Abstract
Saffron is one of the most expensive species of Chinese herbs and has been subjected to various types of adulteration because of its high price and limited production. The present study introduces a loop-mediated isothermal amplification (LAMP) technique for the differentiation of saffron from its adulterants. This novel technique is sensitive, efficient and simple. Six specific LAMP primers were designed on the basis of the nucleotide sequence of the internal transcribed spacer 2 (ITS2) nuclear ribosomal DNA of Crocus sativus. All LAMP amplifications were performed successfully, and visual detection occurred within 60 min at isothermal conditions of 65 °C. The results indicated that the LAMP primers are accurate and highly specific for the discrimination of saffron from its adulterants. In particular, 10 fg of genomic DNA was determined to be the limit for template accuracy of LAMP in saffron. Thus, the proposed novel, simple, and sensitive LAMP assay is well suited for immediate on-site discrimination of herbal materials. Based on the study, a practical standard operating procedure (SOP) for utilizing the LAMP protocol for herbal authentication is provided.
Human cultivation and use of saffron spans more than 3,500 years and extends across cultures, continents, and civilizations1 with documentation appearing as early as the 7th-century BC in an Assyrian botanical reference compiled under Ashurbanipal2. Saffron is a spice derived from the dried stigmas of the plant C. sativus that has remained among the world’s most expensive substances throughout history3. The apocarotenoid-rich saffron has been used as seasoning, fragrance, dye, and medicine, owing to its bitter taste, hay-like fragrance, and slightly metallic notes. It has been traded over the course of four millennia and has been used as treatment for certain disorders, such as cramps, asthma, menstruation disorders, liver disease, and pain for more than 2,500 years2. Chemical analysis of saffron extracts has revealed that its characteristic compounds are ds-crocins and crocetin, including carotenoid, crocetin, crocins, and monoterpene aldehydes of picrocrocin and safranal1,3,4,5. These compounds have medicinal attributes, such as strong antioxidant effects and free radical scavenging properties, as well as potential anti-carcinogenic, anti-mutagenic, anti-depressant, and memory-enhancing features that improve mood3,4. Thus, given saffron’s limited production and high price, many types of contaminants and imitations have been used to mimic saffron over the centuries. Although these imitations have similar appearances to saffron, they are cheaper and do not have the same medicinal value, thus making it difficult for consumers to immediately distinguish them. According to previous reports, the plant materials most frequently used to adulterate saffron are Carthamus tinctorius petals (safflower)6, Curcuma longa powdered rhizomes (turmeric)6, Calendula officinalis6, Daucus carota6, Zea mays6, and Nelumbo nucifera6. Previous methods used to authenticate saffron have involved physical, chemical, and molecular fields, such as 1H NMR metabolite fingerprinting7, two-dimensional gas chromatography8, SCAR markers9,10,11, and melting curve analysis6. These methods can identify saffron and its adulterants more quickly and accurately than traditional morphological detection. However, these processes require professional operation and correlation analysis instruments; therefore, a simple, immediate, and sensitive method for identifying adulterated saffron is required.
The loop-mediated isothermal amplification (LAMP) method is a novel nucleic acid amplification technology that is quick, simple, and highly specific. LAMP was established by Notomi in 200012 and is based on a complex methodology requiring four to six different primers that are specifically designed to recognize six to eight precise gene sequences. DNA amplification is accomplished by a DNA polymerase with strand-displacing activity13,14, thus obviating the need for a thermal denaturation step to obtain single-stranded DNA. Thus, strand-displacing activity allows for amplification under isothermal conditions, in contrast to PCR (polymerase chain reaction), in which a thermal denaturation step is essential. The use of isothermal conditions in the LAMP technique allows for reactions to occur in less time because no temperature changes are required. Compared with that of PCR, the detection limit of LAMP is much lower, and only a few copies are needed15. Additionally, there is no need for expensive and complicated equipment, such as a thermal cycler. Because of its sensitivity, specificity, efficiency, rapidity, and simplicity, the LAMP method is an ideal molecular technique for identifying certain infectious diseases16,17 and conducting food hygiene quality control18,19, inspection, and quarantine20,21. A number of reports have demonstrated that LAMP may be useful for the detection of some plants and medicinal herbs22,23,24,25; however, a standard operating procedure (SOP) for on-site molecular authentication of Chinese herbs has not yet been established. DNA barcoding is currently an effective and widely used tool that enables the accurate identification of plant species. The ITS2 region has proven to be a core DNA barcode for authentication of a broader range of medicinal plant taxa with high variability and species-discrimination26. These characteristics make ITS2 a good bar-code gene choice for LAMP-specific primer design27. This study sought to establish a simple, rapid, and sensitive method to distinguish saffron from its adulterants by using LAMP based on the ITS2 sequence and to provide a practical SOP model for herbal medicine detection.
Results
Multiple alignments of C. sativus with other species’ ITS2 sequences
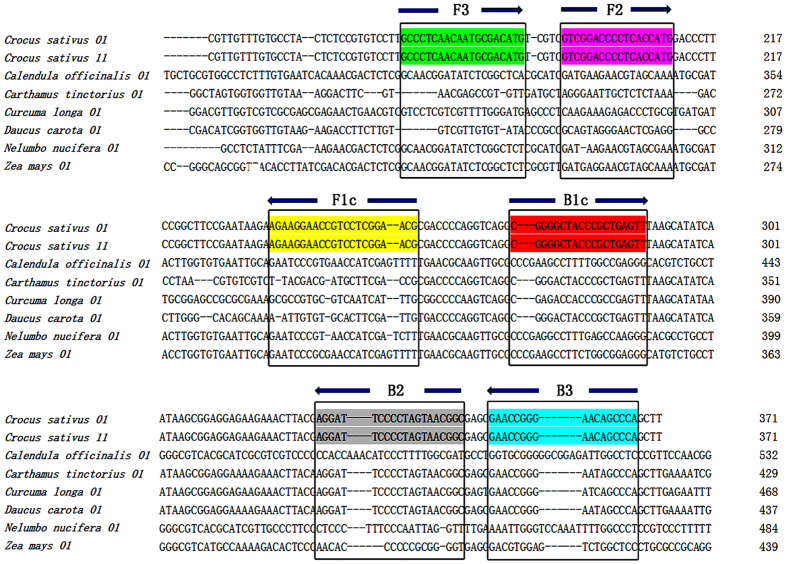
Our results showed that no variation at the site of the ITS2 sequence between the original plant C. sativus and the dry herb saffron. The interspecies variation of ITS2 among saffron and its adulterants ranged from 22.7% to 45.6%. On the basis of the interspecies and intraspecies sequence variation of ITS2, six specific LAMP primers were designed, F3, B3, FIP, BIP, LF, and LR (Table 1). The sequences and directions of F3, B3, F1c, and B1c are shown in Fig. 1. The sequence of FIP was combined from F3 to F1c, and BIP was linked from B3 to B1c. LF and LR were constructed according to F3, B3, F1c, and B1c.
Table 1. Primers used in this study.
| Primer Name | Type | Length (bp) | Sequence (5′-3′) |
|---|---|---|---|
| ITS2 | 2F | ATGCTATACTTGGTGTGAAT | 20 |
| ITS2 | 3R | GACGCTTCTCCAGACTACAAT | 21 |
| F3 | F3 | GCCCTCAACAATGCGACATG | 20 |
| B3 | B3 | TGGGCTGTTCCCGGTTC | 17 |
| FIP | F2 + F1c | CGTTCCGAGGACGGTTCCTTCTGTCGGACCCCTCACCATG | 40 |
| BIP | B2 + B1c | CGGGGCTACCCGCTGAGTTGCCGTTACTAGGGGAATCCT | 39 |
| LF | LF | TTCGGAAGCCGGAAGGGTC | 19 |
| LR | LR | ATAAGCGGAGGAGAAGAAACTTAC | 24 |
Figure 1. Sequence alignment of the ITS2 of C. sativus and its adulterants.
The six ITS2 sequences of the adulterants are aligned against C. sativus. The sequences in coloured boxes indicate the regions that share the specifically designed LAMP primer sequences. The arrow symbols indicate the direction of DNA polymerization from the LAMP primers.
Authentication by LAMP amplification using specific primers
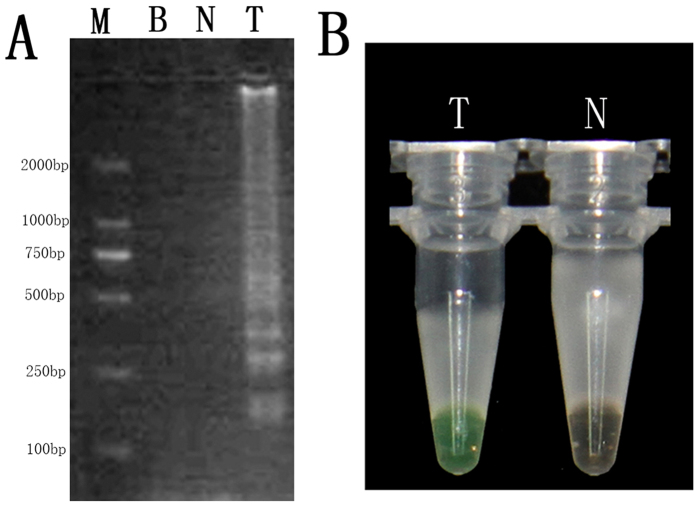
The LAMP products that were detected are shown in Fig. 2. The LAMP product showed the presence of a specific smear pattern of DNA in the gel electrophoresis in lane T. However, no such smear pattern emerged in the CK lane (LAMP product without template DNA, lane N) or the blank (lane B) (Fig. 2A). The LAMP product was directly visualized in tube T, owing to the detectable colour change to green after amplification, whereas no colour change occurred for CK (tube N) (Fig. 2B). These results indicated that the saffron LAMP primers designed were suitable for rapidly amplifying the target nucleic acids of saffron under isothermal conditions.
Figure 2.
Detection of the LAMP products of C. sativus by using DNA electrophoresis (A) and calcein dyeing (B). The LAMP reaction was performed with the specifically designed primers. Lane B, lane N, and lane T in (A) represent the blank lanes, the CK line (LAMP product without template DNA), and the positive amplification lane (LAMP product with template DNA), respectively. Lane M had 2 kb of DNA ladder marker; Tube N (without template DNA) and Tube T (with template DNA) in (B) represent the LAMP products stained with calcein dye.
Figure 3A indicates that the LAMP products (tubes 1–4) were initially detected within the first 15 minutes of the process and increased in concentration thereafter to 60 minutes. By the end of the reaction, the turbidity of the products of tube 1 to tube 4 reached 0.9 to 1, whereas the non-LAMP reactions (tubes 5–11) did not produce amplification curves. Figure 3B indicates that samples of original saffron plant matter and dried saffron both changed from orange to green with the addition of the templates, whereas the other tubes with non-targeted adulterant DNA remained orange after amplification for 60 minutes. Our results indicated that the LAMP method with the specific primers designed in this study can be used to rapidly discriminate saffron from its common adulterants under isothermal conditions within 60 min with the addition of calcein.
Figure 3. Specificity of the LAMP assay for the authentication of saffron and its adulterants.
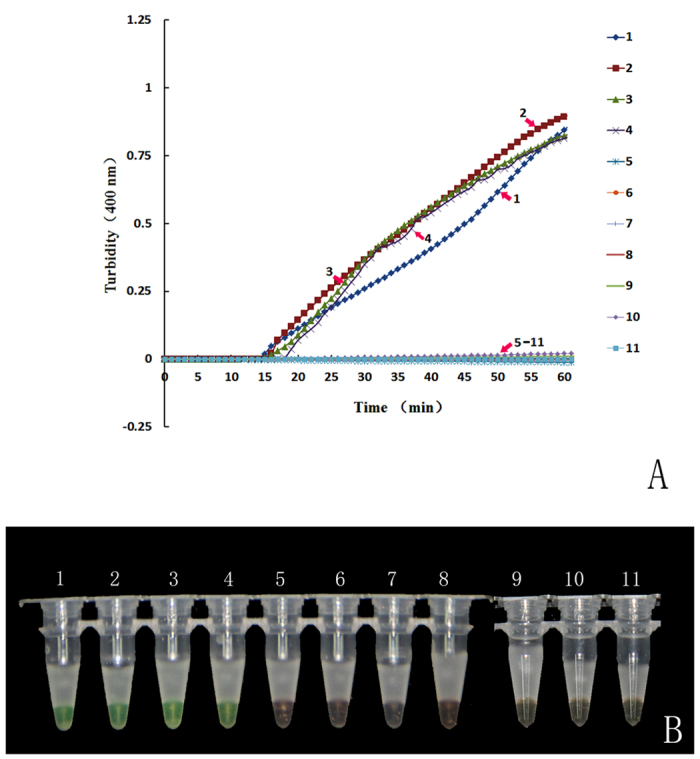
(A) Turbidity was monitored with a Loopamp real-time turbidimeter at 400 nm every 6 s. (B) A visual colour change detection method was compared. 1 μl of calcein (a fluorescent detection reagent) was added to 25 μl of the LAMP reaction mixture before the LAMP reaction. Lines and tubes: 1, C. sativus 01; 2, C. sativus 11; 3, C. sativus 12; 4, C. sativus 13; 5, C. officinalis 01; 6, C. tinctorius 01; 7, C. longa 01; 8, D. carota 01; 9, N. nucifera 01; 10, Z. mays 01; and 11, Neg (negative control, ddH2O).
Sensitivity of LAMP for authentication of saffron
Figure 4 shows the results of the LAMP test that used saffron-specific primers with different template concentrations. The sensitivity results in Fig. 4A show that the LAMP products with template DNA concentrations of 100 ng to 1 pg (lines 1–6) began to accumulate between 9 and 14 minutes into the process. However, the template DNA concentration of 100 fg (line 7) began to amplify later, starting at 19 minutes. By the end of the test, the accumulated products of 100 fg DNA reached 0.75 turbidity. A calcein assay (Fig. 4B) supported the findings from the turbidity curves: no colour change was observed for LAMP reactions with less than 100 fg (line 8) of template DNA. The minimum amount of template used in the LAMP process to test saffron was 100 fg, thus indicating that LAMP is more sensitive than normal PCR, which typically requires a minimum template amount of 10 ng.
Figure 4. Sensitivity of the LAMP reaction for the detection of saffron.
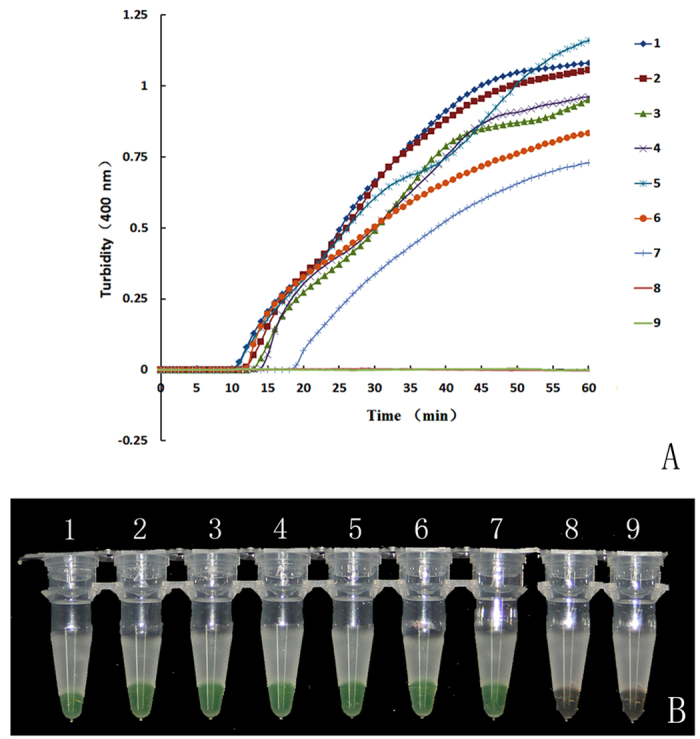
The pure genomic DNA extracted from saffron was diluted in a serial 10-fold dilution. Tubes and lanes: 1, 100 ng/μl; 2, 10 ng/μl; 3, 1 ng/μl; 4, 100 pg/μl; 5, 10 pg/μl; 6, 1 pg/μl; 7, 100 fg/μl; 8, 10 fg/μl; 9, Neg. (A) Turbidity was monitored with a Loopamp real-time turbidimeter at 400 nm every 6 s; (B) A visual colour change detection method was compared. 1 μl of calcein was added to 25 μl of LAMP reaction mixture before the LAMP reaction.
A practical SOP for rapid and immediate detection of herbal medicine using LAMP
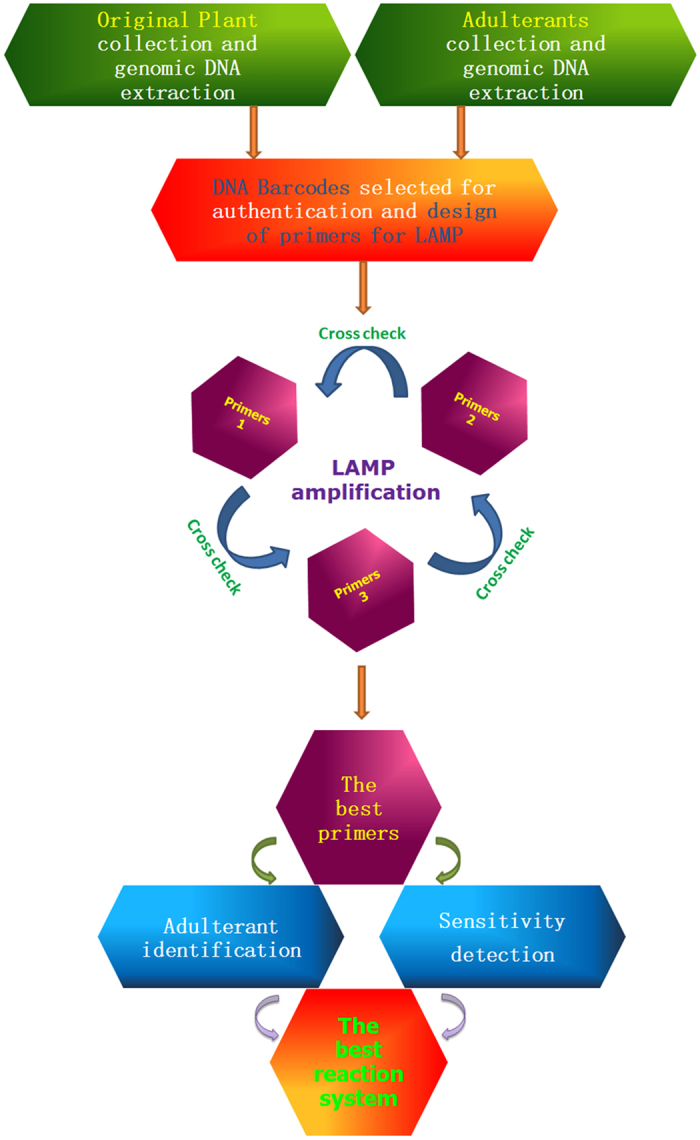
The LAMP method has been used as a valid molecular technique in many fields, such as the identification of infectious diseases, testing of poisonous foods, and authentication of herbs, primarily because of its sensitivity, specificity, high efficiency, rapidity, and simplicity. The need for an immediate method of identification is urgent in the Chinese herb market. To address this, we propose a practical SOP for Chinese herb authentication utilizing the LAMP protocol (Fig. 5). First, genomic DNA from both the authentic medicinal plant and the adulterants needs to be extracted. The extracted DNA is then prepared for use as templates for PCR primers with barcodes. Second, the sections of the selected barcodes are amplified with different LAMP-specific primers designed from different barcoding sequences (i.e., ITS2, psbA-trnH, rbcl, etc.). Third, LAMP amplification is performed with different groups of primers, and then crosschecks are completed. The best group of primers that are both efficient and stable are then screened. Fourth, a preliminary test is performed to detect adulterants by using the LAMP method with specific primers for detection of the sensitivity of LAMP amplification. This fourth step focuses on the sensitivity and the identification of the authentic herb from the adulterants with each of the specific primers. Finally, the best reaction system can be constructed through the aforementioned steps.
Figure 5. A proposed SOP for herb authentication utilizing a LAMP protocol.
Discussion
This study sought to develop a rapid on-site method of discriminating the rare herb saffron (C. sativus) from its adulterants. Previous studies have found that saffron can be authenticated through several methods, such as barcoding-based melting curve analysis6, SCAR markers9,10, or 1H NMR metabolite fingerprinting7. Although these methods precisely identify species, they require professional expertise, statistical analysis, and precise instruments. Among the many methods described in previous studies, none are an efficient means for immediate authentication or can be performed by research staff without knowledge of these complex methods. From the many methods used to identify saffron from its adulterants, our results indicated that LAMP may be a superior process. Although the results of the present paper are encouraging, some problems may remain in the methodology and need to be addressed. First, because the selection of a good barcode sequence is the basis of the primer design, the fragment must be conserved, and the sequence length should be long enough to contain all of the regions adapted to designing LAMP primers. For this reason, not all common barcodes result in highly efficient LAMP primers. Therefore, we recommend designing several groups of LAMP primers based on different barcodes, such as ITS2, psbA-trnH, and rbcl. By screening the amplification efficiency of the chosen fragments, the best candidates for primers can be selected. Second, to ensure the accuracy and credibility of the test, the original plants, rather than the dry herbs, were chosen as templates to amplify the ITS2 gene, which was the target DNA for designing the specific primers for LAMP. Third, owing to the high sensitivity of the LAMP test, the occurrence of false-positive results is likely with the current experimental set-up28,29. To prevent false-positive results, we recommend using low-melting point paraffin wax to decrease the probability of contamination. In addition, spatial separation of the reagent and test samples is needed; this can be accomplished by analysing the turbidity curve in combination with the visual colour change instead of using only the DNA electrophoresis test. Finally, saffron and its adulterants can be discriminated rapidly by visual detection, but the adulterant species are not identified. To understand whether these adulterants produce toxic side effects, the traceability of the adulterants must be determined by using BLAST through the TCM barcode platform30, which is a part of the Traditional Chinese Medicine Database (TCMD) accessible at http://www. tcmbarcode.cn/en/31.
In summary, this study provided a novel molecular technique for rapid on-site authentication of saffron. Based on the high sensitivity of the LAMP amplification, this method is applicable for the authentication of dry herbs as well as for identification of the presence of saffron in health products. Thus, we recommend that this specific, sensitive, and convenient method be used for the commercial identification of herbal products to meet the growing need for immediate herbal authentication.
Methods
Plant and herbal sample collection
As shown in Table 2, we collected 31 plant samples for our analysis. 10 of these samples were pure saffron collected from Tibet, Xinjiang and Hubei province, and 21 were common adulterants collected from the Hubei and Jiangsu province and other locations. The ITS2 region is a conserved section in nuclear ribosomal DNA and is normally used as a barcode for discriminating fresh plants and dry herbs. In this work, the ITS2 sequence of these plant samples were used to design LAMP specific primers. We also purchased dried commercial herbal materials from herb markets and Taobao.com for the analysis (Table 2). Both the plant samples and the dry herb samples were used for the LAMP test. All plant samples were identified by taxonomist Wei Sun from the Chinese Material Medica, China Academy of Chinese Medical Science. All materials were stored in the herbarium of the Institute of Medicinal Plant Development of the Chinese Academy of Medical Sciences in Beijing, China.
Table 2. The species origin, collection area and identifiers of saffron (C. sativus) and its common adulterants in this study.
| Samples | Species | Plant (portion) | Crude drug (portion) | Origin | Identifier |
|---|---|---|---|---|---|
| CS01 | Crocus sativus | Y/filament | N | Tibet China | Wei Sun |
| CS02 | C. sativus | Y/filament | N | Tibet China | Wei Sun |
| CS03 | C. sativus | Y/filament | N | Tibet China | Wei Sun |
| CS04 | C. sativus | Y/filament | N | Tibet China | Wei Sun |
| CS05 | C. sativus | Y/filament | N | Tibet China | Wei Sun |
| CS06 | C. sativus | Y/filament | N | Tibet China | Wei Sun |
| CS07 | C. sativus | Y/filament | N | Xinjiang China | Wei Sun |
| CS08 | C. sativus | Y/filament | N | Xinjiang China | Wei Sun |
| CS09 | C. sativus | Y/filament | N | Hubei China | Wei Sun |
| CS10 | C. sativus | Y/filament | N | Hubei China | Wei Sun |
| DC01 | Daucus carota | Y/leaf | N | Hubei China | Wei Sun |
| DC02 | D. carota | Y/leaf | N | Hubei China | Wei Sun |
| DC03 | D. carota | Y/leaf | N | Hubei China | Wei Sun |
| DC04 | D. carota | Y/leaf | N | Hubei China | Wei Sun |
| DC05 | D. carota | Y/leaf | N | Hubei China | Wei Sun |
| DC06 | D. carota | Y/leaf | N | Hubei China | Wei Sun |
| CL01 | Curcuma longa | Y/root | N | Guangdong China | Wei Sun |
| CL02 | C. longa | Y/root | N | Jiangsu China | Wei Sun |
| CL03 | C. longa | Y/root | N | Fujian China | Wei Sun |
| NU01 | Nelumbo nucifera | Y/root | N | Beijing China | Wei Sun |
| NU02 | N. nucifera | Y/root | N | Jilin China | Wei Sun |
| NU03 | N. nucifera | Y/root | N | Jilin China | Wei Sun |
| ZM01 | Zea mays | Y/leaf | N | Jilin China | Wei Sun |
| ZM02 | Z. mays | Y/leaf | N | Jilin China | Wei Sun |
| ZM03 | Z. mays | Y/leaf | N | Jilin China | Wei Sun |
| CO01 | Calendula officinalis | Y/leaf | N | Yunnan China | Wei Sun |
| CO02 | C. officinalis | Y/leaf | N | Sichuan China | Wei Sun |
| CO03 | C. officinalis | Y/leaf | N | Yunnan China | Wei Sun |
| CT01 | Carthamus tinctorius | Y/petal | N | Yunnan China | Wei Sun |
| CT02 | C. tinctorius | Y/petal | N | Yunnan China | Wei Sun |
| CT03 | C. tinctorius | Y/petal | N | Sichuan China | Wei Sun |
| CS11 | saffron | N | Y/dry filament | TongRenTang pharmacy | |
| CS12 | saffron | N | Y/dry filament | Anguo herb market, Hebei | |
| CS13 | saffron | N | Y/dry filament | Taobao.com | |
| DC07 | D. carota | N | Y/fresh root | Taobao.com | |
| DC08 | D. carota | N | Y/fresh root | Anguo herb market | |
| DC09 | D. carota | N | Y/fresh root | Bozhou herb market | |
| NU04 | N. nucifera | N | Y/dry stigmas | Taobao.com | |
| NU05 | N. nucifera | N | Y/dry stigmas | Anguo herb market | |
| NU06 | N. nucifera | N | Y/dry stigmas | Bozhou herb market | |
| ZM04 | Z. mays | N | Y/dry stigma maydis | Taobao.com | |
| ZM05 | Z. mays | N | Y/dry stigma maydis | Anguo herb market | |
| ZM06 | Z. mays | N | Y/dry stigma maydis | Bozhou herb market | |
| CO04 | C. officinalis | N | Y/dry flowers | Taobao.com | |
| CO05 | C. officinalis | N | Y/dry flowers | Taobao.com | |
| CO06 | C. officinalis | N | Y/dry flowers | Taobao.com | |
| CT04 | C. tinctorius | N | Y/dry stigmas | TongRenTang pharmacy | |
| CT05 | C. tinctorius | N | Y/dry stigmas | YiDeTang pharmacy | |
| CT06 | C. tinctorius | N | Y/dry stigmas | JiXiang pharmacy | |
| CL04 | C. longa | N | Y/dry root | TongRenTang pharmacy | |
| CL05 | C. longa | N | Y/dry root | YiDeTang pharmacy | |
| CL06 | C. longa | N | Y/dry root | JiXiang pharmacy |
Y = Yes and N = No.
Genomic DNA extraction and candidate sequencing
We cut all of the plant samples and medicinal materials into small pieces of approximately 0.5 cm. We took 30 mg from each silica gel-dried plant sample and rubbed the samples for two minutes at a frequency of 30 times/second in a Mixer Mill MM400 (Retsch GmbH, Haan, Germany). The total genomic DNA was isolated using a Plant Genomic DNA kit (Tiangen Biotech Co, Ltd., Beijing, China) according to the manufacturer’s instructions. We determined the DNA concentration and purity by measuring the absorbance of a diluted DNA solution at 260 nm and 280 nm using Qubit® 2.0 (Life Tech, Invitrogen, USA). To estimate the sensitivity of the LAMP assay, we prepared genomic DNA from C. sativus by serial 10-fold dilutions to produce concentrations ranging from 100 ng/μl to 10 fg/μl. To design the LAMP primers for the authentication of saffron, the ITS2 regions of the nuclear ribosomal DNA of C. sativus, C. longa, D. carota, N. nucifera, Z. mays, C. officinalis, and C. tinctorius were obtained using PCR amplification. We used the extracted genomic DNA of each sample as templates for amplifying the ITS2 sequence. The reaction mixture contained 12.5 μL 2 × Taq PCR Master Mix (Aidlab Biotechnologies Co., China), 1 μL of each primer (2.5 μmol/L), 8.5 μL ddH2O, and 2 μL template DNA. The conditions used for the PCR were 94 °C for 3 min, followed by 37 cycles of 94 °C for 1 min, 56 °C for 30 sec, and 72 °C for 1 min, and a final extension cycle at 72 °C for 10 min. We examined 4 μL of PCR products using 1.2% agarose gel electrophoresis. We purified the PCR products with a TIANgel Midi Purification Kit (Tiangen Biotech Co., China); then these purified PCR products were sequenced on an ABI3730XL sequencer (Applied Biosystems Inc.) using amplification primers. Sequence assembly and the generation of consensus sequences were performed with CodonCode Aligner v5.1 (CodonCode Co, USA).
Saffron-specific primer design for LAMP
We constructed multiple alignments of the ITS2 sequence between C. sativus and adulterants to design saffron-LAMP-specific primers (F3, B3, FIP, BIP, LF, and LB). From the alignment results, we were able to design the LAMP-specific primers for C. sativus by using the online software Primer Explorer version 4.0 (http://primerexplorer.jp/elamp4.0.0/index.html). Conditions were as follows: (1) the length of F1c/B1c was 18–23 bp, whereas the lengths of F2/B2 and F3/B3 were 18–22 bp; (2) the melting temperature (Tm) for F1c/B1c was 60–65 °C, whereas the Tm for F2/B2 and F3/B3 was 58–61 °C; (3) the distance between F2 and B2 was 50–150 bp, whereas the distance between F1c and B1c was 0–50 bp. The sequences and target positions of the primers are shown in Table 1 and Fig. 1, respectively.
LAMP amplification and product detection
The LAMP reaction was performed by using a DNA amplification kit (Eiken China Co., Ltd., Shanghai, China) in a mixture at a final volume of 25 μl, including 12.5 μl of 2× Reaction Mix (RM); 1 μl of Bst DNA polymerase; 1.6 μM of each inner primer FIP and BIP; 0.2 μM of each outer primer F3 and B3 (50 mmol); 0.8 μM of each loop primer LF and LR; 1 μl of fluorescent detection reagent (Loopamp, Eiken China Co., Ltd., Shanghai, China), and double distilled water (ddH2O) added to bring the reaction mix to volume. We added 1 μl of genomic DNA from each sample to each LAMP reaction as the template. The C. sativus 01 DNA was added as the template, and ddH2O was included as CK for the LAMP amplification. The mixtures were incubated at 65 °C for 60 min and 80 °C for 10 min to inactivate the Bst DNA polymerase in a loop real-time turbidimeter (LA-230; Eiken Chemical Co., Ltd., Tochigi, Japan). We monitored turbidity by recording the optical density at 400 nm every 6 s, and the addition of 1 μl of calcein (Fluorescence Detection Reagent; Loopamp, Eiken China Co., Ltd., Shanghai, China) allowed for the detection of the colour change by visual inspection. We used 2% agarose gel electrophoresis for verification of the first LAMP reaction mixture. To assess the specificity of LAMP, the saffron genomic DNA and genomic DNA from other adulterant plants were used as templates. From all of the DNA extracted, the original plant of saffron named C. sativus 01, the dry herbs of saffron named C. sativus 11, C. sativus 12, C. sativus 13, and C. officinalis 01, C. tinctorius 01, C. longa 01, D. carota 01, N. nucifera 01, and Z. mays 01 were chosen as templates for LAMP detection. To evaluate the sensitivity of the LAMP assay to detect saffron in different concentrations from its adulterants, varying amounts of extracted saffron DNA were utilized as template DNA. The amounts of extracted saffron DNA ranged from 100 ng to 10 fg.
Additional Information
How to cite this article: Zhao, M. et al. Rapid authentication of the precious herb saffron by loop-mediated isothermal amplification (LAMP) based on internal transcribed spacer 2 (ITS2) sequence. Sci. Rep. 6, 25370; doi: 10.1038/srep25370 (2016).
Acknowledgments
This research was supported by the National High Tech R&D Program of China (863 Program) (No. 2012AA021602) and the Major Scientific and Technological Special Project for “Significant New Drugs Creation” (No. 2014ZX09304307).
Footnotes
Author Contributions M.Z., W.S. and S.C. conceived the study and participated in its design. M.Z., Y.S., L.G., C.X. and S.Y. contributed samples and performed the experiments. M.Z., W.L. and W.S. analysed the data and drafted the manuscript. All authors have read and approved the final manuscript.
References
- Moghaddasi M. S. Saffron chemicals and medicine usage. J Med Plants Res. 4, 427–430 (2010). [Google Scholar]
- Kianbakht S. & Ghazavi A. Immunomodulatory Effects of Saffron: A Randomized Double-Blind Placebo-Controlled Clinical Trial. Phytother Res. 25, 1801–1805 (2011). [DOI] [PubMed] [Google Scholar]
- Tarantilis P. A., Beljebbar A., Manfait M. & Polissiou M. FT-IR, FT-Raman spectroscopic study of carotenoids from saffron (Crocus sativus L.) and some derivatives. Spectrochim Acta Part A. 54, 651–657 (1998). [Google Scholar]
- Abdullaev F. & Espinosa-Aguirre J. Biomedical properties of saffron and its potential use in cancer therapy and chemoprevention trials. Cancer Detect prev. 28, 426–432 (2004). [DOI] [PubMed] [Google Scholar]
- Gohari A. R., Saeidnia S. & Mahmoodabadi M. K. An overview on saffron, phytochemicals, and medicinal properties. Pharmacogn Rev. 7, 61 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C. et al. Barcoding Melting Curve Analysis for Rapid, Sensitive, and Discriminating Authentication of Saffron (Crocus sativus L.) from Its Adulterants. BioMed Res Int. Aricle ID 809037, 10 pages (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrakis E. A., Cagliani L. R., Polissiou M. G. & Consonni R. Evaluation of saffron (Crocus sativus L.) adulteration with plant adulterants by 1H NMR metabolite fingerprinting. Food Chem. 173, 890–896 (2015). [DOI] [PubMed] [Google Scholar]
- Jiang M., Kulsing C., Nolvachai Y. & Marriott P. J. Two-Dimensional Retention Indices Improve Component Identification in Comprehensive Two Dimensional Gas Chromatography of Saffron. Anal Chem. 87 5753–5761 (2015). [DOI] [PubMed] [Google Scholar]
- Torelli A., Marieschi M. & Bruni R. Authentication of saffron (Crocus sativus L.) in different processed, retail products by means of SCAR markers. Food Control. 36, 126–131 (2014). [Google Scholar]
- Babaei S., Talebi M. & Bahar M. Developing an SCAR and ITS reliable multiplex PCR-based assay for safflower adulterant detection in saffron samples. Food Control. 35, 323–328 (2014). [Google Scholar]
- Marieschi M., Torelli A. & Bruni R. Quality control of saffron (Crocus sativus L.): development of SCAR markers for the detection of plant adulterants used as bulking agents. J Agr Food Chem. 60, 10998–11004 (2012). [DOI] [PubMed] [Google Scholar]
- Notomi T. et al. Loop-mediated isothermal amplification of DNA. Nucl Acids Res. 28, e63–e63 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T., Mori Y., Tomita N. & Kanda H. Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. J Microb. 53, 1–5 (2015). [DOI] [PubMed] [Google Scholar]
- Ushikubo H. Principle of LAMP method-a simple and rapid gene amplification method. Uirusu. 54, 107–112 (2004). [DOI] [PubMed] [Google Scholar]
- Kaneko H., Kawana T., Fukushima E. & Suzutani T. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J Biochemical Bioph Meth. 70, 499–501 (2007). [DOI] [PubMed] [Google Scholar]
- Mori Y. & Notomi T. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J Infect Chemoth. 15, 62–69 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph D. L., Sullivan V., Owen S. M. & Curtis K. A. Detection of Acute HIV-1 Infection by RT-LAMP. Plos One. 10(5), e0126609 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Kudo Y. et al. Detection of verotoxigenic Escherichia coli O157 and O26 in food by plating methods and LAMP method: a collaborative study. Int J Food Microbiol. 122, 156–161 (2008). [DOI] [PubMed] [Google Scholar]
- Niessen L., Luo J., Denschlag C. & Vogel R. F. The application of loop-mediated isothermal amplification (LAMP) in food testing for bacterial pathogens and fungal contaminants. Food Microbiol. 36, 191–206 (2013). [DOI] [PubMed] [Google Scholar]
- Lun Z., Youping Y., Yujia W. & Zhongkang W. Regular LAMP and fast LAMP for the detection of Xanthomonas axonopodis pv. citri. Plant Prot. 3, 023 (2013). [Google Scholar]
- Liu D. F. et al. Establishment of reverse transcription loop-mediated isothermal amplification for rapid detection and differentiation of canine distemper virus infected and vaccinated animals. Infect Genet Evol. 32, 102–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. et al. Application of novel loop-mediated isothermal amplification (LAMP) for rapid authentication of the herbal tea ingredient Hedyotis diffusa Willd. Food Chem. 141, 2522–2525 (2013). [DOI] [PubMed] [Google Scholar]
- Sasaki Y., Komatsu K. & Nagumo S. Rapid detection of Panax ginseng by loop-mediated isothermal amplification and its application to authentication of ginseng. Biol Pharm Bull. 31, 1806–1808 (2008). [DOI] [PubMed] [Google Scholar]
- Sasaki Y. & Nagumo S. Rapid identification of Curcuma longa and C. aromatica by LAMP. Biol Pharm Bull. 30, 2229–2230 (2007). [DOI] [PubMed] [Google Scholar]
- Chaudhary A. A., Mohsin M. & Ahmad A. Application of loop-mediated isothermal amplification (LAMP)-based technology for authentication of Catharanthus roseus (L.) G. Don. Protoplasma 249, 417–422 (2012). [DOI] [PubMed] [Google Scholar]
- Chen S. et al. A renaissance in herbal medicine identification: from morphology to DNA. Biotechnol Adv. 32, 1237–1244 (2014). [DOI] [PubMed] [Google Scholar]
- Li X. et al. Plant DNA barcoding: from gene to genome. Biol Rev. 90, 157–166 (2015). [DOI] [PubMed] [Google Scholar]
- Li X. et al. Rapid detection of Trichinella spiralis larvae in muscles by loop-mediated isothermal amplification. Int J Parasitol. 42, 1119–1126 (2012). [DOI] [PubMed] [Google Scholar]
- Liu W. et al. Sensitive and rapid detection of the new Delhi metallo-beta-lactamase gene by loop-mediated isothermal amplification. J Clin Microbiol. 50, 1580–1585 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. et al. A renaissance in herbal medicine identification: from morphology to DNA. Biotechnol Adv. 32, 1237–1244 (2014). [DOI] [PubMed] [Google Scholar]
- Han J. et al. An authenticity survey of herbal medicines from markets in China using DNA barcoding. Sci Rep. 6, 18723 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]