Abstract
RNase H1-dependent antisense oligonucleotides (ASOs) are chemically modified to enhance pharmacological properties. Major modifications include phosphorothioate (PS) backbone and different 2′-modifications in 2–5 nucleotides at each end (wing) of an ASO. Chemical modifications can affect protein binding and understanding ASO-protein interactions is important for better drug design. Recently we identified many intracellular ASO-binding proteins and found that protein binding could affect ASO potency. Here, we analyzed the structure-activity-relationships of ASO-protein interactions and found 2′-modifications significantly affected protein binding, including La, P54nrb and NPM. PS-ASOs containing more hydrophobic 2′-modifications exhibit higher affinity for proteins in general, although certain proteins, e.g. Ku70/Ku80 and TCP1, are less affected by 2′-modifications. We found that Hsp90 protein binds PS-ASOs containing locked-nucleic-acid (LNA) or constrained-ethyl-bicyclic-nucleic-acid ((S)-cEt) modifications much more avidly than 2′-O-methoxyethyl (MOE). ASOs bind the mid-domain of Hsp90 protein. Hsp90 interacts with more hydrophobic 2′ modifications, e.g. (S)-cEt or LNA, in the 5′-wing of the ASO. Reduction of Hsp90 protein decreased activity of PS-ASOs with 5′-LNA or 5′-cEt wings, but not with 5′-MOE wing. Together, our results indicate Hsp90 protein enhances the activity of PS/LNA or PS/(S)-cEt ASOs, and imply that altering protein binding of ASOs using different chemical modifications can improve therapeutic performance of PS-ASOs.
INTRODUCTION
Antisense oligonucleotides (ASOs) that direct RNase H1-mediated cleavage of complementary RNAs have been widely used as research tools and therapeutic agents to regulate gene expression (1–3). After decades of development, the new generation RNase H1-dependent ASO drugs are composed of 14–20 nucleotides in length and are constructed as a chimeric ‘gapmer’ structure. The gapmer ASOs usually contain 10 (or fewer) deoxynucleotides in the central region to support RNase H1 activity, flanked at both ends (wings) by several nucleotides modified at the 2′ position of the ribose with different chemical groups. Commonly used 2′-modifications include, but are not limited to, 2′-O-methyl (Me), 2′-O-methoxyethyl (MOE), locked-nucleic-acid (LNA), constrained-ethyl-bicyclic-nucleic-acid ((S)-cEt, hereafter referred to as cEt) and 2′-fluoro (F) (1,4,5). To increase nuclease resistance and the pharmacological features of ASOs, phosphodiester (PO) linkages of nucleotides are replaced with phosphorothioate (PS).
PS-ASOs were shown to bind plasma proteins (e.g. albumin, α2-macroglobulin) that prevent ASOs from being rapidly discarded through urinary excretion, and enhance cellular uptake both in vitro and in vivo as compared with PO-ASOs (6,7). PS-ASOs enter cells largely through endocytic pathways and can be released from endocytic particles into cytosol/nucleus to act on complementary RNAs by base-pairing (8–10). In addition to the PS backbone modification, various 2′-modifications can also affect ASO activity, likely by increasing ASO/RNA binding affinity. For example, it has been demonstrated that LNA or cEt modified gapmer PS-ASOs (referred to as PS/LNA or PS/cEt ASOs, respectively) are typically more potent compared with 2′-MOE ASOs (designated as PS/MOE ASOs) (11–13). LNA and cEts can increase melting temperature (Tm) ∼3.5°C per modification, whereas MOE increases ∼1–2°C per modification (14,15), suggesting a better affinity of PS/LNA ASOs to target RNAs in contrast to PS/MOE ASOs. This increased ASO/RNA affinity not only increases potency but increases the number of sites in a target RNA that are accessible to binding by ASOs (16). However, increasing Tm seems to not always be beneficial, since ASOs with five LNA modified nucleotides at both wings flanking a 10-deoxynucleotides portion (5–10–5) appeared less active than a 3–10–3 LNA ASO (15). These results suggest that other factors, in addition to binding affinity with RNA target, also contribute to ASO activity. These factors may include the properties of the modified ASOs that affect uptake, release from endocytic pathways and protein binding.
Compared with PO-ASOs, PS-ASOs can bind many more extracellular or intracellular proteins, including plasma proteins such as albumin and some growth factors, and intracellular proteins such as nucleic acid binding proteins (3,17–19). Due to the physicochemical difference between sulfur and oxygen atom in the PO backbone, such as van der Waal's radius and electronegativity, the sulfur in PS-ASO can participate in stronger hydrogen bonding than the equivalent PO-ASO (20), allowing binding of PS-ASOs to many proteins (21).
Proteins that bind ASOs may affect ASO potency in many ways, e.g. by altering ASO distribution in vivo, subcellular localization, ability to base-pair with target RNA and recruitment of RNase H1 protein for cleavage. Recently we identified dozens of intracellular proteins that bind PS-ASOs, and showed that some proteins can affect ASO activity. For example, La, NPM1, ANXA2, PC4 and TCP1 proteins enhance ASO activity, whereas P54nrb, PSPC1, PSF, Ku70 and Ku80 proteins can inhibit ASO activity (21–23). These proteins may influence ASO action through different mechanisms. For instance, Ku70 and P54nrb proteins can compete with RNase H1 for binding to ASO/RNA duplex, thus reducing target RNA cleavage (21). On the other hand, La and NPM1 proteins may facilitate nuclear ASO retention, thus enhancing ASO activities in the nucleus (21). Some other proteins, such as chaperone protein TCP1β and paraspeckle proteins P54nrb, PSF and PSPC1, can co-localize with ASOs in certain nuclear bodies and may thus regulate ASO availability (22,23). In addition, we also found that 2′-Fluoro-modified ASOs can reduce levels of some paraspeckle proteins, leading to cell death (24). Moreover, Hsp70 protein has been shown to interact with ASO/RNA duplexes and inhibit RNase H1 cleavage (25). These results clearly indicate that ASO binding proteins can play important roles in modulating ASO activity. However, information about ASO-protein interactions is still very limited.
In addition to the PS modification, other chemical modifications with different physicochemical properties may also affect protein binding to the ASOs. Indeed, we recently found that while the binding of Ku70 to PS-ASOs were less affected by the types of 2′-modifications, several other ASO binding proteins, including TCP1β and P54nrb, bound more tightly to PS-ASOs containing 2′-F, cEt or LNA modified wings, relative to 2′-MOE modified wings (22,23). Since 2′-F, cEt and LNA modifications are more hydrophobic than 2′-MOE modification, it is likely that the hydrophobicity of 2′-modifications affects protein binding. However, it is not clear how the 2′-modifications affect protein binding at a global level and whether some protein(s) interact specifically with ASOs containing certain 2′-modifications.
In the current study, we analyzed the effect of PS backbone and 2′-modifications on protein binding, and found that in general, significant protein binding requires 10 or more PS-backbone nucleotides, and that 2′-modifications also significantly affect protein binding. PS-ASOs containing more hydrophobic 2′-modifications bind more tightly and more promiscuously to cellular proteins. Interestingly, we found that Hsp90 protein interacts with PS-ASOs containing LNA or cEt modified wings, but not MOE modified wings. Mutational analyses revealed that the mid-domain of the Hsp90α protein binds to ASOs, and Hsp90 binding requires 5′-cEt wing, and not 3′-cEt wing of a PS-ASO. Consistently, Hsp90 enhances the antisense activity of PS-ASOs containing 5′-cEt wings, but not 5′-MOE wing. To our knowledge, this is the first study showing that a protein can bind specifically to PS-ASOs with certain 2′-modifications. Together, our results indicate that the 2′-modifications of ASOs can significantly affect protein binding, which in turn, can affect the antisense activity of ASOs.
MATERIALS AND METHODS
Antibodies, siRNAs, ASOs and primer probe sets used for qRT-PCR are listed in Supplementary Data
Construction
Full length Hsp90α (NM_005348.3) cDNA was purchased from GENECOPOEIA (EX-M0233-M06) and used as a template for PCR-based cloning. Plasmids expressing different Hsp90α domains containing HA-tag at C-terminus were established as followings: N-terminal domain was amplified by PCR using primers XL222 (5′-CGCGGATCCATGCCTGAGGAAACCCAGACCCA-3′) and XL223 (5′-GCACTCGAGTTAATAATCTGGAACATCATATGGATATTCAGCCT CATCATCGCTTACTTC-3′). Mid-domain was amplified using primers XL224 (5′- CGC GGATCCATGGCTGAAGAAAAGGAAGACAAAGAAGAAG-3′) and XL225 (5′-GCACTCGAG TTAATAATCTGGAACATCATATGGATATGCCATGTAACCCATTGTTGAG-3′). C-terminal domain was amplified using primers XL226 (5′-CGCGGATCCATGGCAGCAAAGAAACA CCTGGAGA-3′) and XL227 (5′- GCACTCGAGTTAATAATCTGGAACATCATATGGATA GTCTACTTCTTCCATGCGTGATGT-3′). The full length Hsp90α coding region was amplified using primers XL222 and XL227. The PCR products were digested with BamH I and XhoI and ligated into vector pcDNA4 (Invitrogen), generating expression plasmids for different domains of Hsp90α. The sequence-confirmed plasmids were transfected into HeLa cells (2 μg/15 cm dish) using Effectene (Qiagen), based on manufacturer's instruction. Whole cell lysate was prepared 48 h after transfection and used for Western analysis or affinity selection with biotinylated ASOs.
Cell culture, siRNA and ASO transfection
HeLa or A431 cells were grown as described elsewhere. One day before transfection, cells were re-seeded at ∼50–70% confluency in complete Dulbecco's modified Eagle's medium (DMEM) without antibiotics. Transfection of siRNA and ASOs was carried out as described previously (21). Briefly, siRNA was transfected at 3 nM final concentration using Lipofectamine RNAiMax (Life Technologies), based on manufacturer's instructions. Eight or 24 h after transfection, cells were reseeded at ∼60% confluency in 24-well plates and incubated for 16 h. Next, RNase H1-dependent gapmer ASOs were transfected into cells using Lipofectamine 2000 (Life Technologies), at different final concentrations as indicated in Figures 6 and 7. Four hours after ASO transfection, cells were harvested and RNA or protein was prepared for subsequent analyses. For free uptake, ASOs were directly added to culture medium for 12–18 h. For Hsp90α over-expression, an empty vector (pcDNA4) or the plasmid expressing the full length Hsp90α was transfected into HeLa cells (0.8 μg/10 cm dish) using Effectene. After 36 h, cells were reseeded in 24-well plates, and incubated for 12 h. Next, PS/MOE or PS/cET ASOs were transfected using Lipofectamine 2000 for 4 h, and cells were collected for RNA preparation and qRT-PCR analyses.
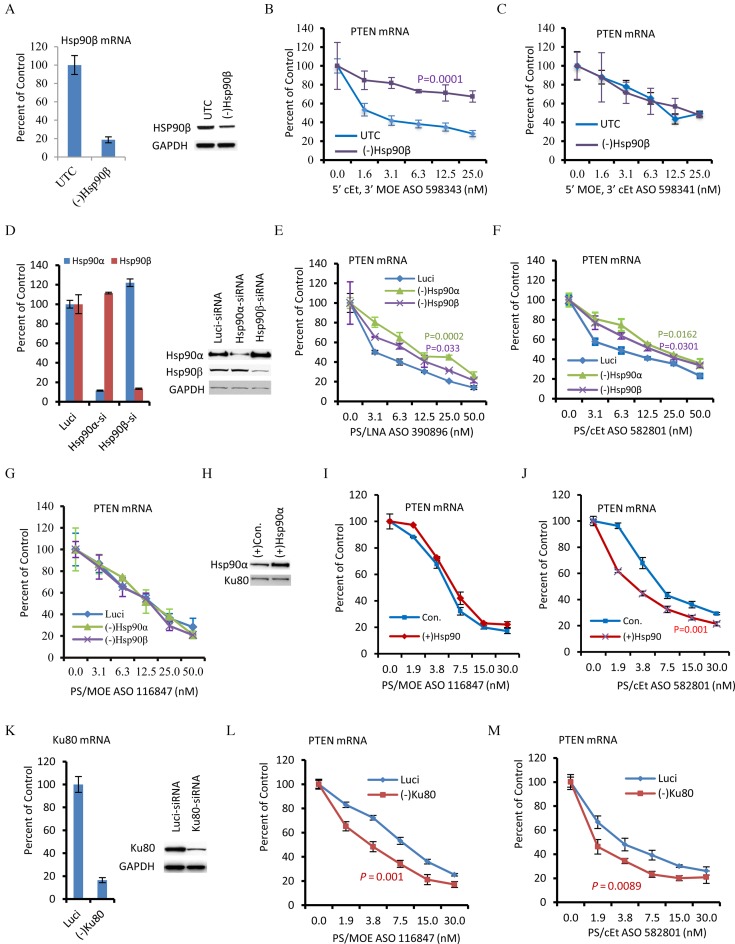
Figure 6.
Hsp90 protein enhances antisense activity of PS-ASOs. (A) The levels of Hsp90β mRNA and protein were reduced in HeLa cells by siRNA (S7001) treatment. Left panel, qRT-PCR analyses for Hsp90β mRNA in mock transfected cells (UTC) or cells treated for 24 h with a Hsp90β specific siRNA. Right panel, Western analyses for Hsp90β protein in control and siRNA treated cells. GAPDH was detected and served as a loading control. (B) qRT-PCR for PTEN mRNA levels in control (UTC) or Hsp90β reduced ((-)Hsp90β) cells transfected for 4 h with different concentrations of a 5–10–5 PS/cEt-MOE gapmer ASO (598343) targeting PTEN mRNA. (C) qRT-PCR for PTEN mRNA levels in cells transfected with a 5–10–5 PS/MOE-cEt ASO (598341) targeting the PTEN mRNA, as in panel B. (D) The mRNA and protein levels of Hsp90α and Hsp90β were specifically reduced by the treatment with corresponding siRNAs (S6995 for Hsp90α, 284645 for Hsp90β), as compared with cells treated with a control luciferase siRNA (Luci). qRT-PCR and Western analysis were performed as in panel A. (E–G) qRT-PCR for PTEN mRNA levels in different cells transfected with PTEN-targeting 5–10–5 gapmer PS/LNA, PS/cEt, and PS/MOE ASOs, respectively. (H) Western analyses for Hsp90α protein in cells transfected with the expression plasmid [(+)Hsp90α] or an empty vector [(+)Con.]. Ku80 was detected and served as a control for loading. (I) qRT-PCR analyses for PTEN mRNA levels in different test cells transfected with ASO116847 for 4 h. (J) qRT-PCR analyses for PTEN mRNA levels in test cells transfected with ASO582801 for 4 h. (K) siRNA-mediated reduction of Ku80 mRNA and protein in HeLa cells was confirmed by qRT-PCR (left panel) and western analyses (right panel). Luci, cells transfected with a control luciferase siRNA. GAPDH was detected and used as a loading control for Western analysis. (L–M) qRT-PCR for PTEN mRNAs in different cells transfected with 5–10–5 PS/MOE (Panel L) or 5–10–5 PS/cEt (Panel M) ASOs, respectively. The experiments were repeated more than 3 times, and representative results are given. In all panels, the error bars represent standard deviation from three independent experiments. The P-values were calculated using Prism with F-test (curve comparison).
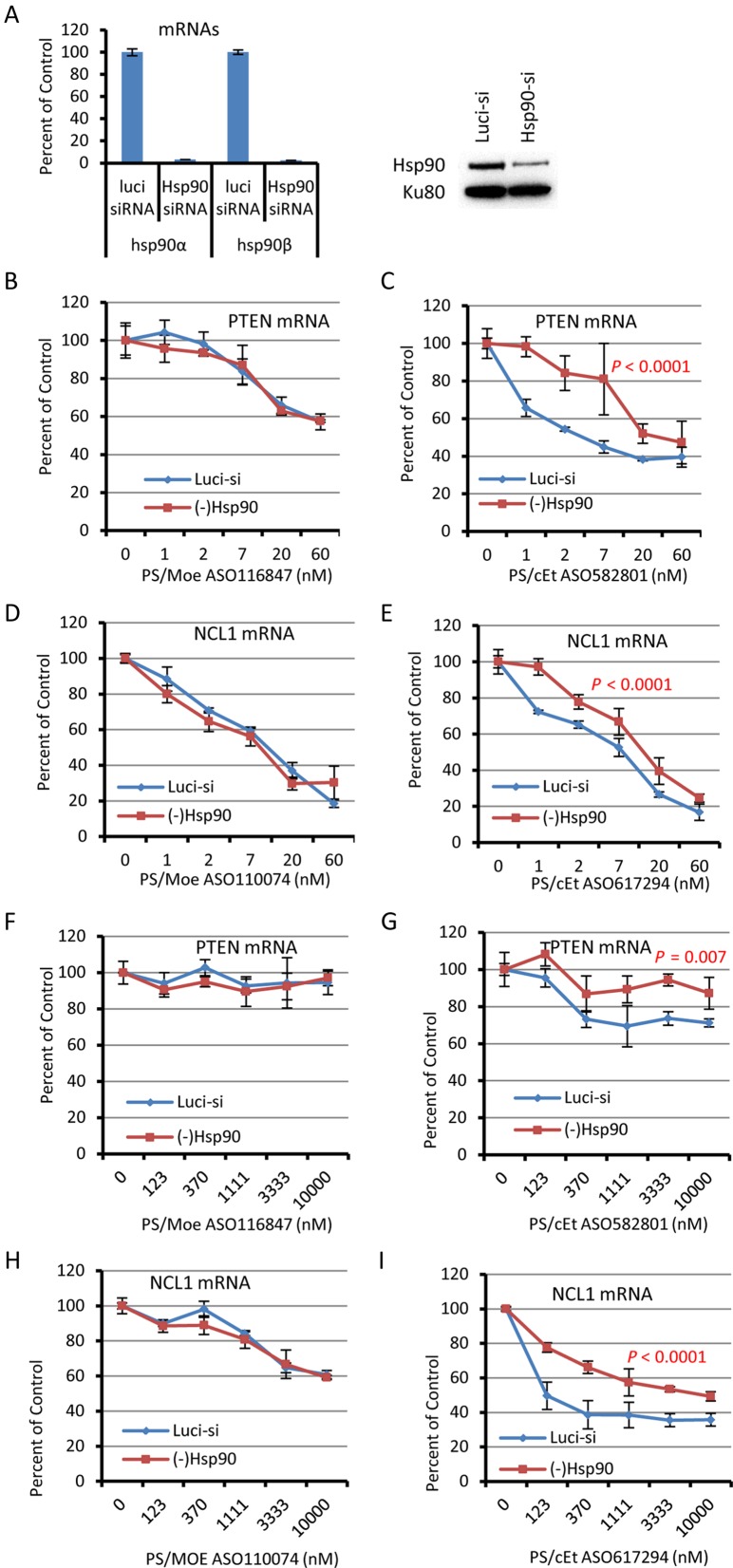
Figure 7.
Reduction of Hsp90 in A431 cells reduced the activity of PS/cEt, but not PS/MOE ASOs with different sequences. (A) Reduction of Hsp90 by siRNA treatment in A431 cells. Left panel, qRT-PCR analyses were performed for the levels of Hsp90 mRNAs in A431 cells treated by 10 nM Luciferase siRNA (luci-siRNA) or by combination of 5 nM Hsp90α siRNA and 5 nM Hsp90β siRNA for 48 h. Right panel, Western assay for Hsp90 proteins using an antibody recognizes both Hsp90α and Hsp90β. Ku80 was detected and served as a loading control. (B and C) qRT-PCR results for PTEN mRNA levels in control or Hsp90 reduced cells that were transfected for 4 h with the PTEN specific PS/MOE ASO (Panel B) or PS/cEt ASO (Panel C), respectively. (D and E) qRT-PCR analyses for NCL1 mRNA levels in control or Hsp90 reduced cells that were transfected for 4 h with NCL1 specific ASOs with PS/MOE (panel D) or PS/cEt (Panel E) modifications. (F and G) The PTEN mRNA levels were determined by qRT-PCR in control or Hsp90 reduced cells that were treated for 16 h by free uptake with PTEN PS/MOE (Panel F) or PS/cEt (panel G) ASOs, respectively. (H and I) qRT-PCR results for NCL1 mRNA in control or Hsp90 reduced cells that were treated by free uptake with NCL1 PS/MOE (Panel H) or PS/cEt (Panel I) ASOs, respectively. In all panels, the error bars represent standard deviation from three independent experiments. The P-values were calculated using Prism with F-test (curve comparison).
Quantitative RT-PCR (qRT-PCR)
qRT-PCR using TaqMan prime probe sets was performed essentially as described previously (26), except using AgPath-ID Onestep RT-PCR kit (Ambion). Briefly, ∼50 ng total RNA in 5 μl water was mixed with 0.3 μl primer probe sets containing forward and reverse primers (10 μM of each), and fluorescently labeled probe (3 μM), 0.3 μl RT enzyme mix (Qiagen), 4.4 μl RNase-free water and 10 μl of 2x PCR reaction buffer in a 20 μl reaction. Reverse transcription was performed at 48°C for 10 mins, and 40 cycles of PCR reaction were conducted at 94°C for 20 s and 60°C for 20 s within each cycle, using Stepone Plus RT-PCR system (Applied Biosystems). The mRNA levels were normalized to the total RNA present in each reaction as determined for duplicate RNA samples by Ribogreen assay (Invitrogen).
Affinity selection of ASO-binding proteins
Affinity selection of ASO-binding proteins was performed as described previously (22). Briefly, 400 μl agarose neutravidin beads (Thermo Scientific) were incubated with 400 μl of 200-μM biotinylated ASO ISIS586183 at 4°C for 2 h in W-100 buffer [50 mM Tris–HCl (pH 7.5), 100 mM KCl, 5 mM EDTA, 0.1% NP-40, 0.05% SDS]. Beads were then incubated for 30 mins in block buffer [10 mg/ml BSA, 1.2 mg/ml glycogen and 0.2 mg/ml yeast tRNA in W-100]. After three washes with W-100, ASO-coated beads were incubated at 4°C for 3 h with 2 mg HeLa cell extract prepared in buffer A [25-mM Tris-HCl pH 8.0, 5-mM MgCl2, 150-mM KCl, 10% glycerol, 0.5-mM PMSF, 5-mM β-mercaptoethanol and one tablet of Protease Inhibitor Cocktail/50 ml (Roche)]. After 7 washes with 1 ml W-300 buffer [50-mM Tris-HCl (pH 7.5), 300-mM KCl, 5-mM EDTA, 0.1% NP-40, 0.05% SDS], 8 aliquots were taken from the beads, and bound proteins for each aliquot were eluted by incubation for 30 mins at room temperature with 100 μl of 50-μM ASO containing different modifications, as indicated in figures. The eluted proteins were precipitated, separated on 4–12% polyacrylamide gel and visualized by silver staining, or analyzed by Western assay. Interested protein bands were excised and identified by mass spectrometry (Alphalyse, CA, USA). Briefly, the samples were digested by trypsin, and the digested peptides were concentrated and analyzed on a Bruker Autoflex Speed MALDI TOF/TOF instrument.
MALDI MS/MS was conducted on 15 peptides for peptide fragmentation analysis. The MS and MS/MS spectra were combined and used to search database with the Mascot software. Alternatively, aliquots of isolated proteins were analyzed by Western blotting.
ASO-Protein binding analyses
In general, ASO-binding proteins were isolated by affinity selection using biotinylated PS-ASOs containing different 2′-modifications, as described above. After binding and washing, bound proteins were either eluted by competition using non-biotinylated ASOs (Figure 1B and C; Figure 2A and B; Figure 3D, E and F; and Figure 4), or directly analyzed by SDS-PAGE by boiling the samples in SDS-buffer (Figure 2C, D, E and G; Figure 3A, B and C). To determine protein binding on ASO/RNA duplex, Biotinylated PS/cEt ASO was annealed with complementary PO-backbone, 2′-O-methylated oligonucleotide XL279, in a 1:1.2 ratio. The annealed duplex was used in affinity selection and isolated proteins were directly eluted using SDS-buffer and analyzed by Western assay. To determine the effects of PS backbone on protein binding, affinity selection was performed essentially as described above, except that 120-μl beads were pre-coated with 120 μl of 200-μM ASO 586183. After binding and washing, beads in W-100 buffer were equally separated into four aliquots and transferred to 1-ml columns. After removal of W-100 by centrifugation, proteins were eluted with 100 μl of 50-μM ASOs with different lengths or different numbers of PS modified nucleotides, as indicated in figures. Eluted proteins were precipitated and analyzed by Western blotting.
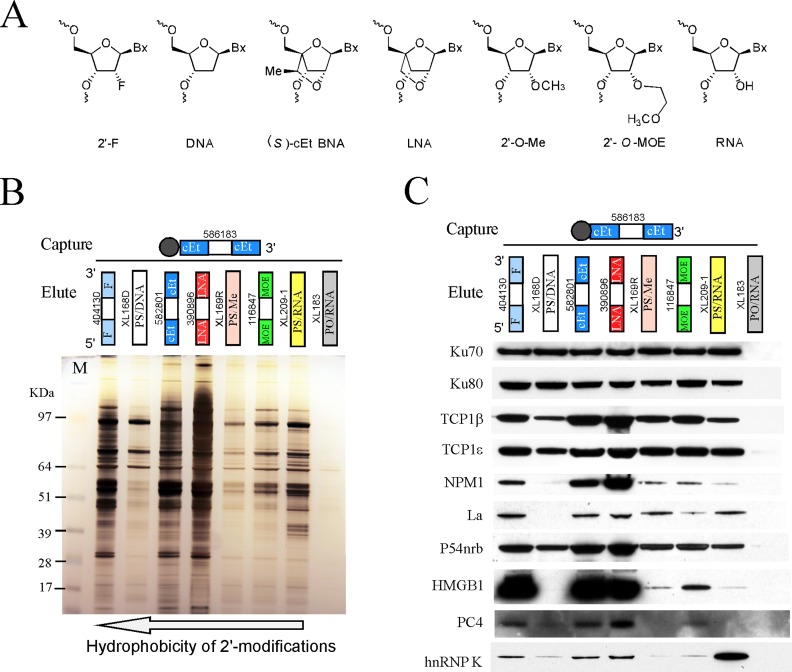
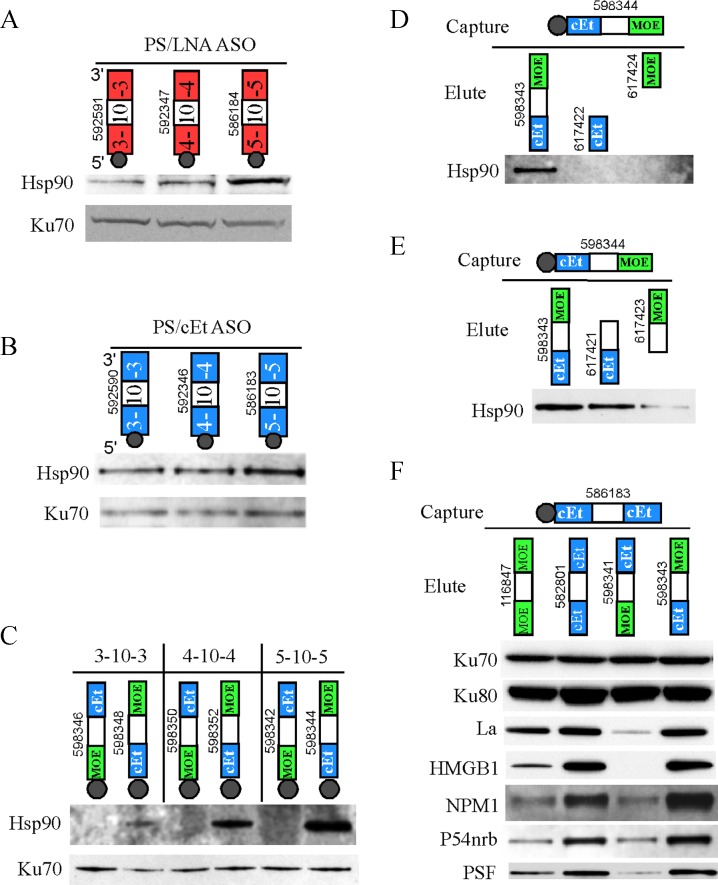
Figure 1.
2′-modification of PS-ASOs significantly affects protein binding. (A) The structures of different 2′-modifications. Bx, base of a nucleotide. Me, 2′-O-methyl. (B) Affinity selection of ASO binding proteins was performed as described in Material and Methods. Bound proteins were separated in 4–12% gradient SDS-PAGE, and visualized by silver staining. The ID numbers of ASOs used for affinity selection and elution are shown. The portions of ASOs with different modifications are indicated by colored boxes. The white box in the middle of gapmer ASOs indicates deoxynucleotides. The dark gray circle at the end of ASOs represents biotin. The experiments were repeated more than three times and representative results are shown. (C) Aliquots of isolated ASO-binding proteins as used in panel B was analyzed by Western analyses. The same blot was probed sequentially with antibodies against different ASO-binding proteins.
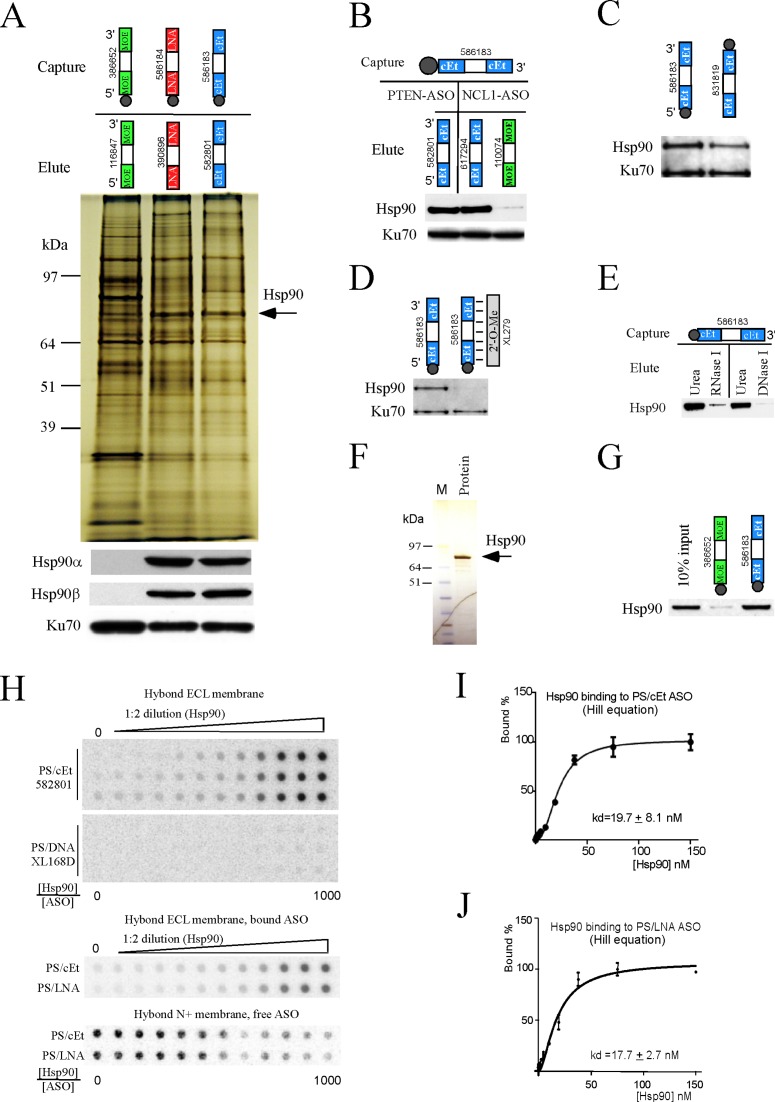
Figure 2.
Hsp90 protein binds PS/cEt or PS/LNA ASOs but not PS/MOE ASO. (A) Affinity selection was performed using biotinylated gapmer PS-ASOs with different 2′-modifications in the wings. Proteins were eluted using non-biotinylated PS-ASOs with the same chemical composition. Isolated proteins were separated by SDS-PAGE, and visualized by silver staining (upper panel), or detected by Western assay for a duplicate gel (lower panel) using antibodies specific to Hsp90α or HSP90β. Ku70 protein served as a control for loading. The protein band containing Hsp90 is indicated for the silver stained gel. (B) Western analysis for Hsp90 protein co-selected using a biotinylated PS/cEt ASO, and eluted by competition using PS/cEt ASOs with two different sequences that target PTEN or NCL1 mRNAs, or using a PS/MOE ASO targeting NCL1. The Hsp90 antibody recognizes both α and β isoforms. Ku70 protein was detected and served as a control. (C) Affinity selection using PS/cEt ASOs that have the same sequence and chemistry but tagged with biotin at either 5′ or 3′ end. Co-isolated Hsp90 and Ku70 were detected by western assay. (D) Affinity selection was carried out using either a single stranded PS/cEt ASO or a duplex formed using the same ASO and a complementary 2′-O-methylated oligonucleotide. Co-isolated proteins were directly separated on SDS-PAGE and analyzed by Western assay for Hsp90 and Ku70 proteins. (E) Affinity selection was conducted using either a PS/MOE ASO 386652 or a PS/cEt ASO 586183. After washing, bound proteins were first eluted by RNase I (5 U/μl) or DNase I (5 U/μl) treatment for 30 min at RT, followed by elution using 9M Urea from the same beads for 30 min at RT. The eluted proteins were precipitated and analyzed by Western assay for Hsp90 protein. (F) Silver staining of one μg of the recombinant Hsp90α protein (Abcam, ab80369). (G) Interaction of the recombinant Hsp90α protein with PS/cEt ASO was determined by affinity selection using biotinylated PS-ASOs and 3 μg of purified Hsp90 protein. After wash, the beads were directly loaded on a 4–12% SDS-PAGE, and Hsp90 protein was detected by Western assay. The ASO ID numbers in each panel are shown. The above experiments were repeated at least three times and representative results are shown. (H) Membrane binding assay for Hsp90α protein and different PS-ASOs, as described in Materials and Methods. For each ASO, triplicate binding reactions were performed. The molar ratio between Hsp90a protein and ASO was given below the wells. An example of a double-filter binding for PS/cEt and PS/LNA ASOs is shown in the middle panel for protein-bound ASOs captured using a Hybond ECL membrane, and the lower panel for unbound ASOs captured using a Hybond-N+ nylon membrane. The signal intensity for the ASOs was quantified and the binding curves for (I) PS/cEt and (J) PS/LNA ASOs were plotted using Prism. The calculated kds are given. The error bars represent standard deviations from three experiments.
Figure 3.
Hsp90 protein binds to the 5′-cEt wing of PS-ASOs. (A) Affinity selection was performed using biotinylated PS/LNA gapmer ASOs containing different lengths of LNA modified wings. Co-selected proteins were directly separated on SDS-PAGE for Western analyses. (B) Affinity selection and Western analyses were performed as in panel A, except that PS/cEt gapmer ASOs with different wing lengths were used. (C) Affinity selection was performed as in panel A, using biotinylated gapmer PS-ASOs with mixed wings, either 5′-cEt + 3′-MOE or vice versa. Isolated proteins were analyzed by Western assay for Hsp90 and Ku70 proteins. (D) Co-selected proteins using a biotinylated 5–10–5 PS-ASO containing 5′-cEt + 3′-MOE mixed wings were eluted by competition using either the same 5–10–5 ASO but without biotin, or with 5-nt oligonucleotides representing 5′-cEt or 3′-MOE wings. The eluted Hsp90 protein was detected by Western assay. (E) Affinity selection and Western assay were performed as in panel D, except that different elution ASOs were used. (F) Co-selected proteins using a 5–10–5 PS/cEt ASO were eluted by competition using 5–10–5 gapmer PS-ASOs with different wing modifications, and analyzed by Western assay. The same membrane was probed sequentially for different proteins. The ASO ID numbers in each panel are shown. The above experiments were repeated at least three times and representative results are shown.
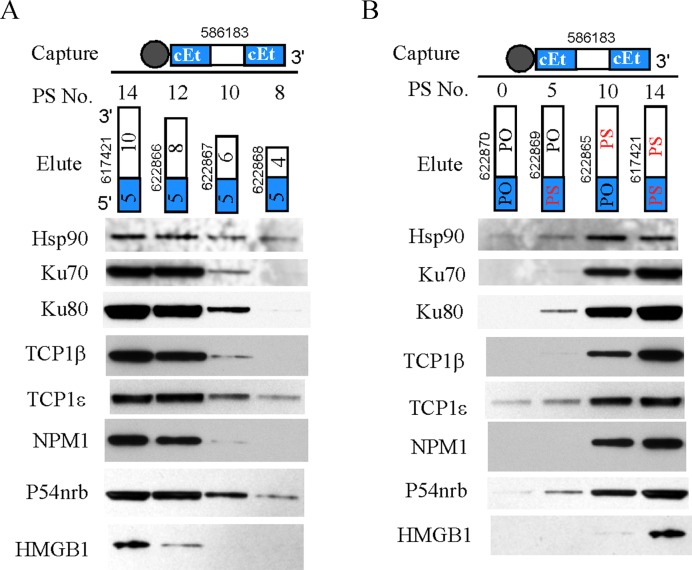
Figure 4.
Significant protein binding requires 10 or more PS-modified nucleotides. (A) Affinity selection was conducted using the 5–10–5 PS/cEt ASO, and bound proteins were eluted using different PS-ASOs containing only the 5′-cEt wing and different numbers of deoxynucleotides, as indicated. Eluted proteins were separated by SDS-PAGE and sequentially analyzed by Western assay for different proteins. The numbers of PS modified nucleotides are shown above each elution ASO. The numbers in the boxes of elution ASOs represent the length of the portion. (B) Affinity selection and Western analyses were performed as in panel A, except different elusion ASOs were used. The elution ASOs are all 15-mer (5–10) containing 5-nt 5′-cEt wing and 10 deoxynucleotides. These ASOs have different PS/PO backbone combinations, as indicated. The numbers of PS modified nucleotides are given above each elution ASO. The blue and white boxes indicate cEt modified wing and deoxynucleotides, respectively. The experiments were repeated twice and representative results are shown.
Measuring the binding constant (kd) of Hsp90 to PS-ASOs
ASO/protein binding was performed in 100 μl binding buffer (20 mM Tris.Cl, pH 7.5; 150 mM NaCl; 1 mM DTT; 10% Glycerol). Each reaction contained increasing amounts of purified recombinant Hsp90α protein (Abcam, ab80369), from 0 to 75 nM with 1:2 dilution in 12 reactions, and 0.075 nM of 5′-32P-labeled ASO582801 (PS/cEt), or ASO390896 (PS/LNA), or XL168D (PS/DNA, as a negative control). After 1 h incubation at 37°C, the samples were loaded on a Hybond ECL nitrocellulose membrane (GE Healthcare) pre-treated for 1 h with wash buffer (20 mM Tris.Cl, pH 7.5; 1 M NaCl) in a 96-well dot-blot apparatus (Bio-Rad Bio-Dot), and the protein-bound ASOs were transferred to the membrane by applying vacuum. Unbound ASOs were removed by washing the membrane 3 times with 500 μl of wash buffer each time. For double-filter binding experiments, the binding reaction was performed as described above. The samples were loaded on the Hybond ECL nitrocellulose membrane to trap proteins and ASO–protein complexes, and the free ASOs were trapped by a Hybond-N+ nylon membrane under the nitrocellulose membrane. After washing, the membranes were then air-dried, and exposed to a PhosphorImager plate. The signal intensity was quantified using ImageJ and the results from three independent experiments were fitted as Hill Slope and plotted, and the kds were calculated using Prism.
Western analysis
Whole cell extracts (∼20 μg) or proteins affinity selected using ASOs were separated in 4–12% gradient SDS-PAGE gels. Proteins were transferred to Nitrocellulose membrane using iBlot gel transfer system (Life Technologies). The membranes were blocked for 1 h using block buffer (5% dry milk in 1x TBS), and incubated with primary antibodies against different proteins (1:500–1:2000) for 2 h at room temperature, or overnight at 4°C. After three 5-min washes with wash buffer (1x TBS, 0.1% Tween-20), membranes were incubated with anti-mouse or anti-rabbit secondary antibody (1:2000) in block buffer at room temperature for 1 h. After three washes, proteins were detected using ECL (Abcam). For subsequent detection of other proteins from the same blot, the membrane was stripped out using Re-blot Plus Strong Solution (Millipore) at 4°C for 30 mins, and re-probed with additional antibodies.
RESULTS
2′-modification can significantly affect the binding of PS-ASOs to intracellular proteins
To determine how 2′-modifications generally influence the binding of PS-ASOs to intracellular proteins, we performed affinity selection using a biotinylated 5–10–5 gapmer PS-ASO modified with cEt in the wings (ISIS586183), as described previously (22). Isolated proteins were eluted from the beads by competition using non-biotinylated PS-ASOs that have the same sequence as ISIS586183, but with different 2′-chemistries (Figure 1A), or using a PO-backbone oligoribonucleotide. The eluted proteins were separated using SDS-PAGE and visualized by silver staining (Figure 1B). Consistent with previous observations that PS-ASOs bind many more proteins than PO-ASOs (17,21), the PO-oligoribonucleotide eluted only a trace amount of proteins from the capture PS/cEt ASO, whereas significant amounts of proteins were eluted by different PS-ASOs. The low efficiency of the PO-oligoribonucleotide to elute proteins was not due to lower stability of this oligonucleotide compared with the PS-ASOs, as we previously demonstrated that the PO-oligoribonucleotide could survive the elution process (22).
In general, 5–10–5 PS-ASOs containing 2′-F, cEt, or LNA modifications in the wing nucleotides eluted many more proteins than the 5–10–5 PS/MOE ASO. Similarly, protein binding to PS/Me or PS/oligoribonucleotide ASOs was also weaker than to PS/cEt, PS/LNA or PS/F ASOs. 2′-F, cEt, and LNA are more hydrophobic than MOE and Me modifications. These results suggest that PS-ASOs with more hydrophobic 2′-functional groups bind more proteins. However, this is not always the case. The uniform PS-oligodeoxynucleotide that contains 2′-H in the ‘wing’ nucleotides bound fewer proteins than PS/F, PS/cEt and PS/LNA ASOs. Since 2′-H is more hydrophobic compared with cEt, the differences in protein binding may also be attributed to other properties of ASOs, such as the structures related to the modifications.
Although most ASO-binding proteins preferentially associate with PS/F, PS/cEt, and PS/LNA ASOs, similar to what we observed previously for TCP1β and P54nrb proteins (22,23), a few protein bands (∼64, 75 and 95 kDa) appeared to be less affected by 2′-modifications. It is therefore likely that changing 2′-modifications may have different impacts on the binding of different proteins. To evaluate this possibility, and to determine how 2′-modifications influence the binding of some proteins that affect ASO activity, as we previously showed (21–23), aliquots of isolated proteins used in panel B were analyzed by western assay (Figure 1C). In addition to Ku70, TCP1β, TCP1ϵ and P54nrb that we reported previously but were included here for comparison purpose (22,23), Ku80, La, NPM1, PC4, hnRNP K and HMGB1 were probed sequentially with the same Western blot.
Similar to what we observed for Ku70 (22), binding of Ku80 protein was also less sensitive than many proteins to changes in 2′-modifications of PS-ASOs, including 5–10–5 gapmer ASOs (PS/F, PS/cEt, PS/LNA or PS/MOE) and uniformly modified PS-ASOs (e.g. PS/oligodeoxynucleotide, PS/oligoribonucleotide or PS/Me ASOs). This is in agreement with the fact that Ku70 and Ku80 exist as a heterodimeric complex (27). However, NPM1, La, PC4, hnRNP K, and HMGB1 all showed stronger binding to PS/F, PS/cEt, and PS/LNA ASOs, compared to PS/MOE ASOs. This is similar to the binding properties of P54nrb and TCP1β proteins (22,23). Although 2′-F, cEt, and LNA modifications are more hydrophobic than MOE or Me, the difference in protein binding is unlikely determined solely by hydrophobicity, as described above. The uniform PS/oligodeoxynucleotide ASO, which has higher hydrophobicity than the PS/cEt ASO, failed to bind significantly to NPM1, La, and HMGB1 proteins. In addition, the degree of variation in binding of NPM1 (and HMGB1) to PS/LNA and to PS/Me ASOs was much greater than the binding difference of La protein to these ASOs, suggesting that different proteins can be affected differentially by 2′-modifications. This is further supported by the observations that both La and hnRNP K proteins bound more tightly to PS/oligoribonucleotide ASOs compared to PS/MOE ASOs, while the opposite effect was found for NPM1 and HMGB1 proteins. Together, these results showed that in general, PS-ASOs with more hydrophobic 2′-modifications tend to bind more proteins, and different proteins can have different responses to changes of 2′-modifications.
Hsp90 protein binds PS/cEt and PS/LNA ASOs but not PS/MOE ASOs
PS/cEt and PS/LNA gapmer ASOs exhibit higher potencies than the PS/MOE gapmer ASOs (13). Since many proteins preferentially bind PS/cEt or PS/LNA ASOs rather than PS/MOE ASOs, we wondered if some proteins specifically interact with PS-ASOs containing certain types of 2′-modifications, and thus contribute to the observed differences in antisense activity. For this purpose, affinity selection was performed using biotinylated gapmer PS-ASOs containing 2′-MOE, cEt or LNA modified wings, and isolated proteins were eluted using corresponding PS-ASOs without biotin. These modifications were tested because they are most commonly used in gapmer ASOs.
The co-selected proteins were separated by SDS-PAGE, and visualized by silver staining (Figure 2A, upper panel). Aliquots of proteins were separated in a duplicate gel for Western analysis (Figure 2A, lower panel). One protein band, at ∼80–90 kDa, was enriched in samples isolated using PS/cEt or PS/LNA ASOs, although more proteins were loaded in the PS/MOE ASO selected samples. This band was identified by mass spectrometry to contain several proteins, including Hsp90α and Hsp90β, two isoforms of Hsp90 protein. The mass spectrometry result was confirmed by Western analysis (lower panel). These Hsp90 proteins were detected using specific antibodies in samples isolated with PS/cEt or PS/LNA ASOs, but not with the PS/MOE ASO. As a control, Ku70 was detected in all samples, consistent with previous observations that Ku70 binding is less affected by 2′-modifications (Figure 1C). These results indicate that Hsp90 protein specifically interacts with PS/cEt and PS/LNA ASOs, but not significantly with PS/MOE ASO. Although a protein band at the same size as the Hsp90 protein was detected by silver staining in the PS/MOE ASO isolated sample, that band must represent other ASO-binding proteins, such as Ku80 (22), which was also detected by mass spectrometry to be present in the same protein band containing Hsp90 in the PS/cEt and PS/LNA ASO isolated samples (data not shown).
The binding of Hsp90 to PS/cEt and not to PS/MOE gapmer ASOs were also confirmed using ASOs with different sequences (Figure 2B). After affinity selection using ASO 586183, Hsp90 protein was successfully eluted by PS/cEt ASO 582801 and PS/cEt ASO 617294, but not by PS/MOE ASO 110074, as determined by Western blotting using an antibody that recognizes both Hsp90α and Hsp90β. The latter two 5–10–5 gapmer PS-ASOs share the same sequence targeting NCL1 mRNA, but are different from the sequence of 582801 that targets PTEN mRNA. This result suggests that the binding of Hsp90 is not unique to a particular ASO sequence.
Next, we evaluated the interaction properties between Hsp90 protein and ASOs using affinity selection and Western analyses. The results showed that Hsp90 protein was able to be co-selected with PS/cEt ASOs tagged with biotin at either 5′ or 3′ end, suggesting that the binding was not blocked by biotin positions (Figure 2C). In addition, Hsp90 protein bound single stranded PS/cEt ASO, but not duplexes since an PS/cEt ASO/RNA duplex failed to co-select Hsp90 protein (Figure 2D), whereas Ku70 protein was isolated using either a single stranded ASO or a duplex, consistent with our previous observations (21). Moreover, the physical interaction between PS/cEt ASOs and Hsp90 protein appeared not to be linked by DNA or RNA, as Hsp90 protein could not be significantly eluted from beads using either DNase or RNase treatment, but was efficiently eluted using 9M urea buffer (Figure 2E), suggesting a protein-ASO interaction.
Hsp90 protein interacts with many co-chaperone proteins (28). It is therefore possible that the ASO-Hsp90 protein interaction may be mediated by other partner proteins. To investigate this possibility, affinity selection was performed using a purified recombinant Hsp90 protein, which is more than 90% pure, as determined by silver staining (Figure 2F). Affinity selection and Western analysis showed that, in the absence of cell lysate, the recombinant Hsp90 protein was also significantly co-selected with the PS/cEt ASO, but not with the PS/MOE ASO (Figure 2G), suggesting a direct interaction between Hsp90 and ASOs. We note that trace amounts of Hsp90 protein were detected in the isolated sample with PS/MOE ASO; however, this is very likely due to incomplete washing during affinity selection using purified Hsp90 protein, which may weakly interact with PS/MOE ASOs, but such interaction may be inhibited in cells by other ASO-binding proteins.
To determine the binding constant of Hsp90 to ASOs, PS/cEt or PS/LNA ASOs were radio-labeled and incubated with different concentrations of purified Hsp90α protein. The protein-bound ASOs were transferred to a nitrocellulose membrane and the signal intensity was determined (Figure 2H). Consistent with the ASO co-selection results (Figure 2A), the binding affinity of Hsp90α determined by the membrane-binding assay also appears comparable for PS/cEt (kd = 19.7 nM) and PS/LNA (kd = 17.7 nM) ASOs (Figure 2H). As a control, no significant binding was found for a PS/DNA ASO. The specific binding of protein-associated ASO to the nitrocellulose membrane was confirmed using a double-filter binding assay for PS/cEt and PS/LNA ASOs. The unbound ASOs were attracted using a Hybond-N+ Nylon membrane under the Hybond ECL nitrocellulose membrane (Figure 2H, middle and lower panels). The bound-ASO signal intensity was quantified and plotted using Prism (Figure 2I and J), and the binding constant to PS/cEt and PS/LNA was determined to be 19.7 nM and 17.7 nM, respectively. Together, these results indicate that Hsp90 protein directly interacts with PS/cEt and PS/LNA ASOs, and that such an interaction is not unique to a particular ASO sequence.
Hsp90 protein recognizes the 5′-cEt wing of PS-ASOs
Since the 5–10–5 PS/cEt, PS/LNA, and PS/MOE gapmer ASOs used in affinity selection differ only at the 2′-modifications of the 5′ and 3′ wing nucleotides, it is conceivable that Hsp90 protein recognizes and/or interacts with the modified wings. Indeed, Hsp90 binding appears to be affected by the length of ASO wings. PS/LNA ASOs with shorter modified wings, such as 3–10–3 and 4–10–4 gapmer ASOs, bound less tightly to Hsp90 protein relative to the 5–10–5 gapmer ASO (Figure 3A). As a control, the binding of Ku70 protein was not significantly affected by shorting the wing length. Similar results were also observed for PS/cEt ASOs. Again, significantly weaker binding of Hsp90 protein was found for ASOs with shorter wings compared with the 5–10–5 ASO (Figure 3B), whereas the binding of Ku70 to PS/cEt ASOs of different wing lengths was comparable.
To determine which wing, 5′ or 3′ or both, interacts with Hsp90 protein, we designed gapmer PS-ASOs containing mixed wings, i.e. 5′-cEt + 3′-MOE (PS/cEt-MOE) or 5′-MOE + 3′-cEt (PS/MOE-cEt) wings. Since Hsp90 binding also depends on the wing length, ASOs were synthesized in the forms of 3–10–3, 4–10–4, and 5–10–5 gapmers. Affinity selection was performed using the biotinylated PS-ASOs with mixed wings, and co-isolated proteins were released by boiling in SDS-buffer and analyzed by Western assay (Figure 3C). Interestingly, Hsp90 protein was co-selected only with PS-ASOs containing a 5′-cEt wing, but not a 3′-cEt wing, in a wing-length dependent manner. Hsp90 binding was significantly increased with ASOs containing longer 5′-cEt wings. No Hsp90 binding was detected for PS/MOE-cEt ASOs, regardless of the wing length. As a control, the binding of Ku70 protein to these various ASOs was comparable. Together, these results indicate that Hsp90 protein requires 5′-cEt wing of PS-ASOs for binding.
The Hsp90 protein binding appears to depend on not only the 5′-cEt wing, but also the central deoxynucleotide gap region, since a 5-nt PS/cEt oligonucleotide representing the 5′-cEt wing of gapmer ASOs failed to elute Hsp90 protein by competition, similar to a 5-nt PS/MOE oligonucleotide representing the 3′-MOE wing (Figure 3D). However, a 15-mer PS-ASO containing 5-nt 5′-cEt wing and 10 deoxynucleotide region (referred as 5–10) eluted Hsp90 protein as efficiently as the 5–10–5 gapmer PS/cEt-MOE ASO (Figure 3E). On the other hand, Hsp90 protein binds poorly to a 15-mer PS-ASO containing 10 deoxynucleotides and a 5-nt 3′-MOE wing (referred as 10–5). These results suggest that Hsp90 protein recognizes 5′-cEt wing, but also requires some downstream PS-deoxynucleotides for binding.
Next, we analyzed if other proteins also prefer to bind to 5′-cEt wing of ASOs. Affinity selection was conducted using the 5–10–5 PS/cEt ASO 586183, and bound protein was eluted with different 5–10–5 PS-ASOs of the same sequence but different 2′-modifications. Western assay results showed that some ASO-binding proteins, such as La, NPM1, HMGB1 and P54nrb, bound preferentially to PS-ASOs containing 5′-cEt wing (Figure 3F). However, the binding of Ku70 and Ku80 was not significantly affected by the position and type of 2′-modifications. Unlike Hsp90 protein, which does not bind 5′-MOE ASOs, La, HMGB1, NPM1, P54nrb and PSF proteins bind 5′-MOE ASOs, but less tightly relative to 5′-cEt ASOs. These results suggest that many ASO-binding proteins may preferentially interact with the 5′-cEt wing of PS-ASOs.
The number of PS moieties in ASOs affects Hsp90 protein binding
Hsp90 protein significantly binds the PS-ASO containing 5′-cEt wing and 10-nt DNA (5–10). Next, we determined the effect of shortening the deoxynucleotide portion downstream from the 5′-cEt wing. Four PS-ASOs were synthesized that contain 5-nt 5′-cEt wing and 4, 6, 8 or 10 deoxynucleotides downstream, referred to 5–4, 5–6, 5–8 and 5–10, respectively. Proteins were affinity selected using the 5–10–5 PS/cEt ASO 586183, and bound proteins were then eluted via competition using the ASOs of different lengths and analyzed by Western assay (Figure 4A). The results showed that the 5–8 and 5–10 PS-ASOs bound much more significantly to Hsp90 protein than the 5–6 and 5–4 ASOs, suggesting that shortening the DNA portion to less than 6 (totally 10 PS) can significantly reduce Hsp90 protein binding. In addition, the membrane was also probed sequentially for other ASO-binding proteins, including NPM1, Ku80 and HMGB1. We note that the results for Ku70, P54nrb and TCP1 proteins were recently published, but included here for comparison purpose (22,23). Consistent with our previous results, the majority of the tested ASO-binding proteins, including NPM1 and HMGB1, bound much more weakly to 5–6 and 5–4 ASOs, as compared to the 5–8 and 5–10 ASOs. Although the binding affinity of these proteins to shorter ASOs can be different, our results suggest that at least 6-nt PS-deoxynucleotide portion or in total 10 PS modified nucleotides, of the cEt ASO are required for significant binding of Hsp90 and some other proteins.
To determine if the reduced protein binding of ASOs with fewer PS backbones was due to shorter ASO length, or due to reduced PS numbers, we performed similar experiments using 5–10 ASOs that all contain 5-nt cEt wing followed by 10 deoxynucleotides. These ASOs contain different numbers of PS modifications, from 0, 5, 10, to 14, at different positions in the ASOs. Significant Hsp90 protein binding was again found for ASOs with 10 or 14 PS modified nucleotides, but not with ASOs containing 0 or 5 PS modified nucleotides (Figure 4B). Similar trends were also observed for the majority of other tested ASO binding proteins, including Ku70, P54nrb and TCP1 proteins that we reported previously (21). In addition to the number of PS, however, the ASO length also plays a role in protein binding, since ASO 622865, a 5–10 ASO containing 10 PS modified nucleotides, bound more proteins than ASO 622867, a 5–6 ASO that also contains 10 PS modified nucleotides (Figure 4A). Importantly, we note that the binding properties of different proteins to these ASOs may vary, since some proteins, such as TCP1ϵ and P54nrb, can be less affected by the number of PS or the length of ASOs, whereas other proteins are more sensitive to changes in these factors, such as HMGB1. Together, these and our previous results indicate that for 5′-cEt ASOs, significant binding of Hsp90 and many proteins requires 10 or more PS modified nucleotides.
The mid-domain of Hsp90 protein binds PS/cEt and PS/LNA ASOs
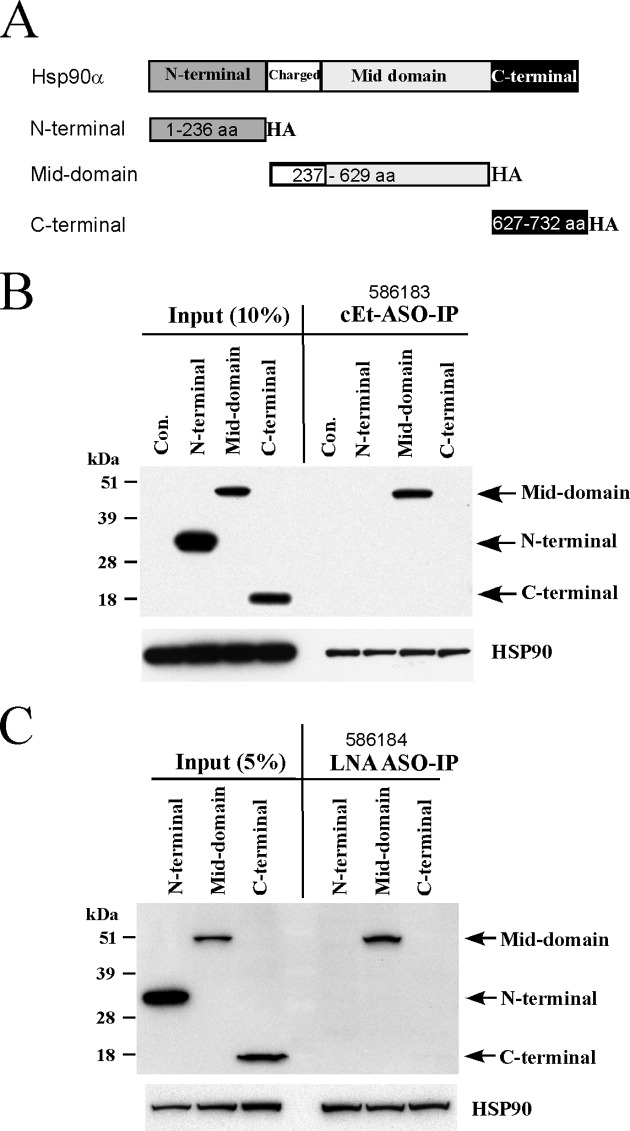
As a molecular chaperon, Hsp90 protein functions as a dimer in the folding of client proteins. Hsp90 contains three domains, the N-terminal domain, mid-domain and C-terminal domain (29). It has been shown that the N-terminal domain binds ATP, mid-domain is involved in client protein binding, and c-terminal domain is required for protein–protein interaction for dimer formation, although all the three domains can be involved in binding of different client proteins (30). To determine which domain(s) of Hsp90 interacts with PS-ASOs, three constructs were established to express N-terminal (amino acids 1–236), mid-domain (237–629) and C-terminal domains (627–732) of Hsp90α protein, respectively (Figure 5A). These truncated domains are HA-tagged at the C-terminus. The plasmids were transfected into HeLa cells, and the transient expression of truncated proteins was confirmed by Western analyses using an anti-HA antibody (data not shown and Figure 5B).
Figure 5.
The mid-domain of Hsp90 protein binds to ASOs. (A) Schematic representation of Hsp90α domains. The amino acid positions and C-terminal HA-tags in the truncated proteins are shown. (B) HeLa cells were transfected with plasmids expressing different domains of Hsp90α protein, or a control plasmid (Con.). Two days after transfection, cell lysate was prepared and affinity selection was performed using the 5–10–5 PS/cEt ASO 586183. Co-isolated proteins and ∼10% cell lysate used in affinity selection were separated by SDS-PAGE and analyzed by Western assay using an anti-HA antibody. The same membrane was subsequently probed for endogenous Hsp90 protein. (C) Plasmid transfection, affinity selection, and Western analyses were performed as in panel B, using a 5–10–5 PS/LNA ASO. The truncated Hsp90α proteins and endogenous Hsp90 protein are indicated. These experiments were repeated twice and representative results are shown.
Next, affinity selection was performed using the biotinylated PS/cEt ASO 582183 and cell lysates prepared from cells expressing different domains of Hsp90α protein, or from control cells transfected with an empty vector. Co-selected proteins were analyzed by Western assay using an anti-HA antibody. Interestingly, only the mid-domain was co-selected with the PS/cEt ASO (Figure 5B). Note that the three truncated domains were all significantly expressed in cells, as detected from the input materials, suggesting that other two domains do not significantly interact with the ASO. As a control for successful affinity selection, endogenous Hsp90α protein was equally co-isolated from all samples (Figure 5B, lower panel). Together, these results indicate that the mid-domain of Hsp90α binds PS/cEt ASOs.
Since PS/LNA ASOs bind Hsp90 protein like PS/cEt ASOs, we next ascertained if PS/LNA ASOs also interacts with the mid-domain of Hsp90α. Affinity selection was performed using a biotinylated PS/LNA ASO 586184 with lysates prepared from cells transiently expressing different domains of Hsp90α protein, as in panel B. Western assay results showed that the mid-domain of Hsp90 protein was co-isolated with the PS/LNA ASO (Figure 5C), consistent with the observation for the PS/cEt ASO. Our results therefore indicate that the mid-domain of Hsp90α contains the site(s) that recognizes and interacts with the PS/LNA and PS/cEt ASOs.
Reduction of Hsp90 protein inhibits the antisense activity of PS/cEt and PS/LNA ASOs
The binding of proteins to ASOs can affect the ASO-directed RNase H1 cleavage activity (21). To determine if Hsp90 protein influences the antisense activity of ASOs, we first analyzed the effect of reducing Hsp90β protein, which is constitutively expressed in cells (31). Both Hsp90β mRNA (Figure 6A, left panel) and protein (Figure 6A, right panel) were significantly reduced by siRNA treatment in HeLa cells, as determined by qRT-PCR and Western analysis, respectively. Next, control or Hsp90β reduced cells were transfected with PS-ASOs with 5′-cEt+3′-MOE (ASO 598343) or 5′-MOE+3′-cEt (ASO 598341) wings, as used in affinity selection (Figure 3F). These ASOs have the same sequence complementary to a site in PTEN mRNA, a target that we have been using as a reporter in our ASO activity assay (21–23). Four hours after ASO transfection, total RNA was prepared and PTEN mRNA levels were detected by qRT-PCR. The results showed that reduction of Hsp90β significantly inhibited the antisense activity of ASO 598343 (Figure 6B), but not ASO 598341 (Figure 6C), as indicated by less reduction of the PTEN mRNA in Hsp90β reduced cells. This is consistent with the binding property: Hsp90 protein interacts with ASO 598343 that contains a 5′-cEt wing, but not with ASO 598341 that harbors a 5′-MOE wing (Figure 3).
To further confirm the observation that Hsp90 protein affects the antisense activity of ASOs that it binds, and to exclude the possibility of potential off-target effects of the Hsp90β siRNA, we treated HeLa cells using a different siRNA targeting Hsp90β. In addition, the Hsp90α isoform was also reduced by siRNA treatment, to determine if the effect is common to both Hsp90 isoforms. To control for potential side-effects related to siRNA transfection (32,33), control cells were transfected with a luciferase siRNA. As expected, the levels of Hsp90α and Hsp90β mRNAs (Figure 6D, left panel) and the corresponding proteins (Figure 6D, right panel) were significantly reduced by specific siRNA treatment. Different cells were then transfected with PS/LNA ASO 390896 or PS/cEt ASO 582801, which can bind Hsp90 proteins or transfected with a PS/MOE ASO 116847, which does not bind significantly to Hsp90 protein. All these 5–10–5 gapmer ASOs have the same sequence targeting PTEN mRNA, and differ only in the 2′-modifications in the wings. qRT-PCR analyses indicate that reduction of Hsp90β using a different siRNA diminished the antisense activity of PS/LNA (Figure 6E) and PS/cEt (Figure 6F) ASOs, as compared with that in control siRNA treated cells. Not surprisingly, reduction of Hsp90α also decreased the activity of PS/LNA and PS/cEt ASOs. However, reduction of either Hsp90 protein had no significant effect on the activity of the PS/MOE ASO (Figure 6G). These results also indicate that the reduced activity for PS/cEt or PS/LNA ASOs was not due to unexpected effects of siRNA treatment.
To further confirm that Hsp90 protein enhances ASO activity, Hsp90α was transiently over-expressed by transfection of an expression plasmid. As expected, The Hsp90α protein level was increased ∼2.6-fold, as compared with transfection of an empty vector (Figure 6H). Over-expression of the Hsp90α protein had no significant effect on the activity of the PS/MOE ASO (Figure 6I), however, the activity of the PS/cEt ASO was significantly increased (Figure 6J), as compared with that in cells transfected with an empty vector. As a positive control, siRNA-mediated reduction of Ku80 (Figure 6K), which binds both PS/MOE and PS/cEt ASOs and inhibits ASO activities (21), increased the antisense activity of both PS/MOE (Figure 6L) and PS/cEt (Figure 6M) ASOs. Together, these results indicate that Hsp90 proteins can enhance antisense activity of PS-ASOs containing 5′-cEt or LNA wings, but not PS/MOE ASO that does not bind Hsp90 proteins.
The Hsp90 effect on ASO activity is not unique to specific ASO sequence or a specific cell type
To evaluate if the effect of Hsp90 protein on ASO activity observed in HeLa cells applies to other cell types, and to different ASO sequences, siRNAs targeting Hsp90α and Hsp90β were co-transfected into A431 cells for 48 h, and ASOs targeting PTEN or NCL1 mRNAs were transfected for an additional 4 h. As expected, Hsp90α and Hsp90β mRNAs were reduced by more than 90%, as determined by qRT-PCR (Figure 7A, left panel), and the protein level was also significantly reduced (Figure 7A, right panel). Consistent with what was observed in HeLa cells, reduction of Hsp90 proteins did not affect the activity of the PS/MOE ASO116847 targeting PTEN mRNA (Figure 7B), whereas the activity of PS/cEt ASO582801 was dramatically decreased, as determined by qRT-PCR assay (Figure 7C). Importantly, a similar effect was observed with a different ASO sequence targeting NCL1 mRNA. Again, a PS/MOE ASO110074 showed comparable activity in control and Hsp90 reduced cells (Figure 7D), while the activity of a PS/cEt ASO617294 was significantly decreased upon Hsp90 reduction (Figure 7E), indicating that the effect of Hsp90 on PS/cEt ASO activity is not unique to a specific sequence or a specific cell type.
Next, we investigated the effect of Hsp90 reduction on ASO activity upon free uptake, namely incubation of ASOs with cells in the absence of transfection reagent. Control or Hsp90 reduced cells were incubated with ASOs for 16 h, and the levels of ASO-targeted mRNAs were analyzed by qRT-PCR. Although, the PS/MOE ASO116847 targeting PTEN mRNA was not active under this condition (Figure 7F), therefore it is difficult to judge the effect of Hsp90 reduction, decreased activity was observed for the PS/cEt ASO582801 in Hsp90 reduced cells (Figure 7G). However, the PS/MOE ASO110074 targeting NCL1 mRNA appeared to be active under free uptake, and reduction of Hsp90 had no significant influence on the activity of this ASO (Figure 7H). On the contrary, Hsp90 depletion dramatically reduced the activity of the PS/cEt ASO617294 targeting NCL1 (Figure 7I). Together, these results indicate that Hsp90 protein can affect the activity of PS/cEt, but not PS/MOE, ASOs with different sequences in different cell types, and under different ASO delivery approaches.
DISCUSSION
It is conceivable that intracellular proteins that bind PS-ASOs could have an impact on the ASO activity (1). Indeed, we have recently identified and characterized more than 50 intracellular proteins, many of which could affect ASO antisense activity, either positively or negatively, via different mechanisms (21). However, how these and other unidentified proteins interact with ASOs remains largely unknown. In the current study, we analyzed the effects of ASO modification on protein binding, focusing on the PS backbone and 2′-modifications in the ribose.
Using affinity selection and competition assays that we established previously for the identification of ASO-binding proteins (21,22), we showed that the PS backbone modification plays dominant roles in protein binding. Compared with PO-backbone, PS-modified ASOs bind more proteins and more tightly, consistent with previous observations (17,21–23). Using ASOs with different lengths and/or PS numbers, we found that the number of PS modified nucleotides dramatically affects the binding of many proteins. ASOs containing less than 10 PS modified nucleotides bound proteins significantly weaker compared to ASOs with greater PS numbers (Figure 4). In addition to the PS number, the length of ASOs also contributes to protein binding, since a 15-mer ASO containing 10 PS bind stronger to many proteins than an 11-mer ASO that also contains 10 PS nucleotides. This PS-number requirement for significant binding of intracellular proteins is consistent with early observations that PS-ASO binding to plasma proteins, mainly albumin and growth factors, also depended on the length of ASOs (3,6,34–37).
In addition to the PS backbone, the 2′-modifications in the wings of gapmer ASOs significantly affect protein binding. Our results showed that in general, ASOs with more hydrophobic 2′-modifications, such as 2′-F, cEt or LNA, tend to bind more proteins than ASOs with more hydrophilic 2′-modifications, such as Me and MOE. Consistent with our previous observations for TCP1β and P54nrb proteins (22,23), many other proteins, including La, NPM1, HMGB1, PC4 and hnRNP K also exhibited binding preference to ASOs with more hydrophobic 2′-modifications. These observations indicate that the ASO/protein interaction can be significantly affected by the nature of the 2′-modifications. This is not surprising, since for single-stranded ASOs, the modified 2′-moiety can be exposed to the surface (38,39), thus may participate in protein binding.
Although the hydrophobicity of 2′-modifications may play an important role in protein binding, other factors, such as the ASO local structure related to the 2′-modification or steric crowding of 2′-modifications, can also influence protein binding. This is supported by the observation that PS/DNA nucleotides have higher hydrophobicity than the cEt modified nucleotides, yet protein binding to the PS/DNA ASO was much weaker than to the PS/cEt ASO. In addition, 2′-modifications can have different impact on the binding of different proteins. For example, interactions of Ku70, Ku80 and TCP1ϵ proteins with ASOs are less significantly affected by 2′-modifications. On the other hand, the binding of other proteins, such as HMGB1 and NPM1, is more sensitive to changes in the 2′-modifications.
The finding that Hsp90 protein binds PS/cEt and PS/LNA ASOs, but not PS/MOE ASOs is interesting. Although Hsp90 is an abundant protein, co-isolation of Hsp90 with ASOs was not due to contamination, since PS/MOE ASOs failed to elute this protein in the same experiment. In addition, the physical interaction of Hsp90 protein with PS-ASOs is not mediated by DNA, RNA or other protein(s), rather, Hsp90 protein can directly bind to ASOs. These findings together indicate the ASO/Hsp90 interaction is specific and direct. Hsp90 protein has been shown to bind RNA (40), suggesting a versatile role of these chaperone proteins. Recently we demonstrated that TCP1 proteins, another group of chaperone proteins, interact and co-localize with PS-ASOs in nuclear PS-bodies and can enhance ASO activity (22). In addition, Hsp70 protein has been found to inhibit RNase H1 cleavage by preventing the binding of RNase H1 to ASO/mRNA duplex (25). These observations, together with the current finding for Hsp90 protein, suggest that chaperone proteins play important roles in modulating PS-ASO action in cells.
Hsp90 protein interacts with many client proteins mainly through hydrophobic interactions (41). The three domains, i.e. N- terminal, C-terminal and Mid-domains, are all involved in binding of client proteins (30). Interestingly, we found that PS/cEt and PS/LNA ASOs interact with the mid-domain of Hsp90α protein. The mid-domain of Hsp90 protein has previously been shown to interact with the 3′ UTR of the Bamboo mosaic virus RNA in plant (40), suggesting a RNA/DNA binding ability of this domain. A recent study also demonstrated that recombinant mammalian Hsp90 protein could interact with norovirus RNA (42). The mid-domain of Hsp90 protein is composed of two αβα motifs that are connected by α-helices. In addition, a hydrophobic patch and amphipathic protrusion in the mid-domain may play important roles in client protein interaction (28,29). Since Hsp90 protein prefers binding to PS-ASOs with more hydrophobic modifications, it is possible that the ASO-protein interaction may involve the hydrophobic patch of Hsp90 protein. However, the ASO/Hsp90 interaction may be different from the RNA/Hsp90 interaction, since the Hsp90 protein recognizes the cEt and LNA modifications of the ASOs, which are not present in natural RNAs. Understanding the detailed mechanism of ASO/protein interaction awaits further investigation, especially by solving the crystal structure for the protein/ASO complex.
Hsp90 protein recognizes and interacts with the 5′-cEt wing and a portion of downstream DNA nucleotides within an ASO (Figures 3 and 4). How Hsp90 distinguishes the direction of a 5–10–5 gapmer ASO is still an enigma. It seems the binding does not require the recognition of 5′ hydrogen or phosphate moiety, since the protein was isolated using ASOs tagged with biotin at either 5′ or 3′ end. It is likely that Hsp90 protein recognizes a cluster of cEt or LNA modified nucleotides; however, downstream PS-DNA nucleotides are also required to form a docking site for Hsp90 protein binding. Intriguingly, several proteins, such as La, NPM1, P54nrb, PSF and HMGB1, also prefer to bind 5′-cEt wing of PS-ASOs (Figure 3D), suggesting that this protein binding property may contribute to the higher activity for PS/cEt ASOs as compared with PS/MOE ASOs (12,13). Since the 5′ half or even an entire 20-mer ASO is relatively short, it is highly unlikely that so many proteins bind to the same ASO molecule. Rather, it is possible that each of these proteins interacts with a sub-population of ASOs (22). This view is supported by the observations that reduction of a particular ASO-binding protein caused only modest effect on ASO activity (21).
Consistent with the ASO binding properties, Hsp90 protein can enhance the antisense activity of PS/cEt or PS/LNA ASOs, but not PS/MOE ASOs with different sequences and in different cell types, as confirmed using both reduction and over-expression of the protein. The effects of Hsp90 on ASO activity were modest, consistent with our observations for several other ASO-binding proteins that we reported recently (21–23). This is not unexpected, however, since PS-ASOs can interact with many different intracellular proteins, likely in a competitive manner (21–24). Thus, reduction of a particular ASO-binding protein may not have dramatic effect on ASO activity, as each individual protein may bind a small portion of cellular ASOs, and the reduction effect of an individual protein can also be partially compensated by other ASO-binding protein(s), as we discussed previously (21–22).
Currently it is not clear how Hsp90 protein enhances ASO activity. Unlike TCP1 proteins that co-localize with ASOs in cells (22), no co-localization of Hsp90 protein and PS/cEt ASO was detected by confocal imaging (data not shown). Hsp90 does not interact with the ASO/RNA duplex, unlike the Hsp70 protein reported previously (25). Although it is possible that Hsp90 may facilitate ASO/RNA annealing by, e.g. preventing the formation of intra- or inter-molecular structures, we do not favor this possibility. This is because the recombinant Hsp90 protein, which binds PS/cEt ASOs (Figure 2G), does not enhance ASO/RNA hybridization, as determined using biotinylated ASOs and a radio-labeled complementary RNA in a beads-mediated precipitation assay (data not shown). However, Hsp90 may enhance ASO activity in several ways. For example, it is possible that binding of this protein may prevent ASOs from being trapped by other inhibitory ASO-binding proteins, thus increasing the local availability of ASOs to the target mRNAs. In addition, Hsp90 has been shown to be involved in nuclear import of virus RNA polymerase (43), therefore it is possible that this protein may facilitate ASO nuclear-cytoplasmic transport. Although no significant difference was observed for ASO subcellular distribution in Hsp90 reduced cells using confocal imaging or cellular fractionation (data not shown), a role of Hsp90 protein in ASO nuclear-cytoplasmic shuttling cannot be excluded. Subtle change in the level of ASOs in different subcellular compartments might be difficult to be convincingly detected, due to the fast diffusion of ASOs. Understanding the underlying mechanisms by which Hsp90 proteins affect ASO activity thus requires further investigation.
Supplementary Material
Acknowledgments
We wish to thank Timothy Vickers, Shiyu Wang, Sebastien Burel, Eric Swayze, and Frank Bennett for discussions. We thank Tori Kniss for editing. This work was supported by an internal funding from IONIS Pharmaceuticals.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
IONIS Pharmaceuticals. Funding for open access charge: IONIS Pharmaceuticals.
Conflict of interest statement. None declared.
REFERENCES
- 1.Crooke S.T., Vickers T.A., Lima W.F., Wu H.-J. In: Antisense Drug Technology - Principles, Strategies, and Applications. 2nd edn. Crooke ST, editor. Boca Raton: CRC Press; 2008. pp. 3–46. [Google Scholar]
- 2.Kurreck J. RNA interference: from basic research to therapeutic applications. Angew Chem. Int. Ed. Engl. 2009;48:1378–1398. doi: 10.1002/anie.200802092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dias N., Stein C.A. Antisense oligonucleotides: basic concepts and mechanisms. Mol. Canc. Therap. 2002;1:347–355. [PubMed] [Google Scholar]
- 4.Swayze E.E., Bhat B. In: Antisense Drug Technology - Principles, Strategies, and Applications. 2nd edn. Crooke ST, editor. Boca Raton: CRC Press; 2008. pp. 143–182. [Google Scholar]
- 5.Kurreck J. Antisense technologies. Improvement through novel chemical modifications. Europ. J. Biochem./FEBS. 2003;270:1628–1644. doi: 10.1046/j.1432-1033.2003.03555.x. [DOI] [PubMed] [Google Scholar]
- 6.Bennett C.F., Swayze E.E. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmac. Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 7.Geary R.S. Antisense oligonucleotide pharmacokinetics and metabolism. Expert Opin. Drug Metab. Toxicol. 2009;5:381–391. doi: 10.1517/17425250902877680. [DOI] [PubMed] [Google Scholar]
- 8.Juliano R.L., Carver K., Cao C., Ming X. Receptors, endocytosis, and trafficking: the biological basis of targeted delivery of antisense and siRNA oligonucleotides. J. Drug Targeting. 2013;21:27–43. doi: 10.3109/1061186X.2012.740674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juliano R.L., Ming X., Nakagawa O. Cellular uptake and intracellular trafficking of antisense and siRNA oligonucleotides. Bioconjugate Chem. 2012;23:147–157. doi: 10.1021/bc200377d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koller E., Vincent T.M., Chappell A., De S., Manoharan M., Bennett C.F. Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic Acids Res. 2011;39:4795–4807. doi: 10.1093/nar/gkr089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simoes-Wust A.P., Hopkins-Donaldson S., Sigrist B., Belyanskaya L., Stahel R.A., Zangemeister-Wittke U. A functionally improved locked nucleic acid antisense oligonucleotide inhibits Bcl-2 and Bcl-xL expression and facilitates tumor cell apoptosis. Oligonucleotides. 2004;14:199–209. doi: 10.1089/oli.2004.14.199. [DOI] [PubMed] [Google Scholar]
- 12.Murray S., Ittig D., Koller E., Berdeja A., Chappell A., Prakash T.P., Norrbom M., Swayze E.E., Leumann C.J., Seth P.P. TricycloDNA-modified oligo-2′-deoxyribonucleotides reduce scavenger receptor B1 mRNA in hepatic and extra-hepatic tissues–a comparative study of oligonucleotide length, design and chemistry. Nucleic Acids Res. 2012;40:6135–6143. doi: 10.1093/nar/gks273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seth P.P., Siwkowski A., Allerson C.R., Vasquez G., Lee S., Prakash T.P., Kinberger G., Migawa M.T., Gaus H., Bhat B., et al. Design, synthesis and evaluation of constrained methoxyethyl (cMOE) and constrained ethyl (cEt) nucleoside analogs. Nucleic Acids Symposium Series. 2008;52:553–554. doi: 10.1093/nass/nrn280. [DOI] [PubMed] [Google Scholar]
- 14.Freier S.M., Altmann K.H. The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 1997;25:4429–4443. doi: 10.1093/nar/25.22.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stanton R., Sciabola S., Salatto C., Weng Y., Moshinsky D., Little J., Walters E., Kreeger J., DiMattia D., Chen T., et al. Chemical modification study of antisense gapmers. Nucleic Acid Ther. 2012;22:344–359. doi: 10.1089/nat.2012.0366. [DOI] [PubMed] [Google Scholar]
- 16.Kurreck J., Wyszko E., Gillen C., Erdmann V.A. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 2002;30:1911–1918. doi: 10.1093/nar/30.9.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown D.A., Kang S.H., Gryaznov S.M., DeDionisio L., Heidenreich O., Sullivan S., Xu X., Nerenberg M.I. Effect of phosphorothioate modification of oligodeoxynucleotides on specific protein binding. J. Biol. Chem. 1994;269:26801–26805. [PubMed] [Google Scholar]
- 18.Weidner D.A., Valdez B.C., Henning D., Greenberg S., Busch H. Phosphorothioate oligonucleotides bind in a non sequence-specific manner to the nucleolar protein C23/nucleolin. FEBS Lett. 1995;366:146–150. doi: 10.1016/0014-5793(95)00517-d. [DOI] [PubMed] [Google Scholar]
- 19.Abdul-Manan N., Williams K.R. hnRNP A1 binds promiscuously to oligoribonucleotides: utilization of random and homo-oligonucleotides to discriminate sequence from base-specific binding. Nucleic Acids Res. 1996;24:4063–4070. doi: 10.1093/nar/24.20.4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eckstein F. Phosphorothioate oligodeoxynucleotides: what is their origin and what is unique about them. Antisense Nucleic Acid Drug Dev. 2000;10:117–121. doi: 10.1089/oli.1.2000.10.117. [DOI] [PubMed] [Google Scholar]
- 21.Liang X.H., Sun H., Shen W., Crooke S.T. Identification and characterization of intracellular proteins that bind oligonucleotides with phosphorothioate linkages. Nucleic Acids Res. 2015;43:2927–2945. doi: 10.1093/nar/gkv143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang X.H., Shen W., Sun H., Prakash T.P., Crooke S.T. TCP1 complex proteins interact with phosphorothioate oligonucleotides and can co-localize in oligonucleotide-induced nuclear bodies in mammalian cells. Nucleic Acids Res. 2014;42:7819–7832. doi: 10.1093/nar/gku484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen W., Liang X.H., Crooke S.T. Phosphorothioate oligonucleotides can displace NEAT1 RNA and form nuclear paraspeckle-like structures. Nucleic Acids Res. 2014;42:8648–8662. doi: 10.1093/nar/gku579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen W., Liang X.H., Sun H., Crooke S.T. 2′-Fluoro-modified phosphorothioate oligonucleotide can cause rapid degradation of P54nrb and PSF. Nucleic Acids Res. 2015;43:4569–4578. doi: 10.1093/nar/gkv298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vickers T.A., Crooke S.T. Antisense oligonucleotides capable of promoting specific target mRNA reduction via competing RNase H1-dependent and independent mechanisms. PLOS One. 2014;9:e108625. doi: 10.1371/journal.pone.0108625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang X.H., Crooke S.T. Depletion of key protein components of the RISC pathway impairs pre-ribosomal RNA processing. Nucleic Acids Res. 2011;39:4875–4889. doi: 10.1093/nar/gkr076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin S., Weaver D.T. Double-strand break repair by Ku70 requires heterodimerization with Ku80 and DNA binding functions. EMBO J. 1997;16:6874–6885. doi: 10.1093/emboj/16.22.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taipale M., Jarosz D.F., Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 29.Li J., Soroka J., Buchner J. The Hsp90 chaperone machinery: conformational dynamics and regulation by co-chaperones. Biochim. Biophys. Acta. 2012;1823:624–635. doi: 10.1016/j.bbamcr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Jackson S.E. Hsp90: structure and function. Topics Curr. Chem. 2013;328:155–240. doi: 10.1007/128_2012_356. [DOI] [PubMed] [Google Scholar]
- 31.Sreedhar A.S., Kalmar E., Csermely P., Shen Y.F. Hsp90 isoforms: functions, expression and clinical importance. FEBS Lett. 2004;562:11–15. doi: 10.1016/s0014-5793(04)00229-7. [DOI] [PubMed] [Google Scholar]
- 32.Liang X.H., Hart C.E., Crooke S.T. Transfection of siRNAs can alter miRNA levels and trigger non-specific protein degradation in mammalian cells. Biochim. Biophys. Acta. 2013;1829:455–468. doi: 10.1016/j.bbagrm.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 33.Liang X.H., Crooke S.T. RNA helicase A is not required for RISC activity. Biochim. Biophys. Acta. 2013;1829:1092–1101. doi: 10.1016/j.bbagrm.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 34.Krieg A.M., Stein C.A. Phosphorothioate oligodeoxynucleotides: antisense or anti-protein. Antisense Res. Dev. 1995;5:241. doi: 10.1089/ard.1995.5.241. [DOI] [PubMed] [Google Scholar]
- 35.Stein C.A., Tonkinson J.L., Zhang L.M., Yakubov L., Gervasoni J., Taub R., Rotenberg S.A. Dynamics of the internalization of phosphodiester oligodeoxynucleotides in HL60 cells. Biochem. 1993;32:4855–4861. doi: 10.1021/bi00069a022. [DOI] [PubMed] [Google Scholar]
- 36.Watanabe T.A., Geary R.S., Levin A.A. Plasma protein binding of an antisense oligonucleotide targeting human ICAM-1 (ISIS 2302) Oligonucleotides. 2006;16:169–180. doi: 10.1089/oli.2006.16.169. [DOI] [PubMed] [Google Scholar]
- 37.Bennett C.F. In: Antisense Drug Technology - Principles, Strategies, and Applications. Crooke ST, editor. Boca Raton: CRC Press; 2006. pp. 273–304. [Google Scholar]
- 38.Teplova M., Minasov G., Tereshko V., Inamati G.B., Cook P.D., Manoharan M., Egli M. Crystal structure and improved antisense properties of 2′-O-(2-methoxyethyl)-RNA. Nat. Struc. Biol. 1999;6:535–539. doi: 10.1038/9304. [DOI] [PubMed] [Google Scholar]
- 39.Pallan P.S., Allerson C.R., Berdeja A., Seth P.P., Swayze E.E., Prakash T.P., Egli M. Structure and nuclease resistance of 2′,4′-constrained 2′-O-methoxyethyl (cMOE) and 2′-O-ethyl (cEt) modified DNAs. Chem. Commun. 2012;48:8195–8197. doi: 10.1039/c2cc32286b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang Y.W., Hu C.C., Liou M.R., Chang B.Y., Tsai C.H., Meng M., Lin N.S., Hsu Y.H. Hsp90 interacts specifically with viral RNA and differentially regulates replication initiation of Bamboo mosaic virus and associated satellite RNA. PLOS Pathog. 2012;8:e1002726. doi: 10.1371/journal.ppat.1002726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu Z., Horwich A.L., Sigler P.B. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature. 1997;388:741–750. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- 42.Vashist S., Urena L., Gonzalez-Hernandez M.B., Choi J., de Rougemont A., Rocha-Pereira J., Neyts J., Hwang S., Wobus C.E., Goodfellow I. Molecular chaperone Hsp90 is a therapeutic target for noroviruses. J. Virol. 2015;89:6352–6363. doi: 10.1128/JVI.00315-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naito T., Momose F., Kawaguchi A., Nagata K. Involvement of Hsp90 in assembly and nuclear import of influenza virus RNA polymerase subunits. J. Virol. 2007;81:1339–1349. doi: 10.1128/JVI.01917-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.