Abstract
The Musashi-2 (Msi2) RNA-binding protein maintains stem cell self-renewal and promotes oncogenesis by enhancing cell proliferation in hematopoietic and gastrointestinal tissues. However, it is unclear how Msi2 recognizes and regulates mRNA targets in vivo and whether Msi2 primarily controls cell growth in all cell types. Here we identified Msi2 targets with HITS-CLIP and revealed that Msi2 primarily recognizes mRNA 3′UTRs at sites enriched in multiple copies of UAG motifs in epithelial progenitor cells. RNA-seq and ribosome profiling demonstrated that Msi2 promotes targeted mRNA decay without affecting translation efficiency. Unexpectedly, the most prominent Msi2 targets identified are key regulators that govern cell motility with a high enrichment in focal adhesion and extracellular matrix-receptor interaction, in addition to regulators of cell growth and survival. Loss of Msi2 stimulates epithelial cell migration, increases the number of focal adhesions and also compromises cell growth. These findings provide new insights into the molecular mechanisms of Msi2's recognition and repression of targets and uncover a key function of Msi2 in restricting epithelial cell migration.
INTRODUCTION
Post-transcriptional gene regulation mediated by RNA-binding proteins (RBPs) is a versatile mechanism that modulates the localization, stability and translation of many protein coding genes (1). Originally identified in Drosophila for controlling sensory organ development (2), the Musashi (Msi) RBPs are now implicated to have important functions in the neural, hematopoietic and gastrointestinal systems in many species including worm, fly, mouse and human (3–8). In vertebrates, the Msi family consists of the paralogs, Msi1 and Msi2, which share a high degree of similarity in their RNA binding domains (RBDs) but also show divergence in their carboxyl terminal (C-terminal) domains (9). Loss of Msi2 in hematopoietic or gastrointestinal systems results in stem cell depletion and compromises normal tissue functions (5,8). Conversely, overexpression of Msi2 drives pathologic cellular proliferation in these systems and is associated with poor prognosis in patients with malignant leukemia (3,5).
To elucidate the molecular underpinning responsible for the oncogenic activities of Msi2, recent works have begun to utilize genomic tools to identify Msi2 targets globally and characterize the regulatory network governed by Msi2 (6,8). Transcriptome-wide binding assays, such as HIgh-Throughput Sequencing of RNA isolated by CrossLinking ImmunoPrecipitation (HITS-CLIP), were used to identify a number of Msi2 targets involved in cell cycle control in human leukemia cell lines and murine intestinal progenitor cells. Although these studies attempted to identify Msi2 targeted mRNAs in vivo, the primary sequence motif recognized by Msi2 is unclear as numerous divergent sequence motifs were identified (8). In contrast, biochemical and structural analysis of Msi binding sites in experimentally identified targets, Numb and Jag1, and in vitro target selection experiments have identified a tripartite nucleotide sequence, UAG, as the prominent recognition motif for both Msi1 and Msi2 (4,9,10). Thus, the discrepancy between HITS-CLIP identified in vivo targets and biochemically examined in vitro binding sites highlights the importance of further investigation into Msi2-recognized targets in vivo and warrants experimental identification of Msi2 targets in a cell context-specific manner.
The Msi proteins are suggested to regulate translation of their targeted mRNAs primarily based on studies of Msi1. A region in the C-terminal of Msi1 interacts with poly(A) binding protein (PABP) and competes with eIF4G for the interaction, which results in inhibition of translation (11). Consistent with this result, a recent study using RNA-seq and ribosome profiling (Ribo-seq) showed that the overexpression of Msi1 inhibits translational efficiency (TE) without causing significant changes to mRNA levels in cultured neural stem cells isolated from mouse embryos (4). However, due to the lack of confidently identified Msi2 targets and the apparent differences in the C-terminal sequences between Msi1 and Msi2, it remains unclear if this mode of gene silencing is also the predominant mechanism for Msi2. Additionally, since Msi2 is broadly expressed in many tissue types including epithelial and neural tissues, this suggests that Msi2 may regulate additional processes distinct from cell growth and stem cell dynamics.
In this study, we identify novel targets and cellular processes regulated by Msi2 by mapping the transcriptome-wide RNA targets and binding sites in primary mouse keratinocytes. Using HITS-CLIP to capture Msi2 associated RNAs in intact cells, we show that the in vivo binding motifs of Msi2 are highly enriched for both single and clustered, multiple copies of UAG in the 3′UTR of mRNAs. RNA-seq and Ribo-seq analyses demonstrate that Msi2 primarily promotes mRNA decay without significantly altering TE. Importantly, we detect novel Msi2 targets that are involved in regulation of focal adhesion (FA), extracellular matrix (ECM)-receptor interaction and the actin cytoskeleton, in addition to regulators of cell proliferation and survival. Guided by these findings, we show that the loss of Msi2 increases the migration of keratinocytes, at least in part, by regulating FA while reducing proliferation by inhibiting cell cycle progression and inducing apoptosis. In support of these results, in wounded skin Msi2 is strongly downregulated in the epidermal stem/progenitor cells, located at the leading edge of the wound. These findings provide new insights into the molecular mechanisms of Msi2-mediated gene repression in mammalian cells and define Msi2 as a novel regulator of epithelial migration and growth.
MATERIALS AND METHODS
RNA-stability measurements
Approximately 300 000 shRNA producing keratinocytes were plated into five 6 cm dishes and allowed to grow in E-Low Media as previously described (13). Once cells reached ∼80% confluency media was supplemented with 5 ug/ml Actinomycin D (Thermo Fisher). The zero time point was marked starting 5 min after Actinomycin D addition. Cellular RNA was harvested using Trizol (Thermo Fisher) at time 0, 2, 4, 6 and 8 h and used in qPCR for targets (Supplementary Table S2 for qPCR primer sequences). Relative expression was computed and normalized to the 0 h time point for each target from four technical replicates using ΔΔCq method normalized to Hprt and Gapdh values with error bars denoting standard error of the mean. RNA half-lives were calculated from linear regression of log transformed expression values for each target as described previously (12). ANCOVA analysis was performed on the resulting regression lines to assess statistical significance.
Msi2 HITS-CLIP
Msi2 HITS-CLIP was performed as previously described for Ago2-HITS-CLIP with minor modifications (13). Briefly, 15 cm dishes of mouse keratinocytes were irradiated with 200 mJ/cm with 254 nm UVC light, harvested by scraping and stored at −80°C. After lysis, the lysates were then treated with 10 μl per ml lysate Turbo DNase (Thermo Fisher), 5 μl per ml lysate RNase OUT (Thermo Fisher), and partially digested with 10 μl of either a 1:1, 1:20, 1:50 or a 1:75 dilutions of an RnaseA/T1 mix (Sigma/Ambion 1x mix = 3.33 μl Rnase-A (2 μg/μl) with 6.66 μl Rnase-T1 (1 U/μl). Crosslinked Msi2 was immunoprecipitated for 2 h at 4°C using 5μg of an anti-Msi2 antibody (Supplementary Table S2) complexed with Protein-G Dynabeads (Thermo Fisher). After end-labelling, 5′adaptor ligation, and phosphatase treatment Msi2-RNA complexes were resolved on a 10% Novex Bis-Tris gel (Thermo Fisher) and transferred to a nitrocellulose membrane (GE Healthcare Life Sciences). The nitrocellulouse was subsequently exposed to X-ray film and a phosphor screen overnight. Protein–RNA complexes migrating between 40 and 60 kDa for the 1:1 and 1:20 RNase dilutions or 70–160+ kDa for the 1:20, 1:50 and 1:75 RNase dilutions were isolated from the nitrocellulose. RNA was extracted by Proteinase-K treatment followed by acidic Phenol-Chloroform extraction and ethanol precipitation. After ligation and reverse transcription, the cDNA were subject to 20 cycles of PCR, purified on a 10% native PAGE gel and subject to 1 × 100 sequencing on an Illumina HiSeq 2000.
Ribosome profiling and RNA-seq library preparation
Ribosome profiling was performed on scrambled shRNA and Msi2 knockdown keratinocytes using the ART-Seq Ribosome profiling kit (Illumina). Briefly, cultured cells were grown to ∼80% confluency on a 15 cm plate and treated with 50 ug/ml cyclohexamaide for 1 min before they were lysed, aliquoted and digested with RNase. Ribosomes and associated RNA were isolated using illustra™ MicroSpin™ S-400 HR Columns (Illumina). RNA was extracted from the isolate and rRNA was depleted once using Ribo-Zero Gold™ kit (Illumina). RNA fragments 28–32 nts long were isolated via denaturing PAGE gel, ligated to a 3′ adapter, reverse transcribed, circularized using CircLigase, depleted for rRNA a second time and PCR amplified following the provided protocol. RNA-seq was performed using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England BioLabs) with minor modifications using lysate matched to the ribosome profiling samples. Briefly, mRNA was isolated from total RNA using Dynabeads mRNA DIRECT Micro Purification Kit (Thermo Fisher) and fragmented for 15 min at 94°C. First strand synthesis, second strand synthesis, end repair, adapter ligation and PCR were performed as described in provided protocol. All PCR products were sequenced on an Illumina HiSeq 2000 using 1 × 100 sequencing.
RESULTS
Msi2-HITS-CLIP identifies Msi2 associated RNA targets
Msi2 is highly expressed in skin progenitor and stem cells of both interfollicular and hair follicle lineages in intact mouse skin and in cultured keratinocytes (Supplementary Figure S1). In addition, using previously published RNA-seq data from total epidermis (14), we found that Msi2 is more than 9-fold more abundant than Msi1, a paralog of Msi2, suggesting that Msi2 is the dominant Msi protein in the epidermis (Supplementary Figure S1F). Furthermore, the same dominant expression of Msi2 over Msi1 was also detected in RNA-seq and ribosome profiling (Ribo-seq) data from keratinocytes (see below), in which Msi2 is ∼13-fold more abundant than Msi1 (Supplementary Figure S1F). Lastly, in our previous study, we generated genome-wide proteomics data from neonatal murine epidermis (15). These mass spectrometry data demonstrated that Msi2 protein is detectable, whereas Msi1 is at undetectable levels (Supplementary Figure S1F). Altogether, these data document that Msi2 is the dominant Msi protein expressed in the epidermis.
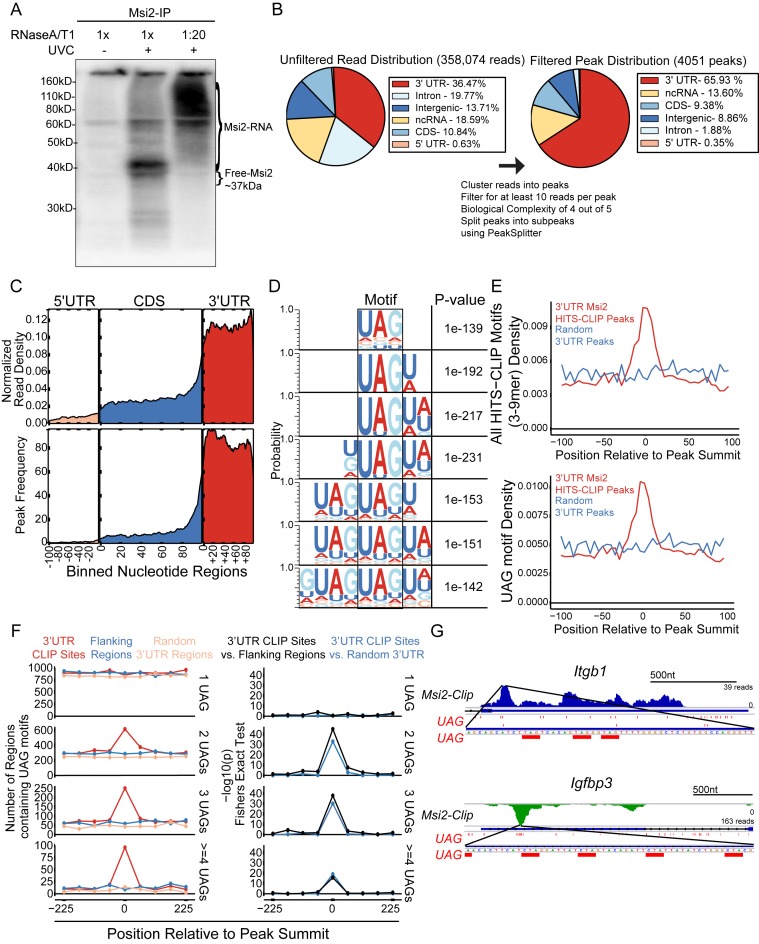
To understand how Msi2 recognizes its targets in intact cells, we first identified Msi2 target mRNAs by performing Msi2-HITS-CLIP on mouse keratinocytes (Supplementary Figure S2A). RNA fragments crosslinked to Msi2 by UV radiation were isolated from lysates treated with different concentrations of RNase A/T1. High RNase treatment resulted in a relatively uniform band with an apparent molecular weight of ∼40 kD, slightly larger than the anticipated 37 kD of Msi2, and low RNase treatment resulted in a broad smear between ∼60 and 110 kD, confirming capture of an RNase-sensitive Msi2-RNA complex (Figure 1A). Of note, partial RNase digestion and recovery of the longer RNA fragments protected by Msi2 would enhance our ability to identify Msi2 binding sites (16). We sequenced five libraries with varying sized Msi2-associated RNA fragments (Supplementary Figure S2B and C). Thirty-four million reads were recovered with ∼350 000 unique reads that were unambiguously mapped to the mouse genome. Overall, the largest portion of the mapped reads aligned to 3′UTRs (36.47%), followed by intronic regions, noncoding RNAs, intergenic regions and coding regions (CDS) (Figure 1B). Overlapping reads were merged together to generate Msi2 binding peaks. We then defined a set of high confidence peaks by requiring at least four out of five libraries to have at least one read per peak and the peak to contain a minimum of 10 total reads. Peaks located in 3′UTRs (65.93%) stood out again as the most dominant region after peak filtering whereas peaks covering intronic and intergenic regions were reduced to 8.86% and 1.88% respectively after filtering, suggesting that intronic and intergeneic binding events may be the result of spurious binding or alignment artifacts (Figure 1B). Overall, 75.66% of the Msi2-HITS-CLIP peaks were located in mRNAs, with 87.14% of those peaks located in the 3′UTR. Metagene plots demonstrated that the read and peak distribution across mRNA transcripts were strongly enriched in the 3′UTR region (Figure 1C). Within the 3′UTR, neither the reads distribution nor peak distribution showed a strong preference towards either the stop codon or the 3′ terminus of the transcript (Figure 1C). These data provide a genome-wide view of Msi2's strong preference to 3′UTR of mRNAs.
Figure 1.
Msi2-HITS-CLIP identifies direct Msi2 targets in keratinocytes. (A) Autoradiogram of 32P-labelled Msi2-RNA complexes treated with different RNase concentration resolved on a 10% Bis-Tris gel. (B) Pie chart of the genomic locations of the aligned reads and filtered peaks before (left panel) and after (right panel) the filtering processes, respectively. (C) Metagene of exonic coverage along a scaled mRNA for aligned reads (top panel) and filtered peaks (bottom panel). Reads densities are normalized for library sizes. Peak and reads densities are averaged along all detectable transcripts based on RNA-seq (see ‘Materials and Methods’). (D) Msi2 recognized motifs are identified from de novo motif search for 3–9 mers in the 3′UTR peaks. The top motif identified for each N-mer search is displayed and positioned to highlight the shared UAG motif. (E) Motif occurrences are tabulated in a +/- 100-nucleotide window surrounding the peak summits for all 3–9mer motifs displayed in panel D (top panel) or UAG (bottom panel). (F) Number of UAG motif (1–4) occurrences in a +/- 225-nucleotide window around the peak summit in CLIP sites, flanking region or random 3′UTR background (left panel), with Fisher Exact Test showing motif enrichment in CLIP sites over the flanking region or 3′UTR background (right panel). (G) Gene tracks of Msi2 HITS-CLIP reads for Msi2 bound transcripts. Reads from all libraries were combined and positive strand reads are coloured blue whereas the negative strand reads are coloured green. The UAG motifs and its reverse complement, CUA, in a window around the peak summits are highlighted in red.
To probe Msi2 recognized binding sites in vivo, we performed de novo motif searching to detect sequence-specific signatures of Msi2 binding within 3′UTR, 5′UTR, intronic and CDS regions. We found enrichment for a set of motifs in the 3′UTRs that contained a 3-nucleotide UAG core element (Figure 1D), supporting previous in vitro results obtained from SELEX as well as structural and biochemical studies (9,10,17). In addition, only peaks located in 3′UTRs generated a confidently enriched and coherent set of motifs, indicating that Msi2 primarily binds the 3′UTR of mRNA transcripts by recognizing UAG containing motifs (Figure 1D, Supplementary Figure S3A–C).
The relative position of the UAG core motif or all identified 3–9mer motifs in aggregate showed a significant enrichment around the summit of the HITS-CLIP peaks, defined as the middle of the region in a peak with the greatest read coverage over the background, further supporting that our HITS-CLIP approach detected mRNA regions directly recognized and protected by Msi2 (Figure 1E, Supplementary Figure S4A). Because both Msi2 and Msi1 contain two RNA recognition motifs (RRMs) and each is believed to interact with a UAG motif (9) and because the 7–9mer motifs identified consisted of repeated adjacent UAG sequences, we wanted to determine if the RNA regions recognized by Msi2 were enriched for multiple UAGs. We observed a significant enrichment of two or more UAG motifs within a 50 nt window surrounding the peak summit of the Msi2 binding sites, in contrast to randomized 3′UTR regions or regions flanking the binding sites (Figure 1F). This suggests that multiple UAGs may be required for Msi2 recognition. These multiple clustered UAG motifs within a short distance probably provide a mechanism for Msi2 to distinguish bona fide binding sites from random UAG sequences that are prevalent in the genome. Indeed, inspection of a representative set of targets demonstrated widespread existence of the UAG motif in the 3′UTRs and multiple UAGs were enriched within a portion of the HITS-CLIP identified Msi2 binding regions (Figure 1G and Supplementary Figure S5). However, it was clear, based on the widespread distribution of UAGs within these individual examples, that UAG alone is not sufficient for predicting Msi2 binding due to the large number of non-clipped UAGs. Furthermore, the lack of UAG was also observed in some peaks, indicating that additional elements or features may be used for Msi2 recognition (Supplementary Figure S5).
In addition, because the RRMs are highly similar between Msi1 and Msi2 (9), we also assessed the positional distribution of the Msi1 consensus motif, (G/A)U(n)AGU (n = 1–3), previously identified by SELEX (17). Notably, the GUAGU motif showed more robust enrichment near the peak summit than the GUUAGU motif did whereas the GUUUAGU or AU1-3AGU motifs were not enriched (Supplementary Figure S4B). These data suggest that GU1-2AGU motifs may also be bound by Msi2 in vivo. Altogether, our Msi2-HITS-CLIP reveals characteristics of Msi2 binding and identifies a trinucleotide UAG core Msi2 recognition motif in intact cells.
Loss of Msi2 increases targeted mRNA abundance
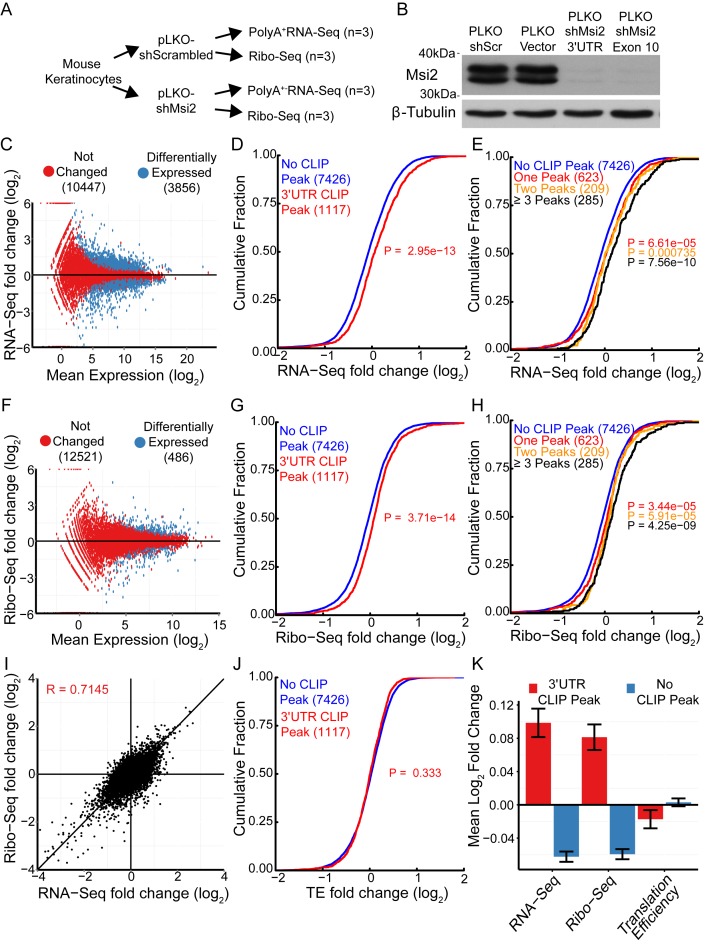
One thousand one hundred and ninety-nine mRNAs were identified as Msi2 recognized targets with at least one Msi2 HITS-CLIP peak located in the 3′UTR. To determine molecular effects of Msi2 binding to these targeted transcripts, we knocked down Msi2 in cultured mouse keratinocytes using two short hairpin RNAs (shRNA) and measured steady-state mRNA abundance and ribosome associated mRNAs by RNA-seq and ribosome profiling (Ribo-seq) (18), respectively (Figure 2A). Knockdown of Msi2 using both shRNAs, one targeting the 3′ UTR and the other targeting the coding regions, achieved highly efficient reduction of protein levels (Figure 2B). All subsequent experiments utilized the 3′UTR targeting shRNA (shMsi2). Additionally, the Ribo-seq data showed the expected phasing of reads in the CDS, consistent with capturing actively translating messages (Supplementary Figure S6).
Figure 2.
Loss of Msi2 increases targeted mRNA abundance without affecting TE. (A) Flow-chart of experimental design. (B) Western blot against Msi2 on keratinocytes infected with indicated shRNA lentiviral construct. (C, F) Log2 fold change plotted against Log2 mean expression for RNA-seq (C) and Ribo-seq (F) libraries. Significantly changed genes with FDR < 0.05 plotted in red. (D, G) Cumulative distributions of changes in mRNA abundance (RNA-seq) (D) and ribosome occupancy (Ribo-seq) (G) after Msi2 knockdown. Genes with 3′UTR Msi2-HITS-CLIP peaks are plotted in red, genes without 3′UTR Msi2-HITS-CLIP peaks are plotted in blue. The number of genes in each category is indicated in parenthesis. (E, H) Cumulative distributions as in panels D and G, with genes with one, two or three or more 3′UTR Msi2-HITS-CLIP peaks plotted separately. (I) Comparison of log2 fold-changes for RNA-seq and Ribo-seq. Pearson correlation coefficient displayed. (J) Cumulative distributions of changes in translation efficiency for genes containing or not containing 3′UTR Msi2-HITS-CLIP peaks. (Kolmogorov-Smirnov Test one-sided). (K) Mean log2 fold-changes for RNA-seq, Ribo-seq and translation efficiency for genes containing or not containing 3′UTR Msi2-HITS-CLIP peaks. Standard error of the mean is displayed.
RNA-seq detected 3856 dysregulated genes whereas Ribo-seq detected 486 differentially expressed genes in the Msi2 knockdown cells (FDR <0.05) (Figure 2C and F). The differences were likely due to lower sequencing coverage in the Ribo-seq dataset. Of the 1199 target mRNAs identified through HITS-CLIP, 1117 were detectable by both RNA-seq and Ribo-seq after filtering out genes with low coverage (minimum baseMean of 10 in shScr libraries). A cumulative distribution function was then used to compare the RNA-seq and Ribo-seq fold change of the 1117 targeted mRNAs versus the non-targeted mRNAs to assess Msi2's effects in target mRNA levels and ribosome occupancy. Msi2 bound mRNAs were more likely to be upregulated and additionally have more ribosome occupancy upon reduction of Msi2 levels (Figure 2D and G). Furthermore, the trend toward increased mRNA and ribosome occupancy for Msi2 targets was positively correlated with increasing number of detected Msi2 binding sites, reflecting a dosage response of Msi2 targets to Msi2 binding (Figure 2E and H). These data are consistent with Msi2 acting as a repressive regulatory RBP.
A recent study suggested that Msi2 functions to regulate TE of individual targets when overexpressed (4). However, our global measurements with Msi2 knockdown suggested that the change in ribosome occupancy and mRNA abundance for Msi2 targets was highly correlated (Figure 2I). These results raised a possibility that Msi2 regulates target mRNA stability in addition to modulating ribosome occupancy. To investigate this mechanism, we examined the impact that loss of Msi2 had on TE by calculating the ratio of normalized ribosome protected fragments to normalized RNA-seq reads per transcript. Overall, the TE was unchanged for Msi2 targets upon reduction of Msi2 (Figure 2J and Supplementary Figure S7A), as illustrated by the cumulative distribution of TE changes, or by comparing the mean fold RNA-seq, Ribo-seq or TE change of Msi2 targets versus non-targets (Figure 2K). This effect was also evident for individual genes when we analyzed the previously identified Msi2 target Jag1 and found mRNA and ribosome occupancy were both significantly increased upon Msi2 knockdown (Supplementary Figure S7B and C).
In addition to binding to 3′UTR regions, Msi2 binding was also detected in 5′UTR, coding and intronic regions at a much lower frequency (Figure 1B). The number of genes containing 5′UTR, CDS or intron binding events were very low (8, 44 and 33 genes respectively), compared to the 3′UTR binding events. However, in addition to 3′UTRs, genes with Msi2 CDS binding events were more likely to be upregulated when Msi2 was depleted, in contrast to 5′UTR and intron binding events, which did not demonstrate any discernible trend (Supplementary Figure S8A–C). Taken as a whole, these data support the notion that Msi2 promotes targeted mRNA decay primarily through association with the 3′UTR and, to a lesser extent, the CDS, similar to what has been reported for miRNAs, another class of post-transcriptional regulators (19,20).
Msi2 regulates target mRNA stability
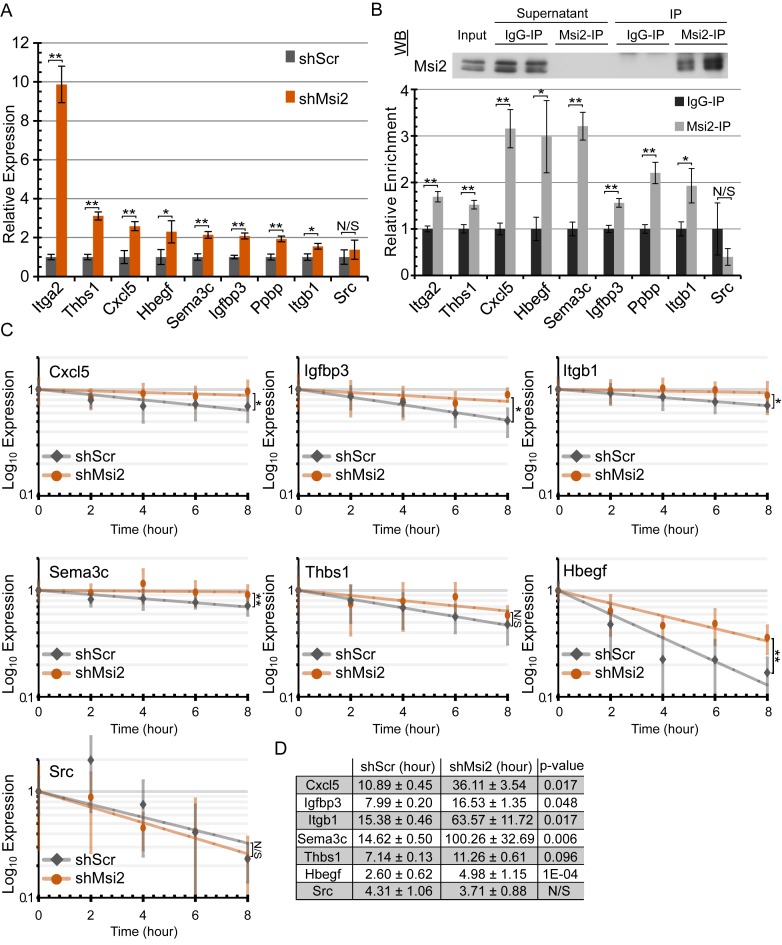
To independently validate that Msi2 regulates RNA stability, we selected a subset of targets and assayed their steady-state transcript levels by qPCR and the rate of mRNA decay upon Actinomycin D treatment. Eight genes (Itga2, Thbs1, Hbegf, Cxcl5, Sema3c, Igfbp3, Ppbp and Itgb1) that exhibited mRNA abundance increases and increased ribosome occupancy in the absence of Msi2 were selected for validation in addition to a nontargeted and unchanged gene, Src, for comparison (Supplementary Figure S7D). The steady-state mRNA levels of all eight selected Msi2 targets were found to be upregulated in independently generated Msi2 knockdown cell lines while Src was not significantly changed (Figure 3A). Additionally, we performed Msi2 immunoprecipitation followed by qPCR and found that the eight targets were enriched in Msi2 immunoprecipitates over control genes Gapdh and Hprt, confirming their association with Msi2 proteins, while Src showed no enrichment (Figure 3B). To determine the stability of the target mRNAs, RNA decay curves for the eight targets and Src were assayed using an Actinomycin D time course in shScr and shMsi2 keratinocytes followed by qPCR. Unfortunately, Itga2 was too lowly expressed in shScr to accurately assess RNA stability. Additionally, Ppbp suffered from poor qPCR detection and normalization and was excluded from the analysis. Nevertheless, all other target mRNAs except Thbs1 were significantly more stable in the absence of Msi2 (Figure 3C). While Thbs1 changes did not achieve statistical significance, it also trended toward being more stable in Msi2 knockdown samples. In support of this observation, RNA half-lives were calculated from the decay curves and found to be anywhere from 2–7-fold higher in Msi2 knockdown cells over the scrambled control counterparts (Figure 3D). These data demonstrate that Msi2 likely functions to repress target genes at least in part by de-stabilizing the mRNA transcript.
Figure 3.
Msi2 functions by promoting target mRNA decay. (A) qPCR detection of the expression of selected Msi2 targets in scrambled control and Msi2 knockdown keratinocytes. (n = 4 biological replicates, **P < 0.01; *P < 0.05). (B) RNA-immunoprecipitation of Msi2 complexes with Msi2 or control rabbit IgG antibody, followed by qPCR detection of the selected Msi2 targets using Gapdh and Hprt as non-targeting controls. Relative enrichment was calculated by enrichment over IgG control. Western blot validation of successful Msi2 immunoprecipitation is shown. (**P < 0.01; *P < 0.05). (C) RNA stability curves plotted using qPCR expression versus time. ANCOVA analysis was used in determining statistical significance. Standard error mean is displayed for each time point. (**P < 0.01; *P < 0.05). (D) RNA half-lives in hours calculated from the stability curves. ANCOVA P-values displayed. **P < 0.01; *P < 0.05) (ANCOVA). N.S. not significant.
Msi2 targets are highly enriched for genes involved in migration and proliferation
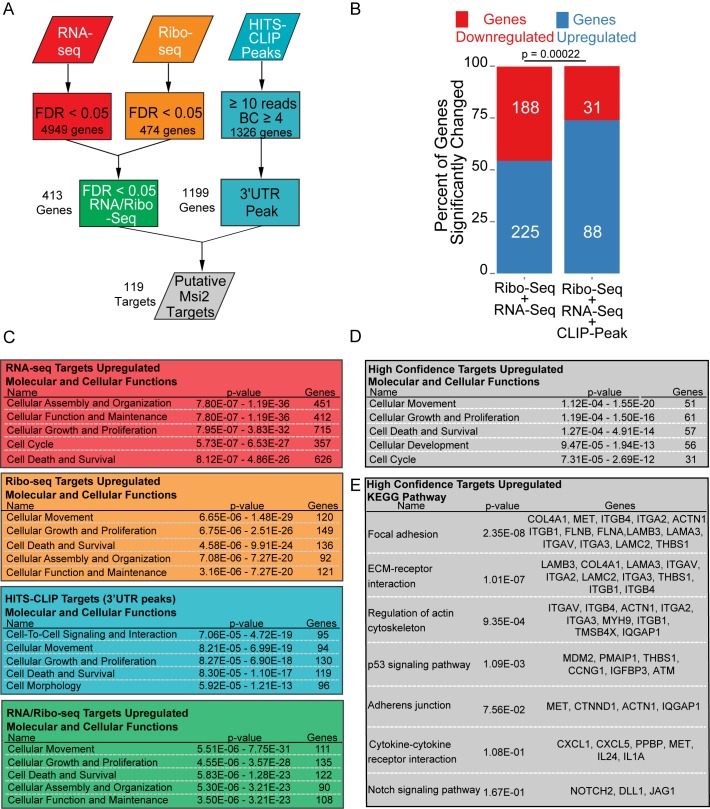
Our identification of direct Msi2 targets through HITS-CLIP combined with transcriptome and ribosome occupancy measurements provided a platform to identify molecular pathways regulated by Msi2. A list of regulated Msi2 targets with Msi2 binding sites was generated as follows: genes were filtered for FDR < 0.05 in both Ribo-seq and RNA-seq as calculated by DEseq and cross-referenced with the high-confidence 3′UTR HITS-CLIP peaks (Figure 4A). Putative targets in this list were largely upregulated in the absence of Msi2 (88 out of 119, 74%), consistent with Msi2 being a negative regulator of gene expression (Figure 4B). We deemed these 88 genes exhibiting upregulation in both RNA-seq and Ribo-seq upon Msi2 knockdown as high confidence Msi2 targets (Supplementary Table S1).
Figure 4.
Msi2 targets are highly enriched for regulators of migration and proliferation. (A) Schematic depicting analysis method to extract high-confidence Msi2 targets. (B) Proportion of genes upregulated in combined RNA-seq/Ribo-seq background data, or the identified high-confidence putative Msi2 targets. P value was assessed with Chi-Squared test. (C) Ingenuity Pathway Analysis of genesets derived from panel A. The top terms for molecular and cellular functions are displayed. Note that only genes upregulated upon loss of Msi2 were selected for analysis. (D) Ingenuity Pathway analysis of 88 high confidence Msi2 targets. (E) KEGG Pathway enrichment of 88 high confidence Msi2 targets.
In order to identify a common group of pathways that are regulated by Msi2, each parental dataset (HITS-CLIP, RNA-seq and Ribo-seq) was analyzed using Ingenuity Pathway Analysis (IPA). Genes involved in cell proliferation, survival and cell cycle were among the top pathways enriched in all datasets, consistent with previously reported functions of Msi2 in promoting cell growth (3–5,8) (Figure 4C). Surprisingly, a significant enrichment for targets involved in cell movement was also commonly detected in all datasets, indicating a widespread effect of Msi2 on cell migration (Figure 4C). Importantly, when the high confidence targets were analyzed with IPA, cellular movement was identified as the most enriched pathway, followed by proliferation and survival (Figure 4D). In particular, KEGG pathway analysis highlighted genes involved in FA, ECM-receptor interaction and the actin cytoskeleton (Figure 4E). These data suggest a possibility that Msi2 controls cellular movement by regulating expression levels of regulators in these pathways.
Within the list of Msi2 targets are many genes that have functions in cell migration, including Flnb/a, Plec, Lama3, and many of the targets selected for qPCR validation and RNA-stability measurements, Itgb1, Itga2, Hbegf, Cxcl5, Sema3c, Igfbp3 and Ppbp (Supplementary Table S1). Furthermore, consistent with Msi's known functions in regulating the Notch signaling pathway (17,21), Dll1, Jag1 and Notch2 are found among the high-confidence Msi2 targets, providing additional evidence for Msi2's role in the Notch pathway (Figure 4E, Supplementary Table S1).
We next compared the HITS-CLIP identified targets in our study to previously published results for Msi2 in the blood and intestine (Supplementary Figure S9A). A number of common targets (221) were identified in addition to a large number of mRNAs unique to each experiment. Interestingly, when GO term analysis was performed for the common and cell type-specific targets, we found GO terms related to cell migration were most prominently enriched in our dataset (Supplementary Figure S9B and C), while the commonly identified 221 genes were not strongly enriched for any GO term related to cell migration (Supplementary Figure S9D). These results demonstrate that Msi2 regulates both common and distinct mRNA targets in different cell-types, and additionally suggests that Msi2 may regulate cellular pathways in a cell-type specific manner.
Msi2 promotes cell proliferation and survival in keratinocytes
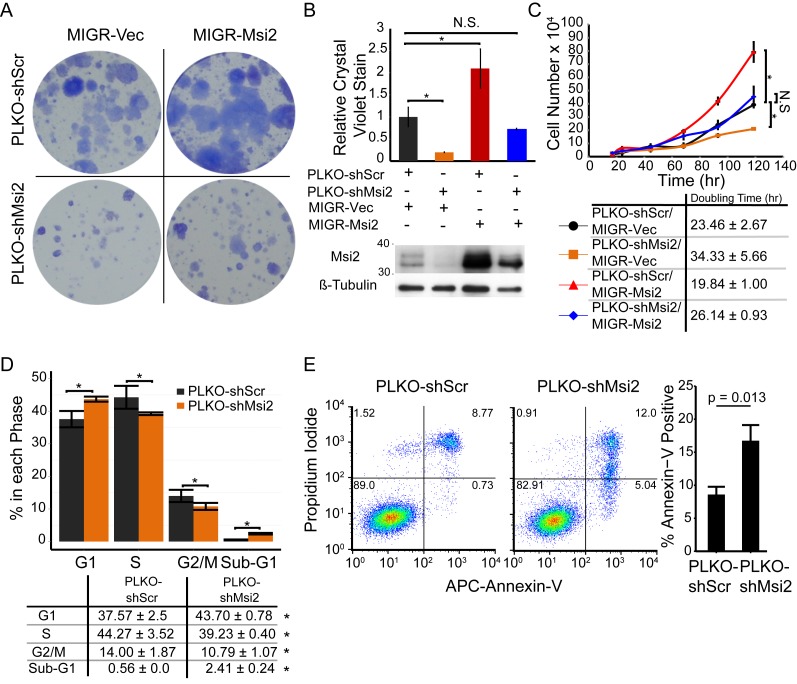
Msi2 has previously been shown to play a critical role in regulating cellular proliferation in many systems (3,4,8). Interestingly, in our high confidence targets we detected negative regulators of cell-cycle progression and pro-apoptotic factors including the DNA damage checkpoint regulator Atm, and the pro-apoptotic inducer Pmaip1. We thus assessed the role of Msi2 in regulating keratinocyte proliferation and survival. Keratinocytes with reduced Msi2 demonstrated reduced colony forming ability in contrast to Msi2 overexpression, which resulted in enhanced colony forming ability (Figure 5A). Additionally, introduction of an shRNA resistant Msi2 cDNA into the knockdown cells rescued the impaired colony forming phenotype (Figure 5A and B). To further characterize this phenotype, the doubling times of shScr, shMsi2, overexpression and rescued keratinocytes were examined. Loss of Msi2 reduced the doubling time by ∼50%. Whereas, overexpression of Msi2 increased doubling time by ∼15% (Figure 5C). The rescue of Msi2 expression in shMsi2 cell lines returned the doubling time to near control levels, validating that the cell growth phenotype is a result of loss of Msi2.
Figure 5.
Msi2 promotes keratinocyte proliferation. (A) Colony formation assay of keratinocytes with control (MIGR-Vec/shScrCtrl), Msi2 overexpression (MIGR-Msi2/shScrCtrl), Msi2 knockdown (MIGR-Vec/shMsi2) or co-infected (MIGR-Msi2/shMsi2) to rescue Msi2 levels in the knockdown condition. Results are representative of two independent biological samples each assayed in triplicate. (B) Crystal violet quantification of colony forming and western blot of keratinocytes infected with the indicated lentiviral and retroviral constructs to knockdown or overexpress Msi2, respectively. Standard error mean displayed. (C) Growth curves of keratinocytes. Results are representative of n = 2 independent biological replicated plated in duplicate. (D) Cell cycle analysis of EdU pulsed keratinocytes with Ctrl and knockdown of Msi2. Representative chart of cell populations shown with quantification of n = 5 biological replicates. *P < 0.05. (Student T-Test two-way). N.S.: not significant. (E) Propidium iodide and Annexin V flow cytometry analysis performed in triplicate to identify the population of apoptotic cells. Representative result displayed from n = 3 independent experiments (Student T-test two-way).
To explore the mechanism by which Msi2 promotes cell growth, we performed cell cycle analysis by EdU incorporation. Loss of Msi2 results in an increased G1 population and reduced S and G2/M populations, suggesting that Msi2 regulates the G1-S transition (Figure 5D). Additionally, the sub-G1 population, reflective of apoptotic or necrotic cells, was significantly increased in the Msi2 knockdowns, indicating that the observed growth differences are caused by defects in both cell proliferation and survival (Figure 5D). To measure apoptosis induced by Msi2 knockdown, Annexin-V/PI staining was performed. The knockdown cells showed ∼2-fold increased levels of Annexin-V positive apoptotic cells when compared to control scrambled cells (Figure 5E). Together, these results provide evidence that Msi2 regulates cell proliferation by controlling cell cycle progression and survival in keratinocytes.
Msi2 inhibits keratinocyte migration
Unexpectedly, our analyses identified many targets of Msi2 involved in the regulation of cell migration. Indeed, the mRNA targets selected for qPCR validation are all involved in cell migration, as either structural adhesion molecules or regulators of cell migration processes. Among Msi2 targets, Itga2, Itgav and Itgb1 are integrin subunits that form heterodimeric transmembrane protein complexes. These proteins mediate cell–cell or cell–ECM attachment and have regulatory effects on cell migration (22). Similarly, the glycoprotein Thbs1, also known as TSP-1, mediates cell-to-cell or cell-matrix interactions through binding a multitude of receptors and can act to simulate or inhibit migration depending on the cellular context (23). Cxcl5 and Ppbp are members of the secreted chemokine CxC ligand family that stimulate cell migration in addition to providing chemoattractant cues (24,25). Additionally, Sema3c, a member of the semaphorin family, stimulates cell migration in diverse tissues (26). Lastly, Igfbp3, an insulin growth factor binding protein, can regulate cell migration in a cellular context dependent manner (27,28).
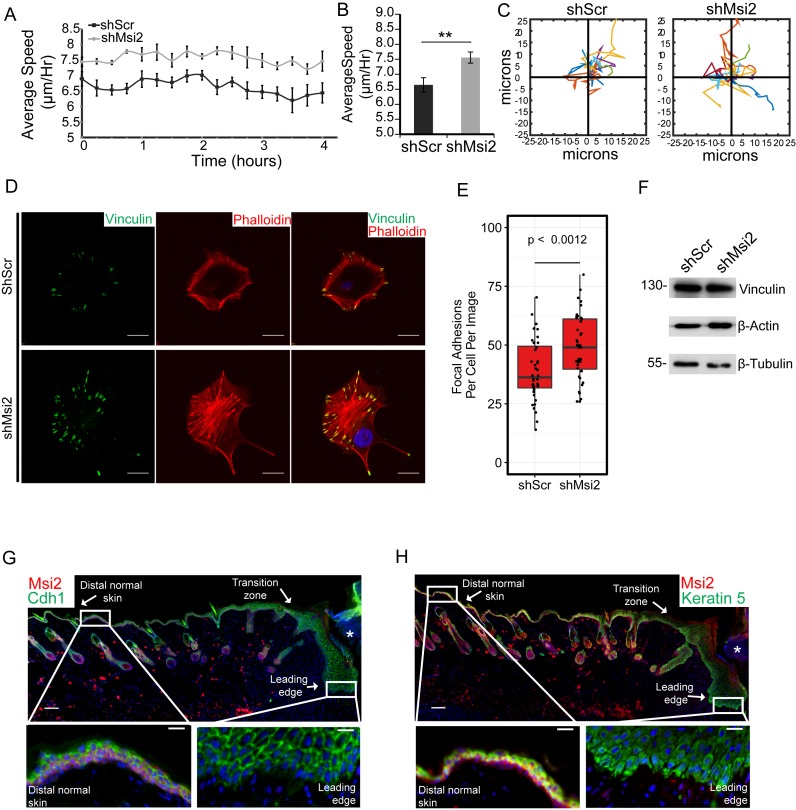
To investigate the role of Msi2 in cell migration, individual cells were tracked using live-cell imaging permitting the measurement of cell migration speed and direction (29). This approach allowed us to investigate cellular migration differences between control and Msi2 knockdown cells independent of cell proliferation rates. Consistent with the identification of numerous Msi2 targets involved in cell migration, loss of Msi2 significantly perturbed keratinocytes migration. Msi2 knockdown cells demonstrated enhanced migration speed compared to scrambled control (Figure 6A and B). Migratory tracks of individual cells revealed no tendency for changed migratory patterns upon Msi2 knockdown (Figure 6C).
Figure 6.
Msi2 inhibits keratinocyte migration and is downregulated during skin wound healing. (A) Cellular migration speed is measured over time for the control and Msi2 knockdown keratinocytes. Data shown are representative of two independent experiments. (B) Average migration speed for the control and Msi2 knockdown keratinocytes. (C) Example cell migration tracks for the control and Msi2 knockdown keratinocytes. (D) Immunofluorescence of a FA marker, Vinculin (green), and an actin marker, Phalloidin (red), for shScr and shMsi2 keratinocytes. Scale bars = 20 μm. (E) Quantification of FA numbers per cell per image for 40 images for shScr and shMsi2 keratinocytes. (F) Western blot for Vinculin, beta-Actin and beta-Tubulin in shScr and shMsi2 cell lines. (G, H) Immunofluorescence of Msi2 (red) and an epithelial cell marker, E-Cadherin (green) or Msi2 (red) and a basal cell marker, Krt5 (green), in a 7-day old skin wound on mouse backskin. Insertions represent regions at the distal or the leading edge of the wounded site. Asterisks (*) represent the granulation tissue at the wounded site. Nuclei are shown in blue. Scale bars = 100 μm (inset = 20 μm). *P < 0.05, **P < 0.01 (Student two-way T-Test).
To establish a mechanistic link between Msi2-mediated gene expression regulation and cell migration, we turned our focus to the pathways that are most enriched for Msi2 targeted genes - regulators of FA and the integrin molecules (Figure 4E). Extensive studies have linked FA and integrins to cell migration (22,30,31). However, it is not clear how expression levels of individual components affect FA formation and cell migration. Immunofluorescence and imageJ quantification for Vinculin and Phalloidin to mark FAs and actin, respectively, showed significantly increased numbers of FAs in the Msi2 knockdown cells without a change in total cellular levels of Vinculin or actin (Figure 6D–F). These data suggest that elevated expression and/or utilization of FA components and integrins in the absence of Msi2 correlates with increased FA formation, likely leading to increased cell migration in these cells.
To further corroborate the role for Msi2 in inhibiting cell migration in a physiologically relevant condition, we examined Msi2 expression during wound healing in adult mouse skin. We used Keratin 5 staining to mark the basal layer, which consists of the basal progenitor cells, and E-cadherin to mark all epidermal keratinocytes in order to localize Msi2. In both the basal epidermal progenitors and the hair follicle stem cells in intact skin regions, Msi2 was readily detectable (Figure 6G and H). Strikingly, within the epidermis at the leading edge of the wound adjacent to the granulation tissue (marked with an asterisks), Msi2 was nearly undetectable, yet its expression recovered gradually with increasing distance away from the wound site (Figure 6G and H). This observation suggests an intriguing possibility that the downregulation of Msi2 is a prerequisite step to promote cell migration in keratinocytes at the leading edge of the wound and that precise control of Msi2 abundance may be required for proper wound healing processes.
DISCUSSION
In this study, we provide a critical link between the UAG motif that has been determined biochemically in vitro and the Msi2: mRNA binding events occurring in vivo. We identified Msi2 targeted mRNAs using an HITS-CLIP approach, which directly crosslinks Msi2 and its associated RNAs and allows transcriptome-wide identification of both Msi2 bound mRNAs and the specific Msi2 binding site. Consistent with the role of Msi2 as a post-transcriptional regulator, we found that the main regions recognized by Msi2 are 3′UTRs (66%) (Figure 1B and C). We also identified a core UAG motif as the critical element for Msi2 recognition, consistent with the previously identified Msi1 motif using biochemical and structural studies (Figure 1D). This is in line with the observation that the RBDs of Msi1 and Msi2 are highly conserved. However, we found that two or more closely positioned UAG motifs are most significantly enriched within the Msi2 HITS-CLIP peaks instead of a single UAG (Figure 1F and G). It also suggests that the two RRMs of Msi2 may each bind to a UAG motif within a short distance for target recognition, providing a mechanism for Msi2 to distinguish its bona fide targets among all UAG sequences that are pervasive in the genome. Such a mechanism was also reported for hnRNPA1, sex-lethal and other RBPs. Indeed, Msi1's RRMs, and by extrapolation, Msi2's RRMs can bind UAG sequences independently, with the linker between the RRMs providing structural guidance and allowing UAGs of varying distance apart to be recognized (9).
We present several lines of evidence that Msi2 in keratinocytes primarily controls abundance of targeted mRNAs, likely by promoting mRNA decay, without affecting TE. To determine the molecular consequences of Msi2 binding to their targets, we depleted Msi2 and measured mRNA abundance and ribosome occupancy by RNA-seq and Ribo-seq, respectively. Recent studies show that Msi1 functions by regulating translation rather than steady-state RNA levels through the interaction with PABP and competing with eIF4G (4,11). However, in our study of Msi2 in keratinocytes, we found that the changes in RNA abundance for Msi2 targets are very similar to and largely explain the changes in ribosome occupancy (Figure 2). Thus, the effect of Msi2 mediated post-transcriptional regulation is similar to what is ascribed to miRNAs (32), another prominent class of post-transcriptional regulators, which regulate mRNAs through interaction with DDX6 and the CCR4-NOT complex (20). We note that Msi2 differs considerably from Msi1 in the C-terminal domain, which is likely responsible for recognition and recruitment of downstream effectors for regulating mRNAs. Our finding raises a possibility that Msi1 and Msi2 evolve different mechanisms for regulating mRNA abundance and translation. Future investigations that distinguish interacting proteins of Msi1 and Msi2 will be required to delineate precise regulatory mechanisms of Msi1 and Msi2.
Our data reveal a new role of Msi2 in regulating migration of keratinocytes. Using our HITS-CLIP data together with RNA-seq and Ribo-seq, we generated a list of high-confidence targets that are directly bound and regulated by Msi2 (Figure 4). Unexpectedly, we found a large number of regulators of FA, ECM-receptor interaction and the actin cytoskeleton, in addition to cell proliferation and survival, as Msi2 targets. Interestingly, many of these targets appear to be cell type-specific when we compared our Msi2 results to what were found in blood and intestine (Supplementary Figure S9). Guided by these findings, we demonstrated that the loss of Msi2 significantly increases cell motility of keratinocytes (Figure 6A–C). Furthermore, we documented increased FA numbers in the absence of Msi2, likely a molecular underpinning for the enhanced motility. Thus, the collective suppression of FA regulators and integrins by Msi2 may partly explain the increased cell migration observed in Msi2 depleted keratinocytes. Altogether, our findings reveal that Msi2 has dual functions in promoting cell proliferation and inhibiting cell migration in the epidermis.
The role of Msi2 in migration and its regulation suggest its downregulation could play important roles in normal development and wound healing. Msi2 appears to be downregulated in the outer root sheath (ORS) progenitors in the normal skin (Supplementary Figure S1B and C). Intriguingly, long-range migration and downwards extension of these ORS progenitors are recently reported to fuel hair follicle growth (33). We also documented significantly suppressed Msi2 expression in the leading edge of the wounded skin (Figure 6G and H). These data suggest that Msi2 also regulates migration in vivo and Msi2 downregulation may be an important prerequisite for stimulating cell migration in these cellular contexts.
Additionally, we also identified many targeted signaling pathways in our data sets, including integrin signaling and the Notch signaling pathway (Figure 4E, Supplementary Table S1). In our high-confidence Msi2 targets, we identified five integrin subunits including Itgb1, Itgb4, Itga2, Itga3 and Itgav as well as Flnb/a, Met and Iqgap1, key regulators of FA and actin dynamics (34,35). These data provide a molecular basis to investigate Msi2's role in cell adhesion and motility in epithelial cells. We also identified new components of the Notch signaling pathway including Notch2 and Dll1 as Msi2 targets. This suggests that the regulation of the Notch pathway by Msi2 is more widespread than currently appreciated. It will also be critical to investigate whether Msi2's regulation of proliferation and migration is linked through these signaling pathways. Future studies using both gain- and loss-of-function mouse models will begin to answer some of these important questions. In conclusion, our study has provided new insights into Msi2 target recognition in intact cellular context, molecular mechanisms of target repression and cellular functions in stratified epithelia. These findings provide a foundation to further examine Msi2 functions in normal development, tissue homeostasis, wound healing and tumorigenesis.
ACCESSION NUMBERS
Raw sequencing and analyzed data are available as a GEO super series with accession number GSE71333.
Supplementary Material
Acknowledgments
We thank R. Parker for comments, B. Gao, K. Diener, J. Kershner for Illumina sequencing. We thank members of the Yi laboratory for discussion and critical reading of this manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Health (NIH) [AR059697 and AR066703 to R.Y.]. Funding for open access charge: NIH - NIAMS (R01AR059697 and R01AR066703).
Conflict of interest statement. None declared.
REFERENCES
- 1.Gerstberger S., Hafner M., Tuschl T. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014;15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakamura M., Okano H., Blendy J.A., Montell C. Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron. 1994;13:67–81. doi: 10.1016/0896-6273(94)90460-x. [DOI] [PubMed] [Google Scholar]
- 3.Ito T., Kwon H.Y., Zimdahl B., Congdon K.L., Blum J., Lento W.E., Zhao C., Lagoo A., Gerrard G., Foroni L., et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466:765–768. doi: 10.1038/nature09171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katz Y., Li F., Lambert N.J., Sokol E.S., Tam W.-L., Cheng A.W., Airoldi E.M., Lengner C.J., Gupta P.B., Yu Z., et al. Musashi proteins are post-transcriptional regulators of the epithelial-luminal cell state. eLife. 2014;3:e03915. doi: 10.7554/eLife.03915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kharas M.G., Lengner C.J., Al-Shahrour F., Bullinger L., Ball B., Zaidi S., Morgan K., Tam W., Paktinat M., Okabe R., et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat. Med. 2010;16:903–908. doi: 10.1038/nm.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park S.-M., Deering R.P., Lu Y., Tivnan P., Lianoglou S., Al-Shahrour F., Ebert B.L., Hacohen N., Leslie C., Daley G.Q., et al. Musashi-2 controls cell fate, lineage bias, and TGF-β signaling in HSCs. J. Exp. Med. 2014;211:71–87. doi: 10.1084/jem.20130736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakakibara S., Nakamura Y., Yoshida T., Shibata S., Koike M., Takano H., Ueda S., Uchiyama Y., Noda T., Okano H. RNA-binding protein Musashi family: roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proc. Natl. Acad. Sci. U. S. A. 2002;99:15194–15199. doi: 10.1073/pnas.232087499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang S., Li N., Yousefi M., Nakauka-Ddamba A., Li F., Parada K., Rao S., Minuesa G., Katz Y., Gregory B.D., et al. Transformation of the intestinal epithelium by the MSI2 RNA-binding protein. Nat. Commun. 2015;6:6517. doi: 10.1038/ncomms7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohyama T., Nagata T., Tsuda K., Kobayashi N., Imai T., Okano H., Yamazaki T., Katahira M. Structure of Musashi1 in a complex with target RNA: the role of aromatic stacking interactions. Nucleic Acids Res. 2012;40:3218–3231. doi: 10.1093/nar/gkr1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zearfoss N.R., Deveau L.M., Clingman C.C., Schmidt E., Johnson E.S., Massi F., Ryder S.P. A conserved three-nucleotide core motif defines Musashi RNA binding specificity. J. Biol. Chem. 2014;289:35530–35541. doi: 10.1074/jbc.M114.597112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawahara H., Imai T., Imataka H., Tsujimoto M., Matsumoto K., Okano H. Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP. J. Cell Biol. 2008;181:639–653. doi: 10.1083/jcb.200708004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C.-Y.A., Ezzeddine N., Shyu A.-B. Messenger RNA half-life measurements in mammalian cells. Methods Enzymol. 2008;448:335–357. doi: 10.1016/S0076-6879(08)02617-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riemondy K., Wang X., Torchia E.C., Roop D.R., Yi R. MicroRNA-203 represses selection and expansion of oncogenic Hras transformed tumor initiating cells. eLife. 2015;4:e07004. doi: 10.7554/eLife.07004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beronja S., Janki P., Heller E., Lien W.-H., Keyes B.E., Oshimori N., Fuchs E. RNAi screens in mice identify physiological regulators of oncogenic growth. Nature. 2013;501:185–190. doi: 10.1038/nature12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang D., Zhang Z., O'Loughlin E., Lee T., Houel S., O'Carroll D., Tarakhovsky A., Ahn N.G., Yi R. Quantitative functions of Argonaute proteins in mammalian development. Genes Dev. 2012;26:693–704. doi: 10.1101/gad.182758.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kishore S., Jaskiewicz L., Burger L., Hausser J., Khorshid M., Zavolan M. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat. Methods. 2011;8:559–564. doi: 10.1038/nmeth.1608. [DOI] [PubMed] [Google Scholar]
- 17.Imai T., Tokunaga A., Yoshida T., Hashimoto M., Mikoshiba K., Weinmaster G., Nakafuku M., Okano H. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol. Cell. Biol. 2001;21:3888–3900. doi: 10.1128/MCB.21.12.3888-3900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingolia N.T., Ghaemmaghami S., Newman J.R.S., Weissman J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jonas S., Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015;16:421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 21.Okabe M., Imai T., Kurusu M., Hiromi Y., Okano H. Translational repression determines a neuronal potential in Drosophila asymmetric cell division. Nature. 2001;411:94–98. doi: 10.1038/35075094. [DOI] [PubMed] [Google Scholar]
- 22.Huttenlocher A., Horwitz A.R. Integrins in cell migration. Cold Spring Harb. Perspect. Biol. 2011;3:a005074. doi: 10.1101/cshperspect.a005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Streit M., Velasco P., Riccardi L., Spencer L., Brown L.F., Janes L., Lange-Asschenfeldt B., Yano K., Hawighorst T., Iruela-Arispe L., et al. Thrombospondin-1 suppresses wound healing and granulation tissue formation in the skin of transgenic mice. EMBO J. 2000;19:3272–3282. doi: 10.1093/emboj/19.13.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu M., Berk R., Kosir M.A. CXCL7-mediated stimulation of lymphangiogenic factors VEGF-C, VEGF-D in human breast cancer cells. J. Oncol. 2010;2010 doi: 10.1155/2010/939407. Article ID 939407, 10 pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuo P.-L., Chen Y.-H., Chen T.-C., Shen K.-H., Hsu Y.-L. CXCL5/ENA78 increased cell migration and epithelial-to-mesenchymal transition of hormone-independent prostate cancer by early growth response-1/snail signaling pathway. J. Cell. Physiol. 2011;226:1224–1231. doi: 10.1002/jcp.22445. [DOI] [PubMed] [Google Scholar]
- 26.Goshima Y., Ito T., Sasaki Y., Nakamura F. Semaphorins as signals for cell repulsion and invasion. J. Clin. Invest. 2002;109:993–998. doi: 10.1172/JCI15467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang K.-H., Chan-Ling T., McFarland E.L., Afzal A., Pan H., Baxter L.C., Shaw L.C., Caballero S., Sengupta N., Calzi S.L., et al. IGF binding protein-3 regulates hematopoietic stem cell and endothelial precursor cell function during vascular development. Proc. Natl. Acad. Sci. 2007;104:10595–10600. doi: 10.1073/pnas.0702072104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gribben L., Baxter R.C., Marsh D.J. Insulin-like growth factor binding protein-3 inhibits migration of endometrial cancer cells. Cancer Lett. 2012;317:41–48. doi: 10.1016/j.canlet.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 29.Chapnick D.A., Jacobsen J., Liu X. The development of a novel high throughput computational tool for studying individual and collective cellular migration. PLoS ONE. 2013;8:e82444. doi: 10.1371/journal.pone.0082444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Case L.B., Waterman C.M. Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat. Cell Biol. 2015;17:955–963. doi: 10.1038/ncb3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ridley A.J., Schwartz M.A., Burridge K., Firtel R.A., Ginsberg M.H., Borisy G., Parsons J.T., Horwitz A.R. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 32.Eichhorn S.W., Guo H., McGeary S.E., Rodriguez-Mias R.A., Shin C., Baek D., Hsu S.-H., Ghoshal K., Villén J., Bartel D.P. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol. Cell. 2014;56:104–115. doi: 10.1016/j.molcel.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rompolas P., Deschene E.R., Zito G., Gonzalez D.G., Saotome I., Haberman A.M., Greco V. Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration. Nature. 2012;487:496–499. doi: 10.1038/nature11218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacquemet G., Morgan M.R., Byron A., Humphries J.D., Choi C.K., Chen C.S., Caswell P.T., Humphries M.J. Rac1 is deactivated at integrin activation sites through an IQGAP1-filamin-A-RacGAP1 pathway. J. Cell Sci. 2013;126:4121–4135. doi: 10.1242/jcs.121988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen S.-Y., Chen H.-C. Direct interaction of Focal Adhesion Kinase (FAK) with met is required for fak to promote hepatocyte growth factor-induced cell invasion. Mol. Cell. Biol. 2006;26:5155–5167. doi: 10.1128/MCB.02186-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.