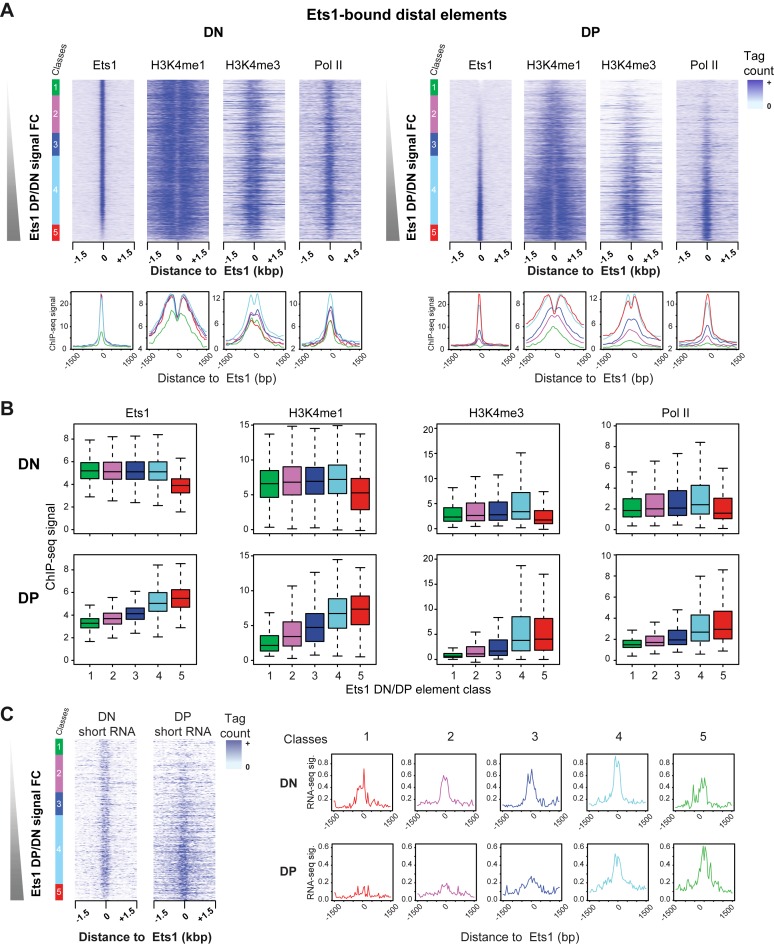
Figure 3.
Core hallmarks of transcription activation mirror Ets1 binding between DN and DP stages. (A) Dynamic recruitment of Ets1 and core transcriptional hallmarks at distal Ets1 binding sites. Heatmaps of Ets1, H3K4me1, H3K4me3 and Pol II signals [−1500; +1500 bp] around distal Ets1-bound sites sorted by increasing Ets1 DP/DN fold change (top) in DN and DP stages (left, right). Average profiles for each class is shown with its corresponding color (bottom). (B) Quantitatively differential recruitment of Ets1 and core transcriptional hallmarks at distal Ets1 binding sites. Boxplots of signals for each mark for all classes are shown in both DN and DP. (C) Mirroring of short RNA signals and Ets1-fold change. Left: heatmaps of DN and DP short RNA-seq tag counts sorted by increasing Ets1 DP/DN fold change in distal Ets1 DN + DP sites. Classes are indicated left of the heatmaps. Right: average profiles for all classes in DN and DP.
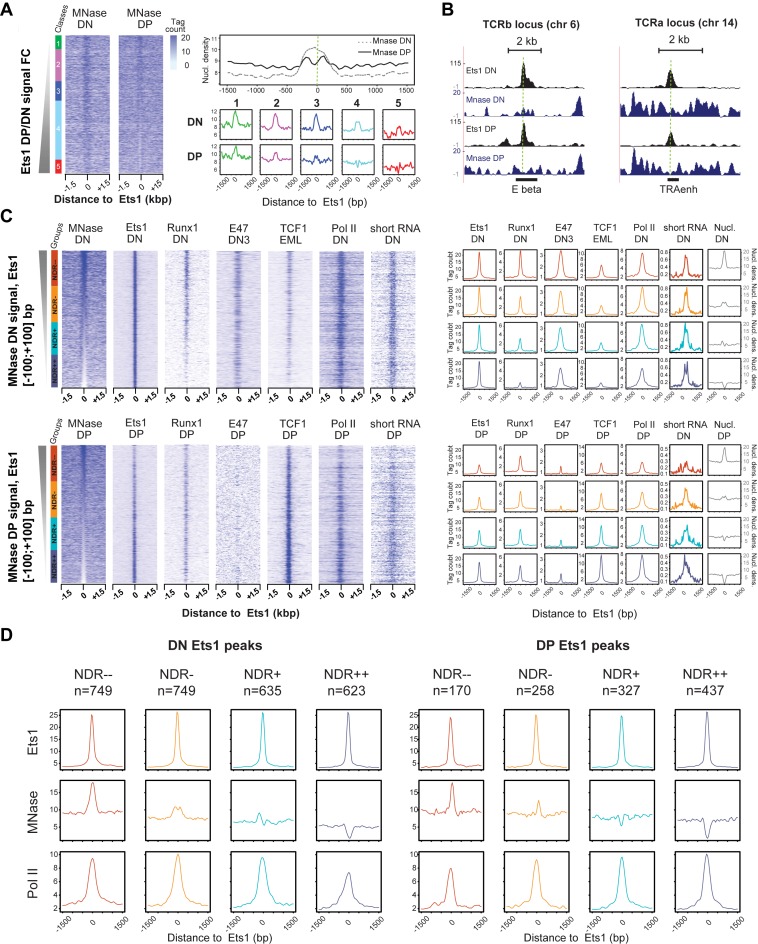
Figure 5.
Stage-specific and co-association-mediated NDRs around Ets1-bound sites. (A) Stronger presence of NDRs in DP Ets1-bound sites. Left: MNase DN and DP heatmaps in Ets1-bound distal elements sorted by increasing Ets1 DP/DN fold change. Right: global DN and DP average profiles for MNase-seq tag counts in all Ets1 DN + DP sites (top), and DN and DP average profiles for MNase tag counts in all classes (bottom). (B) Example screenshots of DP-specific NDR in the TCRb and Tcra enhancers. (C) Stage-specific enrichments for Ets1, partner TFs as well as Pol II in NDRs and nucleosome-occupied regions (NORs) in DN + DP Ets1 peaks. Heatmaps of tag counts of MNase-seq, ChIP-seq for Ets1, partner TFs and Pol II sorted by increasing DN (top) and DP (bottom) MNase-seq. Corresponding average ChIP-seq profiles for each class of NDRs are shown on the right (colored), as well as average nucleosomal density in each case (gray). The numbers of Ets1 peaks for NDR−−, NDR−, NDR+ and NDR++ classes are indicated above each graph. (D) Altered Pol II recruitment in Ets1-bound nucleosome -occupied and -depleted regions between the DN and DP stages. Average profiles for Ets1, MNase and Pol II high-throughput sequencing of Ets1-bound classes of nucleosome occupancy levels at the DN and DP stages (left, right).
To further investigate the transcriptional activity and potential eRNAs associated with Ets1 distal regulatory regions, we performed short RNA-seq (15–50 nt) in DN cells and compared it with our previously described DP dataset (38). This protocol, based on size selection of RNAs, allows for enriching for paused transcripts found both at promoters and enhancers. We found that eRNAs essentially recapitulated Pol II patterns during DN to DP transition as well as those of Ets1 in DP only, suggesting highest enhancer activity in class 4 at the DN stage and classes 4–5 at the DP stage (Figure 3C). Thus, distal Ets1 enhancers broadly recruit Pol II that is able to generate paused transcripts.
Altogether, our results suggest that at distal sites, rather than at promoters, core transcriptional epigenetic hallmarks globally correlate with Ets1 binding in a dynamic, stage-specific (DP) manner, although there are some notable exceptions whereby recruitment of active transcriptional marks (H3K4me3 and Pol II) in DP is preceded by Ets1 binding in DN accompanied by high H3K4me1 levels. Importantly, these results also pointed to higher enhancer activity in DN-DP shared and DP-specific sites.
Ets1 is dynamically co-associated with increasingly T-cell specific motifs and TFs during the DN to DP transition
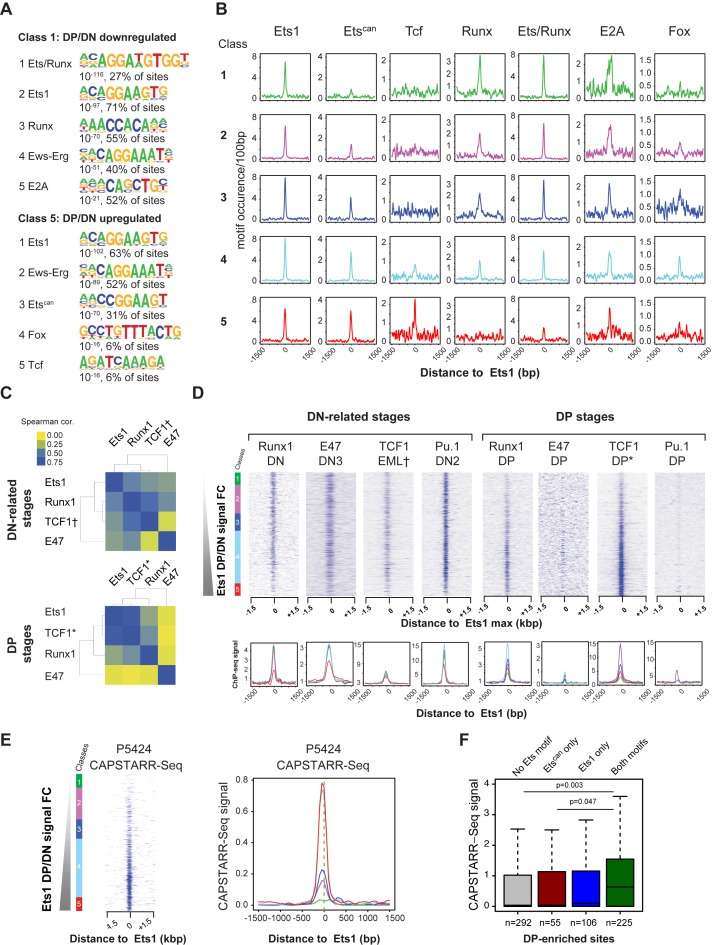
Several co-factors of Ets1 were previously described, such as Runx1 (22), or even itself via homo-dimerization (62). To gain further insights as to whether Ets1 alone, or via co-association, is responsible for switching gene expression programs and if co-association with any TF could explain the observed differential properties of Ets1, we first proceeded to identify potential co-factors using motif discovery in different classes of distal Ets1 DN + DP peaks. Enriched motifs appeared differentially represented when comparing DN- and DP-specific sites (classes 1 and 5), with the Ets/Runx (composite), Pu1-like Ets1 (AGGAAG, Ets1), Runx, Erg and E-box (E2A) motifs over-represented in class 1, and Ets1, Erg, a second, distinct canonical Ets motif (CGGAAG, Etscan), Fox and Tcf (HMG) motifs in class 5 (Figure 4A).
Figure 4.
Differential motif and TF co-association of Ets1 in DN and DP. (A) Specific co-occurrence of Ets1 with Runx motifs in DN and Tcf motifs in DP in Ets1 DN + DP sites. Motif discovery in DN- and DP-specific distal Ets1 sites (classes 1 and 5) showing differential motif content. (B) Decrease of DN-specific and increase of DP-specific motif occurrences with increasing Ets1 DP/DN fold change. Average profiles of motif frequencies by increasing Ets1 DP/DN fold change class. All Ets1 DN + DP classes are shown with a different color, [−1500; +1500 bp] around the Ets1 summit for each motif in each class. (C) Specific co-association of Ets1 with Runx1 in DN and TCF1 in DP in Ets1 DN + DP sites. Spearman correlation clustering of Ets1, Runx1, TCF1 and E47 ChIP signals in Ets1 DN + DP sites in DN (top) and DP (bottom). Datasets from EML cells are marked by a dagger sign (†). Datasets from total thymus are indicated by an asterisk (*). (D) Ets1-dictated occupancies of Runx1, E47, TCF1 and Pu.1 in Ets1 DN + DP sites. Top: heatmaps of ChIP-seq tag counts for Runx1, E47, TCF1, Pu.1 in DN (left) and DP (right) sorted by increasing Ets1 DP/DN fold change. All Ets1 DN + DP classes are shown with a different color. Bottom: average binding profiles [−1500; +1500 bp] around the Ets1 summit for each factor in each class. (E) P5424 CAPSTARR-Seq signal heatmaps and average profiles by increasing Ets1 DP/DN fold change. (F) P5424 CAPSTARR-Seq enhancer activity of Ets1 DP-specific sites with various Ets motif combinations in Ets1 DP-specific sites.
We next sought to characterize the occurrence of these motifs throughout all Ets1 DN + DP classes. To this end, we looked for DN- and DP-specific (class 1 and 5) enriched motifs around Ets1 summits, and plotted hits sorted by increasing Ets1 DP/DN fold change (Figure 4B, Supplementary Figure S6A). We noted that while the main Ets1 motif expectedly showed no differential representation, the Etscan as well as Tcf motifs displayed gradually higher frequencies with increased Ets1 DP/DN fold change. Conversely, Runx and the composite Ets/Runx motifs showed reverse trends, with motifs for Ets/Runx (composite) nearly absent and Runx completely absent in DP-specific sites. Since these over- and under-representations may well be independent, we performed motif co-occurrence clustering to ascertain that these hits could theoretically correspond to enriched transcription factor binding sites (TFBS) of factors associated to the same location. Interestingly, we did notice very strong co-association for the Ets and Runx motifs in DN but much lower in DP, where Ets1 motifs mostly co-localized with Etscan, FoxO1 and Tcf motifs (Supplementary Figure S6B). E-box motifs (E2A) seemed to cluster at equal frequency with Ets1 in both stages.
To study whether the Etscan motif could potentially correspond to a second, distinct TFBS for an additional Ets1 protein (or other Ets family member), we measured DP Ets1 ChIP-seq signal subdividing de novo sites according to their different Ets motif content, as previously performed for other TFs (29). We found that DP-enriched sites containing the Etscan motif alone showed no significantly higher signal than sites with no Ets motifs (Supplementary Figure S6C, P = 0.74), but that sites with the Ets1 motif only showed significantly higher binding than those with no motifs and Etscan only (P = 5.2 × 10−3 and P = 0.04, respectively). We also found that sites with both Ets1 and Etscan motifs had higher ChIP-Seq signal compared to sites with no motif, Etscan only and Ets1 only (P = 2.5 × 10−8, P = 3.2 × 10−4 and P = 6 × 10−6 and respectively). This indicates that sites with both motifs likely correspond to two binding events and thus a higher propensity of Ets1 to homo- or hetero-dimerize with another Ets protein bound to a secondary, distinct motif in de novo DP sites. Interestingly, Etscan motifs exhibited preferential positioning at −4 bp of Ets1 motifs, resulting in a CGGAAGGAAG composite motif for 10 sites, of which 8 were class 4 peaks associated with critical T-cell genes such as Lef1 and Cstad (Supplementary Figure S6D and Table S6).
To confirm whether Runx1, E47 and TCF1, the predominant TFs expressed at both stages actually co-localized with Ets1 in DN and DP, we performed ChIP-seq for Runx1 in DN, E47 in DP and retrieved publicly available data for E47 in DN3 (54), TCF1 in EML cells, a model related to ETPs (51,63) as well as whole thymus (representing essentially DP cells, typically 80% CD4+/CD8+ pure) TCF1 (52,53) and Runx1 in DP (49). We subsequently obtained corresponding tag counts around DP/DN sorted Ets1 summits. Hierarchical clustering of tag-count correlations revealed that Runx1 signal more closely mirrored that of Ets1 in DN than that of TCF1, although this trend was reversed in DP (Figure 4C), in agreement with our earlier observations. E47 seemed distantly related at both stages, albeit less in DN. We also confirmed a lower number of common Ets1/Runx1 peaks in DP, both as percentage of Ets1 and Runx1 peaks (Supplementary Figure S6E). Heatmap representation of tag counts around Ets1 summits also revealed that Runx1 binding was mostly restricted to classes 1–4 in DN, and to class 4 in DP, with signal lower in class 5 where few Ets/Runx or Runx motifs were present (Figure 4B and D). TCF1 signal was also higher and closely followed Ets1 sites in DP as compared to DN. We further validated the dynamic recruitment of TCF1 and Runx1 by ChIP-qPCR at the Tcra enhancer, which is bound by similar levels of Ets1 in both DN and DP thymocytes (see Figure 5B). Indeed, activation of the Tcra locus in DP thymocytes correlates with specific recruitment of TCF1 at this stage and reduced binding of Runx1 (Supplementary Figure S6F). As for E47, while its signal followed a pattern broadly similar to Ets1 in DN, we found it to be restricted to very few sites and thus near-absent in DP. Additionally, having noticed that our main Ets1 motif closely resembled that of Pu.1, and since Pu.1 expression is abolished in DN3, we wished to determine whether Ets1 could potentially take over DN2 Pu.1-bound sites in DN3. For this purpose, we also retrieved DN2 and DP Pu.1 ChIP-seq datasets (11). DN2 Pu.1 signal very closely matched that of DN Ets1, whereby more than one in four DN Ets1 site was occupied by Pu.1 in DN2, a stage anterior to β-selection (Figure 4D, Supplementary Figure S6G). Examples of stage-specific loci, such as Ptcra, featuring high Ets1, Runx1, E47, DN2 Pu.1 but not TCF1 in DN and background signal in DP, as well as Bambi-ps1 (a DP-specific gene), featuring background signal in DN and high Ets1, TCF1 but not Runx1 nor E47 signals in DP are shown in Supplementary Figure S6H. At these sites, eRNA signal was also detected, indicating that they are active regulatory elements. Since we had observed signs for higher transcriptional activity at the DP stage (Figure 3), we wanted to determine whether these sites, characterized by enrichment for distinctive motifs, could correspond to enhancer regions activating the thymus and T-cell gene expression program. To relate Ets1 binding to actual enhancer activity, we retrieved enhancer scores from a high-throughput reporter assay, CAPSTARR-Seq, previously performed in the DP-related P5424 thymic cell line (50). These enhancer scores were ranked by increasing DP/DN Ets1-fold change. A direct parallel between Ets1-fold change and enhancer activity was observed, with class 4 and 5 sites showing the highest enhancer activity in P5424 cells (Figure 4E). The impact of the presence of Etscan on enhancer activity was also investigated. To retrieve enough motifs, we isolated the top 10% DP-enriched sites and split peaks containing (i) no Ets motif, (ii) Etscan only, (iii) Ets1 only and (iv) both motifs. We found that overall, only those peaks containing both motifs showed noticeable enhancer activity compared to peaks with no Ets motif or bearing only Etscan (Figure 4F, P = 0.0032 and P = 0.0476, respectively). This suggests a critical role for the co-association of Etscan and Ets1 motifs at the DP stage.
Based on previous reports that the Etscan motif was found at promoters (22,25), we further wanted to determine whether its over-representation patterns there as well as other motifs described in the manuscript followed the trends observed at distal sites. The densities of all candidate motifs including Etscan were plotted as in Supplementary Figure S7A. Interestingly, while we saw no differential representation of Etscan between DN-up, shared and DP-up sites, the distal Ets motif seemed to be under-represented in DN-specific promoters and increasingly enriched in shared and DP-specific promoters. The Ets-Runx motif was lost in DP-specific sites, while other motifs were not over-represented in promoter regions. Intriguingly, since this motif is composed of a CpG dinucleotide, the question as to whether these sites are found in intergenic CpG islands arose. When plotting the occurrence of CpG islands by increasing DP/DN Ets1-fold change, we did not notice over-representation of those in classes where the Etscan motif was found (Supplementary Figure S7B). Only 76 occurrences of Ets1 peaks overlapped with annotated CpG islands and only one significantly enriched motif was found, an Ets/Runx composite motif (Supplementary Figure S7C). This suggests that these motifs are protected from DNA methylation outside of CpG islands by successive occupancy, at least for DN-DP shared sites, by e.g. Pu.1 at previous stages (Figure 4, Supplementary Figure S6F) or other TFs such as Fli1 at even earlier stages of hematopoietic development (64).
Taken together, these results show differential co-association of Ets1 in DN and DP, with (i) Runx1, E47 in sites from classes 1–3 in DN only, (ii) Runx1, TCF1 in class 4 sites in both DN and DP, with E47 binding only in DN and (iii) Etscan and to TCF1. This suggests that all these TFs are likely part of a same complex in DN and DP, with Runx1 and TCF1 in turn binding directly to stage-specific enhancers, except for E47, which is lost in DP.
Differential properties of Ets1 nucleosomal occupancy at regulatory elements
Since we had observed dynamic recruitment of transcriptional hallmarks, as well as a change of co-factors between DN and DP, we wondered whether Ets1 could by itself be acting as or associate itself with a pioneering factor in DP, given that Ets1 was previously described to be capable to bind nucleosomal DNA (65). We also wished to determine whether chromatin accessibility was identical in DN-specific, DN-DP shared and DP-specific classes, hypothesizing that it would not in light of the differential enhancer activity that was observed for these classes. To address this question, we performed genome-wide nucleosome mapping via MNase-seq in DN and DP thymocytes. Strikingly, upon plotting MNase tag densities around Ets1 summits sorted by DP/DN fold change, a well-positioned nucleosome was visible as a dominant feature at distal Ets1-bound DN sites, while conversely a dominant NDR was instead observed at distal Ets1-bound DP sites (Figure 5A). Well characterized such examples include the Tcrb and Tcra enhancers (Figure 5B). Interestingly, a nucleosome was visible in all Ets1 distal element classes in DN thymocytes, as well as in Ets1-unbound classes in DP (classes 1–2), with the NDR only present in classes 3–5 at this stage (Figure 4A, bottom right panel).
To gain insights with regards to these differences, we consequently wished to identify which TFs, if any, were more associated with Ets1-bound nucleosome-depleted and NORs. Since some of the (Ets1 DP/DN-sorted) average nucleosomal profiles could be resulting from a combination of mixed NDR and NOR profiles, we first identified NDRs in both DN and DP, by ranking the union of DN + DP Ets1 sites by decreasing nucleosome density (Figure 5C). We subsequently identified which distal Ets1 DN + DP sites were NDRs and found that these represented around 50% of sites in both cases (DN: 2044 NORs, 1856 NDRs; DP: 2072 NORs, 1828 NDRss). Thus, this analysis reveals that Ets1 carries the ability to bind either NORs or NDRs at both differentiation stages.
NDR-sorted peaks were further divided into equally-sized classes: 2 NORs classes, and 2 NDRs-associated classes. By plotting corresponding signals for all TFs, this revealed that (i) Ets1 indeed binds both NDR and NORs sites in DN and DP but is more restricted to NDRs in DP, (ii) Runx1 is restricted to NORs in DN, (iii) TCF1 is mainly found in DP NDRs and (iv) NDRs are less transcriptionally active than NORs in DN since they display less Pol II signal, and vice versa in DP, whereby NDRs displayed increased Pol II occupancy. In DN, Ets1 equally efficiently bound both NDR and nucleosomal regions, while in DP Ets1 binding is contingent on nucleosome depletion. This strongly suggests a dynamic switch in the chromatin binding properties of Ets1 during the DN to DP transition. We further extended our analyses to H3K4me1/3 epigenetic marks, using the same analytical set-up (Supplementary Figure S8A). Consistent with increased Ets1, TCF1 and Pol II in NDRs (specifically in DP cells), both methylation marks correlated with decreased nucleosome occupancy in DP, but not in DN, where only H3K4me1 decreased with nucleosome occupancy. Finally, gene expression of genes adjacent to Ets1 distal regions followed the same trend (Supplementary Figure S8B), further strengthening the possibility that Ets1 relates more directly to enhancer activity in DP than it does in DN.
Since DN NDRs showed lower Pol II signal than DN NORs (Figure 5C), we wished to determine whether this held also true only for Ets1-bound NDRs. We computed average profiles of Ets1, Pol II and nucleosome densities around Ets1 sites bound either in DN or DP. This analysis revealed a switch in the properties of Ets1 binding to distal sites as Ets1 associated predominantly to NDRs in DP (65% of sites) whereas it was slightly more associated to NORs in DN (55%, Figure 5D). Interestingly, we observed that Pol II bound more NORs in DN and more NDRs in DP emphasizing another transition at Ets1-bound sites. This entails a switch in the transcriptional properties between DN and DP with regards to nucleosome occupancy and Pol II recruitment.
In light of the dual ability of Ets1 to bind both nucleosome-occupied and depleted regions, we next set out to establish whether or not Ets1 could be a pioneer TF. First, we tested if Ets1 bound-sites in T-cells were bound by nucleosomes in vitro (32) or in another primary cell type (mESC) (38). We observed that the union of Ets1 DN + DP target sites was nucleosome-occupied in both cases (Supplementary Figure S9A). This suggests that by default, these sites correspond to a closed chromatin conformation, which is seemingly opened during early T-cell differentiation. To further determine whether Ets1 is responsible for this mechanism, we isolated four classes from DN-DP shared Ets1 sites, corresponding to trends in nucleosome occupancy changes (DN NDR → DP NDR, DN NDR → DP NOR, DN NOR → DP NDR and DN NOR → DP NOR). If Ets1 is a pioneer TF, gain of active epigenetic and transcriptional marks should be observed at enhancers associated with appearance of NDR (class DN NOR → DP NDR, Supplementary Figure S9B). While we observed enhanced nucleosome phasing in this class, we also noted pre-existing Pol II recruitment in DN and no change in Pol II levels between both stages. This was also true for H3K4me1 and H3K4me3 (data not shown). The observed Pol II signal was also the strongest as compared to the other classes, suggesting high transcriptional activity for these sites. This result was supported by CAPSTARR-Seq levels closely matching those of Pol II in Ets1-bound, DP NOR and NDR regions (Figure 5D, Supplementary Figure S9C), differential enhancer activity between Ets1-bound DN NOR and DP NDR regions. Further, since we had seen that Ets1 binding was more related to its potential enhancer activity in DP NDR regions (with higher levels of Pol II, Figure 5D), we wished to identify a potential mechanism. Ets1 could potentially be bringing in the transcriptional machinery without necessarily engaging Pol II in DN. We plotted the distance of Pol II peaks relative to the Ets1 motif orientation in both stages and noticed that while Pol II was centered on Ets1 motifs in DN, it was slightly downstream of Ets1 motifs in DP, suggesting that Pol II is engaged by Es1 in DP, where its enhancer activity is fully achieved (Supplementary Figure S9D). According to this model, Pol II would be stalled against a nucleosome in DN but not in DP and recruitment of Pol II by Ets1 could be directional, in agreement with results pertaining to pioneer TFs and directional chromatin opening (66). We conclude that Ets1 is able to bind nucleosome-bound DNA together with the transcription machinery, suggesting that it might prime transcription activation by recruiting Pol II on nucleosomes.
In summary our analyses reveal that Ets1 can bind both NORs and NDRs at both differentiation stages. IN DP, increased TCF1 and Pol II are found associated with Ets1 at NDRs as well as active epigenetic marking by H3K4me1 and me3, overall indicative of more active enhancers. It is also remarkable that Runx1 shows a clear preference for nucleosome-bound Ets1 in DN but not in DP. This observation is also true to a lesser extent for TCF1 sites in EML cells (more related to DN). Altogether, our data suggest that Ets1 properties as regard to gene activation and chromatin remodeling have dramatically changed between the DN to DP transition, allowing this TF to become a more crucial and direct transcriptional activator.
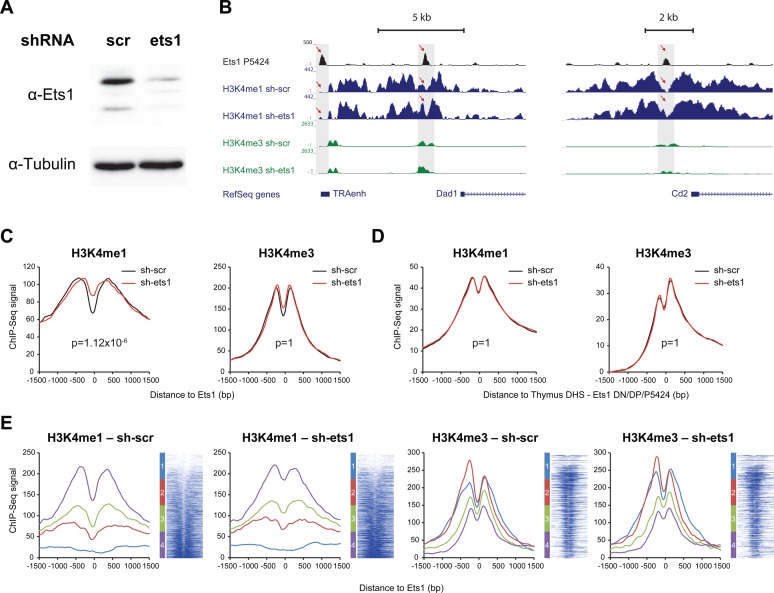
Ets1 induces chromatin remodeling of H3K4me1-marked nucleosomes
To test whether Ets1 does actually induce chromatin remodeling, we performed knockdown of Ets1 in the P5424 thymic cell line using shRNA (sh-ets1), alongside with a scramble shRNA control (sh-scr). After 48 h, marked decreases of Ets1 protein levels could be observed in P5424 cells having been transfected with sh-ets1 (Figure 6A). We next conducted ChIP-Seq experiments for H3K4me1 and H3K4me3 in both conditions, as well as ChIP-Seq for Ets1 in the untransfected P5424 cell line. We detected 3565 Ets1 peaks in P5424 cells. Increases of H3K4me1 were observed by visual inspection at several Ets1 sites, including at the Tcra and Cd2 loci (Figure 6B). To determine whether this finding held true genome-wide, we isolated distal Ets1 sites and retrieved signals for H3K4me1 and H3K4me3 around Ets1 summits. Our results show global increases only of H3K4me1 nucleosomes around Ets1 sites (Figure 6C, Supplementary Figure S10A) together with an essentially unaffected H3K4me3 signal. Critically, we showed that this effect was specific to Ets1-bound sites since no differences were observed using control sites, comprising ENCODE thymus DHSs depleted of Ets1 peaks in DN, DP and P5424 cells (Figure 6D). By defining four classes based on increasing sh-scr H3K4me1 [−500, +500 bp] around Ets1, we determined that this effect was homogenous, apart for sites originally depleted in H3K4me1 (Figure 6E). This effect also coincided where Ets1 signal was maximal (Supplementary Figure S10B). Importantly, Ets1 binding patterns in P5424 cells mirrored those of DP Ets1 (Supplementary Figure S10C), while increases of H3K4me1 could also be observed in the union of DN + DP peaks, suggesting that these findings can be extended to primary cells (Supplementary Figure S10D). Thus, while its loss does not appear to completely erase epigenetic patterns at enhancers, Ets1 does however contribute to chromatin remodeling at DP-related stages, possibly by triggering NDRs and precise positioning within H3K4me1 nucleosomes.
Figure 6.
Loss of Ets1 induces chromatin remodeling via increased H3K4me1 signal genome-wide in the thymic P5424 cell line. (A) Western blot showing reduction of Ets1 protein levels following knockdown in the P5424 cell line. (B) UCSC genome browser screenshot of the Tcra (left) and Cd2 loci (right) showing increased H3K4me1 signal at the Ets1 binding site following knockdown in the P5424 cell line. (C and D) Total average profiles of H3K4me1 (left), H3K4me3 (right) with and without Ets1 knockdown (red and black, respectively) in the thymic P5424 cell line at (C) P5424 Ets1 sites and (D) control, ENCODE thymus DHS overlapping with Ets1 peaks neither in DN, DP nor P5424. P-values adjusted with Bonferroni correction for multiple testing. (E) Average profiles per class of H3K4me1 NDR and heatmaps for H3K4me1 (left), H3K4me3 (right) with and without Ets1 knockdown (red and black, respectively) in the thymic P5424 cell line at P5424 Ets1 sites, ranked by increasing P5424 sh-scr H3K4me1 signal [−500; +500 bp] around the Ets1 site. Classes of H3K4me1 NDR status are indicated left of heatmaps.
DISCUSSION
In this work, we performed an extensive analysis of Ets1 role during thymic differentiation, focusing on the DN to DP transition. Through various genome-wide approaches, including by using knock-out and knockdown experiments, we identified differentially Ets1-bound regulatory elements and Ets1-regulated genes. Converging evidence also indicate that while it still regulates many hematopoietic genes in DN, Ets1 plays an important role in activating thymic specific in DP cells. Interestingly, we further point out differential co-association of Ets1 with Runx1 in DN and with TCF1 in DP at specific subsets of genes. Our data also argues for a dual co-association to both nucleosomal and accessible DNA in vivo at both differentiation stages with more association to NORs in DN and to NDRs in DP. Finally our knockdown experiments indicate a role for Ets1 in H3K4me1 chromatin remodeling in a DP-related cell line.
Based on our observations, we thus distinguished two main modes of action related to Ets1 binding: (i) at DN-specific sites where it entails little or no evidence of enhancer activity and (ii) at DN-DP shared sites or DP-specific sites where it correlates with enhancer function. In DN cells, Ets1 binds DN-specific sites with high H3K4me1 and lower H3K4me3 compared to DN-DP shared or DP-specific sites. These sites were also marked with lower eRNA signal, suggesting less enhancer activity. The fact that genes associated with these sites in DN are expressed in the absence of Ets1 in DP also suggests that Ets1 confers them little enhancer activity. A possible explanation is that Ets1 takes over Pu.1 as a placeholder for a substantial part of these sites as Pu.1 expression is largely diminished at the DN3 stage (11) that become repressed at the next stage, concomitantly or following Ets1 recruitment. This model is supported by the nucleosome-occupied nature of these sites at both the DN and DP stages. The other mode of action entails direct enhancer activity, which encompasses DN-DP shared sites at both stages and DP-specific sites at the DP stage. At these sites, high H3K4me3 and eRNA levels were observed, together with a high enhancer activity (using CAPSTARR-Seq data). In this model, DN-DP shared sites are bound by Ets1 in DN and would become even more active in DP through a still unknown mechanism involving chromatin remodeling. Finally, the loss of expression of genes associated with the deletion of Ets1 suggests direct involvement of Ets1 in their enhancer activity.
Ets1-bound enhancers also exhibited differential TF co-associations in DN and DP cells; Ets1 was more associated with Runx1 and E47 in DN thymocytes, while it seemingly homodimerized/heterodimerized with another Ets family member in DP cells, with a higher co-association with TCF1. For Runx1, these results concur with a previous study showing reduced roles for this TF at the DP stage as a requirement for the DN to DP transition (67). Importantly, we showed that sites bearing two distinct Ets motifs, Ets1 (AGGAAG) and Etscan (CGGAAG) exhibited high enhancer activity. This class of sites could correspond to ‘redundant’ Ets binding sites for which multiple members of the Ets family compete (25). The fact that we also observed different ontologies between classes generally only enriched in the Ets1 motif and those enriched in both motifs also extends in a stage-specific manner, to distal elements, previous findings that different ontologies of genes are bound by Ets transcription factors with different binding affinities at promoters (25). Further, we showed higher Runx1 binding in NORs in DN and conversely, higher TCF1 motif presence and binding in NDRs in DP cells. This suggests a chromatin-state based transcriptional switch resulting in TCF1 binding in DP NDRs. Moreover, we showed that distal NDRs are more transcriptionally active than NORs in DP, a situation that is not observed in DN thymocytes.
It is remarkable that Ets1 carries the ability to bind both NORs and NDRs and is able to induce chromatin remodeling. This observation is in agreement with previous reports attributing nucleosome-binding properties to Ets1 based on in vitro characterization (33,34). We also noted that in DP, Ets1 clearly occupied more NDRs, which is reflected by its mode of action. This also raises the question as to whether this mechanism is specific to Ets1. A recent ENCODE work on 119 TFs suggested that a large majority of TFs, including Ets1, are associated with NDRs in various cell lines (68). This study however only considered metagene analyses to draw this conclusion and it would be of interest to extend more detailed future investigation to this data set by discriminating possible subclasses of nucleosome occupancies. A recent study in macrophage has shown that Pu.1, another Ets family member, essentially binds to NDRs in vivo, while the same genomic locations tend to be nucleosome-occupied in vitro or in other lineages (32). This indicates that the dichotomy of Ets1 association to NORs/NDRs in T-cells does not stand for all Ets family members, although bound nucleosomes in DN could potentially be enriched for the H2A.Z variant and thus be labile. Conversely, other TFs, such as FoxA2 in the liver, were described to bind to both nucleosomes and NDRs. In this case however, no difference in both classes of TFBS (nucleosome-occupied/NDR) were observed for gene expression regulation (69). Thus Ets1, at least in early differentiating T-cells, seems to carry a somewhat specific dichotomy in its binding ability to nucleosomes and NDRs.
Our results also confirm, at the genome-wide scale, previous studies describing impaired DN to DP transition upon Ets1 knockout (4), arguing for a role whereby Ets1 facilitates the transition by switching from distal elements of DN to DP-specific genes. This likely happens as DP-specific genes are lost as a result of the deletion of Ets1, and that commitment to the T-cell lineage is somewhat reduced in light of the up-regulation of genes from other lineages in the absence of Ets1. However, because the block in DN is not complete (5,6), this might suggest that for critical DP genes, other factors may compensate for the loss of Ets1 in bound enhancers. Recent studies have also argued for dramatic changes in the transcriptional landscape of DP cells between the DP blast stage (DPbl) and small DP stage (DPsm), whereby many housekeeping genes are shut down (70). Thus, changes in the transcriptional landscape of thymocytes observed in this study could be related with the repression of many housekeeping genes. Additionally, we also noted downregulation of non-T lineage specific yet hematopoietic genes between DN and DP cells. The implication of Ets1-mediated transactivation of such genes in DN hints at a role in alternative lineage commitment for Ets1 at this stage, as was described for Pu.1 at earlier stages of differentiation (71). For certain lineage-specific genes, the observed differential in numbers of peaks hints at the involvement of Ets1 in a fine tuning of gene expression via minute activation of certain enhancer or silencer elements, as for Cd4 or Cd8. By showing direct Ets1 binding in the Cd8 locus, we extend findings from (72) who emphasized the importance of Ets1 during CD8+ T-cell differentiation. Importantly, we also found Ets1 to bind distal elements of the Runx3 gene (Supplementary Table S4), in line with reports that Ets1 contributes also toward CD8+ single positive T-cell commitment via Runx3 transactivation (73).
Such an Ets1-mediated DN to DP transition might occur via differential TF co-association or chromatin remodeling, but also by change of motif specificity between these stages. We identified both previously and independently described motifs for Ets1 (AGGAAG), present in DN-specific and shared classes, and Etscan (CGGAAG), present in DP-specific and shared classes. This change of motif specificity, possibly linked with homodimerization/heterodimerization to other Ets proteins in DP, could indicate a post-translational modification at this stage. The MAPK/ERK and PKC pathways are for example known to increase the transcriptional activity of Ets1 via phosphorylation of threonine 38 and exon VII respectively (74,75), which could suggest that Ets1 relates to its cognate enhancer activity only following TCR stimulation which occurs from the DN3b stage onward thus not in Rag2−/− mice). Runx1 is also known to activate Ets1 by binding to exon VII thus blocking its autoinhibitory domain (76). In the absence of Runx1 as we observed in DP-specific classes, this role could be achieved by homodimerization by binding at separate locations within one turn of nucleosomal DNA, occupied or not (77). These modifications and co-associations could thus potentially alter the ability to interact with other TFs and/or to promote nucleosome exclusion locally, thus providing a wider TFs/DNA landing platform and ultimately leading to a more direct role in gene transactivation. The presence of Ets1 on DN-shared NORs may also correspond to Ets1 acting as a placeholder for the transcriptional machinery at this stage, whereby distal elements would be opened-up by activation-induced recruitment of other TFs and/or remodelers in DP thymocytes. This mode of activation could also be extended to later stages of T-cell activation with recruitment of TFs in NORs (78).
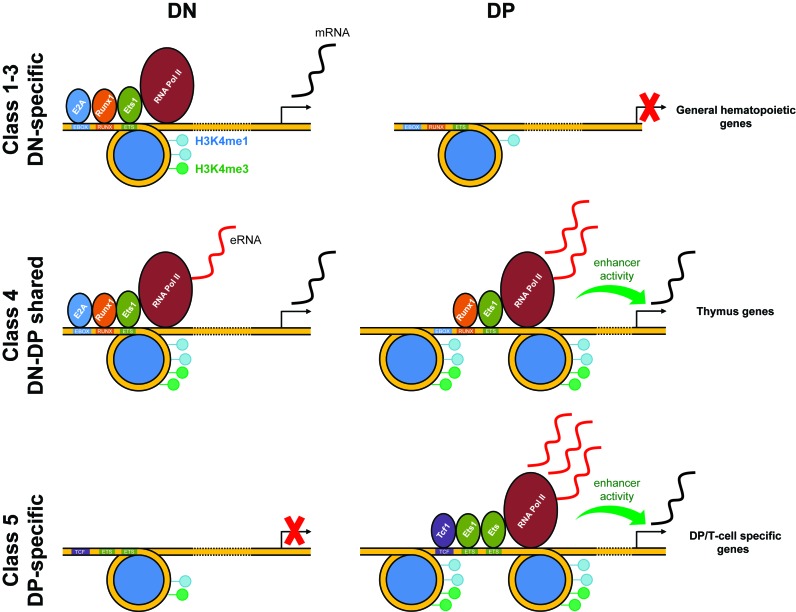
In summary, our analyses reveal (i) Ets1-mediated chromatin remodeling between the DN and DP stages at these sites, (ii) differential, maturational stage and chromatin-state specific TF occupancy. Ets1 binds NDRs and NORs in DN but more NDRs in DP cells. Runx1 and TCF1 were enriched in NORs and NDRs in DN and DP cells, respectively, (iii) increased Pol II recruitment and eRNAs transcription were found in NDRs in DP but not in DN, suggesting enhancer activity mostly at the DP stage and (iv) that this dual mode of action corresponds to the specific upregulation of the DP gene expression program but negatively regulates the DN and other-lineage hematopoietic gene expression programs during the DN to DP transition. These findings are summed up as a model in Figure 7. Our results thus provide an original model whereby one transcription factor changes its binding ability to nucleosomal DNA during differentiation with consequences on its cognate enhancer activity.
Figure 7.
Ets1-promoted lineage commitment by dynamic co-association and chromatin remodeling. Model summing up transactivation mechanics of class 1 (DN-specific), 4 (DN-DP shared) and 5 (DP-specific) Ets1 targets. For class 1, Ets1 binds to NORs in DN, recruiting E2A, Runx1 partners and Pol II, leading to downstream activation of DN-specific, early hematopoietic genes. eRNA is produced, however at modest quantities. These sites are lost in DP. For class 4, Ets1 binds to NORs in DN, recruiting E2A, Runx1 partners and Pol II, transactivating general hematopoietic genes. High quantities of eRNA are produced. These sites are maintained and nucleosome-free in DP, with E2A being lost. In class 5 sites, these sites are nucleosome-occupied and devoid of TFs in DN, with the nearest gene being repressed. In DP, Ets1 binds as a homodimer, opens up the chromatin, recruiting TCF1 and Pol II, resulting in upregulation of DP/T-cell lineage-specific genes, as well as high eRNA production.
ACCESSION NUMBERS
All datasets produced for this work (DP expression data, DN, DP ChIP-seq datasets, DN, DP MNase-seq datasets and DN short RNA dataset) were submitted to the Gene Expression Omnibus (GEO) database as superseries GSE56395. This superseries was subdivided by experiment type, namely DP expression data (GEO accession GSE56357), DN short RNA-seq (GSE56358), DN and DP MNase-seq (GSE56360), as well as DN and DP ChIP-seq (GSE56393). Ets1−/− and WT DP expression microarray datasets were deposited under GEO series accession GSE73849. WT P5424 Ets1, sh-scr, sh-ets1 H3K4me1 and H3K4me3 ChIP-Seq datasets were deposited under GEO series accession GSE74042.
Supplementary Material
Acknowledgments
We are grateful to Nadine Obier, Peter Cockerill and Constanze Bonifer for critical reading of the manuscript as well as to Matthew Ingham from the CNAG lab for sequencing quality controls.
Footnotes
Present address: Maria A Santana, Facultad de Ciencias, Universidad Autónoma del Estado de Morelos. Av. Universidad 1001, Chamilpa, Cuernavaca, Morelos, Mexico.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
CNRS; ‘Agence Nationale de la Recherche’ (ANR); Fondation pour la Recherche Medicale (FRM starting grant) in the JCA lab; A*MIDEX project N° [ANR-11-IDEX-0001-02 to S.S.]; INCa (to P.C.); Fondation pour la Recherche Médicale (to P.C.); Genopole (to P.C.); ‘amorcage jeunes équipes’ [FRM AJE20130728183 to M.A.M.]; Genopole (to J.Z.); European Union's FP7 Program [agreement n° 282510-BLUEPRINT to L.V.]; ‘Chromatin Plasticity’ Marie Curie RTN and Association pour la Recherche sur le Cancer (to F.K.); Genopole (to R.F.); CNRS (to R.F.); CONACYT (to M.A.S.); UAEM (to M.A.S.); Inserm; GIS IBiSA; Aix-Marseille Université; ANR [10-INBS-0009-10]. Funding for open access charge: CNRS; “Agence Nationale de la Recherche" (ANR); Fondation pour la Recherche Medicale.
Conflict of interest statement. None declared.
REFERENCES
- 1.Anderson M.K. At the crossroads: diverse roles of early thymocyte transcriptional regulators. Immunol. Rev. 2006;209:191–211. doi: 10.1111/j.0105-2896.2006.00352.x. [DOI] [PubMed] [Google Scholar]
- 2.Barton K., Muthusamy N., Fischer C., Ting C.N., Walunas T.L., Lanier L.L., Leiden J.M. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity. 1998;9:555–563. doi: 10.1016/s1074-7613(00)80638-x. [DOI] [PubMed] [Google Scholar]
- 3.Bhat N.K., Komschlies K.L., Fujiwara S., Fisher R.J., Mathieson B.J., Gregorio T.A., Young H.A., Kasik J.W., Ozato K., Papas T.S. Expression of ets genes in mouse thymocyte subsets and T cells. J. Immunol. 1989;142:672–678. [PubMed] [Google Scholar]
- 4.Eyquem S., Chemin K., Fasseu M., Bories J.C. The Ets-1 transcription factor is required for complete pre-T cell receptor function and allelic exclusion at the T cell receptor beta locus. Proc. Natl. Acad. Sci. U.S.A. 2004;101:15712–15717. doi: 10.1073/pnas.0405546101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grenningloh R., Kang B.Y., Ho I.C. Ets-1, a functional cofactor of T-bet, is essential for Th1 inflammatory responses. J. Exp. Med. 2005;201:615–626. doi: 10.1084/jem.20041330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grenningloh R., Miaw S.C., Moisan J., Graves B.J., Ho I.C. Role of Ets-1 phosphorylation in the effector function of Th cells. Eur. J. Immunol. 2008;38:1700–1705. doi: 10.1002/eji.200738112. [DOI] [PubMed] [Google Scholar]
- 7.Moisan J., Grenningloh R., Bettelli E., Oukka M., Ho I.C. Ets-1 is a negative regulator of Th17 differentiation. J. Exp. Med. 2007;204:2825–2835. doi: 10.1084/jem.20070994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sacchi N., Watson D.K., Guerts van Kessel A.H., Hagemeijer A., Kersey J., Drabkin H.D., Patterson D., Papas T.S. Hu-ets-1 and Hu-ets-2 genes are transposed in acute leukemias with (4;11) and (8;21) translocations. Science. 1986;231:379–382. doi: 10.1126/science.3941901. [DOI] [PubMed] [Google Scholar]
- 9.Hashiya N., Jo N., Aoki M., Matsumoto K., Nakamura T., Sato Y., Ogata N., Ogihara T., Kaneda Y., Morishita R. In vivo evidence of angiogenesis induced by transcription factor Ets-1: Ets-1 is located upstream of angiogenesis cascade. Circulation. 2004;109:3035–3041. doi: 10.1161/01.CIR.0000130643.41587.DB. [DOI] [PubMed] [Google Scholar]
- 10.Charafe-Jauffret E., Ginestier C., Monville F., Finetti P., Adelaide J., Cervera N., Fekairi S., Xerri L., Jacquemier J., Birnbaum D., et al. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene. 2006;25:2273–2284. doi: 10.1038/sj.onc.1209254. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J.A., Mortazavi A., Williams B.A., Wold B.J., Rothenberg E.V. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 2012;149:467–482. doi: 10.1016/j.cell.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rothenberg E.V., Taghon T. Molecular genetics of T cell development. Annu. Rev. Immunol. 2005;23:601–649. doi: 10.1146/annurev.immunol.23.021704.115737. [DOI] [PubMed] [Google Scholar]
- 13.Winandy S. Regulation of chromatin structure during thymic T cell development. J. Cell Biochem. 2005;95:466–477. doi: 10.1002/jcb.20391. [DOI] [PubMed] [Google Scholar]
- 14.Sudo T., Nishikawa S., Ohno N., Akiyama N., Tamakoshi M., Yoshida H. Expression and function of the interleukin 7 receptor in murine lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 1993;90:9125–9129. doi: 10.1073/pnas.90.19.9125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Godfrey D.I., Kennedy J., Suda T., Zlotnik A. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3-CD4-CD8- triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J. Immunol. 1993;150:4244–4252. [PubMed] [Google Scholar]
- 16.von Boehmer H. Developmental biology of T cells in T cell-receptor transgenic mice. Annu. Rev. Immunol. 1990;8:531–556. doi: 10.1146/annurev.iy.08.040190.002531. [DOI] [PubMed] [Google Scholar]
- 17.Komine O., Hayashi K., Natsume W., Watanabe T., Seki Y., Seki N., Yagi R., Sukzuki W., Tamauchi H., Hozumi K., et al. The Runx1 transcription factor inhibits the differentiation of naive CD4+ T cells into the Th2 lineage by repressing GATA3 expression. J. Exp. Med. 2003;198:51–61. doi: 10.1084/jem.20021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woolf E., Xiao C., Fainaru O., Lotem J., Rosen D., Negreanu V., Bernstein Y., Goldenberg D., Brenner O., Berke G., et al. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc. Natl. Acad. Sci. U.S.A. 2003;100:7731–7736. doi: 10.1073/pnas.1232420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prosser H.M., Wotton D., Gegonne A., Ghysdael J., Wang S., Speck N.A., Owen M.J. A phorbol ester response element within the human T-cell receptor beta-chain enhancer. Proc. Natl. Acad. Sci. U.S.A. 1992;89:9934–9938. doi: 10.1073/pnas.89.20.9934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho I.C., Bhat N.K., Gottschalk L.R., Lindsten T., Thompson C.B., Papas T.S., Leiden J.M. Sequence-specific binding of human Ets-1 to the T cell receptor alpha gene enhancer. Science. 1990;250:814–818. doi: 10.1126/science.2237431. [DOI] [PubMed] [Google Scholar]
- 21.Hollenhorst P.C., Shah A.A., Hopkins C., Graves B.J. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 2007;21:1882–1894. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hollenhorst P.C., Chandler K.J., Poulsen R.L., Johnson W.E., Speck N.A., Graves B.J. DNA specificity determinants associate with distinct transcription factor functions. PLoS Genet. 2009;5:e1000778. doi: 10.1371/journal.pgen.1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verger A., Duterque-Coquillaud M. When Ets transcription factors meet their partners. Bioessays. 2002;24:362–370. doi: 10.1002/bies.10068. [DOI] [PubMed] [Google Scholar]
- 24.Wei G.H., Badis G., Berger M.F., Kivioja T., Palin K., Enge M., Bonke M., Jolma A., Varjosalo M., Gehrke A.R., et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29:2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Odrowaz Z., Sharrocks A.D. ELK1 uses different DNA binding modes to regulate functionally distinct classes of target genes. PLoS Genet. 2012;8:e1002694. doi: 10.1371/journal.pgen.1002694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei G., Abraham B.J., Yagi R., Jothi R., Cui K., Sharma S., Narlikar L., Northrup D.L., Tang Q., Paul W.E., et al. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity. 2011;35:299–311. doi: 10.1016/j.immuni.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dore L.C., Chlon T.M., Brown C.D., White K.P., Crispino J.D. Chromatin occupancy analysis reveals genome-wide GATA factor switching during hematopoiesis. Blood. 2012;119:3724–3733. doi: 10.1182/blood-2011-09-380634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chlon T.M., Dore L.C., Crispino J.D. Cofactor-mediated restriction of GATA-1 chromatin occupancy coordinates lineage-specific gene expression. Mol. Cell. 2012;47:608–621. doi: 10.1016/j.molcel.2012.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samstein R.M., Arvey A., Josefowicz S.Z., Peng X., Reynolds A., Sandstrom R., Neph S., Sabo P., Kim J.M., Liao W., et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151:153–166. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raney B.J., Cline M.S., Rosenbloom K.R., Dreszer T.R., Learned K., Barber G.P., Meyer L.R., Sloan C.A., Malladi V.S., Roskin K.M., et al. ENCODE whole-genome data in the UCSC genome browser (2011 update) Nucleic Acids Res. 2011;39:D871–D875. doi: 10.1093/nar/gkq1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghisletti S., Barozzi I., Mietton F., Polletti S., De Santa F., Venturini E., Gregory L., Lonie L., Chew A., Wei C.L., et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 32.Barozzi I., Simonatto M., Bonifacio S., Yang L., Rohs R., Ghisletti S., Natoli G. Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers. Mol. Cell. 2014;54:844–857. doi: 10.1016/j.molcel.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh A.K., Swarnalatha M., Kumar V. c-ETS1 facilitates G1/S-phase transition by up-regulating cyclin E and CDK2 genes and cooperates with hepatitis B virus X protein for their deregulation. J. Biol. Chem. 2011;286:21961–21970. doi: 10.1074/jbc.M111.238238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu J., Pazin M.J., Ravid K. Properties of ets-1 binding to chromatin and its effect on platelet factor 4 gene expression. Mol. Cell. Biol. 2004;24:428–441. doi: 10.1128/MCB.24.1.428-441.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J., Zhuang J., Iyer S., Lin X., Whitfield T.W., Greven M.C., Pierce B.G., Dong X., Kundaje A., Cheng Y., et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012;22:1798–1812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mombaerts P., Iacomini J., Johnson R.S., Herrup K., Tonegawa S., Papaioannou V.E. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 37.Koch F., Fenouil R., Gut M., Cauchy P., Albert T.K., Zacarias-Cabeza J., Spicuglia S., de la Chapelle A.L., Heidemann M., Hintermair C., et al. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat. Struct. Mol. Biol. 2011;18:956–963. doi: 10.1038/nsmb.2085. [DOI] [PubMed] [Google Scholar]
- 38.Fenouil R., Cauchy P., Koch F., Descostes N., Cabeza J.Z., Innocenti C., Ferrier P., Spicuglia S., Gut M., Gut I., et al. CpG islands and GC content dictate nucleosome depletion in a transcription-independent manner at mammalian promoters. Genome Res. 2012;22:2399–2408. doi: 10.1101/gr.138776.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pekowska A., Benoukraf T., Zacarias-Cabeza J., Belhocine M., Koch F., Holota H., Imbert J., Andrau J.C., Ferrier P., Spicuglia S. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 2011;30:4198–4210. doi: 10.1038/emboj.2011.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benoukraf T., Cauchy P., Fenouil R., Jeanniard A., Koch F., Jaeger S., Thieffry D., Imbert J., Andrau J.C., Spicuglia S., et al. CoCAS: a ChIP-on-chip analysis suite. Bioinformatics. 2009;25:954–955. doi: 10.1093/bioinformatics/btp075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cauchy P., James S.R., Zacarias-Cabeza J., Ptasinska A., Imperato M.R., Assi S.A., Piper J., Canestraro M., Hoogenkamp M., Raghavan M., et al. Chronic FLT3-ITD signaling in acute myeloid leukemia is connected to a specific chromatin signature. Cell Rep. 2015;12:821–836. doi: 10.1016/j.celrep.2015.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kreher S., Bouhlel M.A., Cauchy P., Lamprecht B., Li S., Grau M., Hummel F., Kochert K., Anagnostopoulos I., Johrens K., et al. Mapping of transcription factor motifs in active chromatin identifies IRF5 as key regulator in classical Hodgkin lymphoma. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E4513–E4522. doi: 10.1073/pnas.1406985111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saldanha A.J. Java Treeview–extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 44.Zhu L.J., Gazin C., Lawson N.D., Pages H., Lin S.M., Lapointe D.S., Green M.R. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Hoon M.J., Imoto S., Nolan J., Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 47.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 48.Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lepoivre C., Belhocine M., Bergon A., Griffon A., Yammine M., Vanhille L., Zacarias-Cabeza J., Garibal M.A., Koch F., Maqbool M.A., et al. Divergent transcription is associated with promoters of transcriptional regulators. BMC Genomics. 2013;14:914. doi: 10.1186/1471-2164-14-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vanhille L., Griffon A., Maqbool M.A., Zacarias-Cabeza J., Dao L.T., Fernandez N., Ballester B., Andrau J.C., Spicuglia S. High-throughput and quantitative assessment of enhancer activity in mammals by CapStarr-seq. Nat. Commun. 2015;6:6905. doi: 10.1038/ncomms7905. [DOI] [PubMed] [Google Scholar]
- 51.Wu J.Q., Seay M., Schulz V.P., Hariharan M., Tuck D., Lian J., Du J., Shi M., Ye Z., Gerstein M., et al. Tcf7 is an important regulator of the switch of self-renewal and differentiation in a multipotential hematopoietic cell line. PLoS Genet. 2012;8:e1002565. doi: 10.1371/journal.pgen.1002565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li L., Zhang J.A., Dose M., Kueh H.Y., Mosadeghi R., Gounari F., Rothenberg E.V. A far downstream enhancer for murine Bcl11b controls its T-cell specific expression. Blood. 2013;122:902–911. doi: 10.1182/blood-2012-08-447839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dose M., Emmanuel A.O., Chaumeil J., Zhang J., Sun T., Germar K., Aghajani K., Davis E.M., Keerthivasan S., Bredemeyer A.L., et al. beta-Catenin induces T-cell transformation by promoting genomic instability. Proc. Natl. Acad. Sci. U.S.A. 2014;111:391–396. doi: 10.1073/pnas.1315752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miyazaki M., Rivera R.R., Miyazaki K., Lin Y.C., Agata Y., Murre C. The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naive fate of T cells. Nat. Immunol. 2011;12:992–1001. doi: 10.1038/ni.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shay T., Kang J. Immunological Genome Project and systems immunology. Trends Immunol. 2013;34:602–609. doi: 10.1016/j.it.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Geimer Le Lay A.S., Oravecz A., Mastio J., Jung C., Marchal P., Ebel C., Dembele D., Jost B., Le Gras S., Thibault C., et al. The tumor suppressor Ikaros shapes the repertoire of notch target genes in T cells. Sci. Signal. 2014;7:ra28. doi: 10.1126/scisignal.2004545. [DOI] [PubMed] [Google Scholar]
- 57.Ichiyama K., Chen T., Wang X., Yan X., Kim B.S., Tanaka S., Ndiaye-Lobry D., Deng Y., Zou Y., Zheng P., et al. The methylcytosine dioxygenase Tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity. 2015;42:613–626. doi: 10.1016/j.immuni.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin C.C., Bradstreet T.R., Schwarzkopf E.A., Sim J., Carrero J.A., Chou C., Cook L.E., Egawa T., Taneja R., Murphy T.L., et al. Bhlhe40 controls cytokine production by T cells and is essential for pathogenicity in autoimmune neuroinflammation. Nat. Commun. 2014;5:3551. doi: 10.1038/ncomms4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang S., Fujikado N., Kolodin D., Benoist C., Mathis D. Immune tolerance. Regulatory T cells generated early in life play a distinct role in maintaining self-tolerance. Science. 2015;348:589–594. doi: 10.1126/science.aaa7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harker N., Garefalaki A., Menzel U., Ktistaki E., Naito T., Georgopoulos K., Kioussis D. Pre-TCR signaling and CD8 gene bivalent chromatin resolution during thymocyte development. J. Immunol. 2011;186:6368–6377. doi: 10.4049/jimmunol.1003567. [DOI] [PubMed] [Google Scholar]
- 61.Painter M.W., Davis S., Hardy R.R., Mathis D., Benoist C. Transcriptomes of the B and T lineages compared by multiplatform microarray profiling. J. Immunol. 2011;186:3047–3057. doi: 10.4049/jimmunol.1002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lamber E.P., Vanhille L., Textor L.C., Kachalova G.S., Sieweke M.H., Wilmanns M. Regulation of the transcription factor Ets-1 by DNA-mediated homo-dimerization. EMBO J. 2008;27:2006–2017. doi: 10.1038/emboj.2008.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kutlesa S., Zayas J., Valle A., Levy R.B., Jurecic R. T-cell differentiation of multipotent hematopoietic cell line EML in the OP9-DL1 coculture system. Exp. Hematol. 2009;37:909–923. doi: 10.1016/j.exphem.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lichtinger M., Ingram R., Hannah R., Muller D., Clarke D., Assi S.A., Lie A.L.M., Noailles L., Vijayabaskar M.S., Wu M., et al. RUNX1 reshapes the epigenetic landscape at the onset of haematopoiesis. EMBO J. 2012;31:4318–4333. doi: 10.1038/emboj.2012.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Steger D.J., Workman J.L. Stable co-occupancy of transcription factors and histones at the HIV-1 enhancer. EMBO J. 1997;16:2463–2472. doi: 10.1093/emboj/16.9.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sherwood R.I., Hashimoto T., O'Donnell C.W., Lewis S., Barkal A.A., van Hoff J.P., Karun V., Jaakkola T., Gifford D.K. Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat. Biotechnol. 2014;32:171–178. doi: 10.1038/nbt.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wong W.F., Nakazato M., Watanabe T., Kohu K., Ogata T., Yoshida N., Sotomaru Y., Ito M., Araki K., Telfer J., et al. Over-expression of Runx1 transcription factor impairs the development of thymocytes from the double-negative to double-positive stages. Immunology. 2010;130:243–253. doi: 10.1111/j.1365-2567.2009.03230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kundaje A., Kyriazopoulou-Panagiotopoulou S., Libbrecht M., Smith C.L., Raha D., Winters E.E., Johnson S.M., Snyder M., Batzoglou S., Sidow A. Ubiquitous heterogeneity and asymmetry of the chromatin environment at regulatory elements. Genome Res. 2012;22:1735–1747. doi: 10.1101/gr.136366.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Z., Schug J., Tuteja G., White P., Kaestner K.H. The nucleosome map of the mammalian liver. Nat. Struct. Mol. Biol. 2011;18:742–746. doi: 10.1038/nsmb.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mingueneau M., Kreslavsky T., Gray D., Heng T., Cruse R., Ericson J., Bendall S., Spitzer M.H., Nolan G.P., Kobayashi K., et al. The transcriptional landscape of alphabeta T cell differentiation. Nat. Immunol. 2013;14:619–632. doi: 10.1038/ni.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dionne C.J., Tse K.Y., Weiss A.H., Franco C.B., Wiest D.L., Anderson M.K., Rothenberg E.V. Subversion of T lineage commitment by PU.1 in a clonal cell line system. Dev. Biol. 2005;280:448–466. doi: 10.1016/j.ydbio.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 72.Clements J.L., John S.A., Garrett-Sinha L.A. Impaired generation of CD8+ thymocytes in Ets-1-deficient mice. J. Immunol. 2006;177:905–912. doi: 10.4049/jimmunol.177.2.905. [DOI] [PubMed] [Google Scholar]
- 73.Zamisch M., Tian L., Grenningloh R., Xiong Y., Wildt K.F., Ehlers M., Ho I.C., Bosselut R. The transcription factor Ets1 is important for CD4 repression and Runx3 up-regulation during CD8 T cell differentiation in the thymus. J. Exp. Med. 2009;206:2685–2699. doi: 10.1084/jem.20092024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang B.S., Hauser C.A., Henkel G., Colman M.S., Van Beveren C., Stacey K.J., Hume D.A., Maki R.A., Ostrowski M.C. Ras-mediated phosphorylation of a conserved threonine residue enhances the transactivation activities of c-Ets1 and c-Ets2. Mol. Cell. Biol. 1996;16:538–547. doi: 10.1128/mcb.16.2.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lindemann R.K., Braig M., Ballschmieter P., Guise T.A., Nordheim A., Dittmer J. Protein kinase Calpha regulates Ets1 transcriptional activity in invasive breast cancer cells. Int. J. Oncol. 2003;22:799–805. [PubMed] [Google Scholar]
- 76.Kim W.Y., Sieweke M., Ogawa E., Wee H.J., Englmeier U., Graf T., Ito Y. Mutual activation of Ets-1 and AML1 DNA binding by direct interaction of their autoinhibitory domains. EMBO J. 1999;18:1609–1620. doi: 10.1093/emboj/18.6.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Babayeva N.D., Baranovskaya O.I., Tahirov T.H. Structural basis of Ets1 cooperative binding to widely separated sites on promoter DNA. PLoS One. 2012;7:e33698. doi: 10.1371/journal.pone.0033698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Johnson B.V., Bert A.G., Ryan G.R., Condina A., Cockerill P.N. Granulocyte-macrophage colony-stimulating factor enhancer activation requires cooperation between NFAT and AP-1 elements and is associated with extensive nucleosome reorganization. Mol. Cell. Biol. 2004;24:7914–7930. doi: 10.1128/MCB.24.18.7914-7930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.