Abstract
Accurate recognition of splice sites is essential for pre-messenger RNA splicing. Mammalian 5′ splice sites are mainly recognized by canonical base-pairing to the 5′ end of U1 small nuclear RNA, yet we described multiple noncanonical base-pairing registers by shifting base-pair positions or allowing one-nucleotide bulges. By systematic mutational and suppressor U1 analyses, we prove three registers involving asymmetric loops and show that two-nucleotide bulges but not longer can form in this context. Importantly, we established that a noncanonical uridine-pseudouridine interaction in the 5′ splice site/U1 helix contributes to the recognition of certain 5′ splice sites. Thermal melting experiments support the formation of noncanonical registers and uridine-pseudouridine interactions. Overall, we experimentally validated or discarded the majority of predicted noncanonical registers, to derive a list of 5′ splice sites using such alternative mechanisms that is much different from the original. This study allows not only the mechanistic understanding of the recognition of a wide diversity of mammalian 5′ splice sites, but also the future development of better splice-site scoring methods that reliably predict the effects of disease-causing mutations at these sequences.
INTRODUCTION
Splicing initiates by the recognition of the three essential cis-acting splicing elements, which are the 5′ and 3′ splice sites (5′ss and 3′ss) at either end of introns as well as the branch point sequence (BPS). In major or U2-type introns, which usually have GT-AG boundaries and comprise over 99% of all mammalian introns (1), these elements are respectively recognized by binding of U1 small nuclear ribonucleoprotein (snRNP), U2-auxiliary factor (U2AF) heterodimer and U2 snRNP. These factors induce the assembly of the macromolecular and highly dynamic complex that catalyzes splicing known as the major spliceosome (2). A parallel spliceosome with mostly different snRNPs excises the minor or U12-type introns, which are ∼0.36% of the total (3). In addition, exonic or intronic splicing enhancer or silencer sequences on the pre-mRNA activate or repress splicing respectively by recruiting RNA binding proteins (4). Splicing is a crucial process in mammals, as virtually all mRNAs are regulated by alternative splicing (5,6), 10–50% of all human disease-causing mutations do so by affecting splicing, and ∼10% of all deleterious mutations map to splice sites (7,8). The algorithmic prediction of the effects of mutations on splicing is still largely unreliable, partially because these tools do not include all the mechanistic variations involved in splice-site selection (9,10).
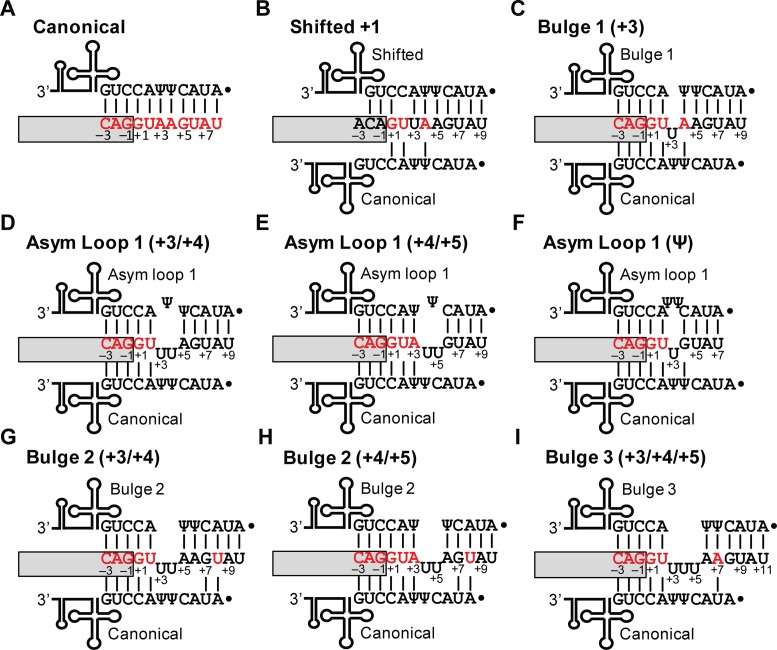
A critical step in splicing is the recognition of U2-type 5′ss via base-pairing to the 5′ end of the U1 small nuclear RNA (snRNA), which is the noncoding RNA moiety that composes the U1 snRNP along with 10 polypeptides (11–13). The 5′ss motif includes the last 3 exonic and first 8 intronic nucleotides, which can establish up to 11 base pairs with U1 snRNA for the consensus 5′ss in the most common, canonical register (14) (Figure 1A). Despite their lack of conservation, the seventh and eighth nucleotides in the intron (positions +7 and +8) can contribute to 5′ss selection in mammals but perhaps not in budding yeast (15–17). Natural 5′ss are remarkably diverse, as >9000 distinct sequences function as bona fide 5′ss in the human transcriptome. Indeed, it is not understood how a single trans-acting factor as U1 snRNP manages to recognize such a wide diversity of sequences, and the observation that several mismatches are allowed in 5′ss/U1 helices only explains in part this conundrum. Importantly, we showed that the 5′ss/U1 interaction allows for multiple noncanonical base-pairing registers in which the position of the 5′ss nucleotides that base-pair to the same U1 nucleotide is different. These schemes include the shifted register in which all base pairs of the 5′ss/U1 helix are ‘shifted’ by one position (10) (Figure 1B), and registers involving bulged nucleotides at either the 5′ss or the U1 strand (14,16) (Figure 1C).
Figure 1.
Noncanonical base-pairing registers between 5′ss and U1 snRNA. (A–I) Boxes represent exons. 5′ss consensus and nonconsensus nucleotides are shown in red and black, respectively. 5′ss positions are numbered according to convention, with −3 to −1 at the end of the exon, and +1 to +8 at the beginning of the intron. U1 snRNA is schematically depicted above and below the 5′ss, with its 5′ end spanning A1 to G11. In each case the canonical base-pairing scheme is shown below for comparison with the noncanonical register. Ψ, pseudouridine; Dot, 2,2,7-trimethylguanosine cap at the 5′ end of U1. Vertical lines between nucleotides represent base pairs. (A) Mammalian consensus 5′ss using canonical base-pairing. (B) 5′ss that uses the shifted register (10). (C) 5′ss that uses a register with a bulge at position +3 (or +2) (16). (D) 5′ss predicted to use asymmetric-loop (Asym loop) 1 (+3/+4) register. (E) 5′ss predicted to use asymmetric loop 1 (+4/+5). (F) 5′ss predicted to use asymmetric loop Ψ. (G) 5′ss predicted to use a register with a bulge of two nucleotides at +3 and +4. (H) 5′ss predicted to use a register with a bulge of two nucleotides at +4 and +5. (I) 5′ss predicted to use a register with a bulge of three nucleotides at +3, +4 and +5.
Later in the reaction, U1 must be replaced by U6 snRNA, which forms a few base pairs to the 5′ss and directly catalyzes the two trans-esterification reactions (18–21). There are a few examples in which U1 base-pairs at some distance from the actual 5′ss, with the cleavage site being subsequently determined by the site of U6 base-pairing (22–24). There is also a report of a natural human U2-type intron that is U1 snRNA-independent (25). Because U1 is dispensable for catalysis and likely only marks the 5′ss for subsequent U6 binding, the 5′ss/U1 helix is indeed highly flexible, allowing for a diversity of base-pairing registers.
Here, we refine our knowledge on the contribution of noncanonical 5′ss/U1 registers to splicing. We demonstrate the usage of three new registers involving asymmetric internal loops at different nucleotide positions (Figure 1D–F), and show that bulges of 2 nucleotides but not longer (Figure 1G–I) can contribute to 5′ss selection, but so far only appear to do so in artificial substrates. Importantly, we also found a pattern that restricts the formation of noncanonical base-pairing helices, but that in turn results in the formation of a noncanonical base pair between a uridine at the 5′ss and a pseudouridine in U1 (U–Ψ base pair) earlier reported for budding yeast 5′ss (26). Pseudouridines (Ψ) are C5-glycoside isomers of uridines, and this modification confers additional stability to RNA helices (27,28). We found that one of the U1 pseudouridines establishes such a noncanonical interaction that contributes to mammalian splicing. This work should enhance our understanding of noncanonical mechanisms of 5′ss selection in human cells.
MATERIALS AND METHODS
Universal minigene vector (UMV) construction and minigene cloning
We designed a ‘Universal Minigene Vector’ (UMV) to facilitate the cloning and testing of many 5′ss in their native exonic context. The original plasmid included an acyl-Coenzyme A dehydrogenase, C-4 to C-12 straight chain (ACADM) minigene cloned into pcDNA+3.1 plasmid using the HindIII and XhoI sites. This minigene consists of exons 8 to 10 of the ACADM gene with internally-deleted intervening introns, each retaining 250 nt of their native 5′ and 3′ ends. We performed PCR mutagenesis on the minigene to remove the middle exon plus 250 nt of flanking intronic sequences, and to introduce KpnI and EcoRI restriction sites in the primers (all oligonucleotide sequences available upon request). We digested the plasmid with KpnI and EcoRI (New England Biolabs, USA), gel purified and then ligated it to two 5′-phosphorylated oligonucleotides with compatible overhangs to introduce the multiple cloning site (MCS). We confirmed this and all constructs by sequencing (1st BASE, Singapore).
We used a previous list of naturally-occurring human 5′ss predicted to use noncanonical registers (16) by the UNAFold hybrid tool (29), which is built from experimental thermodynamic parameters (30). We amplified by high fidelity PCR the selected DNA fragments consisting of test exons together with 300 nt of flanking intronic sequences. We used human genomic DNA (Promega, USA) as template, PrimeSTAR® Max DNA Polymerase Premix (Takara Bio, Japan), and primers bearing restriction sites on their 5′ ends. We purified the products with Qiaquick® PCR Purification Kit (Qiagen, Germany) and digested them along with UMV plasmids with the respective pair of restriction enzymes (New England Biolabs, USA). We gel-extracted the resultant DNA fragments with the Qiaquick Gel Extraction Kit (Qiagen, Germany), and ligated them to vector at 8:1 ratio with T4 DNA Ligase (New England Biolabs, USA) to obtain the UMV splicing minigenes.
Testing 5′ss in SMN1/2 heterologous context
The SMN1/2 minigenes were used extensively in previous work (10,16,31). The SMN1/2 minigenes consist of exons 6 to 8 as well as a truncated intron 6 and a full-length intron 7 of the SMN1/2 genes cloned into the pCI vector. SMN1 exhibits nearly complete exon 7 inclusion, but this exon is predominantly skipped in SMN2 by virtue of a C-to-T transition at the sixth nucleotide in SMN2 exon 7 that affects splicing element/s (32). Therefore, by replacing the native 5′ss of exon 7 in the SMN1/2 minigenes with test 5′ss sequences via PCR mutagenesis, we examined the efficiency of the test 5′ss in these two heterologous contexts.
PCR mutagenesis
We performed PCR mutagenesis using HiFi PCR (Kapa Biosystems, USA) kits to incorporate the test 5′ss or introduce point mutations into the minigenes. For each 5′ss we used a common reverse primer whose 3′ end is complementary to a specific forward primer bearing the mutation. We removed the template DNA in the PCR products with DpnI (New England Biolabs) before bacterial transformation. We made minipreps with E.Z.N.A.® Plasmid Mini Kit (Omega Bio-tek, USA) and midipreps with PureLink® HiPure Plasmid Midiprep Kit (Invitrogen, USA).
Cell culture and transfection
We cultured HEK293T cells in Hyclone Dulbecco′s Modified Eagle′s medium (DMEM) (Thermo Scientific, USA) with 10% (v/v) fetal bovine serum (FBS) and antibiotics (100 U ml−1 penicillin and 100 mg ml−1 streptomycin). We transfected ∼50% confluent HEK293T cells in 12-well plates with 1 μg of DNA per well using 3 μl of X-tremeGENE 9 DNA Transfection Reagent (Roche, Switzerland) diluted in 100 μl of Hyclone Opti-MEM (Thermo Scientific, USA). We mixed test constructs (UMV or SMN1/2 minigenes) with control plasmids at 1:11 ratio. For suppressor experiments we mixed test constructs with suppressor U1 and control plasmids at 1:10:1.
RNA extraction, reverse transcription and PCR
We harvested cells 48 h after transfection and extracted total RNA with PureLink® RNA Mini Kit (Life Technologies, USA). We eliminated residual DNA by RQ1 RNase-Free DNaseI (Promega, USA). We reverse transcribed 1 μg of RNA with Moloney Murine Leukemia Virus Reverse Transcriptase (New England Biolabs, USA) and oligo-dT (18T).
We amplified cDNAs derived from expression of UMV (with pcDNA3.1+ backbone) or SMN1/2 (with pCI) constructs via semi-quantitative PCR, using primer pairs pcDNA.F-R, or pCI.FwB-Rv, respectively (10). These primers anneal to the transcribed portion of the plasmids upstream of the 5′ exon and downstream of the 3′ exon. We 5′-end-radio-labeled 10 pmol of the forward primer using T4 PolyNucleotide Kinase (New England Biolabs, USA) and γ-32P-ATP (Perkin-Elmer, USA) at a final concentration of 90 μCi/μl. We purified the labeled primer using MicroSpin G-25 columns (GE Healthcare, USA). We performed 23 cycles of semi-quantitative PCR with GoTaq DNA polymerase in Colourless GoTaq reaction buffer (Promega, USA). The annealing temperature was 58°C for pCI-FwB and pCI-Rv, and 54°C for pcDNA F and pcDNA R. With 23 cycles the PCR amplification remains within the exponential phase, ensuring that amplimer abundances correspond to those of their templates. We identified PCR products by agarose gel-extraction followed by sequencing (1st BASE, Singapore) with one of the PCR primers.
We separated the PCR products by 6% native PAGE at 10 V/cm for 6 h in TBE. The gels were subsequently vacuum-dried with a Model 583 gel-dryer (Bio-Rad, USA). We scanned the exposed phosphorimaging screens with Typhoon Trio, and quantified bands with ImageQuant TL software (all GE Healthcare Life Sciences, USA). Exon inclusion percentages from three experimental replicas (RT-PCRs of total RNA acquired from three independent transfections) allowed us to derive means and standard deviations (SD). SDs at or below 5% indicate that the exon-inclusion percentages are highly reproducible between experiments. We deemed values as different if the inclusion percentages between two experiments are distinct enough so that the means and standard deviations do not overlap. We generated the Figures by exposing Medical X-ray Film General Purpose Green (Kodak, USA) to the radioactive gels at −80°C and developing them with a Kodak Model 2000 X-Ray Film Processor.
Thermal melting experiments
The RNA oligonucleotides were synthesized and underwent RNase-free HPLC purification at IDT (Integrated DNA Technologies). We mixed equimolar amounts of the 5′ss and U1 oligonucleotides to a final total concentration of 8 μM, in a buffer containing 1 M NaCl, 20 mM HEPES, and 0.1 mM EDTA at a pH of 7.3. We measured the absorbance of each sample at 260 nm as a function of temperature using a Shimadzu UV-2550 UV-Vis Spectrophotometer. For each measurement, we cooled the samples from 85°C to 20°C, and then reheated them back to 85°C, at a rate of ±0.5°C/min. We performed at least three such measurements for each mixture. We derived the melting point temperature (Tm) and the free-energy (ΔGexp) of the duplexes based on a 2-state model via the Meltwin 3.5 software (33). Finally, we also derived melting curves for each oligonucleotide in isolation to ascertain that the derived Tm for each pair was not caused by self-dimers or intramolecular base-pairing.
RESULTS
This section contains three sets of data: (i) asymmetric internal-loop registers between 5′ss and U1, (ii) registers with bulges longer than one nucleotide and (iii) the contribution of a noncanonical U–Ψ interaction to 5′ss selection in human cells.
Asymmetric-loop registers in 5′ss/U1 helices
In addition to the one-nucleotide bulge registers that were already validated (Figure 1C), our previous data set included predictions of asymmetric-loop registers (Figure 1D–F) (16). Asymmetric loops have an uneven number of unpaired nucleotides on both sides of the helix which are flanked by base pairs, and the difference in number of unpaired nucleotides is used as the loop number (which is 1). By mutational analyses and suppressor U1 experiments, we demonstrate three asymmetric-loop 1 registers (10,16).
Asymmetric loop 1 (+3/+4)
In this register, nucleotides at 5′ss positions +3 and +4 as well as Ψ6 in U1 form a loop in the helix (Figure 1D). Using UNAFold (29), a total of 349 human 5′ss sequences were predicted to base-pair to U1 more stably via this noncanonical register.
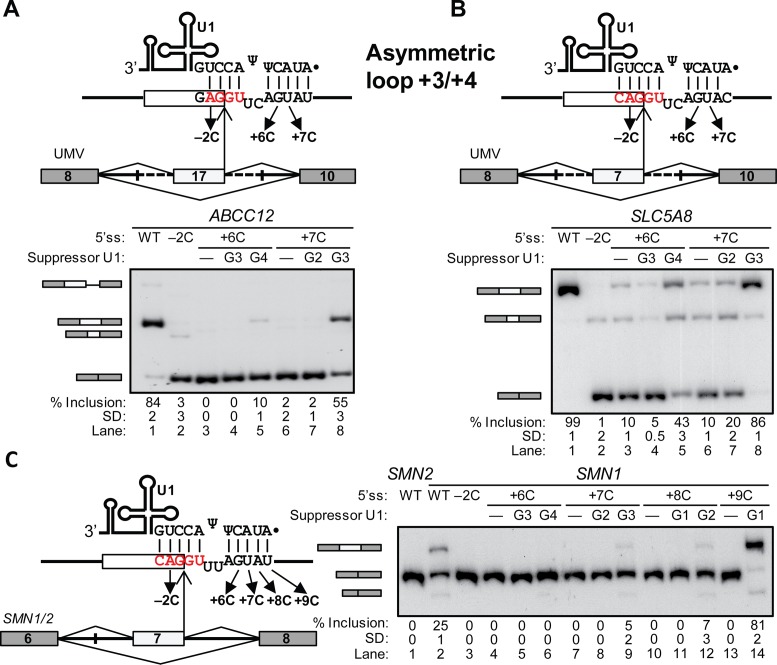
We tested this new register in three naturally-occurring 5′ss: ATP-Binding Cassette Sub-Family C Member 12 (ABCC12) exon 17, Solute Carrier Family 5 Member 8 (SLC5A8) exon 7 and Poly-ADP-Ribose Polymerase Family Member 14 (PARP14) exon 9 (Figure 2A and B; Supplementary Figures S1A and S2). We constructed UMV hybrid minigenes with the corresponding exons containing the test 5′ss and flanking intronic sequences (see Materials and Methods). We analyzed the splicing patterns of the minigenes by radioactive RT-PCR upon HEK293T cell transfection, so that exon inclusion reflects recognition of the test 5′ss (10,16). We derived mean percentage of inclusion and standard deviation from three experimental replicates per mutant (three independent transfections).
Figure 2.
Asymmetric loop 1 (+3/+4). (A–C) Schematics depict the minigenes with gray boxes as flanking exons and lines as introns, highlighting the middle exons (white box) and flanking intronic sequences (dashed lines) with the test 5′ss. U1 snRNA is schematically drawn and its 5′ end is shown base paired in the asymmetric loop 1 (+3/+4). Arrows show the tested point mutations. Bottom gel images show the radioactive RT-PCR results of transfections with the mutant minigenes and suppressor U1s indicated above each lane. The identity of the various spliced mRNAs is indicated on the left. In all gel panels, the mean percentage and Standard Deviation (SD) of middle exon inclusion are shown below each lane, which were derived from three experimental replicates (samples from independent transfections). Exon inclusion percentages are different between two experiments if the means and standard deviations do not overlap. (A) ABCC12 UMV minigene. From large to small, bands represent use of a cryptic 5′ss at position 76, correct exon 17 inclusion, use of a cryptic 5′ss at 20 nt upstream of the test 5′ss, and exon skipping. (B) SLC5A8 UMV minigene. From large to small, Bands represent correct exon 7 inclusion, use of a cryptic 5′ss at 52 nt upstream of the test 5′ss, and exon skipping. (C) SMN1 minigene. Bands represent correct exon 7 inclusion, exon skipping and use of a cryptic 5′ss 50 nucleotides upstream of the normal 5′ss of SMN1/2 exon 6 (10).
Both ABCC12 and SLC5A8 wild-type minigenes exhibited predominant test exon inclusion (Figure 2A and B, lanes 1). As expected, the −2C point mutations, which break one base pair in both registers, almost completely abolished correct exon inclusion, resulting in exon skipping and cryptic 5′ss activation (Figure 2A and B, lanes 2). The +6C and +7C, which affected only the noncanonical register, also caused strong or complete loss of exon inclusion (Figure 2A and B, lanes 3 and 6), suggesting that these 5′ss are recognized via the asymmetric loop. Mutations or deletions that eliminate looped nucleotides were not tested as these would introduce the canonical or other new registers.
We next performed suppressor U1 snRNA experiments to further test the noncanonical register hypothesis (10–13,16). For +6C mutants, exon inclusion was rescued by the suppressor U1 that restores a base pair in the asymmetric loop, which is U1 with the G4 mutation (Figure 2A and B, lanes 5; diagrams in Supplementary Figure S2A and S2C). The suppressor G3, rescuing a base pair in the canonical register for this mutant, did not restore correct splicing at all (Figure 2A and B, lanes 4). Similarly for +7C, exon inclusion was rescued by the asymmetric-loop suppressor G3, while the canonical suppressor G2 was not so effective (Figure 2A and B, lanes 7 and 8). Overall, the suppressor U1 acting via the canonical register did not restore the correct splicing patterns, or performed much less effectively versus their asymmetric-loop counterparts. We considered as rescue any suppressor-U1 driven increase in percentage of inclusion which is separate enough such that the corresponding exon inclusion means with standard deviations did not overlap. PARP14 minigene analysis also supports 5′ss recognition via asymmetric loop (Supplementary Figure S1A and S2B).
To test the general applicability of this noncanonical register, the 5′ss was analyzed in a heterologous context, the Survival of Motor Neuron 1 and 2 minigenes (SMN1/2, (10,16)). SMN1 and SMN2 are paralogous genes in which a single-nucleotide divergence in the sixth nucleotide of exon 7 results in full inclusion in SMN1 and predominant skipping in SMN2 (34). We replaced the natural exon 7 5′ss in minigenes with a representative sequence for the asymmetric loop (Figure 2C, left diagram). SMN2 transcripts with the test 5′ss displayed complete exon skipping while SMN1 retained some inclusion (Figure 2C, lanes 1 and 2). Henceforth, we used only the SMN1 minigene for mutational analysis. As above, −2C resulted in complete exon skipping, and all intronic mutations, only affecting the asymmetric loop, caused complete exon skipping (Figure 2C, lanes 3, 4, 7, 10 and 13). The +7C and +8C (but not +6C) mutations were rescued by the asymmetric loop and not the canonical suppressors, albeit to a lesser extent than in the other minigenes (Figure 2C, lanes 4–12). Finally, +9C was strongly rescued by the only possible suppressor G1, which acts via the noncanonical register (Figure 2C, lanes 13–14). All this evidence demonstrates that the test 5′ss are recognized via the asymmetric-loop register.
Other asymmetric-loop registers
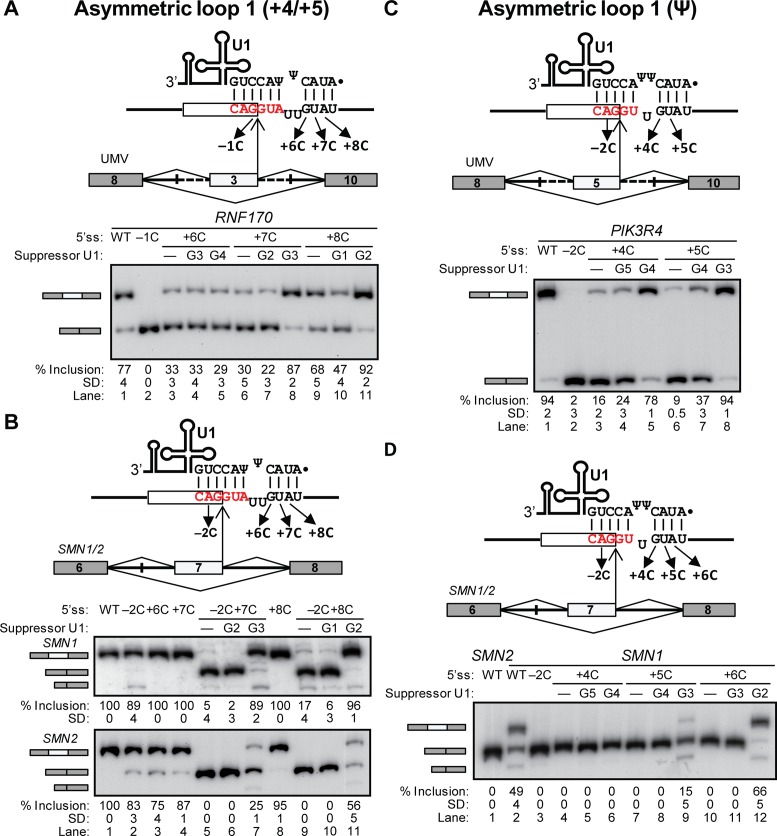
Asymmetric loop 1 (+4/+5) involves a loop with the indicated 5′ss positions and U1 Ψ5 (Figure 1E), and could account for recognition of 654 human 5′ss. The Ring Finger Protein 170 (RNF170) exon 3 minigene in UMV produced transcripts with 77% test exon inclusion, which was reduced by the +6C through +8C mutations affecting only the noncanonical register (Figure 3A, lanes 1–3, 6 and 9). Suppressor U1 snRNAs in the asymmetric loop but not in the canonical register strongly rescued inclusion in +7C and +8C (Figure 3A, lanes 6–11; diagrams in Supplementary Figure S3A). Mutational analyses and suppressor U1 experiments on the Kinase insert Domain Receptor (KDR) exon 27 and F-Box and Leucine-Rich Repeat Protein 13 (FBXL13) exon 5 minigenes gave consistent results (Supplementary Figure S1B and S1C; diagrams in Supplementary Figure S3B and S3C).
Figure 3.
Asymmetric loop 1 (+4/+5). (A–D) Schematics of the UMV (A and C) and SMN1/2 (B and D) splicing minigenes as in Figure 2, indicating the 5′ss/U1 base-pairing in the asymmetric-loop 1 (+4/+5) register (A and B) and in the asymmetric-loop Ψ register (C and D). Bottom gel images show the radioactive RT-PCR results of transfections with the mutant minigenes and suppressor U1s indicated above each lane. The identity of the various spliced mRNAs is indicated on the left. In all gel panels, the mean percentage and SD of middle exon inclusion are shown below each lane, which were derived from three experimental replicates (samples from independent transfections). (A) RNF170 UMV minigene. Bands on the left indicate exon 3 inclusion and skipping. (B) SMN1/2 minigene analysis of asymmetric loop 1 (+4/+5). Bands are labeled as in Figure 2C. (C) PIK3R4 UMV minigene. Bands on the left indicate exon 5 inclusion and skipping. (D) SMN1/2 minigene analysis of asymmetric loop Ψ. Bands are labeled as in Figure 2C.
The corresponding test 5′ss inserted in the SMN1/2 minigenes resulted in full exon 7 inclusion. While −2C slightly reduced inclusion in both minigenes, +6C through +8C only did so in SMN2 (Figure 3B, lanes 1–4, and 8). In both minigenes, the double mutants −2C+7C and −2C+8C showed a significant loss in exon inclusion that was much larger than that of the single mutants combined (Figure 3B, lanes 5 and 9). The noncanonical suppressor U1s strongly rescued inclusion in these mutants while the canonical suppressors did not (Figure 3B, lanes 5–7 and 9–11). All this evidence demonstrates that this test 5′ss are recognized via the asymmetric loop 1 (+4/+5).
In the asymmetric loop 1 Ψ, the longer side of the loop involves the two U1 Ψs (Figure 1F), and accounts for 115 predicted human 5′ss. The Phosphoinositide-3-kinase regulatory subunit 4 (PIK3R4) exon 5 minigene in UMV produced transcripts with almost complete test exon inclusion, which was fully abrogated by the −2C mutation, and partially with +4C and +5C (Figure 3C, lanes 1–3 and 6). Inclusion in the latter two mutants was more efficiently rescued by asymmetric loop than by canonical suppressors (Figure 3C, lanes 3–8; diagrams in Supplementary Figure S4A). Analysis of UMV minigenes containing DNA-directed Polymerase theta (POLQ) exon 20 and 5′-Nucleotidase Domain Containing 3 (NT5DC3) exon 12, as well as SMN1/2 minigenes gave consistent results (Figure 3D and Supplementary Figure S1D and S1E; diagrams in Supplementary Figure S4B and S4C). We conclude that the test 5′ss are recognized via the predicted noncanonical register.
Registers with bulges longer than 1 nucleotide
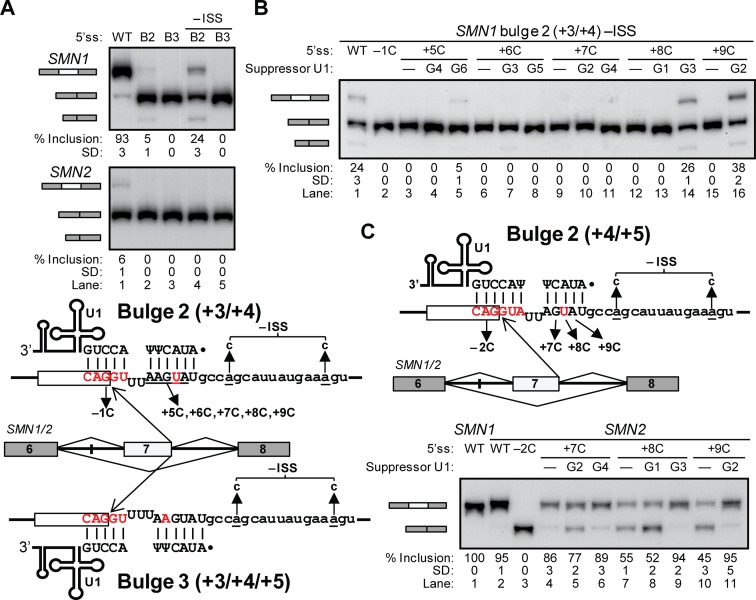
Our previous study predicted that ∼3000 5′ss base-pair to U1 by forming longer bulges on the 5′ss strand, ranging from 2 to 8 nucleotides (16). About half of these correspond to bulges of 2 or 3 nucleotides (‘bulge 2’ or ‘bulge 3’ registers) (Figure 1G–I). Test 5′ss for the bulge 2 (at +3 and +4) (Figure 1G) and bulge 3 (Figure 1I) registers in SMN1/2 were extremely inefficient (Figure 4A, lanes 2–3), with only traces of middle exon inclusion for bulge 2 in SMN1. By mutational inactivation of a known Intronic Splicing Silencer (ISS) in intron 7 (35), exon inclusion was increased in SMN1 bulge 2 (Figure 4A, -ISS, lanes 4–5) but remained undetectable in the other three minigenes. Therefore, we did not further pursue bulges longer than 2 nucleotides.
Figure 4.
Bulges of 2 or more nucleotides in heterologous SMN1/2 context. (A–C) Gel images of the RT-PCR results with different SMN1/2 minigenes and suppressor U1s as indicated on top of each lane. Splicing products with exon 7 inclusion or skipping are schematically shown on the left as in Figure 1. In all gel panels, the mean percentage and SD of middle exon inclusion are shown below each lane, which were derived from three experimental replicates (samples from independent transfections). Exon inclusion percentages are different between two experiments if the means and standard deviations do not overlap. (A) SMN1/2 minigenes with the bulge 2 (+3/+4) (b2) and bulge 3 (+3/+4/+5) (b3) model 5′ss are shown in the bottom schematics, with the base-pairing to U1 above and below the 5′ss, respectively. The two A to C transversions to inactivate the SMN1/2 intronic silencer (-ISS) as well as the 5′ss point mutations are indicated by arrows. Gel images on top show very low or no exon inclusion. (B) Gel image of the mutational analysis for the bulge 2 (+3/+4) in SMN1-ISS context. (C) Base-pairing schematic and gel image for the analysis of the bulge 2 (+4/+5) 5′ss in SMN2.
We performed mutational analysis and suppressor U1 experiments in the SMN1 bulge 2 -ISS minigene. The point mutations only breaking a base pair in the bulge register resulted in complete exon 7 skipping (Figure 4B, lanes 3, 6 and 15). The bulge but not the canonical suppressors rescued exon 7 inclusion for the +5C and +8C mutants, albeit weakly, and no suppressor rescued +6C or +7C (Figure 4B, lanes 3–14). +9C was strongly rescued by the only possible suppressor in bulge 2 (Figure 4B, lanes 1,15,16). The rescue by bulge suppressors demonstrate that the bulge 2 register is used in this artificial context. We similarly verified the register with a 2-nucleotide bulge at positions +4 and +5 in SMN2 (Figures 1H and 4C).
For bulge 2, all 5′ss tested so far in their natural exonic context showed use of the canonical register instead (Table 1, Supplementary Figure S5 and S6). Most of these 5′ss sequences deviate from the tested 5′ss, and several have a G at position +5 (see next section). We conclude that registers with bulges of 2 nucleotides can contribute to 5′ss selection but they do not appear to do so in natural 5′ss.
Table 1. 5′ss that unexpectedly show recognition by U1 via the canonical register.
| Gene | Exon | Sequence | Predicted Register | ΔΔGa UNAFold (kcal/mol) | ΔΔGa RNAstructure (kcal/mol) |
|---|---|---|---|---|---|
| RPS6KC1 | 5 | AAG/GUUUGGUAGb | Asymmetric loop 1 (+3/+4) | −4.5 | −1.6 |
| DNAI1 | 9 | CAG/GUUUGGUGUc | Asymmetric loop 1 (+3/+4) | −4.2 | −2 |
| CCDC132 | 15 | CAG/GUUUGGUUU | Asymmetric loop 1 (+3/+4) | −2.6 | 0 |
| DHODH | 6 | GAG/GUUUGAGUCG | Bulge 2 (+3/+4) | −1.8 | −1 |
| ARHGAP12 | 4 | GAG/GUAUGAGUAA | Bulge 2 (+3/+4) or (+4/+5) | −2.1 | −1.8 |
| ATG2B | 3 | CAG/GUUUGGUGU | Asymmetric loop 1 (+3/+4) | −4.2 | −2 |
aΔΔG, mean energetic advantage of the noncanonical over the canonical register in 1 M NaCl at 37°C.
b+5G highlighted in bold, and +4U is underlined.
cBase-pairing in both noncanonical and canonical registers is shown in Figure 5A.
A noncanonical +4U-Ψ5 interaction is critical for the recognition of certain 5′ss
While testing new registers, a subset of candidate 5′ss unexpectedly displayed canonical base-pairing, despite a strong energetic advantage predicted for the noncanonical registers (Table 1). All these 5′ss had a common feature, which was a G at position +5 flanked by nonconsensus nucleotides. UNAfold predicted that this +5G would not base-pair to C4 in U1 snRNA. Consistently, RNAstructure (36,37) predicts that the base-pairing profile with +5G-C4 usually confers a lesser stability than the noncanonical register. The thermodynamic instability of this base pair might be due to the lack of hydrogen bonding in the flanking mispairs being unable to overcome the electrostatic repulsion of the phosphates on the 5′ss and U1 strands (38).
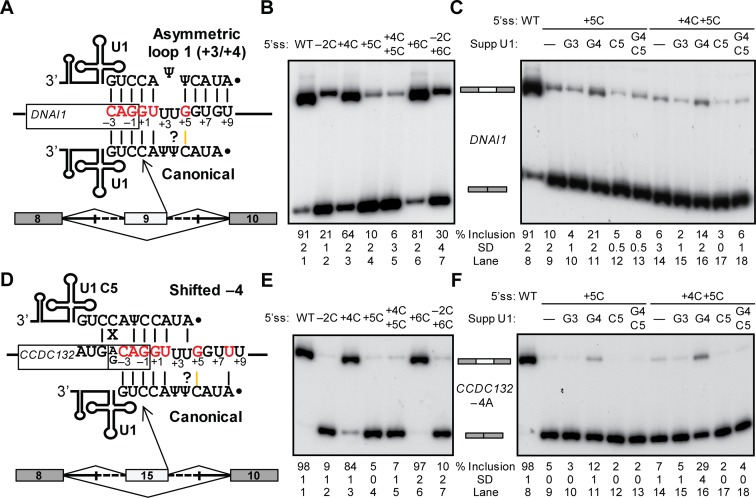
Our data demonstrates that this base pair forms in 5′ss/U1 helices. The 5′ss in exon 9 of Dynein Axonemal Intermediate Chain 1 (DNAI1), which was predicted to use an asymmetric-loop register (Figure 5A), resulted in 91% exon inclusion in UMV (Figure 5B, lane 1). Interestingly, the +5C mutation, which only disrupts a weak G–Ψ base pair in the asymmetric loop had a much stronger effect than +6C, which breaks a strong G–C base pair in this register (Figure 5B, lanes 4 and 6). The double mutant −2C+6C did not further disrupt exon inclusion when compared to −2C alone (Figure 5B, lanes 2 and 7), strongly suggesting that the contribution of +6 is negligible. The suppressor G4 rescued exon inclusion in the +5C mutant (Figure 5C, lane 11), demonstrating that +5G base-pairs to C4 in U1 in the absence of flanking Watson–Crick base pairs. These data indicate that the DNAI1 exon 9 5′ss base-pairs to U1 in the canonical register, thus contradicting the thermodynamic predictions.
Figure 5.
Contribution of a noncanonical U–Ψ interaction to 5′ss selection. (A and D) Schematics of the indicated potential base-pairing registers for the 5′ss in the DNAI1 (A) and CCDC132 -4A (D) minigenes. Yellow lines indicate the +5G−C4 base pair in canonical register, and question marks represent the potential +4U−Ψ5 interaction. (B,C,E,F) Gel images of the RT-PCR results for the mutational analyses (B and E) and suppressor U1 experiments (C and F) with different UMV DNAI1 (B and C) and CCDC132 -4A (E and F) minigenes and suppressor U1s as indicated on top of each lane. Splicing products with middle exon inclusion or skipping are schematically shown between panels. In all gel panels, the mean percentage and SD of middle exon inclusion are shown below each numbered lane, which were derived from three experimental replicates (samples from independent transfections). Exon inclusion percentages are different between two experiments if the means and standard deviations do not overlap.
Next, we hypothesized that a +4U−Ψ5 interaction stabilizes the adjacent +5G−C4, which otherwise might be too weak to form. This U–Ψ interaction could correspond to a base pair with two hydrogen bonds (39) and/or base-stacking that stabilize the adjacent base pair. Non-canonical base pairs are those that are not Watson–Crick or classical wobble (G–U or Inosine based). The non-canonical +4U−Ψ5 was reported in budding yeast 5'ss, whose consensus sequence has a U at +4 (26).
As first indirect evidence of the +4U−Ψ5 interaction, the +4C mutation in DNAI1 reduced exon inclusion much more than +6C (Figure 5B, lanes 3 and 6), strongly suggesting that position +4 is involved in the recognition of this 5′ss.
Suppressor U1 snRNA experiments further supported the formation of +4U−Ψ5. Typically, suppressor U1 experiments are used to confirm Watson–Crick base pairs, as these are disrupted and restored by compensatory mutations on the other strand. In this case, suppression of the Watson–Crick base pair at +5 was tested in the presence or absence (C mutations) of the adjacent +4U or Ψ5, individually or combined. The DNAI1 +5C was partially rescued by G4, but neither by the unrelated control suppressor G3, nor by the G4C5 which lacks the adjacent pseudouridine (Figure 5C, lanes 9–13; diagrams in Supplementary Figure S7A). The double mutant +4C+5C, which lacks the U at +4, was rescued to a lesser extent by suppressor G4 but neither by G3 nor by G4C5 (Figure 5C, lanes 14–18). These results indicate that, for suppression of the +5C mutation in this minigene, the adjacent Ψ5 is essential, and the U at +4 is important, thus supporting the contribution of the noncanonical +4U−Ψ5 interaction.
We further tested this interaction in the Coiled-Coil Domain Containing 132 gene (CCDC132) exon 15 minigene in UMV. In this minigene, the control suppressor C5 activated a spurious cryptic 5′ss 4 nucleotides upstream of the natural 5′ss (Supplementary Figure S8A–C). Thus, we inactivated this confounding 5′ss by mutating the −4G to A (Figure 5D). Mutational analysis of the CCDC132 5′ss showed consistent results with those of DNAI1, except that +6C had no effect, and +4C had smaller yet clear reduction in inclusion (Figure 5E). Importantly, +5C almost completely disrupted inclusion, which was rescued by suppressor G4 but neither with the control nor with the suppressor lacking Ψ5 (Figure 5F, lanes 8–13; diagrams in Supplementary Figure S9). Here the suppressor G4 (but not G4C5) better rescued the +5C mutation when accompanied with a C at +4 (Figure 5F, lanes 14–18), suggesting that a C4−Ψ5 forms in some instances (see Discussion). Analogous data with the ATG2B exon 3 minigene (Supplementary Figure S7B and S8D–F) also supported the contribution of noncanonical +4U−Ψ5 interactions in 5′ss/U1 helices. Lastly, U1 decoy experiments (10,16) demonstrated that the test 5′ss are recognized by U1 snRNA in cells (Supplementary Figure S10).
Thermal melting experiments support noncanonical registers and U–Ψ interactions
We carried out oligonucleotide thermal melting experiments to provide further evidence for the noncanonical mechanisms of 5′ss selection. These thermodynamic measurements reflect the intrinsic potential of 5′ss sequences to adopt distinct base-pairing conformations with U1 in the absence of proteins like the U1C polypeptide, a component of the U1 snRNP (40,41). As before (16), we designed two types of RNA oligonucleotides (Table 2): two 11-nt oligonucleotides mimicking the 5′ end of U1, with or without pseudouridines (U1–1 and U1–2); and 23 oligonucleotides with different 5′ss sequences, which include calibration controls and pairs of oligonucleotides to test three asymmetric loops, several bulge registers, and the formation of U–Ψ interactions (5′ss-1 to 5′ss-23). The four calibration oligonucleotides included the consensus 5′ss with 11 base pairs to U1 (5′ss-1), and 5′ss with 9 and 7 base pairs to U1 (5′ss-2 and 5′ss-3, respectively), as well as a very weak 5′ss that showed no specific binding (5′ss-20). The test pairs included a 5′ss predicted to base-pair to U1 with noncanonical register (NC oligonucleotide), and a control with one-nucleotide difference that abolishes the noncanonical register by disrupting a Watson–Crick base pair (C oligonucleotide right after NC).
Table 2. Summary of thermal melting data.
| Oligonucleotide | Name | Sequence (5′ to 3′) | ΔGa RNAst (kcal/mol) | ΔGUb (kcal/mol) | TmUb (°C) | ΔGΨc (kcal/mol) | TmΨc (°C) |
|---|---|---|---|---|---|---|---|
| U1 unmodified | U1–1 | AUACUUACCUG | – | – | – | – | |
| U1 modified | U1–2 | AUACΨΨACCUG | – | – | – | – | |
| Consensus | 5′ss-1 | ACAGGUAAGUAUA | −15.2d | −14.7 | 66.5 | −14.5 | 68.9 |
| Control consensus | 5′ss-2 | ACAGGUAAGUCCA | −12.6 | −13.2 | 56.4 | −13.4 | 59.4 |
| Control consensus +5C | 5′ss-3 | ACAGGUAACUCCA | −8.5 | −8.4 | 38.5 | −9.4 | 45.3 |
| Shifted +1 (NC) | 5′ss-4 | ACAGUCAAGUAUA | −6.5 | −6.8 | 29.4 | −7.6 | 34.2 |
| Shifted +1 +6C (C) | 5′ss-5 | ACAGUCAACUAUAe | −1.6 | NDTf | NDT | NDT | NDT |
| Bulge -1 (NC) | 5′ss-6 | ACAGAGUAAGUAUA | −11.4 | −10.6 | 47.4 | −11.2 | 50.3 |
| Bulge -1 -2U (C) | 5′ss-7 | ACAUAGUAAGUAUA | −8.2 | −9.1 | 40.7 | −9.3 | 43.3 |
| Asymmetric loop 1 +3/+4 (NC) | 5′ss-8 | ACAGGUUUAGUAUA | −10.0 | −10.1 | 44.7 | −11.2 | 48.7 |
| Asymmetric loop 1 +3/+4 +6C (C) | 5′ss-9 | ACAGGUUUACUAUA | −6.1 | NDT | NDT | NDT | NDT |
| Asymmetric loop 1 +4/+5 (NC) | 5′ss-10 | ACAGGUAUUGUAUA | −10.0 | −9.7 | 44.9 | −11.1 | 49.3 |
| Asymmetric loop 1 +4/+5 +6C (C) | 5′ss-11 | ACAGGUAUUCUAUA | −7.7 | −8.0 | 36.1 | −8.7 | 40.8 |
| Asymmetric loop 1 Ψ (NC) | 5′ss-12 | ACAGGUUGUAUA | −8.7 | −8.2 | 44.3 | −8.5 | 46.5 |
| Asymmetric loop 1 Ψ +6C (C) | 5′ss-13 | ACAGGUUGUCUA | −6.1 | −7.4 | 31.1 | −7.5 | 32.7 |
| Bulge 2 +3/+4 (NC) | 5′ss-14 | ACAGGUCUAAGUAUA | −10.1 | −10.6 | 46.2 | −10.9 | 48.3 |
| Bulge 2 +3/+4 +7C (C) | 5′ss-15 | ACAGGUCUAACUAUA | −6.1 | NDT | NDT | NDT | NDT |
| Bulge 2 +4/+5 (NC) | 5′ss-16 | ACAGGUAUUAGUAUA | −10.5 | −9.9 | 44.3 | −10.8 | 47.6 |
| Bulge 2 +4/+5 +7C (C) | 5′ss-17 | ACAGGUAUUACUAUA | −7.7 | −7.9 | 35.6 | −8.8 | 40.9 |
| Bulge 3 +3/+4/+5 (NC) | 5′ss-18 | ACAGGUUCUAAGUAUA | −9.7 | −10.0 | 43.8 | −10.1 | 44.7 |
| Bulge 3 +3/+4/+5 +8C (C) | 5′ss-19 | ACAGGUUCUAACUAUA | −6.1 | NDT | NDT | NDT | NDT |
| Control weak | 5′ss-20 | ACAGGUUUUUUUU | −6.1 | NDT | NDT | NDT | NDT |
| U–Ψ | 5′ss-21 | ACAGGUUUGGUUA | −7.4g | −8.9 | 40.2 | −9.9 | 44.9 |
| U–Ψ +4C | 5′ss-22 | ACAGGUUCGGUUA | −7.4h | −8.4 | 38.2 | −8.8 | 40.4 |
| U-Ψ +5C | 5′ss-23 | ACAGGUUUCGUUA | −6.1i | −7.6 | 34.5 | −8.1 | 36.9 |
aFree energy predicted by RNAstructure (RNAst) in 1 M NaCl at 37°C. All the experimental free energy values shown were measured in 1 M NaCl at 37°C.
bValues obtained by pairing the corresponding 5′ss oligonucleotide with the unmodified U1–1.
cValues obtained by pairing the corresponding 5′ss oligonucleotide with the pseudouridylated U1–2.
dValues are averages derived from three separate measurements.
eNucleotide difference between canonical (C) versus noncanonical (NC) oligonucleotides is underlined.
fNDT, No Defined Transition.
gPredicted ΔG for both asymmetric loop and for canonical register with +5G−C4 are −7.4 kcal/mol.
hPredicted ΔG for canonical register with +5G−C4 drops to −6.5 kcal/mol.
iThis predicted ΔG is for canonical register with only 5 base pairs and no +5G−C4.
We obtained thermal melting curves for each 5′ss and U1 oligonucleotide duplex, and derived experimental Tm and ΔG (ΔGexp, which can be ΔGU or ΔGΨ) (Table 2). The reliability of our measurements was substantiated by the small standard deviation of the values for three thermal melting curves, and by the correlation between ΔGexp and the predicted ΔG by RNAstructure (R = 0.96 for the unmodified U1–1). A total of 5 out of 23 5′ss mixed with U1 oligonucleotides did not show a cooperative hyperchromic shift as a function of temperature, consistent with these pairs of RNAs not base-pairing under our conditions (Table 2; Supplementary Figure S11A and S11B). The Tm measurements for duplexes with pseudouridylated U1 were on average 3.1°C (±1.9) higher than those for the unmodified oligonucleotide (Table 2; Supplementary Figure S11A and S11B), which is almost identical to the 3.4°C (±1.6) previously obtained by us with other oligonucleotides (16), and close to the 2°C measured by others (42).
When we compare the Tms and ΔGexp for each pair of NC versus C oligonucleotides, in four cases the NC oligonucleotide would base-pair with a substantially higher Tm and more favorable ΔGexp (Table 2; Supplementary Figure S11A and S11B), and this difference ranged from 6.7°C and −0.8 kcal/mol to 13.7°C and −2.4 kcal/mol. In the other four cases, the NC oligonucleotide resulted in a measurable Tm and ΔGexp, whereas the C RNA did not show a cooperative transition, and hence no values were derived. In all eight comparisons, the NC oligonucleotide base-paired to U1 more strongly than the C oligonucleotide. These results are consistent with the formation of the three asymmetric loops (5′ss-8 to 5′ss-13), two bulge 2 (5′ss-14 to 5′ss-17) and one bulge 3 register (5′ss-18 and 5′ss-19), as well as the previously published shifted register (5′ss-4 and 5′ss-5, (10)), and the predicted bulge 1 (-1) (5′ss-6 and 5′ss-7) (see Discussion).
The last four oligonucleotides tested the contribution of noncanonical U–Ψ interactions adjacent to a potential +5G−C4 base pair in 5′ss/U1 helices. The 5′ss-21 oligonucleotide is nearly identical to the CCDC132 5′ss (Figure 5D), and has a +5G−C4 base pair stabilized by +4U−Ψ5 with a canonical register, in addition to the predicted asymmetric-loop register, both with identical stability according to RNAstructure (Table 2). Consistent with both registers, 5′ss-21 binds to U1–1 and U1–2 much more strongly than the 5′ss-20 control, which shows no cooperative transition. In addition, 5′ss-22 with the +4C mutation would not affect the asymmetric loop, but would replace the U–Ψ by a weaker C–Ψ base pair (39). The lower Tm for 5′ss-22 (compared to 5′ss-21) with both U1 oligonucleotides strongly suggests formation of the +5G−C4 base pair stabilized by +4U−Ψ5 (Supplementary Figure S11C and 11D). In turn, 5′ss-23 has a C at +5 which breaks the strong +5G−C4 base pair and also the asymmetric loop, thus lowering the Tm even more. The contribution of the pseudouridine modification to the noncanonical interaction is not clear, as the unmodified U1–1 oligonucleotide with 5′ss-21 also shows high Tm, thus reflecting a U–U interaction (43–45). Nevertheless, the comparison of 5′ss-21 versus the single point mutations at +4 or +5 is consistent with the noncanonical +4U−Ψ5 stabilizing the adjacent +5G−C4 base pair, thus supporting the splicing data in this protein-free binding assay.
Revised list of 5′ss that potentially use noncanonical registers and U–Ψ interactions
This study generates a curated list of 5′ss that are predicted to use noncanonical base-pairing registers (Table 3). In previous work (10,16), out of a total of 201 541 bona-fide human 5′ss, we projected 10 248 (5.1% of the total) to use noncanonical registers with bulges or asymmetric loops. This list was based on the minimum free-energy of 5′ss/U1 base-pairing in the noncanonical (ΔG1, in kilocalories per mole) over the corresponding free-energy in the canonical register (ΔG2), by using UNAFold hybrid (29). For 5877 5′ss, the energetic difference between the noncanonical and canonical register was substantial (ΔΔG ≤ −1 kcal/mol, where ΔΔG = ΔG1 − ΔG2) (16). We experimentally demonstrated several registers with single-nucleotide bulges.
Table 3. Numbers and distribution of predicted non-canonical 5′ss.
| Register | Position | Noncanonical 5′ssa | 5′ss with +5Gb | Updated noncanonical 5′ssc | Average ΔΔGd | 5'ss with ΔΔG ≤ −1e |
|---|---|---|---|---|---|---|
| Total | 10 248 | 4766 | 4270 | −1.32 | 2703 | |
| Reliable | 3176 | −1.35 | 2073 | |||
| Validated | 3119 | −1.35 | 2019 | |||
| Bulge 1 | All | 5766 | 3386 | 2358 | −1.24 | 1448 |
| Ψh,i | 501 | 0 | 501 | −0.63 | 75 | |
| –1 | 2913 | 2891 | 0 | NAj | 0 | |
| +2 | 1 | 0 | 1 | −4.9 | 1 | |
| +2/+3k | 68 | 14 | 54 | −2.21 | 45 | |
| +3 | 51 | 15 | 36 | −1.65 | 28 | |
| +3/+4/+5 | 579 | 0 | 579 | −1.16 | 368 | |
| +4 | 1118 | 466 | 652 | −1.66 | 463 | |
| +5 | 535 | 0 | 535 | −1.25 | 468 | |
| Asymmetric loop 1 | All | 1179 | 361 | 818 | −1.69 | 625 |
| Ψ | 115 | 0 | 115 | −2.27 | 111 | |
| +3/+4 | 349 | 297 | 52 | −2.33 | 36 | |
| +4/+5 | 654 | 59 | 595 | −1.52 | 425 | |
| +3/+4/+5 | 52 | 0 | 52 | −1.73 | 51 | |
| With +2l | 9 | 5 | 4 | −0.78 | 2 | |
| Bulge 2 | All | 1067 | 389 | 678 | −1.15 | 395 |
| +3/+4 | 320 | 252 | 68 | −1.09 | 29 | |
| +4/+5 | 656 | 70 | 586 | −1.19 | 365 | |
| Othersm | 91 | 67 | 24 | −0.35 | 1 | |
| Asymmetric loop 2 | All | 472 | 56 | 416 | −1.37 | 235 |
| Bulge/Asymmetric loop 3–8 | All | 1764 | 574 | 0 | NA | 0 |
aPredictions from (16).
b5′ss from the original predictions with a G at +5, which would base pair in canonical register.
cCurated list in which we removed the +5G 5′ss, the bulge 1 (−1) and bulges and asymmetric loops longer than 2.
dPredicted ΔΔG = ΔG1 − ΔG2 (in 1 M NaCl at 37°C); mean energetic advantage of the bulge over the canonical register.
ePredicted ΔΔG ≤ −1, (in 1 M NaCl at 37°C); number of 5′ss in which the bulge register confers a substantial advantage over the canonical register.
fReliable list excludes the provisional registers with long bulges and asymmetric loops.
gValidated list includes only registers directly proven by experiments.
hPseudouridine at either position 5 or 6 of the 5’ end of U1 snRNA.
iBold indicates registers with experimental validation here or in (16).
jNA, not applicable.
k(+2/+3) Either 5′ss position +2 or +3 is bulged.
lGroup of asymmetric-loop 1 registers for GC 5′ss whereby the loop involves position +2 and others.
mThree registers with bulges at either both U1 pseudouridines, at 5′ss positions +2/+3 or at +5/+6.
The updated list (Table 3) includes the asymmetric-loop registers that are now experimentally validated, with the bulges of 2 nucleotides on a ‘provisional’ status. Most importantly, this list is very different from the original, because many 5′ss were removed on the basis of our data. First, we excluded 4766 5′ss with a G at +5, as +5G would likely always form canonical base-pairing. Second, we eliminated the register with bulge at position −1, as candidate 5′ss did not pass the mutational analyses (see Discussion). Third, we removed the bulges and asymmetric loops longer than two nucleotides, as our experiments disproved formation of bulge 3 in SMN1/2. Consequently, the curated list of 5′ss that potentially use noncanonical registers retains 4270 5′ss, which comprise 2.1% of the total (Supplementary Data File 1). The list is reduced to 3176 (1.6%) by excluding the provisional bulge 2 (and asymmetric loop) categories. Of these, 3119 (1.5%) belong to registers with experimental validation, and of these 2019 (1%) confer a substantial energetic advantage (ΔΔG ≤ −1 kcal/mol). These numbers strongly argue that the revised list is much more reliable than the original.
For the U–Ψ noncanonical interaction, as many as 26 048 (12.9%) of human 5′ss have a U at +4, and 16 892 of these have a G at +5. Around half of such +4U+5G 5′ss (7807, 3.9% of all) do not have a U at +6 which would base pair to A3 in U1 snRNA, so that the U–Ψ would be crucial to stabilize the +5G−C4 base pair. We conclude that the +4U−Ψ5 noncanonical interaction should be crucial to stabilize the adjacent +5G−C4 base pair in almost 4% of all 5′ss.
DISCUSSION
We present evidence for new noncanonical registers which complement yet largely modify the experimentally-verified list from our previous study (16). The new registers consist of three asymmetric loops within 5′ss/U1 helices. In addition, this work demonstrates that 2-nucleotide bulges can occur in the context of 5′ss/U1 duplexes, yet these registers were only proven in artificial SMN1/2 substrates. Importantly, our study also proved that not all predicted registers can form, so we expunged from the list more than half of the originally predicted cases. The 5′ss that are predicted to use proven bulge or asymmetric loops (without bulges of 2 or longer) represent 3119 or 1.5% of 201 541 human 5′ss (Table 3). At this stage, there are only a few 5′ss with predicted registers awaiting verification (1151 or 0.6% of all 5′ss), so we now have a clearer picture of the global impact of these mechanisms. In any case, the estimated 1.5% of 5′ss to use validated noncanonical registers still exceeds the percentage of GC-AG 5′ss (0.9%), U12-type introns (0.36%) and U2-type AT–AC splice sites with totals barely in the double digits (1,3,46). We conclude that noncanonical 5′ss/U1 base-pairing registers account for the recognition for a small but important fraction of all human 5′ss.
We discarded the register with a single-nucleotide bulge at position −1 because the candidate 5′ss did not pass the mutational analysis in UMV and SMN1/2 (16). Nevertheless, our thermal melting experiment was consistent with formation of 5′ss/U1 helices with a bulge at −1 in this simplified system. Our data suggest that a factor in or outside the U1 snRNP prevents the bulge at −1, and potentially this could be U1C (40,41,47).
We only proved the bulge 2 registers in heterologous but not in natural contexts and with the best possible 5′ss sequences, suggesting a ‘weak’ contribution of these registers to 5′ss/U1 base-pairing stability. Nevertheless, this demonstration leaves open the possibility that some 5′ss from humans or other species use this mechanism. Bulges of two or more nucleotides entail severe energetic penalties due to the loss of base-stacking (48), and the increasing angle of the helix kink as a function of bulge length (49,50). In addition, we found evidence against the contribution to splicing by bulges of 3 nucleotides. However, our thermal melting experiments argued for the formation of bulge 3 register, indicating that the disagreement between the prediction and our splicing data in cells is not merely due to energetic overestimations, but perhaps to other factors involved in 5′ss selection.
The validated asymmetric loops involve 5′ss positions +3 through +5, and the two U1 pseudouridines, which also allow single-nucleotide bulges and are restricted to the innermost positions of the helix (16). Mutations adjacent to the loop were often not rescued by suppressor U1s (such as +6C in Figure 3A), likely because these mutations also disrupted a strong G–C base pair. As previously documented (10,16), suppressor U1s may fail to rescue 5′ss recognition because of the competing and very abundant endogenous U1. Expression differences between suppressors may account for different extents of splicing rescue, yet this is minimized by using the same set of suppressors (G1–G6) throughout this study. As particular suppressors showed strong rescue in some minigenes but less or no effects in others, suppression also depends on the exonic context of the 5′ss (for instance, G3 in Figure 2A and B lanes 8 versus 2C lane 9). Besides, the new asymmetric loops are equivalent to one-nucleotide bulges followed or preceded by a mismatch. Many of the asymmetric-loop 5′ss have a U opposed to a Ψ in the loop, thus possibly forming a noncanonical U–Ψ interaction. In the context of the BPS/U2 snRNA helix, an A–Ψ base pair helps place the adjacent bulged adenosine in extrahelical conformation (51,52), but a U–Ψ base pair next to a bulge might have different effects. Finally, as for the 5′ss using the shifted register (10), the diversity of 5′ss/U1 registers might not necessarily be followed by a diversity of 5′ss/U6 helices, due to the much more limited base-pairing potential between 5′ss and U6 snRNA, and the role of U6 in splicing catalysis (18).
The contribution of the noncanonical registers to splicing implies that certain 5′ss might be longer. For instance, for asymmetric loops or two-nucleotide bulges the 5′ss might use up to +9 or +10 (the ninth or tenth intronic nucleotide) as opposed to +8 for the canonical register. Most 5′ss scoring methods only consider 6 intronic nucleotides (53–55) and others 8 (9), thereby missing 5′ss position/s that might play a role in their recognition. Nevertheless, the 5′ss/U1 base-pairing by noncanonical registers might also be favored or competed by proteins and/or by internal pre-mRNA structures (56–58). Finally, this diversity of 5′ss/U1 registers also increases the number of potential pseudo 5′ss, which are sequences—typically intronic—that resemble 5′ss but that are not apparently functional in splicing (59). Recently such sequences have been proposed to play a role in the excision of long introns by recursive splicing or related mechanisms (60), repression of pseudoexons (61) and repression of intronic polyadenylation (62,63). Indeed, a recent high-throughput mapping of U1-binding sites in the transcriptome revealed that U1 binds along introns outside the classical 5′ss (64), so it is possible that many of these intronic binding sites base-pair to U1 via noncanonical registers.
We also establish the formation of the noncanonical U–Ψ interaction between 5′ss position +4 and Ψ5 in U1 snRNA, and for the first time its contribution to mammalian splicing. Even if the +4U nucleotide also base-pairs to A43 in within the invariant U6 ACAGAG box, our suppressor U1 experiments show that the contribution of +4U to splicing is at least in large part mediated by U1. A U–Ψ noncanonical base pair can occur in either of two different conformations, each contributing two hydrogen bonds (39), in addition to base-stacking without hydrogen bonding. The +4U–Ψ5 interaction was already described in budding yeast (26), in which the very highly conserved consensus 5′ss has a +4U instead of +4A in mammals (1). We propose that the functional significance of this interaction might be different between yeast and mammals. +4U forms a weaker base pair with U1 nucleotide Ψ5 than +4A, and in yeast 5′ss such interaction facilitates the U1-off rate from the 5′ss, and thus its replacement by U6 during spliceosome assembly (26), which is rate limiting in this species (17). We disfavor the U1/U6 replacement role of the noncanonical 4U–Ψ5 in mammalian 5′ss, because extended U1/5′ss helices are non-detrimental for mammalian splicing (15). Instead, +4U interacting with Ψ5 might facilitate the recognition of a highly divergent set of mammalian 5′ss sequences, by promoting the formation of a +5G–C4 base pair in the absence of flanking canonical base pairs (our study).
Pseudouridine is the most common modified nucleotide in RNA with usually higher base-pairing stability than uridine (28,65). Recent studies are establishing the thermodynamic contributions of pseudouridines to the structural stability of RNAs (39,66), but these values are not yet incorporated into thermodynamic tools like UNAFold or RNAstructure. The role of the two pseudouridines at positions 5 and 6 of the 5′ end of U1 (67) is not clear, yet A–Ψ base pairs confer slightly higher stability than A–U (42), a trend that we also observed (Table 2). Consistent with the splicing analyses, our thermal melting results provided evidence for the +4U−Ψ5 interaction, even though it also formed in the absence of pseudouridines. Furthermore, our data supports a contribution of C–Ψ base pairs to splicing which is weaker than that of U–Ψ, like a prior thermodynamic study (39). However, the C–Ψ interaction in CCDC132 appears to be more stable than the U–Ψ, so the relative energetic parameters of these base pairs might be context-dependent. By analogy to C–U mismatches, C–Ψ may be structurally compatible with the potential adjacent +3U−6Ψ (43–45,68). Future work should elucidate the importance of noncanonical base pairs in 5′ss selection in mammals, and the role of the U1 pseudouridines in splicing.
Supplementary Material
Acknowledgments
We thank all members of the Roca lab for advice.
Author contributions: J.T., X.R. and G.C. designed the experiments. J.T., J.X.J.H. and S.F.L. conducted the splicing assays in cells. J.T. and X.R. analyzed and interpreted the splicing results with help from J.X.J.H. and S.F.L. J.T. and Z.Z. carried out the thermal melting experiments and the corresponding data analysis under supervision of G.C.. X.R. wrote the manuscript with the main help by J.T. and G.C., and all other authors read and approved the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Singapore's Ministry of Education (MoE) [Academic Research Fund (AcRF) Tier 1 grant (RG 20/11)]; SBS-NTU Startup grant 2011. Funding for open access charge: Academic Research Fund (AcRF) Tier 1 grant (RG 33/15) from Singapore Ministry of Education (MoE).
Conflict of interest statement. None declared.
REFERENCES
- 1.Sheth N., Roca X., Hastings M.L., Roeder T., Krainer A.R., Sachidanandam R. Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res. 2006;34:3955–3967. doi: 10.1093/nar/gkl556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wahl M.C., Will C.L., Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Turunen J.J., Niemela E.H., Verma B., Frilander M.J. The significant other: splicing by the minor spliceosome. Wiley Interdiscip. Rev. RNA. 2013;4:61–76. doi: 10.1002/wrna.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Busch A., Hertel K.J. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip. Rev. RNA. 2012;3:1–12. doi: 10.1002/wrna.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nilsen T.W., Graveley B.R. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krawczak M., Thomas N.S., Hundrieser B., Mort M., Wittig M., Hampe J., Cooper D.N. Single base-pair substitutions in exon-intron junctions of human genes: nature, distribution, and consequences for mRNA splicing. Hum. Mutat. 2007;28:150–158. doi: 10.1002/humu.20400. [DOI] [PubMed] [Google Scholar]
- 8.Lopez-Bigas N., Audit B., Ouzounis C., Parra G., Guigo R. Are splicing mutations the most frequent cause of hereditary disease? FEBS Lett. 2005;579:1900–1903. doi: 10.1016/j.febslet.2005.02.047. [DOI] [PubMed] [Google Scholar]
- 9.Hartmann L., Theiss S., Niederacher D., Schaal H. Diagnostics of pathogenic splicing mutations: does bioinformatics cover all bases? Front. Biosci. 2008;13:3252–3272. doi: 10.2741/2924. [DOI] [PubMed] [Google Scholar]
- 10.Roca X., Krainer A.R. Recognition of atypical 5′ splice sites by shifted base-pairing to U1 snRNA. Nat. Struct. Mol. Biol. 2009;16:176–182. doi: 10.1038/nsmb.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seraphin B., Kretzner L., Rosbash M. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5′ cleavage site. EMBO J. 1988;7:2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siliciano P.G., Guthrie C. 5′ splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev. 1988;2:1258–1267. doi: 10.1101/gad.2.10.1258. [DOI] [PubMed] [Google Scholar]
- 13.Zhuang Y., Weiner A.M. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell. 1986;46:827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- 14.Roca X., Krainer A.R., Eperon I.C. Pick one, but be quick: 5′ splice sites and the problems of too many choices. Genes Dev. 2013;27:129–144. doi: 10.1101/gad.209759.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freund M., Hicks M.J., Konermann C., Otte M., Hertel K.J., Schaal H. Extended base pair complementarity between U1 snRNA and the 5′ splice site does not inhibit splicing in higher eukaryotes, but rather increases 5′ splice site recognition. Nucleic Acids Res. 2005;33:5112–5119. doi: 10.1093/nar/gki824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roca X., Akerman M., Gaus H., Berdeja A., Bennett C.F., Krainer A.R. Widespread recognition of 5′ splice sites by noncanonical base-pairing to U1 snRNA involving bulged nucleotides. Genes Dev. 2012;26:1098–1109. doi: 10.1101/gad.190173.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Staley J.P., Guthrie C. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol. Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- 18.Fica S.M., Tuttle N., Novak T., Li N.S., Lu J., Koodathingal P., Dai Q., Staley J.P., Piccirilli J.A. RNA catalyses nuclear pre-mRNA splicing. Nature. 2013;503:229–234. doi: 10.1038/nature12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kandels-Lewis S., Seraphin B. Involvement of U6 snRNA in 5′ splice site selection. Science. 1993;262:2035–2039. doi: 10.1126/science.8266100. [DOI] [PubMed] [Google Scholar]
- 20.Lesser C.F., Guthrie C. Mutations in U6 snRNA that alter splice site specificity: implications for the active site. Science. 1993;262:1982–1988. doi: 10.1126/science.8266093. [DOI] [PubMed] [Google Scholar]
- 21.Wassarman D.A., Steitz J.A. Interactions of small nuclear RNA's with precursor messenger RNA during in vitro splicing. Science. 1992;257:1918–1925. doi: 10.1126/science.1411506. [DOI] [PubMed] [Google Scholar]
- 22.Brackenridge S., Wilkie A.O., Screaton G.R. Efficient use of a ‘dead-end’ GA 5′ splice site in the human fibroblast growth factor receptor genes. EMBO J. 2003;22:1620–1631. doi: 10.1093/emboj/cdg163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen J.B., Snow J.E., Spencer S.D., Levinson A.D. Suppression of mammalian 5′ splice-site defects by U1 small nuclear RNAs from a distance. Proc. Natl. Acad. Sci. U.S.A. 1994;91:10470–10474. doi: 10.1073/pnas.91.22.10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hwang D.Y., Cohen J.B. U1 snRNA promotes the selection of nearby 5′ splice sites by U6 snRNA in mammalian cells. Genes Dev. 1996;10:338–350. doi: 10.1101/gad.10.3.338. [DOI] [PubMed] [Google Scholar]
- 25.Fukumura K., Taniguchi I., Sakamoto H., Ohno M., Inoue K. U1-independent pre-mRNA splicing contributes to the regulation of alternative splicing. Nucleic Acids Res. 2009;37:1907–1914. doi: 10.1093/nar/gkp050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Libri D., Duconge F., Levy L., Vinauger M. A role for the Psi-U mismatch in the recognition of the 5′ splice site of yeast introns by the U1 small nuclear ribonucleoprotein particle. J. Biol. Chem. 2002;277:18173–18181. doi: 10.1074/jbc.M112460200. [DOI] [PubMed] [Google Scholar]
- 27.Arnez J.G., Steitz T.A. Crystal structure of unmodified tRNA(Gln) complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry. 1994;33:7560–7567. doi: 10.1021/bi00190a008. [DOI] [PubMed] [Google Scholar]
- 28.Charette M., Gray M.W. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49:341–351. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- 29.Markham N.R., Zuker M. UNAFold: software for nucleic acid folding and hybridization. Methods Mol. Biol. 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
- 30.Mathews D.H., Sabina J., Zuker M., Turner D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 31.Cartegni L., Hastings M.L., Calarco J.A., de Stanchina E., Krainer A.R. Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2. Am. J. Hum. Genet. 2006;78:63–77. doi: 10.1086/498853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monani U.R., Lorson C.L., Parsons D.W., Prior T.W., Androphy E.J., Burghes A.H., McPherson J.D. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 33.McDowell J.A., Turner D.H. Investigation of the structural basis for thermodynamic stabilities of tandem GU mismatches: solution structure of (rGAGGUCUC)2 by two-dimensional NMR and simulated annealing. Biochemistry. 1996;35:14077–14089. doi: 10.1021/bi9615710. [DOI] [PubMed] [Google Scholar]
- 34.Lorson C.L., Hahnen E., Androphy E.J., Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. U.S.A. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hua Y., Vickers T.A., Okunola H.L., Bennett C.F., Krainer A.R. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet. 2008;82:834–848. doi: 10.1016/j.ajhg.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mathews D.H., Disney M.D., Childs J.L., Schroeder S.J., Zuker M., Turner D.H. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. U.S.A. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reuter J.S., Mathews D.H. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinform. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gutell R.R. Comparative analysis of the higher-order structure of RNA. In: Russell R, editor. Biophysics for the Life Sciences, Biophysics of RNA folding series. Vol. 3. Springer Publishing; 2012. pp. 11–22. [Google Scholar]
- 39.Kierzek E., Malgowska M., Lisowiec J., Turner D.H., Gdaniec Z., Kierzek R. The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res. 2014;42:3492–3501. doi: 10.1093/nar/gkt1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bringmann P., Luhrmann R. Purification of the individual snRNPs U1, U2, U5 and U4/U6 from HeLa cells and characterization of their protein constituents. EMBO J. 1986;5:3509–3516. doi: 10.1002/j.1460-2075.1986.tb04676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hinterberger M., Pettersson I., Steitz J.A. Isolation of small nuclear ribonucleoproteins containing U1, U2, U4, U5, and U6 RNAs. J. Biol. Chem. 1983;258:2604–2613. [PubMed] [Google Scholar]
- 42.Hall K.B., McLaughlin L.W. Properties of a U1/mRNA 5′ splice site duplex containing pseudouridine as measured by thermodynamic and NMR methods. Biochemistry. 1991;30:1795–1801. doi: 10.1021/bi00221a010. [DOI] [PubMed] [Google Scholar]
- 43.SantaLucia J. Jr., Kierzek R., Turner D.H. Stabilities of consecutive A.C, C.C, G.G, U.C, and U.U mismatches in RNA internal loops: Evidence for stable hydrogen-bonded U.U and C.C.+ pairs. Biochemistry. 1991;30:8242–8251. doi: 10.1021/bi00247a021. [DOI] [PubMed] [Google Scholar]
- 44.Wu M., McDowell J.A., Turner D.H. A periodic table of symmetric tandem mismatches in RNA. Biochemistry. 1995;34:3204–3211. doi: 10.1021/bi00010a009. [DOI] [PubMed] [Google Scholar]
- 45.Zhong Z., Soh L.H., Lim M.H., Chen G. A U.U pair-to-U.C pair mutation-induced RNA native structure destabilisation and stretching-force-induced RNA misfolding. ChemPlusChem. 2015;80:1267–1278. doi: 10.1002/cplu.201500144. [DOI] [PubMed] [Google Scholar]
- 46.Kubota T., Roca X., Kimura T., Kokunai Y., Nishino I., Sakoda S., Krainer A.R., Takahashi M.P. A mutation in a rare type of intron in a sodium-channel gene results in aberrant splicing and causes myotonia. Hum. Mutat. 2011;32:773–782. doi: 10.1002/humu.21501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kondo Y., Oubridge C., van Roon A.M., Nagai K. Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5′ splice site recognition. Elife. 2015;4 doi: 10.7554/eLife.04986. doi:10.7554/eLife.04986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Znosko B.M., Silvestri S.B., Volkman H., Boswell B., Serra M.J. Thermodynamic parameters for an expanded nearest-neighbor model for the formation of RNA duplexes with single nucleotide bulges. Biochemistry. 2002;41:10406–10417. doi: 10.1021/bi025781q. [DOI] [PubMed] [Google Scholar]
- 49.Bhattacharyya A., Murchie A.I., Lilley D.M. RNA bulges and the helical periodicity of double-stranded RNA. Nature. 1990;343:484–487. doi: 10.1038/343484a0. [DOI] [PubMed] [Google Scholar]
- 50.Gohlke C., Murchie A.I., Lilley D.M., Clegg R.M. Kinking of DNA and RNA helices by bulged nucleotides observed by fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. U.S.A. 1994;91:11660–11664. doi: 10.1073/pnas.91.24.11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin Y., Kielkopf C.L. X-ray structures of U2 snRNA-branchpoint duplexes containing conserved pseudouridines. Biochemistry. 2008;47:5503–5514. doi: 10.1021/bi7022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Newby M.I., Greenbaum N.L. Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine. Nat. Struct. Biol. 2002;9:958–965. doi: 10.1038/nsb873. [DOI] [PubMed] [Google Scholar]
- 53.Sahashi K., Masuda A., Matsuura T., Shinmi J., Zhang Z., Takeshima Y., Matsuo M., Sobue G., Ohno K. In vitro and in silico analysis reveals an efficient algorithm to predict the splicing consequences of mutations at the 5′ splice sites. Nucleic Acids Res. 2007;35:5995–6003. doi: 10.1093/nar/gkm647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Senapathy P., Shapiro M.B., Harris N.L. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- 55.Yeo G., Burge C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 56.Eperon I.C., Makarova O.V., Mayeda A., Munroe S.H., Caceres J.F., Hayward D.G., Krainer A.R. Selection of alternative 5′ splice sites: role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol. Cell. Biol. 2000;20:8303–8318. doi: 10.1128/mcb.20.22.8303-8318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Erkelenz S., Mueller W.F., Evans M.S., Busch A., Schoneweis K., Hertel K.J., Schaal H. Position-dependent splicing activation and repression by SR and hnRNP proteins rely on common mechanisms. RNA. 2013;19:96–102. doi: 10.1261/rna.037044.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Warf M.B., Berglund J.A. Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem. Sci. 2010;35:169–178. doi: 10.1016/j.tibs.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun H., Chasin L.A. Multiple splicing defects in an intronic false exon. Mol. Cell. Biol. 2000;20:6414–6425. doi: 10.1128/mcb.20.17.6414-6425.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sibley C.R., Emmett W., Blazquez L., Faro A., Haberman N., Briese M., Trabzuni D., Ryten M., Weale M.E., Hardy J., et al. Recursive splicing in long vertebrate genes. Nature. 2015;521:371–375. doi: 10.1038/nature14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Buratti E., Baralle D. Novel roles of U1 snRNP in alternative splicing regulation. RNA Biol. 2010;7:412–419. doi: 10.4161/rna.7.4.12153. [DOI] [PubMed] [Google Scholar]
- 62.Berg M.G., Singh L.N., Younis I., Liu Q., Pinto A.M., Kaida D., Zhang Z., Cho S., Sherrill-Mix S., Wan L., et al. U1 snRNP determines mRNA length and regulates isoform expression. Cell. 2012;150:53–64. doi: 10.1016/j.cell.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vorlova S., Rocco G., Lefave C.V., Jodelka F.M., Hess K., Hastings M.L., Henke E., Cartegni L. Induction of antagonistic soluble decoy receptor tyrosine kinases by intronic polyA activation. Mol. Cell. 2011;43:927–939. doi: 10.1016/j.molcel.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Engreitz J.M., Sirokman K., McDonel P., Shishkin A.A., Surka C., Russell P., Grossman S.R., Chow A.Y., Guttman M., Lander E.S. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. 2014;159:188–199. doi: 10.1016/j.cell.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Davis D.R. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995;23:5020–5026. doi: 10.1093/nar/23.24.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hudson G.A., Bloomingdale R.J., Znosko B.M. Thermodynamic contribution and nearest-neighbor parameters of pseudouridine-adenosine base pairs in oligoribonucleotides. RNA. 2013;19:1474–1482. doi: 10.1261/rna.039610.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reddy R., Henning D., Busch H. Pseudouridine residues in the 5′-terminus of uridine-rich nuclear RNA I (U1 RNA) Biochem. Biophys. Res. Commun. 1981;98:1076–1083. doi: 10.1016/0006-291x(81)91221-3. [DOI] [PubMed] [Google Scholar]
- 68.Theimer C.A., Finger L.D., Trantirek L., Feigon J. Mutations linked to dyskeratosis congenita cause changes in the structural equilibrium in telomerase RNA. Proc. Natl. Acad. Sci. U.S.A. 2003;100:449–454. doi: 10.1073/pnas.242720799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.