Abstract
Most budding yeast introns exist in the many duplicated ribosomal protein genes (RPGs) and it has been posited that they remain there to modulate the expression of RPGs and cell growth in response to stress. However, the mechanism by which introns regulate the expression of RPGs and their impact on the synthesis of ribosomal proteins remain unclear. In this study, we show that introns determine the ratio of ribosomal protein isoforms through asymmetric paralog-specific regulation of splicing. Exchanging the introns and 3′ untranslated regions of the duplicated RPS9 genes altered the splicing efficiency and changed the ratio of the ribosomal protein isoforms. Mutational analysis of the RPS9 genes indicated that splicing is regulated by variations in the intron structure and the 3′ untranslated region. Together these data suggest that preferential splicing of duplicated RPGs provides a means for adjusting the ratio of different ribosomal protein isoforms, while maintaining the overall expression level of each ribosomal protein.
INTRODUCTION
Ribosomes are highly conserved ribonucleoprotein complexes that are required for protein synthesis (1). In eukaryotes, the ribosomes are composed of two subunits, the 60S and 40S. The 60S subunit contains three rRNAs (28/25S, 5.8S and 5S) and ∼49 proteins while the 40S subunit contains a single rRNA (18S) and ∼33 proteins (2). Ribosome assembly is a major undertaking that occurs in the nucleus and requires coordinated expression of the different RNA and protein components (3,4). The biogenesis of ribosomes involves hundreds of rDNA repeats and ribosomal protein genes (RPGs) and also requires ∼76 small nucleolar RNAs (snoRNAs) and > 200 different assembly factors (5). This complex process has to meet the high demand for protein synthesis, without compromising quality or wasting energy. Accordingly, perturbing the expression of one or more components of the ribosome often impacts the ensemble of genes required for ribosome biogenesis and leads to defects at different levels of the assembly pipeline (6).
In the yeast Saccharomyces cerevisiae, the majority of ribosomal proteins (RPs) are produced from duplicated genes or ohnologs created by whole genome duplication (7). These ohnologs are believed to be functional paralogs preserved during evolution (8,9), but the reason for their preservation is not entirely clear. There is still ongoing debate about whether duplicated RPGs (dRPGs) are preserved to ensure constant dose (10,11) or to increase the functional diversity of RPs (8,12). Most experimental deletions of duplicated genes generated copy-specific phenotypic effects, arguing against redundant functions. For example, the deletion of RPL7A, RPL12B, RPL22A and RPS18B and not their paralog affected the localized translation of Ash1 mRNA (8). Indeed, we have recently shown that the deletion of the minor and not the major copy of several dRPGs may affect growth under stress without major effect on the global amount of RP (12,13). Deletions of duplicated genes were also shown to generate different transcription profiles and affect growth in a copy-specific manner, suggesting the dRPGs may serve different functions (8). Consistently, most paralogs are expressed at different levels and have different localization and assembly patterns suggesting that the expression of RPGs is functionally regulated in a copy-specific manner (8).
Systematic deletions of introns from dRPGs indicated that they might play an important role in defining the expression pattern of dRPGs (12). The majority of intron deletions affected the expression of the dRPGs in a paralog-specific manner. The presence of introns did not always lead to inhibition of gene expression, as would be expected from the obligate delay in gene expression caused by splicing. Instead, introns induced the expression of about half of the genes and inhibited the expression of the other half indicating that introns may function as modular gene-specific regulators of gene expression (12). Deletion of RPGs introns affected not only the expression of the host gene (intragenic regulation) but also altered the expression of its paralog in trans (intergenic regulation). This interplay between two splicing events in dRPGs provides a means to alter the specific ratio of similar but non-identical mRNAs, thus allowing modification of the constitution of ribosomes in certain growth conditions. Indeed, exposing cells to stress altered the expression ratio of the dRPGs in an intron-dependent manner and intron deletions altered sensitivity to drugs (12). For example, while exposing cells to caffeine favors the expression of the B form of the small subunit protein RPS9, exposure to NaCl favors the expression of the A form. Deleting the intron of RPS9A blocked the caffeine-dependent modulation of the paralog ratio by constitutively increasing the expression of RPS9A and preventing the induction of RPS9B. Similarly, exposing cells to staurosporine favored the expression of RPS9A and the deletion of its intron rendered the cell sensitive to staurosporine (12). Therefore, while deletion of the RPG introns demonstrated their importance as regulators of RP synthesis, the mechanism by which introns regulate the expression of duplicated genes and their impact on ribosome production remains unclear.
In this study, we examined the mechanism by which introns regulate the expression of dRPGs and evaluated their impact on the synthesis of RPs. Mutational analysis of the genes coding for the S9 protein indicated that the expression levels of these dRPGs, which produce two protein isoforms that differ by five amino acids (14), are defined by their different intron structures and the different nature of their 3′ end. The minor paralog (least expressed copy, RPS9A) has a long 3′ UTR and has an intronic two-way helical structure that inhibits splicing. Replacing these elements with the corresponding sequences of the predominant paralog (most expressed copy, RPS9B) enhanced splicing and increased the expression of RPS9A. Surprisingly, increasing the levels of the S9 proteins preferentially inhibited the splicing of the minor paralog. This suggests that the dose of dRPGs is regulated by an asymmetric negative feedback loop that maintains the correct level of expression of duplicated genes. Indeed, we show here that Rps9B differentially binds to the intron of RPS9A in the presence of the intronic element that is required for inhibiting the expression of RPS9A. Together our data provide an explanation for the preservation of introns in RPGs and they highlight the importance of splicing for controlling gene expression in yeast.
MATERIALS AND METHODS
Strains and plasmids
Yeast strains were grown and manipulated using standard procedures (15). Strains carrying chromosomal copies of the 6 His-tagged version of RPS9 were grown at 30°C in YEP media supplemented with 2% dextrose. Strains carrying plasmids expressing different versions of RPS9 were grown at 30°C in synthetic media without leucine (16). Tagging the chromosomal copies of RPS9 genes was achieved using a standard PCR based pop-in / pop-out method (17). RPS9A or RPS9B genes were replaced by the nutritional marker gene URA3 (18) and the resulting knockout strains (rps9aΔ::URA3 or rps9bΔ::URA3) were transformed by PCR fragments containing the tagged versions of RPS9 genes (19). The primers used for the two-step PCR reactions are listed in Supplementary Table S1. Cells in which URA3 was replaced by the tagged constructs were selected in media containing 5-FOA and the integration of the marker was verified using PCR. The tagged genes were introduced in the intron deletion strains described in Figure 1 by mating and sporulation of the diploid strains (17). Mutations of the RPS9 genes were generated by gene synthesis (Bio Basic Canada, Markham, ON), while the majority of sequence substitutions were achieved by standard cloning techniques using silent restriction sites introduced by gene synthesis (Bio Basic Canada, Markham, ON). The detailed description and sources of the different plasmids and strains used in this study are listed in Supplementary Tables S2 and S3, respectively. Plasmids were named using the standard nomenclature scheme where P indicated plasmids, H indicates Histidine tag, and each region of the gene (promoter, 5′ UTR, ORF and 3′ UTR) is indicated by letter A for RPS9A and letter B for RPS9B. US indicates the upper stem structure in the intron of RPS9A and M indicates mutations.
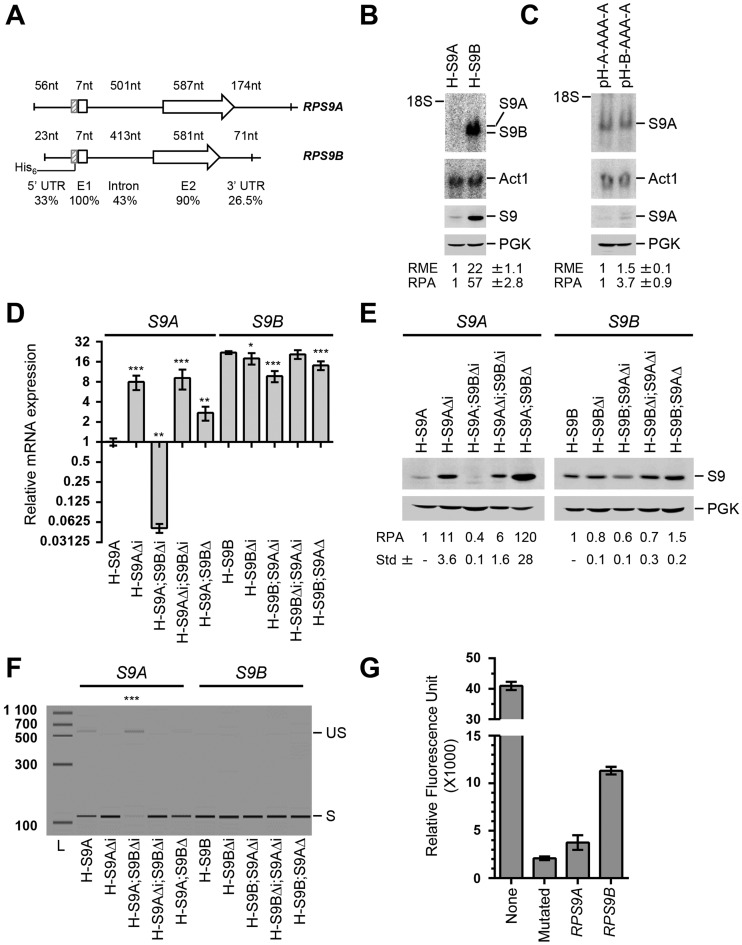
Figure 1.
Synthesis of the ribosomal protein S9 is regulated by asymmetric splicing of duplicated genes. (A) Schematic representation of the RPS9 paralogs. The sizes of the 5′ untranslated region (5′ UTR), first exon (E1), intron, second exon (E2) and 3′ untranslated region (3′ UTR) are shown above each gene. The position of the 6-histidine tag (His6) used for the detection of RPS9 mRNA and Rps9 proteins is indicated on the left. The percent homology between the different regions of the RPS9 paralogs is shown below. (B) The expression of RPS9A is repressed under normal growth conditions. The RNA and proteins were extracted from cells expressing either a tagged version of either RPS9A (H-S9A) or RPS9B (H-S9B) and visualized using Northern blot (upper panel) and western blot (lower panel), respectively. The RNA was hybridized to probes complementary to the His6-tag sequence, while the protein was detected using antibodies against the tag. The position of RPS9 mRNA and proteins is shown on the right. ACT1 mRNA was used as loading control for the Northern blot and Pgk1 was used as loading control for the western blot. The relative mRNA expression (RME) and protein amounts (RPA) were calculated from three independent experiments and are indicated at the bottom along with the standard deviation (Std) for each data point. (C) The expression of RPS9A is repressed post-transcriptionally. Northern blots and Western blots were performed using the His6-tag as described in B to compare the expression of RPS9A mRNA and protein before and after the substitution of the RPS9A promoter and 5′ UTR with that of RPS9B (pH-B-AAA-A). (D) Introns asymmetrically regulate the expression of Rps9 mRNA. RNA was extracted from strains expressing normal or mutated versions of tagged RPS9A (S9A) or RPS9B (S9B) and the expression of the tagged RNA was detected using quantitative RT-PCR. H-S9 indicates mRNA extracted from strains carrying a tagged chromosomal copy of either RPS9A or RPS9B. Strains carrying intron deletions are indicated by Δi and those with complete gene deletions are indicated by Δ. The data are the average of three independent experiments. The asterisks indicate statistically significant differences between the mRNA levels detected in the mutated and non-mutated strains (**P-value < 0.01 and ***P < 0.001). (E) Introns preferentially repress the production of Rps9A proteins. Proteins were extracted from the strains indicated in D and subjected to Western blot analysis using antibodies against the His6-tag as described in (B). (F) Expression of RPS9B inhibits the splicing of RPS9A. The splicing efficiency of RPS9A and RPS9B was evaluated by comparing the relative amount of the spliced (S) and unspliced (US) mRNA using end-point RT-PCR. The bands were detected using capillary electrophoresis and the relative migration of the PCR amplicon were simulated in the forms of bands. The size marker is shown on the left. The splicing levels of the tagged version of each RPG are listed in Supplementary Table S4. (G) RPS9A is spliced less efficiently than RPS9B pre-mRNA. Cells expressing a previously established yellow fluorescent protein (YFP) dependent synthetic splicing reporter carrying either the RPS9A or RPS9B intron were grown in synthetic media and the relative fluorescent units are presented in the bar graph. An intronless reporter (none) was used as a positive control and a reporter carrying a mutated or splice defective intron (mutated) was used as negative control. The data is an average of three independent experiments and the standard deviation is indicated as error bars.
RNA extraction and analysis
Total RNA was extracted from exponentially growing cultures using glass beads (425–600 μm, Sigma-Aldrich, St. Louis, MO) and Northern blot analysis was performed as previously described (20). Briefly, 15 μg of total RNA was separated on a 1.2% denaturing agarose gel and transferred to nylon membrane (Hybond N+, GE Healthcare, Mississauga, ON). The RNA was visualized by autoradiography using a labelled oligonucleotide complementary to the six His-tag or labeled probes corresponding to ACT1 mRNA as control. Images were obtained using a phosphorimager (Storm 860, GE Healthcare, Mississauga, ON). The primers used to generate the probes are listed in Supplementary Table S1.
PCR and splicing index analysis
Total RNA was extracted, treated with DNase (Qiagen, Toronto, ON) and quantitative RT-PCR was performed as previously described (12). End-point PCR (30 cycles) was performed using Platinum Taq (Invitrogen Life Technologies, Burlington, ON) and 10 ng of cDNA as previously described (21). The sequences of the primers used for the PCR amplifications are shown in Supplementary Table S1.
Protein extraction and analysis
Proteins were extracted from exponentially growing cells using glass beads (425–600 μm; Sigma-Aldrich, St. Louis, MO) in lysis buffer (Tris 20 mM pH 8; NaCl 150 mM; Triton X-100 0.1%, PMSF 1 mM; Roche Protease inhibitor Cocktail tablets 1×, Roche Canada, Mississauga, ON). Cells were broken using five cycles of 30 s at 5000 rpm in a Precellys® homogenizer (Laboratory Supply Network, Atkinson, NH) and 40 μg of total proteins were separated on 15% SDS-polyacrylamide gel and transferred to nitrocellulose membrane (Hybond-ECL; GE Healthcare, Mississauga, ON). Membranes were incubated in a hybridization solution (TBS-Tween 0.1%; milk 5%) with antibodies against the six His-tag (dilution 1:500; Santa Cruz Biotechnology, Santa Cruz, CA) or against Pgk1p, as loading control (dilution 1:10 000; Invitrogen Life Technologies, Burlington, ON). The proteins were visualized by chemiluminescence (Lightning Plus-ECL Chemiluminescence kit, Perkin Elmer, Woodbridge, ON) using secondary anti-mouse antibodies conjugated to horseradish peroxidase (dilution 1:2000; GE Healthcare, Mississauga, ON). The protein images were captured using LAS 4000 (GE Healthcare, Mississauga, ON) and the relative protein amounts were calculated using Quantity One software (Bio-Rad, Saint-Laurent, QC).
Growth and fluorescence measurements
Cells expressing yellow fluorescent protein (YFP) (22) were grown in synthetic media to a final concentration of 2 × 106 cells/ml and 100 μl of the cell suspension of the different strains examined was transferred to a 96-well microplate to monitor either growth or fluorescence using the PowerWave XS and FLx800 microplate readers, respectively (BioTek Instruments, Winooski, VT). The relative fluorescence units (RFU) were calculated after ∼2.5 generations by subtracting the fluorescence units (FU) generated by a strain that does not express YFP from that generated by strains carrying different versions of the reporter gene.
Chromatin immunoprecipitation (ChIP) assays
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (23). Briefly, cells were grown to an OD600 of 0.5–0.6. Formaldehyde solution was added into the culture to 1% final concentration and incubated for 20 min at room temperature. The fixation was quenched with glycine (0.36 M final) for 5 min. The cells were washed twice with cold Tris-buffered saline (20 mM Tris–HCl pH 7.5, 150 mM NaCl) and pellets were resuspended in FA lysis buffer (50 mM HEPES–KOH pH 7.5, 150mM NaCl, 1 mM EDTA pH 8, 1% Triton X-100, 0.1% sodium deoxycholate, protease inhibitors). Cells were disrupted using six cycles of 10 s at 6500 rpm in a Precellys® homogenizer (Laboratory Supply Network, Atkinson, NH) and chromatin was sheared by 12 pulses of 10 s at 20% using Branson digital sonifier (Branson Ultrasonics, Danbury, CT). Whole-cell extracts were treated with 7.5 U RNase A (Affymetrix, Santa Clara, CA) and 300U RNase T1 (Sigma Aldrich, St. Louis, MO) for 30 min at 15°C. Untreated samples were also incubated under the same condition in the absence of ribonucleases as control. Immunoprecipitation was performed by incubating the different samples with HA antibody (1.5 μg; 12CA5; Sigma-Aldrich, St. Louis, MO) coupled to magnetic beads (Dynabeads Pan Mouse IgG, Invitrogen Life Technologies, Burlington, ON). The beads were washed and the eluates were collected. The eluates were decrosslinked by incubation in TE–SDS 1% buffer overnight at 65°C. The DNA was purified and detected using quantitative PCR as described (23). The sequences of the primers used for the PCR amplifications are shown in Supplementary Table S1.
RESULTS
Synthesis of Rps9 protein isoforms is regulated by asymmetric paralog specific splicing
The fact that intron deletions differentially affect the expression of dRPGs suggests that differences in intron sequence may play an important role in controlling and coordinating the expression of RPGs (12). However, the intronic effects on RP synthesis and the mechanism by which they affect the expression of dRPGs remain unclear. To evaluate the effects of introns and other regulatory sequences on protein production, we generated tagged versions of a model dRPGs pair. RPS9A and RPS9B were chosen as the model for this study (Figure 1A) because introns of RPS9 were retained during evolution (24) and their deletions altered growth under stress (12). The tagged copies of RPS9 were created by inserting a six His-tag after the start codon of RPS9A (H-S9A) or RPS9B (H-S9B) to produce strains expressing one tagged paralog at a time. The tag inclusion did not affect growth and had no significant effect on RNA expression levels (Supplementary Table S4). By using one epitope in two different strains, we were able to directly compare the expression levels of RPS9 paralogs using a single probe or antibody. As shown in Figure 1B, RPS9B produced 20 times more RNA and almost 60 times more proteins than RPS9A, indicating that RPS9B is the most expressed copy, or the ‘housekeeping’ gene, that produces the majority of S9 protein.
To understand the source of the great difference in the amount of proteins produced by the RPS9 genes, we evaluated the effects of the intron and promoter sequence on gene expression. Promoters are considered important regulators of RPGs expression (25,26) and thus were included in the assay as control. Surprisingly, the substitutions of the promoter and 5′ UTR of RPS9A with those of RPS9B in Figure 1C (pH-B-AAA-A; our convention of naming constructs states the origin of each part of the gene according to the system pHis- 5′ UTR - first exon / intron / second exon – 3′ UTR) did not substantially increase the levels of RPS9A mRNA, indicating that the promoter identity is not responsible for the difference in the RNA levels of these two paralogs (Figure 1C, upper panel). This is also consistent with the RPS9A and RPS9B RNAPII and nucleosome association profiles, which indicate that RPS9B is not more transcribed than RPS9A (Supplementary Figure S1).
Despite the small difference in mRNA quantities detected upon the promoter substitutions, the presence of the RPS9B promoter and 5′ UTR increased Rps9A protein production by 3.7 times (Figure 1C, lower panel). This suggests that the sequence upstream of the coding sequence of RPS9 may influence the translation of its paralog's mRNAs. In contrast, intron deletion substantially increased both the RNA and protein levels of RPS9A but not of RPS9B (Figure 1D and E). This clearly indicates that the intron is the main cause of the preferential repression of RPS9A. Unexpectedly, while deletion of the RPS9B gene increased the expression of RPS9A, deletion of the RPS9B intron inhibited the expression of RPS9A (Figure 1D and E). This suggests that whereas the presence of RPS9B represses the expression of RPS9A, its intron alleviates the repression of RPS9A, perhaps by competing for potential inhibitors in trans. We conclude that RPS9B is preferentially expressed over RPS9A due to asymmetric regulation by their respective introns.
To determine the mechanism by which introns influence the expression of RPS9 paralogs, we compared the splicing efficiency of RPS9A and RPS9B pre-mRNAs and evaluated the effects of their introns on the splicing of a reporter gene. The amount of spliced and unspliced transcripts was detected by end-point RT-PCR using primers complementary to the region surrounding the splice site of each paralog. As indicated in Figure 1F, RPS9A generated a detectable quantity of unspliced RNA, while RPS9B did not. Deleting the 5′-3′ exoribonucleases that normally degrade unspliced pre-mRNA increased the amount of the paralog pre-mRNA and made it possible to detect the precursor of RPS9B (Supplementary Figure S2). However, even in the absence of ribonuclease we still detected more pre-mRNA for RPS9A than RPS9B (Supplementary Figure S2), consistent with the diminished splicing efficiency of RPS9A. To directly compare the splicing efficiency of the RPS9 introns, we monitored their effect on the maturation of same reporter gene (22). As indicated in Figure 1G, the splicing reporter produced much more yellow fluorescent protein (YFP) with the intron of RPS9B than with that of RPS9A. This result confirms that the difference between the splicing efficiency of RPS9A and RPS9B is mostly determined by the intron sequence.
Deletion of the complete RPS9B gene enhanced the splicing (Figure 1F) and increased the expression of RPS9A (Figure 1D and E). However, the deletion of the RPS9B intron, which does not increase the amount of S9B proteins (Figure 1E), resulted in strong inhibition of RPS9A splicing (Figure 1D and F) and expression (Figure 1D and E). This suggests that the introns of these two gene copies may communicate independently of the amount of proteins produced from their coding regions. Deletion of RPS9A (H-S9B;S9AΔ) and its intron (H-S9B;S9AΔi) had minor effect on protein levels and no effect on the splicing of RPS9B (Figure 1E–F). We conclude that the introns of RPS9 paralogs encode different splicing programs that define the expression level of their host genes.
Splicing efficiency is controlled by paralog specific intronic structure
Comparison of the splicing efficiency of RPS9 paralogs indicated that RPS9A is less efficiently spliced than RPS9B (Figure 1F and G). However, the reason behind this difference in the paralogs splicing efficiency is unclear. It was previously shown that intronic secondary structure might regulate splicing in yeast (27–29). Therefore, we compared the secondary structure of the RPS9A and RPS9B introns with the goal of identifying possible paralog-specific structural elements that might explain the difference in splicing efficiency. The intronic sequence of both RPS9A and RPS9B were folded using the nucleic acid folding and hybridization prediction program mfold (30) and consensus structures of each paralog found in other Saccharomyces species were retained and compared. As indicated in Figure 2A, both introns formed extended long range secondary structures that bring the 5′ and 3′ ends into close proximity, which seems to be a general feature of yeast pre-mRNA (31). Differences in the structure of the paralogs’ introns were found in two regions, the first near the paired termini defining the splice sites and the second near the upper stem (US) loop structure (Figure 2A). To verify whether these variations in structures are indeed the reason for the differences in the splicing efficiency of RPS9A and RPS9B, we substituted the intron of RPS9A with that of RPS9B and monitored the effect on the splicing and expression of RPS9A mRNA. As indicated in Figure 2B, intron substitution (pH-A-ABA-A) significantly increased the amount of the mature mRNA generated by RPS9A, which confirms that differences in intron structure may influence paralog expression levels. The increase in the expression of pH-A-ABA-A raised the amount of the Rps9A protein without greatly affecting the expression of RPS9B (Figure 2C and D). Remarkably, intron substitution did not change the ratio of RPS9A spliced and unspliced mRNA (Figure 1B). Therefore, while differences between RPS9A and RPS9B introns are sufficient to induce the expression of RPS9A, additional elements may influence the preferential stability of RPS9A pre-mRNA.
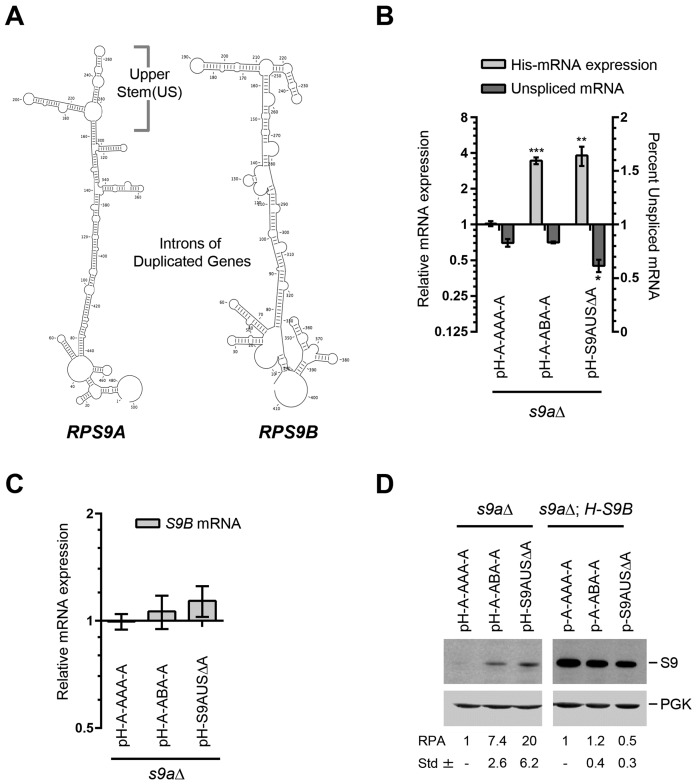
Figure 2.
The differential splicing of RPS9 pre-mRNAs is generated by paralog specific intronic elements. (A) The introns of RPS9A and RPS9B have distinct structural elements. The secondary structures of the RPS9A and RPS9B introns were folded using mfold and the most likely conserved structures found in related Saccharomyces species are shown. (B) The intron sequence defines the splicing efficiency of the RPS9 paralogs. The relative mRNA expression of tagged RPS9A (pH-A-AAA-A), RPS9A carrying the intron of RPS9B (pH-A-ABA-A) and RPS9A lacking the upper stem structure that differentiates introns between the RPS9A and B (pH-S9AUSΔA) were determined using quantitative RT-PCR. The splicing efficiency was determined as described in Figure 1F and presented in the form of a bar graph. The experiments were conducted in triplicates and statistically significant differences in expression and splicing efficiency are indicated by asterisks (*P-value <0.05; **P-value < 0.01 and ***P-value < 0.001). (C) Modification of theRPS9A intron has little effect on the expression of RPS9B mRNA. The levels of RPS9B were detected using quantitative RT-PCR in the different RPS9A strains described in (B). (D) The intron sequence regulates the expression of S9 proteins. The mutations described in B were expressed with (pH-A-AAA-A, pH-A-ABA-A and pH-S9AUSΔA) or without (p-A-AAA-A, p-A-ABA-A and p-S9AUSΔA) the His6-tag in strains lacking RPS9A (s9aΔ) or cells lacking RPS9A and expressing His-tagged RPS9B (s9aΔ; H-S9B) and the protein was extracted and visualized using antibodies against the His6-tag. The position of the S9 protein and loading control Pgk1 is shown on the right. The relative protein amount calculated from three independent experiments and their respective standard deviations are shown at bottom.
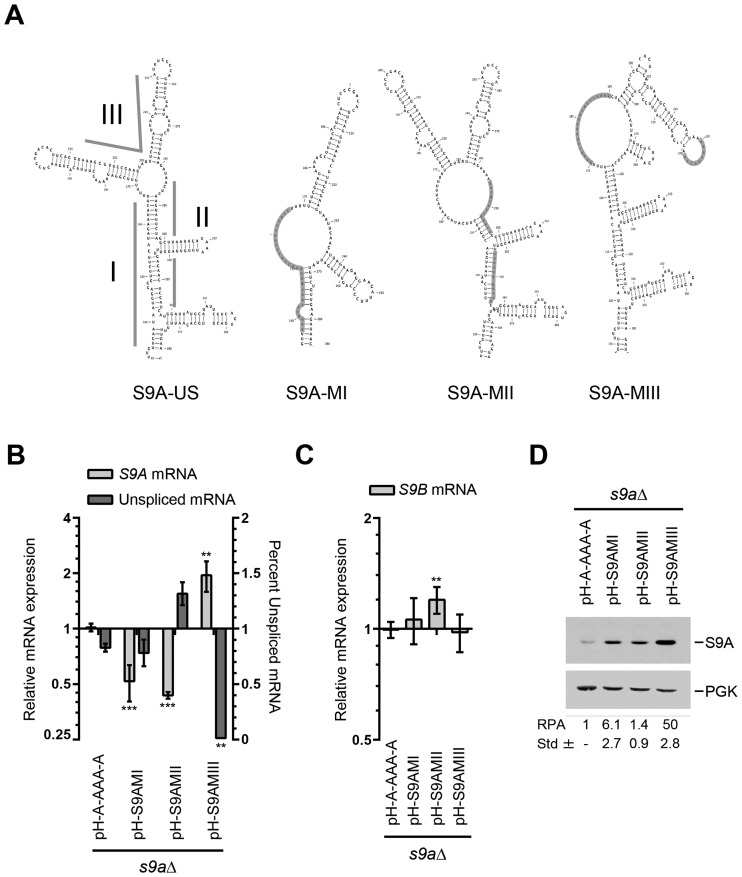
To better characterize the intronic elements inhibiting the expression of RPS9A, we deleted the US structure (Figure 2A), which harbors most of the differences between the RPS9 paralogs, and examined its effect on expression, splicing and protein synthesis. As predicted, the deletion of the RPS9A US (pH-S9AUSΔA) resulted in a significant increase in the RPS9A mRNA and reduced the amount of unspliced pre-mRNA (Figure 2B). This increase in mRNA induced the expression of Rps9A protein and inhibited the expression of Rps9B protein without significantly decreasing the levels of RPS9B mRNA (Figure 2C and D). These data indicate that the ratio of the RPS9 paralogs is defined by intronic structural elements that preferentially attenuate the splicing and inhibit the production of the RPS9A mRNA. Indeed, replacing the US loop structure of A with that of B is sufficient to enhance the splicing and increase the expression of RPS9A (Supplementary Figure S3). To identify the specific structural element that regulates the splicing of RPS9 paralogs, we divided the US loop structure of RPS9A into three regions and examined the effect of each mutated region on expression and splicing efficiency (Figure 3A). As indicated in Figure 3B, mutations of regions I and II (S9A-MI and S9A-MII), which are located on each side of the lower region of the US structure, did not increase the levels of mature RPS9A mRNA or enhance splicing. S9A-MII did not affect protein production, while S9A-MI resulted only in a moderate increase in protein production (Figure 3D). In contrast, mutations in region III (S9A-MIII), which includes the putative two-way helical structure, enhanced splicing and increased expression of RPS9A significantly (Figure 3B and D). Despite the substantial increase in S9A-MIII proteins, the amount of RPS9B mRNA did not change (Figure 3C). In any case, the data clearly indicate that while different mutations may influence gene expression only mutations disrupting the two-way helical structure (S9A-MIII) enhance splicing. We conclude that the splicing of RPS9A is inhibited by a putative two-way helical structure that differs in junction size and stem orientation between paralogs.
Figure 3.
Splicing of RPS9A is inhibited by a putative two-way helical structure. (A) Schematic representation of the RPS9A upper stem structure (S9A-US) required for the inhibition of RPS9A splicing and its mutated derivatives. The roman numbers indicate the three subsections of S9A-US targeted for mutation. The mutated regions are highlighted and identified by roman number (I–III). (B) Disruption of the upper stem two-way junction promotes the splicing and increases the expression of RPS9A. The relative mRNA expression and splicing efficiency were determined using PCR and presented in the form of a bar graph. The experiments were conducted in triplicates and statistically significant differences are indicated by asterisks (**P-value < 0.01 and ***P-value < 0.001). (C) Disruption of RPS9A upper stem structure has limited effect on the expression of RPS9B. The expression of RPS9B in the different RPS9A mutations was determined by quantitative RT-PCR and presented in the form of bar graph. (D) Disruption of the putative two-way helical structure induced the expression of the Rps9A protein. The relative protein amounts (RPA) and standard deviations were calculated using three independent experiments and shown at bottom.
Splicing of RPS9B is promoted by multiple elements ensuring efficient splicing and translation
To determine whether the intron of RPS9A is sufficient for inhibiting the expression of its host gene and to understand how the expression of RPS9B is regulated, we substituted the non-coding sequences of RPS9B with those of RPS9A and determined the impact on expression (Figure 4A). As indicated in Figure 4B, the intron substitution (pH-B-BAB-B) greatly inhibited the expression of RPS9B, confirming the capacity of RPS9A intron to independently inhibit gene expression. The substitution of both the RPS9B promoter and intron sequences with those of RPS9A (pH-A-BAB-B) did not decrease the expression level of RPS9B (Figure 4B). This suggests that RPS9A is normally more transcribed than RPS9B. Indeed, ChIP-Seq data indicate that RNAPII associates more with RPS9A than with RPS9B and more nucleosomes are found near the promoter of RPS9B than that of RPS9A (Supplementary Figure S1). Therefore, while RPS9A might be more transcribed than RPS9B, the relative amount of the mature mRNA available for translation is influenced by differences in the splicing efficiency of these two duplicated genes.
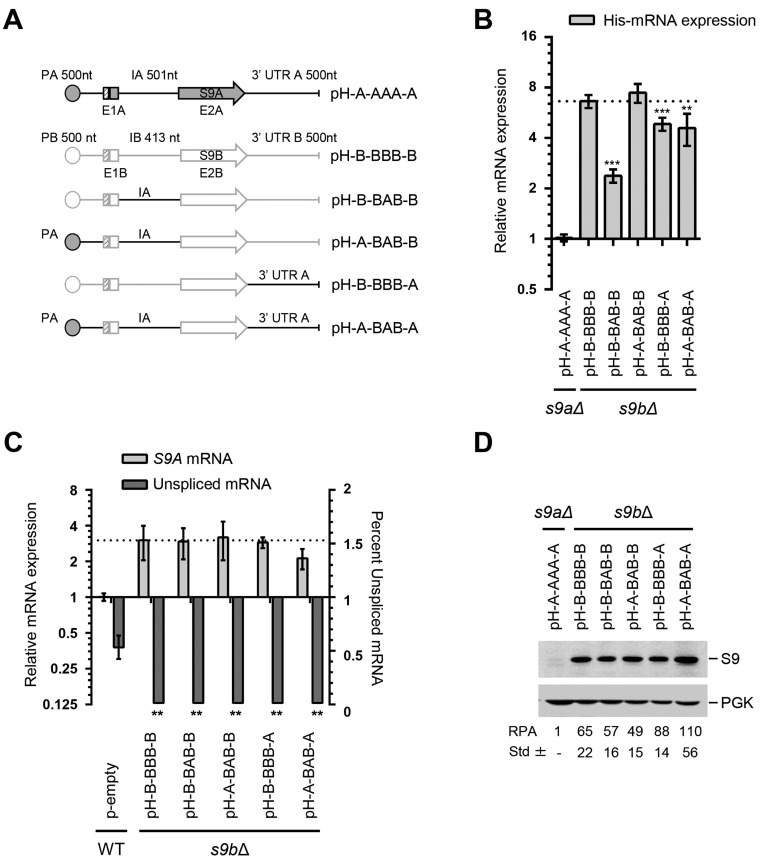
Figure 4.
Expression of RPS9B is promoted by both intron and 3′ UTR sequences. (A) Schematic representation of the RPS9 genes and its associated RPS9B mutations. The size of the promoter (P), exons (E), introns (I) and 3′ UTR is indicated above each gene in nucleotides (nt). The name of each mutant is indicated on the right. The sequence of RPS9A is outlined in black while that of RPS9B outlined in gray. (B) The intron and 3′ UTR of RPS9A inhibit the expression of RPS9B. The expression of the different mutations of RPS9B indicated in A was detected using quantitative RT-PCR and shown in the form of a bar graph. The experiments were conducted in triplicates and statistically significant differences in expression are indicated by asterisks (** P-value < 0.01 and ***P-value < 0.001). (C) Inclusion of the RPS9A intron and 3′ UTR in RPS9B has little effect on the expression of RPS9A. The splicing of RPS9A and the level of mature mRNA were detected using RT-PCR and indicated in the form of a bar graph. (D) Changes in the levels of RPS9B mRNA have limited effect on the expression of Rps9B protein. The effect of the different mutations on protein levels was monitored using a His6-tag specific antibody. The expected position of the S9 and the control protein Pgk1 is shown on the right. The relative protein amounts and the standard deviation were determined using three independent experiments and indicated at the bottom.
The substitution of the 3′ UTR of RPS9B with that of RPS9A (pH-B-BBB-A) reduced the expression of RPS9B but not to the same level as the intron substitution (pH-B-BAB-B) (Figure 4B). Substitution of all non-coding sequences of RPS9B with those of RPS9A within the same construct did not further reduce the expression of RPS9B (pH-A-BAB-A) (Figure 4B). Additionally, the unspliced mRNA was not detected for any of the constructs containing the coding sequence of RPS9B under normal growth condition (Supplementary Table S4). However, the mutant RPS9B containing the intron of RPS9A (pH-B-BAB-B) had increased accumulation of pre-mRNA in the absence of the ribonucleases that normally degrade unspliced pre-mRNA (Supplementary Figure S2). Therefore, while the intron and the 3′ UTR may alter the levels of RPS9B mRNA (pH-B-BAB-B and pH-B-BBB-A), other sequences embedded within the coding sequence contribute to the instability of RPS9B pre-mRNA (Supplementary Figure S2). The introduction of RPS9A regulatory elements in RPS9B did not greatly affect the expression of RPS9A because none of the mutations greatly reduced the amount of Rps9B protein under the conditions tested (Figure 4C and D). The amount of Rps9B proteins produced by the different substitutions (e.g. pH-A-BAB-B and pH-A-BAB-A) did not correlate directly with the amount of mRNA produced from each construct. This suggests that unlike RPS9A, the expression of RPS9B might also be regulated at the level of translation (Figure 4B and D). Comparison of the paralog codon usage frequency indicated that RPS9B mRNA contains more codons with higher usage frequency than RPS9A (Supplementary Figure S4), which may promote the translation of the Rps9B isoform over its Rps9A counterpart. Overall these observations indicate that while differential repression of dRPGs is achieved by a simple modification of mRNA levels, the preferential expression of the dominant copy can be achieved by multiple factors at several levels of gene expression.
The 3′ UTR of RPS9 influences splicing in an intron dependent manner
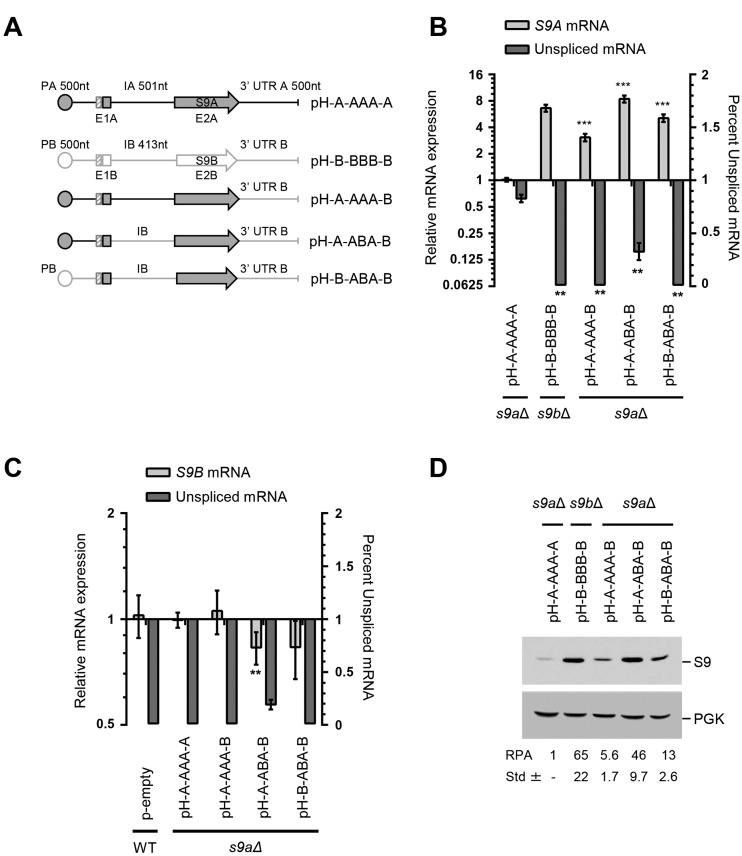
The capacity of RPS9A 3′ UTR to reduce the expression of RPS9B (Figure 4B) suggests that elements other than the intron might also modulate the expression of RPS9A. To verify this possibility, we substituted the promoter and 3′ end of the RPS9A with those of RPS9B and monitored the effect on expression (Figure 5A). Surprisingly, the substitution of RPS9A 3′ UTR with that of RPS9B (pH-A-AAA-B) not only resulted in a significant increase in the expression of RPS9A, but also reduced the proportion of unspliced mRNA (Figure 5B). Substitution of both the 3′ end and the intron of RPS9A (e.g. pH-A-ABA-B) reduced the amount of unspliced mRNA and further increased the expression of RPS9A mRNA to levels comparable to those observed with the intact RPS9B (Figure 5B). Substitution with the RPS9B promoter (pH-B-ABA-B) did not increase the expression any further suggesting that both the intron and the 3′ UTR collaborate to inhibit the expression of RPS9A regardless of the promoter used for transcription. The substitution of the intron and 3′ UTR of RPS9A (pH-A-ABA-B) also inhibited the expression and reduced the splicing of RPS9B in trans (Figure 5C). This decrease in the expression of RPS9B is not necessarily due to increased production of Rps9A protein (Figure 5D) since other mutations that also increase the amount of Rps9A protein (e.g. pH-S9AMIII, Figure 3D) did not reduce the amount of RPS9B mRNA (Figure 3C). Instead, we propose that the introduction of extra copies of the RPS9B intron and 3′ end sequesters factors necessary for the expression of the chromosomal copy. As observed with mRNA (Figure 5B), the highest increase in the level of the Rps9A protein was observed with the substitution of both intron and 3′ UTR (pH-A-ABA-B, Figure 5D). However, a significant increase in protein expression was also observed with the substitution of the 3′ UTR alone (pH-A-AAA-B), which confirms that both the intron and the 3′ UTR can independently regulate protein production by altering the splicing efficiency. We conclude that the ratio of the dRPGs, at least in the case of RPS9, is determined mostly post-transcriptionally through differences in introns and the 3′ end sequences.
Figure 5.
RPS9A 3′ UTR mediates intron dependent attenuation of splicing. (A) Schematic representation of the RPS9 genes and its associated RPS9A mutations. The size of the promoter (P), exons (E), introns (I) and 3′ UTR is indicated above each gene in nucleotides (nt). The name of each mutant is indicated on the right. The sequence of RPS9A is outlined in black with that of RPS9B in gray. (B) The 3′ UTR of RPS9B induces the splicing and increases the expression of RPS9A. The splicing and the expression of the different mutations of RPS9A indicated in A were detected using RT-PCR and are shown in the form of a bar graph. The experiments were conducted in triplicates and statistically significant differences are indicated by asterisks (**P-value < 0.01 and ***P-value < 0.001). (C) Inclusion of both the RPS9B intron and its 3′ UTR in the RPS9A gene repress the expression of RPS9B in trans. The splicing of RPS9B and the level of mature mRNA were detected using RT-PCR and are indicated in the form of a bar graph. (D) The intron and 3′ end sequences collaborate to repress the expression of Rps9A protein. The effect of the different mutations on protein levels was monitored using antibodies specific to the His6-tag. The expected positions of the S9 and the control protein Pgk1p are shown on the right. The relative protein amounts and the standard deviation were determined using three independent experiments and indicated at the bottom.
The S9 protein differentially interacts with the intronic sequence of the duplicated RPS9 genes
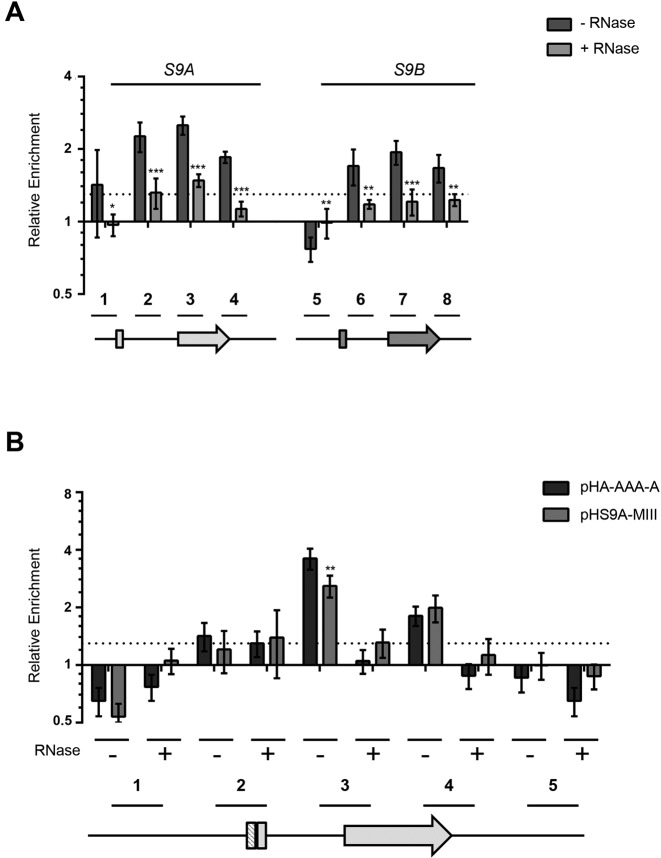
The two-way helical structure (US, Figure 2A) required for inhibiting the splicing of RPS9A features a central bulge similar to that required for the binding of the Rps9 protein to 18S rRNA (32). Therefore, we hypothesized that Rps9B may inhibit the expression of RPS9A by binding to the intron and inhibiting its splicing. However, testing the interaction between Rps9 protein and the introns of the RPS9 genes is difficult since very little unspliced mRNA is detected for these two genes (Figure 1F). Alternatively, since most pre-mRNAs are believed to be spliced co-transcriptionally (33,34), we reasoned that Rps9 protein might be present at the chromatin of the duplicated RPS9 genes in an intron-dependent manner. To verify this possibility, we immunoprecipitated an HA-tagged version of the RPS9 paralogs before and after RNase treatment. As indicated in Figure 6A, DNA fragments corresponding to the intron, the beginning of exon 2 and the 3′ UTR of RPS9A were enriched by the immunoprecipitation of Rps9B protein. In contrast, mock precipitation in the absence of antibodies and amplification using primers against unrelated RPGs did not enrich any of the DNA fragments. The biggest enrichment was detected in fragments within the intron and around the exon 2 splice site (fragments 2 and 3). Enrichment was also observed in fragments corresponding to the RPS9B intron and exon 2 (fragments 6 and 7) but to a lesser extent. Furthermore, unlike RPS9A, no significant enrichment was observed in the intron of RPS9B when compared with its 3′ UTR. All observed enrichments were lost after ribonuclease treatment as would be expected if the interaction between Rps9B protein and the chromatin of the duplicated genes is mediated by RNA. Together these data indicate that while Rps9B protein may interact with different parts of the duplicated transcripts it has higher affinity to the intron and exon 2 splice site of RPS9A. Mutation of the two-way helical structure that inhibits the splicing of RPS9A (Figure 3A) reduced the interaction of the intronic sequence with Rps9B to a similar level to that observed with the intron of RPS9B and this had little effect on the capacity of Rps9B to interact with the 3′ end sequence (Figure 6B). Therefore, while Rps9B may be recruited to chromatin by multiple sequences within the Rps9A and B mRNAs, its recruitment is enhanced in the presence of the RPS9A-specific helical structure. Together these data suggest that the differential inhibition of the RPS9 paralogs is achieved through differences in the introns’ affinity for Rps9 protein.
Figure 6.
Rps9B protein binds to the intron of RPS9A in the presence of the two-way helical structure required for inhibiting the splicing of RPS9A pre-mRNA. (A) Rps9B protein differentially interacts with the chromatin of the RPS9 paralogs in an RNA dependent manner. ChIP assays were performed using an HA tagged version of Rps9B isoform before (dark gray bars) or after (light gray bars) RNase treatment and the co-precipitated DNA was detected using paralog specific primers. The enrichment of the different PCR amplicons relative to an untranscribed region in chromosome V is presented in the form of a bar graph. The position of the primers used is shown relative to the paralog gene's structures under the graph. The untranslated regions and introns are shown as lines and exons as boxes. The experiments were conducted in triplicates and the standard deviations are shown as error bars. DNA enrichments were considered significant when greater than 30% relative to the untranscribed control (dotted line). Statistically significant differences between RNase treated and untreated samples are indicated by asterisks (*P-value <0.05, **P-value < 0.01 and ***P-value < 0.001). (B) The Rps9B interaction with RPS9A is enhanced in the presence of the two-way helical structure required for the inhibition of RPS9A expression. The interaction of Rps9B with DNA originating from plasmids carrying wild type RPS9A (pH-A-AAA-A) or RPS9A with mutated two-way helical structure (pHS9A-MIII) was assayed before (–) or after (+) treatment with RNase using ChIP as described in (A). The position of the primers is shown below the graph. Significant differences between the enrichment patterns of the wild type gene and its mutated versions are indicated by asterisks (**P-value < 0.01).
Introns define the expression hierarchy of many dRPGs
To evaluate the generality of the RPS9 mode of inter-paralog gene regulation, we compared the expression levels of the dRPGs to the splicing efficiency of their introns and determined the number of genes where low expression levels correlated with reduced splicing efficiency. The expression ratio of dRPG paralogs was calculated by comparing the expression levels of each gene as determined by single molecule sequencing (35), while the relative splicing efficiency of each gene pair was calculated by comparing the amount of spliced mRNA generated by a reporter gene carrying the introns of the different dRPGs (22). As indicated in Supplementary Table S5 and Supplementary Figure S5A, 71% (27 out of 38) of dRPGs tested were both under expressed and their intron less spliced when compared to their paralog. This indicates that, in most cases, splicing determines the paralog relative expression levels of the paralogs. Indeed, in 83% of the cases deleting the under-spliced intron from the minor dRPG copy led to increased expression of the host gene (Supplementary Table S5 and Supplementary Figure S5B). Strikingly, in 57% of the cases the deletion of the minor copy intron completely changed the paralog expression hierarchy also gave dominance to the otherwise minor copy. As in the case of RPS9 (Figure 5), 12 out of these 15 genes with a strict intron-dependent expression hierarchy also have longer 3′ UTRs than their counterparts (Supplementary Table S5). This is consistent with a role of the UTR length in enforcing the splicing inhibition of the minor dRPG copies. We conclude that the expression hierarchy of several dRPGs is determined in large part post-transcriptionally by the intron identity assisted at least in some cases by other elements like the 3′ UTR length.
DISCUSSION
Previous studies indicated that introns influence the expression of dRPGs in a paralog specific manner (12). However, the mechanism by which introns regulate the expression of dRPGs and their impact on the accumulation of RPs remained unclear. In this study, we show that differences in intron structure determine the expression levels of dRPGs by influencing their splicing efficiency. The study of RPS9A and RPS9B indicated that the expression hierarchy of these duplicated genes is determined by copy specific intronic structures that differentially interact with the Rps9 protein. The presence of a putative two way helical structure in the intron of the minor copy (RPS9A) increased binding to the Rps9 protein and reduced splicing to favor the expression of the major copy (RPS9B) (Figures 3 and 6). Mutational analysis indicated that while the repression of RPS9A was primarily based on the nature of intron and 3′ UTR the promotion of RPS9B expression requires additional elements embedded within the coding sequence (Figures 4 and 5). Together, the data indicate that the expression hierarchy of dRPGs is not simply achieved by accidental repression of the minor paralog during evolution, but rather represents a deep-rooted post-transcriptional regulatory program that promotes the expression of one copy over the other.
Most yeast introns are found in genes coding for RPs, which begs the question as to whether introns make a special contribution to the expression and function of RPGs (36). Previous studies using RNA as a marker for gene expression indicated that half of the RPGs are repressed in the presence of introns, while the other half is induced in their absence (12). In addition, studies of the large subunit protein gene RPL30 and the small subunit protein gene RPS14B indicated that the intron structure play an important role in regulating the splicing efficiency and expression level of these genes (28,37). However, the impact of these introns on the synthesis of RPs and the mechanism by which they regulate the expression of dRPGs remained unclear. In this study, we show that introns may directly affect the production of RPs by modifying the splicing of the dRPGs in a paralog specific manner. Comparison of the duplicated genes producing the small subunit protein Rps9 indicated that introns carry paralog-specific splicing structures that permit the inhibition of one copy and induce the expression of the other. This mode of differential regulation is not limited to RPS9 but extend to other gene pairs (Supplementary Figure S5 and Supplementary Table S5). Indeed, a well-defined structure in the intron of RPS14B, and not RPS14A, was shown to inhibit the splicing of its host gene in response to changes in the levels of Rps14p (37). Accordingly, we propose that introns may function as codes for gene expression that help cells to program mini-circuits of inter-gene regulation. These codes would permit the coordination of the duplicated gene's expression without interfering with the general transcription program that control ribosome production.
Together, our data suggest a model (Figure 7) in which intron structure and 3′ UTR size determine the relative expression levels of dRPGs. This mode of asymmetric gene expression permits the adjustments of the paralogs expression levels without interfering with the promoter-dependent regulatory network at the root of ribosome synthesis control (25). In this model, the two RPG copies are not necessarily preserved to maintain constant dose of RPs but instead serve as tools to broaden the regulatory spectrum of RPGs. This notion is supported by the fact that the minor copy of the dRPGs does not fully compensate for the loss of its paralog (Supplementary Table S4) and increasing the amount of RP (e.g. expression of extra copies of RPS9, Supplementary Table S6) does not inhibit the expression of the paralog producing the majority of the RP (RPS9B). Instead, it is the minor copy (RPS9A) that is preferentially repressed by the Rps9 protein, perhaps through direct interaction between the Rps9 protein and the intron of RPS9A (Figure 6).
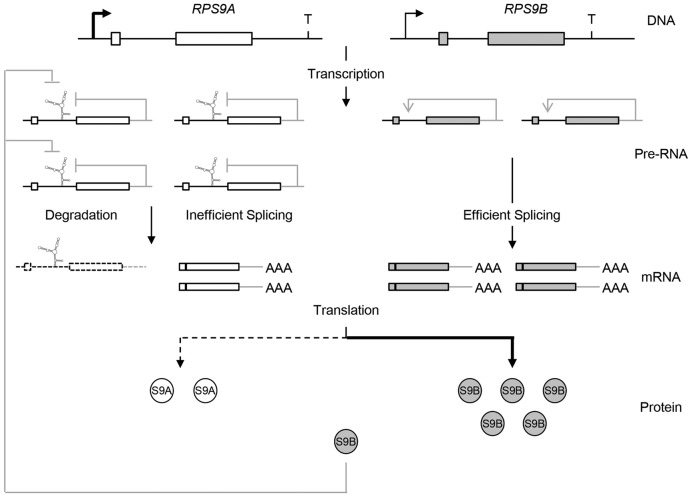
Figure 7.
Schematic representation of the multi level RPS9 regulatory circuit. The differential regulation of RPS9A and RPS9B is illustrated at different levels of gene expression. The coding regions are shown in the form of boxes and non-coding sequence (introns and UTRs) are shown as lines. The transcription start sites are illustrated by arrows. The secondary structure within the RPS9A intron represents the structure required for the inhibition of RPS9A splicing. Steady state inhibition and enhancement of splicing are illustrated by gray lines.
The ratio of the RPS9 paralogs is maintained under normal growth conditions regardless of the amount of protein produced (Supplementary Table S4) but their relative expression is dramatically altered under stress (12). Exposure to osmotic stress reverses the ratio of RPS9 paralogs by increasing the expression of RPS9A and repressing the expression of RPS9B (12). Deleting the introns perturbs the expression of both paralogs and results in a constitutive induction of RPS9A and conceals the response to stress. Therefore, it appears that the minor copy of the RPG pairs is preserved to adjust expression under different growth conditions and perhaps to produce alternative protein isoforms that are better suited for growth under stress. The RPS9 protein isoforms vary by 5 amino acids, which slightly increases the charge of the C-terminal portion of the minor copy (14). Deleting the region harboring these alternative amino acids altered the ribosome association with many mRNAs (14). However, it remains to be seen if these amino acid differences are sufficient to explain the requirement of RPS9A for growth under stress. In all cases, the models presented in this study provide an explanation of the preservation of introns in RPGs and highlight the importance of splicing for gene regulation in yeast.
Supplementary Material
Acknowledgments
We thank Dr Maya Schuldiner for the YFP reporter constructs. The authors would like to thank Mustafa Malik for determining the codon usage frequency used in Supplementary Figure S4 and for reading the manuscript. We thank Mélodie Berthoumieux for the analysis of the ChIP-seq data presented in Supplementary Figure S1. We are indebted for Dr Jonathan Warner for critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natural Sciences and Engineering Research Council of Canada (NSERC); S.A. is Canada Research Chair in RNA Biology and Cancer Genomics. Funding for open access charge: NSERC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ramakrishnan V. The ribosome emerges from a black box. Cell. 2014;159:979–984. doi: 10.1016/j.cell.2014.10.052. [DOI] [PubMed] [Google Scholar]
- 2.Perry R.P. Balanced production of ribosomal proteins. Gene. 2007;401:1–3. doi: 10.1016/j.gene.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 4.Xiao L., Grove A. Coordination of ribosomal protein and ribosomal RNA gene expression in response to TOR signaling. Curr. Genomics. 2009;10:198–205. doi: 10.2174/138920209788185261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woolford J.L., Jr, Baserga S.J. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013;195:643–681. doi: 10.1534/genetics.113.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kressler D., Hurt E., Bassler J. Driving ribosome assembly. Biochim. Biophys. Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 7.Langkjaer R.B., Cliften P.F., Johnston M., Piskur J. Yeast genome duplication was followed by asynchronous differentiation of duplicated genes. Nature. 2003;421:848–852. doi: 10.1038/nature01419. [DOI] [PubMed] [Google Scholar]
- 8.Komili S., Farny N.G., Roth F.P., Silver P.A. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131:557–571. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evangelisti A.M., Conant G.C. Nonrandom survival of gene conversions among yeast ribosomal proteins duplicated through genome doubling. Genome Biol. Evol. 2010;2:826–834. doi: 10.1093/gbe/evq067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kafri R., Levy M., Pilpel Y. The regulatory utilization of genetic redundancy through responsive backup circuits. Proc. Natl. Acad. Sci. U.S.A. 2006;103:11653–11658. doi: 10.1073/pnas.0604883103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ihmels J., Collins S.R., Schuldiner M., Krogan N.J., Weissman J.S. Backup without redundancy: genetic interactions reveal the cost of duplicate gene loss. Mol. Syst. Biol. 2007;3:86. doi: 10.1038/msb4100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parenteau J., Durand M., Morin G., Gagnon J., Lucier J.F., Wellinger R.J., Chabot B., Elela S.A. Introns within ribosomal protein genes regulate the production and function of yeast ribosomes. Cell. 2011;147:320–331. doi: 10.1016/j.cell.2011.08.044. [DOI] [PubMed] [Google Scholar]
- 13.Parenteau J., Lavoie M., Catala M., Malik-Ghulam M., Gagnon J., Abou Elela S. Preservation of gene duplication increases the regulatory spectrum of ribosomal protein genes and enhances growth under stress. Cell Rep. 2015;13:2516–2526. doi: 10.1016/j.celrep.2015.11.033. [DOI] [PubMed] [Google Scholar]
- 14.Pnueli L., Arava Y. Genome-wide polysomal analysis of a yeast strain with mutated ribosomal protein S9. BMC Genomics. 2007;8:285. doi: 10.1186/1471-2164-8-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rose M.D., Winston F., Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. NY: Cold Spring Harbor; 1990. [Google Scholar]
- 16.Guthrie C., Fink G.R. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic Press; 1991. [Google Scholar]
- 17.Parenteau J., Durand M., Veronneau S., Lacombe A.A., Morin G., Guerin V., Cecez B., Gervais-Bird J., Koh C.S., Brunelle D., et al. Deletion of many yeast introns reveals a minority of genes that require splicing for function. Mol. Biol. Cell. 2008;19:1932–1941. doi: 10.1091/mbc.E07-12-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gietz R.D., Woods R.A. Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods Mol. Biol. 2006;313:107–120. doi: 10.1385/1-59259-958-3:107. [DOI] [PubMed] [Google Scholar]
- 20.Abou Elela S., Igel H., Ares M., Jr RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996;85:115–124. doi: 10.1016/s0092-8674(00)81087-9. [DOI] [PubMed] [Google Scholar]
- 21.Klinck R., Bramard A., Inkel L., Dufresne-Martin G., Gervais-Bird J., Madden R., Paquet E.R., Koh C., Venables J.P., Prinos P., et al. Multiple alternative splicing markers for ovarian cancer. Cancer Res. 2008;68:657–663. doi: 10.1158/0008-5472.CAN-07-2580. [DOI] [PubMed] [Google Scholar]
- 22.Yofe I., Zafrir Z., Blau R., Schuldiner M., Tuller T., Shapiro E., Ben-Yehezkel T. Accurate, model-based tuning of synthetic gene expression using introns in S. cerevisiae. PLoS Genet. 2014;10:e1004407. doi: 10.1371/journal.pgen.1004407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghazal G., Gagnon J., Elela S.A. Detection and characterization of transcription termination. Methods Mol. Biol. 2012;809:593–607. doi: 10.1007/978-1-61779-376-9_38. [DOI] [PubMed] [Google Scholar]
- 24.Plocik A.M., Guthrie C. Diverse forms of RPS9 splicing are part of an evolving autoregulatory circuit. PLoS Genet. 2012;8:e1002620. doi: 10.1371/journal.pgen.1002620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bosio M.C., Negri R., Dieci G. Promoter architectures in the yeast ribosomal expression program. Transcription. 2011;2:71–77. doi: 10.4161/trns.2.2.14486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goncalves P.M., Griffioen G., Minnee R., Bosma M., Kraakman L.S., Mager W.H., Planta R.J. Transcription activation of yeast ribosomal protein genes requires additional elements apart from binding sites for Abf1p or Rap1p. Nucleic Acids Res. 1995;23:1475–1480. doi: 10.1093/nar/23.9.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li B., Vilardell J., Warner J.R. An RNA structure involved in feedback regulation of splicing and of translation is critical for biological fitness. Proc. Natl. Acad. Sci. U.S.A. 1996;93:1596–1600. doi: 10.1073/pnas.93.4.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vilardell J., Warner J.R. Ribosomal protein L32 of Saccharomyces cerevisiae influences both the splicing of its own transcript and the processing of rRNA. Mol. Cell. Biol. 1997;17:1959–1965. doi: 10.1128/mcb.17.4.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.AbuQattam A., Gallego J., Rodriguez-Navarro S. An intronic RNA structure modulates expression of the mRNA biogenesis factor Sus1. RNA. 2016;22:75–86. doi: 10.1261/rna.054049.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Howe K.J., Ares M., Jr Intron self-complementarity enforces exon inclusion in a yeast pre-mRNA. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12467–12472. doi: 10.1073/pnas.94.23.12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ben-Shem A., Jenner L., Yusupova G., Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 33.Neugebauer K.M. On the importance of being co-transcriptional. J. Cell Sci. 2002;115:3865–3871. doi: 10.1242/jcs.00073. [DOI] [PubMed] [Google Scholar]
- 34.Aitken S., Alexander R.D., Beggs J.D. Modelling reveals kinetic advantages of co-transcriptional splicing. PLoS Comput. Biol. 2011;7:e1002215. doi: 10.1371/journal.pcbi.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lipson D., Raz T., Kieu A., Jones D.R., Giladi E., Thayer E., Thompson J.F., Letovsky S., Milos P., Causey M. Quantification of the yeast transcriptome by single-molecule sequencing. Nat. Biotechnol. 2009;27:652–658. doi: 10.1038/nbt.1551. [DOI] [PubMed] [Google Scholar]
- 36.Neuveglise C., Marck C., Gaillardin C. The intronome of budding yeasts. C. R. Biol. 2011;334:662–670. doi: 10.1016/j.crvi.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 37.Li Z., Paulovich A.G., Woolford J.L., Jr Feedback inhibition of the yeast ribosomal protein gene CRY2 is mediated by the nucleotide sequence and secondary structure of CRY2 pre-mRNA. Mol. Cell. Biol. 1995;15:6454–6464. doi: 10.1128/mcb.15.11.6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.