Abstract
The transcription factor (TF) SOX18 drives lymphatic vessel development in both embryogenesis and tumour-induced neo-lymphangiogenesis. Genetic disruption of Sox18 in a mouse model protects from tumour metastasis and established the SOX18 protein as a molecular target. Here, we report the crystal structure of the SOX18 DNA binding high-mobility group (HMG) box bound to a DNA element regulating Prox1 transcription. The crystals diffracted to 1.75Å presenting the highest resolution structure of a SOX/DNA complex presently available revealing water structure, structural adjustments at the DNA contact interface and non-canonical conformations of the DNA backbone. To explore alternatives to challenging small molecule approaches for targeting the DNA-binding activity of SOX18, we designed a set of five decoys based on modified Prox1-DNA. Four decoys potently inhibited DNA binding of SOX18 in vitro and did not interact with non-SOX TFs. Serum stability, nuclease resistance and thermal denaturation assays demonstrated that a decoy circularized with a hexaethylene glycol linker and terminal phosphorothioate modifications is most stable. This SOX decoy also interfered with the expression of a luciferase reporter under control of a SOX18-dependent VCAM1 promoter in COS7 cells. Collectively, we propose SOX decoys as potential strategy for inhibiting SOX18 activity to disrupt tumour-induced neo-lymphangiogenesis.
INTRODUCTION
SRY-related box 18 (SOX18) belongs to the SOX transcription factor (TF) family comprising 20 paralogous members in the human genome with critical but diverse roles in specifying and maintaining cellular identities throughout development (1–4). DNA recognition is accomplished by a 79 amino acid high mobility group (HMG) box. The HMG box is ubiquitously found in a large number of DNA binding proteins that bind DNA either specifically or non-specifically (5). The HMG box of SOX proteins confers sequence specificity and all members bind to a short (A/T)2T3T4G5T6 core sequence with high affinity (6,7). Yet, high-throughput studies suggest subtle variations within the core and flanking regions that could demarcate preferred recognition sequences of individual SOX family members (6,8). The Sox DNA motif is short and degenerated and therefore highly abundant in the human genome. Thus, it remains an open question how individual SOX proteins select specific sets of target genes to dictate cell fate decisions. An in depth understanding how target DNA selectivity is achieved would also inform efforts for the design of nucleic acid-based drugs. To aid these goals we explored in this study the DNA binding mechanism of SOX18 for its purported role in the metastasis of solid tumours.
SOX18 regulates angiogenesis (9), lymphangiogenesis (10) and hair follicle differentiation (11). In mice, SOX18 is expressed at 9 days post coitum (dpc) in a subpopulation of endothelial cells from the anterior cardinal vein and induces a lymphatic endothelial cell (LEC) fate which subsequently gives rise to the initial lymphatic vascular plexus (10). SOX18 binds to and causes the transactivation of the Prospero homeobox 1 (Prox1) gene, the hallmark of LEC identity. In Sox18 as well as Prox1 loss of function scenarios, deficient mice are devoid of lymphatic vessels, develop severe generalized edema and die in utero at around 14.5 dpc (10,12). The regulatory cascade from SOX18 to Prox1 is therefore regarded as a key regulatory switch conferring lymphatic cell specification (reviewed in (13,14)). Reminiscent to its role in the early embryo, SOX18 also initiates the formation of new lymphatic vasculature (neo-lymphangiogenesis) in mouse melanoma models (15). Gene targeted deletion of Sox18 has been shown to interfere with tumour-induced lymphangiogenesis and SOX18 was therefore proposed as a novel molecular target to prevent the metastatic spread of solid tumours (16). In human, SOX18 was implicated as prognostic marker of poor patient outcome for solid tumours such as gastric cancer, breast adenocarcinoma and non-small cell lung cancer (17–19). With the view to manipulating SOX18 activity in a therapeutic setting, here we explore a way to inhibit the function of this TF. As the DNA binding domains of TF proteins are widely deemed ‘undruggable’ using small-molecules (20,21), alternative strategies need to be considered to make this target class accessible for cancer therapy (22). We therefore set out to structurally analyse the DNA recognition by SOX18 and to utilize these insights to design SOX DNA decoys for its selective inhibition of gene transactivation.
Published structural data of SOX/DNA complexes are available for SRY (23), SOX2 (24,25), SOX17 (26) and SOX4 (27) greatly improved our understanding of SOX DNA recognition. Yet, these structures were built using data at intermediate resolution regimes with a 2.4Å structure of the SOX4/DNA complex providing the highest resolution dataset. To further analyse how SOX TFs in general and SOX18 in particular engage their target genes, we solved the crystal structure of SOX18-HMG bound to a 16 bp regulatory region derived from an intron of its target gene Prox1 using crystals diffracting to 1.75Å resolution. We used the Prox1-DNA to design SOX decoys and tested their potency, selectivity, stability as well as their effects on the expression of SOX18 target genes in cell culture. This approach led to the identification of a pan-SOX inhibitory decoy. This SOX-HMG family specific decoy shows prolonged resistance to nuclease digestion, degradation in mouse serum and thermal denaturation. Further, it inhibits the expression of SOX18 dependent reported genes.
MATERIALS AND METHODS
Production of SOX18-HMG for crystallization
The pENTR-mSOX18-HMG entry clone encoding 79 amino acids of the HMG box of mouse SOX18 (accession BC006612) was generated by GATEWAY BP (Invitrogen) using DNA oligos GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAAACCTGTATTTTCAGGGCTTGCGCATTCGGCGGCC and GGGGACCACTTTGTACAAGAAAGCTGGGTTTATCACTGTTTTTTGCGGCGAGGCC (gene-specific portion underlined) containing attB sites and a 5′ tobacco etch virus (TEV) protease cleavage site. The SOX18-HMG box was recombined by GATEWAY LR into the pDESTHisMBP plasmid and heterologously expressed in BL21 (DE3) cells and purified as described previously (28). In brief, the fusion protein was purified using an immobilized metal affinity chromatography step, tag cleavage achieved using TEV protease followed by ion exchange and size exclusion chromatography (Figure 1A).
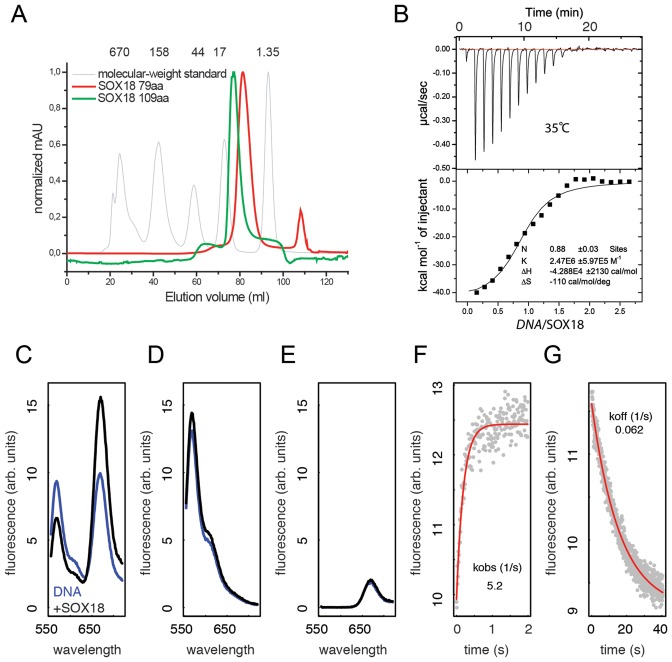
Figure 1.
Calorimetry, DNA deformation and kinetics of SOX18-HMG/Prox1-DNA complex formation. (A) Calibrated size exclusion chromatogram of 79 aa and 109 aa versions of the SOX18-HMG showing that the proteins elute as symmetrical peaks corresponding to the molecular weight of the respective monomers. The grey curve is a molecular weight standard with weights indicated in kDa. (B) Representative ITC titration of 70 μM 15-bp Prox1-DNA in the syringe and 10 μM SOX18-HMG (79 aa) in the cell. The lower panel shows the equilibrium isotherm and the inset the parameters obtained using single site binding models and the Origin software. (C) FRET spectra of 15 bp Prox1-DNA labelled with 5′ cy3 at the forward and 5′ cy5 at the reverse strand before (blue) and after (black) addition of the SOX18-HMG. Spectra were recorded from 555–720 nm after excitation at 535 nm for double labelled (C) or single labelled Prox1 elements (D and E). The Association (F) and the dissociation (G) kinetics of SOX18 with Prox1 were measured by recording the cy5 (FRET acceptor) emission at 668 nm. Red curves are mono-exponential fits to the data.
Crystallization and data collection
A solution of polyacrylamide gelelectrophoresis (PAGE)-purified 16 bp Prox1-DNA elements with CG-overhangs (5′-CACTAGCATTGTCTGGG-3′) was obtained from Sigma-Proligo at 1 mM and adjusted to pH 8.0 using Tris–HCl. Annealing was carried out by first combining equimolar amounts of complementary DNA, followed by heating to a temperature of 95°C for 5 min and cooling to 4°C at the rate of 0.5°C/s using a polymerase chain reaction (PCR) thermocycler (Bio-Rad). The protein/DNA complex was prepared by mixing SOX18-HMG (79 aa) and Prox1-DNA at a molar ratio of 1:1.2. The complex was concentrated to 15 mg/ml using a Vivaspin centrifugal filter column with molecular weight cut-off of 3 kDa (Sartorius). A buffer containing 0.1 M DL-Malic acid, MES monohydrate, Tris (MMT), pH 5.0, 25% (w/v) PEG1500 was used to grow crystals at ambient temperature in a hanging–drop vapour diffusion setting. A drop volume of 1 μl was soaked with 1 μl of soaking–solution (1 % dimethyl sulfoxide (DMSO), 0.62 mM compound, 0.1 M MMT buffer pH 5.0, 25% (w/v) PEG1500). The cryoprotectant used consisted of 0.1 M MMT buffer pH 5.0, 25% (w/v) PEG1500 plus 15% (w/v) glycerol. Crystals were flash frozen in liquid nitrogen. A 1.75Å resolution data set was collected at the X29 beamline of the National Synchrotron Light Source (Upton NY) and processed using the HKL–2000 software (29) (Supplementary Table S1).
Structure solution and refinement
The search model for molecular replacement was prepared using the coordinates of the crystal structure of the SOX4/Lama1-DNA complex (PDB ID: 3U2B (27)). Molecular replacement was performed with phaser (30) using data from 50Å to 1.75Å searching for one protein/DNA complex in the asymmetric unit by including space groups P64 and P62. Phaser found one solution in space group P64 and a first Fourier difference electron density map revealed new interpretable features, confirming the correctness of the solution. The model was built manually using Coot (31) by interpreting 2Fo-Fc and Fo-Fc electron density maps. Refinement was performed with Phenix.refine (32) including a simulated annealing run at the beginning of refinement. 5% of the reflections were marked for cross-validation throughout the refinement. Water molecules were built using Phenix.refine. The final rounds included translation-libration-screw refinement.
Isothermal titration calorimetry (ITC)
A 15-bp Prox1-DNA element (5′- CTAGCATTGTCTGGG; Life Technologies) was annealed in 1× annealing buffer (20 mM Tris–HCl, pH 8.0; 50 mM MgCl2; 50 mM KCl) as described above. The isothermal titration calorimetry (ITC) experiment was carried out in degassed ITC buffer containing 10 mM potassium phosphate, 150 mM KCl, pH 7.0. Prox1-DNA was extensively dialysed against 2× 1 l of degassed ITC buffer with 3.5K MWCO Snakeskin Dialysis Tubing (Life Technologies) at 4°C. The dialysate was replaced after 2 h and dialysis was continued overnight. For buffer exchange of SOX18-HMG (79 aa), a batch of freshly purified protein was applied to a Superdex S75 16/60 column equilibrated in ITC buffer and peak fractions were concentrated up to 70 μM, flash-frozen and stored at −80°C. Reactants were concentrated with Nanosep 3K MWCO OMEGA spin tubes (Pall Corporation) at 5000 g at 4°C for 20 min followed by a 10 min centrifugation at 13 000 rpm at 4°C. The concentrations were measured with a Nanodrop 2000 (Thermo Fisher) using the dialysis buffer as blank. The ITC experiment was performed using a MicroCal iTC200 (Malvern Instruments) at 35°C with 10 μM of protein in the 200 μl sample cell and 40 μl of 70 μM Prox1-DNA in the syringe. The protein concentration was adjusted to 10 μM using the Prox1-DNA dialysate after overnight dialysis. The titration was initiated with a 0.4 μl initial injection to minimize equilibration artifacts followed by 19 × 2 μl injections in 80 s intervals. Experimental data were corrected for the heat of dilution of the Prox1-DNA into the solvent and analysed using Origin™ software. The concentration of SOX18-HMG (79 aa) was corrected to 5.4 μM of active protein, determined by an active fraction experiment (Supplementary Figure S1).
Fluorescence resonance energy transfer (FRET) assay
To study SOX18-HMG box-induced DNA bending we adopted procedures previously reported for the TBP/TATA system (33). The SOX18-HMG (79 aa) and several dye-modified 15-bp Prox1-DNA elements were prepared including a donour-only control (DO, single 5′ cy3 label), acceptor-only control (AO, single 5′ cy5 label) and a 5′ cy3/cy5 double labelled (DL) element. Every measurement was done in triplicates and included a buffer control and six reactions: (i) 60 nM DO DNA with and (ii) without 500 nM SOX18-HMG, (iii) 60 nM AO DNA with and (iv) without 500 nM SOX18-HMG, (v) 60 nM DL DNA with and (vi) without 500 nM SOX18-HMG in 30 μl 1× fluorescence resonance energy transfer (FRET) buffer (10 mM Tris–HCl pH 8.0, 5 mM MgCl2, 50 mM KCl and 10% glycerol). Reactions were set up in NUNC 384-well black bottom plates (Thermo Fisher) and incubated at RT for 1 h. Spectra were recorded using a Varioskan LUX Platereader (Thermo Fisher) and two measurements were made: (i) donor excitation at 535 nm with 555–720 nm emission scan and (ii) acceptor excitation at 635 nm with 655–720 nm emission scan. Dye-to-dye distances were calculated using single point measurements at 567 nm for the donor and 668 nm for the acceptor emission (33). The association kinetic was measured using 668 nm acceptor emission and immediately after the protein (500 nM final concentration) was added to the DNA (60 nM final). Dissociation kinetic was measured by adding 1800 nM unlabelled Prox1-DNA to the reaction. The dead time of the experiment was 20 ms and readings were taken in 10 ms (association) or 40 ms (dissociation) intervals. On-rates were calculated using equation kon = (kobs-koff)/cprot where kobs is the observed association rate, koff the observed dissociation rate per second and cprot the molar protein concentration. To measure kobs and koff averaged association and dissociation data from 40 independent readings were modelled as single exponential rise or decay as described (34). The equation Kd = koff/kon. was used to calculate the binding constant (Kd).
Decoy DNA elements used
A 15-bp element was used for decoy design CTAGCATTGTCTGGG (SOX motif underlined) including a mutant control CTAGCAGGCTCTGGG. A circular SOX decoy version where hexaethylene glycol linkers covalently connect the 3′ and 5′ ends of the dsDNA (‘CIRC’), a decoy with phosphorothioated internucleotide linkages at the three terminal nucleotide pairs on either end of the DNA element (‘PS’), a decoy version where the PS modification and the circularization are combined (‘PSCIRC’) and a decoy variant where all nucleotides contain a 2′ O-methyl base (‘MEO’) were selected. PS mutant and PSCIRC mutant contain the same motif as the native ('NAT') mutant control. CIRC, PSCIRC, PSCIRC mutant, MEO (Integrated DNA Technologies), NAT, PS, NAT mutant, PS mutant (Life technologies) were obtained as powder, re-suspended in DNase and RNase-free deionized water (TIANGEN) and annealed in 1× annealing buffer as described above. The CIRC and PSCIRC decoys were supplied with internal nick leaving a free 3′-OH and 5′-monophosphate between the central TT dinucleotide and ligation was carried out after annealing using a T4 DNA ligase (NEB). Ligation mix was incubated at 16°C overnight, the enzyme was deactivated at 60°C for 1 h and pelleted by centrifugation at 13 000 rpm in a bench-top centrifuge for 10 min. Decoys were stored at −20°C until usage.
Electrophoretic mobility shift assays (EMSAs)
5′-cy5 labelled DNA was used for electrophoretic mobility shift assays (EMSAs). In preparation for the decoy competition assays the minimal protein concentrations required to ensure complete supershifts of DNA were determined for SOX-familiy proteins purified as described previously (28) and other unrelated TFs produced for previous studies along with their cognate DNA elements: PAX9 (35), OCT6 (36), ESRRB (37) (Supplementary Table S2). Proteins were used at varying concentrations (16.6, 31.3, 62.5, 125, 250, 500, 1000 nM). The following concentrations were used for decoy competition experiments: 100 nM of mouse SOX2-HMG (79 aa), 100 nM of mouse SOX4-HMG (79 aa), 200 nM of his-tagged mouse SOX6-HMG (79 aa), 50 nM mouse SOX18-HMG (109 aa or 79 aa), 80 nM of mouse SOX10-HMG (79 aa), 100nM of mouse SOX17-HMG (79 aa), 100 nM of mouse ESRRB, 200 nM of mouse OCT6-POU or BRN2-POU domain and 1 μM of mouse PAX9. All reactions were incubated for 1.5 h at 4°C with 20 nM cy5-labelled DNA in EMSA buffer (10 mM Tris–HCl pH 8.0, 0.1 mg/ml bovine serum albumin, 50 μM ZnCl2, 100 mM KCl, 10% Ultrapure Glycerol, 0.10% NP-40, 2 mM BME). Next, 20 μl samples were loaded onto 15% (w/v) 1× Tris-glycine (TG) polyacrylamide gels pre-run at 100 V in 1× TG buffer (25 mM Tris, pH 8.3, 192 mM glycine) and electrophoresed at 200 V for 30 min at 4°C in the dark. The gels were visualized using a FLA-7000 image reader (FUJIFILM). The ImageQuant TL Software (GE Healthcare Life Sciences) was used to quantify the fraction of bound DNA and IC50 values were estimated using non-linear regression methods and custom R scripts.
Fluorescence polarization
Fluorescence polarization (FP) competition experiments were carried out with a 5′-6-carboxyfluorescein (FAM) labelled Prox1-DNA (5′-GGTTCCCCCGCCCCCAGACAATGCTAGTTTGCATACAAAG, core 15 bp element underlined). A total of 2 nM FAM-dsDNA was mixed with 160 nM SOX18-HMG box (109 aa) and decoys were added at different concentrations (3.9, 7.8, 15.6, 31.3, 62.5, 125, 250, 500 nM) in reaction buffer (20 mM Tris–HCl pH 8.0, 100nM NaCl, 0.01% NP-40) to a final reaction volume of 30μl in a black 384-well plate (NUNC, Yuwei). After incubating for 1 h at 25°C plates were centrifuged at 1800 g for 1 min and readings were taken using the Wallac 1420 multilabel counter (Perkin Elmer) with 480/535 nm wavelengths. Readings were performed in triplicates.
Thermal stability assay
The decoy concentrations were adjusted to 1 μM in TE buffer (10 mM Tris pH 8.0, 1 mM ethylenediaminetetraacetic acid (EDTA)). The solutions were placed into a 1 ml Varian quartz cuvette at a volume of 500 μl. All five decoys and a TE buffer blank were analysed side-by-side using a temperature regulated Cary 100 UV-Vis spectrophotometer (Varian, South East Chemicals & Instrument Ltd). The absorbance at 260 nm was monitored from 25°C to 90°C (1°C/min heating rate).
Serum stability assay
Mouse serum was obtained from mouse orbital blood by allowing the blood to clot at RT for 15 to 30 min followed by centrifugation at 1500 g for 10 min at 4°C. The supernatant was transferred into a PCR tube and stored at −20°C until further use. A total of 6 μl of each decoy stock was added to 44 μl mouse serum to a final decoy concentration of 0.05 μg/μl. The reaction mix was incubated at 37°C in a water bath. At different time points (0, 1, 2, 4, 6, 8, 10, 36 h), 3 μl were removed and placed into a 1.5 ml tube containing 3 μl of 2 × formamide loading buffer (90% formamide, 0.5% EDTA, 0.1% Xylene cyanol, 0.1% Bromphenol Blue). The samples were kept on ice until loading onto the gel. After heating the samples to 60°C for 10 min they were loaded onto a pre-run denaturating 15% polyacrylamid gel containing 7 M urea and electrophoresed at 60 V for 2 h. The gel was stained for 10 min with 1× SYBR Gold nucleic acid gel stain (Invitrogen) in the dark and visualized under UV light (Tanon 1600 Gel Imaging System).
DNase I resistance assay
A total of 0.05 μg/μl of each decoy was incubated at 37°C in a final reaction volume of 50 μl containing 0.1 U DNase I in 1× DNase I buffer (NEB). At different time points (0, 2, 4, 6, 10, 20, 40, 60, 120 and 180 min), 3 μl of the reaction mix were combined with 3 μl 2× formamide loading buffer, heated and electrophoresed as described above.
VCAM-1 luciferase reporter assay
COS7 cells were grown in Dulbecco's Modified Eagle Medium (DMEM, Life Technologies) with 10% fetal bovine serum (FBS, Biowest) in 24-well plates and grown for 24 h to 80–90% confluency. Decoys, 200 ng VCAM1 Luciferase Reporter plasmid (38), 50 ng mouse SOX18 expression vector (38) and 1 ng pRL-SV40 Renilla expression plasmid dissolved in OptiMEM (Invitrogen) were transfected using X-tremeGENE 9 DNA Transfection Reagent (Roche). As a negative control one set of samples was transfected with the reporter and the pRL-SV40 plasmid only. After incubating at 37°C, 5% CO2 for 48 h the cells were carefully washed with Dulbecco's Phosphate Buffered Saline (Gibco) and lysed in 100 μl 1× Passive Lysis Buffer (Promega). The lysate was stored at −80°C until measurements. Luciferase and Renilla activity were measured using Dual Luciferase Reporter System (Promega) and a Veritas Microplate Luminometer (Turner Biosystems) following the manufacturer's protocols.
Quantitative reverse transcription PCR
COS7 cells were grown in DMEM + 10% FBS for 24 h to 60% confluency in a 48-well plate and then transfected with X-tremeGENE 9 DNA transfection reagent (Roche) with 1 μM PSCIRC decoy or with water, 2 ng pRL-SV40 (Renilla plasmid), 50 ng Sox18 expression vector and 200 ng VCAM reporter plasmid. RNA were collected after 48 h of transfection with TRIzol® Reagent (Life Technologies) then extracted using isopropyl alcohol precipitation. RNA concentration was measured by Nanodrop 2000 (Thermo), 2 μg of RNA was reverse transcribed using ReverTra Ace (Toyobo) for 60 min at 42°C and 99°C for 5 min. cDNA was diluted 1:80 for each experiment and qPCR was performed using a CFX96 device (Bio-Rad) with iTaq™ Universal SYBR® Green Supermix (Bio-Rad). The primers are listed in Supplementary Table S3. Raw data were transformed using the 2−ΔCt method with GAPDH as normalization control and log transformation.
RESULTS
In solution analysis of SOX18/Prox1-DNA interaction
To study DNA recognition by SOX18, 79 and 109 amino acid SOX18-HMG box protein constructs were overexpressed and purified to homogeneity. Both constructs eluted from size exclusion columns at a volume consistent with monomeric proteins (Figure 1A). For binding assays a 15-bp Sox18-binding site regulating the expression of the Prox1 gene was selected. It has previously been reported that sequence-specific HMG boxes are partially unfolded and refold upon DNA complex formation accompanied by a considerable heat effect (39). These heat effects can be measured calorimetrically and we therefore performed ITC with SOX18-HMG (79 aa) and Prox1-DNA. As expected, we observed strong exothermic signals at 35°C with a ΔH = −42.9 kcal/mol, indicating HMG box refolding and effective SOX18/Prox1 complex formation (Figure 1B). Surprisingly, the association constant Ka derived from our ITC measurements was only 2.47 × 106 ± 5.97 × 105 M−1, which deviates from previously reported affinity measurements carried out with alternative methods (26,39–40). This is likely because these measurements do not distinguish between heat effects upon HMG box refolding and heat effects of association (39). We therefore went on to study SOX18/Prox1-DNA binding using optical methods that are not sensitive to temperature changes. The DNA bending by HMG boxes can be detected in solution by monitoring changes of FRET signals using end-labelled DNA (39). We established this assay for the SOX18/Prox1-DNA binding system and observed a substantial increase of the acceptor emission with concurrent decrease of the donor emission when SOX18 was added to Prox1-DNA (Figure 1C). This observation is due to FRET and not caused by FRET-independent changes of the fluorescence intensities upon SOX18 binding (Figure 1D and E). This finding indicates a pronounced reduction of the distance between the complementary ends of the Prox1 element upon SOX18 binding from 57.8 to 49.0Å. Next, the FRET assay was adopted to study the kinetics of the SOX18/Prox1 interaction. The complex formation is driven by fast association kinetics with an association constant kon 1.02 ± 0.06 × 107 (M−1 × s−1) (Figure 1F). By incubating with an excess of unlabelled competitor, the dissociation constant koff was measured to be 0.062+/− 0.001 (s−1) corresponding to a binding constant Kd = koff/kon = 6.1 × 10−9M (Figure 1G). This value is in good agreement with previously measured affinities for SOX-HMG boxes using EMSAs (26) and SRY binding to its cognate DNA binding element by FRET (40). Collectively, these experiments indicate a fast, stable and functional interaction of SOX18 with the Prox1-DNA fragment accompanied by pronounced DNA bending.
Overall structure of the SOX18/Prox1-DNA complex
To investigate the SOX18/Prox1-DNA interactions at atomic resolution, extensive screening for suitable crystallization conditions was carried out. A 16-bp Prox1-DNA element containing CG overhangs and the SOX18-HMG box (79 aa) construct was found to be most suitable for the growth of well-diffracting crystals (Figure 2A and B, Supplementary Figure S2). A high resolution 1.75 Å dataset was collected, the structure was determined by molecular replacement and could be refined to R/Rfree factors of 17.4/19.9% (Supplementary Table S1).
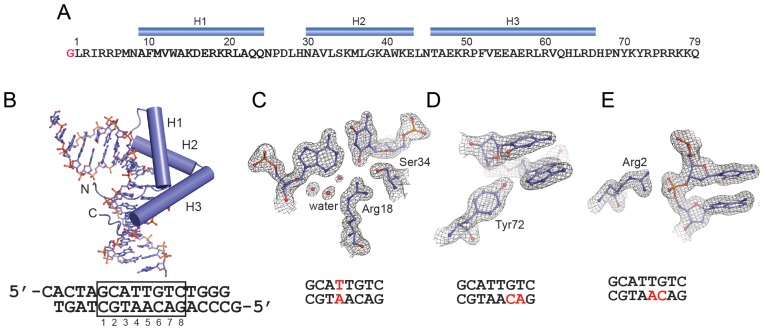
Figure 2.
Structure of the SOX18-HMG/Prox1-DNA complex. (A) Sequence of the SOX18-HMG construct used. Numbering follows the convention used for HMG boxes (1) and does not refer to the full length protein. The N-terminal glycine marked in red is a relict of the TEV protease site and H1-H3 denote the three alpha-helices. (B) Final model of the SOX18/Prox1-DNA complex with DNA shown as ball-and-sticks and the protein as cartoon. Helices H1-H3 are shown as cylinders and N-and C-termini are marked. The sequence of the Prox1 element with CG overhangs used for crystallization is depicted below. The core Sox binding site is boxed. (C–E) Snapshots of the final 2Fo-Fc electron density contoured at 1.5 σ (C and D) or 1.0 (E) and carved at 1.8 Å around the selected atoms. The position of the displayed nucleotides within the Prox1 element is marked with red letters. (C and D) show specific base contacts including interface waters (C) and (E) shows the interaction of Arg2 with a backbone phosphate adopting a non-canonical conformation.
The SOX18-HMG box consists of a three-helix bundle exhibiting an L-shaped structure with the long arm composed of helix 3 and the short arm of helices 1 and 2 (Figure 2A and B). The N-terminus (residue 1–9, amino acid numbering according to HMG conventions (1)) and the C-terminus (residue 66–75) adopt extended conformations and interlace approximately orthogonally. Binding occurs exclusively in the minor groove of the DNA. The DNA is located in the concave face of the HMG domain and the conformation of the DNA follows the contours of the concave binding surface. In this article we will refer to the following numbering scheme of the core SOX motif G1C2A3T4T5G6T7C8 and define the reverse complement sequences in a 3′-5′ direction as C’1G2’T3’A4’A5’C6’A7’G8’ (Figure 2B). The C-terminal amino acids of the SOX18-HMG 76–79 and the 3′ ‘T’ of the reverse strand could not be traced in the electron density map and were omitted from the final model. The remainder of the complex shows electron density of high quality (Figure 2C–E). In particular the protein–DNA contact interface is well defined and reliably reveals details including water molecules (Figure 2C) and unambiguously allows the placement of side-chains that were only moderately well defined in previous lower resolution structures, e.g. Tyr72 (Figure 2D, Supplementary Figure S3). Moreover, the phosphodiester backbone at A5’ is found to adopt a non-canonical gauche+/trans configuration having the α angle (O3*(-1)-P-O5*-C5* = 65°) and the γ torsion angle (O5*-C5*-C4*-C3* = 174°) (Figure 2E). The majority of nucleotides adopt a gauche−/gauche+ configuration but gauche+/trans is found in less than 2.4% of analysed nucleotides (41). A5’ is contacted by Arg2 which likely abets the gauche+/trans conformation. Therefore, the conformation of the A5′C6′ backbone could contribute to an ‘indirect readout’ mechanism and indicate that selective binding at this position is dictated by DNA shape rather than by complementary hydrogen bonding between interface amino acids and nucleotides (42).
Comparing DNA bending and binding by different SOX TFs
The topology of SOX/DNA complexes could affect the assembly of regulatory complexes and thereby influence gene expression. We thus compared the structure of the DNA elements co-crystallized with SOX18 and four other SOX proteins. We first estimated the total bending angle using Curves+ (43) (Supplementary Table S4). Globally, the DNA co-crystallized with SOX2, SOX4, SOX17, SOX18 and SOX9 exhibits very similar bending angles in the narrow range of 60–70° (Figure 3A). Locally, C2A3T4 show the strongest deviations from the base-pair parameters of canonical B-DNA (Figure 3B and C). In particular the roll angle is increased at the site of the intercalation of Phe10Met11 (between T4 and T5) but also at the two preceding base-pairs. Concurrently, unwinding of the double helix is reflected in the reduced twist angles (Figure 3C). The pattern and the magnitude of the changes to inter base-pair parameters are very similar for all five SOX/DNA complexes. Therefore, the DNA architecture induced by SOX proteins is not profoundly affected by the exact sequence of the DNA binding site nor by amino acid variations between paralogous SOX proteins. Further, DNA structure is not markedly changed despite different crystallization conditions, the presence of co-factor as in the ternary SOX2/OCT1/DNA complex and different crystal packing interactions that are mostly involving the DNA ends. Collectively, this analysis suggests that the mechanism of DNA bending is conserved throughout the SOX family and rather insensitive to variations in DNA sequence.
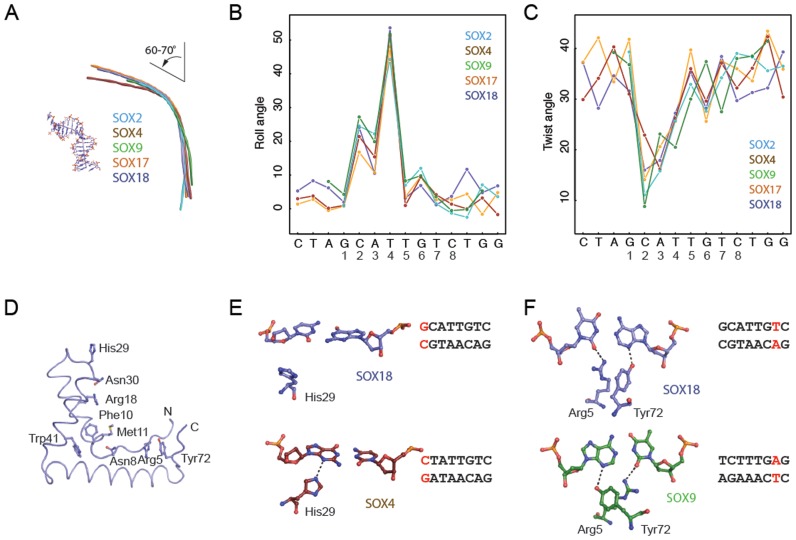
Figure 3.
SOX proteins induce conserved bends and rearrange to accommodate variant DNA elements. (A) Helical axis calculated by Curves+ (43) used to estimate the total bend for SOX2 (PDB ID: 1gt0), SOX4 (3u2b), SOX9 (4euw), SOX17 (3f27) and SOX18 (this study). All five SOX/DNA complexes show a similar overall bend between 60° and 70° (Supplementary Table S4). The inter base-pair parameters roll angle (B) and twist angle (C) were calculated using Curves+ (43) and plotted against the DNA sequence. The x-axis shows the forward strand of the Prox1-DNA sequence co-crystallized with the SOX18-HMG. (D) Tube representation of the SOX18-HMG highlighting the conserved set of DNA-binding residues present in all 20 SOX TFs. DNA was omitted and orientation was changed with respect to Figure 2B for optimized visibility. (E) Comparison of the conformation of His29 in SOX18 (slate blue) and SOX4 (ruby red) and the G1C’1 base-pair (SOX18) which is converted into C1G1’ in the SOX4/DNA complex. (F) Conformational switch of Arg5 and Tyr72 in SOX18 (slate blue) and SOX9 (green) to accommodate the different environment provided by T7A7’ (SOX18) or A7T7’ (SOX9), respectively. H-bonds are indicated with dashed lines. Displayed base pairs are highlighted in red in the core Prox1 elements used.
We next inspected the DNA contact interface of the five SOX proteins to identify structural difference. SOX HMG boxes possess a core set of DNA contact residues that are invariantly conserved amongst paralogues (Arg5, Asn8, Phe10, Met11, Arg18, His29, Asn30, Ser34, Trp41 and Tyr72, Figure 3D). We have previously noted that the side-chains of Arg18 and Asn30 re-orient in concert to accommodate different DNA sequences (27). Here we found some additional variability in the way conserved amino acids engage DNA in the various SOX/DNA complexes. For example, His29 forms a H-bond with N3 of G1′ in the SOX4/Lama1 structure. However, this contact is lost in SOX18 presumably because G1′ is replaced by C1’ in the Prox1 sequence providing a less favourable chemical environment (Figure 3E). Further, in SOX18 Tyr72 forms an H-bond with the N3 of A7’ and Arg5 with the O2 of the reverse complementary T7. By contrast, when the bases are inverted, as in the DNA element co-crystallized with SOX9, the side chains of Tyr72 and Arg5 concurrently reorient to now interact with A7 or T7’, respectively (Figure 3F). We conclude that the observed minor variations at SOX/DNA contact interfaces reflect structural adjustment to accommodate different DNA sequences and are not indicative of binding modes specific for individual SOX proteins (Supplementary Figure S4A–C).
Decoy oligonucleotides potently inhibit the SOX18/Prox1-DNA interactions
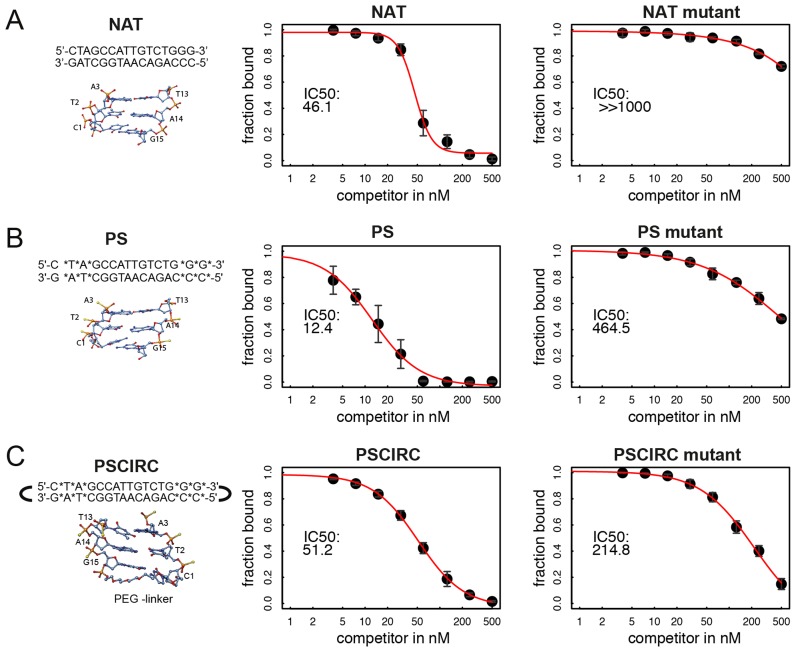
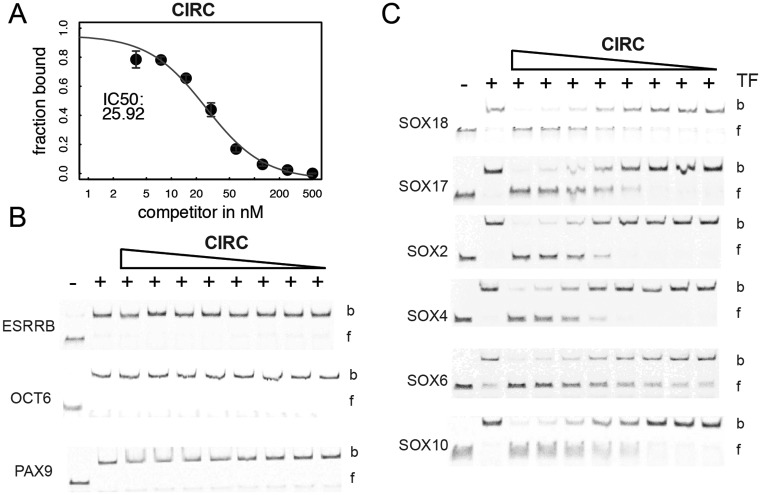
We have previously shown that polyanionic polyoxomethalates (POM) potently interfere with the DNA binding activity of SOX proteins but even POMs with organic modifications were found to be rather unselective and inhibited a wide range of TF classes (44,45). We therefore decided to explore whether DNA decoys can be designed to inhibit the DNA binding activity of SOX18 with a better selectivity profile. First, we chose to design a circular SOX decoy version in which hexaethylene glycol linkers covalently connect the 3′ and 5′ ends of the DNA (‘CIRC’). A similar modification was used to design STAT3 decoys for phase 0 clinical trials (46). Secondly, decoys with phosphorothioated internucleotide linkages at the three terminal nucleotide pairs on either end of the DNA element were considered (‘PS’ modification (47)). Third, we opted for a decoy variant where all nucleotides contain a 2′ O-methyl base (‘MEO’). The MEO modification is widely used to stabilize RNA based therapeutics (48). Forth, the PS and the CIRC modifications were used in combination (‘PSCIRC’). Modified DNA decoy elements were designed in silico using a 15-bp DNA and modelled onto the SOX18/Prox1-DNA crystal structure. From these models the modifications are not expected to directly impair the binding by SOX18 (Figure 4). All decoys were tested for their capacity to interfere with the binding activity of the SOX18-HMG box to a cy5-labelled Prox1-DNA in EMSAs (Figure 4, Supplementary Figure S5). As expected, the unlabelled and unmodified ‘NAT’ decoy competed effectively with the labelled reporter whereas a mutant element (‘NAT mutant’) barely showed inhibitory effects (Figure 4A). The PS, PSCIRC (Figure 4B and C) and CIRC decoys (Figure 5A) inhibited reporter DNA binding similarly as the NAT decoy (see also Supplementary Figure S5). When mutations of the SOX binding sites were tested in the context of the PS and the PSCIRC modifications, the IC50 increased by 40- or 4-fold, respectively (Figure 4B and C). This suggests that the modifications relax the requirements of SOX18 for its consensus-binding site.
Figure 4.
Modified SOX decoys potently inhibit DNA binding of SOX18. The potency to inhibit binding of 50 nM SOX18-HMG to 20 nM cy5-labelled Prox1-DNA by three decoys was assessed by EMSAs using decoy concentrations 500, 250, 125, 60, 30, 15, 7.5, 3.75 nM. The left panel shows a scheme of the decoys and a structural cartoon of the termini to illustrate the modifications. (A) Unmodified decoy ‘NAT’, (B) ‘PS’ decoy where asterisk and yellow spheres indicate phosphorothioate modification sites and (C) ‘PSCIRC’ containing ‘PS’ modifications and an additional PEG linker connecting 3′ and 5′ ends. Central and right panels show quantifications of competition EMSAs with the fraction of bound cy5-labelled reported DNA on the y-axis plotted against decoy concentration. IC50 values were estimated using non-linear curve fitting and a 4-parametric model. The IC50 value is shown and the modelled curve is depicted in red. For all mutants the central TTG was replaced by a GGC sequence. Data points in the plots represent mean +/− standard deviation of triplicate experiments. Representative EMSA gel images used for quantification are shown in Supplementary Figure S5.
Figure 5.
Decoys inhibit SOX but not unrelated TFs. (A) IC50 fit to competition EMSA data using the ‘CIRC’ decoy and the SOX18-HMG carried out as for ‘NAT’, ‘PS’ and ‘PSCIRC’ in Figure 4 with gel images in Supplementary Figure S5. Binding of ESRRB, OCT6 and PAX9 to their cy5-labelled cognate DNA binding site (B) or of the SOX family members SOX18, SOX17, SOX2, SOX4, SOX6 and SOX10 (C) to the Prox1 DNA was challenged using the CIRC decoy. The triangle above the EMSA images denotes 2-fold dilution series for the CIRC decoy (1000, 500, 250, 125, 62.5, 31.3, 15.6, 7.8). b and f denote bound and free DNA, respectively. See Supplementary Figure S7 for PSCIRC data.
The MEO modification leads to a drastic reduction of the inhibitory potential and was excluded from further assays (Supplementary Figure S5). FP assays using FAM-labelled reporter DNA showed an inhibitory profile consistent with the EMSAs for all tested decoy modifications (Supplementary Figure S6). Apparent IC50 values vary between EMSA and FP assays as different reporter DNA and SOX18-HMG protein concentrations were used to ensure satisfactory signal-to-noise ratios of both assays. Collectively, PS, CIRC and PSCIRC decoys represent DNA modifications that retain high-affinity interactions with the SOX18-HMG box.
Modified decoys specifically interfere with DNA binding of the SOX transcription factor family
We have previously encountered difficulties in identifying small molecule compounds that show a satisfactory selectivity profile (44). To probe whether SOX decoys bind to other TFs, we tested the inhibition of several non-SOX TFs that represent a set of structurally diverse DNA binding domains. The orphan nuclear receptor ESRRB, the paired domain protein PAX9 and the highly homologous POU domains of OCT6 and BRN2 were selected. We used CIRC and PSCIRC decoys that effectively bind to SOX18 for these experiments (Figures 4C and 5A). CIRC and PSCIRC do not inhibit DNA binding to cognate DNA elements by ESRRB, PAX9 and OCT6 at concentrations up to 500 nM while unlabelled competitors for these proteins encoding their cognate binding elements effectively compete at such concentrations (Figure 5B, Supplementary Figure S7). Next, decoy binding to members of the SOX family was tested using members from all major SOX subfamilies including SOX2 (SOXB1), SOX4 (SOXC), SOX6 (SOXD), SOX10 (SOXE) and SOX17 which as SOX18 belongs to the SOXF family. Results show that the CIRC and PSCIRC decoys effectively inhibits DNA binding by all SOX factors (Figure 5C, Supplementary Figure S7).
Modified SOX decoys enhance stability and interference with transcriptional regulation
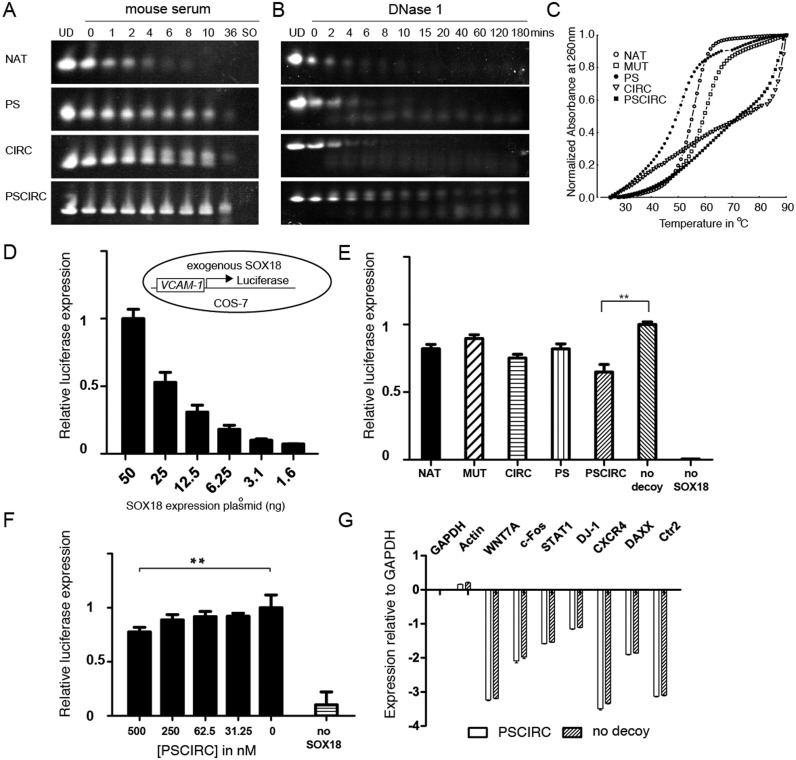
A key limitation for the application of decoys as research tools or therapeutics is their rapid degradation in a cellular environment or in the blood. We therefore conducted a series of experiments to assess the stability of the SOX decoys. First, the decoys were incubated in mouse serum and their time dependent integrity was monitored using urea-PAGE and SYBR Gold staining (Figure 6A). We found that all tested modifications increased the resistance to degradation in mouse serum with PSCIRC remaining intact even after 36 h of serum exposure. Second, their stability in the presence of DNase I was tested (Figure 6B). The PSCIRC decoy was observed to be most resistant to DNase I degradation whereas other decoys showed only marginal stability improvements against this endonuclease activity. Third, by measuring the absorbance at 260 nm versus temperature the thermal stability of the decoys was assessed. We observed that the PS modification caused a reduction in the thermal stability compared to the unmodified DNA (Figure 6C). By contrast, the CIRC decoy substantially delayed thermal denaturation. Moreover, the circularization rescued the heat sensitivity of the PS modification illustrated by the postponed melting of the PSCIRC variant.
Figure 6.
Modified SOX decoys show enhanced stability and repress reporter gene transactivation. (A) Decoys were incubated in mouse serum and their integrity was assessed by urea-PAGE and SYBR gold nucleic acid staining as a function of incubation time. The ‘0’ lane marks a sample taken immediately after mixing with serum and numbers denote time in hours. UD: undigested decoys. SO: serum only. (B) Decoys were incubated with DNase I and analysed as in (A). (C) The melting of decoys was monitored by heating 1 μM DNA to 90°C at 1°C/min and recording the absorbance at 260 nm. (D) SOX18 exogenously expressed in COS-7 cells activates the expression of a luciferase reporter under control of a regulatory region derived from the VCAM-1 gene (38) in a dose-dependent manner. The inlet illustrates the assay set-up. (E) The effect of decoys at 1000 nM on the VCAM-1 reporter activity was compared 48 h post-transfection using 50 ng of SOX18 plasmid. (F) Effects of the PSCIRC on luciferase expression at different concentrations. (G) Expression of selected genes detected by RT-qPCR in the absence or presence of the PSCIRC. The mean and standard deviation from three experiments each carried out in triplicates is shown. The asterisks (**) denote P < 0.001 (t-test).
We next studied whether SOX decoys can disrupt SOX18-mediated transcriptional activity in a cellular environment using a reporter assay where the luciferase gene is under the control of an 1889-bp promoter region of the vascular cell adhesion molecule-1 (VCAM-1) gene to which the SOX18 protein binds (Figure 6D) (38). The COS7 cell line was chosen for these experiments, as SOX18 is not normally expressed in this cell type. We first reaffirmed that SOX18 is necessary for the transcriptional upregulation of the reporter in a dose-dependent manner (Figure 6D). Next, we tested the inhibitory effects using 50 ng of SOX18 plasmid of the studied decoys side-by-side at 1 μM concentration 48 h post-transfection (Figure 6E). All decoys were found to exert some noticeable inhibitory activity. Yet, the PSCIRC inhibited transcription most potently and reduced the luciferase expression by ∼40% as compared to the vehicle control (no decoy). The inhibitory effect was also observed at lower PSCIRC concentrations (Figure 6F). The expression of control genes was not perturbed upon PSCIRC treatment indicating that PSCIRC does not globally interfere with transcription (Figure 6G). This finding is in accordance with the stability assays and suggests that the stability-enhanced PSCIRC decoy possesses the best properties to interfere with SOX18 activity in a cellular environment.
DISCUSSION
SOX proteins act as master regulators mediating cellular reprogramming and dictate multiple cell fate decisions. They often maintain multipotential progenitor states and stem cell-like phenotypes, e.g. SOX2. These properties likely underlie the competence of many SOX proteins to drive tumorigenesis. For example, SOX4 contributes to acute myeloid leukemia (49) and prostate cancer (50). Further, SOX2 reprograms cancer cells and maintains their stem-like properties (51,52) and SOX10 initiates and maintains melanomas (53). Thus, technologies that enable direct interference with SOX proteins in such pathological situations would be hugely beneficial not only as potential therapy but also as research tools. However, the DNA-binding HMG box is thus far the only structurally analysed portion of SOX proteins and while regions outside the box have been functionally annotated as transcriptional activators and dimerization elements (2), it is unclear whether such regions represent globular domains that fold independently of interacting proteins. Therefore, the HMG box currently represents the only tractable structural target for therapeutic intervention. However, targeting the DNA recognition interface of TFs using conventional small molecules is notoriously challenging, as hydrophobic pockets that could easily accommodate such compounds are virtually absent.
Here, we explored a DNA decoy approach as an alternative avenue for SOX inhibition. We focused on SOX18 because of its role in initiating tumour-induced neo-lymphangiogenesis paving the way for subsequent tumour metastasis (16). TF decoys are short double stranded DNA sequences encoding cognate binding sequences of TFs (47,54). This way, TFs are sequestered in the nucleus or the cytoplasm preventing them from regulating their targets and gene expression programs leading to tumorigenesis are thereby suppressed. First proof-of-principle studies were carried out with decoys targeting the TF E2F (55–57). To develop nucleic acid into therapeutics a number of roadblocks are to be removed: challenging pharmacokinetics, delivery to the target tissue and cellular uptake. Nevertheless, antisense drugs based on modified RNAs that received regulatory approval for systemic delivery provide tailwinds for nucleic acid based therapeutics (58). Moreover, DNA decoys targeting different classes of TF proteins have been advanced into different stages of clinical trials including decoys targeting E2F to prevent bypass vein graft failures (56,59), EGR1 to treat various forms of pain (60), STAT3 in head and neck tumours (46) and the anti-inflammatory NF-κB protein (61,62).
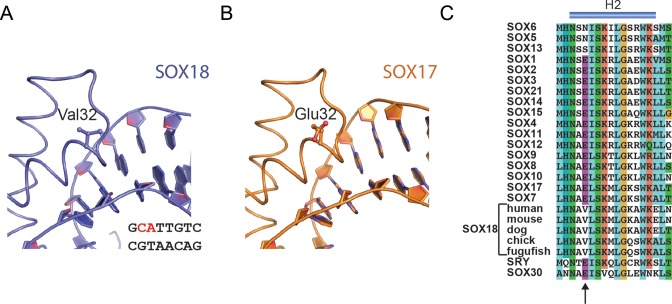
In this study, we present a high-resolution crystal structure and used the structural models to design SOX decoys based on the DNA sequence of the cognate SOX18 binding site controlling the expression of Prox1. A novel innovative DNA circularization strategy developed for STAT3 decoys enabled systemic delivery in human (46). We combined this circularization approach with the more classical phosphorothioation and demonstrated that SOX decoys potently inhibit the DNA binding of all SOX proteins but not of unrelated TFs. Encouragingly, the stability-enhanced PSCIRC decoy interferes with SOX18-mediated transcriptional regulation. Collectively, we suggest the SOX decoy as a viable approach to target SOX18 and other SOX proteins. One remaining challenge remains to invent decoy modifications that selectively bind to only SOX18 but not to other SOX HMG boxes. It has recently been reported that decoys that discriminate between the closely related paralogous TFs STAT1 and STAT3 could be designed with rational nucleotide substitutions (63). However, our structural analysis revealed a virtually identical set of DNA contacting amino acids shared by all SOX proteins (Figure 3D). We therefore inspected the SOX18/Prox1-DNA structure in search of structural elements not directly involved in DNA recognition that could permit the design of specific SOX18 decoys. We reasoned that Val32, at the start of helix 2, provides the most promising interfacial residue to design SOX18-selective decoys (Figure 7A). First, Val32 is located close to the phosphodiester bond connecting C2 and A3 (Figure 7A). Second, only SOX18 contains an aliphatic valine at position 32 whereas most other SOX proteins, including the closely related SOXF factors SOX7 and SOX17, encode an acidic glutamate (Figure 7B and C). Third, Val32 is highly conserved amongst orthologous SOX18 proteins suggesting functional relevance (Figure 7C). Thus, we propose Val32 as a selectivity hotspot that could provide an avenue to design SOX18-selective decoys. As Val32 is positioned too remote from actual nucleobases, nucleotide substitutions are unlikely to contribute to selectivity improvements. Rather, derivatization of the phosphate backbone itself with amino acids and other organic moieties via a phosphotriester linkage presents a promising strategy. The general feasibility of this approach has recently been demonstrated for RNAi prodrugs where neutral phosphotriester groups augmented cellular delivery (64). To further substantiate this idea we performed structural modelling and covalently attached a proline derivative (2-pyrrolidinecarbothionic S-acid) to the phosphate backbone proximal to Val32 (Supplementary Figure S8). After energy minimization, the proline residue makes a favourable hydrophobic contact with Val32. By contrast, this modification causes the acidic side-chain of Glu32 present at equivalent positions in SOX17 or SOX4 to reorient suggesting an impairment of the interaction. We therefore suggest that modification of the decoy backbone with organic moieties present a promising strategy to engineer SOX18-selective decoys. Collectively, we propose SOX decoys as a potential strategy for inhibiting SOX18 activity, an approach which upon further development could open up new therapeutic avenues to counteract tumour-induced neo-lymphangiogenesis and associated cancer metastasis.
Figure 7.
Engineering a SOX18-selective decoy. (A) Valine 32 of SOX18 maps to a region proximal to the phosphodiester connecting nucleotides C2 and A3. Only SOX18 possesses an aliphatic valine at position 32 but SOX17 and many other SOX proteins contain an acidic glutamate (B and C). Valine 32 is evolutionarily conserved amongst SOX18 orthologs but unique within the SOX family (C).
ACCESSION NUMBER
Structure factors and coordinates have been deposited to the protein data bank with accession number 4Y60.
Supplementary Material
Acknowledgments
The authors are grateful to Howard Robinson for data collection and processing. We thank Colyn Crane-Robinson, University of Portsmouth, UK, for advise and critical reading of the manuscript. We are grateful to Jucheng Gong for administrative support.
Author contribution statement: N.P. purified proteins, performed crystallization and structural analysis. M.K. performed EMSA, FP and decoy stability experiments with the help of Y.H.Y. and L.H. M.G. performed EMSAs and ITC. M.K. and X.W. performed the luciferase reporter and RT-qPCR assays. Y.H.Y. performed FRET experiments. Y.S. performed structural modelling of decoy modifications. K.N., P.K. and M.F. contributed to data analysis, study and assay design. R.J. designed the study, analyzed data and wrote the paper. All authors contributed to manuscript writing, data analysis and approved the final manuscript.
Footnotes
Present address: Nina Prokoph, Asia & Emerging Markets iMed, AstraZeneca, Zhangjiang Hi-Tech Park, 199 Liangjing Road, 201203 Shanghai, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
2013 MOST China-EU Science and Technology Cooperation Program [2013DFE33080 to R.J.]; National Natural Science Foundation of China [31471238 to R.J.]; Chinese Academy of Sciences 100 talent award (to R.J.); Science and Technology Planning Project of Guangdong Province, China [2014B030301058 to R.J.]; Diffraction data were collected at beamline X29 of the National Synchrotron Light Source (NSLS) supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences; German Academic Exchange Service (DAAD) ( to M.K., N.P., M.G.]; Deutsche Gesellschaft fuer Internationale Zusammenarbeit (GIZ) GmbH (to N.P.); Project grants from Australian Research Council [DP14010048 to M.F.]; Chinese Government Scholarship (CGS) (to Y.S.); University of the Chinese Academy of Sciences (UCAS) (to Y.S.). Funding for open access charge: Chinese Academy of Sciences.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bowles J., Schepers G., Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev. Biol. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- 2.Kamachi Y., Kondoh H. Sox proteins: regulators of cell fate specification and differentiation. Development. 2013;140:4129–4144. doi: 10.1242/dev.091793. [DOI] [PubMed] [Google Scholar]
- 3.Pevny L.H., Lovell-Badge R. Sox genes find their feet. Curr. Opin. Genet. Dev. 1997;7:338–344. doi: 10.1016/s0959-437x(97)80147-5. [DOI] [PubMed] [Google Scholar]
- 4.Wegner M. From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res. 1999;27:1409–1420. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malarkey C.S., Churchill M.E. The high mobility group box: the ultimate utility player of a cell. Trends Biochem. Sci. 2012;37:553–562. doi: 10.1016/j.tibs.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Badis G., Berger M.F., Philippakis A.A., Talukder S., Gehrke A.R., Jaeger S.A., Chan E.T., Metzler G., Vedenko A., Chen X., et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mertin S., McDowall S.G., Harley V.R. The DNA-binding specificity of SOX9 and other SOX proteins. Nucleic Acids Res. 1999;27:1359–1364. doi: 10.1093/nar/27.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jolma A., Yan J., Whitington T., Toivonen J., Nitta K.R., Rastas P., Morgunova E., Enge M., Taipale M., Wei G., et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 9.Duong T., Koltowska K., Pichol-Thievend C., Le Guen L., Fontaine F., Smith K.A., Truong V., Skoczylas R., Stacker S.A., Achen M.G., et al. VEGFD regulates blood vascular development by modulating SOX18 activity. Blood. 2014;123:1102–1112. doi: 10.1182/blood-2013-04-495432. [DOI] [PubMed] [Google Scholar]
- 10.Francois M., Caprini A., Hosking B., Orsenigo F., Wilhelm D., Browne C., Paavonen K., Karnezis T., Shayan R., Downes M., et al. Sox18 induces development of the lymphatic vasculature in mice. Nature. 2008;456:643–647. doi: 10.1038/nature07391. [DOI] [PubMed] [Google Scholar]
- 11.Pennisi D., Gardner J., Chambers D., Hosking B., Peters J., Muscat G., Abbott C., Koopman P. Mutations in Sox18 underlie cardiovascular and hair follicle defects in ragged mice. Nat. Genet. 2000;24:434–437. doi: 10.1038/74301. [DOI] [PubMed] [Google Scholar]
- 12.Wigle J.T., Oliver G. Prox1 function is required for the development of the murine lymphatic system. Cell. 1999;98:769–778. doi: 10.1016/s0092-8674(00)81511-1. [DOI] [PubMed] [Google Scholar]
- 13.Albrecht I., Christofori G. Molecular mechanisms of lymphangiogenesis in development and cancer. Int. J. Dev. Biol. 2011;55:483–494. doi: 10.1387/ijdb.103226ia. [DOI] [PubMed] [Google Scholar]
- 14.Alitalo K. The lymphatic vasculature in disease. Nat. Med. 2011;17:1371–1380. doi: 10.1038/nm.2545. [DOI] [PubMed] [Google Scholar]
- 15.Duong T., Proulx S.T., Luciani P., Leroux J.C., Detmar M., Koopman P., Francois M. Genetic ablation of SOX18 function suppresses tumor lymphangiogenesis and metastasis of melanoma in mice. Cancer Res. 2012;72:3105–3114. doi: 10.1158/0008-5472.CAN-11-4026. [DOI] [PubMed] [Google Scholar]
- 16.Duong T., Koopman P., Francois M. Tumor lymphangiogenesis as a potential therapeutic target. J. Oncol. 2012;2012:204946. doi: 10.1155/2012/204946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Azhikina T., Kozlova A., Skvortsov T., Sverdlov E. Heterogeneity and degree of TIMP4, GATA4, SOX18, and EGFL7 gene promoter methylation in non-small cell lung cancer and surrounding tissues. Cancer Genet. 2011;204:492–500. doi: 10.1016/j.cancergen.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 18.Bidkhori G., Narimani Z., Hosseini Ashtiani S., Moeini A., Nowzari-Dalini A., Masoudi-Nejad A. Reconstruction of an integrated genome-scale co-expression network reveals key modules involved in lung adenocarcinoma. PLoS One. 2013;8:e67552. doi: 10.1371/journal.pone.0067552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eom B.W., Jo M.J., Kook M.C., Ryu K.W., Choi I.J., Nam B.H., Kim Y.W., Lee J.H. The lymphangiogenic factor SOX 18: a key indicator to stage gastric tumor progression. Int. J. Cancer. 2012;131:41–48. doi: 10.1002/ijc.26325. [DOI] [PubMed] [Google Scholar]
- 20.Darnell J.E., Jr Transcription factors as targets for cancer therapy. Nat. Rev. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 21.Yan C., Higgins P.J. Drugging the undruggable: transcription therapy for cancer. Biochim. Biophys. Acta. 2013;1835:76–85. doi: 10.1016/j.bbcan.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fontaine F., Overman J., Francois M. Pharmacological manipulation of transcription factor protein-protein interactions: opportunities and obstacles. Cell Regen. (Lond) 2015;4:2. doi: 10.1186/s13619-015-0015-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy E.C., Zhurkin V.B., Louis J.M., Cornilescu G., Clore G.M. Structural basis for SRY-dependent 46-X,Y sex reversal: modulation of DNA bending by a naturally occurring point mutation. J. Mol. Biol. 2001;312:481–499. doi: 10.1006/jmbi.2001.4977. [DOI] [PubMed] [Google Scholar]
- 24.Remenyi A., Lins K., Nissen L.J., Reinbold R., Scholer H.R., Wilmanns M. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 2003;17:2048–2059. doi: 10.1101/gad.269303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams D.C., Jr, Cai M., Clore G.M. Molecular basis for synergistic transcriptional activation by Oct1 and Sox2 revealed from the solution structure of the 42-kDa Oct1.Sox2.Hoxb1-DNA ternary transcription factor complex. J. Biol. Chem. 2004;279:1449–1457. doi: 10.1074/jbc.M309790200. [DOI] [PubMed] [Google Scholar]
- 26.Palasingam P., Jauch R., Ng C.K., Kolatkar P.R. The structure of Sox17 bound to DNA reveals a conserved bending topology but selective protein interaction platforms. J. Mol. Biol. 2009;388:619–630. doi: 10.1016/j.jmb.2009.03.055. [DOI] [PubMed] [Google Scholar]
- 27.Jauch R., Ng C.K., Narasimhan K., Kolatkar P.R. Crystal structure of the Sox4 HMG/DNA complex suggests a mechanism for the positional interdependence in DNA recognition. Biochem. J. 2012;443:39–47. doi: 10.1042/BJ20111768. [DOI] [PubMed] [Google Scholar]
- 28.Ng C.K., Li N.X., Chee S., Prabhakar S., Kolatkar P.R., Jauch R. Deciphering the Sox-Oct partner code by quantitative cooperativity measurements. Nucleic Acids Res. 2012;40:4933–4941. doi: 10.1093/nar/gks153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enz. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 30.McCoy A.J., Grosse-Kunstleve R.W., Storoni L.C., Read R.J. Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 31.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 32.Afonine P.V., Grosse-Kunstleve R.W., Adams P.D. The Phenix refinement framework. https://www.phenix-online.org/papers/ccp4_july_2005_afonine.pdf CCP4 Newsl. 2005;42 [Google Scholar]
- 33.Blair R.H., Goodrich J.A., Kugel J.F. Using FRET to monitor protein-induced DNA bending: the TBP-TATA complex as a model system. Methods Mol. Biol. 2013;977:203–215. doi: 10.1007/978-1-62703-284-1_16. [DOI] [PubMed] [Google Scholar]
- 34.Pollard T.D., De La Cruz E.M. Take advantage of time in your experiments: a guide to simple, informative kinetics assays. Mol. Biol. Cell. 2013;24:1103–1110. doi: 10.1091/mbc.E13-01-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narasimhan K., Pillay S., Huang Y.H., Jayabal S., Udayasuryan B., Veerapandian V., Kolatkar P., Cojocaru V., Pervushin K., Jauch R. DNA-mediated cooperativity facilitates the co-selection of cryptic enhancer sequences by SOX2 and PAX6 transcription factors. Nucleic Acids Res. 2015;43:1513–1528. doi: 10.1093/nar/gku1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jauch R., Choo S.H., Ng C.K., Kolatkar P.R. Crystal structure of the dimeric Oct6 (POU3f1) POU domain bound to palindromic MORE DNA. Proteins. 2011;79:674–677. doi: 10.1002/prot.22916. [DOI] [PubMed] [Google Scholar]
- 37.Sun W., Hu X., Lim M.H., Ng C.K., Choo S.H., Castro D.S., Drechsel D., Guillemot F., Kolatkar P.R., Jauch R., et al. TherMos: estimating protein-DNA binding energies from in vivo binding profiles. Nucleic Acids Res. 2013;41:5555–5568. doi: 10.1093/nar/gkt250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hosking B.M., Wang S.C., Downes M., Koopman P., Muscat G.E. The VCAM-1 gene that encodes the vascular cell adhesion molecule is a target of the Sry-related high mobility group box gene, Sox18. J. Biol. Chem. 2004;279:5314–5322. doi: 10.1074/jbc.M308512200. [DOI] [PubMed] [Google Scholar]
- 39.Dragan A.I., Read C.M., Makeyeva E.N., Milgotina E.I., Churchill M.E., Crane-Robinson C., Privalov P.L. DNA binding and bending by HMG boxes: energetic determinants of specificity. J. Mol. Biol. 2004;343:371–393. doi: 10.1016/j.jmb.2004.08.035. [DOI] [PubMed] [Google Scholar]
- 40.Racca J.D., Chen Y.S., Maloy J.D., Wickramasinghe N., Phillips N.B., Weiss M.A. Structure-function relationships in human testis-determining factor SRY: an aromatic buttress underlies the specific DNA-bending surface of a high mobility group (HMG) box. J. Biol. Chem. 2014;289:32410–32429. doi: 10.1074/jbc.M114.597526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varnai P., Djuranovic D., Lavery R., Hartmann B. Alpha/gamma transitions in the B-DNA backbone. Nucleic Acids Res. 2002;30:5398–5406. doi: 10.1093/nar/gkf680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oguey C., Foloppe N., Hartmann B. Understanding the sequence-dependence of DNA groove dimensions: implications for DNA interactions. PLoS One. 2010;5:e15931. doi: 10.1371/journal.pone.0015931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lavery R., Moakher M., Maddocks J.H., Petkeviciute D., Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves+ Nucleic Acids Res. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Narasimhan K., Micoine K., Lacote E., Thorimbert S., Cheung E., Hasenknopf B., Jauch R. Exploring the utility of organo-polyoxometalate hybrids to inhibit SOX transcription factors. Cell Regen. (Lond) 2014;3:10. doi: 10.1186/2045-9769-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Narasimhan K., Pillay S., Bin Ahmad N.R., Bikadi Z., Hazai E., Yan L., Kolatkar P.R., Pervushin K., Jauch R. Identification of a Polyoxometalate Inhibitor of the DNA Binding Activity of Sox2. ACS Chem. Biol. 2011;6:573–581. doi: 10.1021/cb100432x. [DOI] [PubMed] [Google Scholar]
- 46.Sen M., Thomas S.M., Kim S., Yeh J.I., Ferris R.L., Johnson J.T., Duvvuri U., Lee J., Sahu N., Joyce S., et al. First-in-human trial of a STAT3 decoy oligonucleotide in head and neck tumors: implications for cancer therapy. Cancer Discov. 2012;2:694–705. doi: 10.1158/2159-8290.CD-12-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bielinska A., Shivdasani R.A., Zhang L.Q., Nabel G.J. Regulation of gene expression with double-stranded phosphorothioate oligonucleotides. Science. 1990;250:997–1000. doi: 10.1126/science.2237444. [DOI] [PubMed] [Google Scholar]
- 48.Majlessi M., Nelson N.C., Becker M.M. Advantages of 2′-O-methyl oligoribonucleotide probes for detecting RNA targets. Nucleic Acids Res. 1998;26:2224–2229. doi: 10.1093/nar/26.9.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang H., Alberich-Jorda M., Amabile G., Yang H., Staber P.B., Di Ruscio A., Welner R.S., Ebralidze A., Zhang J., Levantini E., et al. Sox4 is a key oncogenic target in C/EBPalpha mutant acute myeloid leukemia. Cancer Cell. 2013;24:575–588. doi: 10.1016/j.ccr.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scharer C.D., McCabe C.D., Ali-Seyed M., Berger M.F., Bulyk M.L., Moreno C.S. Genome-wide promoter analysis of the SOX4 transcriptional network in prostate cancer cells. Cancer Res. 2009;69:709–717. doi: 10.1158/0008-5472.CAN-08-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boumahdi S., Driessens G., Lapouge G., Rorive S., Nassar D., Le Mercier M., Delatte B., Caauwe A., Lenglez S., Nkusi E., et al. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 2014;511:246–250. doi: 10.1038/nature13305. [DOI] [PubMed] [Google Scholar]
- 52.Suva M.L., Rheinbay E., Gillespie S.M., Patel A.P., Wakimoto H., Rabkin S.D., Riggi N., Chi A.S., Cahill D.P., Nahed B.V., et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157:580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shakhova O., Zingg D., Schaefer S.M., Hari L., Civenni G., Blunschi J., Claudinot S., Okoniewski M., Beermann F., Mihic-Probst D., et al. Sox10 promotes the formation and maintenance of giant congenital naevi and melanoma. Nat. Cell Biol. 2012;14:882–890. doi: 10.1038/ncb2535. [DOI] [PubMed] [Google Scholar]
- 54.Mann M.J., Dzau V.J. Therapeutic applications of transcription factor decoy oligonucleotides. J. Clin. Invest. 2000;106:1071–1075. doi: 10.1172/JCI11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morishita R., Gibbons G.H., Horiuchi M., Ellison K.E., Nakama M., Zhang L., Kaneda Y., Ogihara T., Dzau V.J. A gene therapy strategy using a transcription factor decoy of the E2F binding site inhibits smooth muscle proliferation in vivo. Proc. Natl. Acad. Sci. U.S.A. 1995;92:5855–5859. doi: 10.1073/pnas.92.13.5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mann M.J., Whittemore A.D., Donaldson M.C., Belkin M., Conte M.S., Polak J.F., Orav E.J., Ehsan A., Dell'Acqua G., Dzau V.J. Ex-vivo gene therapy of human vascular bypass grafts with E2F decoy: the PREVENT single-centre, randomised, controlled trial. Lancet. 1999;354:1493–1498. doi: 10.1016/S0140-6736(99)09405-2. [DOI] [PubMed] [Google Scholar]
- 57.Tomita N., Kim J.Y., Gibbons G.H., Zhang L., Kaneda Y., Stahl R.A., Ogborn M., Venderville B., Morishita R., Baran D., et al. Gene therapy with an E2F transcription factor decoy inhibits cell cycle progression in rat anti-Thy 1 glomerulonephritis. Int. J. Mol. Med. 2004;13:629–636. [PubMed] [Google Scholar]
- 58.Sinha G. Antisense battles small molecule for slice of rare lipid disorder market. Nat. Biotechnol. 2013;31:179–180. doi: 10.1038/nbt0313-179. [DOI] [PubMed] [Google Scholar]
- 59.Ehsan A., Mann M.J., Dell'Acqua G., Dzau V.J. Long-term stabilization of vein graft wall architecture and prolonged resistance to experimental atherosclerosis after E2F decoy oligonucleotide gene therapy. J. Thorac. Cardiovasc. Surg. 2001;121:714–722. doi: 10.1067/mtc.2001.111204. [DOI] [PubMed] [Google Scholar]
- 60.Mamet J., Klukinov M., Yaksh T.L., Malkmus S.A., Williams S., Harris S., Manning D.C., Taylor B.K., Donahue R.R., Porreca F., et al. Single intrathecal administration of the transcription factor decoy AYX1 prevents acute and chronic pain after incisional, inflammatory, or neuropathic injury. Pain. 2014;155:322–333. doi: 10.1016/j.pain.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 61.Egashira K., Suzuki J., Ito H., Aoki M., Isobe M., Morishita R. Long-term follow up of initial clinical cases with NF-kappaB decoy oligodeoxynucleotide transfection at the site of coronary stenting. J. Gene Med. 2008;10:805–809. doi: 10.1002/jgm.1192. [DOI] [PubMed] [Google Scholar]
- 62.De Stefano D. Oligonucleotides decoy to NF-kappaB: becoming a reality? Discov. Med. 2011;12:97–105. [PubMed] [Google Scholar]
- 63.Souissi I., Ladam P., Cognet J.A., Le Coquil S., Varin-Blank N., Baran-Marszak F., Metelev V., Fagard R. A STAT3-inhibitory hairpin decoy oligodeoxynucleotide discriminates between STAT1 and STAT3 and induces death in a human colon carcinoma cell line. Mol. Cancer. 2012;11:12. doi: 10.1186/1476-4598-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meade B.R., Gogoi K., Hamil A.S., Palm-Apergi C., van den Berg A., Hagopian J.C., Springer A.D., Eguchi A., Kacsinta A.D., Dowdy C.F., et al. Efficient delivery of RNAi prodrugs containing reversible charge-neutralizing phosphotriester backbone modifications. Nat. Biotechnol. 2014;32:1256–1261. doi: 10.1038/nbt.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.