Abstract
Staufen2 (Stau2) is an RNA-binding protein involved in cell fate decision by controlling several facets of mRNA processing including localization, splicing, translation and stability. Herein we report that exposure to DNA-damaging agents that generate replicative stress such as camptothecin (CPT), 5-fluoro-uracil (5FU) and ultraviolet radiation (UVC) causes downregulation of Stau2 in HCT116 colorectal cancer cells. In contrast, other agents such as doxorubicin and ionizing radiation had no effect on Stau2 expression. Consistently, Stau2 expression is regulated by the ataxia telangiectasia and Rad3-related (ATR) signaling pathway but not by the DNA-PK or ataxia telangiectasia mutated/checkpoint kinase 2 pathways. Stau2 downregulation is initiated at the level of transcription, independently of apoptosis induction. Promoter analysis identified a short 198 bp region which is necessary and sufficient for both basal and CPT-regulated Stau2 expression. The E2F1 transcription factor regulates Stau2 in untreated cells, an effect that is abolished by CPT treatment due to E2F1 displacement from the promoter. Strikingly, Stau2 downregulation enhances levels of DNA damage and promotes apoptosis in CPT-treated cells. Taken together our results suggest that Stau2 is an anti-apoptotic protein that could be involved in DNA replication and/or maintenance of genome integrity and that its expression is regulated by E2F1 via the ATR signaling pathway.
INTRODUCTION
Chromosomal DNA is constantly exposed to endogenous and exogenous mutagens (1) that induce DNA damage with attendant genotoxic consequences including cell death, mutagenesis and carcinogenesis (2). Therefore, to maintain genomic integrity, eukaryotic cells have evolved a finely-tuned global response, termed the ‘DNA damage response’ (DDR), consisting of DNA damage detection leading to activation of signal transduction cascades that mediate reversible periods of cell cycle arrest and DNA repair (3,4). Alternatively, when repair pathways fail or become overwhelmed, or if cells are able to re-enter the growth cycle before repair is completed, mechanisms of irreversible growth arrest (senescence) or programmed cell death (apoptosis) are initiated (3). Senescence and apoptosis constitute powerful tumor-suppressive mechanisms that, respectively, completely forestall proliferation of, or destroy, severely genetically-damaged cancer-prone cells.
DDR pathways involve a preeminent contribution by the phosphoinositide 3-kinase related kinases, including ataxia telangiectasia mutated (ATM), ataxia telangiectasia and Rad3-related (ATR) and DNA-activated protein kinase (DNA-PK) (1,2). During genotoxic stress these enzymes phosphorylate hundreds of substrates either alone, or through the intermediacy of the downstream effector kinases checkpoint kinase 1 (CHEK1) and checkpoint kinase 2 (CHEK2) activated primarily by ATR and ATM, respectively. Among other effects, this culminates in stimulation of transcription factors such as p53, E2F1 and NF-κB which in turn positively and/or negatively regulate DDR gene expression. The DDR is differentially regulated depending on the type of DNA damage sustained by cells (1,2,5). Specifically, DNA double-strand breaks (DSBs) engender rapid activation of the ATM and DNA-PK pathways (6) whereas DNA adducts that induce replicative stress by blocking the progression of DNA polymerases trigger rapid activation of the ATR pathway (7). Moreover, stalled replication forks may eventually collapse leading to DSB formation, and thus initial activation of ATR signaling can be followed by activation of ATM a number of hours later (8). Similarly, DSB formation initially sensed by ATM signaling is followed later during the repair process by DNA end resection, which generates RPA-coated single stranded overhangs leading to ATR activation (1,2,6). In any case, the mechanisms by which cells ‘decide’ to induce programs leading to either cell cycle arrest/DNA repair or senescence/apoptosis are not entirely clear; however the balance between levels of pro- and anti-apoptotic proteins, mediated in large part by transcription factors such as p53, E2F1 and NF-κB, lie at the heart of the decision (3,9–12). For example, E2F1-mediated activation of p53 results primarily in p53-dependent apoptosis rather than growth arrest (13–15). Indeed, certain critical proteins, many of which are transcription factors, can integrate diverse signals modulated by levels of DNA damage thereby finely tuning the equilibrium of pro- versus anti-apoptotic protein expression.
High-throughput genomic/proteomic approaches have revealed RNA-binding proteins, as well as proteins implicated in RNA processing and post-transcriptional mRNA regulation, as putative novel regulators of the DDR (16–19). We thus became interested in the possibility of a potential role for Stau2 in the DDR. Stau2 is a double-stranded RNA-binding protein that associates with RNA secondary structures (20,21). The Stau2 gene, through differential splicing, generates at least four isoforms varying at their N- and/or C-termini. Stau2 is a component of ribonucleoprotein complexes (20,22,23) involved in mRNA transport (20,21,24), differential splicing (25), translation (26,27) and mRNA decay (28). In mammals, downregulation of this protein impairs mRNA transport to neuronal dendrites, causes dendritic spine defects and prevents long-term depression of hippocampal neurons (21,24,26). In zebrafish, Stau2 is required for survival and migration of primordial germ cells (29), while in Xenopus it participates in anterior endodermal organ formation (30). Interestingly, in chicken, Stau2 downregulation engenders small eye development as a consequence of reduced cell proliferation, with no evidence of necrosis or apoptosis (31). Similarly, in rat neural stem cells, Stau2 regulates the balance of stem cell maintenance versus differentiation (32). In the latter case, downregulation of the protein induces cell differentiation, whereas over-expression induces proliferation (33).
We recently employed a genome-wide approach to identify Stau2-bound mRNAs in HEK293T cells (34), which revealed prevalent groups of transcripts involved in ubiquitination, catabolism, splicing and intracellular transport (34). Interestingly 49 Stau2-bound mRNAs encode proteins specifically involved in the DDR, and 150 encode ones related to cell death or apoptotic pathways. Moreover, a previous high-throughput siRNA screen aimed at identifying proteins that normally protect against apoptosis uncovered 73 and 72 pro-survival kinases and phosphatases respectively (35). We note that, strikingly, mRNAs coding for 6 kinases and 10 phosphatases within this collection are among those bound by Stau2. Herein, we show that during genotoxic stress, Stau2 is downregulated at the promoter level in an ATR- and E2F1-dependent manner, leading to increased levels of DNA damage and apoptosis.
MATERIALS AND METHODS
Plasmids, antibodies and reagents
Plasmids coding for HA-ER-E2F1 and HA-ER were previously described (36). Stau259-FLAG3 was generated by the insertion of the full length cDNA (Clone MGC 12191-ATCC) into the pCMV-Sport plasmid (Invitrogen). Three copies of the FLAG sequence were then introduced at the NotI site. Promoter segments of 4320, 1445 and 394 bp were cloned into the pGL3 vector after digestion of a bacterial artificial chromosome (BAC) with restriction enzymes SdaI-MreI, Nde1-MreI and PstI-MreI, respectively. Proximal deletions were generated with the restriction enzymes SdaI-NdeI and SdaI-SacII to create Δ1447 and Δ860, respectively. Shorter promoter fragments were synthesized (Life Technologies Inc.) and cloned into the pGL3 vector. Sequences are given in Supplementary Table SI.
Antibodies against, Stau2, β-Actin (Ac-74), FLAG (M2), HA (rabbit polyclonal) were purchased from Sigma; anti-PARP1, anti-E2F1, anti-H2AX and anti-γH2AX were obtained from New England Biolabs. Monoclonal antibodies against Stau2 (20), HA (37) and Stau1 (38) were previously described. ATM inhibitor (118 500), ATR inhibitor (118 510), CHEK1 inhibitor (681 637) and CHEK2 inhibitor (220 486) were purchased from EMD Millipore, ATR inhibitor (VE821) and DNA-PK inhibitor (NU7441) from Selleckchem. Inhibitor specificities are listed in Supplementary Table SII. Camptothecin (CPT), 5-fluoro-uracil, doxorubicin and DMSO were purchased from Sigma. SMARTpool ON-TARGETplus Stau2 siRNA or ON-TARGETplus non-targeting siRNA pool were purchased from Dharmacon.
For ultraviolet radiation (UVC) irradiation, a monochromatic 254-nm G25T8 germicidal lamp (Philips) was used at a fluence of 1 J/m2/s measured using a Spectroline DRC 100× digital radiometer equipped with DIX-254 sensor. In the case of ionizing radiation, cells were treated with a 137Cs source (Gamma Cell; Atomic Energy Canada) at a dose rate of 6.3 × 10−2 Gy/s.
Cell culture
The human cell lines hTert-RPE1and HEK293T were cultured in Dulbecco modified Eagle's medium (DMEM, Invitrogen) and the human cell line HCT116 cells was maintained in McCoy's medium (Invitrogen) supplemented with 10% fetal bovine serum (Wisent), 100 μg/ml streptomycin and 100 units/ml penicillin (Wisent) (hereafter referred to as complete DMEM or McCoy's). Cells were cultured at 37°C under a 5% CO2 atmosphere. When required, drugs were added to the medium for the indicated periods before harvesting the cells for western blotting.
DNA transfection and infection
For transient expression, cells were transfected with lipofectamine 2000 (Invitrogen) or Mirus (Mirus Bio. LLC) according to the manufacturer's instructions. When needed, cells were selected with 3 μg/ml puromycin for 2 days. For infection, ecotropic Phoenix cells (2 × 106) were transfected by using lipofectamine 2000, with 10 μg of retroviral plasmids (pBabe-puromycin), either control (pBabe empty) or a construct encoding Stau259-FLAG3. Forty-eight hours after transfection, supernatants from those infected cells were collected, filtered (0.2 μm) and added along with polybrene (8 μg/ml) to 4 × 105 HCT116 cells. Forty-eight hours post infection, HCT116 cells were treated for five days with puromycin (2 μg/ml).
Western blot analysis
Total-cell extracts were prepared in lysis buffer (50 mM Tris–HCl pH 7.5, 15 mM EDTA, 0.5% Triton X-100, 100 mM NaCl, 1 mM dithiothreitol [DTT] and a protease inhibitor cocktail [Roche]), and protein concentrations were determined by BCA assays. Cell extracts (10–20 μg) were analyzed by western blotting. Data were collected either on X-ray films (Fujifilm) or with the ChemiDoc MP Imaging System (Bio-Rad Laboratories) and the western blot signals were quantified with the ImageLab (Bio-Rad Laboratories) software.
RNA isolation and RT-qPCR
To determine the steady state level of endogenous Stau2 expression in synchronized cells, RNA was isolated from cell extracts using the TRIZOL Reagent (Invitrogen) according to the manufacturer's procedure. RNA was resuspended in 50 μl water and digested with DNase using the TURBO DNA-free kit (Ambion). Reverse transcription reactions were done with 1 μg of RNA, RevertAid H Minus First Stand cDNA Synthesis kit (Thermo Scientific), according to the manufacturer's procedure. Resulting cDNAs were qPCR amplified using the LightCycler 480 SYBR Green I Master kit (Roche) and the LightCycler 480 instrument (Roche). Sense and antisense sequences of the primer pairs used for qPCR amplification are shown in Supplementary Table SIII.
Luciferase assay
Different truncated regions of Stau2 promoter were subcloned into the pGL3-basic luciferase reporter plasmid (Promega, Madison, WI, USA). Then, cells were transiently cotransfected with the constructs and with YFP plasmid (a normalizing control) using Mirus X2 (Biolynx). Twenty-four hours later, cells treated with or without hydroxyltamoxifen (OHT) (500 nM) were harvested, and luciferase activity was determined by the Luciferase Assay System (Promega) on Fusion α-FP apparatus (Perkin-Elmer). Each experiment was done in duplicate. Results were shown as relative firefly luciferase activities, which were obtained by normalizing to YFP luciferase activities.
Chromatin immunoprecipitation (ChIP)
HCT116 cells were stably transfected with a plasmid coding for HA-ER-E2F1 and incubated in the presence or absence of 4-OHT for 24 h. Cells were then treated with or without CPT (300 nM) for 4 h. Cells were cross-linked with a crosslinking mix (11% formaldehyde, 100 mM NaCl, 0.5 mM EGTA, 50 mM HEPES pH 8.0), then lysed (0.1% SDS, 5 mM EDTA pH 8.0, 50 mM Tris–HCl pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate) and sonicated using the Sonic Dismembrator Model 100 (Fisher Scientific, Ottawa, Canada). Immunoprecipitation was performed by overnight incubation at 4°C with the beads (anti-HA monoclonal beads and using anti-Flag monoclonal beads as negative control - Sigma). Protein–DNA complexes were washed and eluted with the elution buffer (1% SDS, 100 mM NaHCO3). Reverse cross-links were carried out, followed by treatment by proteinase K, phenol–chloroform extractions and DNA precipitation. Real-Time polymerase chain reaction (PCR) was performed to analyze DNA fragments by LightCycler 480 SYBR GreenIMaster (Roche Applied Sciences, Basel, Switzerland) on the LightCycler 480 (Roche Applied Sciences, Basel, Switzerland). Input DNA was analyzed simultaneously and used as normalization. Primers HMBS were used as negative control.
RESULTS
Downregulation of Stau2 in response to camptothecin
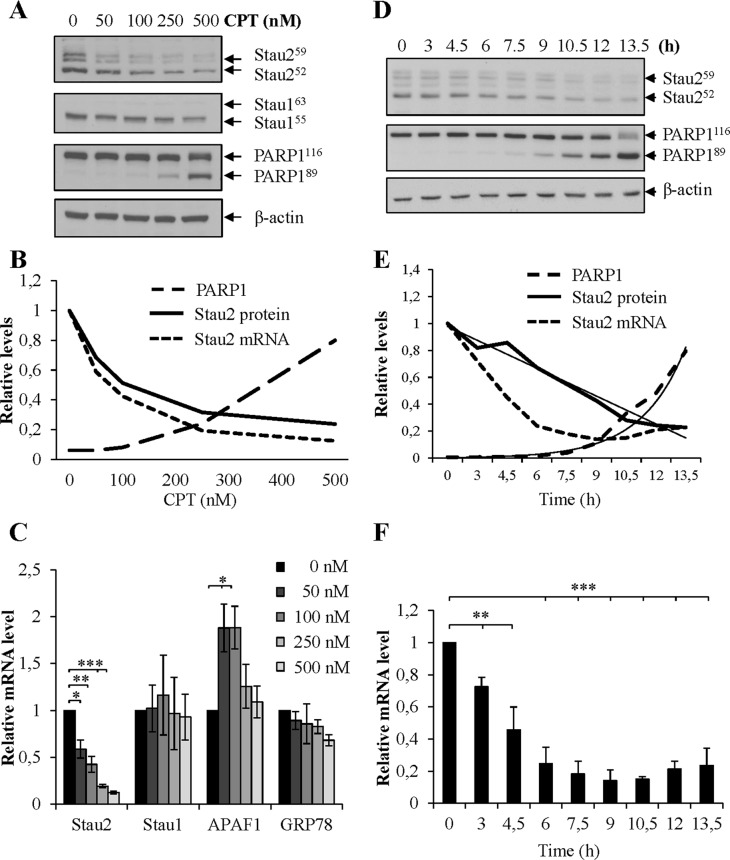
Since Stau2 binds many mRNAs coding for proteins involved in DNA repair and/or apoptosis (34), we evaluated whether Stau2 expression might be modulated following DNA damage. We initially monitored Stau2 expression as a function of dose in response to the toposisomerase 1 inhibitor CPT (39) using the model colorectal carcinoma cell line HCT116. Cells were incubated for 24 h with increasing CPT concentrations and analyzed by Western blotting. Our results show that Stau2 protein levels decreased in a dose-dependent manner in response to CPT (Figure 1A and B). All Stau2 isoforms were affected similarly. Interestingly, the decrease was apparent even at a dose of CPT (50 nM) that did not induce apoptosis as determined by PARP1 cleavage, indicating that CPT-induced reduction of Stau2 expression is not a consequence of apoptosis. We then employed RT-qPCR to quantify steady-state levels of Stau2 mRNA in response to increasing doses of CPT in HCT116 cells. Figure 1B and C clearly show that Stau2 mRNA levels were downregulated in response to CPT, and the decrease observed at 24 h in mRNA levels paralleled that in protein levels. To test the specificity of the response, i.e. to rule out the possibility that attenuation of general transcription might explain our findings, we analyzed the expression of Stau1, a Stau2 paralog that also binds double-stranded RNAs and controls RNA fate. Stau1 protein and mRNA levels remained unchanged by drug treatment. As further controls, APAF1 and GRP78 mRNA, known to be upregulated and not to be regulated, respectively, following DNA damage (40) responded as expected (Figure 1C).
Figure 1.
Decrease of Stau2 expression in response to camptothecin (CPT) treatment. (A–C) HCT116 cells were treated for 24 h with increasing doses of CPT as indicated and cell extracts were analyzed by western blotting (A) and RT-qPCR (C). (B) Quantification of Stau252 protein and mRNA (all isoforms) levels and of PARP1 cleavage (PARP189/PARP1116) in the representative experiment. The western blot is representative of three independently preformed experiments. PARP1 cleavage was used as a measure of apoptosis and β-actin as a loading control. The RT-qPCR data represent the means and standard deviation of three independently performed experiments. The ratio of specific gene mRNAs on GAPDH mRNA in DMSO-treated cells (0 nM) was arbitrary fixed to 1. Stau1, Staufen 1 (an RNA-binding protein—paralog of Stau2); APAF1, apoptotic peptidase activating factor 1; GRP78, glucose-related protein 78. APAF1 mRNAs, known to be upregulated in apoptotic cells, was used as positive controls. GRP78 was used as a negative control. Statistical analyses (Student's t-test) are indicated when significant. ***P-value ≤ 0.001; **P-value ≤ 0.01; *P-value ≤ 0.05. (D–F) HCT116 cells were incubated in constant amounts of CPT (300 nM) for increasing periods of time. Cells were lysed and Stau2 expression was analyzed by western blotting (D) and RT-qPCR (F). (E) Quantification of Stau2 protein and mRNA levels and of PARP1 cleavage (PARP189/PARP1116) over times in the representative experiment. The thin black lines represented the best fit on the curves. The western blot is representative of four independently performed experiments. Stau2 decline and PARP1 cleavage were normalized to their values at time 0. The RT-qPCR data represent the means and standard deviation of four independently performed experiments. The ratio of specific gene mRNAs on GAPDH mRNA at time 0 was arbitrary fixed to 1.
To more precisely analyze the consequence of CPT treatment on Stau2 expression, HCT116 were treated with 300 nM of the drug for increasing time periods and analyzed as above by western blotting and RT-qPCR. Stau2 protein expression declined in a linear fashion as a function of time (Figure 1D and E). A reduction in Stau2 expression was also observed at the mRNA level, although this downregulation appeared to occur more rapidly than for the protein (Figure 1E and F) suggesting that Stau2 mRNA decrease precedes that of the protein. Interestingly, cleavage of PARP1 was detected at later time points than Stau2 decrease (Figure 1E), suggesting again that Stau2 degradation is not a consequence of apoptosis.
Stau2 expression is downregulated by CPT in other transformed and untransformed cell lines
To rule out the possibility that Stau2 downregulation is cell-type specific, we treated transformed human embryo kidney HEK293T cells (Supplementary Figure S1A–C) and untransformed human hTert-RPE1 cells (Supplementary Figure S1D–F) with different doses of CPT. As observed for HCT116, exposure for 24 h to high doses of CPT caused a decrease in Stau2 expression in both cell lines (Supplementary Figure S1A,D). Under these conditions, PARP1 was not cleaved indicating that apoptosis was not induced. We also incubated both cell lines with a lower dose of CPT for different time periods (Supplementary Figure S1B,E). At 50 nM CPT, Stau2 expression decreased slightly at 3 h in hTert-RPE1, and much more profoundly in both lines by 24 h. Stau2 decrease was also observed with 100 and 250 nM CPT at 24 h. The treatment caused DNA damage as early as 3 h post treatment, as evaluated by the induction of the phospho-histone γH2AX. At the RNA level, 50 nM CPT was sufficient to significantly downregulate Stau2 mRNA in h-TERT-RPE1 (Supplementary Figure S1F). Although not statistically significant (P = 0.06 at 3 and 24 h), the same tendency was observed in HEK293T cells (Supplementary Figure S1C). When compared at 24 h at 50 nM CPT, Stau2 mRNA downregulation in HEK293T (0.77 ± 0.06) is slightly lower than in hTert-RPE1 (0.50 ± 0.04) or HCT116 cells (0.58 ± 0.09).
Stau2 downregulation occurs in a mutagen-specific manner
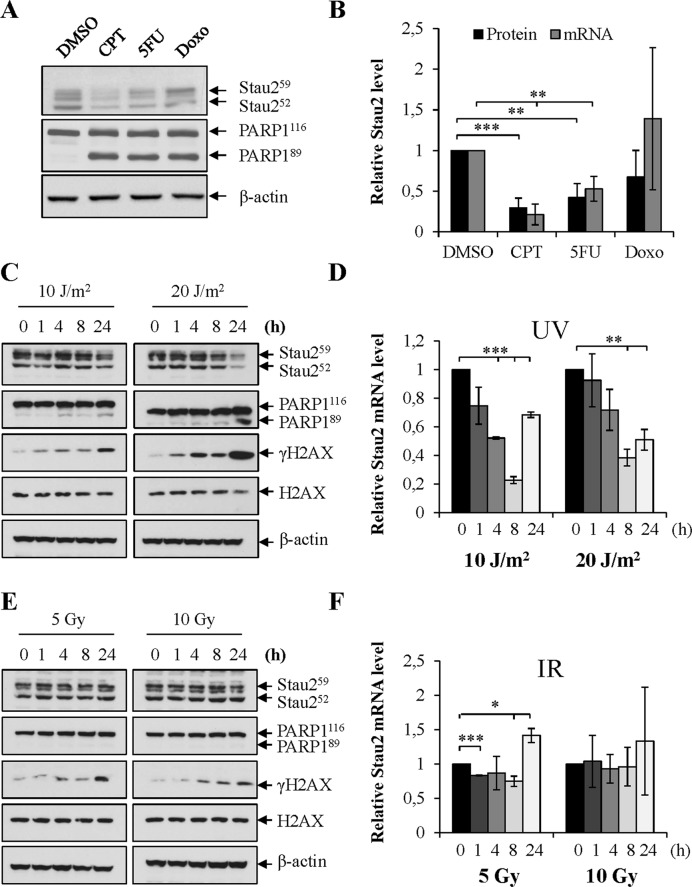
HCT116 cells were treated for 24 h with various DNA damaging agents (Figure 2A and B), such as doxorubicin (Doxo), an inhibitor of topoisomerase II (41) and the base-analog 5-fluoro-uracil (5FU) (42). As observed in Figure 2A and B, 5FU treatment significantly reduced Stau2 expression both at the protein and mRNA levels whereas Doxo had no significant effect. This indicates that Stau2 downregulation is induced by specific DNA damaging agents.
Figure 2.
Stau2 is downregulated in response to other DNA damaging agents. (A and B) HCT116 cells were incubated in the presence of different DNA damaging agents for 24 h. DMSO, dimethyl sulfoxide (vehicle used as control); CPT (300 nM); 5FU, 5-fluoro-uracile (3 μM); Doxo, doxorubicin (6 μM). (C–F) HCT116 cells were irradiated with different doses of UVC (C and D) or IR (E and F). Cells were collected at different time points post-treatment and cell extracts were analyzed by western blotting (A, C and E) and RT-qPCR (B,D and F). (A, C and E) The western blots are representative of three independently performed experiments. Induction of apoptosis was monitored by the cleavage of PARP1 and DNA damage by the presence of γH2AX. β-actin antibody was used as a loading control. Stau2 protein quantification is shown in (B) and in the Supplementary Figure S2. (B, D and F) Stau2 mRNA expression was normalized to that of GAPDH mRNA, the ratio in mocked-treated cells being fixed to 1. Data represent the means and standard deviation of three independently performed experiments. Statistical analyses (Student's t-test) are indicated when significant. ***P-value ≤ 0.001; **P-value ≤ 0.01; *P-value ≤ 0.05.
HCT116 cells were then treated with UVC or ionizing radiation (IR), which directly induce cyclobutane pyrimidine dimers (replication stress) or DNA double-stranded breaks, respectively. Irradiation with UVC (10 or 20 J/m2) clearly generated a decrease in the amount of Stau2 protein 24 h post-treatment (Figure 2C, Supplementary Figure S2A). Decline at the mRNA level was faster i.e. observed as soon as 4 h post-treatment (Figure 2D). Both apoptosis (PARP1 cleavage) and DNA damage (γH2AX) were triggered by these treatments (Figure 2C). HCT116 cells were also irradiated with 5 and 10 Gy IR and Stau2 protein and mRNA levels quantified at different time points. IR exerted only a modest (if any) effect on Stau2 expression both at the protein (Figure 2E, Supplementary Figure S2B) and mRNA level (Figure 2F). A small but significant reduction in Stau2 mRNA was observed post-treatment with 5 Gy but not 10 Gy of IR (Figure 2F). Both treatments induced DSBs (γH2AX) (Figure 2E). Altogether, these results suggest that replication stress promotes Stau2 decrease, i.e. in this respect, replication-blocking DNA adducts are more efficient than DNA double stranded breaks.
Stau2 mRNA expression is regulated by the ATR/CHEK1 pathways
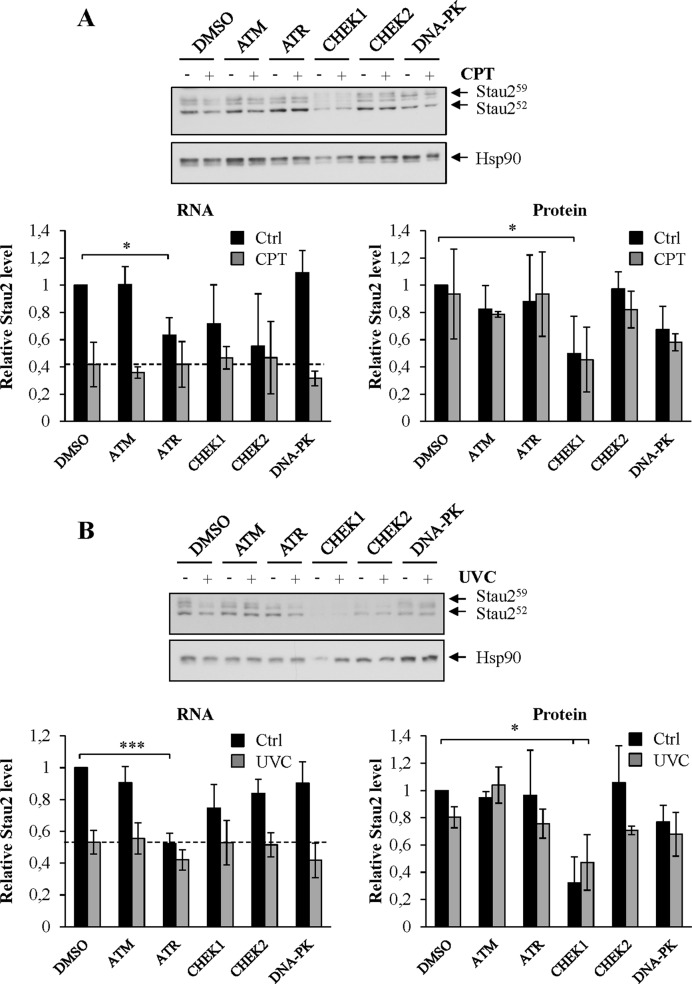
To obtain clues regarding the pathways that link DNA damaging agents to Stau2 expression, we tested a series of pharmacological agents known to inhibit various kinases involved in the DDR including ATM, CHEK2, ATR, CHEK1 and DNA-PK (Supplementary Table SII). HCT116 cells were treated with inhibitor for 4 h and then exposed to CPT in the presence of the inhibitor for an additional 4 h (Figure 3A). The vehicle DMSO was used as control. Cells were collected and the expression of Stau2 was quantified by RT-qPCR and western blotting. Unexpectedly, none of these inhibitors prevented the CPT-mediated mRNA Stau2 expression decline. However, the ATR inhibitor caused a significant decrease in Stau2 mRNA expression in the absence of CPT indicating that Stau2 expression is regulated by the ATR pathway in unstressed cells. Similar results were obtained with a different ATR inhibitor (Supplementary Figure S3). In contrast, DNA-PK and ATM inhibitors had no effect on Stau2 expression both in untreated and CPT-treated cells. CHEK1 and CHEK2 inhibitors caused intermediate responses in Stau2 mRNA expression in unstressed cells that are not statistically significant. Consistent with data in Figure 1, Stau2 expression was only marginally reduced at the protein level when HCT116 cells were incubated for 4 h in CPT (Figure 3A). Interestingly, Stau2 protein expression was however strongly reduced in CHEK1-inhibitor-treated cells. Inhibition of other DDR kinases had no effect on Stau2. Our results indicate that Stau2 expression is under the control of the ATR/CHEK1 pathway in untreated cells.
Figure 3.
Signaling pathways involved in Stau2 downregulation. HCT116 cells were incubated in the presence of different kinase inhibitors for 4 h, then with the inhibitors and CPT (300 nM) (A) for another 4 h, or (B) incubated in the presence of different kinase inhibitors for 4 h, then irradiated at 10 J/m2 and re-incubated for 4 h in the presence of inhibitors. Stau2 protein expression was analyzed by western blotting while Stau2 mRNA levels were quantified by RT-qPCR. Stau2 expression was normalized to that of Hsp90 protein or GAPDH mRNA, the ratio in DMSO-treated cells without inhibitors being fixed to 1. Data represent the means and standard deviation of three independently performed experiments. Statistical analyses (Student's t-test) are indicated when significant. ***P-value ≤ 0.001; *P-value ≤ 0.05. DMSO, dimethyl sulfoxide; ATM (20 μM); ATR (20 μM); CHEK1 (20 μM); CHEK2 (20 μM); DNA-PK (10 μM).
To expand on these results, HCT116 cells were treated with the kinase inhibitors for 4 h, irradiated with 10 J/m2 of UVC and incubated for an additional 4 h in the presence of the inhibitors. As observed above with CPT, UVC also caused a reduced expression of Stau2 mRNA even in the presence of the inhibitors (Figure 3B). However, the ATR inhibitor completely repressed Stau2 mRNA expression while CHEK1 inhibitor reduced Stau2 protein level in non-irradiated cells indicating again a role for ATR/CHEK1 in Stau2 expression.
To get more insight into the molecular mechanism, we prepared cell extracts from CPT- and UVC-treated cells in the presence or absence of specific inhibitors and analyzed the induction of p53 (Supplementary Figure S4A and B). Both CPT and UVC induced p53 accumulation as compared to untreated cells. In both cases, the ATR inhibitor prevented p53 induction, indicating that ATR is involved in CPT- and UVC-mediated p53 expression. Other inhibitors had no effect on p53 induction. Interestingly, although CHEK1 inhibitor did not prevent p53 increase in CPT-treated cells, it caused p53 induction in unstressed cells. This is likely due to the high level of spontaneous DNA damage that is generated when CHEK1 is inhibited (18,43,44), as also seen on western blot with γH2AX (not shown). We next compared p53 induction in Doxo-treated cells as a means to compare pathways that are activated with a drug that does not cause a decrease in Stau2 expression (Supplementary Figure S4C). Inhibitors of the ATM, CHEK2 and ATR pathways prevented p53 accumulation in Doxo-treated cells. Inhibitors of CHEK1 and DNA-PK had no effect. All together, these results indicate that CPT and UVC activate the ATR pathway while Doxo activates ATM/CHEK2 as well as ATR in HCT116 cells.
Stau2 downregulation occurs at the transcriptional level
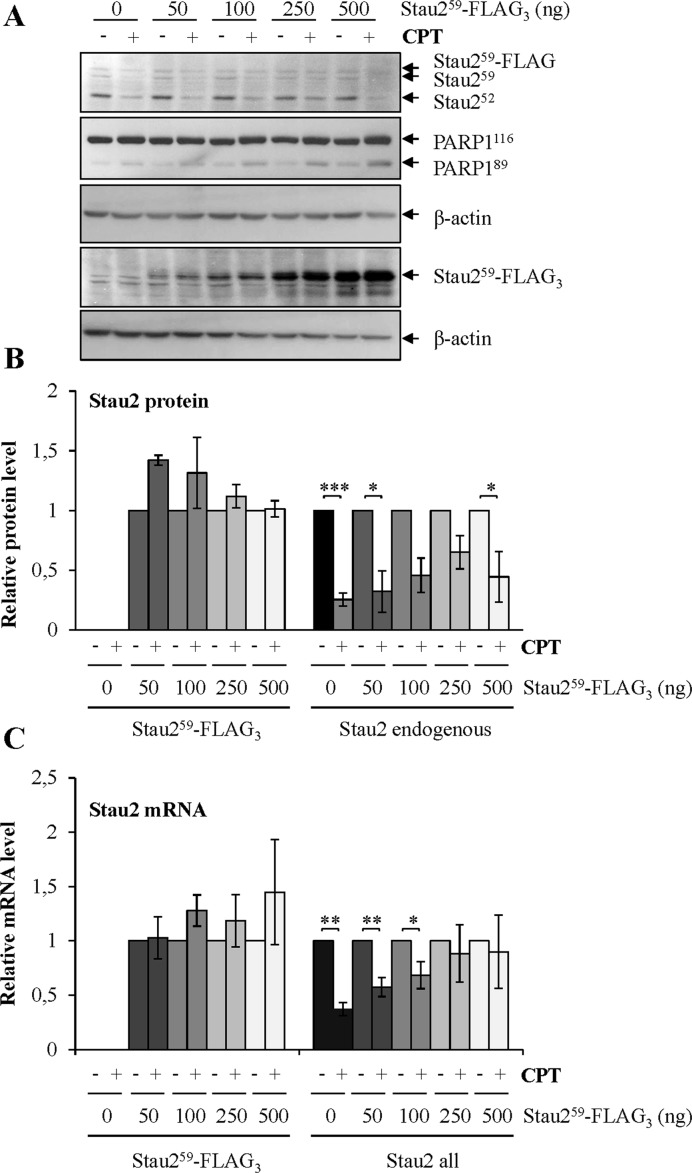
To determine if the decrease in Stau2 mRNA levels in response to CPT is due to a transcriptional or a post-transcriptional mechanism, we constructed a full length FLAG-tagged Stau259 isoform (containing the 5′- and 3′-UTRs) under the control of a retroviral LTR promoter (MSCV). HCT116 cells were transfected with increasing concentrations of plasmid and then treated with 300 nM CPT. We maintained expression of the exogenous protein at lower levels relative to the endogenous counterpart. (Figure 4A). Under these conditions, whereas the expression of endogenous Stau2 was downregulated as expected in the presence of CPT, that of exogenous Stau259-FLAG was not affected (Figure 4A and B). This was also the case at the mRNA level (Figure 4C). These data suggested that RNA stability and post-translational-mediated degradation of Stau2 protein are not involved in Stau2 downregulation, and that the mechanism is likely transcriptional.
Figure 4.
Stau2-FLAG expressed from a viral promoter is not downregulated in response to CPT treatment. HCT116 cells were transfected with the empty vector (0) or with increasing concentrations of a plasmid coding for Stau259-FLAG3. Cells were then incubated in 300 nM CPT for 24 h. (A) Cells were lysed and Stau2 expression was analyzed by western blotting using anti-Stau2 and anti-FLAG antibodies. PARP1 cleavage was used as an indicator of apoptosis. Representative data of three independently performed experiments. (B) Quantification of endogenous Stau2 and of transfected Stau259-FLAG3. The ratio of Stau2 on β-actin in cells without CPT (−) was arbitrary fixed to 1. (C) mRNAs isolated from HCT116 cells (as in (A)) were quantified by RT-qPCR using oligonucleotides that only recognized Stau259-FLAG3 or that recognized both Stau2 endogenous and Stau259-FLAG3 (Stau2 all). Data represent the means and standard deviation of three independently performed experiments. The ratio of specific gene mRNAs on GAPDH mRNA in vector-transfected cells without CPT (−) was arbitrary fixed to 1. Statistical analyses (Student's t-test) are indicated when significant. ***P-value ≤ 0.001; **P-value ≤ 0.01; *P-value ≤ 0.05.
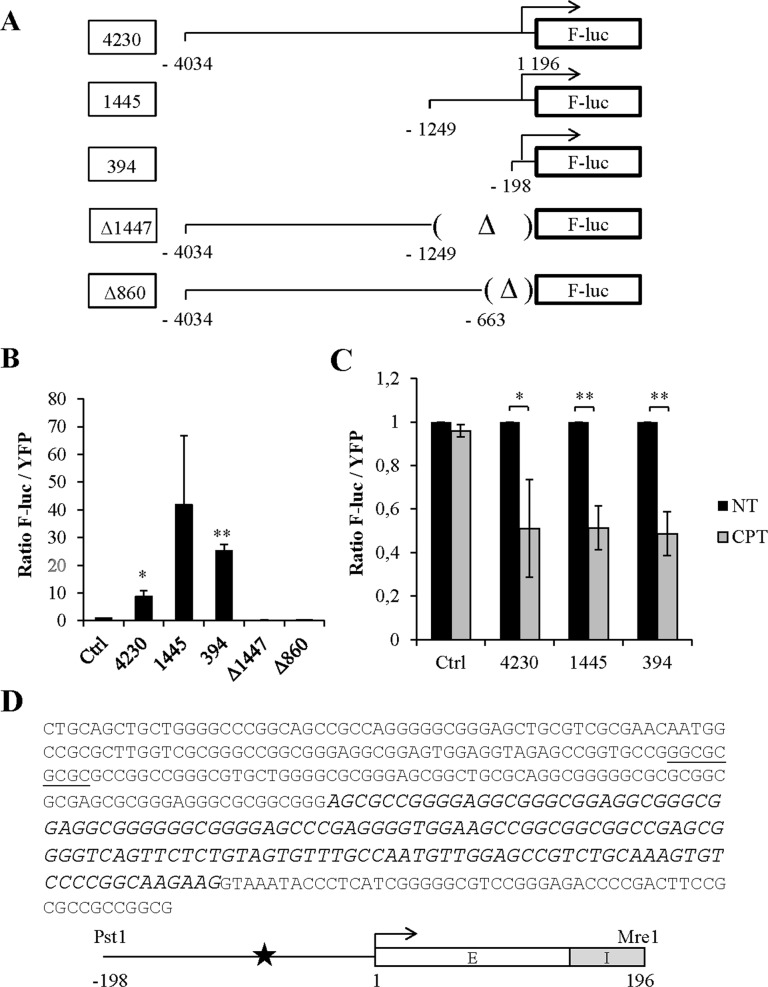
To confirm the transcriptional modulation of Stau2 following CPT treatment, we first identified and characterized its promoter region. Since nothing was previously known about transcriptional regulation of Stau2, we started with a long 4230 bp genomic fragment isolated from the 5′ end of the gene. This fragment was cloned upstream of the F-Luc coding region (Figure 5A). Shorter clones with deletions at either the 5′ or 3′ end were also generated to identify a functional promoter region. HEK293T cells were transfected with these plasmids along with a plasmid coding for YFP as transfection normalization control. Analysis of F-Luc activity indicated that the 3′ most 394 bp fragment of the Stau2 promoter was sufficient and necessary to drive F-Luc expression (Figure 5B). Accordingly, deletion of the proximal 860 bp completely abolished F-Luc expression.
Figure 5.
Identification of the endogenous Stau2 promoter. (A) Schematic representation of plasmids coding for the F-Luciferase gene (F-luc) under the control of different fragments isolated from the putative promoter region of the Stau2 gene. Arrows indicated the position of the transcription start site. (Δ), deletion. (B) HEK293T cells were transfected with plasmids coding for YFP and the promoter-less F-luc construct (Ctrl) or co-transfected with plasmids coding for F-luc as described in (A) and YFP (as a transfection normalization marker). Expression of F-luc and YFP was quantified and the ratio of luciferase activity on the YFP signal was calculated. The ratio from YFP-transfected cells was arbitrary fixed to 1. Data represent the means and standard deviation of three independently performed experiments. (C) HCT116 cells that expressed YFP (Ctrl) or YFP along with different F-luc constructs as indicated were incubated or not in 300 nM CPT for 24 h. Expression of F-luc and YFP was quantified and the ratio of luciferase activity on the YFP signal was calculated. The ratio from untreated cells (NT) was arbitrary fixed to 1. Data represent the means and standard deviation of three independently performed experiments. Statistical analyses (Student's t-test) are indicated when significant. **P-value ≤ 0.01; *P-value ≤ 0.05. (D) Sequence and schematic representation of the minimal 394 bp Stau2 promoter sequence. In the sequence, the position of the putative E2F1 binding site is underlined whereas nucleotides of the first exon are in italics. Schematic representation of the 198 nt of promoter sequence, with the position of the E2F1 binding site (star), the position of the transcription start site (arrow) and the 196 bp exon (E)/intron (I) region of the Stau2 gene. Restriction enzymes used to clone this fragment are indicated.
These results indicate that we successfully isolated a functional Stau2 promoter. We next tested if this region is sufficient to respond to CPT treatment as does the endogenous promoter. HCT116 cells were transfected as above with the functional F-Luc and YFP constructs and F-Luc activity monitored in the presence vs absence of CPT. Interestingly, F-Luc expression was downregulated in CPT-treated cells expressing a functional promoter region (Figure 5C), even with the minimal 394 bp region, suggesting that the cis-acting sequence involved in Stau2 regulation by CPT lies within this latter region. The sequence of the minimal functional promoter indicates that the 394 bp clone contains 198 bp of upstream promoter sequence, the first exon, and part of the first intron of Stau2 (Figure 5D). The minimal promoter has no TATA or CAT box sequences but is GC-rich and, interestingly, contains a putative E2F1 transcription factor binding site. Therefore we reasoned that E2F1, known to be involved in DNA repair and apoptosis (11,12), may be crucial for regulation of Stau2.
The transcription factor E2F1 enhances Stau2 expression
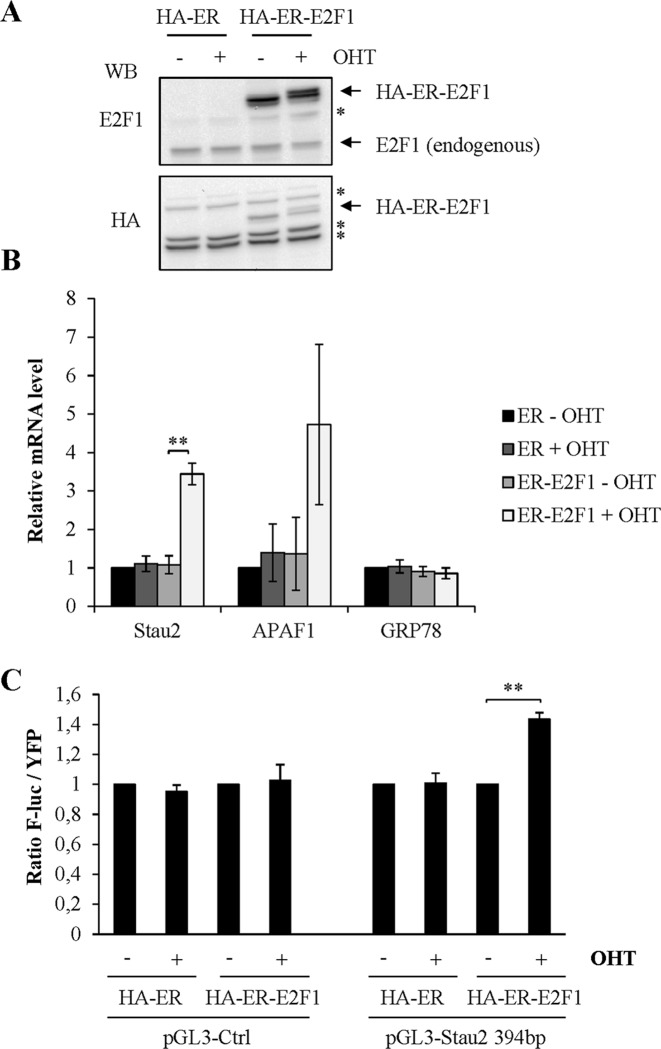
Consistent with the prediction of a putative E2F1 binding site in the functional promoter region of the Stau2 gene, the ENCODE consortium (http://www.genome.ucsc.edu/encode/) which compiles results of ChIP experiments on the human genome, indeed reported that E2F1 associates with the Stau2 promoter. This is interesting because E2F family members (i) are known to regulate expression of genes involved in G1-S transition, DNA replication and DNA repair (i.e. double-strand break repair and nucleotide- and base-excision repair) during genotoxic stress (11) and (ii) are well known to be upregulated following DNA-damage (45,46). We hypothesized that induction of E2F1 by DNA-damaging agents and subsequent binding to the Stau2 promoter may negatively regulate Stau2 gene expression. Therefore, we tested whether overexpression of E2F1 can modulate Stau2 expression. We used an inducible vector in which E2F1 was fused to the estrogen receptor (ER), allowing E2F1 to be imported into the nucleus in the presence of 4-OHT. Western blot analysis indicated that the plasmids are well expressed when transfected in HCT116 cells (Figure 6A). Interestingly, upon incubation of HCT116 with OHT, the Stau2 levels increased by 3.7-fold (Figure 6B), indicating that E2F1 upregulates Stau2 expression. As controls, we showed that E2F1 enhanced, as expected, the expression of APAF1, a known E2F1 target (40) but did not modulate the expression of GRP78.
Figure 6.
The transcription factor E2F1 upregulates Stau2 transcription. (A) E2F1 is fused to the estrogen receptor-binding domain tagged with HA (HA-ER). The ER-fusion protein is expressed in the cytosol and translocates to the nucleus in the presence of the ER ligand 4-hydroxytamoxifen (OHT). HCT116 cells were transfected with plasmids coding for HA-ER or HA-ER-E2F1 and incubated in the presence (+) or absence (−) of OHT (500 nM). Expression of the proteins was detected by western blotting. *: non-specific bands. (B) Expression of known (APAF1) and putative (Stau2) endogenous targets of E2F1 was analyzed by RT-qPCR and normalized on that of GAPDH mRNA. Expression in the absence of OHT and E2F1 (ER-OHT) was arbitrarily normalized to 1. Data represent the means and standard deviation of three independently performed experiments. (C) HA-ER- and HA-ER-E2F1-expressing HCT116 cells were co-transfected with plasmids coding for YFP and F-luc under the control of the minimal Stau2 promoter (pGL3 Stau2 -394) or without any promoter region (pGL3 control). Cells were then incubated in the absence or presence of OHT. A ratio of F-luc activity on YFP signal was calculated in each condition. The ratio in the absence of OHT was arbitrary fixed to 1. Data represent the means and standard deviation of three independently performed experiments. Statistical analyses (Student's t-test) are indicated when significant. **P-value ≤ 0.01.
To determine if E2F1 has the same effect on the minimal promoter region, HCT116 cells were transfected with the F-Luc and YFP constructs. Addition of OHT enhanced F-Luc expression (Figure 6C), as observed above with the endogenous promoter. This indicates that E2F1 influences Stau2 promoter expression and may be linked to the observed CPT response.
Positive correlation between the E2F1 binding site and Stau2 regulation in the absence or presence of CPT
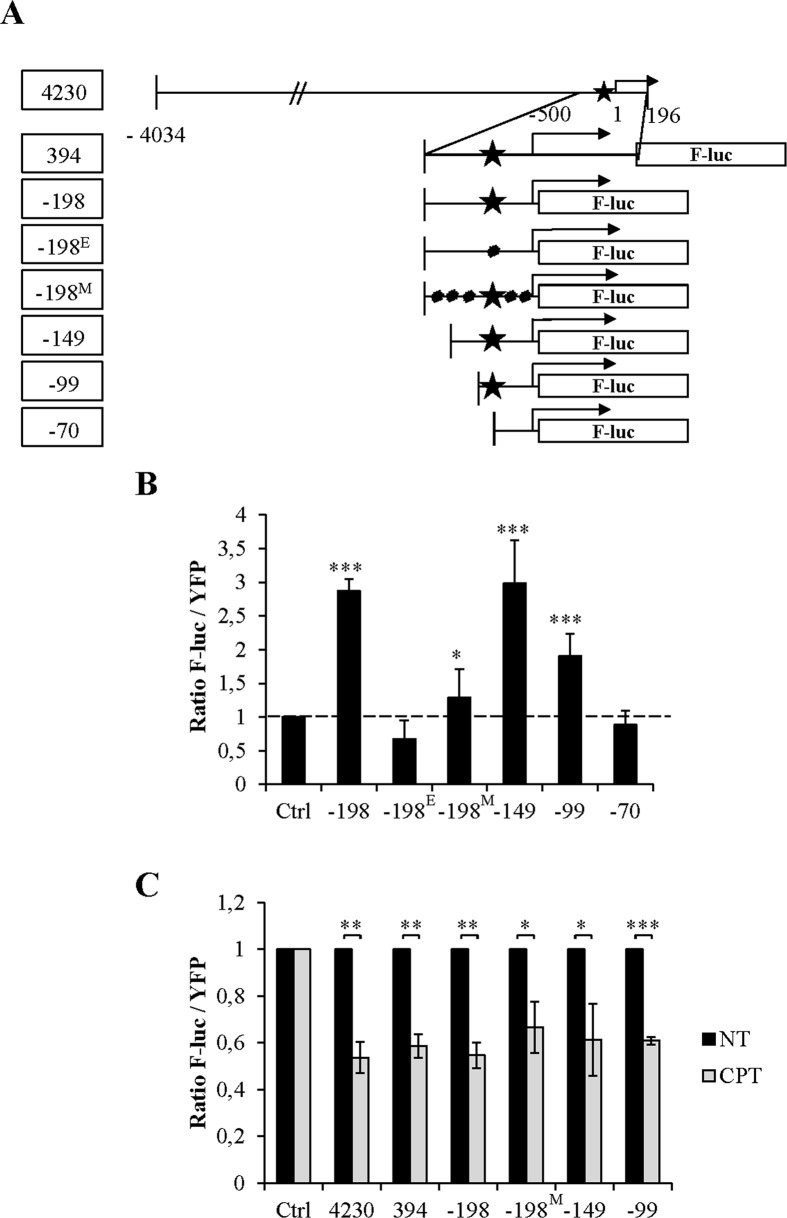
To confirm that E2F1 is involved in Stau2 expression, we generated a series of mutated promoters (Figure 7A). We first deleted the Stau2 exon/intron region (clone -198) and showed that the resulting clone was functional, although it was seven times less efficient than the -394 clone. In this truncated clone, we then introduced point mutations to alter the putative E2F1 binding site (clone -198E). Alternatively, we mutated the GC-rich region of the promoter region but not the putative E2F1 binding site (clone -198M), or progressively deleted the distal part of the promoter to eventually generate a construct that lacks the E2F1 binding site. Expression of these F-Luc constructs along with YFP indicated that the removal of the Stau2 exon/intron region (clone -198) did not abrogate F-Luc expression (Figure 7B). Similarly, mutations (clone -198M) or deletions of parts of the promoter (clones -149 and -99) did not completely abolish its activity as long as the E2F1 binding site was still present. Consistently, mutation (clone -198E) or removal (clone -70) of this site completely abrogated promoter activity. Therefore we have established a strong correlation between the presence of the putative E2F1 binding site and Stau2 promoter expression.
Figure 7.
Correlation between the E2F1-binding site in the Stau2 promoter and Stau2 expression. (A) Schematic representation of the mutated Stau2 promoters fused to F-luc. Star, putative E2F1-binding site; dot, point mutations; -198E, mutation in the E2F1-binding site; -198M, several mutations in the promoter but not in the E2F1-binding site; Arrow, transcription start site. (B) HCT116 cells were co-transfected with plasmids coding for YFP and F-luc under the control of a Stau2 promoter. Expression of F-luc and YFP was quantified and the ratio of luciferase activity on the YFP signal was calculated. The ratio from cells transfected with the control plasmid (Ctrl) was arbitrary fixed to 1. Data represent the means and standard deviation of three independently performed experiments. The dashed line indicates the level of expression of the control F-luc plasmid that has no promoter sequence. Statistical values are relative to the expression of the control plasmid. Statistical analyses (Student's t-test) are indicated when significant. *P-value ≤ 0.05; ***P-value ≤ 0.001. (C) Transfected HCT116 cells (as in B) were incubated in the absence (NT) or presence (CPT) of 300 nM CPT for 24 h. The ratio from untreated cells was arbitrary fixed to 1. Data represent the means and standard deviation of three independently performed experiments. ***P-value ≤ 0.001; **P-value ≤ 0.01; *P-value ≤ 0.05.
Although we cannot directly determine if the E2F1 binding site is also involved in CPT-mediated Stau2 downregulation (since promoters lacking this site have no activity), we nevertheless evaluated whether CPT influences F-Luc activity when the gene is driven by minimal functional promoters containing mutations and/or deletions. As seen in Figure 7C, removal of the Stau2 exon/intron region or of the distal part of the promoter did not abolish Stau2 promoter downregulation in the presence of CPT. Similarly, downregulation of F-Luc activity driven by the promoter containing multiple mutations (that did not alter the E2F1 binding site) was still observed. These results suggest that the E2F1 binding site may be involved in the Stau2 gene response to CPT.
ATR inhibition reduces E2F1 expression in unperturbed cells
We showed above (Figure 3) that Stau2 mRNA levels are downregulated when cells are treated with the ATR inhibitor. To look for a possible link between signaling pathways and Stau2 transcription, we monitored Stau2 and E2F1 expression in CPT-, UVC- and Doxo-treated cells, in the presence of kinase inhibitors. Strikingly, E2F1 expression was abolished in ATR- and CHEK1-inhibitor-treated cells in the presence and absence of DNA-damaging agents (Supplementary Figure S5A). While Stau2 expression was not decreased in ATR-inhibitor-treated cells, it was almost completely abolished in CHEK1-inhibitor-treated cells (Figure 3). Inhibition of other DDR kinases had no effect on Stau2 or E2F1 expression. These data suggested that E2F1 expression requires ATR/CHEK1 activity and that the decrease in Stau2 expression may be a downstream consequence of a reduction in E2F1 levels.
CPT treatment displaces E2F1 from the Stau2 promoter
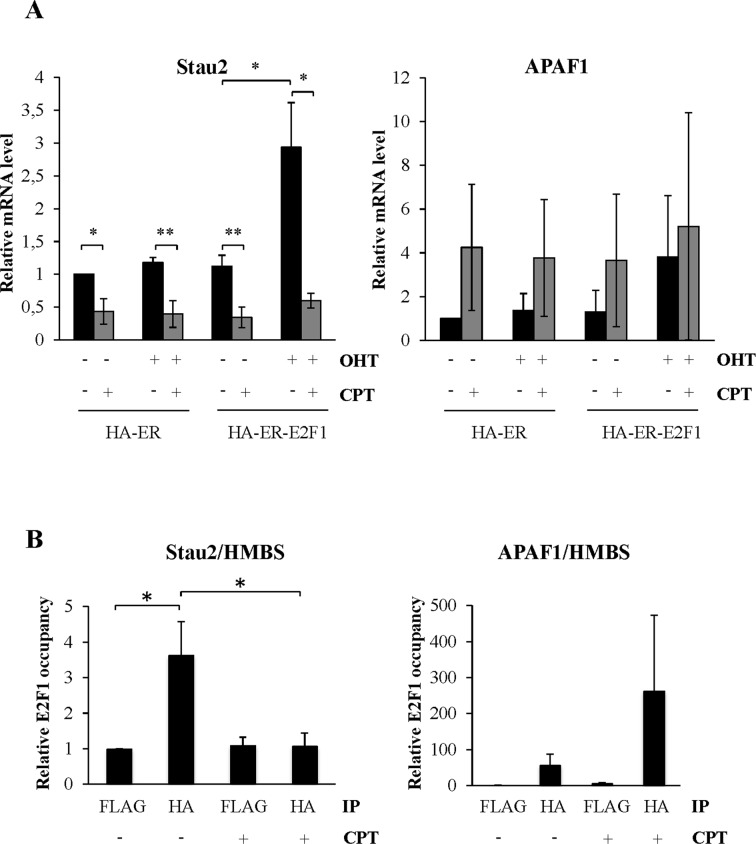
To elucidate the relationship between E2F1 and CPT in relation to Stau2 expression, we overexpressed E2F1 and quantified Stau2 expression in the presence or absence of CPT in a manner similar to that described in Figure 6B. Overexpression of E2F1 increased Stau2 expression in HCT116 cells both at the RNA (Figure 8A) and protein (Supplementary Figure S6) levels; moreover, interestingly, incubation with CPT not only abolished E2F1-mediated stimulation of Stau2 but also decreased Stau2 expression to the level observed in CPT-treated cells. As control, both CPT treatment and E2F1 overexpression increased APAF1 expression as expected.
Figure 8.
CPT displaces E2F1 from the Stau2 promoter. (A) HCT116 cells were transfected with plasmids coding for HA-ER or HA-ER-E2F1 as described in Figure 6A. Cells were further incubated in the absence or presence of CPT (300 nM) for 24 h. Expression of Stau2 mRNA and as control of APAF1 was analyzed by RT-qPCR and normalized on that of GAPDH mRNA. Expression in the absence of OHT, E2F1 (ER -OHT) and CPT was arbitrarily normalized to 1. Expression data represent the means and standard deviation of three independently performed experiments. Statistical analyses (Student's t-test) are indicated when significant. *P-value ≤ 0.05; **P-value ≤ 0.01. Note that similar profiles of APAF1 expression were observed in the three experiments although the levels of APAF1 induction varied from one experiment to the others. (B) ChIP assay. HA-ER-E2F1-expressing cells were incubated in the presence or absence of OHT (500 nM) for 24 h and then treated or not with CPT (300 nM) for 3 h. DNA was immunoprecipitated with anti-HA or anti-FLAG (as control) antibodies. Resulting DNA was qPCR-amplified with primers located in the Stau2 promoter as well as in the APAF1 promoter as positive control and HMBS as negative control. Ratios of the amount of immunoprecipitated DNA in the Stau2 or APAF1 promoters over that in the control region were calculated. Data represent the ratios of E2F1 occupancy in HA-ER-transfected cells over that in HA-ER-E2F1-transfected cells. Data represent the means and standard deviation of three independently performed experiments. *P-value ≤ 0.05.
To reinforce the link between E2F1 and Stau2, we studied E2F1 interaction with the Stau2 promoter by ChIP. HA-ER-E2F1-expressing HCT116 cells were treated or not with OHT for 24 h and then exposed to CPT or not for an additional 3 h. Cells were fixed and the sonicated DNA immunoprecipitated with anti-HA antibody, or anti-FLAG antibody as negative control. The DNA was qPCR-amplified with primers located in the Stau2 promoter or in a neighboring intron (as negative control). As a positive control, we also amplified the promoter of APAF1, a known target of E2F1. In the absence of OHT, HA-ER-E2F1 was not found in any precipitates (not shown). However in the presence of OHT, HA-ER-E2F1 was detected in HA- but not FLAG-precipitates, for both the Stau2 and APAF1 promoters (Figure 8B). Interestingly, after CPT incubation, HA-ER-E2F1 was no longer associated with the Stau2 promoter although it was still present within the APAF1 promoter. These results indicate that E2F1 is bound to the Stau2 promoter in untreated cells and that it is displaced following DNA damage by CPT, consistent with the expression of Stau2 in the presence or absence of CPT.
Downregulation of Stau2 expression increases DNA-damage and facilitates apoptosis
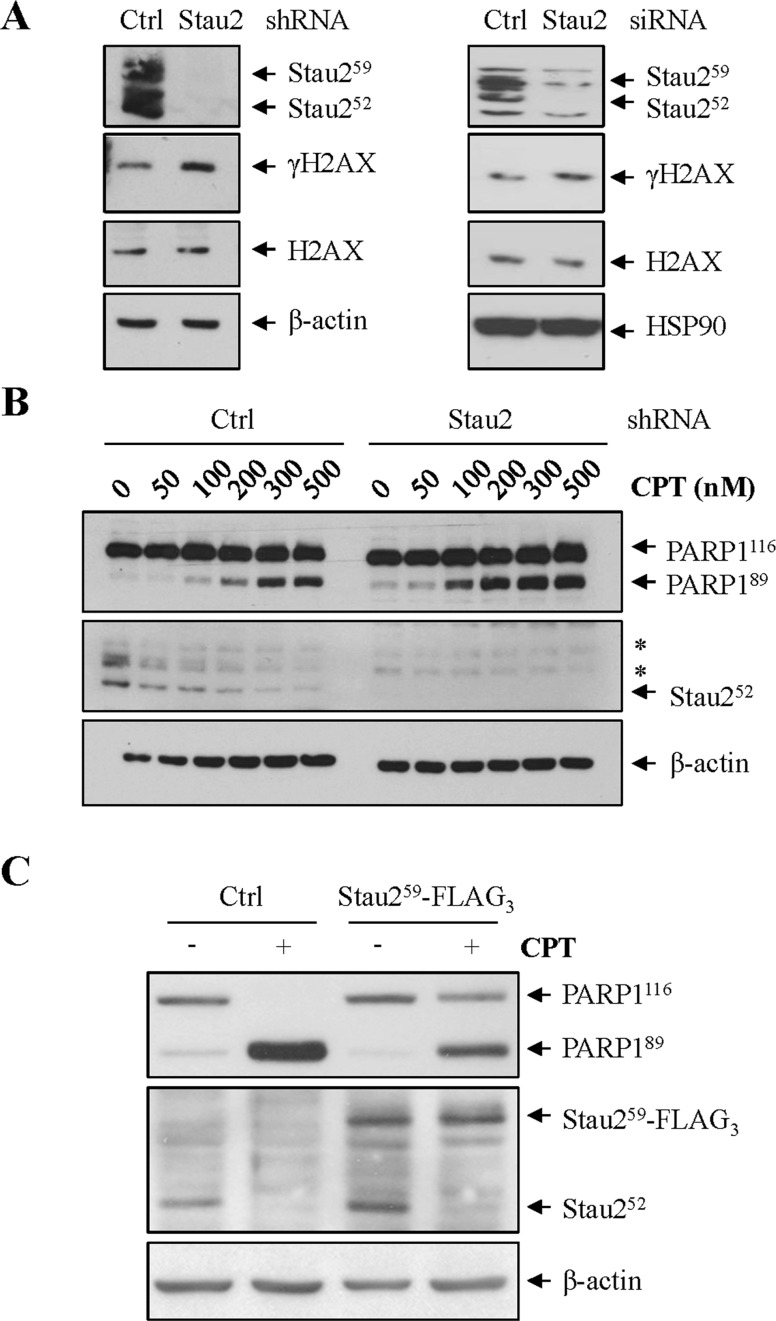
Our data indicated that Stau2 expression is modulated in unstressed cells by ATR. Therefore, we determined whether Stau2, as a downstream effector in the ATR pathway, might contribute to DNA repair. HCT116 cells were infected with viruses expressing control or shRNA against Stau2 and analyzed at 24 h post-infection by western blotting. Alternatively, cells were transfected with control or siRNA against Stau2. Depletion of Stau2 protein was observed in both cases and generated an increase in DSB formation as evaluated by increased levels of γH2AX (Figure 9A). These results in untreated cells indicate that Stau2 downregulation causes an accumulation of DSBs and therefore suggest that it may promote apoptosis.
Figure 9.
Stau2 downregulation facilitates apoptosis and increases DNA damage. (A) HCT116 cells were either infected with viruses expressing a non-targeting shRNA (Ctrl) or a shRNA against Stau2. Cells were selected on puromycin for 48 h and then cell extracts were analyzed by western blotting for Stau2 expression, DNA damage (γH2AX) and loading (β-actin). Alternatively, cells were transfected with a non-targeting (Ctrl) or an siRNA against Stau2 and analyzed as above. (B) HCT116 cells were infected with a retrovirus expressing a non-targeting shRNA (Ctrl) or a shRNA against Stau2. Cells were selected on puromycin for 48 h and then treated or not with increasing concentrations of CPT for 24 h. Proteins were analyzed by western blotting as above. Quantification of PARP1 cleavage is shown in the Supplementary Figure S7. *, non-specific bands. (C) HCT116 cells were transfected with plasmids coding for the empty vector (Ctrl) or Stau259-FLAG3, selected on puromycin for 48 h and treated or not with CPT (300 nM) for 24 h. Cells extracts were prepared and analyzed by western blotting for apoptosis (PARP1 cleavage), Stau2 expression and loading (β-actin). In Stau259-FLAG3-expressing cells, a 5.8-fold reduction in PARP1 cleavage (PARP189/PARP1116) was observed as compared to vector-transfected cells.
To test this latter possibility, we downregulated Stau2 expression with shRNA and then treated with increasing concentrations of CPT. Apoptosis was evident at CPT concentrations lower than those required for cells transfected with a non-targeting shRNA, indicating that apoptosis is promoted by Stau2 depletion (Figure 9B; Supplementary Figure S7). Similar results were obtained following siRNA-mediated downregulation of Stau2 (data not shown). To confirm this observation, we expressed Stau259-FLAG3 in HCT116 cells and 24 h later incubated the cells with or without CPT. We maintained Stau259-FLAG3 expression at the same level as the endogenous counterpart (Figure 9C). As shown earlier (Figure 4), whereas endogenous Stau2 protein levels decreased as a consequence of CPT-treatment, that of ectopically-expressed Stau259-FLAG3 did not (Figure 9C). Interestingly, under conditions where the overall amount of the Stau2 protein in CPT-treated cells was kept at the level of untreated cells via expression of Stau259-FLAG3, CPT-induced apoptosis was reduced, indicating that Stau2 downregulation in response to CPT promotes apoptosis.
DISCUSSION
In this manuscript we show that Stau2 expression is rapidly inhibited in response to DNA damaging agents. This response is initially transcriptional, causing a subsequent decrease in the amount of Stau2 protein. Our data indicate that E2F1 is involved in regulation of Stau2 expression. In unstressed cells, this transcription factor binds to the Stau2 promoter and activates Stau2 expression. In CPT-treated cells, E2F1 is displaced from the promoter causing the observed downregulation of Stau2 expression. Interestingly, Stau2 downregulation increases DNA damage and promotes apoptosis. We propose that Stau2 downregulation participates in an E2F1-mediated process of cell fate decision contributing to apoptosis following DNA damage.
Downregulation of Stau2 in response to genotoxic stress
Stau2 downregulation is initiated by specific types of DNA damaging agents such as CPT, 5FU and UVC radiation. CPT, through its binding to DNA topoisomerase I, causes the covalent association of Topo I with DNA, leading to the formation of stable complexes on DNA that accumulate at the site of cleavage (39). In addition, CPT prevents the ligation step of the cleavage/ligation reaction of DNA topoisomerase I thus generating single-strand breaks. CPT has indeed been shown to activate the ATR pathway (47,48). Similarly, UVC directly induces cyclobutane pyrimidine dimers (1). CPT-mediated complexes and cyclobutane pyrimidine dimers interfere with DNA replication and transcription by blocking the progression of DNA and RNA polymerases. 5FU metabolites are incorporated into RNA and DNA and/or inhibit thymidylate synthase causing impairment of DNA synthesis and transcription (42). Eventually, in response to CPT, UVC and 5FU, resulting stalled replication forks may collapse, leading to DSB formation. In contrast, Stau2 expression is not regulated by Doxo or IR. Doxo, an inhibitor of topoisomerase II, traps topoisomerase II onto DNA and inhibits DNA re-ligation after the formation of a DNA DSB (49–52). Doxo activates the ATM/CHEK2 pathway (53–56). Similarly, IR directly causes DNA double-stranded breaks and is known to trigger the ATM and DNA-PK pathways (2,6). Both Doxo and IR also induce reactive oxygen species that cause redox imbalance and oxidative DNA damage (1,54). Considering that Doxo and IR both play marginal role in Stau2 expression, our results suggest that Stau2 downregulation via the ATR/CHEK1 signaling pathway is a consequence of early replication and/or transcription stresses rather than subsequent DSB formation that induces the ATM pathway.
In unstressed cells, Stau2 expression is controlled by the ATR and CHEK1 signaling pathways. Both kinases are essential for cell viability in unperturbed dividing cells (57). They are activated at each S phase of the cell cycle and are required for the regulation of replication origin firing (43,44). While ATR is involved in the control of early firing origins, CHEK1 regulates firing of late ones following activation by DNA-PK (58). ATR and CHEK1 are also required for the maintenance of genome integrity (43,44,58). When replication forks stall as a consequence of endogenous (spontaneous) or exogenous DNA damage, ATR stabilizes and repairs the forks, thereby preventing their collapse into DSBs (57). CHEK1 recognizes DNA strand instability during replication and negatively regulates cell cycle progression (43,44). Downregulation of CHEK1 by siRNA (18) or its inhibition with specific drugs (data not shown) strongly induce phosphorylation of H2AX, a marker of double strand breaks. The control of Stau2 expression by ATR and CHEK1 in unstressed cells is interesting because it suggests that Stau2 may be a downstream effector of these signaling pathways and that it may play a role in genome maintenance. Indeed, an increase of spontaneous DNA damage was observed following Stau2 downregulation by RNAi (Figure 9) (18).
Our results identify the transcription factor E2F1 as the most plausible effector. First, we show that its expression is downregulated in the presence of ATR or CHEK1 inhibitors (Supplementary Figure S5), concomitant with reduced expression of Stau2 both at the mRNA and/or protein levels (Figure 3). Indeed, E2F1 binds to the Stau2 promoter to trigger transcription of Stau2 mRNA and the downregulation of Stau2 in response to DNA damage correlates with displacement of E2F1 from the promoter (Figure 8). Second, E2F1 is normally rapidly degraded during the S and G2 phases of the cell cycle by the SKP2-mediated ubiquitination and proteasome pathway (59). However, activation of ATR in S phase induces E2F1 phosphorylation on Ser31 causing its dissociation from SKP2 and thereby stabilizing E2F1 (45,57). Similarly, CHEK1 activation stabilizes E2F1 (60). It is obvious that other proteins and/or post-translational modifications are also required as cofactors since E2F1 can be stabilized by several other kinases not involved in Stau2 expression (43,61). Interestingly, whereas ATR inhibition for 8h clearly reduces Stau2 mRNA level concomitant with a modest effect at the protein level, inhibition of CHEK1 almost completely eliminates Stau2 protein despite an intermediate decrease at the mRNA level. The simplest explanation would be that CHEK1 can phosphorylate and stabilize Stau2. Indeed two putative phosphorylation sites are found in the Stau2 sequence and T294 is found 3 amino acids downstream of the observed ubiquitinated K297 (62). It will be interesting to test if T294 is phosphorylated by CHEK1 and if this modification prevents Stau2 ubiquitination.
Transcriptional regulation of Stau2
Our data clearly indicate that Stau2 downregulation in response to DNA damaging agents is transcriptional. Consequently, we identified the Stau2 promoter region and delimited its activity within a 198 bp fragment. This fragment is necessary and sufficient for Stau2 expression in untreated cells and for its downregulation in response to CPT. Our results are consistent with a model in which E2F1 binds the Stau2 promoter and upregulates its expression in cells in the absence of applied genotoxic stress. However, following mutagen exposure, E2F1 is displaced from the Stau2 promoter thus downregulating the latter's expression. An apparent paradox concerns why ATR-dependent activation of E2F1 during the S phase of unstressed cells allows Stau2 expression, while ATR-dependent activation of E2F1 during genotoxic stress inhibits Stau2 expression. The answer probably resides in the molecular mechanisms of E2F1 activation and of its interacting partners. In unstressed cells, E2F1 is known to be activated at the G1/S phase of the cell cycle by dissociation of its inhibitory partner Rb (63). Its release from Rb regulates the transcription of genes involved in cell proliferation and DNA replication/repair (64). Its subsequent phosphorylation by ATR in S phase prevents its degradation (45,57). Following genotoxic stress, E2F1 is upregulated via its stabilization by phosphorylation by several kinases (12). The function of E2F1 then changes from the control of cell proliferation to the induction of apoptosis. Pro-apoptotic genes are transcribed and anti-apoptotic genes are silenced. Furthermore, it was recently shown that ATR phosphorylates different targets in unstressed and DNA damage cells (65), suggesting that alternative downstream responses may be controlled by signaling thresholds.
In addition to Stau2, E2F1 was previously shown to repress several genes following DNA damage. The anti-apoptotic Mcl-1 and BCl-2 genes are downregulated via direct transcriptional repression by E2F1 in a p53 independent manner (66,67). In both cases, the DNA-binding domain of E2F1 is required for repression, suggesting that an E2F1-containing inhibitory complex may be formed on the promoter. Cofactor proteins are likely involved in the modulation of E2F1 functions. For example, it was shown that E2F1 activates transcription when linked to P/CAF (68) whereas it inhibits transcription when associated with TopBP1 (69). For Stau2 downregulation, these E2F1-inhibitory complexes may not be involved since we did not detect E2F1 on the Stau2 promoter after CPT treatment (Figure 8). Alternatively, competing transcription factors may be activated to displace E2F1 from the promoter in a CPT-dependent manner. Proteins such as p53, E2F4 and Sp1 were previously shown to antagonize E2F1 activity (12,70–73).
DNA damage and apoptosis
Why is Stau2 expression downregulated in response to DNA damage? For cells not challenged with DNA damaging agents, our results suggest that Stau2 is an anti-apoptotic protein that could be involved in genome maintenance. On the other hand, during periods of genotoxic stress, Stau2 downregulation may facilitate E2F1-mediated apoptosis. We propose that Stau2 may be part of a decision pathway that modulates the precarious equilibrium between pro-survival and pro-apoptotic proteins to promote cell death. The post-transcriptional functions of Stau2 could complement the transcriptional activity of E2F1 by stabilizing or degrading mRNAs coding for pro- and anti-apoptotic genes respectively. Stau2 is indeed an important regulator of post-transcriptional gene expression (34) and it will be important to determine if expression of at least some of these genes is regulated by Stau2 during genotoxic stress. Its downregulation following DNA damage potentially represents a novel facet of post-transcriptional DDR regulation, required to fine-tune gene expression in order to precisely link protein synthesis to specific cellular requirements with acute precision (74–76). For example, following treatment with IR, the RNA-binding protein HuR is phosphorylated by p38MAPK, allowing HuR binding to p21Cip1/WAF mRNA and subsequent p21 mRNA stabilization/protein accumulation (77,78). HuR is also phosphorylated by CHEK2 causing the dissociation of the HuR/SIRT1 mRNA complex and the degradation of the released mRNA, thus contributing to apoptosis (79). Alternatively, we do not exclude the possibility that Stau2 interacts with nuclear proteins to protect against apoptosis. Although mainly cytoplasmic, Stau2 was also shown to transit in the nucleus (80) and to associate with nuclear proteins in an RNA-independent manner (22,81,82).
Many anti-cancer drugs and radiotherapy target DNA, and their therapeutic efficacy primarily depends on the induction of apoptosis. Unfortunately, many tumor cells can disrupt apoptotic pathways allowing them to evade the cytotoxic action of anti-cancer treatments. Thus, elucidation of the molecular functions of Stau2 in DNA repair and apoptosis may contribute to our understanding of how pro-survival functions may be subverted in neoplastic cells, providing new strategies for therapeutic interventions in cancer.
Supplementary Material
Acknowledgments
We thank François Belanger for his help with the UVC and IR experiments, Louise Cournoyer for help with cell culture, and Celine Frechina, Natacha Dozois, Evelyn Leblanc and Othmane Ennajih for technical assistance.
Footnotes
Present address: Wildriss Viranaicken, PIMIT, Processus Infectieux en Milieu Insulaire Tropical, I2T team, Université de La Réunion.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institute for Health Research of Canada (CIHR) [MOP-229979 to L.D.G.]; Natural Science and Engineering Research Council of Canada (NSERC) [41596-09 to L.D.G.]. NSERC Studentships (to V.T.). Funding for open access charge: Canadian Institute of Health Research [MOP-229979].
Conflict of interest statement. None declared.
REFERENCES
- 1.Roos W.P., Kaina B. DNA damage-induced cell death: from specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett. 2013;332:237–248. doi: 10.1016/j.canlet.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Ciccia A., Elledge S.J. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reinhardt H.C., Schumacher B. The p53 network: cellular and systemic DNA damage responses in aging and cancer. Trends Genet. 2012;28:128–136. doi: 10.1016/j.tig.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Medema R.H., Macurek L. Checkpoint control and cancer. Oncogene. 2012;31:2601–2613. doi: 10.1038/onc.2011.451. [DOI] [PubMed] [Google Scholar]
- 5.Aziz K., Nowsheen S., Pantelias G., Iliakis G., Gorgoulis V.G., Georgakilas A.G. Targeting DNA damage and repair: embracing the pharmacological era for successful cancer therapy. Pharmacol. Ther. 2012;133:334–350. doi: 10.1016/j.pharmthera.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 6.Hiom K. Coping with DNA double strand breaks. DNA Repair (Amst) 2010;9:1256–1263. doi: 10.1016/j.dnarep.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 7.Guo Z., Kumagai A., Wang S.X., Dunphy W.G. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 2000;14:2745–2756. doi: 10.1101/gad.842500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stiff T., Walker S.A., Cerosaletti K., Goodarzi A.A., Petermann E., Concannon P., O'Driscoll M., Jeggo P.A. ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J. 2006;25:5775–5782. doi: 10.1038/sj.emboj.7601446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bieging K.T., Attardi L.D. Deconstructing p53 transcriptional networks in tumor suppression. Trends Cell Biol. 2012;22:97–106. doi: 10.1016/j.tcb.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mirzayans R., Andrais B., Scott A., Murray D. New insights into p53 signaling and cancer cell response to DNA damage: implications for cancer therapy. J. Biomed. Biotechnol. 2012:170325. doi: 10.1155/2012/170325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engelmann D., Putzer B.M. The dark side of E2F1: in transit beyond apoptosis. Cancer Res. 2012;72:571–575. doi: 10.1158/0008-5472.CAN-11-2575. [DOI] [PubMed] [Google Scholar]
- 12.Biswas A.K., Johnson D.G. Transcriptional and nontranscriptional functions of E2F1 in response to DNA damage. Cancer Res. 2012;72:13–17. doi: 10.1158/0008-5472.CAN-11-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murray-Zmijewski F., Slee E.A., Lu X. A complex barcode underlies the heterogeneous response of p53 to stress. Nat. Rev. Mol. Cell Biol. 2008;9:702–712. doi: 10.1038/nrm2451. [DOI] [PubMed] [Google Scholar]
- 14.Chen D., Padiernos E., Ding F., Lossos I.S., Lopez C.D. Apoptosis-stimulating protein of p53–2 (ASPP2/53BP2L) is an E2F target gene. Cell Death Differ. 2005;12:358–368. doi: 10.1038/sj.cdd.4401536. [DOI] [PubMed] [Google Scholar]
- 15.Hershko T., Chaussepied M., Oren M., Ginsberg D. Novel link between E2F and p53: proapoptotic cofactors of p53 are transcriptionally upregulated by E2F. Cell Death Differ. 2005;12:377–383. doi: 10.1038/sj.cdd.4401575. [DOI] [PubMed] [Google Scholar]
- 16.Fan J., Yang X., Wang W., Wood W.H., Becker K.G., Gorospe M. Global analysis of stress-regulated mRNA turnover by using cDNA arrays. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10611–10616. doi: 10.1073/pnas.162212399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R., 3rd, Hurov K.E., Luo J., Bakalarski C.E., Zhao Z., Solimini N., Lerenthal Y., et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 18.Paulsen R.D., Soni D.V., Wollman R., Hahn A.T., Yee M.C., Guan A., Hesley J.A., Miller S.C., Cromwell E.F., Solow-Cordero D.E., et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol. Cell. 2009;35:228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilker E.W., van Vugt M.A., Artim S.A., Huang P.H., Petersen C.P., Reinhardt H.C., Feng Y., Sharp P.A., Sonenberg N., White F.M., et al. 14–3–3sigma controls mitotic translation to facilitate cytokinesis. Nature. 2007;446:329–332. doi: 10.1038/nature05584. [DOI] [PubMed] [Google Scholar]
- 20.Duchaine T.F., Hemraj I., Furic L., Deitinghoff A., Kiebler M.A., DesGroseillers L. Staufen2 isoforms localize to the somatodendritic domain of neurons and interact with different organelles. J. Cell Sci. 2002;115:3285–3295. doi: 10.1242/jcs.115.16.3285. [DOI] [PubMed] [Google Scholar]
- 21.Tang S.J., Meulemans D., Vazquez L., Colaco N., Schuman E. A role for a rat homolog of staufen in the transport of RNA to neuronal dendrites. Neuron. 2001;32:463–475. doi: 10.1016/s0896-6273(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 22.Maher-Laporte M., Berthiaume F., Moreau M., Julien L.A., Lapointe G., Mourez M., DesGroseillers L. Molecular composition of staufen2-containing ribonucleoproteins in embryonic rat brain. PLoS One. 2010;5:e11350. doi: 10.1371/journal.pone.0011350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mallardo M., Deitinghoff A., Muller J., Goetze B., Macchi P., Peters C., Kiebler M.A. Isolation and characterization of Staufen-containing ribonucleoprotein particles from rat brain. Proc. Natl. Acad. Sci. U.S.A. 2003;100:2100–2105. doi: 10.1073/pnas.0334355100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goetze B., Tuebing F., Xie Y., Dorostkar M.M., Thomas S., Pehl U., Boehm S., Macchi P., Kiebler M.A. The brain-specific double-stranded RNA-binding protein Staufen2 is required for dendritic spine morphogenesis. J. Cell Biol. 2006;172:221–231. doi: 10.1083/jcb.200509035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Leary D.A., Sharif O., Anderson P., Tu B., Welch G., Zhou Y., Caldwell J.S., Engels I.H., Brinker A. Identification of small molecule and genetic modulators of AON-induced dystrophin exon skipping by high-throughput screening. PLoS One. 2009;4:e8348. doi: 10.1371/journal.pone.0008348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lebeau G., Miller L.C., Tartas M., McAdam R., Laplante I., Badeaux F., DesGroseillers L., Sossin W.S., Lacaille J.C. Staufen 2 regulates mGluR long-term depression and Map1b mRNA distribution in hippocampal neurons. Learn. Mem. 2011;18:314–326. doi: 10.1101/lm.2100611. [DOI] [PubMed] [Google Scholar]
- 27.Miki T., Kamikawa Y., Kurono S., Kaneko Y., Katahira J., Yoneda Y. Cell type-dependent gene regulation by Staufen2 in conjunction with Upf1. BMC Mol. Biol. 2011;12:48. doi: 10.1186/1471-2199-12-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park E., Gleghorn M.L., Maquat L.E. Staufen2 functions in Staufen1-mediated mRNA decay by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity. Proc. Natl. Acad. Sci. U.S.A. 2013;110:405–412. doi: 10.1073/pnas.1213508110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramasamy S., Wang H., Quach H.N., Sampath K. Zebrafish Staufen1 and Staufen2 are required for the survival and migration of primordial germ cells. Dev. Biol. 2006;292:393–406. doi: 10.1016/j.ydbio.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 30.Bilogan C.K., Horb M.E. Xenopus staufen2 is required for anterior endodermal organ formation. Genesis. 2012;50:251–259. doi: 10.1002/dvg.22000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cockburn D.M., Charish J., Tassew N.G., Eubanks J., Bremner R., Macchi P., Monnier P.P. The double-stranded RNA-binding protein Staufen 2 regulates eye size. Mol. Cell. Neurosci. 2012;51:101–111. doi: 10.1016/j.mcn.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 32.Vessey J.P., Amadei G., Burns S.E., Kiebler M.A., Kaplan D.R., Miller F.D. An asymmetrically localized Staufen2-dependent RNA complex regulates maintenance of mammalian neural stem cells. Cell Stem Cell. 2012;11:517–528. doi: 10.1016/j.stem.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 33.Kusek G., Campbell M., Doyle F., Tenenbaum S.A., Kiebler M., Temple S. Asymmetric segregation of the double-stranded RNA binding protein Staufen2 during mammalian neural stem cell divisions promotes lineage progression. Cell Stem Cell. 2012;11:505–516. doi: 10.1016/j.stem.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Furic L., Maher-Laporte M., DesGroseillers L. A genome-wide approach identifies distinct but overlapping subsets of cellular mRNAs associated with Staufen1- and Staufen2-containing ribonucleoprotein complexes. RNA. 2008;14:324–335. doi: 10.1261/rna.720308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacKeigan J.P., Murphy L.O., Blenis J. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat. Cell Biol. 2005;7:591–600. doi: 10.1038/ncb1258. [DOI] [PubMed] [Google Scholar]
- 36.Vigo E., Muller H., Prosperini E., Hateboer G., Cartwright P., Moroni M.C., Helin K. CDC25A phosphatase is a target of E2F and is required for efficient E2F-induced S phase. Mol. Cell. Biol. 1999;19:6379–6395. doi: 10.1128/mcb.19.9.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wickham L., Duchaine T., Luo M., Nabi I.R., DesGroseillers L. Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol. Cell. Biol. 1999;19:2220–2230. doi: 10.1128/mcb.19.3.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martel C., Dugre-Brisson S., Boulay K., Breton B., Lapointe G., Armando S., Trepanier V., Duchaine T., Bouvier M., Desgroseillers L. Multimerization of Staufen1 in live cells. RNA. 2010;16:585–597. doi: 10.1261/rna.1664210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu L.F., Desai S.D., Li T.K., Mao Y., Sun M., Sim S.P. Mechanism of action of camptothecin. Ann. N.Y. Acad. Sci. 2000;922:1–10. doi: 10.1111/j.1749-6632.2000.tb07020.x. [DOI] [PubMed] [Google Scholar]
- 40.Moroni M.C., Hickman E.S., Lazzerini Denchi E., Caprara G., Colli E., Cecconi F., Muller H., Helin K. Apaf-1 is a transcriptional target for E2F and p53. Nat. Cell Biol. 2001;3:552–558. doi: 10.1038/35078527. [DOI] [PubMed] [Google Scholar]
- 41.Tacar O., Sriamornsak P., Dass C.R. Doxorubicin: an update on anticancer molecular action, toxicity and novel drug delivery systems. J. Pharm. Pharmacol. 2013;65:157–170. doi: 10.1111/j.2042-7158.2012.01567.x. [DOI] [PubMed] [Google Scholar]
- 42.Longley D.B., Harkin D.P., Johnston P.G. 5-fluorouracil: mechanisms of action and clinical strategies. Nat. Rev. Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y., Hunter T. Roles of Chk1 in cell biology and cancer therapy. Int. J. Cancer. 2014;134:1013–1023. doi: 10.1002/ijc.28226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patil M., Pabla N., Dong Z. Checkpoint kinase 1 in DNA damage response and cell cycle regulation. Cell. Mol. Life Sci. 2013;70:4009–4021. doi: 10.1007/s00018-013-1307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin W.C., Lin F.T., Nevins J.R. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 2001;15:1833–1844. [PMC free article] [PubMed] [Google Scholar]
- 46.O'Connor D.J., Lu X. Stress signals induce transcriptionally inactive E2F-1 independently of p53 and Rb. Oncogene. 2000;19:2369–2376. doi: 10.1038/sj.onc.1203540. [DOI] [PubMed] [Google Scholar]
- 47.Helt C.E., Cliby W.A., Keng P.C., Bambara R.A., O'Reilly M.A. Ataxia telangiectasia mutated (ATM) and ATM and Rad3-related protein exhibit selective target specificities in response to different forms of DNA damage. J. Biol. Chem. 2005;280:1186–1192. doi: 10.1074/jbc.M410873200. [DOI] [PubMed] [Google Scholar]
- 48.Bensimon A., Aebersold R., Shiloh Y. Beyond ATM: the protein kinase landscape of the DNA damage response. FEBS Lett. 2011;585:1625–1639. doi: 10.1016/j.febslet.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 49.Wijdeven R.H., Pang B., van der Zanden S.Y., Qiao X., Blomen V., Hoogstraat M., Lips E.H., Janssen L., Wessels L., Brummelkamp T.R., et al. Genome-wide identification and characterization of novel factors conferring resistance to topoisomerase II poisons in cancer. Cancer Res. 2015;75:4176–4187. doi: 10.1158/0008-5472.CAN-15-0380. [DOI] [PubMed] [Google Scholar]
- 50.Pommier Y. Drugging topoisomerases: lessons and challenges. ACS Chem. Biol. 2013;8:82–95. doi: 10.1021/cb300648v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pang B., Qiao X., Janssen L., Velds A., Groothuis T., Kerkhoven R., Nieuwland M., Ovaa H., Rottenberg S., van Tellingen O., et al. Drug-induced histone eviction from open chromatin contributes to the chemotherapeutic effects of doxorubicin. Nat. Commun. 2013;4:1908. doi: 10.1038/ncomms2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang F., Kemp C.J., Henikoffa S. Anthracyclines induce double-strand DNA breaks at active gene promoters. Mutat. Res. 2015;773:9–15. doi: 10.1016/j.mrfmmm.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bakkenist C.J., Czambel R.K., Hershberger P.A., Tawbi H., Beumer J.H., Schmitz J.C. A quasi-quantitative dual multiplexed immunoblot method to simultaneously analyze ATM and H2AX Phosphorylation in human peripheral blood mononuclear cells. Oncoscience. 2015;2:542–554. doi: 10.18632/oncoscience.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kurz E.U., Douglas P., Lees-Miller S.P. Doxorubicin activates ATM-dependent phosphorylation of multiple downstream targets in part through the generation of reactive oxygen species. J. Biol. Chem. 2004;279:53272–53281. doi: 10.1074/jbc.M406879200. [DOI] [PubMed] [Google Scholar]
- 55.Brum G., Carbone T., Still E., Correia V., Szulak K., Calianese D., Best C., Cammarata G., Higgins K., Ji F., et al. N-acetylcysteine potentiates doxorubicin-induced ATM and p53 activation in ovarian cancer cells. Int. J. Oncol. 2013;42:211–218. doi: 10.3892/ijo.2012.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshida M., Shiojima I., Ikeda H., Komuro I. Chronic doxorubicin cardiotoxicity is mediated by oxidative DNA damage-ATM-p53-apoptosis pathway and attenuated by pitavastatin through the inhibition of Rac1 activity. J. Mol. Cell Cardiol. 2009;47:698–705. doi: 10.1016/j.yjmcc.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 57.Cimprich K.A., Cortez D. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buisson R., Boisvert J.L., Benes C.H., Zou L. Distinct but concerted roles of ATR, DNA-PK, and Chk1 in countering replication stress during S phase. Mol. Cell. 2015;59:1011–1024. doi: 10.1016/j.molcel.2015.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marti A., Wirbelauer C., Scheffner M., Krek W. Interaction between ubiquitin-protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat. Cell Biol. 1999;1:14–19. doi: 10.1038/8984. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y.W., Jones T.L., Martin S.E., Caplen N.J., Pommier Y. Implication of checkpoint kinase-dependent up-regulation of ribonucleotide reductase R2 in DNA damage response. J. Biol. Chem. 2009;284:18085–18095. doi: 10.1074/jbc.M109.003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Putzer B.M., Engelmann D. E2F1 apoptosis counterattacked: evil strikes back. Trends Mol. Med. 2013;19:89–98. doi: 10.1016/j.molmed.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 62.Kim W., Bennett E.J., Huttlin E.L., Guo A., Li J., Possemato A., Sowa M.E., Rad R., Rush J., Comb M.J., et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dynlacht B.D. Regulation of transcription by proteins that control the cell cycle. Nature. 1997;389:149–152. doi: 10.1038/38225. [DOI] [PubMed] [Google Scholar]
- 64.Dynlacht B.D., Moberg K., Lees J.A., Harlow E., Zhu L. Specific regulation of E2F family members by cyclin-dependent kinases. Mol. Cell. Biol. 1997;17:3867–3875. doi: 10.1128/mcb.17.7.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bastos de Oliveira F.M., Kim D., Cussiol J.R., Das J., Jeong M.C., Doerfler L., Schmidt K.H., Yu H., Smolka M.B. Phosphoproteomics reveals distinct modes of Mec1/ATR signaling during DNA replication. Mol. Cell. 2015;57:1124–1132. doi: 10.1016/j.molcel.2015.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Croxton R., Ma Y., Song L., Haura E.B., Cress W.D. Direct repression of the Mcl-1 promoter by E2F1. Oncogene. 2002;21:1359–1369. doi: 10.1038/sj.onc.1205157. [DOI] [PubMed] [Google Scholar]
- 67.Eischen C.M., Packham G., Nip J., Fee B.E., Hiebert S.W., Zambetti G.P., Cleveland J.L. Bcl-2 is an apoptotic target suppressed by both c-Myc and E2F-1. Oncogene. 2001;20:6983–6993. doi: 10.1038/sj.onc.1204892. [DOI] [PubMed] [Google Scholar]
- 68.Korah J., Falah N., Lacerte A., Lebrun J.J. A transcriptionally active pRb-E2F1-P/CAF signaling pathway is central to TGFbeta-mediated apoptosis. Cell Death Dis. 2012;3:e407. doi: 10.1038/cddis.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu K., Luo Y., Lin F.T., Lin W.C. TopBP1 recruits Brg1/Brm to repress E2F1-induced apoptosis, a novel pRb-independent and E2F1-specific control for cell survival. Genes Dev. 2004;18:673–686. doi: 10.1101/gad.1180204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu S., Murai S., Kataoka K., Miyagishi M. Cooperative regulation of p73 promoter by Yin Yang 1 and E2F1. Nucleic Acids Symp. Ser. (Oxf) 2007;51:347–348. doi: 10.1093/nass/nrm174. [DOI] [PubMed] [Google Scholar]
- 71.Taura M., Suico M.A., Fukuda R., Koga T., Shuto T., Sato T., Morino-Koga S., Okada S., Kai H. MEF/ELF4 transactivation by E2F1 is inhibited by p53. Nucleic Acids Res. 2011;39:76–88. doi: 10.1093/nar/gkq762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ingram L., Munro S., Coutts A.S., La Thangue N.B. E2F-1 regulation by an unusual DNA damage-responsive DP partner subunit. Cell Death Differ. 2011;18:122–132. doi: 10.1038/cdd.2010.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Racek T., Buhlmann S., Rust F., Knoll S., Alla V., Putzer B.M. Transcriptional repression of the prosurvival endoplasmic reticulum chaperone GRP78/BIP by E2F1. J. Biol. Chem. 2008;283:34305–34314. doi: 10.1074/jbc.M803925200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moore M.J. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 75.Chan C.S., Elemento O., Tavazoie S. Revealing posttranscriptional regulatory elements through network-level conservation. PLoS Comput. Biol. 2005;1:e69. doi: 10.1371/journal.pcbi.0010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Keene J.D. Minireview: global regulation and dynamics of ribonucleic Acid. Endocrinology. 2010;151:1391–1397. doi: 10.1210/en.2009-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lafarga V., Cuadrado A., Lopez de Silanes I., Bengoechea R., Fernandez-Capetillo O., Nebreda A.R. p38 Mitogen-activated protein kinase- and HuR-dependent stabilization of p21(Cip1) mRNA mediates the G(1)/S checkpoint. Mol. Cell. Biol. 2009;29:4341–4351. doi: 10.1128/MCB.00210-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang W., Furneaux H., Cheng H., Caldwell M.C., Hutter D., Liu Y., Holbrook N., Gorospe M. HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 2000;20:760–769. doi: 10.1128/mcb.20.3.760-769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abdelmohsen K., Pullmann R., Jr, Lal A., Kim H.H., Galban S., Yang X., Blethrow J.D., Walker M., Shubert J., Gillespie D.A., et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell. 2007;25:543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Macchi P., Brownawell A.M., Grunewald B., DesGroseillers L., Macara I.G., Kiebler M.A. The brain-specific double-stranded RNA-binding protein Staufen2: nucleolar accumulation and isoform-specific exportin-5-dependent export. J. Biol. Chem. 2004;279:31440–31444. doi: 10.1074/jbc.C400226200. [DOI] [PubMed] [Google Scholar]
- 81.Elvira G., Massie B., DesGroseillers L. The zinc-finger protein ZFR is critical for Staufen 2 isoform specific nucleocytoplasmic shuttling in neurons. J. Neurochem. 2006;96:105–117. doi: 10.1111/j.1471-4159.2005.03523.x. [DOI] [PubMed] [Google Scholar]
- 82.Monshausen M., Gehring N.H., Kosik K.S. The mammalian RNA-binding protein Staufen2 links nuclear and cytoplasmic RNA processing pathways in neurons. Neuromol. Med. 2004;6:127–144. doi: 10.1385/NMM:6:2-3:127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.