Abstract
During HIV-1 reverse transcription, the single-stranded RNA genome is converted into proviral double stranded DNA by Reverse Transcriptase (RT) within a reverse transcription complex composed of the genomic RNA and a number of HIV-1 encoded proteins, including the nucleocapsid protein NCp7. Here, we developed a one-step and one-pot RT polymerization assay. In this in vitro assay, RT polymerization is monitored in real-time by Förster resonance energy transfer (FRET) using a commercially available doubly-labeled primer/template DNA. The assay can monitor and quantify RT polymerization activity as well as its promotion by NCp7. Z-factor values as high as 0.89 were obtained, indicating that the assay is suitable for high-throughput drug screening. Using Nevirapine and AZT as prototypical RT inhibitors, reliable IC50 values were obtained from the changes in the RT polymerization kinetics. Interestingly, the assay can also detect NCp7 inhibitors, making it suitable for high-throughput screening of drugs targeting RT, NCp7 or simultaneously, both proteins.
INTRODUCTION
During HIV-1 reverse transcription, the reverse transcriptase (RT) transcribes (+) strand viral RNA genome to form integration-competent double stranded DNA (1). RT is a p66/p51 heterodimer that exhibits RNA- and DNA-directed DNA polymerase and RNAse H activities. The p66 subunit is enzymatically active and can be structurally divided into thumb, palm, fingers and connection sub-domains and RNase H domain (2,3). Although the p51 subunit shares the same polypeptide chain as the p66 subunit with the exception of the last 120 C-terminal amino acids, its role seems to be purely structural (2–4). Because of its critical role in reverse transcription and thus in retroviral replication, RT is an important target for anti-HIV drugs and two well represented classes of approved RT-inhibitor drugs are available (5–7). The first class called nucleoside reverse transcriptase inhibitors (NRTI) consists of nucleotide analogues such as Zidovudine (AZT), which directly compete with dNTP monomers to block the extension of the primer. The second class called non-nucleoside reverse transcriptase inhibitors (NNRTI) is composed of molecules structurally unrelated to nucleotides that bind non-competitively to RT allosteric sites (8,9). Both of these inhibitor classes, substantially slow down the progression of the virus. However, even when the two classes are used in association, the high prevalence of mutations in the HIV-1 genome results in the selection of drug resistant strains, leading to frequent viral escapes over the long term patient's therapeutic management. In this context, a continuous development of new RT inhibitor drugs targeting the enzymatic activities of RT is needed (10–12). Alternatively, since RT is regulated by a number of other HIV-1 proteins in the reverse transcription complex (RTC), these proteins may constitute additional targets to block the reverse transcription process. For instance, an attractive complementary target could be the HIV-1 nucleocapsid protein NCp7 (13–15), a highly conserved 55-amino acid protein, characterized by two zinc finger motifs, that is quantitatively the main HIV-1 protein in RTC. NCp7 is thought to promote reverse transcription through its nucleic acid chaperone activity (16–21), but also by stabilizing the complex of RT with its substrate (22–24) and/or through direct interaction with RT (25–27).
Multiple assays have been reported to investigate and monitor RT polymerization activity. Most of these assays are usually discontinuous multistep assays, using radioactively or fluorescently labeled nucleotide analogs (28–42) and requiring extensive sample handling. Recently, a number of fluorescence-based approaches have been developed to further characterize the kinetics and mechanism of RT (43–47). Based on these approaches, RT was notably shown to dynamically flip between two opposite orientations on its substrate, namely a polymerase conformation, where the finger sub-domain is toward the primer 3′ end and a RNase H conformation, where the RNase H domain is toward the primer 3′ end, explaining how the binding mode of RT tunes its enzymatic activity (43,46). Based on these data, we recently developed a one-step and one-pot assay using Förster resonance energy transfer (FRET) between fluorescently labeled RT and primer/template (p/t) that allows monitoring RT flipping and polymerization in real time (48). This assay was shown to discriminate non-nucleoside RT inhibitors from nucleoside RT inhibitors and to determine reliably their potency. Though this assay is highly sensitive and convenient, it suffers from the need to produce a RT mutant with a single accessible Cys, to label it by a fluorophore and to purify it, in an enzymatically active state. Therefore, this assay can hardly be used with wild-type RT or with RT mutants resistant to RT inhibitors. In order to overcome these limitations, we proposed here a continuous one-pot FRET-based polymerization assay, using a commercially available doubly-labeled p/t similar to that previously used in single molecule experiments for monitoring RT polymerase activity (46). Through this assay, the RT-mediated extension of the primer as well its promotion by NCp7 could be monitored in real time, by conventional fluorescence techniques. Therefore, we propose here a simple and robust assay for screening and validating anti-RT and anti-NCp7 molecules.
MATERIALS AND METHODS
Proteins
Recombinant, heterodimeric mutant of HIV-1BH10 RT was expressed in Escherichia coli and purified as described previously (49). The expression system and purification protocol allowed the preparation of large quantities of heterodimeric enzyme in a homogeneous form. The prepared RT contains a E478Q mutation in its RNase H domain, but it was previously checked that this mutation does not affect the polymerase activity (50). RT labeled in its p51 thumb subdomain by Alexa 488 was prepared and its activity was tested as described (48). Proteins were stored at −80°C. Protein concentrations were determined using extinction coefficients at 280 nm of 260 450 M−1 cm−1.
Full length NCp7 peptide was synthesized by solid-phase peptide synthesis on a 433A synthesizer (ABI, Foster City, CA, USA) as described (51). Purification by HPLC was carried out on a C8 column (Uptisphere 300 A, 5 μm; 250 × 10, Interchim, France) in 0.05% trifluoroacetic acid (TFA) with a linear gradient of 10–70% of acetonitrile for 30 min. The peptide purity and molecular weight were checked by LC/MS. An absorption coefficient of 5700 M−1 cm−1 at 280 nm was used to determine its concentration.
Nucleotides, RT-inhibitors
The deoxynucleotide (dNTP) mixture was prepared by mixing each of UltraPure dATP, dCTP, dGTP and dTTP, purchased from Sigma Aldrich. Nevirapine (11-Cyclopropyl-5,11-dihydro-4-methyl-6H-dipyrido[3,2-b:2′,3′-e][1,4]diazepin-6-one) was also purchased from Sigma Aldrich and its stock solution was dissolved in dimethyl sulfoxide (DMSO). AZT triphosphate (3′-Azido-3′-Deoxythymidine-5′ Triphosphate) was purchased from Genexxon bioscience (Germany).
Oligonucleotides
Synthetic purified oligodeoxynucleotides were purchased from IBA (Göttingen, Germany) with the necessary modifications and their concentrations were determined by UV absorbance at 260 nm by using the extinction coefficients provided by the supplier. FRET measurements were carried out using a 23-mer (5′- CAG CAG TAC AAA TGG CAG TAT TC) DNA-primer labeled at the T19 position with Cyanine 5 (Cy5), annealed to a 63-mer (3′-TGT CGT CAT GTT TAC CGT CAT AAG TAG GTG TTA CTA GTC CGA TTT CCC CTA GTC CGA CCC ATG)-template labeled at the T2 position with carboxytetramethylrhodamine (TMR). Both TMR and Cy5 were covalently attached by supplier, via a C6 amino link to their respective T residues in the primer and the template. Primer and template oligodeoxynucleotides were annealed by heating equimolar amounts in buffer at 90°C for 2 min, followed by cooling to room temperature over several hours in a heating block. Unless noted otherwise, all experiments were routinely carried out at 20°C in a buffer containing 50 mM Tris–HCl (pH 7.5), 10 mM KCl and 6 mM MgCl2.
Fluorescence measurements
All steady-state and kinetic experiments were performed using the doubly-labeled p/t duplexes, which are mentioned above, in the presence of HIV-1 RT. All measurements were performed at 20°C using either a Fluoromax 4 spectrofluorometer (Jobin-Yvon Horiba) or a stopped-flow apparatus (SF3, Biologics). The FRET donor, TMR, was excited at 540 nm, and its emission was recorded at 580 nm. Nucleotide incorporation kinetics was triggered by addition of deoxyribonucleotides (dNTPs) in excess to a pre-incubated mixture of HIV-RT and p/t duplexes at equimolar concentrations. The kinetics was fast enough to monitor the fluorescence intensities continuously without photobleaching. The annealing kinetic traces were adequately fitted using:
 |
(1) |
where t0 is the dead time, kobs1,2 are the observed kinetic rate constants, a is the amplitude of the fast component, and I0 and IF are the fluorescence intensities before dNTPs addition and at completion of the reaction, respectively. The I0 value was obtained from the steady-state fluorescence spectrum of the doubly labeled p/t duplex in the presence of RT and was thus fixed. All fitting procedures were carried out with Origin™ 8.6 software using nonlinear, least-square methods and the Levenberg-Marquardt algorithm. Analytical analysis was systematically cross-validated using Bayesian inference with Markov Chain Monte Carlo and Gibbs sampling (see Supplementary Data and Supplementary Figures S1 and 2).
Measurements in high-throughput screening (HTS) format
Experiments were performed in 96-well plates. The complex of RT with p/t was formed by mixing 100 nM RT and 100 nM doubly-labeled p/t duplex in a total volume of 200 μl. All measurements were performed at 20°C using a FLX-Xenius plate reader (Safas Monaco). Excitation wavelength was 540 nm and emission was recorded at 580 nm using 10 nm slits. Polymerization kinetics were triggered by addition of 100 μM dNTPs. When kinetics were recorded in the presence of RT-inhibitors, the RT–p/t complex was pre-incubated with inhibitors for at least 5 min, unless otherwise stated. Each set of experiments was performed in three independent triplicates.
RESULTS AND DISCUSSION
Design and validation of the assay
A HIV-1 relevant DNA/DNA p/t sequence was selected from the HIV-1 pol gene. This p/t sequence lies outside any reported pausing-site (52) and thus, can be used to mimic a naturally occurring polymerization step during the plus strand synthesis of the HIV-1 genome. The doubly labeled p/t duplex was prepared by heat-annealing a 63-nt long DNA template labeled close to its 5′ end at its T2 position by TMR, used as a FRET donor, with a 23-nucleotide long DNA primer labeled at its T19 position with Cy5, used as a FRET acceptor (Figure 1A). The TMR label was placed 38 nt away from the 3′ end of the primer and 42 nt away from the Cy5 label and thus should not perturb RT binding.
Figure 1.
Design and validation of the FRET based assay. (A) Schematic representation of the FRET-based assay. The DNA–DNA p/t was labeled with both TMR (green) and Cy5 (red). Addition of RT and dNTP led to extension of the DNA primer, which shifted the two dyes further apart from each other, resulting in a decrease in FRET. (B) Polymerization kinetic trace of 100 nM RT with 100 nM doubly-labeled p/t duplex on addition of 100 μM dNTPs (red trace). The black line corresponds to the fit of the trace by Equation (1), with kobs1 = 19 (±1) × 10−3 s−1, kobs2 = 17 (±3) × 10−4 s−1 and a = 0.35 (±0.1). (Inset) Emission spectra of the doubly-labeled duplex in the absence (black line) and the presence (red line) of RT and after completion of polymerization with the addition of 100 μM dNTPs (blue line). For comparison, the emission spectrum of the duplex with TMR-labeled template and non-labeled primer (pink line) in the presence of RT is given to indicate the emission of the donor alone. Excitation and emission wavelengths for the kinetic trace were 540 and 580 nm, respectively.
Heat-annealed duplexes of labeled primer and template resulted in an FRET efficiency of 65% (Figure 1B, inset). This high value indicates that the average inter-dye distance is very short (∼ 4.5 nm), suggesting that the single-stranded domain of the template is highly flexible, bringing the 5′ end of the template close to the 3′ end of the unextended primer (Figure 1A). Formation of the complex between RT and DNA p/t (hereinafter referred to as E.DNA) led to a marginal decrease in FRET (from 65 to 63%), indicating that RT binding induces marginal changes in the distance between the two chromophores. In contrast, addition of 100 μM dNTPs led to a progressive increase in TMR fluorescence that reached a plateau, corresponding to 16% FRET, in about 45 min (Figure 1B), due to the increase in the distance (from ∼4.5 to ∼7 nm) between the fluorescent dyes as the full ds-duplex forms. As a control, neither the binding of RT nor the extension of the duplex was found to significantly affect the fluorescence of the same duplex labeled only with TMR (Supplementary Figure S3), clearly indicating that the FRET changes observed in Figure 1 were reflecting the polymerization activity of RT.
The progress curve in Figure 1B could be adequately fitted by a biexponential equation (Equation 1) with kobs1 = 19 (±1) × 10−3 s−1 and kobs2 = 17 (±3) × 10−4 s−1. Surprisingly, these rate constants were at least 10-fold slower as compared to the dNTP incorporation rates (0.1–0.4 s−1) reported under similar experimental conditions (43,53–55). To check whether these slower rate constants may be related to the Cy5 label in the primer sequence (Figure 2A), we monitored the polymerization reaction by using RT labeled in its p51 thumb subdomain by Alexa 488 and a duplex where the Cy5-labeled primer is annealed to the template labeled at its 3′ end by TMR (Figure 2B). In this approach, the polymerization is monitored through the changes in FRET between the Alexa 488-labeled RT and the TMR label in the p/t. In line with previous data where a similar approach has been used with a non labeled primer (48), the obtained kobs values (kobs1 = 17 (±3) × 10−2 s−1, kobs2 = 9 (±2) × 10−3 s−1) were similar to the dNTP incorporation rates (46,48,53), indicating that the Cy5 label in the primer does marginally affect RT polymerization. An alternative hypothesis may be that the TMR-containing region in the template is progressively folded during primer extension (Figure 1A), giving complex changes in the distance and thus, in the FRET efficiency between TMR and Cy5, so that the observed fluorescence signal is no more directly related to the number of nucleotides incorporated. To check this hypothesis, we compared the elongation kinetic traces of E.DNA with those obtained with E.DNA + 9 and E.DNA + 21 primers (Figure 2A) that were extended by respectively 9 and 21 nt, in respect with E.DNA (Figure 2C). Interestingly, though the primer was only extended by 9 nt in E.DNA + 9, the initial donor fluorescence was already close that of the final plateau, which as expected, strongly affected the amplitude associated to kobs1 (from 0.35 to 0.85). Moreover, the initial fluorescence of the trace with E.DNA + 21 was even higher than the plateau, indicating that the distance between the two dyes was increased as compared to the fully extended ds-DNA duplex. This increased distance likely results from the about 2-fold increased length per base of single stranded DNA as compared to double stranded DNA (56), so that at least for short segments, higher inter-chromophore distances can be achieved. Both the E.DNA + 9 and E.DNA + 21 data clearly indicate that the fluorescence changes observed as RT elongates the p/t DNA results from complex donor–acceptor distance changes, so that the apparent rate constants retrieved from the experimental curves do not reflect the real catalytic rates of RT.
Figure 2.
Kinetic curves with primers of different lengths. (A) Nucleic acid substrate sequences used in this study. Acceptor and donor fluorophores are attached by a linker to the T residues represented in red and green, in the primer and template sequences, respectively. (B) Polymerization kinetic trace of 100 nM RT labeled in its p51 thumb subdomain by Alexa 488 with 100 nM duplex, where the Cy5-labeled primer is annealed to the template labeled at its 3′ end by TMR, upon addition of 100 μM dNTPs. The black line corresponds to the fit of the trace by Equation (1), with kobs1 = 17 (±3) × 10−2 s−1, kobs2 = 9 (±2) × 10−3 s−1 and a = 0.5 (±0.1). Excitation and emission wavelengths were 480 and 520 nm, respectively. (C) Polymerization kinetic trace of 100 nM E.DNA (gray), 100 nM E.DNA + 9 (red) and E.DNA + 21 (blue) in the presence of 100 μM dNTP. All traces were fitted by Equation (1). Fitting of the gray trace according to Equation (1) provides kobs1 = 19 (±1) × 10−3 s−1, kobs2 = 17 (±3) × 10−4 s−1 and a = 0.35 (±0.1), fitting of the red trace provides kobs1 = 14 (±1) × 10−3 s−1, kobs2 = 13 (±3) × 10−4 s−1 and a = 0.85 (±0.1) and fitting of the blue trace provides kobs1 = 18 (±2) × 10−2 s−1, kobs2 = 63 (±5) × 10−4 s−1 and a = 0.5 (±0.1). Excitation and emission wavelengths were 540 and 580 nm for the kinetic traces, respectively.
Next, we explored the dependence of the kobs values on the dNTP concentrations to further identify the mechanistic steps monitored by this assay (Supplementary Figure S4). From the analysis of this dependence, the assay is thought to monitor the formation of the final extended duplex via two parallel pathways that depend on the initial binding events of RT to the p/t substrate. In this proposed mechanism, the lower pathway corresponds to the primer extension by RT properly bound in polymerase orientation, while the upper and slower pathway is related to either RT flipping or dissociation-rebinding of RT faulty bound on the nucleic acid substrate (Supplementary Figure S4).
Use of the assay to monitor the effect of RT inhibitors
As the assay monitors the RT polymerization, we next evaluated its ability to screen for RT inhibitors. To this end, we monitored the effect of two well-characterized RT inhibitors, Nevirapine and Zidovudine (AZTTP) (5–7,57), on the FRET-based kinetic curves. Inhibitors were serially diluted and pre-incubated at increasing concentrations with the E.DNA complex in cuvettes as well as in 96-well plates. The polymerization reaction was then triggered by adding dNTPs. In the presence of AZTTP, we observed a dramatic dose-dependent decrease of the values of the fluorescence intensity plateau (Figure 3A) and the two kinetic rate constants (Figure 3B). The changes in kobs1 and kobs2 values (Figure 3B) as a function of the inhibitor concentrations, I, were then used to determine IC50 values, according to (58):
 |
(2) |
where kobs0 and kobsf represent the kobs values in the absence and at saturating concentration of inhibitor, respectively. IC50 represents the half maximal inhibitory concentration and p denotes the Hill coefficient. IC50 values determined for AZTTP were found to be 7 (±1) μM and 6 (±1) μM for kobs1 and kobs2, respectively. These values are in good agreement with the values reported in literature (48,59).
Figure 3.
RT polymerization in the presence of RT inhibitors. (A) Representative set of kinetic traces of 100 nM E.DNA in the absence (gray) or in the presence of increasing concentrations of AZTTP (red—1 μM, blue—3 μM, magenta—5 μM, dark green—10 μM, cyan—20 μM) recorded in a 96-well plate. The black lines correspond to the fits of the kinetic traces with Equation (1) and the kobs values given in panel B. (B) Dependence of kobs1 and kobs2 (inset) values on the concentration of AZTTP. The red lines correspond to the best fits of the kinetic traces to Equation (2) and the IC50 values given in text. (C) Representative set of kinetic traces of 100 nM E.DNA in the absence (gray) or in the presence of increasing concentrations of Nevirapine (green—20 nM, magenta—50 nM, blue—70 nM, red—100 nM, cyan—200 nM) recorded in cuvettes. The black lines correspond to the fits of the kinetic traces with Equation (1) and the kobs values given in panel D. (D) Dependence of kobs1 and kobs2 (inset) values on the concentration of Nevirapine. The kobs values were obtained from the fit of the kinetic traces in panel C to Equation (1). The red lines correspond to best fit of the data to Equation (2) and the IC50 values given in the text. Excitation and emission wavelengths were 540 and 580 nm, respectively.
Similar dose-dependent decrease in the values of kobs1 and kobs2 was observed upon addition of Nevirapine to E.DNA complex (Figure 3C and D). IC50 values for Nevirapine calculated from kobs1 (Figure 3D) and kobs2 (Figure 3D, inset) were found to be 71 (±8) nM and 74 (±9) nM, respectively. Once again, these IC50 values were similar to previously reported IC50 values (48,60). Thus, the assay appears highly relevant for quantitatively determining the potency of RT inhibitors.
Finally, in order to evaluate the quality of the assay for high-throughput screening (HTS) applications, we calculated a Z-factor for the kobs1 and kobs2 values at each inhibitor concentration, using Equation (3):
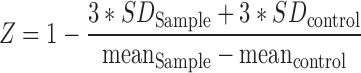 |
(3) |
where mean and SD, corresponds to the mean and standard deviation of the kobs values for three different sets of experiments. The control values corresponded to the kobs values in the absence of inhibitor. The Z factor is a quantitative parameter of the dynamic range and data variation associated with the measured signal in the assay (61). Except for the low inhibitor concentrations, we observed Z-factors in the range of 0.5 to 0.89 for both AZTTP and Nevirapine (Figure 4A and B), showing the excellent sensitivity and robustness of the assay (61).
Figure 4.
Z-factor of the assay. Z-factor values for kobs1 (open squares) and kobs2 (closed disks). These values were calculated for (A) AZTTP and (B) Nevirapine according to Equation (3).
Use of the assay to monitor the concerted activity of NCp7 and RT
Emergence of resistance to highly active antiviral therapy and notably RT inhibitors remains a critical problem in HIV-1 patient management. Development of new antiviral agents targeting multiple HIV-1 proteins could limit cross-resistance observed with the currently used drugs (62–68). A particularly attractive complementary target to RT could be the highly conserved NCp7 protein (13–15), that is thought to promote reverse transcription in the RTC. In this context, the disclosure of hits that would target simultaneously RT and NCp7 could be of utmost interest.
We investigated whether the joint activities of RT and NCp7 could be monitored using our FRET based assay. In the presence of 1 μM of NCp7, we observed a 3-fold increase in both kobs1 and kobs2 values, as well as a small increase in the final fluorescence plateau (Figure 5A, orange trace and Figure 5B), indicating that our assay can sensitively monitor the promotion by NCp7 of the RT polymerization activity. Noticeably, no significant change in the emission of p/t labeled only by the FRET donor was observed in the presence of NCp7 (Supplementary Figure S3), suggesting that no noticeable NCp7-induced aggregation of p/t occurred in these conditions. This is likely the consequence of the high Mg2+ concentration in the polymerization buffer that was shown to strongly reduce the nucleic acid aggregation properties of NCp7 (23). To further validate our assay, and notably its ability to screen for NCp7 inhibitors, we monitored the effect of WDO-217 on RT polymerization in the presence of NCp7. WDO-217 is a phenyl-thiadiazolylidene-amine derivative that binds to NCp7 and efficiently ejects zinc from its two zinc fingers, even when NCp7 is bound to nucleic acids, inhibiting the nucleic acid chaperone activity of NCp7 (69). The addition of WDO-217 led to a full inhibition of the NCp7 effect on RT polymerization, providing the same kinetic parameters and fluorescence plateau as compared to RT alone (Figure 5A, compare magenta and green traces, and Figure 5B). In contrast, WDO-217 showed no effect on RT (Figure 5A, compare green and dark blue traces). The inhibition of NCp7 activity likely results from the previously demonstrated change in its binding mode to nucleic acids, as a result of zinc ejection (69). The binding mode of the zinc-depleted NCp7 is likely dominated by electrostatic interactions with the phosphate groups of the oligonucleotide backbone and is thought to prevent the nucleic acid chaperone properties of NCp7 as well as its productive interaction with RT. This has been verified by replacing NCp7 with NCp7(SSHS), a mutant that is unable to bind zinc (70) but still binds nucleic acids with high affinity (71). This mutant provided similar polymerization profile (Figure 5A, black trace) and kinetic parameters (Figure 5B) as compared to RT alone, mimicking the condition where WDO-217 is added to NCp7. Unfortunately, the additional control using an inhibitor of both proteins could not be performed as no molecule to our knowledge has been reported to be active both on RT polymerization and NCp7.
Figure 5.
RT polymerization in the presence of NCp7 and an NCp7 inhibitor. (A) Kinetic traces of 100 nM E.DNA in the absence (green) and in the presence of 1 μM NCp7 (orange) or 10 μM WDO (blue) or 1 μM NCp7(SSHS) (black). The magenta trace was recorded for 100 nM E.DNA in the presence of both 1 μM NCp7 and 10 μM WDO. All traces were fitted to Equation (1) and the rate constant values provided in panel B. (B) Histogram showing the kobs1 (black bars) and kobs2 (red bars) values extracted from the fits of the kinetic curves of panel A to Equation (1). Excitation and emission wavelengths were 540 and 580 nm, respectively.
Taken together, our data indicate that our direct and continuous assay can monitor the concerted activity of NCp7 and RT, and has the potential to identify molecules that inhibit one of these proteins or both at the same time.
CONCLUSION
Here, we describe an efficient, robust and reproducible real-time assay to screen for RT inhibitors, by monitoring RT polymerization through changes in the FRET signal of a doubly-labeled p/t duplex. In contrast to most existing assays (28–42), this assay is a continuous one-step and one-pot method that does not require cumbersome sample handling. Moreover, it presents the advantage to use commercially available fluorescently labeled oligonucleotides and non-modified RT. This is in contrast to previous assays that require fluorescent or radiolabeled nucleosides (28–42) or the preparation and purification of fluorescently-labeled RT (43–47). Using AZT and Nevirapine as representative examples of NRTI and NNRTI, respectively, it was found that both compounds dramatically decreased the kinetic rate constants and the fluorescence plateau of the FRET-based progress curves. From the dependence of the kinetic rate constants on the inhibitor concentration, reliable IC50 values were obtained for both inhibitors. Moreover, the assay shows high Z-factor values, showing its high sensitivity and suitability for HTS. In addition, the assay proved useful to monitor the promotion by NCp7 of RT polymerization. This promotion was fully reverted by the addition of an NCp7 inhibitor, indicating that this assay can also disclose NCp7 inhibitors. Therefore, the assay appears highly potent, being able to screen for RT and NCp7 inhibitors, as well as for compounds targeting the two proteins. The latter would be of high interest, as RT inhibitors are prone to resistance while due to its high conservation, NCp7 inhibitors are thought to be much less prone to resistance (15,16). Finally, due to its simplicity and ease to use, our one-pot and one-step FRET-based assay will certainly be useful for monitoring the activity of other eukaryotic and prokaryotic polymerases, in order to substitute previous assays using more sophisticated fluorescence-based techniques (72–77) or radiolabeled substrates (78–80). Of course, the assay is expected also to present some limitations, the main one being the interference by the auto-fluorescent compounds in the libraries used for screening. Nevertheless, as we are using TMR and CY5 that absorb above 500 nm, the number of such interfering compounds should be limited. Another limitation will be provided by DNA intercalators that will alter the RT polymerization activity, and thus appear as false positives.
Supplementary Material
Acknowledgments
Agence Nationale de la Recherche (ANR blanc Femtostack) (to Y.M.); European Project THINPAD ‘Targeting the HIV-1 Nucleocapsid Protein to fight Antiretroviral Drug Resistance’ [FP7-Grant Agreement 601969 to Y.M.]. Funding for open access charge: European Project THINPAD ‘Targeting the HIV-1 Nucleocapsid Protein to fight Antiretroviral Drug Resistance’ [FP7-Grant Agreement 601969 to Y.M.].
Conflict of interest statement. None declared.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
REFERENCES
- 1.Le Grice S.F. Human immunodeficiency virus reverse transcriptase: 25 years of research, drug discovery, and promise. J. Biol. Chem. 2012;287:40850–40857. doi: 10.1074/jbc.R112.389056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacobo-Molina A., Ding J., Nanni R.G., Clark A.D., Jr, Lu X., Tantillo C., Williams R.L., Kamer G., Ferris A.L., Clark P. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc. Natl. Acad. Sci. U.S.A. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohlstaedt L.A., Wang J., Friedman J.M., Rice P.A., Steitz T.A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 4.Hachiya A., Shimane K., Sarafianos S.G., Kodama E.N., Sakagami Y., Negishi F., Koizumi H., Gatanaga H., Matsuoka M., Takiguchi M., et al. Clinical relevance of substitutions in the connection subdomain and RNase H domain of HIV-1 reverse transcriptase from a cohort of antiretroviral treatment-naive patients. Antiviral Res. 2009;82:115–121. doi: 10.1016/j.antiviral.2009.02.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barber A.M., Hizi A., Maizel J.V., Jr, Hughes S.H. HIV-1 reverse transcriptase: structure predictions for the polymerase domain. AIDS Res. Hum. Retroviruses. 1990;6:1061–1072. doi: 10.1089/aid.1990.6.1061. [DOI] [PubMed] [Google Scholar]
- 6.Li X., Zhang L., Tian Y., Song Y., Zhan P., Liu X. Novel HIV-1 non-nucleoside reverse transcriptase inhibitors: a patent review (2011–2014) Expert Opin. Ther. Pat. 2014;24:1199–1227. doi: 10.1517/13543776.2014.964685. [DOI] [PubMed] [Google Scholar]
- 7.Paydary K., Khaghani P., Emamzadeh-Fard S., Alinaghi S.A., Baesi K. The emergence of drug resistant HIV variants and novel anti-retroviral therapy. Asian Pac. J. Trop. Biomed. 2013;3:515–522. doi: 10.1016/S2221-1691(13)60106-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Althaus I.W., Chou J.J., Gonzales A.J., Deibel M.R., Chou K.C., Kezdy F.J., Romero D.L., Aristoff P.A., Tarpley W.G., Reusser F. Steady-state kinetic studies with the non-nucleoside HIV-1 reverse transcriptase inhibitor U-87201E. J. Biol. Chem. 1993;268:6119–6124. [PubMed] [Google Scholar]
- 9.Althaus I.W., Chou J.J., Gonzales A.J., Deibel M.R., Chou K.C., Kezdy F.J., Romero D.L., Palmer J.R., Thomas R.C., Aristoff P.A., et al. Kinetic studies with the non-nucleoside HIV-1 reverse transcriptase inhibitor U-88204E. Biochemistry. 1993;32:6548–6554. doi: 10.1021/bi00077a008. [DOI] [PubMed] [Google Scholar]
- 10.Domaoal R.A., Demeter L.M. Structural and biochemical effects of human immunodeficiency virus mutants resistant to non-nucleoside reverse transcriptase inhibitors. Int. J. Biochem. Cell Biol. 2004;36:1735–1751. doi: 10.1016/j.biocel.2004.02.026. [DOI] [PubMed] [Google Scholar]
- 11.Sarafianos S.G., Das K., Hughes S.H., Arnold E. Taking aim at a moving target: designing drugs to inhibit drug-resistant HIV-1 reverse transcriptases. Curr. Opin. Struct. Biol. 2004;14:716–730. doi: 10.1016/j.sbi.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 12.Sarafianos S.G., Hughes S.H., Arnold E. Designing anti-AIDS drugs targeting the major mechanism of HIV-1 RT resistance to nucleoside analog drugs. Int. J. Biochem. Cell Biol. 2004;36:1706–1715. doi: 10.1016/j.biocel.2004.02.027. [DOI] [PubMed] [Google Scholar]
- 13.Darlix J.L., Garrido J.L., Morellet N., Mély Y., de Rocquigny H. Properties, functions, and drug targeting of the multifunctional nucleocapsid protein of the human immunodeficiency virus. Adv. Pharmacol. 2007;55:299–346. doi: 10.1016/S1054-3589(07)55009-X. [DOI] [PubMed] [Google Scholar]
- 14.Goldschmidt V., Jenkins L.M.M., de Rocquigny H., Darlix J.-L., Mély Y. The nucleocapsid protein of HIV-1 as a promising therapeutic target for antiviral drugs. HIV Ther. 2010;4:179–198. [Google Scholar]
- 15.Mori M., Kovalenko L., Lyonnais S., Antaki D., Torbett B.E., Botta M., Mirambeau G., Mely Y. Nucleocapsid protein: a desirable target for future therapies against HIV-1. Curr. Top. Microbiol. Immunol. 2015;389:53–92. doi: 10.1007/82_2015_433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darlix J.L., Godet J., Ivanyi-Nagy R., Fosse P., Mauffret O., Mely Y. Flexible nature and specific functions of the HIV-1 nucleocapsid protein. J. Mol. Biol. 2011;410:565–581. doi: 10.1016/j.jmb.2011.03.037. [DOI] [PubMed] [Google Scholar]
- 17.Godet J., Mely Y. Biophysical studies of the nucleic acid chaperone properties of the HIV-1 nucleocapsid protein. RNA Biol. 2010;7:687–699. doi: 10.4161/rna.7.6.13616. [DOI] [PubMed] [Google Scholar]
- 18.Levin J.G., Mitra M., Mascarenhas A., Musier-Forsyth K. Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biol. 2010;7:754–774. doi: 10.4161/rna.7.6.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas J.A., Gorelick R.J. Nucleocapsid protein function in early infection processes. Virus Res. 2008;134:39–63. doi: 10.1016/j.virusres.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anthony R.M., Destefano J.J. In vitro synthesis of long DNA products in reactions with HIV-RT and nucleocapsid protein. J. Mol. Biol. 2007;365:310–324. doi: 10.1016/j.jmb.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levin J.G., Guo J., Rouzina I., Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 22.Bampi C., Bibillo A., Wendeler M., Divita G., Gorelick R.J., Le Grice S.F., Darlix J.L. Nucleotide excision repair and template-independent addition by HIV-1 reverse transcriptase in the presence of nucleocapsid protein. J. Biol. Chem. 2006;281:11736–11743. doi: 10.1074/jbc.M600290200. [DOI] [PubMed] [Google Scholar]
- 23.Grohmann D., Godet J., Mely Y., Darlix J.L., Restle T. HIV-1 nucleocapsid traps reverse transcriptase on nucleic acid substrates. Biochemistry. 2008;47:12230–12240. doi: 10.1021/bi801386r. [DOI] [PubMed] [Google Scholar]
- 24.Tanchou V., Delaunay T., Bodeus M., Roques B., Darlix J.L., Benarous R. Conformational changes between human immunodeficiency virus type 1 nucleocapsid protein NCp7 and its precursor NCp15 as detected by anti-NCp7 monoclonal antibodies. J. Gen. Virol. 1995;76:2457–2466. doi: 10.1099/0022-1317-76-10-2457. [DOI] [PubMed] [Google Scholar]
- 25.Druillennec S., Caneparo A., de Rocquigny H., Roques B.P. Evidence of interactions between the nucleocapsid protein NCp7 and the reverse transcriptase of HIV-1. J. Biol. Chem. 1999;274:11283–11288. doi: 10.1074/jbc.274.16.11283. [DOI] [PubMed] [Google Scholar]
- 26.Kim J., Roberts A., Yuan H., Xiong Y., Anderson K.S. Nucleocapsid protein annealing of a primer-template enhances (+)-strand DNA synthesis and fidelity by HIV-1 reverse transcriptase. J. Mol. Biol. 2012;415:866–880. doi: 10.1016/j.jmb.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lener D., Tanchou V., Roques B.P., Le Grice S.F., Darlix J.L. Involvement of HIV-I nucleocapsid protein in the recruitment of reverse transcriptase into nucleoprotein complexes formed in vitro. J. Biol. Chem. 1998;273:33781–33786. doi: 10.1074/jbc.273.50.33781. [DOI] [PubMed] [Google Scholar]
- 28.Antoun M.D., Rios Y.R., Mendoza N.T., Proctor G. Reverse transcriptase inhibition as prescreen for potential antiviral bioactives. P.R. Health Sci. J. 1994;13:17–18. [PubMed] [Google Scholar]
- 29.Chang A., Ostrove J.M., Bird R.E. Development of an improved product enhanced reverse transcriptase assay. J. Virol. Methods. 1997;65:45–54. doi: 10.1016/s0166-0934(96)02168-4. [DOI] [PubMed] [Google Scholar]
- 30.Hizi A., Tal R., Shaharabany M., Currens M.J., Boyd M.R., Hughes S.H., McMahon J.B. Specific inhibition of the reverse transcriptase of human immunodeficiency virus type 1 and the chimeric enzymes of human immunodeficiency virus type 1 and type 2 by nonnucleoside inhibitors. Antimicrob. Agents Chemother. 1993;37:1037–1042. doi: 10.1128/aac.37.5.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanyara J.N., Njagi E.N. Anti-HIV-1 activities in extracts from some medicinal plants as assessed in an in vitro biochemical HIV-1 reverse transcriptase assay. Phytother. Res. 2005;19:287–290. doi: 10.1002/ptr.1536. [DOI] [PubMed] [Google Scholar]
- 32.Krebs J.F., Kore A.R. Novel FRET-based assay to detect reverse transcriptase activity using modified dUTP analogues. Bioconjug. Chem. 2008;19:185–191. doi: 10.1021/bc700284y. [DOI] [PubMed] [Google Scholar]
- 33.Lovatt A., Black J., Galbraith D., Doherty I., Moran M.W., Shepherd A.J., Griffen A., Bailey A., Wilson N., Smith K.T. High throughput detection of retrovirus-associated reverse transcriptase using an improved fluorescent product enhanced reverse transcriptase assay and its comparison to conventional detection methods. J. Virol. Methods. 1999;82:185–200. doi: 10.1016/s0166-0934(99)00111-1. [DOI] [PubMed] [Google Scholar]
- 34.Odawara F., Abe H., Kohno T., Nagai-Fujii Y., Arai K., Imamura S., Misaki H., Azuma H., Ikebuchi K., Ikeda H., et al. A highly sensitive chemiluminescent reverse transcriptase assay for human immunodeficiency virus. J. Virol. Methods. 2002;106:115–124. doi: 10.1016/s0166-0934(02)00142-8. [DOI] [PubMed] [Google Scholar]
- 35.Porstmann T., Meissner K., Glaser R., Dopel S.H., Sydow G. A sensitive non-isotopic assay specific for HIV-1 associated reverse transcriptase. J. Virol. Methods. 1991;31:181–188. doi: 10.1016/0166-0934(91)90156-t. [DOI] [PubMed] [Google Scholar]
- 36.Silver J., Maudru T., Fujita K., Repaske R. An RT-PCR assay for the enzyme activity of reverse transcriptase capable of detecting single virions. Nucleic Acids Res. 1993;21:3593–3594. doi: 10.1093/nar/21.15.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki K., Craddock B.P., Kano T., Steigbigel R.T. Colorimetric reverse transcriptase assay for HIV-1. J. Virol. Methods. 1993;41:21–28. doi: 10.1016/0166-0934(93)90159-o. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki K., Craddock B.P., Kano T., Steigbigel R.T. Chemiluminescent enzyme-linked immunoassay for reverse transcriptase, illustrated by detection of HIV reverse transcriptase. Anal. Biochem. 1993;210:277–281. doi: 10.1006/abio.1993.1196. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki K., Craddock B.P., Okamoto N., Kano T., Steigbigel R.T. Poly A-linked colorimetric microtiter plate assay for HIV reverse transcriptase. J. Virol. Methods. 1993;44:189–198. doi: 10.1016/0166-0934(93)90054-u. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki K., Craddock B.P., Okamoto N., Kano T., Steigbigel R.T. Detection of human immunodeficiency virus (HIV) by colorimetric assay for reverse transcriptase activity on magnetic beads. Biotechnol. Appl. Biochem. 1993;18:37–44. [PubMed] [Google Scholar]
- 41.Urabe T., Sano K., Tanno M., Mizoguchi J., Otani M., Lee M.H., Takasaki T., Kusakabe H., Imagawa D.T., Nakai M. A non-radioisotopic reverse transcriptase assay using biotin-11-deoxyuridinetriphosphate on primer-immobilized microtiter plates. J. Virol. Methods. 1992;40:145–154. doi: 10.1016/0166-0934(92)90063-j. [DOI] [PubMed] [Google Scholar]
- 42.Vassiliou W., Epp J.B., Wang B.B., Del Vecchio A.M., Widlanski T., Kao C.C. Exploiting polymerase promiscuity: a simple colorimetric RNA polymerase assay. Virology. 2000;274:429–437. doi: 10.1006/viro.2000.0492. [DOI] [PubMed] [Google Scholar]
- 43.Abbondanzieri E.A., Bokinsky G., Rausch J.W., Zhang J.X., Le Grice S.F., Zhuang X. Dynamic binding orientations direct activity of HIV reverse transcriptase. Nature. 2008;453:184–189. doi: 10.1038/nature06941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kellinger M.W., Johnson K.A. Role of induced fit in limiting discrimination against AZT by HIV reverse transcriptase. Biochemistry. 2011;50:5008–5015. doi: 10.1021/bi200204m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kirmizialtin S., Nguyen V., Johnson K.A., Elber R. How conformational dynamics of DNA polymerase select correct substrates: experiments and simulations. Structure. 2012;20:618–627. doi: 10.1016/j.str.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu S., Abbondanzieri E.A., Rausch J.W., Le Grice S.F., Zhuang X. Slide into action: dynamic shuttling of HIV reverse transcriptase on nucleic acid substrates. Science. 2008;322:1092–1097. doi: 10.1126/science.1163108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schauer G.D., Huber K.D., Leuba S.H., Sluis-Cremer N. Mechanism of allosteric inhibition of HIV-1 reverse transcriptase revealed by single-molecule and ensemble fluorescence. Nucleic Acids Res. 2014;42:11687–11696. doi: 10.1093/nar/gku819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharma K.K., Przybilla F., Restle T., Boudier C., Godet J., Mely Y. Reverse transcriptase in action: FRET-based assay for monitoring flipping and polymerase activity in real time. Anal. Chem. 2015;87:7690–7697. doi: 10.1021/acs.analchem.5b01126. [DOI] [PubMed] [Google Scholar]
- 49.Muller B., Restle T., Weiss S., Gautel M., Sczakiel G., Goody R.S. Co-expression of the subunits of the heterodimer of HIV-1 reverse transcriptase in Escherichia coli. J. Biol. Chem. 1989;264:13975–13978. [PubMed] [Google Scholar]
- 50.Schatz O., Cromme F.V., Gruninger-Leitch F., Le Grice S.F. Point mutations in conserved amino acid residues within the C-terminal domain of HIV-1 reverse transcriptase specifically repress RNase H function. FEBS Lett. 1989;257:311–314. doi: 10.1016/0014-5793(89)81559-5. [DOI] [PubMed] [Google Scholar]
- 51.Shvadchak V., Sanglier S., Rocle S., Villa P., Haiech J., Hibert M., Van Dorsselaer A., Mely Y., de Rocquigny H. Identification by high throughput screening of small compounds inhibiting the nucleic acid destabilization activity of the HIV-1 nucleocapsid protein. Biochimie. 2009;91:916–923. doi: 10.1016/j.biochi.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 52.Klarmann G.J., Schauber C.A., Preston B.D. Template-directed pausing of DNA synthesis by HIV-1 reverse transcriptase during polymerization of HIV-1 sequences in vitro. J. Biol. Chem. 1993;268:9793–9802. [PubMed] [Google Scholar]
- 53.Kati W.M., Johnson K.A., Jerva L.F., Anderson K.S. Mechanism and fidelity of HIV reverse transcriptase. J. Biol. Chem. 1992;267:25988–25997. [PubMed] [Google Scholar]
- 54.Sarafianos S.G., Pandey V.N., Kaushik N., Modak M.J. Site-directed mutagenesis of arginine 72 of HIV-1 reverse transcriptase. Catalytic role and inhibitor sensitivity. J. Biol. Chem. 1995;270:19729–19735. doi: 10.1074/jbc.270.34.19729. [DOI] [PubMed] [Google Scholar]
- 55.Sarafianos S.G., Pandey V.N., Kaushik N., Modak M.J. Glutamine 151 participates in the substrate dNTP binding function of HIV-1 reverse transcriptase. Biochemistry. 1995;34:7207–7216. doi: 10.1021/bi00021a036. [DOI] [PubMed] [Google Scholar]
- 56.Chi Q., Wang G., Jiang J. The persistence length and length per base of single-stranded DNA obtained from fluorescence correlation spectroscopy measurements using mean field theory. Phys. A Stat. Mech. Appl. 2013;392:1072–1079. [Google Scholar]
- 57.Wright K. AIDS therapy. First tentative signs of therapeutic promise. Nature. 1986;323:283. doi: 10.1038/323283a0. [DOI] [PubMed] [Google Scholar]
- 58.Martin L. Benchmark Dose Software (BMDS) Version 2.1 User's Manual Version 2.0. Washington, DC: United States Environmental Protection Agency, Office of Environmental Information; 2009. [Google Scholar]
- 59.Caliendo A.M., Savara A., An D., DeVore K., Kaplan J.C., D'Aquila R.T. Effects of zidovudine-selected human immunodeficiency virus type 1 reverse transcriptase amino acid substitutions on processive DNA synthesis and viral replication. J. Virol. 1996;70:2146–2153. doi: 10.1128/jvi.70.4.2146-2153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Richman D., Rosenthal A.S., Skoog M., Eckner R.J., Chou T.C., Sabo J.P., Merluzzi V.J. BI-RG-587 is active against zidovudine-resistant human immunodeficiency virus type 1 and synergistic with zidovudine. Antimicrob. Agents Chemother. 1991;35:305–308. doi: 10.1128/aac.35.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J.H., Chung T.D., Oldenburg K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 62.Cohen J. HIV suppressed long after treatment. Science. 1997;277:1927. doi: 10.1126/science.277.5334.1927. [DOI] [PubMed] [Google Scholar]
- 63.Cohen J. Novel campaign to test live HIV vaccine. Science. 1997;277:1035. doi: 10.1126/science.277.5329.1035. [DOI] [PubMed] [Google Scholar]
- 64.Cohen J. The daunting challenge of keeping HIV suppressed. Science. 1997;277:32–33. doi: 10.1126/science.277.5322.32. [DOI] [PubMed] [Google Scholar]
- 65.Finzi D., Hermankova M., Pierson T., Carruth L.M., Buck C., Chaisson R.E., Quinn T.C., Chadwick K., Margolick J., Brookmeyer R., et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 66.Gulnik S.V., Suvorov L.I., Liu B., Yu B., Anderson B., Mitsuya H., Erickson J.W. Kinetic characterization and cross-resistance patterns of HIV-1 protease mutants selected under drug pressure. Biochemistry. 1995;34:9282–9287. doi: 10.1021/bi00029a002. [DOI] [PubMed] [Google Scholar]
- 67.Wong J.K., Gunthard H.F., Havlir D.V., Zhang Z.Q., Haase A.T., Ignacio C.C., Kwok S., Emini E., Richman D.D. Reduction of HIV-1 in blood and lymph nodes following potent antiretroviral therapy and the virologic correlates of treatment failure. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12574–12579. doi: 10.1073/pnas.94.23.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wong J.K., Hezareh M., Gunthard H.F., Havlir D.V., Ignacio C.C., Spina C.A., Richman D.D. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 69.Vercruysse T., Basta B., Dehaen W., Humbert N., Balzarini J., Debaene F., Sanglier-Cianferani S., Pannecouque C., Mely Y., Daelemans D. A phenyl-thiadiazolylidene-amine derivative ejects zinc from retroviral nucleocapsid zinc fingers and inactivates HIV virions. Retrovirology. 2012;9:95–109. doi: 10.1186/1742-4690-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Green L.M., Berg J.M. Retroviral nucleocapsid protein-metal ion interactions: folding and sequence variants. Proc. Natl. Acad. Sci. U.S.A. 1990;87:6403–6407. doi: 10.1073/pnas.87.16.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beltz H., Clauss C., Piemont E., Ficheux D., Gorelick R.J., Roques B., Gabus C., Darlix J.L., de Rocquigny H., Mely Y. Structural determinants of HIV-1 nucleocapsid protein for cTAR DNA binding and destabilization, and correlation with inhibition of self-primed DNA synthesis. J. Mol. Biology. 2005;348:1113–1126. doi: 10.1016/j.jmb.2005.02.042. [DOI] [PubMed] [Google Scholar]
- 72.Griep M.A. Fluorescence recovery assay: a continuous assay for processive DNA polymerases applied specifically to DNA polymerase III holoenzyme. Anal. Biochem. 1995;232:180–189. doi: 10.1006/abio.1995.0005. [DOI] [PubMed] [Google Scholar]
- 73.Ma C.B., Comerford L., Wilson J., Puttlitz C.M. Biomechanical evaluation of arthroscopic rotator cuff repairs: double-row compared with single-row fixation. J. Bone Joint Surg. Am. 2006;88:403–410. doi: 10.2106/JBJS.D.02887. [DOI] [PubMed] [Google Scholar]
- 74.Schwartz J.J., Quake S.R. Single molecule measurement of the ‘speed limit’ of DNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 2009;106:20294–20299. doi: 10.1073/pnas.0907404106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Seville M., West A.B., Cull M.G., McHenry C.S. Fluorometric assay for DNA polymerases and reverse transcriptase. Biotechniques. 1996;21:664–672. doi: 10.2144/96214st04. [DOI] [PubMed] [Google Scholar]
- 76.Tveit H., Kristensen T. Fluorescence-based DNA polymerase assay. Anal. Biochem. 2001;289:96–98. doi: 10.1006/abio.2000.4903. [DOI] [PubMed] [Google Scholar]
- 77.Joyce C.M. Techniques used to study the DNA polymerase reaction pathway. Biochim. Biophys. Acta. 2010;1804:1032–1040. doi: 10.1016/j.bbapap.2009.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Horowitz R.W., Zhang H., Schwartz E.L., Ladner R.D., Wadler S. Measurement of deoxyuridine triphosphate and thymidine triphosphate in the extracts of thymidylate synthase-inhibited cells using a modified DNA polymerase assay. Biochem. Pharmacol. 1997;54:635–638. doi: 10.1016/s0006-2952(97)00259-1. [DOI] [PubMed] [Google Scholar]
- 79.Sherman P.A., Fyfe J.A. Enzymatic assay for deoxyribonucleoside triphosphates using synthetic oligonucleotides as template primers. Anal. Biochem. 1989;180:222–226. doi: 10.1016/0003-2697(89)90420-x. [DOI] [PubMed] [Google Scholar]
- 80.Smid K., Van Moorsel C.J., Noordhuis P., Voorn D.A., Peters G.J. Interference of gemcitabine triphosphate with the measurements of deoxynucleotides using an optimized DNA polymerase elongation assay. Int. J. Oncol. 2001;19:157–162. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.







