Abstract
The folding topology of DNA G-quadruplexes (G4s) depends not only on their nucleotide sequences but also on environmental factors and/or ligand binding. Here, a G4 ligand, 3,6-bis(1-methyl-4-vinylpyridium iodide)-9-(1-(1-methyl-piperidinium iodide)-3,6,9-trioxaundecane) carbazole (BMVC-8C3O), can induce topological conversion of non-parallel to parallel forms in human telomeric DNA G4s. Nuclear magnetic resonance (NMR) spectroscopy with hydrogen-deuterium exchange (HDX) reveals the presence of persistent imino proton signals corresponding to the central G-quartet during topological conversion of Tel23 and Tel25 G4s from hybrid to parallel forms, implying that the transition pathway mainly involves local rearrangements. In contrast, rapid HDX was observed during the transition of 22-CTA G4 from an anti-parallel form to a parallel form, resulting in complete disappearance of all the imino proton signals, suggesting the involvement of substantial unfolding events associated with the topological transition. Site-specific imino proton NMR assignments of Tel23 G4 enable determination of the interconversion rates of individual guanine bases and detection of the presence of intermediate states. Since the rate of ligand binding is much higher than the rate of ligand-induced topological conversion, a three-state kinetic model was evoked to establish the associated energy diagram for the topological conversion of Tel23 G4 induced by BMVC-8C3O.
INTRODUCTION
Human telomeric G-rich DNA sequences can form G-quadruplex (G4) structures through Hoogsteen hydrogen bonding in the presence of monovalent cations (1,2). G4 formation can inhibit telomerase activity leading to suppression of cancer cell proliferation. Thus, telomeric G4s are potential therapeutics and targets for drug design (3–6). However, even slight variations in human telomeric sequences can result in different G4 structures. For example, d[TAG3(T2AG3)3] (Tel23) adopts a three-layer hybrid-I G4 topology (7,8), whereas d[G3(T2AG3)3T] (Tel21-T) adopts a two-layer basket G4 topology (9). In addition, different G4 structures can coexist for the same human telomeric sequence, such as those for d[AG3(T2AG3)3] (Tel22) in K+ solution (10,11). Such structural polymorphisms make telomeric G4s challenging not only for structural analysis but also for drug design (12,13). Recently, Phan et al. (14) showed that all different nonparallel forms of human telomeric G4s can be converted into parallel forms in the presence of 40% (v/v) polyethylene glycol (PEG). Despite the crowding condition that resembles cellular environments (14–16), the underlying mechanism of PEG-induced G4 conformational changes is not owing to the crowding effect (17–19).
Inspired by the solvent properties of PEG, we designed a derivative of a previously reported G4 ligand: 3,6-bis(1-methyl-4-vinylpyridinium) carbazole diiodide (BMVC), by covalent attachment of a tetraethylene glycol (8C3O) terminating in a methyl-piperidinium cation; we named this derivative BMVC-8C3O (19). Upon binding to G4s, the BMVC moiety introduced 8C3O into the solvent sphere of the G4 structure and thus caused local dehydration. It is known that in addition to converting non-parallel forms of human telomeric G4s to parallel forms, BMVC-8C3O binding dramatically increases thermal stability (as shown by an increased melting temperature of >45°C). The ability of BMVC-8C3O to induce G4 topological conversions in the μM range implies that the underlying mechanism is not due to a bulk solvent effect, but rather due to a local dehydration effect that disrupts the water structure in the vicinity of the G4. Water depletion favors formation of the parallel form, as shown by X-ray crystallography (20,21). Nonetheless, the mechanism underlying this topological conversion has not been described previously.
A number of reports have described the folding and transition kinetics of human telomeric G4s. Yang et al. (22) suggested that the rapid spectral conversion reflects topological rearrangement of Tel22 from the basket Na+ form to a hybrid K+ form via the opening of the central G-quartet during the Na+/K+ exchange. Based on molecular dynamic simulations, Sugiyama et al. (23) proposed the presence of a triplex intermediate along the G4 folding pathways. Imino proton-based NMR-HDX analysis displays exquisite sensitivity towards G-quartet opening during conformational rearrangements, and we applied such a strategy to establish that Na+/K+ exchange in Tel23 G4 involves neither rapid unfolding nor triplex intermediates as evidenced by the long-lived imino proton resonances corresponding to the middle G-quartet during HDX (24). We further determined the topology of the Na+ form of Tel23 G4 to be the same hybrid form as that of the K+ form of Tel23, which highlights the possibility for G4s to display rapid spectral conversions, for both NMR and far-UV CD spectroscopy, without major conformational changes via unfolding and refolding. Very recently, Schwalbe et al. (25) employed high resolution NMR spectroscopy to delineate the topological change between hybrid-I and hybrid-II of d[T2G3(T2AG3)3A] (Tel24-M), which involves the formation of partially unfolded hairpin-like structures, illustrating the broad range of possibilities for human telomeric G4s to undergo topological interconversions.
In this study, we employed NMR spectroscopy to determine the topology of Tel23 in a K+ solution after topological conversion upon the addition of BMVC-8C3O. The G4 topology observed was the same as that attained by other G4s in the presence of concentrated PEG (14). Furthermore, we employed time-resolved imino proton NMR spectroscopy coupled with HDX analysis to elucidate, at a nucleotide-specific level, the transition pathway and kinetics of topological conversion from an initial hybrid G4 to a final parallel G4 as a result of BMVC-8C3O binding to Tel23. Supplementary Table S1 lists the DNA sequences studied in this work.
MATERIALS AND METHODS
Preparation of reagents and samples
The chemical synthesis of BMVC-8C3O was performed as described previously (19). All unlabeled oligonucleotides were purchased from Bio Basic (Ontario, Canada); the site-specifically 15N-labeled oligonucleotides were synthesized using the solid-phase method as described previously (24,26). DNA concentrations were determined by absorbance at 260 nm using a UV-Vis absorption spectrometer (Nano-Viewer, GE Healthcare). The oligonucleotides were dissolved in a buffer consisting of 10 mM Tris-HCl (pH 7.5) and 150 mM KCl unless otherwise specified, followed by heat denaturation at 95°C for 5 min and slow annealing to 25°C (1 min/°C). The annealed oligonucleotides were stored at 4°C overnight prior to use.
CD spectroscopy
The CD experiments were conducted using a spectropolarimeter (J-815, Jasco, Tokyo, Japan) with a bandwidth of 2 nm, at the scanning speed of 50 nm/min and step resolution of 0.2 nm across the spectral range of 210–350 nm. The sample concentration was 5 μM in the buffer specified above.
Kinetic binding experiment
G4 ligand binding was measured using label-free technology (Bio-Layer Interferometry, BLI) (ForteBio, Inc., Menlo Park, CA). The affinity measurements were performed with the Octet RED96 system equipped with superstreptavidin (SA) biosensor tips (ForteBio, Inc., Menlo Park, CA, USA). The assays were performed at 25°C and 1000 rpm. The tips were loaded with 500 nM of biotinylated G4 sequence for 5 min. The typical capture levels of eight biosensors were in the range of 0.4–0.5 nm. The association (kon) and dissociation (kdis) were then established by dipping the biosensors for 10 min in various concentrations of BMVC-8C3O dispensed in 96-microwell plates (Fisher Scientific, Turnberry Drive, Hanover Park, IL, USA) at a volume of 200 μl per well. The data were processed and analyzed using the Octet data analysis software version 7.0 (ForteBio, Inc., Menlo Park, CA). The binding profile is a comparison of the shift in the interference patterns of light reflected from a reference layer within the biosensor versus molecules as the bind to the biosensor tip. The results were fitted to a two-site binding model, which is presented in supplementary information (Equation S1).
NMR spectroscopy
All NMR experiments were performed on a Bruker AVIII 500 MHz NMR spectrometer equipped with a prodigy probehead and on a Bruker AVIII 800 MHz NMR spectrometer equipped a cryoprobe (Bruker, Karlschurle, Germany). The 1D imino proton NMR spectra were recorded by a WATERGATE or a jump-return pulse sequence for water suppression. The 1D 15N-1H SOFAST-HMQC spectra were used for unambiguous assignment of individual imino proton resonances using a series of site-specifically 15N-labeled NMR samples, where 8% of 15N-labeled guanine was introduced into one of the 11 G-quartet-forming guanine residues as described previously (24,26). In the NMR experiments, the analyte concentrations were typically 100–200 μM for the 1D experiments and 0.5–1.0 mM for the 2D experiments, in buffer consisting of 10% (v/v) D2O, 150 mM KCl and the internal reference 0.1 mM 4,4-dimethyl-4-silapentane-1-sulfonic acid. The resonances of exchange protons were assigned using two-dimensional nuclear Overhauser effect spectroscopy (mixing time 50–400 ms).
NMR-HDX
For these experiments, five equivalents of BMVC-8C3O were added to a G4 without annealing, followed by lyophilization. The lyophilized oligonucleotides were resuspended in 99% D2O at a DNA concentration of 100–200 μM immediately before NMR spectroscopy with a dead time of ∼3 min. The time for recording an individual NMR-HDX spectrum was 8 min.
RESULTS
Topology of a Tel23 G4 after topological conversion induced by BMVC-8C3O
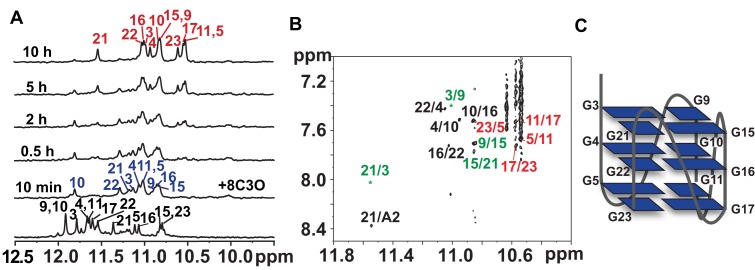
Multidimensional NMR spectroscopy was used to determine the topology of Tel23 G4 induced by BMVC-8C3O. Rapid changes in the imino proton NMR spectra of Tel23 were observed immediately after the addition of five equivalents of BMVC-8C3O, followed by a slower spectral change (Figure 1A). The imino proton NMR assignments of the initial and final forms of BMVC-8C3O-bound Tel23 were achieved by recording one-dimensional 15N-edited SOFAST-HMQC spectra for site-specific assignment of the imino (Supplementary Figure S1) and aromatic proton resonances along with NOE assignment of the cross-peaks between imino and H8 protons (Figure 1B). The end state of the BMVC-8C3O-bound Tel23 consists of three G-quartets with identical hydrogen bonding directionalities, i.e. G-3→G-9→G-15→G-21, G-4→G-10→G-16→G-22 and G-5→G-11→G-17→G-23, resulting in a double-chain-reversal configuration for all connecting loops. The folding topology was confirmed by the sequential H1-H1 NOEs between neighboring guanines, i.e. G-4/G-5, G-10/G-11, G-16/G-17 and G-21/G-22 (Supplementary Figure S2A). The distinct sequential H8-H1′ NOEs from T-1 to G-23 and their patterns further suggested that all guanines adopted an anti-glycosidic conformation (Supplementary Figure S2B). Taken together, our NMR data established that the end state of the BMVC-8C3O-bound Tel23 in K+ solution is in a parallel topology (Figure 1C), which is the same all-parallel topology as previously observed in the crystalline state by X-ray crystallography and under dehydration conditions by solution state NMR spectroscopy (14,20,21).
Figure 1.
A folding topology of a Tel23 G4 induced by BMVC-8C3O. (A) Imino proton NMR spectra of the Tel23 G4 in the presence of 150 mM K+ (bottom) and time-resolved imino proton NMR spectra recorded at 10 min and at 0.5, 2, 5 and 10 h after addition of 5 eq. BMVC-8C3O at 310 K, as indicated in the ascending order. (B) An expanded NOESY spectrum of the Tel23 G4 in complex with BMVC-8C3O; the spectrum was recorded with mixing time 250 ms and showed the H1/H8 correlations. Green, black and red labels correspond to the top, middle and bottom G-quartets, respectively. (C) Schematic representation of the Tel23 G4 topology induced by BMVC-8C3O.
NMR-HDX revealed a novel transition pathway of topological conversion from a hybrid G4 to a parallel G4 of Tel23 induced by BMVC-8C3O and acetonitrile
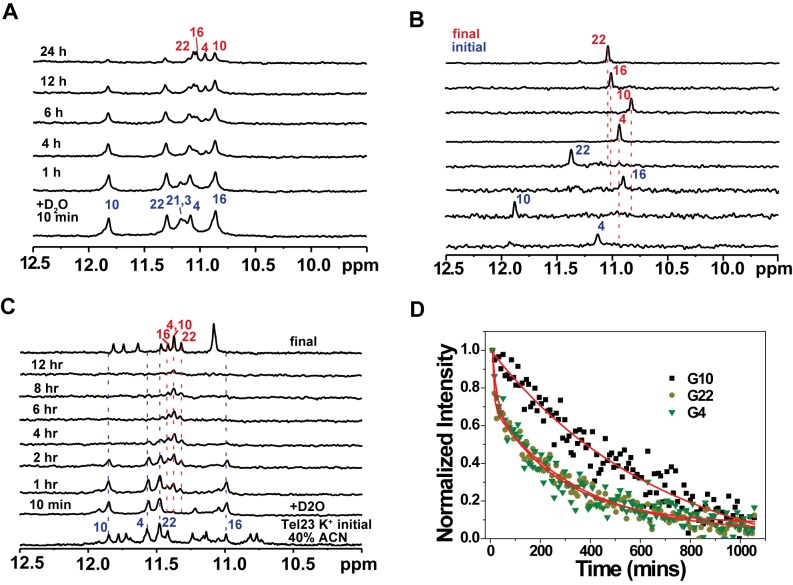
To determine whether such topological conversion involves global or local rearrangements, we analyzed NMR-HDX data to elucidate the mechanism underlying the BMVC-8C3O binding-induced topological transition of Tel23 (Figure 2A). Immediately after the addition of D2O, most of the imino proton signals corresponding to the top and bottom G-quartets, except for that of G-3, disappeared due to fast HDX processes while the imino proton signals of the central G-quartet (G-4, G-10, G-16 and G-22) persisted for several hours, indicating stable hydrogen bonding of the central G-quartet. Concurrently, the persistent imino protons of the central G-quartet exhibited up-field-shifted chemical shift changes during HDX, suggesting conformational rearrangements that do not open the central G-quartet, and will otherwise lead to immediate signal quench due to rapid HDX for unprotected imino protons (Figure 2A) (24).
Figure 2.
NMR spectroscopy with HDX of BMVC-8C3O or acetonitrile induced conformational changes in a Tel23 G4. (A) NMR-HDX spectra of the Tel23 G4 with BMVC-8C3O. At the initial time point, Tel23 G4 sample in the presence of 150 mM K+ was lyophilized after addition of 5 eq. BMVC-8C3O and then dissolved in 99% D2O immediately before NMR measurement, followed by recording of spectra at 25°C at 10 min; at 1, 4, 6, 12 and 24 h, in the ascending order. (B) Site-specific assignment of imino proton resonances in the initial and final forms of the central quartet of Tel23 during interaction with the BMVC-8C3O. The 1D 15N-1H SOFAST-HMQC spectra of 16% 15N-enriched Tel23 G4 samples are shown with the assignment and site-specific labeling that correspond to the labeled sites. (C) Imino proton NMR spectra of the Tel23 G4 in the presence of 150 mM K+ after addition of 40% v/v acetonitrile 10 mins (bottom) and after annealing, and stacked plot with NMR-HDX spectra, in which lyophilized Tel23 G4 sample in the presence of 150 mM K+ dissolved in D2O and 40% v/v acetonitrile-d3, and immediately before NMR measurement, followed by recording of spectra at 25°C at 10 min; at 1, 4, 6, 12 and 24 h. (D) The NMR-HDX curves of G-4, G-10 and G-22; these data were extracted from the imino proton spectra and analyzed by bi-exponential fitting.
Considering the prevalent structural polymorphism in telomeric G4s and the possibility that BMVC-8C3O may preferentially recognize and stabilize a minor conformation of Tel23 that correspond to the long-lived imino proton resonances during HDX, it is important to verify whether the four persistent imino protons during HDX correspond to a pre-existing minor population of Tel23 that corresponds to a parallel form as we deduced from the NMR data as described above. To ensure sufficient signal-to-noise ratio, we used 16% 15N-enriched Tel23 G4 samples to compare the 15N-edited imino proton NMR spectra of Tel23 between the initial state of hybrid form and final state of parallel form (Figure 2B). The well-defined and distinct imino proton resonances in the two different states provided strong evidence that the imino proton resonance intensity decay and concomitant chemical shift changes do reflect a high degree of protection of the central G-quartet of Tel23 from HDX during BMVC-8C3O binding-induced topological conversion. This indicates that there is no evidence of global unfolding during the topological conversion.
To examine whether these persistent imino protons are only induced by ligand binding, NMR-HDX analysis was conducted in the presence of 40% acetonitrile (ACN) to monitor topological conversion of Tel23 into a parallel form under dehydration conditions (Figure 2C) (14,17). The results showed that after the addition of D2O, concurrent decay of the central four imino proton signals of the initial hybrid G4 and emergence of the central four imino proton signals of the final parallel G4 were observed within the first 4 h and eventually all imino proton signals disappeared after 12 h. It appears that the conversion process that exhibits long-lived imino proton signals of the central G-quartet during NMR-HDX is not limited to ligand-induced process but also in solvent induced process. However, the existence of persistent imino proton signals during HDX is likely due to further stabilization of G4 by BMVC-8C3O binding (19).
Detailed comparison of the NMR-HDX data revealed that the rate at which the imino proton signal of G-10 decreases is significantly lower than that of the other three guanines within the central G-quartet (Figure 2D). Because of severe spectral overlap between G-11 and G-4, G-15/G-9 and G-16, and G-21 and G-22, most of HDX kinetic traces of the observed imino proton required multiple exponential fitting to account for the multiple components (Figure 2D). The HDX time constant of G-10 was extracted from single exponential fitting to be 690 min, whereas those of G-4 and G-22 required bi-exponential fitting that resulted in two time constants, 15 min and 300 min; the shorter time constant is attributed to the rapid HDX of G-21 or G-3 that are more exposed to bulk solvent. Differential HDX rates of these central imino proton signals suggested asymmetric local dynamics during the topological conversion from a hybrid G4 to a parallel G4 of Tel23 as a result of G-tract flipping, which is reminiscent of the asymmetric HDX behavior of c-kit2 (27). Collectively, these results suggested that G-10 sustains more stable hydrogen bonding during the topological conversion after the binding to BMVC-8C3O.
NMR-HDX analysis of topological conversion of other telomeric G4s induced by BMVC-8C3O
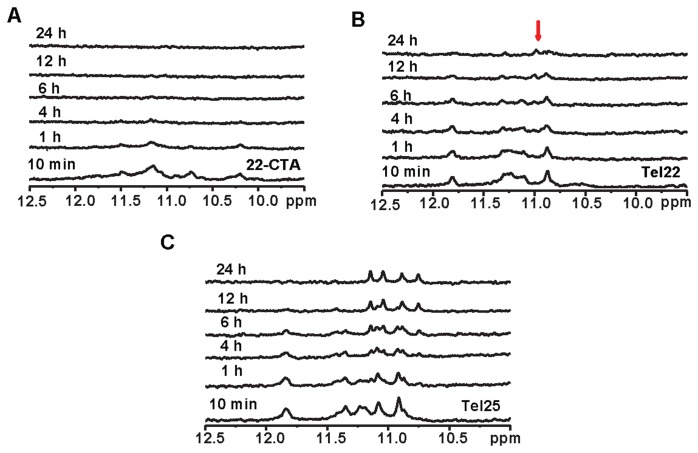
Since BMVC-8C3O binding can induce topological changes of different non-parallel human telomeric G4s into the same parallel topology (19), we examined whether the central imino proton signals of these telomeric G4s are also long-lived during BMVC-8C3O-induced topological conversions, particularly the topological conversion of [AG3(CTAG3)3] (22-CTA), since 22-CTA forms an anti-parallel chair G4 containing two G-quartets and a G•C•G•C quartet in K+ solution (28). Notably, the imino proton NMR spectra of the end topologies of 22-CTA and Tel22 after transition induced by BMVC-8C3O are almost identical, implying that both form similar parallel G4s containing three G-quartets (19). Thus, one would expect that the ligand-induced topological conversion of 22-CTA involves global unfolding. Indeed, the imino proton signals of the final parallel G4 of 22-CTA were not observed during topological conversion (Figure 3A), suggesting the opening of the central G-quartet during the topological conversion. Interestingly, several weak imino proton signals of the final parallel G4 of Tel22 appeared during topological conversion (Figure 3B). Considering that the coexistence of two major G4 topologies of Tel22 in K+ solution have not been determined, it is therefore crucial to define the G4 topologies before and after ligand binding as reference points for the study of topological conversion. Since the previous study suggested that Tel22 could form a hybrid G4 in K+ solution (10,11), these weak signals of the final parallel G4 may result from topological conversion of the initial hybrid G4. NMR-HDX analysis was further applied to test a hybrid-II G4 of d[TAG3(T2AG3)3TT] (Tel25) (8), the central imino proton signals of Tel25 were also long-lived during topological conversion (Figure 3C). Nevertheless, the middle four imino proton NMR signals of these final parallel G4s induced by BMVC-8C3O binding were all long-lived during HDX (Supplementary Figure S3), suggesting that these middle four imino proton signals are valid markers for analysis of the opening of the central G-quartet during topological transition.
Figure 3.
NMR spectroscopy with HDX of BMVC-8C3O or acetonitrile induced conformational changes in human telomeric G4s NMR-HDX spectra of lyophilized (A) 22-CTA, (B) Tel22 and (C) Tel25 G4 sample in the presence of 150 mM K+ after addition of 5 eq. BMVC-8C3O and 99% D2O, followed by recording of spectra at 25°C at 10 min; at 1, 4, 6, 12 and 24 h, in the ascending order.
Investigation of the BMVC-8C3O binding to a hybrid G4 and a parallel G4
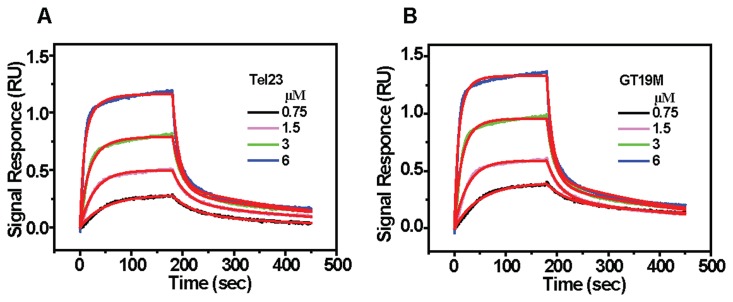
To examine the binding kinetics of BMVC-8C3O to the initial (hybrid) and final (parallel) G4 states, we used interferometry to measure the association and dissociation rate constants as a function of ligand concentrations (Figure 4A). A two-site binding model (Equation S1) was applied to fit the observed binding curves resulting in two association rate constants (ka) 1.1 × 104 M−1 s−1 and 4.4 × 103 M−1 s−1, and two dissociation rate constants (kd) 2.9 × 10−3 s−1 and 4.4 × 10−2 s−1, respectively, resulting in two association constants (Ka) 3.8 × 106 M−1 and 1.0 × 105 M−1 (Supplementary Table S2). If BMVC-8C3O had a higher binding affinity for the parallel G4 over hybrid G4, as a result the topological conversion could be attributed to conformational selection. To verify the possibility of a conformational selection mechanism of BMVC-8C3O preference for G4s, we further examined its binding to a parallel G4, GT19M, d[(TAG3)2TG3TAG3] (29). The binding kinetics of BMVC-8C3O to GT19M are shown in Figure 4B. The results are similar to that of Tel23 with two similar ka values 1.5 × 104 M−1 s−1 and 4.7 × 103 M−1 s−1, and two similar kd values 2.6 × 10−3 s−1 and 4.4 × 10−2 s−1, that yield two association constants, 5.8 × 106 M−1 and 1.1 × 105 M−1, respectively. Similar ligand binding parameters to the hybrid G4 of Tel23 and parallel G4 of GT19M (Supplementary Table S2) suggested that BMVC-8C3O has no specific binding preference to these G4s of distinct topologies.
Figure 4.
Kinetic binding curves for the interaction of BMVC-8C3O with (A) Tel23 and (B) GT19M G4. The kinetic traces as shown on increasing BMVC-8C3O concentration from 0.75 μM (lower curve) to 6 μM (upper curve) were fit to a two-site binding model (red lines) using the built-in software of ForteBio with a correlation coefficient (R2 = 0.99). The experiments were conducted in 10 mM Tris-HCl buffer with 150 mM K+ at 25°C.
Transition kinetics of the topological conversion of Tel23 G4 based on time-resolved imino proton NMR spectroscopy
Kinetic insight into the G4 topological conversion is important not only for a better understanding of the relevant biological functions but also for better understanding of DNA folding dynamics. In this work, ligand binding is the driving force for conformational change. For simplicity, the transition kinetics of ligand induced topological conversion induced by BMVC-8C3O may be briefly described as:
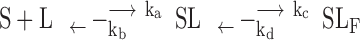 |
where S and L are G4 and ligand, i.e. BMVC-8C3O, and SL and SLF are the initial and final states of the G4-ligand complex, ka is the association rate and kb is the dissociation rate of ligand binding, and kc and kd are the forward and reverse transition rates of topological conversion, respectively. The analytical solutions for a three-state transition model (30) have been described in supplementary information (Equation S2). The individual rate constants were determined experimentally and fit to a three-state transition model to yield the populations of individual states as a function of time (Supplementary Figure S4). The results indicated that the higher reaction rate of ligand binding has no appreciable effect on the lower transition rate of topological conversion.
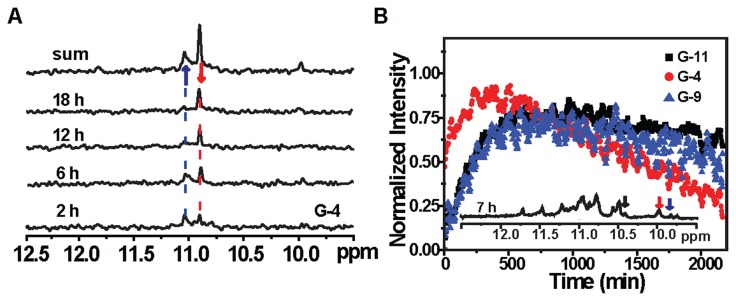
To explore the nature of a potential folding intermediate of Tel23 during topological conversion, we collected a series of time-resolved 15N-edited imino proton NMR spectra with site-selective 15N-labeling to avoid assignment ambiguity resulting from spectral overlap (Figure 5A). For G-4, a weak imino proton signal was observed at ∼10 ppm in addition to those correspond to the initial and final states, and it was tentatively assigned to an intermediate state of G-4. Several intermediate imino proton resonances were also observed for other G4-forming residues (Supplementary Figure S5). Collectively, three well-resolved kinetic traces that correspond to the intermediate populations of G-4, G-9 and G-11 were extracted (Figure 5B). These intermediate states exhibit different reaction kinetics, suggesting that the BMVC-8C3O-induced topological conversion indeed involves multiple steps. In order to construct an energy diagram to describe the BMVC-8C3O binding-induced conformational transition, we subsequently determined the reaction kinetics across a range of temperatures (Figure 6A and Supplementary Figure S6).
Figure 5.
(A) Time-resolved 15N-edited-imino proton spectra of Tel23 G4 after addition of 5 eq. BMVC-8C3O, with site-specific labeling at the G-4 position. The spectra were recorded 2, 6, 12 and 18 h after ligand addition, as shown in the ascending order. (B) Kinetic curves of the G-4, G-9 and G-11 intermediates of Tel23 G4 after addition of 5 eq. BMVC-8C3O at 303 K.
Figure 6.
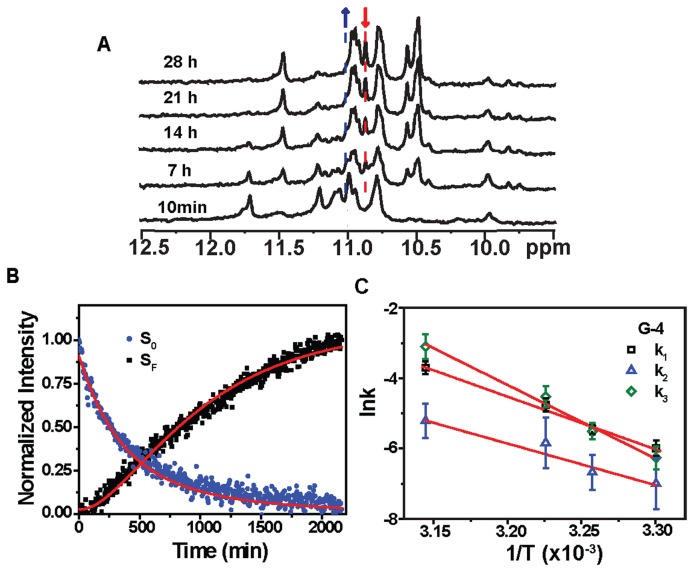
(A) Imino proton NMR spectra of the Tel23 G4 in the presence of 150 mM K+ (bottom) and time-resolved imino proton NMR spectra recorded at 10 min and at 7, 14, 21 and 28 h after addition of 5 eq. BMVC-8C3O at 303K, as indicated in the ascending order. (B) The kinetic trace from the intensity of peaks of the imino proton in G-4. The decay signal of the initial form (blue) and the arising signal of the final form (black) were normalized and then fitted to a linear three-state kinetic model (red lines). (C) Arrhenius plots of the rate constants k1, k2 and k3 of imino proton G-4 against temperature, with the slopes of 30, 24 and 42 kcal/mol, respectively.
Although the intermediate signals of other G4-forming residues were too weak to be quantified, there is evidence to suggest the involvement of multiple steps during the topological conversion given the differential kinetic rates observed for G-4, G-9 and G-11 (Figure 5B). Thus, the transition pathway of topological conversion can be described as:
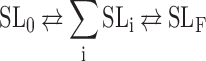 |
where SL0 is the initial state, SLF is the final state, and 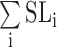 is the sum of a series of intermediate states that form either sequentially or independently along parallel pathways. The spectral characteristics of SL0 and SLF can be established in steady states; detection of the individual intermediates, however, is not feasible in our current experimental setup. We therefore substituted
is the sum of a series of intermediate states that form either sequentially or independently along parallel pathways. The spectral characteristics of SL0 and SLF can be established in steady states; detection of the individual intermediates, however, is not feasible in our current experimental setup. We therefore substituted 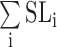 by an apparent intermediate state SLI by a first-order approximation such that the kinetic reaction of the conformational change for an individual guanine is fitted to a three-state transition model (Equation S2) as follows:
by an apparent intermediate state SLI by a first-order approximation such that the kinetic reaction of the conformational change for an individual guanine is fitted to a three-state transition model (Equation S2) as follows:
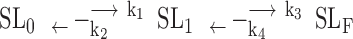 |
where the k1 and k2 are the forward and reverse transition rates associated with the first step between the initial and intermediate states, respectively, while k3 and k4 are the forward and reverse transition rates associated with the second step between the intermediate and final states, respectively.
Using the time-resolved imino proton signals of G-4, which has the highest spectral quality (Figure 6B and Supplementary Figure S6), associated with the initial and final states of individual guanines as input for a three-state kinetic model (Equation S2), we could extract the transition rates, k1, k2 and k3 associated with each step of the topological conversion from the initial hybrid form (SL0) via the folding intermediate (SLI), to the parallel form (SLF) (Figures 6B, Supplementary Figures S6, S7 and Supplementary Table S3). Here, we ignore the contribution from k4 because the uncertainty of k4 parameter is very large during the fitting. Simulation of the three-state kinetic transition with two different k4 values, 0 and 10−4 min−1, shows essentially identical results (Supplementary Figure S8). In addition, the topological conversion from the initial hybrid G4 to the final parallel G4 suggests the k4 rate should be much lower than the k3 rate.
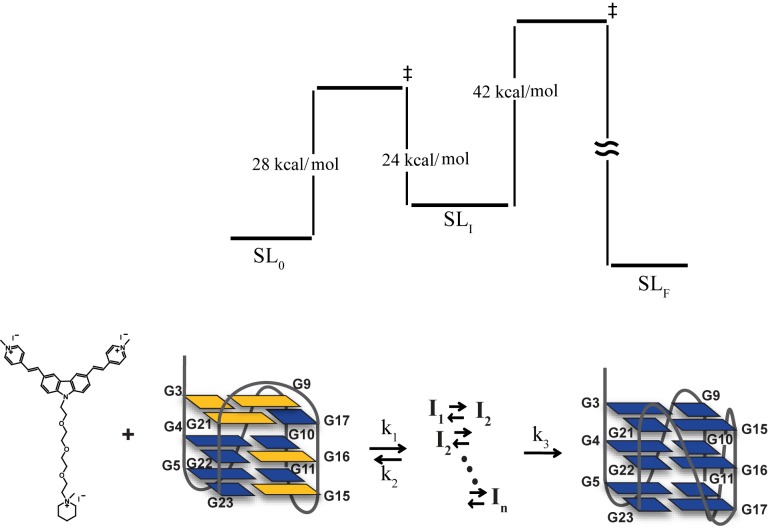
The temperature-dependent kinetic rates were subsequently fit to an Arrhenius equation to yield three different activation energies (Materials and Methods), which correspond to the kinetic barrier from the initial state to the intermediate state, Ea(k1) ≈ 30 kcal/mol, the kinetic barrier from the intermediate state back to the initial state, Ea(k2) ≈ 24 kcal/mol, and the kinetic barrier from the intermediate state to the final state, Ea(k3) ≈ 42 kcal/mol (Figure 6C and Supplementary Table S4). Similar energy parameters could be obtained with uncertainty for G-21 despite the apparent differences in the observed kinetic rates (Supplementary Figure S9 and Supplementary Table S3). Based on these results, an energy diagram was constructed to describe the process of topological conversion of the Tel23 G4 under the influence of BMVC-8C3O (Figure 7).
Figure 7.
A simple energy diagram of topological conversion from a hybrid G4 to a parallel G4 of Tel23 induced by BMVC-8C3O binding.
DISCUSSION
In this work, we first used NMR spectroscopy to determine the end topology of the BMVC-8C3O-bound Tel23 G4 to be the same parallel form as the PEG-induced form in K+ solution. Since the G4 topology of Tel23 has been determined as a hybrid-I form (7,8), it appears that BMVC-8C3O can induce topological conversion of Tel23 from a hybrid-I G4 to a parallel G4 in K+ solution. Previously, Yatsunyk et al. (31) showed that N-methyl mesoporphyrin IX (NMM) can induce the isomerization of Tel22 into a parallel topology in K+ solution because the CD spectra of NMM binding to Tel22 in 5 mM K+ solution showed a spectral conversion from a non-parallel G4 characterized by the 290 nm CD band to a parallel G4 characterized by the 265 nm CD band. In addition, they determined the crystallographic structure of Tel22 binding to NMM as a propeller type parallel G4 (32). It is worth noting that a parallel topology has also been determined by X-ray crystallography for Tel22 in the presence of K+ (20). However, the addition of NMM results in rapid signal loss of the imino proton resonances Tel22 in 5 mM K+ together with the emergence of broad signals that are attributed to NMM (Supplementary Figure S10A). Despite the CD spectral conversion (Supplementary Figure S10B), the poorly resolved NMR resonances suggest the formation of higher order aggregates with undefined topologies, which are difficult to characterize by NMR spectroscopy (33,34). Due to differences in experimental conditions, caution should be taken when using crystal structures to describe the topological changes of G4s in solution (20,35).
The BMVC-8C3O induced topological conversion of Tel23 in 150 mM K+ solution may be entirely different from the effect of NMM binding to Tel22 in 5 mM K+ solution. In this study, instead of creating a dehydrating bulk environment using highly concentrated PEG, we demonstrated that a stoichiometric amount of BMVC-8C3O is sufficient to achieve the ligand binding-induced topological conversion because increase in the local concentration of the ethylene glycol moiety of BMVC-8C3O is capable of generating localized dehydration (19). Therefore, our results suggest a novel strategy for ligand-induced topological switch in G4s, exploiting the local dehydration effect.
NMR-HDX spectroscopy provides valuable insights into the transition pathway at the level of individual guanine bases during the topological conversion. The NMR-HDX results of BMVC-8C3O-induced topological conversion of Tel23 reveal the presence of persistent imino proton signals during HDX that correspond to the central four guanines of Tel23. The gradual decay of initial hybrid G4 signals and the emergence of the final parallel G4 signal take place on a much slower timescale compared to the intrinsic HDX rates for unprotected imino protons, suggesting that the topological conversion does not involve major global unfolding events that would otherwise lead to immediate NMR signal loss during HDX. Although the primary component of Tel23 G4 in K+ solution has been determined as a hybrid-I form, a minor population of Tel23 G4 was observed but not characterized. It is therefore important to examine whether the topological conversion between the two distinct G4 structures can be attributed to conformational selection or induced fit (31,36). Conformational selection is the result of the higher binding preference for final structure, resulting a topological change via stabilizing the thermodynamic favorable complex, while induced fit involves binding to the initial structure followed by conformational rearrangement to the final structure (37,38).
To address this question, we used highly enriched site-specifically 15N-labeled Tel23 G4 to rule out the contribution of a minor component, which may be a parallel form, to the observed NMR signals at the initial state. We also determined the binding kinetics of BMVC-8C3O binding to the Tel23 G4 by interferometry measurement, which showed that BMVC-8C3O exhibits higher association rates (on the timescale of tens of millisecond) and lower dissociation rates (on the timescale of about hundreds of seconds) that are indiscriminative of the types of G4 topologies. Considering that the kinetics of ligand binding is over six orders of magnitudes higher than that of G4 topological conversion (milliseconds versus hundreds of minutes), these two models are not mutually exclusive (37,38). If the topological conversion were a result of conformational selection and the topological conversion were on the slow-exchange regime on an NMR timescale, one would expect the coexistence of the imino proton resonances corresponding to the initial and final states during the time course of topological conversion, which was not the case for Tel23 G4 in the presence of stoichiometric amount of BMVC-8C3O. Therefore, the induced fit model is a more likely scenario for explaining the topological conversion of Tel23 from a hybrid-I G4 to a parallel G4 induced by BMVC-8C3O. A similar conclusion could be drawn for Tel25, which showed distinct imino proton signals, corresponding to the central G-quartet, that converted from the initial hybrid-II G4 to the final parallel G4 under HDX.
Having examined the possible mechanism of ligand-driven G4 structural conversion, we next investigated how G4s undergo topological conversions upon BMVC-8C3O binding by NMR-HDX analyses. Imino-proton-based NMR-HDX provides exquisite sensitivity toward G-quartet opening during conformational rearrangements, as the time window of HDX for duplex DNA is on the timescale of milliseconds (39). Thus, imino protons of an opened guanine tract must be exchanged by deuterium due to slow topological conversion. Hence, the long-lived imino proton signals of the central G-quartet due to transition from a hybrid G4 to a parallel G4 of Tel23 induced by BMVC-8C3O suggests that there is no global unfolding of G-tract involved during topological conversion. Since flipping of the third guanine tract (G-15, G-16 and G-17) is capable of converting a hybrid-I G4 to a parallel G4, we propose that the local rearrangement of flipping the third guanine tract is the major mechanism for topological conversion of Tel23 G4. In addition, the detection of intermediate signals suggests that topological conversion from a hybrid G4 to a parallel G4 is not due to a simple two-state transition by flipping of the G-tract, but involves multiple local rearrangements, due to the fact that some guanines require the anti-syn conversions during topological conversion. A similar mechanism can also account for the topological conversion from hybrid-II G4 to a parallel G4 of Tel25 induced by BMVC-8C3O.
Despite the involvement of one G•C•G•C quartet in 22-CTA G4 in K+ solution, NMR-HDX analysis of 22-CTA, which adopts an anti-parallel chair G4, showed no emergence of its imino proton signals of end parallel G4 during topological conversions. The difference in the NMR-HDX data between Tel23 and 22-CTA suggests the involvement of different transition pathways associated with the topological conversion. It is significant that a single strand flipping can convert a hybrid G4 to a parallel G4 as in the case of Tel23 and Tel25 without exposing the middle four imino protons too much to allow HDX, while a single strand flipping cannot convert an anti-parallel G4 to a parallel G4 as in the case of 22-CTA.
The dynamics and kinetics associated with topological conversions of G4s have been studied by several research groups using molecular dynamics simulations (23), spectroscopic methods (40–42) and single-molecule mechanical analysis (43). On the basis of these findings, Chaires et al. (44) proposed a folding mechanism for Tel22, where a single-stranded oligonucleotide rapidly folds into a U-shaped hairpin, followed by formation of an anti-parallel chair G4, and eventually a much slower conversion step to the final hybrid G4 via formation of a triplex intermediate. In their model, opening the first guanine tract (G-2, G-3 and G-4) can lead to the proposed triplex intermediate and subsequent flipping of this guanine tract can convert the structure into a hybrid-I G4. Very recently, Schwalbe et al. (25) employed high resolution NMR spectroscopy to demonstrate the presence of a slow topological change between hybrid-I and hybrid-II of Tel24-M through a partially unfolded hairpin-like intermediate state. Thus, it is possible that the conversion from an anti-parallel G4 to a parallel G4 may undergo the formation of unfolded intermediate states. Alternatively, the topological conversion from an anti-parallel chair G4 to a parallel G4 could take the transition pathway via a hybrid G4 by flipping the first G-tract and the third G-tract. Nevertheless, the details of the transient intermediates for topological conversion of anti-parallel G4s remain to be elucidated.
The use of highly enriched site-specifically 15N-labeled samples enabled us to unambiguously identify an intermediate signal associated with its initial and final imino proton NMR signals. Since the ligand binding rate is much higher than the rate of ligand-induced topological conversion, a three-state kinetic model was proposed to describe slow conversion of Tel23 G4 induced by BMVC-8C3O. Time-resolved imino proton NMR signals of G-4 and G-21 as a function of temperature was used to derive the associated activation energies. The results showed that the folding intermediate SLI is separated by two kinetic barriers: (i) from the initial state SL and (ii) from the final state SLF, (26–30 kcal/mol for the first forward process, 24 kcal/mol for the reverse process and 41–42 kcal/mol for the second process). Based on CD analysis data in the presence of 40% PEG with data fitting to a two-state model to yield a single kinetic barrier (Ea ≈ 33 kcal/mol), the results are comparable to the kinetic barrier reported by Phan et al. (14). Recently, Schwalbe et al. (25) used a unique imino proton signal to monitor the topological conversion of a hybrid I–hybrid II transition of a DNA G4 by NMR spectroscopy; they determined the associated activation energy of 32–55 kcal/mol, which is comparable with the value that we reported in this study. Our findings indicate that the structural transition can start from the initial hybrid form via multiple intermediate states (which could not be resolved spectroscopically in our study) to the final parallel form with a long time scale (hundreds of minutes). The much higher energy barrier (41–42 kcal/mol) in comparison with the previously determined energy barrier (33 kcal/mol) for PEG-induced conformational transition of G4s can be attributed to steric hindrance of ligand binding.
CONCLUSION
In this study, we determined the final topology of a human telomeric G4, Tel23, in complex with a G4-specific ligand, BMVC-8C3O, to be a parallel topology that is distinct from the initial hybrid topology. NMR-HDX analysis reveals a novel transition pathway via multiple steps of asymmetric local rearrangements for the ligand-induced topological conversion of the hybrid G4s. In contrast, the anti-parallel 22-CTA G4 may undergo a different transition pathway. Thermodynamic studies on topological conversion of these telomeric G4s induced by BMVC-8C3O will be investigated in future studies. Finally, we delineate a three-state free-energy diagram based on time-resolved NMR analysis as a function of temperature. The results revealed two large kinetic barriers that separate the folding intermediate from the initial hybrid form and the final parallel form of the Tel23 G4, which display consistent lower conversion rates in the order of hours at room temperature. Investigation of the ligand-DNA interaction is important not only for drug design but also for better understanding the mechanisms underlying DNA dynamics.
Supplementary Material
Acknowledgments
We thank Dr Shing-Jong Huang and Mrs. Shou-Ling Huang (Instrumentation Center, National Taiwan University) for assistance in obtaining the Bruker AVIII 500 & 800 MHz FT-NMR data and Dr. Shu-Chuan (Chris) Jao (Biophysical Core Facility, Academia Sinica) for assistance on the Bio-Layer Interferometry experiments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Ministry of Science and Technology of the Republic of China [ MOST-103-2113-M-001-017-MY3 to T.C.C.]; Career Development Award [CDA-00025/2010-C] from the International Human Frontier Science Program; Ministry of Science and Technology of the Republic of China [100-2113-M-001-031-MY2, 101-2627-M-001-004 and 102-2113-M-001-017-MY2]; Academia Sinica, Taiwan (to S.T.D.H).
Conflict of interest statement. None declared.
REFERENCES
- 1.Williamson J.R., Raghuraman M.K., Cech T.R. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 2.Sundquist W.I., Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- 3.Zahler A.M., Williamson J.R., Cech T.R., Prescott D.M. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 4.Mergny J.L., Helene C. G-quadruplex DNA: a target for drug design. Nat. Med. 1998;4:1366–1367. doi: 10.1038/3949. [DOI] [PubMed] [Google Scholar]
- 5.Neidle S., Parkinson G. Telomere maintenance as a target for anticancer drug discovery. Nat. Rev. Drug Discov. 2002;1:383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- 6.Rezler E.M., Bearss D.J., Hurley L.H. Telomere inhibition and telomere disruption as processes for drug targeting. Ann. Rev. Pharm. Toxicol. 2003;43:359–379. doi: 10.1146/annurev.pharmtox.43.100901.135733. [DOI] [PubMed] [Google Scholar]
- 7.Luu K.N., Phan A.T., Kuryavyi V., Lacroix L., Patel D.J. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phan A.T., Kuryavyi V., Luu K.N., Patel D.J. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007;35:6517–6525. doi: 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim K.W., Amrane S., Bouaziz S., Xu W., Mu Y., Patel D.J., Luu K.N., Phan A.T. Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers. J. Am. Chem. Soc. 2009;131:4301–4309. doi: 10.1021/ja807503g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai J., Carver M., Punchihewa C., Jones R.A., Yang D. Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007;35:4927–4940. doi: 10.1093/nar/gkm522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray R.D., Petraccone L., Trent J.O., Chaires J.B. Characterization of a K+-induced conformational switch in a human telomeric DNA oligonucleotide using 2-aminopurine fluorescence. Biochemistry. 2010;49:179–194. doi: 10.1021/bi901357r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai J., Carver M., Yang D. Polymorphism of human telomeric quadruplex structures. Biochimie. 2008;90:1172–1183. doi: 10.1016/j.biochi.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phan A.T. Human telomeric G-quadruplex: structures of DNA and RNA sequences. FEBS J. 2010;277:1107–1117. doi: 10.1111/j.1742-4658.2009.07464.x. [DOI] [PubMed] [Google Scholar]
- 14.Heddi B., Phan A.T. Structure of human telomeric DNA in crowded solution. J. Am. Chem. Soc. 2011;133:9824–9833. doi: 10.1021/ja200786q. [DOI] [PubMed] [Google Scholar]
- 15.Miyoshi D., Nakao A., Sugimoto N. Molecular crowding regulates the structural switch of the DNA G-quadruplex. Biochemistry. 2002;41:15017–15024. doi: 10.1021/bi020412f. [DOI] [PubMed] [Google Scholar]
- 16.Xue Y., Kan Z.Y., Wang Q., Yao Y., Liu J., Hao Y.H., Tan Z. Human telomeric DNA forms parallel-stranded intramolecular G-quadruplex in K+ solution under molecular crowding condition. J. Am. Chem. Soc. 2007;129:11185–11191. doi: 10.1021/ja0730462. [DOI] [PubMed] [Google Scholar]
- 17.Miller M.C., Buscaglia R., Chaires J.B., Lane A.N., Trent J.O. Hydration is a major determinant of the G-quadruplex stability and conformation of the human telomere 3' sequence of d(AG3(TTAG3)3) J. Am. Chem. Soc. 2010;132:17105–17107. doi: 10.1021/ja105259m. [DOI] [PubMed] [Google Scholar]
- 18.Hansel R., Lohr F., Foldynova-Trantirkova S., Bamberg E., Trantirek L., Dotsch V. The parallel G-quadruplex structure of vertebrate telomeric repeat sequences is not the preferred folding topology under physiological conditions. Nucleic Acids Res. 2011;39:5768–5776. doi: 10.1093/nar/gkr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z.F., Chang T.C. Molecular engineering of G-quadruplex ligands based on solvent effect of polyethylene glycol. Nucleic Acids Res. 2012;40:8711–8720. doi: 10.1093/nar/gks578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parkinson G.N., Lee M.P.H., Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 21.Parkinson G.N., Cuenca F., Neidle S. Topology conservation and loop flexibility in quadruplex-drug recognition: Crystal structures of inter- and intramolecular telomeric DNA quadruplex-drug complexes. J. Mol. Biol. 2008;381:1145–1156. doi: 10.1016/j.jmb.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 22.Ambrus A., Chen D., Dai J., Bialis T., Jones R.A., Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mashimo T., Yagi H., Sannohe Y., Rajendran A., Sugiyama H. Folding pathways of human telomeric type-1 and type-2 G-quadruplex structures. J. Am. Chem. Soc. 2010;132:14910–14918. doi: 10.1021/ja105806u. [DOI] [PubMed] [Google Scholar]
- 24.Wang Z.F., Li M.H., Hsu S.T., Chang T.C. Structural basis of sodium-potassium exchange of a human telomeric DNA quadruplex without topological conversion. Nucleic Acids Res. 2014;42:4723–4733. doi: 10.1093/nar/gku083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bessi I., Jonker H.R., Richter C., Schwalbe H. Involvement of long-lived intermediate states in the complex folding pathway of the human telomeric G-quadruplex. Angew. Chem. Int. Ed. Engl. 2015;54:8444–8448. doi: 10.1002/anie.201502286. [DOI] [PubMed] [Google Scholar]
- 26.Phan A.T., Patel D.J. A site-specific low-enrichment 15N,13C isotope-labeling approach to unambiguous NMR spectral assignments in nucleic acids. J. Am. Chem. Soc. 2002;124:1160–1161. doi: 10.1021/ja011977m. [DOI] [PubMed] [Google Scholar]
- 27.Hsu S.T., Varnai P., Bugaut A., Reszka A.P., Neidle S., Balasubramanian S. A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics. J. Am. Chem. Soc. 2009;131:13399–13409. doi: 10.1021/ja904007p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim K.W., Alberti P., Guedin A., Lacroix L., Riou J.F., Royle N.J., Mergny J.L., Phan A.T. Sequence variant (CTAGGG)n in the human telomere favors a G-quadruplex structure containing a G:C:G:C tetrad. Nucleic Acids Res. 2009;37:6239–6248. doi: 10.1093/nar/gkp630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu L., Lim K.W., Bouaziz S., Phan A.T. Giardia telomeric sequence d(TAGGG)4 forms two intramolecular G-quadruplexes in K+ solution: effect of loop length and sequence on the folding topology. J. Am. Chem. Soc. 2009;131:16824–16831. doi: 10.1021/ja905611c. [DOI] [PubMed] [Google Scholar]
- 30.Korobov V., Ochkov V. Chemical kinetics with Mathcad and Maple. Vienna: Springer-Verlag/Wien; 2011. pp. 35–72. [Google Scholar]
- 31.Nicoludis J.M., Barrett S.P., Mergny J.L., Yatsunyk L.A. Interaction of human telomeric DNA with N-methyl mesoporphyrin IX. Nucleic Acids Res. 2012;40:5432–5447. doi: 10.1093/nar/gks152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicoludis J.M., Miller S.T., Jeffrey P.D., Barrett S.P., Rablen P.R., Lawton T.J., Yatsunyk L.A. Optimized end-stacking provides specificity of N-methyl mesoporphyrin IX for human telomeric G-quadruplex DNA. J. Am. Chem. Soc. 2012;134:20446–20456. doi: 10.1021/ja3088746. [DOI] [PubMed] [Google Scholar]
- 33.Lin C.T., Tseng T.Y., Wang Z.F., Chang T.C. Structural conversion of intramolecular and intermolecular G-quadruplexes of bcl2mid: the effect of potassium concentration and ion exchange. J. Phys. Chem. B. 2011;115:2360–2370. doi: 10.1021/jp107623n. [DOI] [PubMed] [Google Scholar]
- 34.Kuo M.H., Wang Z.F., Tseng T.Y., Li M.H., Hsu S.T., Lin J.J., Chang T.C. Conformational transition of a hairpin structure to G-quadruplex within the WNT1 gene promoter. J. Am. Chem. Soc. 2015;137:210–218. doi: 10.1021/ja5089327. [DOI] [PubMed] [Google Scholar]
- 35.Li J. Not so crystal clear: the structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005;33:4649–4659. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buscaglia R., Miller M.C., Dean W.L., Gray R.D., Lane A.N., Trent J.O., Chaires J.B. Polyethylene glycol binding alters human telomere G-quadruplex structure by conformational selection. Nucleic Acids Res. 2013;41:7934–7946. doi: 10.1093/nar/gkt440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hammes G.G., Chang Y.C., Oas T.G. Conformational selection or induced fit: a flux description of reaction mechanism. Proc. Natl. Acad. Sci. U.S.A. 2009;106:13737–13741. doi: 10.1073/pnas.0907195106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogt A.D., Di Cera E. Conformational selection or induced fit? a critical appraisal of the kinetic mechanism. Biochemistry. 2012;51:5894–5902. doi: 10.1021/bi3006913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel D.J., Ikuta S., Kozlowski S., Itakura K. Sequence dependence of hydrogen-exchange kinetics in DNA duplexes at the individual base pair level in solution. Proc. Natl. Acad. Sci. U.S.A. 1983;80:2184–2188. doi: 10.1073/pnas.80.8.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gray R.D., Li J., Chaires J.B. Energetics and kinetics of a conformational switch in G-quadruplex DNA. J. Phys. Chem. B. 2009;113:2676–2683. doi: 10.1021/jp809578f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gray R.D., Buscaglia R., Chaires J.B. Populated intermediates in the thermal unfolding of the human telomeric quadruplex. J. Am. Chem. Soc. 2012;134:16834–16844. doi: 10.1021/ja307543z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boncina M., Lah J., Prislan I., Vesnaver G. Energetic basis of human telomeric DNA folding into G-quadruplex structures. J. Am. Chem. Soc. 2012;134:9657–9663. doi: 10.1021/ja300605n. [DOI] [PubMed] [Google Scholar]
- 43.Koirala D., Ghimire C., Bohrer C., Sannohe Y., Sugiyama H., Mao H. Long-loop G-quadruplexes are misfolded population minorities with fast transition kinetics in human telomeric sequences. J. Am. Chem. Soc. 2013;135:2235–2241. doi: 10.1021/ja309668t. [DOI] [PubMed] [Google Scholar]
- 44.Gray R.D., Trent J.O., Chaires J.B. Folding and unfolding pathways of the human telomeric G-quadruplex. J. Mol. Biol. 2014;426:1629–1650. doi: 10.1016/j.jmb.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.