Abstract
Programmable control of the cell cycle has been shown to be a powerful tool in cell-biology studies. Here, we develop a novel system for controlling the bacterial cell cycle, based on binding of CRISPR/dCas9 to the origin-of-replication locus. Initiation of replication of bacterial chromosomes is accurately regulated by the DnaA protein, which promotes the unwinding of DNA at oriC. We demonstrate that the binding of CRISPR/dCas9 to any position within origin or replication blocks the initiation of replication. Serial-dilution plating, single-cell fluorescence microscopy, and flow-cytometry experiments show that ongoing rounds of chromosome replication are finished upon CRISPR/dCas9 binding, but no new rounds are initiated. Upon arrest, cells stay metabolically active and accumulate cell mass. We find that elevating the temperature from 37 to 42°C releases the CRISR/dCas9 replication inhibition, and we use this feature to recover cells from the arrest. Our simple and robust method of controlling the bacterial cell cycle is a useful asset for synthetic biology and DNA-replication studies in particular. The inactivation of CRISPR/dCas9 binding at elevated temperatures may furthermore be of wide interest for CRISPR/Cas9 applications in genomic engineering.
INTRODUCTION
The initiation of the replication of a chromosome is an evolutionary conserved process, and the mechanism of initiation is very similar among different organisms (1–3). In bacteria, replication is initiated when DnaA proteins recognize and bind to specific sequences—DnaA boxes—within the origin of replication locus (oriC). Upon polymerization, they cause adjacent double-stranded DNA to melt, providing a single stranded substrate onto which the helicase loader DnaC loads the replication helicase DnaB (4–6). The process of initiation is tightly regulated in time, and several mechanisms for preventing premature initiation are known in different species. Origin sequestration by SeqA protein, distant DNA tethering to the bacterial membranes, or specific protein regulators interacting with replication initiation machinery are known to act as negative regulators of initiation of the replication (7–9).
Programmable control of the cell cycle has proven to be a powerful tool in cell biology, elucidating many processes involved in replication and metabolism (10,11). The dynamic growth of the field of synthetic biology also strives for robust methods for controlling the rate of bacterial replication which can be integrated into genetic circuits (12). A number of methods are currently available for bacterial cell-cycle control: bacterial cells can be arrested in a pre-replication state using thermosensitive variants of DnaC and DnaA replication-initiation proteins (13), using columns which release newborn cells (14), or using restricted growth conditions (15). None of these methods, however, provide an easy and chemically inducible control over the bacterial cell cycle.
The CRISPR (clustered regularly interspaced short palindromic repeats) system, naturally responsible for bacterial immunity against viruses, has recently been widely engineered and repurposed for many biological applications, from genome engineering and in vitro RNA digestion, fluorescent labeling of genomic positions, or production of antimicrobials (16–21). A simple type II CRISPR system, originating from Streptococcus pyogenes, consists of only two components, a Cas9 protein and small guide RNA (sgRNA). CRISPR/Cas9 system is very efficient in recognizing DNA sequences that are complementary to the specific 20 nucleotides of the sgRNA sequence (22). A modification of Cas9 protein, which abolishes its nuclease activity, allows CRISPR/dCas9 (dCas9 – nuclease-deficient Cas9) to form such a stable complex with the complementary DNA that, if targeted to the promoter region of the gene, excludes RNA polymerase from binding to the promoter sequence. Such an approach was used to regulate gene expression in a wide range of organisms (23–25).
Here, we repurpose a CRISPR/dCas9 system for synchronization of Escherichia coli cells into the pre-replication state. We do so by inactivating the initiation of replication of the bacterial chromosome by targeting CRISPR/dCas9 to the E. coli origin of replication. The replication arrest is found to be highly specific to the origin-of-replication locus and is not observed when dCas9 is targeted to proximal regions of origin of replication. Flow cytometry chromosome-number counting and single-cell fluorescence microscopy show that initiation of replication is blocked very efficiently after the expression of our system in bacterial cells. Furthermore, we show that CRISPR/dCas9 is not active at elevated temperatures, and we exploit that property to recover bacterial cells from the arrested state of replication.
MATERIALS AND METHODS
Strains and culture conditions
All experiments were done with derivatives of E. coli K12 TB28 (MG1655; ΔlacIZYA (26)) with the exception of the serial-dilution plating experiment shown in Figure 1, which was done with E. coli K12 AB1157 strain with chromosomal loci marked with lacO (27). For serial-dilution plating experiments and genetic manipulations, cells were grown in a Lysogeny broth (LB) at 37°C, except for the experiment testing the thermo-sensitive properties of CRISPR/dCas9, where cells were grown in LB media at 30, 37 or 42°C, as specified. Ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml) were added when required. Expressions of dCas9 and dCas9deg were induced with the addition of anhydrotetracycline (aTc, 200 ng/ml). For microscopy and flow-cytometry experiments, cells were grown in M9 media supplemented with 0.2% glucose at 37°C, or at 42°C for the recovery experiments.
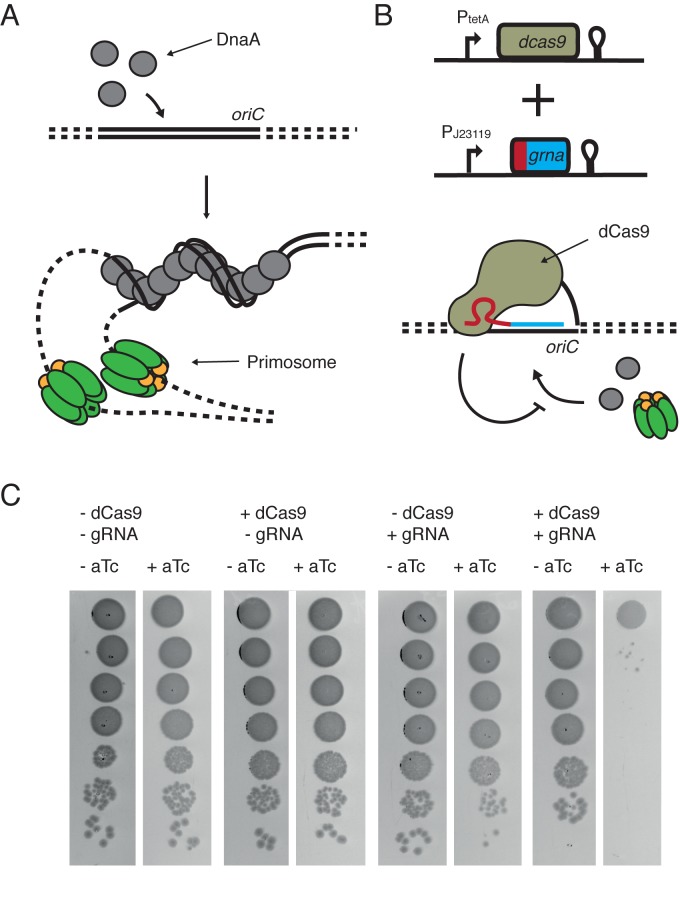
Figure 1.
CRISPR/dCas9 system blocks initiation of replication at the oriC locus. (A) Schematic of initiation of replication by DnaA. Cooperate binding of DnaA proteins to oriC induces unwinding of an adjacent AT-rich region, thus providing single-stranded DNA substrate that is recognized by primosome complexes. Green – helicase DnaB, yellow – helicase loader DnaC. (B) The CRISPR/dCas9 system consists of two plasmids, one coding for dCas9 under the control of an aTc-inducible promoter and the other coding for sgRNA under control of a constitutive promoter. When CRISPR/dCas9 binds to the oriC region, DnaA cannot bind and unwind the DNA, and initiation of replication is blocked. (C) Simultaneous expression of dCas9 and sgRNA has a lethal effect on cells. Serial 10-fold dilutions of liquid bacterial cultures were plated either on the media supplemented (+aTc) or not (−aTc) with 200 ng/ml of aTc. Only in presence of both CRISPR/dCas9 components, cells are not viable.
Plasmid and strain construction
Supplementary Table S1 lists the plasmids and sgRNA targets used in this study. Top10 cells (Thermo Fisher) and the Mix & Go E. coli transformation kit (Zymo Research) were used to transform all cloning reactions. Plasmids pdCas9-bacteria and pgRNA-bacteria were obtained from Addgene (23). Plasmid pdCas9deg was created by restriction digestion and ligation of a PCR fragment obtained by amplification of pdCas9 plasmid backbone with primers Jw098 and Jw099 containing a LAA degradation tag sequence (28) and XhoI restriction sites.
Modifications of sgRNA 20nt sequences were done by PCR amplification of a pgRNA backbone with primers carrying a SpeI restriction site and 20 bp of sgRNA sequence. The PCR fragment was digested and ligated into a circular plasmid. A list of primers used to create pgRNA plasmids can be found in Supplementary Table S2. J23119 constitutive promoter drove the expression of sgRNA.
Plasmid pdCas9deg3 was created by a CPEC reaction (29), by combining pdCas9deg with the sgRNA region of plasmid pgRNA3 with primers Jw121, Jw122, Jw124 and Jw125. In this construct, pdCas9deg was under the control of an aTc-inducible promoter and gRNA3 was placed under the control of a constitutive J23119 promoter.
The strain containing the origin-proximal FROS system and LacI-tagGFP was constructed by P1 phage transduction (as described in (30)) from strain IL01, carrying an origin proximal lacO array (27), and a strain BN1442 carrying a LacI-tagGFP fusion under the control of lactose promoter, into the TB28 strain. Resistances were removed, when possible, using a Flp recombinase expressed from pCP20 (31).
Serial-dilution experiments
For serial-dilution experiments, cells were grown at 37°C in LB with addition of ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml) as needed. Ten-fold serial dilutions were made by diluting 20 μl of cell suspension in 180 μl of LB media in every dilution step. Cells were plated on LB agar plates supplemented with antibiotics, as needed, and incubated at an appropriate temperature for 18 h before imaging. All serial dilution-plating experiments were done at least in two repetitions.
Fluorescence microscopy
Fluorescence microscopy experiments were carried using a Nikon Ti-E microscope with CFI Plan Apochromat λ DM 100× objective, Lumencor Spectra X LED light source, Andor Zyla 4.2 CMOS camera, and a Lumencor SpectraX filter set. Images were collected with Nikon NIS software and analyzed using FIJI software (32), microbeTracker suite (33) and custom Matlab scripts and functions. Sample sizes are as follows: Figure 4C: - aTc t0 = 378 cells, t1 = 402, t2 = 732, t3 = 1100, t4 = 1993, + aTc t0 = 378, t1 = 567, t2 = 379, t3 = 398, t4 = 864. Figure 4D: - aTc t0 = 228, t1 = 140, t2 = 255, t3 = 375, t4 = 544, + aTc t0 = 228, t1 = 224, t2 = 158, t3 = 90, t4 = 170.
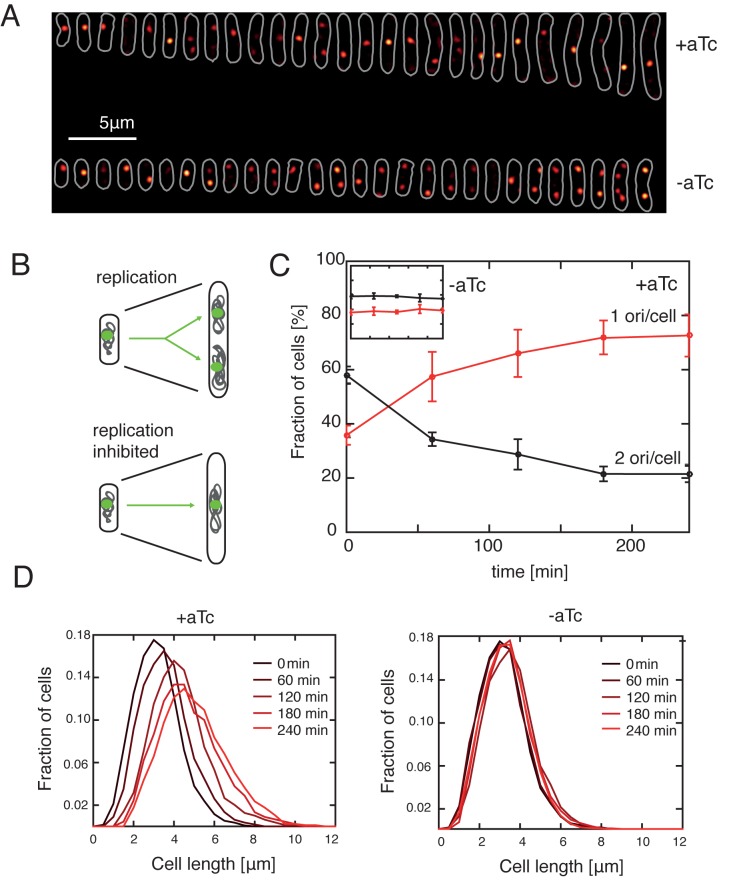
Figure 4.
CRISPR/dCas9deg3 binding to the origin of replication inhibits the initiation of replication but not the cell growth. (A) Representative images of individual cells grown in M9 glucose at 37°C with (top stripe) or without 200 ng/ml of aTc (bottom stripe). All cells images were collected at 240 min after the beginning of induction. White color denotes the cell outlines obtained with microbeTracker software. Red spots denote the oriC loci LacI-tagGFP labels. Cells were lined up from the shortest to the longest and are a representative of the bacterial population. Background has been subtracted. (B) Inhibition of replication leads to an arrested cell phenotype. Arrested cells do not replicate the DNA (hence have one chromosome), but stay metabolically active and increase in size. Green – oriC; gray – DNA. (C) CRISPR/dCas9deg3 system arrests the cell population, yielding cells with one chromosome. After the addition of 200 ng/ml of aTc the population of cells with only one oriC focus is increasing, and saturates to a fraction larger than 70% after 180 min. Population cultured without the CRISPR/dCas9deg3 induction maintains stable level of oriC content. Cells were grown in M9 glucose in 37°C. Error bars indicate SD for three independent experiments for each dataset for each time point, with the exception of t = 240 h, +aTc, where the error bar denotes the mean standard deviation of the dataset. Colors in the inset correspond to those in the main panel. (D) Cell lengths increase after the induction of CRISPR/dCas9deg3-induced arrest. The distribution of lengths of cells in the population is shifting towards longer cells after the induction of arrest. The distribution stabilizes after ∼180 min of induction. Cells are grown as in panels (A) and (C).
Flow cytometry
Cells were grown in M9 glucose at 37°C (or at 42°C for recovery experiments). Induction with 200 ng/ml of aTc was started in the early exponential phase (A600nm ∼ 0.1). Cell samples were prepared as described in (34), and Syto16 (Life Technologies) was used to stain the DNA. Data were collected with the use of a FACSacn flow cytometer (BD Biosciences) and analyzed with custom Matlab scripts. In each experiment 30.000 events were recorded. Raw fluorescent microscopy data were loaded into a Matlab environment using the FCS-data-reader function (http://www.mathworks.com/matlabcentral/fileexchange/9608-fcs-data-reader) and plotted using standard Matlab functions.
RESULTS
CRISPR/dCas9 blocks the initiation of replication at the oriC locus
The initiation of replication in E. coli is restrained to a relatively short (245 bp) and well-defined DNA sequence called oriC (Figure 1A top) (35). Because the earliest step of initiation relies on DnaA binding to specific regions of oriC, called DnaA boxes, it is possible to design a simple CRISPR/dCas9 system that inhibits the cellular replication-initiation machinery by hindering DnaA–oriC interactions (Figure 1B bottom). To engineer such a system, we used a previously described set of two plasmids, one coding for the dCas9 protein, under control of an aTc inducible tetracycline promoter (pdCas9), and the second plasmid coding for the small guide RNA (sgRNA) under the control of a constitutive promoter (Figure 1B) (23). Previous results showed that binding of dCas9 to the promoter region of a gene can efficiently block its transcription (25,23), and targeting it at any protein-recognized DNA sequence may hinder protein–DNA interactions specific to that region. To test if CRISPR/dCas9 binding to the origin of replication can impair the initiation of replication, we constructed and tested a CRISPR/dCas9 that binds to one of the DnaA boxes in the oriC (R1 DnaA box) (36).
Serial-dilution plating shows that cells were not viable if dCas9 and sgRNA were expressed simultaneously: bacteria plated on aTc-containing media showed a drastic reduction in survival (Figure 1C, rightmost row). Expression of guide RNA or dCas9 alone had no significant effect on cell survival in the serial-dilution plating experiments (Figure 1C, left). We suggest that the observed effect results from a competition between DnaA and dCas9 binding to the same DnaA box. Tight binding of CRISPR/dCas9 can prevent DnaA from sequence recognition, thus preventing DnaA-filament formation and the subsequent associated replication-bubble opening that starts replication.
CRISPR/dCas9 inhibition of replication initiation is specific to the origin of replication region
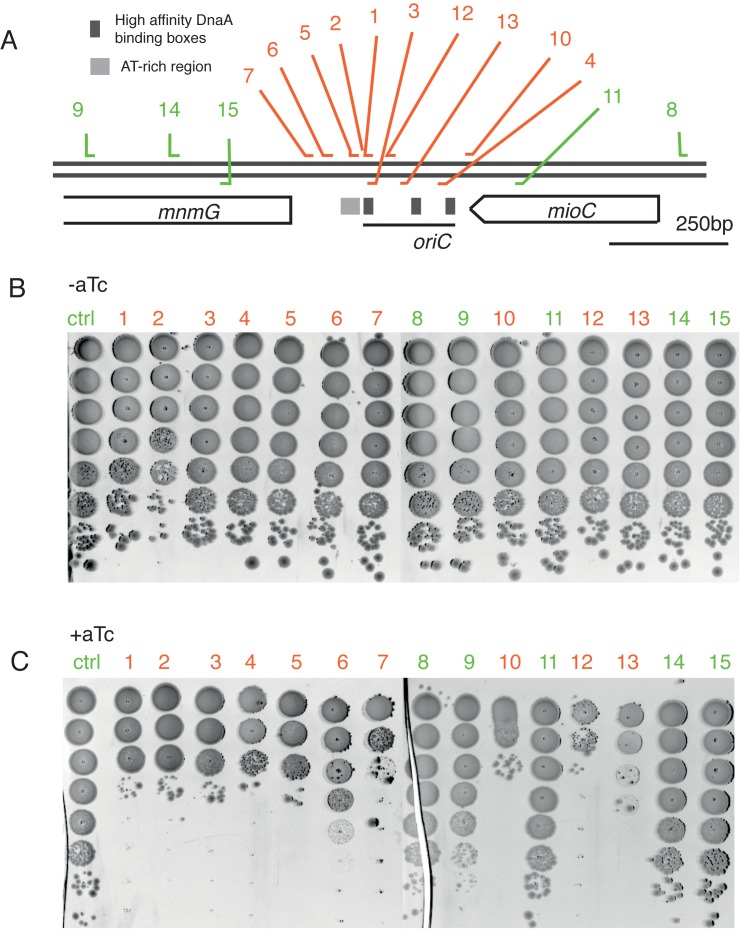
To test if the observed effect is specifically caused by inhibition of replication initiation, we probed the effect of CRISPR/dCas9 binding to many targets throughout the entire origin of replication region, as well as adjacent proximal regions, using a library of sgRNA constructs. The location of all used sgRNA targets is shown on the Figure 2A, overlaid on a genetic map of the oriC region and its surrounding. Targets displayed in red are binding within the origin whereas green targets are binding outside of the origin of replication (Figure 2A).
Figure 2.
CRISPR/dCas9 inhibition of replication initiation is highly specific to the origin of replication region. (A) Genetic map of the oriC region. The 245 bp oriC sequence is marked as well as two adjacent gene-coding regions. Each number corresponds to a different gRNA 20nt target. Targets complementary to origin of replication are shown in red, targets binding outside of origin region are marked in green. Double line represents the dsDNA; sgRNA targets binding the leading strand are represented to bind the upper strand; gRNA targets complementary to the lagging strand are represented to bind the bottom strand. DnaA boxes from left to right: R1, R2, R4. (B and C) CRPSR/dCas9 inhibits cell growth only when targeted to origin of replication. Serial 10-fold dilutions of bacterial cultures were plated on media without the addition (B) or with addition (C) of 200 ng/ml of aTc. Numbers corresponds to gRNA targets shown on (A).
We find that growth inhibition is only observed when CRISPR/dCas9 is binding to targets within the origin or replication. Binding to regions upstream or downstream of the origin site does not affect cell growth after the induction of CRISPR/dCas9 system with aTc (as observed in the green-labeled serial-plate results in Figure 2C). This indeed indicates a high specificity of the CRISPR/dCas9 system that only blocks the initiation of replication when it binds at any spot within the oriC locus.
A potential alternative explanation for the loss of viability of cells is stalling of the replisome during the collision with CRISPR/dCas9 bound to DNA. Indeed, replication-machinery stalling has been reported when the replication fork encountered a repeated sequence of operators that were bound by repressors (37,38). However, we find that only CRISPR/dCas9 with guide RNA that are complementary to sequences within the origin of replication lead to cell death, whereas guide RNA targeting sites outside of origin of replication, or other genomic positions tested in other studies, are not lethal. These observations lead to the conclusion that the system is indeed specifically blocking the replisome formation, and not its progression.
Single-plasmid coding shows a superior performance over a two-plasmid system
To minimize non-specific effects of high levels of CRISPR/dCas9 expression, we fused dCas9 protein to a strong LAA degradation signal tag (39), creating a dCas9deg protein. Next, we combined dCas9deg (dCas9 fused to LAA degradation tag) protein and sgRNA3 (Figure 2) on a single low-copy-number plasmid, called pdCas9deg3 (Supplementary Figure S1A). The LAA tag is promoting an active degradation of a dCas9deg by the ClpXP protease, preventing an accumulation of surplus CRISPR/dCas9 in the cell (28). Such accumulation could potentially trigger a cellular stress by accumulation of inclusion bodies. The dCas9deg is still active in the serial-dilution plating assay, but the LAA tag reduces the aTc sensitivity of the CRISPR/dCas9deg about 10-fold, compared to a original dCas9 protein (Supplementary Figure S1B).
Combination of dCas9deg and pgRNA3 on a single plasmid further improved the performance of the system. A FACS experiment with a two-plasmid system (pdCas9 and pgRNA3) in bacteria, with their DNA stained with the Syto16 fluorescent dye, showed a population of small-sized particles, presumably the remains of dead cells or anucleated cells (Supplementary Figure S2). An identical experiment done with a single-plasmid system showed a well-preserved culture (Figure 3). Both modifications of CRISPR/dCas9 system, introducing the LAA tag and expressing two components from a single plasmid, proved to be necessary to create a viable cell-cycle control system.
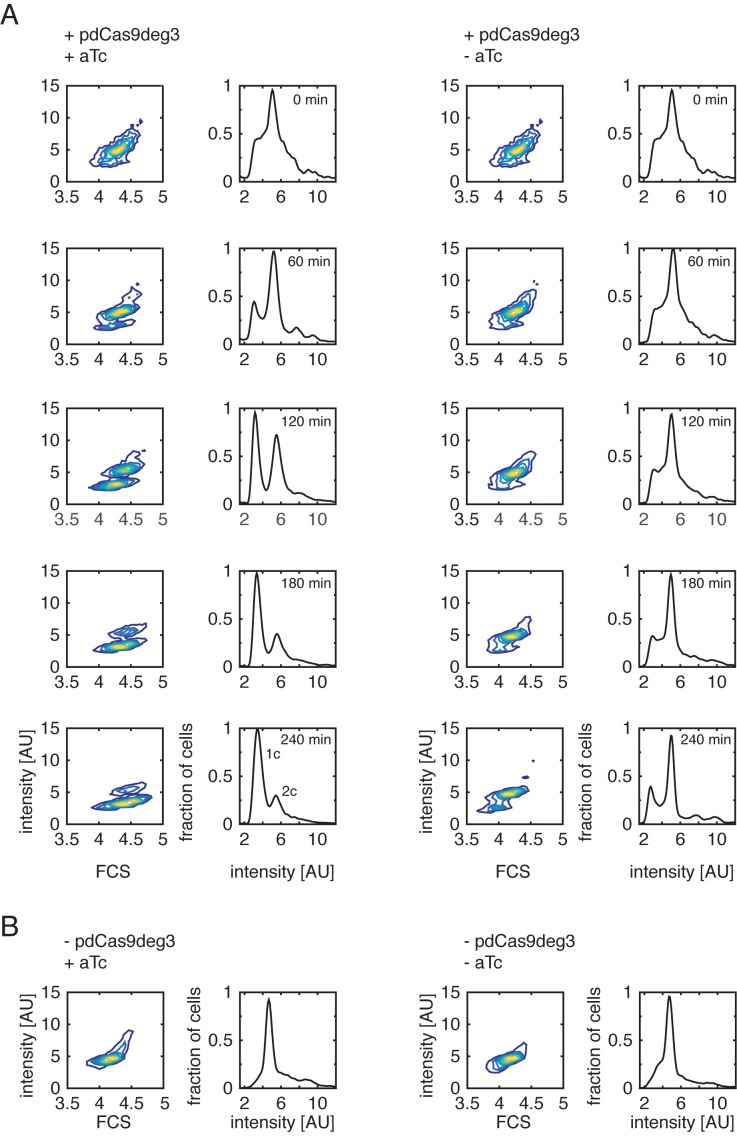
Figure 3.
CRISPR/dCas9deg3 binding to oriC inhibits the initiation of replication. Contour plots of cytograms (left) and fluorescence-intensity histograms (right). (A) Early exponential cell culture (A600nm 0.1-0.2) was divided into two subcultures, where one (left) was induced by addition of 200 ng/ml aTc and the other one (right) was not. Each hour, cells were fixed, DNA was stained with Syto16, and the fluorescence signal was measured. Induced culture showed an arrested chromosome distribution, which saturated at 180 min after induction. Non-induced culture maintained the physiological chromosome content during the entire time of the experiment. The signal equivalent to one or two chromosomes is indicated by ‘1c’ and ‘2c’, respectively, on the 240 min-induced histogram. (B) Addition of 200 ng/ml of aTc had no observed effect on the chromosome content of bacteria lacking CRISPR/dCas9deg3 system.
Chromosome counting verifies that CRISPR/dCas9deg3 system inhibits the initiation of replication
To test the effect of CRISPR/dCas9deg3 binding to the oriC region at the level of individual MG1655 E. coli bacterial cells, we examined the DNA content of cells with flow cytometry. DNA was stained with Syto16, which provides a quantitative measure for the amount of chromosome equivalents as the fluorescent signal scales linearly with DNA content of a cell. At time zero, early exponential cell culture growing in M9 medium supplemented with glucose was split into two smaller cultures, and one of the two was induced with 200 ng/ml of aTc.
Upon induction of the CRISPR/dCas9deg3 system, we could clearly distinguish the development of a population of cells that carry only one copy of the chromosome (Figure 3A, shown as a ‘1c’ population on the bottom-left histogram). While cells in the controls contain on average two chromosomes, the induced arrest of the initiation of replication leads to a reduction of chromosome content to mostly 1 copy per cell. The shift from 2 to 1 chromosomes is visible already ∼60 min after the induction and stabilizes after ∼180 min, and it is clearly visible both on fluorescent intensity histograms and in the appearance of a clearly separated bottom population in the contour plots (Figure 3A). A control experiment with a bacterial culture without the aTc induction shows a typical DNA content distribution of a logarithmic culture in M9 media supplemented with glucose (Figure 3A, right column, mostly two chromosomes per cell). Moreover, DNA content profiles for un-induced culture do resemble the cultures not transformed with pdCas9deg3 plasmid, with, or without addition of aTc (Figure 3B). The data indicate that upon blocking the initiation of replication by CRISPR/dCas9deg3 binding, the ongoing replication is finished and cell division leads to cells with 1 chromosome, which cannot be further replicated. In other words, replication forks that were established before the inhibition of initiation are not interrupted by the CRISPR/dCas9deg3 system, and the observed effect is specific to the initial replisome formation at the oriC.
These results suggest that the CRISPR/dCas9deg3 system specifically and efficiently blocks the initiation of chromosome replication. Importantly, similar experiments exploiting thermosensitive properties of DnaC2 protein yielded a very comparable result for the chromosome content upon inhibition of initiation of replication (13).
Single-cell fluorescence microscopy of CRISPR/dCas9deg3 arrested cells
Next, we investigated the effects of CRISPR/dCas9deg3 binding on cell morphology and chromosome copy number using fluorescence microscopy of MG1655 E. coli cells to which we integrated a fluorescent probe (FROS – fluorescent repressor-operator system) in the proximity of oriC location (27). Because origin regions are segregated within minutes after replication in E. coli, we can thus estimate the number of replicated origins by simply counting the number of origin-proximal foci in each cell. As expected, we find that the bacterial cells in which initiation of replication is inhibited contain mainly a single fluorescent focus (Figure 4A, top), meaning that the region labeled with FROS is not replicated, in contrast to un-induced cells where each round of replication doubles the number of foci (Figure 4A, bottom). Analysis of the origin-proximal foci number thus again indicates that the CRISPR/dCas9deg3 efficiently blocks initiation of replication. At 240 min after the induction, 73 ± 8% cells contain single origin focus, a fraction about twice as large as at the beginning of the experiment where it was 36 ± 4% (Figure 4C, red line) or for a population growing without the presence of aTc which yielded 38 ± 4% (Figure 4C, inset).
CRISPR/dCas9deg3 binding to oriC does not inhibit cell growth
We next tested whether the inhibition of initiation of replication had an effect on cell growth. Cells from previous experiment were found to elongate after the exposure to aTc: At 240 min after the induction, the mean cell length had increased to 4.5 ± 1.5 μm, which is significantly larger than the initial length of 3.1 ± 0.8 μm (see Figure 4A and D). Cells not induced with aTc maintained a stable distribution of cell length throughout the entire length of the experiment (Figure 4D, right). The increased length of the cells suggests that cell division has been inhibited, which can, for example, occur through nucleoid occlusion that can block the formation of the division machinery when E. coli chromosomes are not correctly replicated and segregated (40) (Figure 4D, left). Furthermore, the larger cell sizes indicate that the CRISPR/dCas9deg3 system used in our study is not stopping the cell metabolism, as cells are still able to grow and elongate after the induction. These results demonstrate that our system specifically and efficiently interferes with the initiation of replication, while cells are still well viable and able to increase their mass.
Replication can be restored with the thermo-sensitive properties of CRISPR/dCas9deg
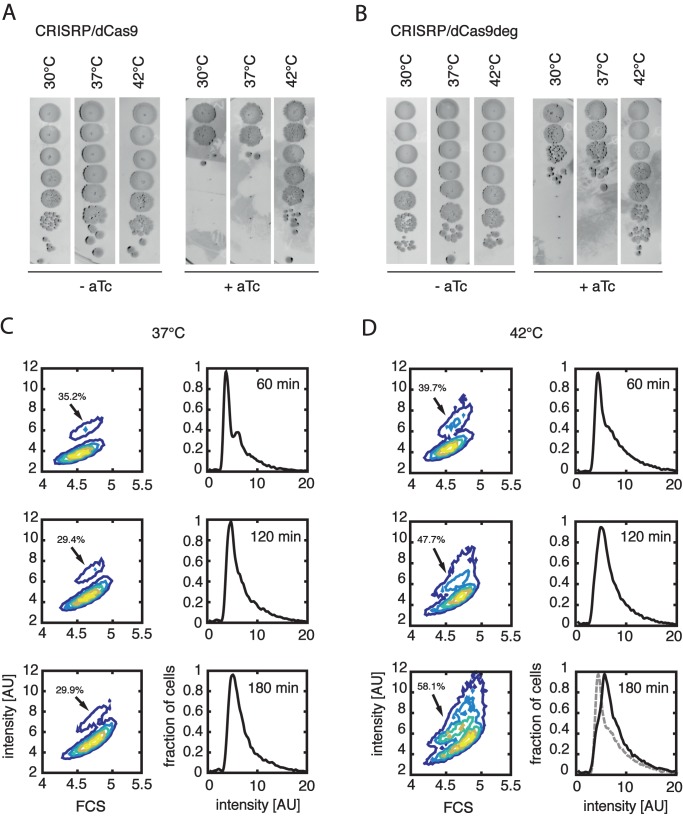
In doing these experiments, we unexpectedly discovered that the CRISPR/dCas9 (and the modified version – CRISRP/dCas9deg) inhibition of replication is not functional at 42°C. Cells plated on a LB agarose media supplemented with aTc are, surprisingly, able to form colonies at 42°C, whereas cells cultured at 37 or 30°C do not proliferate in presence of aTc (Figure 5A and B).
Figure 5.
CRISPR/dCas9 and CRISPR/dCas9deg are thermosensitive and the arrest can be reversed. (A) Effects of temperature on dCas9 supplemented with gRNA3. Cells were cultured in LB media, 10-fold serial dilutions were plated with or without 200 ng/ml of aTc and cultured at different temperatures. In both cases, the system was supplemented with pgRNA3 guide. CRISPR/dCas9 system is not active at elevated temperature and cells are able to form colonies. (B) Same as panel (A), but pdCas9deg was used instead pCas9. CRISPR/dCas9deg is also inactivated at elevated temperature. (C) Cells transformed with pdCas9deg3 were arrested for 150 min in M9 glucose at 37°C, diluted ten times in fresh media without the aTc and grown at either 37°C or 42°C. A population of replicating cells can be observed after 60 min at 42°C, and after 180 min replicating cells are forming a substantial fraction of the whole population. Contour plots show the DNA content (Syto16 intensity) as a function of cell mass (FCS), while the histograms show the distribution of the DNA content (Syto16 intensity) in the population. Arrows are pointing to the population of cells that carry more than one copy of the chromosome. Percentages of cells in population with more that one chromosome are shown at each contour plot. Cells cultured at 37°C are not showing recovery from the arrest. The grey dashed line on the 180 min 42°C histogram shows the intensity histogram for 60 min 42°C, displayed for convenience of direct comparison.
This unexpected property of CRISPR/dCas9 allowed us to reinitiate the previously inhibited replication of chromosomes. Cells transformed with the pdCas9deg3 plasmid were first arrested by the addition of aTc to M9 media supplemented with 0.2% glucose for 2.5 h prior to the start of the temperature-mediated recovery. After arrest, cultures were diluted 10 times in fresh M9 glucose media to lower the cultures OD and provide enough media for proliferation of cells. Two cultures were grown further at 37°C or at 42°C. The cells growing at 37°C did not show signs or recovery during the course of an experiment (Figure 5C). At 42°C, however, first replicating cells were visible already after 60 min of incubation. One hundred eighty minutes after the shift to elevated temperature, a large fraction of replicating cells is clearly visible on the contour plot (Figure 5D, indicated by the arrow). Interestingly, short (15’ or 30’) pules of 42°C incubation, followed by growth in 37°C did not show recovery of replication. This may indicate that the dCas9/dCas9deg that was inactivated by the elevated temperature regains activity after the shift back to 37°C (Supplementary Figure S3). We can thus exploit this newly discovered thermosensitive property of the CRISPR/dCas9 system to reverse effects of inhibition of initiation of replication.
DISCUSSION
This paper reports a novel system for controlling the bacterial cell-cycle stage using programmable inhibition and re-activation of the initiation of replication of the E. coli chromosome. CRISPR/dCas9deg3 efficiently blocks replication at the initiation stage and thus synchronizes a bacterial culture at the pre-replicating state. Arrested cells are still metabolically active, and can restart the replication and proliferation after a switch to 42°C. Our approach is reminiscent of mechanisms based on blocking DnaA from binding to the origin of replication (8,9) by specific proteins (Spo0A in B. subtilis, or CtrA in C. cresentus) and our CRISPR/dCas9 based system mimics such naturally occurring mechanisms.
Interestingly, the DNA-binding footprint of CRISPR/dCas9 (∼20 nucleotides based on the crystallographic studies (41) is much shorter than the oriC sequence (245 bp (35)), and a single CRISPR/dCas9 molecule thus covers only a small fraction of the entire origin. Nevertheless, that single binding process is highly efficient in preventing the initiation of replication, as shown by our local binding data at many (∼10) targets within the oriC. This indicates that the entire origin of replication region is important for DnaA-induced replication-bubble formation. CRISPR/dCas9 binding to regions in close proximity to origin of replication showed, however, no effect on the bacterial viability, also verifying that replication initiation is strictly restrained to the origin region, and that the adjacent DNA does not play a vital role in the process of initiation. The system is also active at temperature of 30°C, at which thermosensitive versions of proteins DnaC and DnaA are able to initiate the replication. Therefore, it can also be used to study processes which require low-temperature growth conditions (13). Moreover our system has an additional advantage, as it requires only one transformation step and addition of the aTc to the media to achieve the replication-arrested phenotype.
In our work, we reported, for the first time to our knowledge, that the system based on CRISPR/dCas9 exhibits thermo-sensitive properties. We used this phenomenon to restart the cell cycle from the arrested phase, but this particular feature of dCas9 systems may find much more general applications. For instance, in Cas9-mediated genome editing, rapid temperature switching may provide control over the dCas9 and likely the wtCas9 activity.
The CRISPR/dCas9deg system controlling the replication can be integrated into any existing genetic circuits. Rational design of genetic circuits lies at the foundation of the rapidly developing field of synthetic biology (42,43). In practical application, our CRISPR/dCas9deg can arrest cell growth after the engineered bacterium served its role (12).
Our results show that CRISPR/dCas9 can be used to efficiently control an important biological process, viz., initiation of DNA replication. It provides a new tool to control the cell cycle, which can be used, for example, in studies of the bacterial metabolism or the bacterial genome. We have also shown that CRISPR/Cas9 systems has thermo-sensitive properties, which adds another level of control to this powerful gene-editing system. In summary, we provide a novel, efficient, and simple method to control a stage of bacterial replication using an engineered CRISPR/dCas9deg system.
Supplementary Material
Acknowledgments
We thank Hugo Snippert for strain BN1442, Amy Rudolph for strain AB1157, Erwin Van Rijn for assistance with FACS experiments, Fabai Wu and Katarzyna Ginda for discussions.
Footnotes
Present address: Christian Lesterlin, Bases Moléculaires et Structurales des Systèmes Infectieux, UMR 5086, Centre National de la Recherche Scientifique, University of Lyon, 69367 Lyon, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Netherlands Organisation for Scientific Research (NWO/OCW) as part of the Frontiers of Nanoscience program; European Research Council NanoforBio No. 247072 (CD) and SynDiv 16 669598 (CD); Wellcome Trust [SIA099204/Z/12Z]; Leverhulme Trust [RP2013-K-017]. Funding for open access charge: NWO/OCW.
Conflict of interest statement. None declared.
REFERENCES
- 1.Leonard A.C., Mechali M. DNA replication origins. Cold Spring Harb. Perspect. Biol. 2013;5:a010116–a010116. doi: 10.1101/cshperspect.a010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sernova N.V., Gelfand M.S. Identification of replication origins in prokaryotic genomes. Brief. Bioinform. 2008;9:376–391. doi: 10.1093/bib/bbn031. [DOI] [PubMed] [Google Scholar]
- 3.Mott M.L., Berger J.M. DNA replication initiation: mechanisms and regulation in bacteria. Nat. Rev. Microbiol. 2007;5:343–354. doi: 10.1038/nrmicro1640. [DOI] [PubMed] [Google Scholar]
- 4.Kornberg A. DNA replication. Biochim. Biophys. Acta BBA-Gene Struct. Expr. 1988;951:235–239. doi: 10.1016/0167-4781(88)90091-7. [DOI] [PubMed] [Google Scholar]
- 5.Yung B.-M., Kornberg A. The dnaA initiator protein binds separate domains in the replication origin of Escherichia coli. J. Biol. Chem. 1989;264:6146–6150. [PubMed] [Google Scholar]
- 6.Robinson A., van Oijen A.M. Bacterial replication, transcription and translation: mechanistic insights from single-molecule biochemical studies. Nat. Rev. Microbiol. 2013;11:303–315. doi: 10.1038/nrmicro2994. [DOI] [PubMed] [Google Scholar]
- 7.Magnan D., Joshi M.C., Barker A.K., Visser B.J., Bates D. DNA replication initiation is blocked by a distant chromosome–membrane attachment. Curr. Biol. 2015;25:2143–2149. doi: 10.1016/j.cub.2015.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katayama T., Ozaki S., Keyamura K., Fujimitsu K. Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat. Rev. Microbiol. 2010;8:163–170. doi: 10.1038/nrmicro2314. [DOI] [PubMed] [Google Scholar]
- 9.Siam R., Marczynski G.T. Cell cycle regulator phosphorylation stimulates two distinct modes of binding at a chromosome replication origin. EMBO J. 2000;19:1138–1147. doi: 10.1093/emboj/19.5.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reyes-Lamothe R., Possoz C., Danilova O., Sherratt D.J. Independent positioning and action of Escherichia coli replisomes in live cells. Cell. 2008;133:90–102. doi: 10.1016/j.cell.2008.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pennington J.M., Rosenberg S.M. Spontaneous DNA breakage in single living Escherichia coli cells. Nat. Genet. 2007;39:797–802. doi: 10.1038/ng2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brophy J.A.N., Voigt C.A. Principles of genetic circuit design. Nat. Methods. 2014;11:508–520. doi: 10.1038/nmeth.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Withers H.L., Bernander R. Characterization of dnaC2 and dnaC28Mutants by flow cytometry. J. Bacteriol. 1998;180:1624–1631. doi: 10.1128/jb.180.7.1624-1631.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bates D., Epstein J., Boye E., Fahrner K., Berg H., Kleckner N. The Escherichia coli baby cell column: a novel cell synchronization method provides new insight into the bacterial cell cycle: The baby cell column. Mol. Microbiol. 2005;57:380–391. doi: 10.1111/j.1365-2958.2005.04693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferullo D.J., Cooper D.L., Moore H.R., Lovett S.T. Cell cycle synchronization of Escherichia coli using the stringent response, with fluorescence labeling assays for DNA content and replication. Methods. 2009;48:8–13. doi: 10.1016/j.ymeth.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen B., Gilbert L.A., Cimini B.A., Schnitzbauer J., Zhang W., Li G.-W., Park J., Blackburn E.H., Weissman J.S., Qi L.S., et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Connell M.R., Oakes B.L., Sternberg S.H., East-Seletsky A., Kaplan M., Doudna J.A. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516:263–266. doi: 10.1038/nature13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang W., Bikard D., Cox D., Zhang F., Marraffini L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bikard D., Euler C.W., Jiang W., Nussenzweig P.M., Goldberg G.W., Duportet X., Fischetti V.A., Marraffini L.A. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat. Biotechnol. 2014;32:1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Citorik R.J., Mimee M., Lu T.K. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat. Biotechnol. 2014;32:1141–1145. doi: 10.1038/nbt.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J., Charpentier E. A programmable dual-RNA guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larson M.H., Gilbert L.A., Wang X., Lim W.A., Weissman J.S., Qi L.S. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 2013;8:2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bikard D., Jiang W., Samai P., Hochschild A., Zhang F., Marraffini L.A. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41:7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernhardt T.G., De Boer P.A. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol. Microbiol. 2003;48:1171–1182. doi: 10.1046/j.1365-2958.2003.03511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lau I.F., Filipe S.R., Søballe B., Økstad O.-A., Barre F.-X., Sherratt D.J. Spatial and temporal organization of replicating Escherichia coli chromosomes: Escherichia coli chromosome dynamics. Mol. Microbiol. 2004;49:731–743. doi: 10.1046/j.1365-2958.2003.03640.x. [DOI] [PubMed] [Google Scholar]
- 28.Keiler K.C., Waller P.R.H., Sauer R.T. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;16:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 29.Quan J., Tian J. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS One. 2009;4:e6441. doi: 10.1371/journal.pone.0006441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu F., van Schie B.G.C., Keymer J.E., Dekker C. Symmetry and scale orient Min protein patterns in shaped bacterial sculptures. Nat. Nanotechnol. 2015;10:719–726. doi: 10.1038/nnano.2015.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sliusarenko O., Heinritz J., Emonet T., Jacobs-Wagner C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol. Microbiol. 2011;80:612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lesterlin C., Pages C., Dubarry N., Dasgupta S., Cornet F. Asymmetry of chromosome replichores renders the DNA translocase activity of FtsK essential for cell division and cell shape maintenance in Escherichia coli. PLoS Genet. 2008;4:e1000288. doi: 10.1371/journal.pgen.1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tabata S., Oka A., Takanami M., Yasuda S., Hirota Y. The 245 base-pair oriC sequence of the E. coli chromosome directs bidirectional replication at an adjacent region. Nucleic Acids Res. 1983;11:2617–2626. doi: 10.1093/nar/11.9.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weigel C., Schmidt A., Rückert B., Lurz R., Messer W. DnaA protein binding to individual DnaA boxes in the Escherichia coli replication origin, oriC. EMBO J. 1997;16:6574–6583. doi: 10.1093/emboj/16.21.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dewar J.M., Budzowska M., Walter J.C. The mechanism of DNA replication termination in vertebrates. Nature. 2015;525:345–350. doi: 10.1038/nature14887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X., Sherratt D.J. Independent segregation of the two arms of the Escherichia coli ori region requires neither RNA synthesis nor MreB dynamics. J. Bacteriol. 2010;192:6143–6153. doi: 10.1128/JB.00861-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prindle A., Selimkhanov J., Li H., Razinkov I., Tsimring L.S., Hasty J. Rapid and tunable post-translational coupling of genetic circuits. Nature. 2014;508:387–391. doi: 10.1038/nature13238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu L.J., Errington J. Nucleoid occlusion and bacterial cell division. Nat. Rev. Microbiol. 2012;10:8–12. doi: 10.1038/nrmicro2671. [DOI] [PubMed] [Google Scholar]
- 41.Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elowitz M.B., Leibler S. A synthetic oscillatory network of transcriptional regulators. J. Biol. Chem. 1999;274:6074–6079. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 43.Cameron D.E., Bashor C.J., Collins J.J. A brief history of synthetic biology. Nat. Rev. Microbiol. 2014;12:381–390. doi: 10.1038/nrmicro3239. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.