Abstract
Using an in vitro reconstituted system in this work we provide direct evidence that the yeast repressor/activator protein 1 (Rap1), tightly bound to its consensus site, forms a strong non-polar barrier for the strand displacement activity of DNA polymerase δ. We propose that relief of inhibition may be mediated by the activity of an accessory helicase. To this end, we show that Pif1, a 5′–3′ helicase, not only stimulates the strand displacement activity of Pol δ but it also allows efficient replication through the block, by removing bound Rap1 in front of the polymerase. This stimulatory activity of Pif1 is not limited to the displacement of a single Rap1 molecule; Pif1 also allows Pol δ to carry out DNA synthesis across an array of bound Rap1 molecules that mimics a telomeric DNA-protein assembly. This activity of Pif1 represents a novel function of this helicase during DNA replication.
INTRODUCTION
During DNA replication the actions of the replicative helicase and nucleosome remodelers/chaperones are thought to lead to destabilization of chromatin, thus facilitating progression of the replication fork (1–5). In addition to the need of dealing with nucleosomes packaged into chromatin, non-histone protein barriers along DNA regulate or hinder the progression of DNA replication (6,7). In this case the sole activity of the replicative helicase and polymerase may not be sufficient for efficient progression of replication across a protein barrier. Indeed, growing experimental evidence points to a role in this process of specialized DNA helicases (8).
In eukaryotes, one example of a non-histone protein barrier regulating DNA replication is the S. cerevisiae Fob1 protein (9,10). Fob1 binds to a strong replication fork barrier site and generates a polar protein block that prevents head-on collisions between the replication and transcription forks (10–12). In S. cerevisiae, telomeres are one additional example of a non-histone protein barrier to progression of replication. At these sites, replication stalls at the repetitive telomeric DNA tracts, both terminal and internal to the chromosome ends (13–17). These regions contain multiple Rap1 binding sites and bound Rap1, rather than the nature of the repetitive sequence itself, was shown to be the cause of replication stalling (13).
These observations suggest that the replicative helicase and polymerase within the replisome are not sufficient for efficient bypass of non-histone protein barriers, and the activity of accessory motor proteins may be needed. In S. cerevisiae, deletion of Rrm3, a 5′–3′ helicase that belongs to the Pif1 subfamily of SF1 helicases, increases replication fork pausing at ∼1400 sites across the genome (14–17). These sites include rDNA, bound by Fob1, and telomeres, bound by Rap1. This has led to the proposal that Rrm3 helicase activity is important for efficient progression across a protein barrier (i.e. displacement of the protein) and provides an example in vivo of an accessory motor protein needed for efficient replication fork progression. Despite the genetic evidence in vivo, direct biochemical support in vitro for this function of Rrm3 is still missing. It is interesting to note that at difference with initial reports (13,16), recently it has been shown that Pif1 may also have a role in removal of bound proteins, facilitating fork progression at telomeric sites (18). Whether this function originates from Pif1 removing bound Rap1 at telomeres or else remains to be determined.
During lagging strand DNA synthesis, Pol δ extends the short Okazaki fragments generated by Pol α (19–21) and catalyzes strand displacement DNA synthesis through the downstream Okazaki fragment. Genome wide analysis of the distribution of Okazaki fragments showed that the ligation junctions map in close proximity to nucleosome dyads (22,23). The same is true for the tightly bound transcription factors Abf1, Reb1 and Rap1 (22,23). On the lagging strand, both nucleosomes and tightly bound proteins appear to control the degree of strand displacement of Pol δ, affecting the position of the ligatable nick generated during maturation (23) and the degree to which the DNA synthesized by the error prone Pol α is removed by the strand displacement activity of Pol δ (22). Therefore, it is reasonable to postulate that the degree to which a tightly bound protein (e.g. Rap1) limits Pol δ activity solely relies on the effect that a protein block would have on strand displacement. To the best of our knowledge this has never been examined for Pol δ in vitro. Moreover, the amount of strand displacement activity by Pol δ needs to be regulated to avoid generating long 5′-flaps that can bind RPA, thus becoming inhibitory to FEN1 cleavage (24). Indeed, a secondary pathway for flap processing has been proposed and it involves Dna2 helicase/nuclease cleavage of flaps that have been extended by the Pif1 helicase (25,26). The mechanism that regulates the transition from short to long flaps is currently not well understood. Whether proteins bound to the downstream duplex to be displaced also affect the activities of Pif1 and/or Dna2 also remains to be established.
In this work we used model DNA substrates and purified proteins to ask two basic questions. First, we asked whether a Rap1 protein, tightly bound to the downstream duplex DNA, poses a block to an incoming Pol δ, thereby impairing its strand displacement activity. Our data show that in a reconstituted system a single bound Rap1 is sufficient to block the strand displacement activity of Pol δ, even when the enzyme is in a complex with its processivity factor PCNA. Second, we asked whether the helicase activity of Pif1 or the helicase/nuclease activity of Dna2 is sufficient to remove the bound Rap1 from the dsDNA, thus allowing Pol δ to catalyze primer extension past the protein barrier. Pif1 stimulates the apparent strand displacement activity of the polymerase by unwinding the downstream duplex DNA. Moreover, in the presence of Pif1, but not Dna2, Rap1 is no longer a block for Pol δ, indicating that the helicase activity of Pif1 is sufficient to remove a protein block from the dsDNA.
MATERIAL AND METHODS
Reagents and buffers
All chemicals used were reagent grade. All solutions were prepared with distilled and deionized Milli-Q water (18 MΩ at 25°C). Oligonucleotides were purchased from Integrated DNA Technology (IDT, Coralville, IA, USA). Annealed substrates were prepared by mixing the template, primer and strand to be displaced at a ratio of 1:1.2:1.1, respectively, in 10 mM Tris-HCl (pH 8.1), 50 mM NaCl, 5 mM MgCl2 and heated at 95°C for 3 min, followed by slow cooling to room temperature. The TeloA sequence is 5′-ACACCCACACACC; RPG is 5′-ACACCCATACATT. To generate the pUC19 substrate with a 18 nt gap (pUC19g18), 2 Nt.BbvCI sites spaced by 11 bp were introduced 3′ to the BamHI site by Quickchange mutagenesis. pUC19g18 was nicked with Nt.BbvCI for 60 min at 37°C, followed by addition of 20-fold excess of a 18 nt oligonucleotide complementary to the nicked strand at 65°C for 20 min and purification on MicroSpin S-400HR (GE Healthcare). The 336 bp cassette, containing 16 TeloA sites spaced by 21 bp and flanked by EcoRI/HindIII, was synthetized (Genescript) and cloned in pUC19. The two sites for Nt-BbvCI were then introduced by Quickchange mutagenesis 50 bp from the first TeloA site and the 18 nt gap generated as describe above.
Purification of proteins
DNA polymerase δ wild-type and D520V (Pol δDV), purified as previously described (27–29). Replication Protein A (RPA), PCNA and Replication Factor C (RFC) were purified from E. coli overproduction strains as described (19,30,31). Untagged, full-length Rap1 was overexpressed and purified from E. coli as described (32). Full-length Pif1, its shorter variant missing the first 237 amino acids and the K264A mutant were purified with a N-terminus His6-tag from E. coli (33).
Strand displacement and replication assay
Strand displacement DNA synthesis reactions were carried out in Buffer TM (20 mM Tris-HCl pH7.8, 8 mM MgAc2, 1 mM DTT, 0.1mg/ml BSA) with 75 mM NaCl (or otherwise indicated). For experiments with PCNA a standard loading protocol was followed (20,34). For simplicity the concentrations reported are the final ones after starting the reaction. RFC (25 nM) was allowed to react with a double-biotinilated DNA substrate (25 nM) in presence of neutravidin (600 nM) and ATP (1 mM) for 5 min at 30°C, followed by the addition of Pol δ (25 nM) and dNTP mix (100 μM). RPA (50 nM) and/or Rap1 (100 nM) were added before Pol δ. Pif1, at the indicated concentrations, was added with Pol δ. The experiments in absence of PCNA a DNA-Pol δDV complex (25 nM) was pre-forming in the absence or presence of Rap1 (100 nM, sufficient to saturate the single site with Rap1 in a canonical DNA-binding mode (32)) and/or Pif1 (25 nM) and the reaction started by addition of 100 μM dNTP. At the indicated times the reactions were stopped by the addition of 80 mM EDTA, 0.08% SDS. After addition of formamide (50% final), the samples were heated at 95°C for 2 min and analyzed on a 12% denaturing polyacrylamide gel, pre-run for 2 h in 0.5x TBE. The gels were scanned using a Typhoon 9400 Variable Mode Imager (GE Healthcare), monitoring the Cy3 fluorescence of the labeled primer. Replication assays were performed at 30°C with 10 nM of the indicated plasmid DNA in Buffer TM. PCNA (15 nM) was loaded onto DNA with RFC (15 nM) and 1 mM ATP by incubation at 30°C for 2 min in the presence of RPA (1 μM) and in the absence or presence of Rap1 (400 nM). The reactions were initiated by addition of Pol δ (15 nM) and 100 μM each of dATP, dGTP, dTTP and 10 μM of [α-32P]-dCTP and Pif1 when indicated. The reactions were stopped with 50 mM EDTA and 0.1% SDS (final concentration) and the products analyzed by electrophoresis on a 1% alkaline agarose gel. The gels were dried and visualized by PhosphorImager analysis (GE Healthcare).
RESULTS
A single Rap1 bound to a high-affinity recognition site is a barrier for DNA polymerase δ
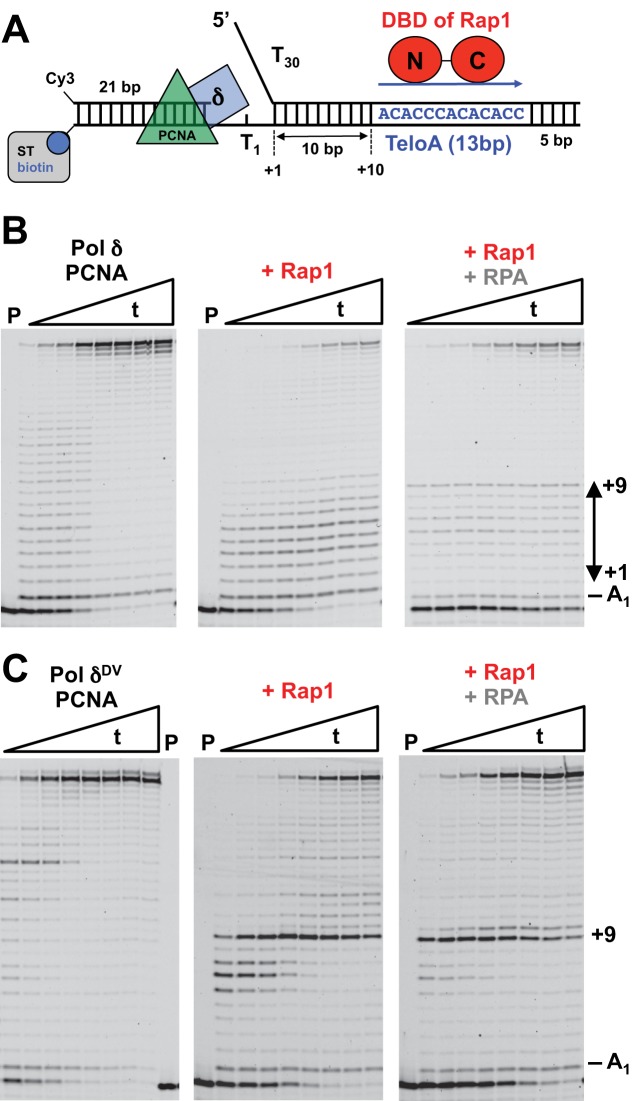
In order to test whether a single Rap1 bound to a downstream duplex is a block to the strand displacement activity of Pol δ, we used a DNA substrate that contains a 21 nt primer labeled at the 5′-end with Cy3, followed by a gap of a single thymine and a 28 bp dsDNA region to be displaced (Figure 1A). The downstream dsDNA region contains a dT30 5′-flap and a high-affinity Rap1 recognition sequence found at telomeres (TeloA) positioned 10 bp from the junction of the 5′-flap. For this orientation of the recognition sequence, Rap1 binds with the N-terminal Myb-like region of its DNA-binding domain (DBD) facing the incoming polymerase and with the majority of the Rap1 contacts with the phosphate backbone of the DNA occurring on the non-template strand (35–38). Finally, the 3′-end of the template strand is biotinylated to allow binding of streptavidin that in combination with the presence of the 5′-flap restricts PCNA binding to the primer region of the substrate.
Figure 1.
A single bound Rap1 is a barrier to the strand displacement activity of Pol δ. (A) Substrate used in the assays. (B) Primer extension assays by PCNA-loaded Pol δ (25 nM) in the absence or presence of Rap1 (100 nM) or RPA (50 nM) and Rap1 (100 nM). The DNA is 25 nM. (C) Same primer extension assays as in (B) but with Pol δDV. Times (t) are: 10”, 20”, 30”, 1’, 2’, 4’, 6’, 10’.
PCNA was loaded on the substrate with RFC and ATP in the absence (Figure 1B, left panel) or presence (Figure 1B, middle panel) of a 4-fold excess of Rap1 over the DNA, the reaction started by addition of Pol δ and dNTPs and monitored by extension of the Cy3-labeled primer. In the presence of Rap1, the amount of full product generated is decreased, suggesting that Rap1 is a block to the strand displacement activity of Pol δ (see Supplementary Figure S1A). Moreover, there is a concomitant increase of intermediate bands between +1 and +9 generated by strand displacement. These intermediate bands likely originate from the 3′–5′ exonuclease activity of Pol δ, which causes the polymerase to idle (reiterative cycles of strand displacement synthesis followed by exonucleolytic degradation) between the nick position and the Rap1 block. Indeed, the presence of a Rap1 block (and disappearance of idling) becomes evident when an exonuclease deficient Pol δ (Pol δDV, D520V) is used (Figure 1C, Supplementary Figure S1B and S1C). In this case, the +9 extension band becomes very pronounced, indicating that the polymerase is halted 1 bp prior to first position of the Rap1 recognition sequence. Taken together these data indicate that Rap1 is a strong block for Pol δ that is bypassed very slowly.
Next, we asked whether binding of RPA to the 5′-flap would allow bypass of Rap1. In the presence of RPA the amount of full-product generated by wild-type Pol δ is more than in its absence (right panel in Figure 1B, and Supplementary Figure S1A). We note that in this case the fraction of substrate utilized during the reaction is also lower, leading to an apparent lower fraction of extended products. It is possible that on this substrate containing a single nucleotide gap, the loading of the polymerase in the presence of RPA and Rap1 is slightly less efficient. Nevertheless, in the presence of RPA there is a clear accumulation of the +9 band. These data indicate that Rap1 is still a block to the strand displacement activity of the polymerase. Because of the lack of exonuclease activity the presence of a +9 extension band becomes evident when Pol δDV is used (right panel in Figure 1C, and Supplementary Figure S1B), again indicating that RPA does not allow bypass of the Rap1 block. However, in the presence of RPA the fraction of polymerase that can extend past the Rap1 block is larger than in its absence, consistent with RPA stimulating the strand displacement activity of Pol δDV.
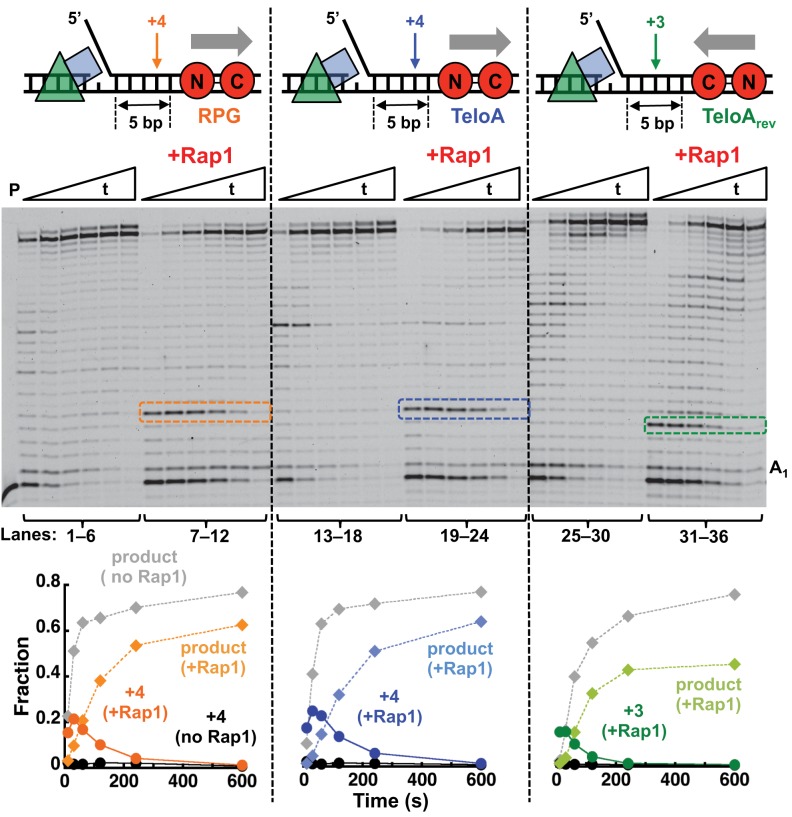
Next, we tested whether the position, orientation, or nature of the Rap1 recognition sequence would affect the ability of Rap1 to hinder Pol δ. Because of the presence of well-defined extension bands, for these experiments we used Pol δDV. Rap1 still blocked Pol δDV when the 13 bp TeloA was moved 5 bp closer to the 5′-flap (Figure 2, lanes 19–24). However, in this case the fraction of Pol δ that can bypass the block is higher than when the TeloA sequence is placed 10 bp downstream. Next, we placed the TeloA sequence in the opposite orientation (Figure 2, lanes 31–36). In this orientation the C-terminal Myb-like region of the DBD of Rap1 faces the incoming polymerase and the majority of the protein contacts with the phosphate backbone of the DNA occur with the template strand (35–38). Rap1 is a block for Pol δDV also when bound in this opposite orientation. Interestingly, now a +3 rather than a +4 extension band is prominent, suggesting an asymmetrical interaction of Rap1 with its recognition sequence. Finally, we also tested a different Rap1 recognition sequence found at the ribosomal protein genes (RPG) (Figure 2, lanes 7–12). Rap1 bound to this alternative recognition sequence is still a block for Pol δDV.
Figure 2.
A single bound Rap1 is a non-polar barrier to the strand displacement activity of Pol δDV. Primer extension assays by PCNA-loaded Pol δDV (25 nM) in the absence or presence of Rap1 (100 nM) bound to the different recognition sequences indicated in the cartoons. The graphs are the quantitation of full-product and the strong pause band observed in the gel (arrow in the cartoons). For reference the same bands were quantitated also for the experiments in the absence of Rap1.
Pif1 stimulates the activity of Pol δ by unwinding the downstream dsDNA
The data in the previous section provide strong evidence that a single tightly bound Rap1 is a block to an incoming Pol δ and therefore it must be removed for strand displacement to occur. The presence of a pre-formed 5′-ssDNA flap or creation of a flap during strand displacement by the polymerase would generate the proper substrate for the activity of a 5′–3′ helicase. Interestingly, it has been shown that Pif1 is involved both in Okazaki fragment maturation and break-induced replication in vivo, processes that involve Pol δ and the presence of 5′-ssDNA available either as a flap or within a D-loop (25,26,39–41). Indeed, in reconstituted reactions excess Pif1 stimulates the strand displacement activity of Pol δ bound to PCNA (25,40). However, we showed that in excess enzyme over the DNA, Pif1 undergoes DNA-induced dimerization (42). Therefore, we tested whether at concentrations where a monomer of Pif1 is favored on the DNA (equimolar to or lower than the DNA), Pif1 would still be able to stimulate Pol δ strand displacement.
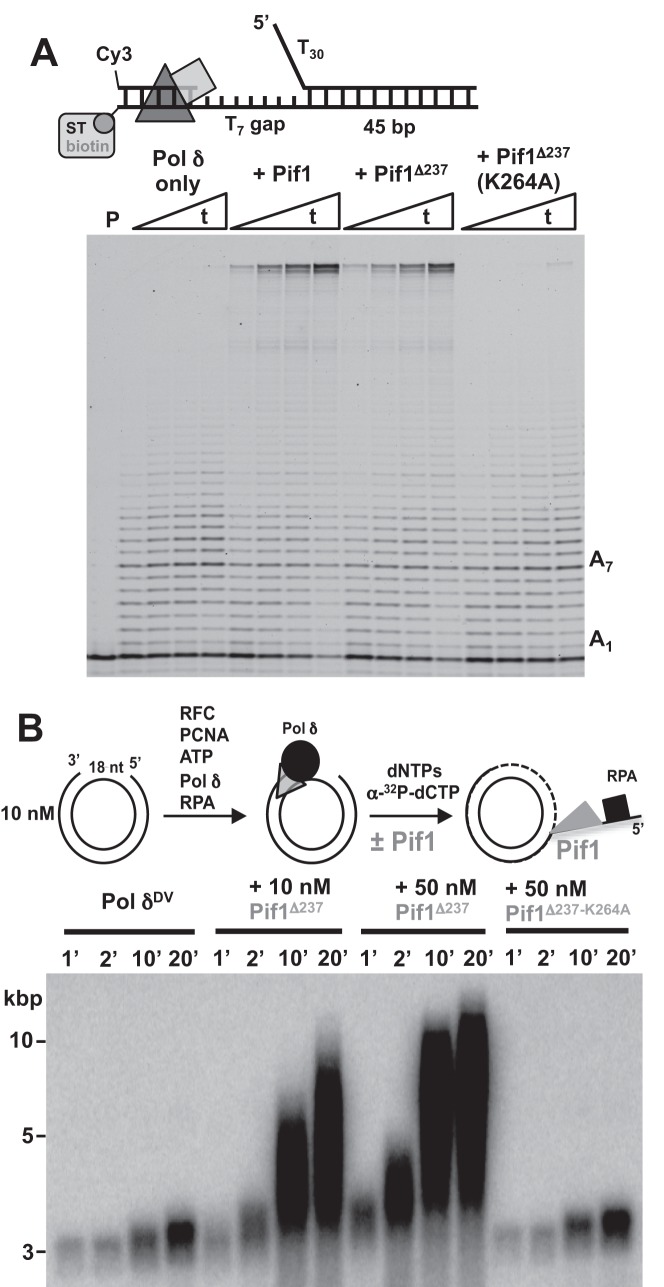
We used a substrate that contains a T7 gap and a longer dsDNA downstream (45 bp) with a T30 5′-flap. When Pol δ is bound to PCNA on the DNA, addition of a low concentration of Pif1 led to higher primer extension activity (Figure 3A). Stimulation of primer extension activity by Pif1 does not require the presence of the first 237 amino acids, suggesting that the helicase core is sufficient, but it requires an active ATPase (K264A variant is inactive (33)). This indicates that at these concentrations binding of Pif1 to the 5′-flap is not sufficient to stimulate strand displacement. Also, we do not find evidence of direct interaction between the helicase and polymerase, as stimulation of primer extension by Pif1 also occurs with the heterologous phage T7 DNA polymerase (33). The stimulation of the primer extension activity of Pol δ originates from multiple turnovers of Pif1 unwinding, with DNA synthesis by the polymerase preventing re-annealing of the template strand.
Figure 3.
Pif1 stimulates the strand displacement activity of Pol δ. (A) Primer extension assays using the indicated DNA (25 nM) and PCNA-loaded Pol δ (25 nM) performed in the absence and presence of different versions of Pif1 (20 nM). Times (t) are: 5”, 15”, 30”, 60”. (B) Replication assays of a 18 nt gapped pUC19 (10 nM) with PCNA-loaded Pol δDV (15 nM) in the absence or presence of the shorter version of Pif1 (10 or 50 nM) or its ATPase inactive variant (50 nM).
Finally, it has been shown that Pif1 in excess over the DNA can stimulate kbps of DNA synthesis when Pol δ extends a D-loop, and this stimulation has been proposed to generate from the activity of Pif1 either in front of the polymerase or at the backend of the D-loop by removing the newly synthesized DNA (40). In order to test whether Pif1 can stimulate Pol δ DNA synthesis over DNA lengths longer than the oligonucleotides used in the previous section, we performed replication assays with pUC19 containing a 18 nt ssDNA gap. With this substrate, only the effect of Pif1 in front of the polymerase is monitored. The data in Figure 3B show that addition of Pif1 stimulates DNA synthesis by Pol δ and this requires the ATPase activity of the helicase. In the presence of Pif1 products longer than the length of the plasmids are generated, indicating that the activity of Pif1 in front of Pol δ is sufficient to lead to synthesis of kbps of DNA.
Pif1 but not Dna2 allows Pol δ to bypass a Rap1 block
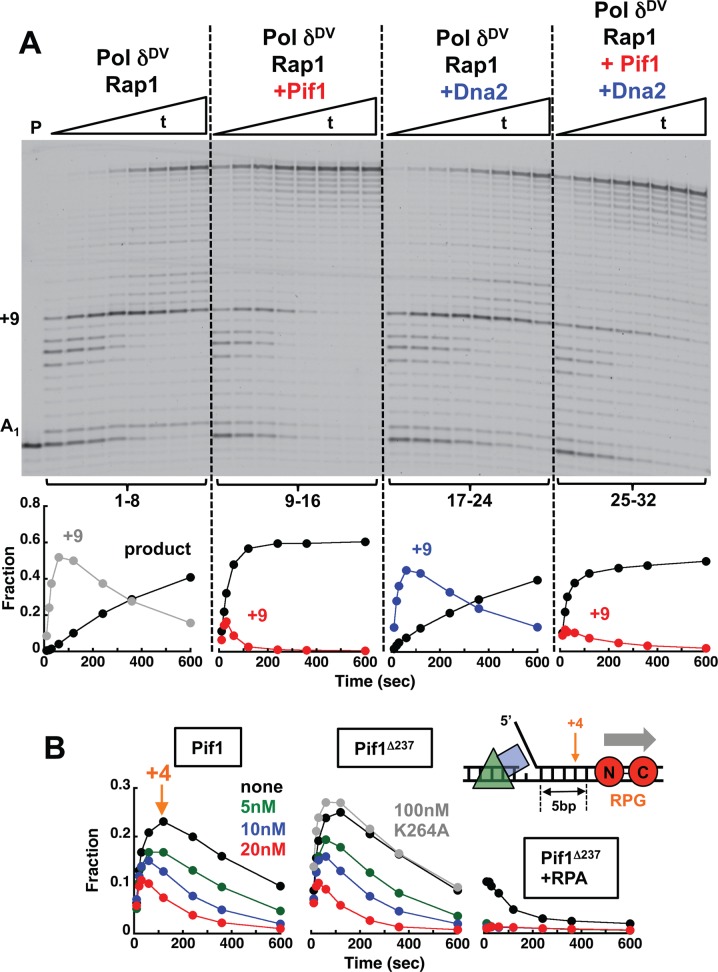
The data in the previous section indicate that Pif1 stimulates polymerase activity of Pol δ by unwinding the downstream dsDNA and suggest that a monomer of Pif1 is sufficient. Next, we asked whether Pif1 would allow Pol δ to catalyze primer extension even across a bound Rap1, indicating that the block has been removed. Figure 4A shows strand displacement reactions with Pol δDV in the presence of bound Rap1 and absence or presence of Pif1. With Pif1 in the reaction (lanes 9–16) a larger amount of full product is formed in shorter times, and also the +9 extension band is less prominent and is cleared faster. This indicates that Pif1 unwinding activity leads to displacement of the Rap1 block, independent of the presence of RPA bound to the 5′-flap (Supplementary Figure S3). Moreover, stimulation by Pif1 of the strand displacement activity of Pol δ and bypass of the Rap1 block were observed also with Pol δ that is not in a complex with PCNA (Supplementary Figure S2), indicating that the reported interaction of Pif1 with PCNA (40) is not required.
Figure 4.
Pif1 allows Pol δ to bypass a Rap1 block. (A) Primer extension assays with PCNA-loaded Pol δDV (25 nM) using the DNA (25 nM) in Figure 1A bound to Rap1 (100 nM). The experiments were performed in the absence (lanes 1–8) or presence of 20 nM Pif1 (lanes 9–16), 20 nM Dna2 (lanes 17–24) or 20 nM Pif1 and 20 nM Dna2 (lanes 25–32). The graphs show quantitation of the full-product and the +9 position in the downstream duplex. (B) Quantitation of full-product and the +4 position using the indicated DNA (25 nM) bound to Rap1 (100 nM). The primer extension assays were performed with PCNA-loaded Pol δDV (25 nM) in the absence (black) or presence of 20 nM (red), 10 nM (blue) or 5 nM (green) of either Pif1 (left) or its variant missing the first 237 amino acids and its ATPase inactive form (middle). The experiments in the right panel were performed in the presence of 50 nM RPA.
In yeast, Dna2 is a 5′–3′ helicase/nuclease that is involved in maturation of Okazaki fragments by cleaving long 5′-flaps in conjunction with Pif1 (25,26,39). In the presence of nuclease activity, it has been shown that Dna2 will preferentially cleave the substrate rather than unwind it (43). Indeed, Dna2 alone does not relieve the Rap1 block (Figure 4A, lanes 17–24). RPA stimulates the activity of Dna2 (43), but its presence did not allow Dna2 helicase to relieve the Rap1 block (Supplementary Figure S3). When Dna2 and Pif1 were added together (Figure 4A, lanes 25–32) the Rap1 block was still relieved, but to a lesser extent than for Pif1 alone, possibly due to the nuclease activity of Dna2 removing the 5′-flap and thus eliminating the entry point for Pif1. These data suggest that if a Rap1 block needs to be dealt with by Pol δ during Okazaki fragment maturation, the activity of Pif1, and not Dna2, is sufficient.
Next, we used a DNA substrate containing the RPG recognition sequence positioned 5 bp from the 5′-flap and performed the experiment at 21°C rather than 30°C to better visualize blocked intermediates. Pif1 displaces Rap1 also when it is bound to this alternative sequence (Figure 4B). The N-terminus region of Pif1 is not required for Rap1 displacement but the presence of an active ATPase is (Figure 4B). Sub-stoichiometric concentrations of Pif1 are sufficient for removal of Rap1, indicating that a monomer of Pif1 unwinds the dsDNA and displaces Rap1. Similar to what observed with the DNA substrate in Figure 1A, in the presence of RPA the fraction of Pol δDV that can bypass a Rap1 bound to the RPG sequence is larger, consistent with RPA stimulating the strand displacement activity of the polymerase. Interestingly, in the presence of RPA even at a concentration of Pif1 4-fold lower than the DNA concentration, Rap1 is efficiently displaced, so much so that no +4 extension band is detected.
A 5′-flap generated during strand displacement is sufficient for Pif1-mediated removal of Rap1 even when bound to an array of sites
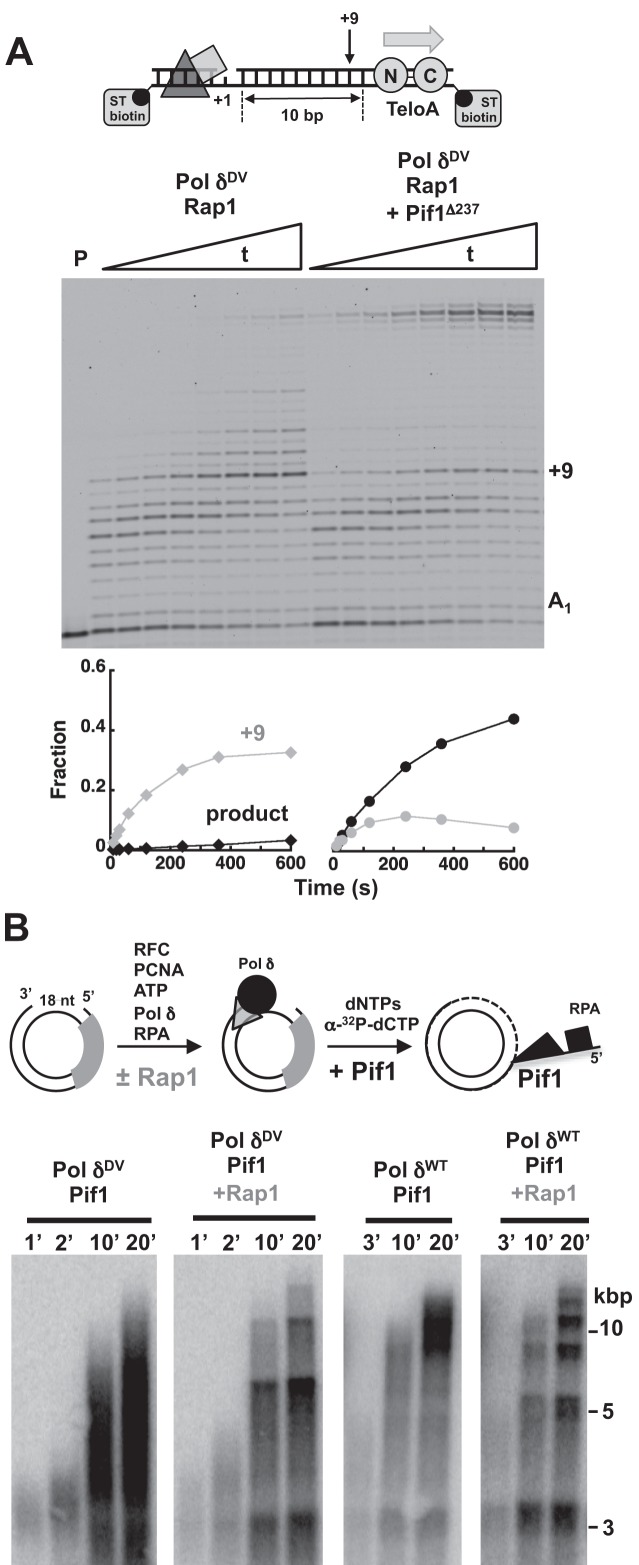
The DNA substrates used in the previous experiments contained a pre-formed 5′-ssDNA flap, however, during Okazaki fragment maturation flaps are formed only transiently. Therefore, we tested whether Pif1 would be able to displace a bound Rap1 when the flap is generated during strand displacement by the polymerase. For this we used a DNA substrate with a T1 gap, the TeloA sequence positioned 10 bp downstream in the dsDNA to be displaced and a doubly biotinylated template bound to streptavidin to prevent PCNA from sliding off the DNA (Figure 5A). Similar to what observed with the same substrate containing a 5′-flap, Rap1 is a block for the incoming Pol δDV. However, addition of Pif1 stimulates formation of full-extension product and the amount of the +9 extension band is reduced. This indicates that generation of a 5′-flap during strand displacement by the polymerase allows for Pif1 to bind to the substrate and displace Rap1.
Figure 5.
Transient generation of a 5′-flap is sufficient for Pif1 to allow Pol δ to bypass a single or multiple Rap1. (A) Primer extension assays with PCNA loaded Pol δDV (25 nM) in the absence and presence of Pif1 (20 nM) with Rap1 (100 nM) bound to a DNA (25 nM) without a 5′-flap. The graphs show quantitation of the full product and the +9 position in the downstream duplex. (B) Replication assays by wild-type and exo-nuclease deficient Pol δ (15 nM) in the absence and presence of Pif1 (10 nM), using a 18 nt gapped pUC19 (10 nM) with an array of 16 Rap1 sites positioned 50 bp from the gap and bound by a 2.5-fold excess of Rap1 relative to the concentration of sites (160 nM).
Next, we asked whether Pif1 could displace multiple Rap1 proteins and thus stimulate DNA synthesis by Pol δ across an array of bound Rap1 that mimics a telomere. Arrays of Rap1 sites with 21 bp spacing are counted in vivo as a normal telomere (44). Thus, we used a 336 bp cassette containing 16 identical sites (TeloA) spaced by 21 bp and in the same orientation. The cassette was cloned into pUC19 and the nicking sites used generate the 18 nt ssDNA gap to initiate DNA synthesis by Pol δ were introduced 50 bp upstream of the cassette. In the presence or the absence of Rap1, the DNA synthesis activity of Pol δDV alone was similar to what observed with pUC19 in Figure 3B (not shown). Similarly, in the absence of Rap1, addition of Pif1 stimulates DNA synthesis by Pol δ and Pol δDV (Figure 5B). In the presence of a 2.5-fold excess of Rap1 relative to the concentration of sites (sufficient for saturation, not shown) Pif1 is still able to stimulate DNA synthesis by Pol δ, indicating that Pif1 displaces multiple bound Rap1 molecules. Indeed, Pif1 displaces Rap1 over multiple cycles of Rap1 dissociation and re-binding, as indicated by the presence of discrete bands spaced according to the size of the plasmid.
DISCUSSION
Despite evidence in vivo of the role of tightly bound proteins in affecting DNA replication, comparatively less is known at the biochemical level using reconstituted proteins. In this work we used S. cerevisiae Rap1 as an example of a well-documented natural protein obstacle (13,15,16,22,23) and showed that a single bound Rap1 is a barrier to the strand displacement activity of Pol δ. The presence of RPA stimulates the strand displacement activity of Pol δ but this is not sufficient to allow bypass of a tightly bound Rap1. Interestingly, for wild-type Pol δ the presence of bound Rap1 leads to an increase in the idling of polymerase in the region preceding the recognition site for Rap1. This suggests that the protein block does not induce dissociation of Pol δ, but rather favors the exo-nuclease activity and backtracking. Bound Rap1 is a barrier independent of the orientation of its recognition site, indicating that Rap1 is not a polar block. Our findings using a reconstituted system, showing that a single Rap1 bound to a high affinity site is sufficient to limit the strand displacement activity of Pol δ, provide strong and direct support to the observation that at the genome-wide level Rap1 sites impart a signature for the boundaries of Okazaki fragment junctions (22,23).
During Okazaki fragment maturation, in addition to the processing by FEN1 of short flaps generated by Pol δ (45,46), a second pathways exists in which long flaps generated by Pif1 are substrates for cleavage by Dna2 (25,26,39). In vitro, excess Pif1 unwinds substrates that mimic an Okazaki fragment (26), thus stimulating the incorporation activity of Pol δ. However, in excess enzyme over the DNA, Pif1 undergoes DNA-induced dimerization (42). The data in this work show that at concentrations that favor binding of a monomer to the DNA, Pif1 stimulates the replication activity of Pol δ (alone or in a complex with PCNA). A monomer of Pif1 binds to the 5′-flap and unwinds the downstream dsDNA, with DNA synthesis by Pol δ preventing re-annealing of the template strand. Surprisingly, Pif1 stimulates the primer extension activity of Pol δ even in the presence of bound Rap1. This provides clear indication that Pif1 can displace bound Rap1 while unwinding the downstream duplex. Removal of Rap1 by Pif1 is independent of the nature of the Rap1 site and its orientation, and requires the ATPase activity of Pif1. The ability of Pif1 to remove Rap1 is not limited to the presence of a pre-formed 5′-flap for initial binding. A short 5′-flap transiently generated by the strand displacement activity of Pol δ is sufficient. RPA stimulates the Rap1 displacement activity of Pif1, likely restricting Pif1 binding to the fork junction. The data in this work show that Pif1 stimulates primer extension activity of Pol δ also when the polymerase is not bound to PCNA. Although we cannot exclude that the reported interaction of Pif1 with PCNA (40) has some role in the removal of a Rap1 block, our data indicate that this interaction is not required. Interestingly, the ability of Pif1 to remove Rap1 in front of Pol δ would suggest that the effect of Rap1 on the distribution of Okazaki fragments and the position of the ligation junctions should not be observed in vivo (22,23). One simple explanation could be that Okazaki fragment maturation is an efficient process and that flap processing by the Dna2/Pif1 pathway is a rare event.
In S. cerevisiae, Rrm3, a second helicase homologue to Pif1, has been proposed to facilitate replication fork progression at specific internal loci and telomeres (13,15,16) and it has been shown in vitro that Rrm3 is a 5′–3′ helicase (16). Surprisingly all of the Rrm3 constructs we generated so far (including a N-terminal truncated version (16)) show ssDNA dependent ATPase activity, yet they posses poor helicase activity even when coupled to the activity of Pol δ (data not shown). We do not currently know the reason for the limited unwinding activity of the Rrm3 constructs. Whether interaction of Rrm3 with the replisome (14) or other factors activate it for unwinding remains to be determined. However, it has been reported that Pif1 may also facilitate replication fork progression at telomeres (18). In support of this novel function of Pif1, we showed that Pif1 displaces multiple Rap1 molecules, allowing Pol δ to replicate across an array of bound Rap1 that mimics a functional telomere.
Protein barriers may pose a problem not only for normal DNA replication but also during break-induced replication (BIR). In BIR, a replication fork is reassembled to allow copying of the template DNA to the end of the chromosome. Depending on the location of the invasion point the activity of the replicative helicase and Pol δ (or Pol ϵ) may not be sufficient to remove tightly bound proteins. Moreover, completion of BIR would be especially problematic at telomeres, which pose a substantial barrier to progression of replication even during normal replication (13–15,17). Interestingly, it has been shown that Rrm3, the helicase proposed to remove proteins bound to DNA (13,15,16), does not have a significant role in BIR; rather, Pif1 does (40). Although the mechanism whereby Pif1 stimulates DNA synthesis from a migrating D-loop in BIR is not fully established, our data show that the activity of Pif1 in front of Pol δ is sufficient to stimulate kbps of DNA synthesis, even across an array of bound Rap1 that mimics a telomere. It is intriguing to speculate that in BIR one of the roles of Pif1 may be to help remove proteins bound to the DNA, especially at telomeres.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM098509 to R.G. and GM032431 to P.B.]. Funding for open access charge: National Institutes of Health [GM098509 to R.G. and GM032431 to P.B.].
Conflict of interest statement. None declared.
REFERENCES
- 1.MacAlpine D.M., Almouzni G. Chromatin and DNA replication. Cold Spring Harb. Perspect. Biol. 2013;5:a010207. doi: 10.1101/cshperspect.a010207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corpet A., Almouzni G. Making copies of chromatin: the challenge of nucleosomal organization and epigenetic information. Trends Cell Biol. 2009;19:29–41. doi: 10.1016/j.tcb.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Groth A., Corpet A., Cook A.J., Roche D., Bartek J., Lukas J., Almouzni G. Regulation of replication fork progression through histone supply and demand. Science. 2007;318:1928–1931. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- 4.Groth A., Rocha W., Verreault A., Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 5.Ransom M., Dennehey B.K., Tyler J.K. Chaperoning histones during DNA replication and repair. Cell. 2010;140:183–195. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mirkin E.V., Mirkin S.M. Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Labib K., Hodgson B. Replication fork barriers: pausing for a break or stalling for time. EMBO Rep. 2007;8:346–353. doi: 10.1038/sj.embor.7400940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruning J.G., Howard J.L., McGlynn P. Accessory replicative helicases and the replication of protein-bound DNA. J. Mol. Biol. 2014;426:3917–3928. doi: 10.1016/j.jmb.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Brewer B.J., Fangman W.L. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- 10.Linskens M.H., Huberman J.A. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 1988;8:4927–4935. doi: 10.1128/mcb.8.11.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi T., Horiuchi T. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells. 1996;1:465–474. doi: 10.1046/j.1365-2443.1996.d01-256.x. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi T. The replication fork barrier site forms a unique structure with Fob1p and inhibits the replication fork. Mol. Cell. Biol. 2003;23:9178–9188. doi: 10.1128/MCB.23.24.9178-9188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makovets S., Herskowitz I., Blackburn E.H. Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol. Cell. Biol. 2004;24:4019–4031. doi: 10.1128/MCB.24.9.4019-4031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Azvolinsky A., Dunaway S., Torres J.Z., Bessler J.B., Zakian V.A. The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev. 2006;20:3104–3116. doi: 10.1101/gad.1478906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivessa A.S., Lenzmeier B.A., Bessler J.B., Goudsouzian L.K., Schnakenberg S.L., Zakian V.A. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol. Cell. 2003;12:1525–1536. doi: 10.1016/s1097-2765(03)00456-8. [DOI] [PubMed] [Google Scholar]
- 16.Ivessa A.S., Zhou J.Q., Schulz V.P., Monson E.K., Zakian V.A. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 2002;16:1383–1396. doi: 10.1101/gad.982902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivessa A.S., Zhou J.Q., Zakian V.A. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell. 2000;100:479–489. doi: 10.1016/s0092-8674(00)80683-2. [DOI] [PubMed] [Google Scholar]
- 18.Anand R.P., Shah K.A., Niu H., Sung P., Mirkin S.M., Freudenreich C.H. Overcoming natural replication barriers: differential helicase requirements. Nucleic Acids Res. 2012;40:1091–1105. doi: 10.1093/nar/gkr836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ayyagari R., Gomes X.V., Gordenin D.A., Burgers P.M. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 AND DNA2. J. Biol. Chem. 2003;278:1618–1625. doi: 10.1074/jbc.M209801200. [DOI] [PubMed] [Google Scholar]
- 20.Jin Y.H., Ayyagari R., Resnick M.A., Gordenin D.A., Burgers P.M. Okazaki fragment maturation in yeast. II. Cooperation between the polymerase and 3′-5′-exonuclease activities of Pol delta in the creation of a ligatable nick. J. Biol. Chem. 2003;278:1626–1633. doi: 10.1074/jbc.M209803200. [DOI] [PubMed] [Google Scholar]
- 21.Maga G., Villani G., Tillement V., Stucki M., Locatelli G.A., Frouin I., Spadari S., Hubscher U. Okazaki fragment processing: modulation of the strand displacement activity of DNA polymerase delta by the concerted action of replication protein A, proliferating cell nuclear antigen, and flap endonuclease-1. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14298–14303. doi: 10.1073/pnas.251193198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reijns M.A., Kemp H., Ding J., de Proce S.M., Jackson A.P., Taylor M.S. Lagging-strand replication shapes the mutational landscape of the genome. Nature. 2015;518:502–506. doi: 10.1038/nature14183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith D.J., Whitehouse I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 2012;483:434–438. doi: 10.1038/nature10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bae S.H., Bae K.H., Kim J.A., Seo Y.S. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature. 2001;412:456–461. doi: 10.1038/35086609. [DOI] [PubMed] [Google Scholar]
- 25.Pike J.E., Burgers P.M., Campbell J.L., Bambara R.A. Pif1 helicase lengthens some Okazaki fragment flaps necessitating Dna2 nuclease/helicase action in the two-nuclease processing pathway. J. Biol. Chem. 2009;284:25170–25180. doi: 10.1074/jbc.M109.023325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pike J.E., Henry R.A., Burgers P.M., Campbell J.L., Bambara R.A. An alternative pathway for Okazaki fragment processing: resolution of fold-back flaps by Pif1 helicase. J. Biol. Chem. 2010;285:41712–41723. doi: 10.1074/jbc.M110.146894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fortune J.M., Stith C.M., Kissling G.E., Burgers P.M., Kunkel T.A. RPA and PCNA suppress formation of large deletion errors by yeast DNA polymerase delta. Nucleic Acids Res. 2006;34:4335–4341. doi: 10.1093/nar/gkl403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stith C.M., Sterling J., Resnick M.A., Gordenin D.A., Burgers P.M. Flexibility of eukaryotic Okazaki fragment maturation through regulated strand displacement synthesis. J. Biol. Chem. 2008;283:34129–34140. doi: 10.1074/jbc.M806668200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koc K.N., Stodola J.L., Burgers P.M., Galletto R. Regulation of yeast DNA polymerase delta-mediated strand displacement synthesis by 5′-flaps. Nucleic Acids Res. 2015;43:4179–4190. doi: 10.1093/nar/gkv260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henricksen L.A., Umbricht C.B., Wold M.S. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 31.Gomes X.V., Gary S.L., Burgers P.M. Overproduction in Escherichia coli and characterization of yeast replication factor C lacking the ligase homology domain. J. Biol. Chem. 2000;275:14541–14549. doi: 10.1074/jbc.275.19.14541. [DOI] [PubMed] [Google Scholar]
- 32.Feldmann E.A., De Bona P., Galletto R. The wrapping loop and Rap1 C-terminal (RCT) domain of yeast Rap1 modulate access to different DNA binding modes. J. Biol. Chem. 2015;290:11455–11466. doi: 10.1074/jbc.M115.637678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh S.P., Koc K.N., Stodola J.L., Galletto R. A monomer of Pif1 unwinds double-stranded DNA and it is regulated by the nature of the non-translocating strand at the 3′-end. J. Mol. Biol. 2016 doi: 10.1016/j.jmb.2016.02.017. doi:10.1016/j.jmb.2016.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burgers P.M., Gerik K.J. Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase delta. J. Biol. Chem. 1998;273:19756–19762. doi: 10.1074/jbc.273.31.19756. [DOI] [PubMed] [Google Scholar]
- 35.Konig P., Giraldo R., Chapman L., Rhodes D. The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell. 1996;85:125–136. doi: 10.1016/s0092-8674(00)81088-0. [DOI] [PubMed] [Google Scholar]
- 36.Taylor H.O., O'Reilly M., Leslie A.G., Rhodes D. How the multifunctional yeast Rap1p discriminates between DNA target sites: a crystallographic analysis. J. Mol. Biol. 2000;303:693–707. doi: 10.1006/jmbi.2000.4161. [DOI] [PubMed] [Google Scholar]
- 37.Matot B., Le Bihan Y.V., Lescasse R., Pérez J., Miron S., David G., Castaing B., Weber P., Raynal B., Zinn-Justin S., et al. The orientation of the C-terminal domain of the Saccharomyces cerevisiae Rap1 protein is determined by its binding to DNA. Nucleic Acids Res. 2012;40:3197–3207. doi: 10.1093/nar/gkr1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le Bihan Y.V., Matot B., Pietrement O., Giraud-Panis M.J., Gasparini S., Le Cam E., Gilson E., Sclavi B., Miron S., Le Du M.H. Effect of Rap1 binding on DNA distortion and potassium permanganate hypersensitivity. Acta Crystallogr. D Biol. Crystallogr. 2013;69:409–419. doi: 10.1107/S0907444912049311. [DOI] [PubMed] [Google Scholar]
- 39.Rossi M.L., Pike J.E., Wang W., Burgers P.M., Campbell J.L., Bambara R.A. Pif1 helicase directs eukaryotic Okazaki fragments toward the two-nuclease cleavage pathway for primer removal. J. Biol. Chem. 2008;283:27483–27493. doi: 10.1074/jbc.M804550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson M.A., Kwon Y., Xu Y., Chung W.H., Chi P., Niu H., Mayle R., Chen X., Malkova A., Sung P., et al. Pif1 helicase and Poldelta promote recombination-coupled DNA synthesis via bubble migration. Nature. 2013;502:393–396. doi: 10.1038/nature12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vasianovich Y., Harrington L.A., Makovets S. Break-induced replication requires DNA damage-induced phosphorylation of Pif1 and leads to telomere lengthening. PLoS Genet. 2014;10:e1004679. doi: 10.1371/journal.pgen.1004679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barranco-Medina S., Galletto R. DNA binding induces dimerization of Saccharomyces cerevisiae Pif1. Biochemistry. 2010;49:8445–8454. doi: 10.1021/bi100984j. [DOI] [PubMed] [Google Scholar]
- 43.Levikova M., Klaue D., Seidel R., Cejka P. Nuclease activity of Saccharomyces cerevisiae Dna2 inhibits its potent DNA helicase activity. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E1992–E2001. doi: 10.1073/pnas.1300390110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grossi S., Bianchi A., Damay P., Shore D. Telomere formation by rap1p binding site arrays reveals end-specific length regulation requirements and active telomeric recombination. Mol. Cell. Biol. 2001;21:8117–8128. doi: 10.1128/MCB.21.23.8117-8128.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garg P., Stith C.M., Sabouri N., Johansson E., Burgers P.M. Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 2004;18:2764–2773. doi: 10.1101/gad.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tishkoff D.X., Filosi N., Gaida G.M., Kolodner R.D. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.