Abstract
The sensory neurons in the olfactory epithelium (OSNs) are equipped with a large repertoire of olfactory receptors and the associated signal transduction machinery. In addition to the canonical OSNs, which express odorant receptors (ORs), the epithelium contains specialized subpopulations of sensory neurons that can detect specific information from environmental cues and relay it to relevant neuronal circuitries. Here we describe a subpopulation of mature OSNs in the main olfactory epithelium (MOE) which expresses CD36, a multifunctional receptor involved in a series of biological processes, including sensory perception of lipid ligands. The Cd36 expressing neurons coexpress markers of mature OSNs and are dispersed throughout the MOE. Unlike several ORs analyzed in our study, we found frequent coexpression of the OR Olfr287 in these neurons, suggesting that only a specific set of ORs may be coexpressed with CD36 in OSNs. We also show that CD36 is expressed in the cilia of OSNs, indicating a possible role in odorant detection. CD36-deficient mice display no signs of gross changes in the organization of the olfactory epithelium, but show impaired preference for a lipid mixture odor. Our results show that CD36-expressing neurons represent a distinct population of OSNs, which may have specific functions in olfaction.
Olfactory sensory neurons (OSNs) in the olfactory epithelium (OE) express different types of olfactory receptors, which participate in the detection of distinct chemosensory stimuli. This information is subsequently transmitted to the olfactory bulb (OB) in the central nervous system1,2. Mature OSNs express the olfactory marker protein (OMP)3 and are functionally and molecularly classified according to the type of olfactory receptor they express, their pattern of axonal projection to olfactory glomeruli in the OB, or presence of specific signal transduction machinery4,5. Most OSNs in the main olfactory epithelium (MOE) express canonical odorant receptors (ORs), which belong to a large family of over a thousand G-protein coupled receptors6 and function in a combinatorial manner to discriminate an enormous number of odorants7. Other OSNs in the MOE express instead trace amine-associated receptors (TAARs) or receptor guanylyl cyclase (GC-D). These receptors are involved in the detection of volatile amines8,9,10 and peptide hormones11 present in urine, respectively. It has also been shown that GC-D expressing neurons detect CO2, which is an important olfactory stimulus for many organisms12.
Distinct subpopulations of OSNs in the olfactory epithelium may also differ in the signaling proteins they express. While the canonical OSNs express the olfactory G protein alpha subunit (Gαolf/Gnal), adenylyl cyclase III (ACIII/Adcy3) and the cyclic nucleotide-gated (Cnga2) channel, other subpopulations express members of the transient receptor potential (Trp) channel family or other subtypes of transduction proteins4.
CD36, also known as fatty acid transporter (FAT), is a single-chain glycoprotein expressed at the cell surface of several mammalian cells types13. Most of the receptor is extracellular; it possesses two transmembrane segments and two short cytoplasmatic portions. Even though its intracellular signaling capacity is apparently limited, CD36 is involved in multiple cellular functions, including lipid binding, lipid uptake and inflammatory signaling13,14. In addition, CD36 functions in the immune system as a scavenger receptor that cooperates with Toll-like receptors (TLRs) to recognize lipids derived from bacterial pathogens15.
Moreover, CD36 is known to play several roles in sensory perception. The sensory neuron membrane protein (SNMP), a D. melanonogaster CD36 homologue, mediates proper responses to the lipid pheromone cis-vaccenyl acetate (cVA) in a population of sensory olfactory neurons16,17. In addition, it was shown that CD36 mediates fatty acid detection in taste cells and preference for lipid ingestion in rodents18,19, suggesting a conserved role for this receptor in lipid sensory recognition.
In previous studies20 we showed that the CD36 gene is expressed in discrete regions of the brain that receive input from both the main and accessory olfactory bulbs21,22,23,24. Here we show that CD36 is expressed in the cilia of a subpopulation of OSNs in the MOE. These neurons coexpress typical markers for mature olfactory sensory neurons, including proteins involved in the canonical OR signal transduction cascade. We found that some of these CD36 expressing neurons also coexpress Olfr287. No coexpression was however observed when other types of ORs were analyzed, indicating that the CD36 expressing neurons may express only a particular subgroup of all possible existing ORs. In addition, we also show that innate preference for lipid odor is impaired in CD36-deficient mice. Altogether, our results suggest that CD36 may play important roles in olfaction.
Results
Cd36 gene expression is abundant in the olfactory epithelium
In order to evaluate the relative abundance of Cd36 transcripts, we conducted real-time RT-PCR using various mouse tissues and two different reference genes (Figs 1a and S1). High levels of Cd36 expression were observed in tissues known to strongly express the gene, such as the heart and adipose tissue (17- fold the expression levels in OE; Fig. 1a), in agreement with the transcript levels originally described for rat tissues14. Cd36 expression in the OE is much higher than in other neural tissues, such as eye or whole brain. Interestingly, Cd36 expression in the dorsal tongue, implicated in fat intake preference19, has not been previously compared to the expression in other tissues. Here we report that Cd36 expression in the OE is comparable to that in the dorsal tongue, both representing epithelial tissues that perform environmental chemical sensing (Fig. 1a).
Figure 1. Cd36 gene is expressed in the olfactory epithelium.
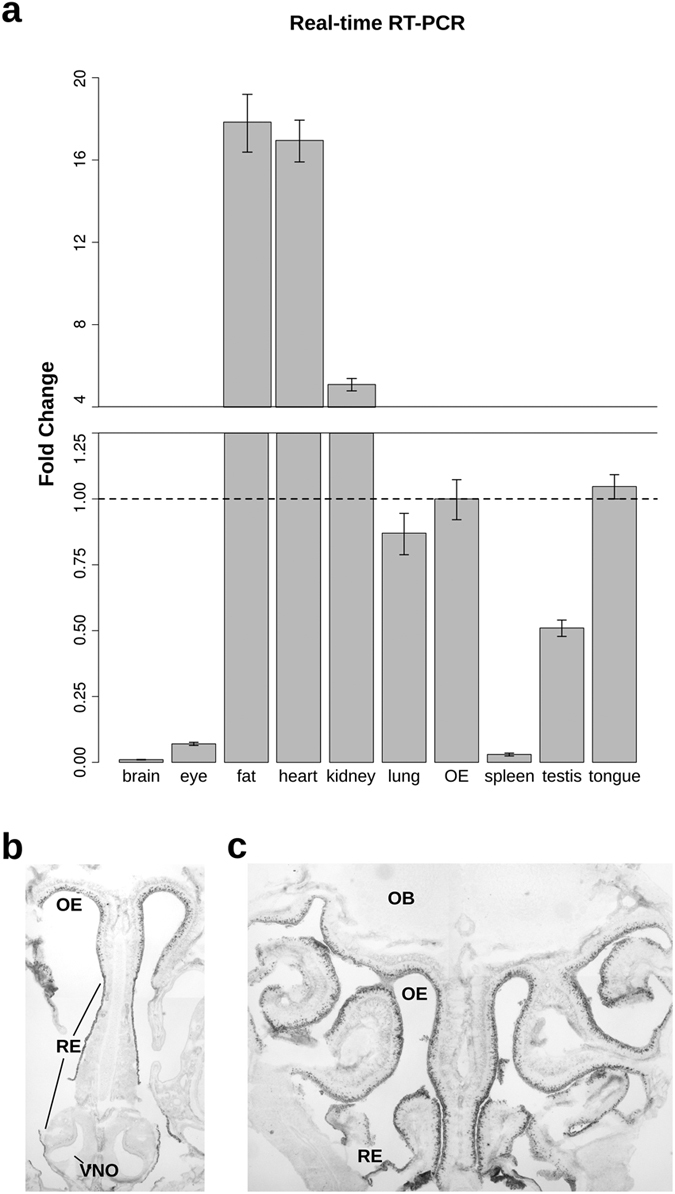
(a) Real-time RT-PCR analysis of Cd36 transcript levels normalized against Actb housekeeping gene for each tissue, represented as fold-change (OE as calibrator; mean of triplicates ± S.D.). Representative microphotographs show widespread pattern of Cd36 mRNA in situ hybridization labeling in rostral (b) and caudal (c) OE cryosections from 4-week-old mice. Note the absence of gene expression in the VNO and olfactory bulb. Abbreviations: OB, olfactory bulb; OE, olfactory epithelium; RE, respiratory epithelium; VNO, vomeronasal organ.
In situ hybridization experiments revealed that Cd36 expression is widely distributed throughout the MOE of adult mice, but absent in the vomeronasal organ (VNO; Fig. 1b) or in the OB (Fig. 1c). The respiratory epithelium also stains positive for the Cd36 message, forming a continuous labeled contour (Fig. 1b). It is remarkable, however, that even though the olfactory epithelium shows intense staining in numerous cells in the neuronal cell layer, only a subset of the cells are stained. These results indicate that Cd36 expression in the MOE is restricted to a subpopulation of neurons.
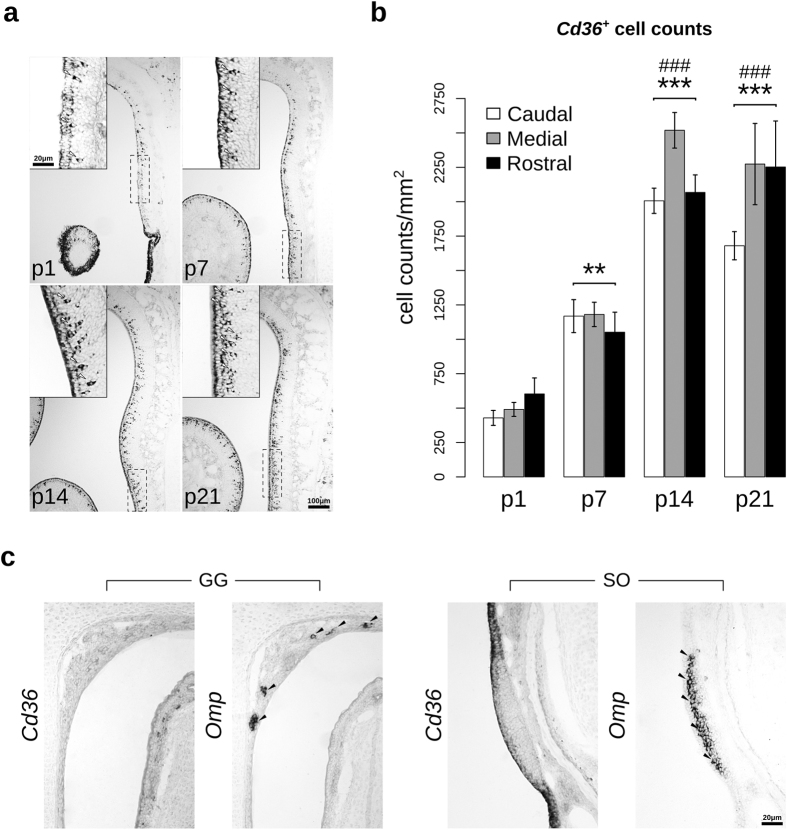
We next examined the expression of Cd36 during the postnatal development of the olfactory epithelium. Even though the staining pattern for Cd36 expression was observed in all developmental stages (Fig. 2a), the number of labeled cells progressively increased until P14, and then stabilized, in all the three levels of the olfactory epithelium measured (rostral, medial and caudal) (Figs 2b and S2). No Cd36 expression was detected in the septal organ (SO) and Grueneberg ganglion (GG) at any tested age (Fig. 2c).
Figure 2. Cellular density of Cd36 mRNA-positive cells increases with postnatal development of the olfactory epithelium.
(a) Cd36 riboprobe signals (white arrowheads) at different postnatal ages (days): p1, p7, p14 and p21. Inserts show higher magnification of the area indicated by dashed rectangles. (b) Quantification of cell density (cell counts/OE area; mean ± S.E.M.) showed in ‘a’, which was performed excluding the stained apical contour cells (supporting cells; SCs). Two-way ANOVA revealed a highly significant main age effect F(3,24) = 73.877, p = 2.91 × 10−12, while the level of the olfactory epithelium (rostral, medial and caudal) effect was only marginal, F(2,24) = 3.367, p = 0.0514. (c) Absence of Cd36 transcripts in the Grueneberg ganglion and septal organ, two noncanonical olfactory subsystems that contain Omp-positive OSNs (black arrowheads). Abbreviations: GG, Grueneberg ganglion; SO, septal organ. **p < 0.0005 and ***p < 0.00001 compared to p1; ###p < 0.00001 compared to p7. Scale bars: as indicated.
Cd36-positive OSNs coexpress OMP, Gαolf, and specific ORs
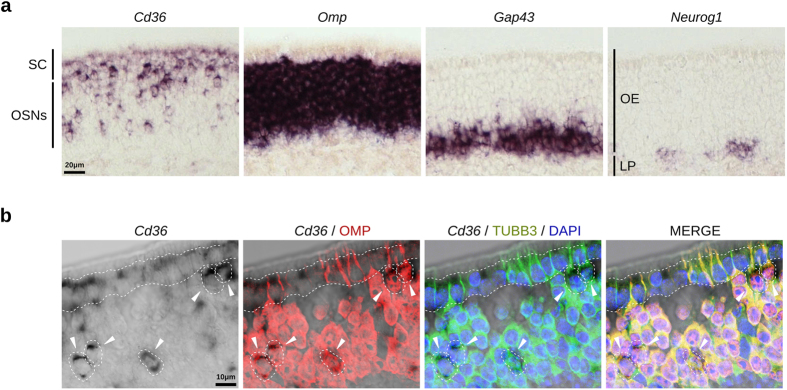
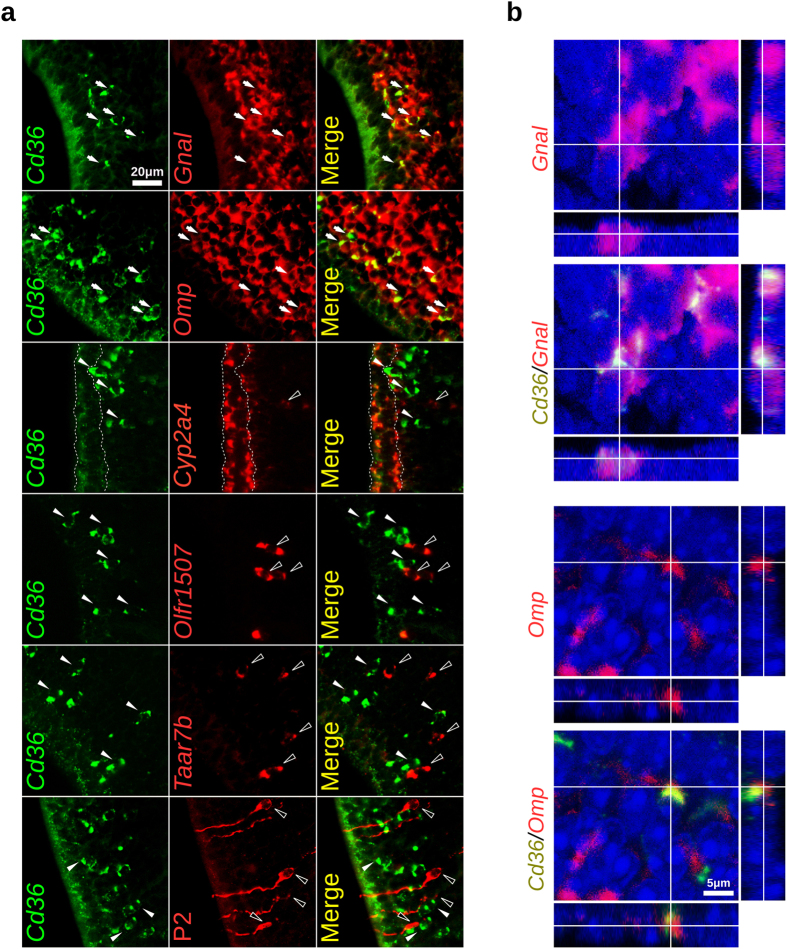
Cd36 mRNA hybridization signals in the olfactory epithelium are observed in two different cell layers, in the supporting cell layer (lighter staining) and in the mature OSNs layer (Fig. 3a). It is noteworthy that cells in the OSNs layer show stronger staining than the ones in the supporting cell layer. Combined in situ hybridization and immunofluorescence staining shows that CD36 expressing cells coexpress both OMP (mature OSNs) and type III β-tubulin (TUBB3; mature and immature OSNs) (Fig. 3b). In addition, two-color fluorescent in situ hybridization experiments revealed that Cd36 is consistently coexpressed with Omp and Gαolf (Gnal), the G protein through which ORs signal (Fig. 4a,b, Table 1). In contrast, duct and supporting cells labeled with the Cyp2a4 riboprobe do not or only weakly express Cd36 (Fig. 4a). Together, these results show that mature OSNs in the MOE are the main source of the transcript.
Figure 3. Cd36 mRNA is expressed in mature OSNs.
(a) Adjacent sections showing the localization of Cd36 expressing cells in the olfactory epithelium from 4-week-old mice compared to Omp (a marker for mature OSNs), Gap43 (a marker for immature OSNs), and Neurog1 (a marker for basal globose cell progenitors) expressing cells. (b) Confocal acquisitions showing coexpression of Cd36 transcripts (chromogenic in situ hybridization) with OMP and TUBB3 proteins (immunofluorescence) in the same OSNs (white arrowheads and dashed lines). Sections from 3-month-old mice; nuclei were stained with DAPI. Supporting cell layer is delimited with dashed lines. Abbreviations: OSNs, olfactory sensory neurons; LP, lamina propria; SC, supporting cells. Scale bars are shown.
Figure 4. Cd36 expressing neurons coexpress canonical OSNs transcripts.
(a) Two-color fluorescent in situ hybridization (FISH) with a Cd36 riboprobe shown in green (white arrowheads) and the other transcripts shown in red (empty arrowheads); arrows indicate coexpression (Omp and Gnal). While a high frequency of coexpression was observed for the canonical OSNs transcripts, no Cd36 coexpression was observed for two ORs (Olfr1507 and P2/Olfr17) or members of the Taar7 subfamily (identified with a Taar7b probe) – please refer to Table 1 for details. In the bottom panel, one-color FISH with a Cd36 riboprobe (green) combined with immunostaining for the GFP protein (red) was performed to identify the P2-IRES-tauGFP expressing neurons. Supporting cells (Cyp2a4 labeled apical cells; delimited with dashed lines) show weaker labeling for Cd36 when compared to OSNs. (b) Confocal acquisitions with orthogonal projections depicting Cd36+ OSNs coexpressing Gnal and Omp transcripts. Nuclei were stained with DAPI (blue). Scale bars are shown.
Table 1. CD36 coexpression with mature OSN markers.
| Transcript or Protein |
Cd36 (mRNA)-positive cells |
|||
|---|---|---|---|---|
| n of cells negative for coexpression | n of cells positive for coexpression (%) | n of cells uncertain for coexpression | Total Cd36+ cells counted | |
| Omp | – | 107 (92%) | 10 | 117 |
| Gnal | 4 | 98 (93%) | 7 | 105 |
| Taar7b | 68 | −(0%)† | – | 312 |
| Olfr1507 | 3321 | −(0%)#† | – | 3209 |
| Tau-EGFP (P2) | 94 | −(0%)† | – | 366 |
| OR pool 1 | 137 | −(0%)† | – | 191 |
| OR pool 2 | 829 | −(0%)#† | – | 1471 |
| OR pool 3 | 615 | −(0%)#† | – | 688 |
| Olfr287 | 40 | 65 (16%)* | – | 407 |
The numbers of Cd36+-OSNs coexpressing the different mature OSN markers are shown. Cell counting was performed for each pair of transcripts using the same microscope fields and restricting Cd36+-OSNs counting to the same OE zone where the ORs are expressed. We only considered for the analysis cells showing defined nuclei. Stained supporting cells were excluded from the analysis. Pool 1: Olfr124 and Olfr1509; pool 2: Olfr646, Olfr1264 and Olfr1372-ps1; pool 3: Olfr78, Olfr569, Olfr638, Olfr691, Olfr692, and Olfr1512). #Significantly smaller than [0.1% x (number of OR genes per probe)] (p < 0.05); *Significantly different than 0.1% (p < 2.2 × 10−16); †Significantly different from coexpression frequency for Olfr287 (p < 1 × 10−10; Bonferroni correction).
Since OR-expressing and TAAR-expressing neurons represent different populations of mature OSNs in the olfactory epithelium, we investigated whether Cd36-positive OSNs coexpress some of the members belonging to these two receptor families. We performed two-color fluorescent in situ hybridization using Cd36 riboprobes together with a riboprobe corresponding to an OR gene which is the most highly expressed OR gene in the mouse olfactory epithelium (Olfr1507, also known as mOR28)25,26 or a Taar7b probe, which should recognize all of the highly related members of the Taar7 subfamily (TAARs 7a, 7b, 7d, 7e, and 7f)9. Neither Olfr1507- nor Taar7b-positive OSNs showed coexpression of Cd36 (Fig. 4a and Table 1). We also used P2-IRES-tauGFP mice, to identify the OSNs expressing the OR P2 (Olfr17)27 and did not observe coexpression of Cd36 with GFP in these neurons (Fig. 4a and Table 1). We therefore analyzed a larger number of OR genes. Probes corresponding to additional 11 OR genes were pooled and used in the two-color fluorescent in situ hybridization together with the probe for Cd36 (pool 1: Olfr124 and Olfr1509; pool 2: Olfr646, Olfr1264 and Olfr1372-ps1; pool 3: Olfr78, Olfr569, Olfr638, Olfr691, Olfr692, and Olfr1512). As shown in Table 1 and figure S3, no coexpression of Cd36 with these OR genes was also observed.
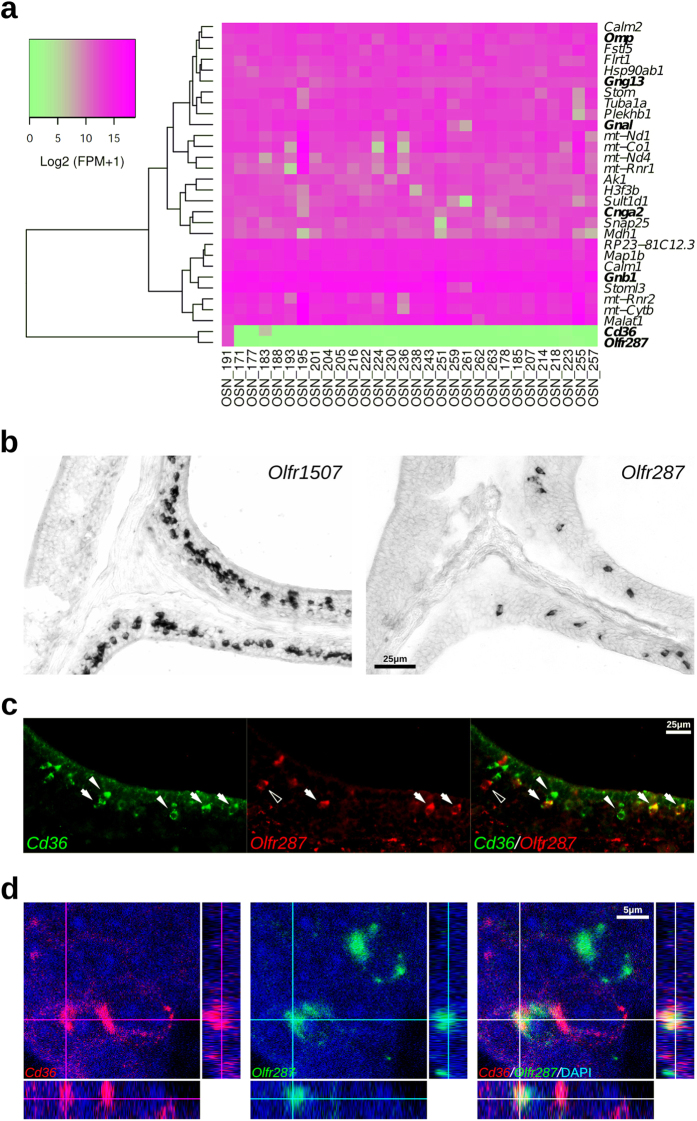
In a recent study, Saraiva and colleagues used single-cell RNA sequencing (RNAseq) experiments to analyze the transcriptomes of individual OSNs28. One of the analyzed neurons (OSN #191) expressed high levels of Cd36. Also highly expressed in this same neuron are the typical markers for mature OSNs (Omp, Gng13, Gnal, Cnga2, Gnb1) and the OR Olfr287 (Fig. 5a). No relevant expression of TAARs, TrpC2, Trpm5 or GC-D was observed in this neuron. Expression of a second OR gene was also detected in this neuron, although at considerably lower levels than Olfr287.
Figure 5. Olfr287 is frequently coexpressed with Cd36 transcripts.
(a) Heatmap from the top thirty expressed genes in a Cd36+-OSN (OSN #191) according to a single-OSN RNAseq annotation data28. Olfr287 and Cd36 are only expressed at high levels in this neuron, which also expresses mature OSN marker transcripts (in bold). Transcript levels are expressed as Log2 (FPM + 1) (FPM: fragments per million). (b) Olfr287 is expressed in the same expression zone in the OE and less frequently than Olfr1507. (c) Two-color fluorescent in situ hybridization (FISH) with a Cd36 riboprobe shown in green (white arrowheads) and Olfr287 shown in red (empty arrowheads); arrows indicate coexpression and arrowheads indicate absence of coexpression in the merged image. (d) Confocal acquisition depicting orthogonal projections for coexpression validation; Cd36 riboprobe shown in red and Olfr287 shown in green. Scale bars: as indicated.
Based on these studies, we next decided to check whether Cd36+-OSNs coexpress Olfr287. In situ hybridization experiments showed that Olfr287+-OSNs are located in the same expression zone as Olfr1507+-OSNs, but are less numerous (Fig. 5b). Two-color FISH revealed substantial coexpression of this receptor and the Cd36 transcript (Fig. 5c,d and Table 1). It should be noted that the Olfr287 probe we used was designed to cover the 3′-untranslated region of the transcript because two other OR genes, Olfr286 and Olfr288, show highly similar coding sequences, and could therefore cross hybridize with the Olfr287 probe. Using this specific probe we found that ~62% of the Olfr287+-OSNs coexpress Cd36, and ~16% of the Cd36+-OSNs coexpress Olfr287 in the same OE zone, indicating that not all Olfr287+-neurons are positive for Cd36 and vice-versa. Nevertheless, Olfr287 coexpression with Cd36 transcript is a frequent event that was not observed for the other OR genes tested in our experiments.
CD36 protein localizes to the ciliary layer of the olfactory epithelium
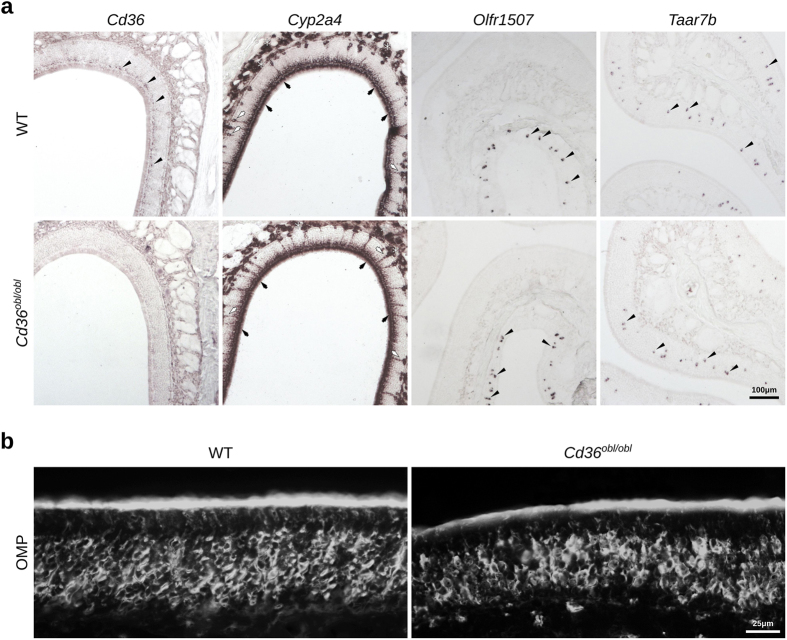
We subsequently investigated whether CD36 is localized to the cilia of OSNs, the site of odorant detection and signal transduction. Double immunostaining and confocal analysis indicate that CD36 colocalizes with acetylated α-tubulin, a marker for cilia, indicating that the CD36 protein is predominantly expressed in the cilia of the OSNs (Fig. 6 and S4). We also analyzed the general morphology of the OE from the Cd36obl/obl mutant mice. These mice carry a nonsense mutation that codes for a premature stop codon within the coding region of the Cd36 gene15, and consequently do not express Cd36 in the olfactory epithelium (Figs 7 and S5). The OE from Cd36obl/obl mice appears normal, and the density and zonal expression profiles of Olfr1507 and Taar7b are unaltered (Fig. 7). In addition, the general distribution of supporting cells, ducts and Bowman’s gland (which express Cyp2a4) and OMP-positive neurons are unaffected in the Cd36 mutant mice (Fig. 7a,b), and no gross changes in the structure of the olfactory bulb of one-year-old CD36-deficient mice are observed (Fig. S6 and Table S1).
Figure 6. CD36 protein is localized to the ciliary layer in the olfactory epithelium.
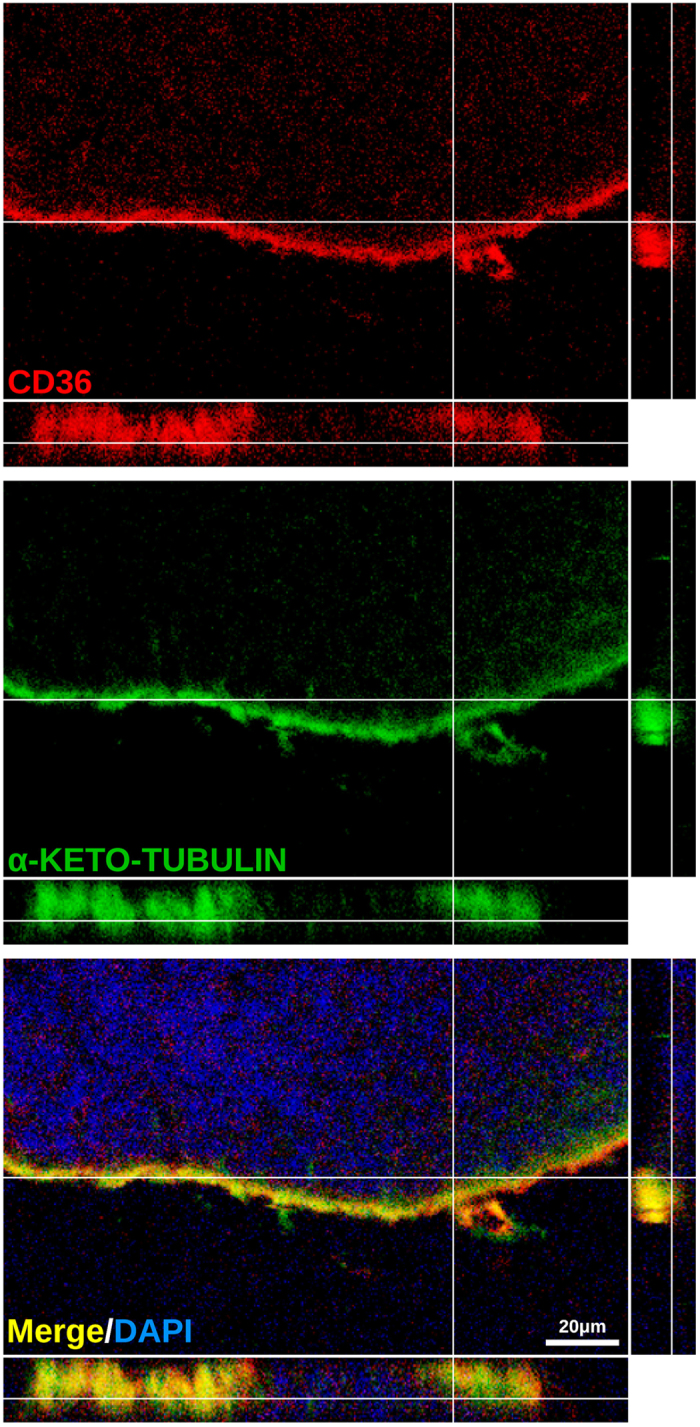
Confocal images of olfactory epithelium sections immunostained with antibodies against CD36 (red) and acetylated α-tubulin (green, α-keto-tubulin), a protein expressed in the cilia of OSNs. Confocal acquisitions with orthogonal projections show localization of the proteins, indicating that the CD36 protein concentrates in the ciliary layer of the olfactory epithelium. Scale bars: as indicated.
Figure 7. The general organization of the olfactory epithelium from CD36-deficient mice is indistinguishable from that of wild-type mice.
(a) In situ hybridization was performed on olfactory epithelium sections from age- and sex-matched C57Bl/6 and Cd36obl/obl mice using riboprobes for Cd36, Olfr1507, Taar7b and Cyp2a4 (which is normally expressed in supporting cells, duct cells and Bowman’s glands). Except for the reduction of Cd36 signals in the mutant mouse, expression patterns for the other probes are similar to the ones in the wild type mouse (WT). (b) OMP immunofluorescence labeling is equivalent between the two genotypes. Black arrowheads indicate labeled OSNs; black arrows indicate supporting cells; white arrows indicate ducts; asterisks indicate Bowman’s gland. Scale bars are shown.
CD36-deficient mice display altered olfactory behavior upon lipid exposure
We next analyzed whether olfactory guided behaviors are perturbed in CD36-deficient mice. Given that CD36 is not expressed in the VNO, it is expected that responses to pheromones or kairomones detected by this organ would not be altered in Cd36obl/obl mice. In fact, we found that defensive behaviors generated upon exposure of mice to predator odors, a response shown to be largely dependent on the VNO29, was equivalent in Cd36obl/obl and wild-type animals (Fig. 8a). In order to assess general olfactory capacity, we conducted the buried food olfactory assay. No significant differences were observed in the performances of wild type and Cd36obl/obl mice in this test, indicating that the loss of CD36 does not affect general odorant detection (Fig. 8b). In addition, no significant changes were observed between the two genotypes when mice were exposed to amyl acetate in an odorant preference test (Fig. S7).
Figure 8. CD36-deficient mice show impaired preference for lipid odorants.
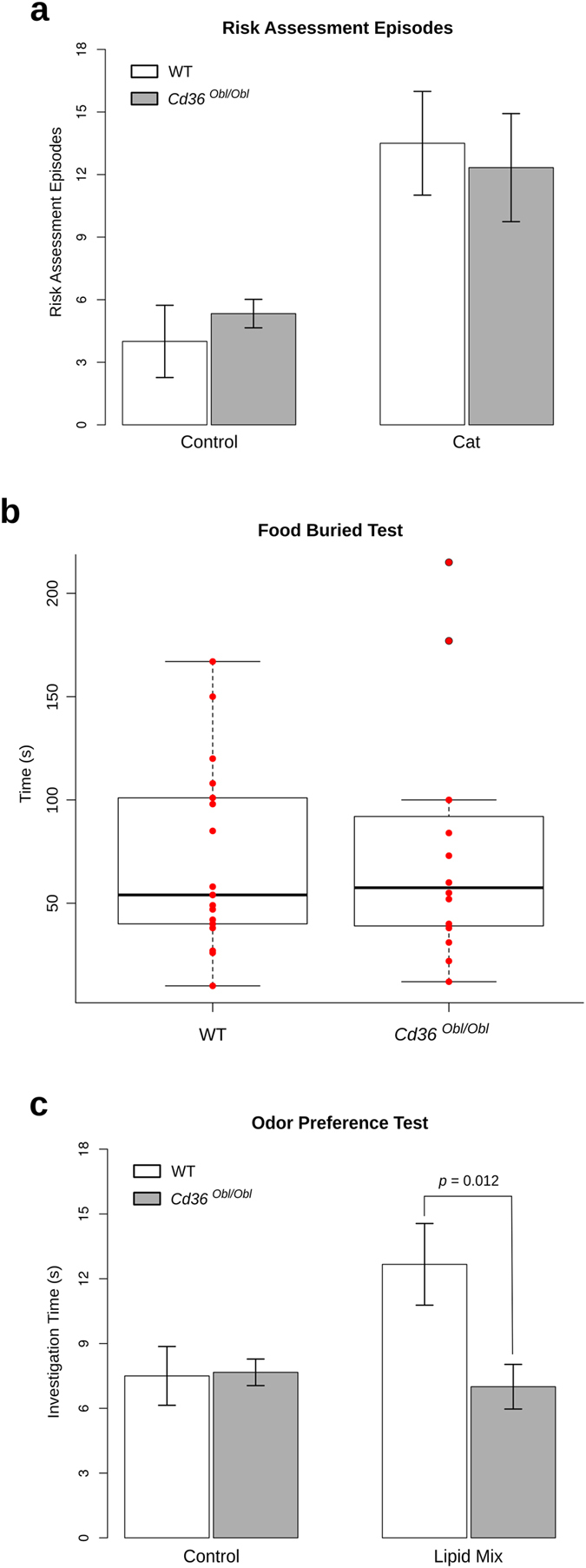
(a) The number of risk assessments was determined in wild-type and Cd36obl/obl mutant mice exposed to gauze rubbed over cat fur. CD36-deficient and wild-type mice showed equivalent responses to the predator kairomones (n = 6 for all groups except for control gauze exposed wild-type; n = 4). (b) Food buried test was performed to evaluate the general olfactory ability of Cd36obl/obl mice. The time required for the animals to find a hidden food pellet was recorded. No statistically significant difference was observed between the two strains. However, innate preference for a filter paper scented with a chemically defined lipid concentrate, evaluated by the investigation time, was lost in Cd36obl/oblmice (n = 6/group) (c). Statistical analysis was performed using two-way ANOVA (a,c) and Welch’s test (b).
Since CD36-deficiency does not lead to aberrant OE morphology or general olfactory behavior impairment, we reasoned that only responses to specific odorants that bind to the receptor should be disturbed in the mutants. Among CD36 known ligands, lipids are candidate molecules, which could induce olfactory responses as environmental cues. In fact, we found that Cd36obl/obl mice animals exhibited significantly decreased investigation time towards a lipid mixture containing saturated and unsaturated long-chain fatty acids (LCFAs), when compared to wild-type mice (Fig. 8c).
Discussion
Previous studies have shown that CD36 and its homologue play a role in chemosensory responses to lipid ligands in insects and mammals16,19. In mice, CD36 is involved in gustatory detection of lipids, and in preferential attraction to lipid-enriched foods and beverages, responses which are abolished in CD36-null mice19,30,31.
Here we show that CD36 is highly expressed in a subset of mature OSNs present in the MOE. These neurons are located in the mature OSN layer and scattered in all zones of the olfactory epithelium. The number of CD36 expressing neurons increases during development, and persists in adult mice. These findings raise important questions regarding the possible functional roles played by CD36 in olfaction.
It has become clear now that the MOE is composed of distinct subpopulations of olfactory neurons. Some of these are well known, like the canonical olfactory sensory neurons, others have been only recently characterized; and there may be other subpopulations, yet to be identified. As mentioned above, the many subpopulations are mainly distinguished by the type of receptor they express: ORs, TAARs, GC-D, and there are also a few olfactory neurons that express V1Rs or V2Rs32,33. Here we identify a new subpopulation of olfactory sensory neurons which express CD36, a receptor that binds lipid ligands and has been previously proposed as a candidate fat taste transducer34.
Some olfactory neurons express members of the Trp channel family, however, it is not clear yet whether they work as receptors or simply as transducing effectors4. A newly identified population of Trpc2-expressing OSNs was recently reported, indicating that previous findings attributed to the VNO Trpc2-positive neurons could also be mediated by MOE cells35. Similarly, our results suggest that, in the olfactory system, the CD36 molecule may have similar functions attributed to these proteins in taste cells.
CD36-positive neurons coexpress Omp and Gnal, initially suggesting that they represent a subpopulation of canonical OSNs. We therefore analyzed whether these cells also express ORs. We did not detect coexpression of Cd36 with 13 different tested ORs; however, we observed a frequent coexpression of Cd36 with Olfr287. Since ~16% of the Cd36+ OSNs in OE zone 4 coexpress Olfr287 we could predict, assuming equivalent proportions, that the remaining Cd36+-OSNs within this same expression zone would express at least additional five OR genes. Also, if Cd36+ OSNs in the other expression zones show similar coexpression frequencies as Cd36+/Olfr287+-OSNs, we would expect that additional eighteen ORs are coexpressed in these cells, totalizing ~24 different OR genes that can be coexpressed with CD36 in the whole OE. The observation that ~40% of Olfr287 cells do not express Cd36 suggests heterogeneity of these neurons. In fact, this is not an unpreceded finding, since Omura and Mombaerts35 showed recently that approximately a third, and not all, of Olfr68/Olfr69+ MOE cells express Trpc2. Altogether, it seems that some OSNs in the MOE are heterogeneous in terms of expression of accessory molecules, even if they have the same Olfr gene identity. Future experiments will be required to determine the complete set of ORs expressed in this subpopulation of OSNs.
Since CD36-OSNs do not represent a major subpopulation of OSNs, one would expect little impact in general olfactory parameters. The general morphology and organization of the CD36-deficient MOE is comparable to wild-type mice, which is also valid for general olfactory behavioral parameters. As mentioned above, CD36 in adipose and taste cells binds LCFAs13. In order to characterize specific olfactory behavior changes possibly related to CD36 loss of function, we tested whether CD36 deficiency impairs preference for a chemically defined lipid concentrate rich in LCFAs. We found that even though Cd36obl/obl mice show normal general olfactory behaviors towards regular odorants, they do not show preference for a lipid mixture when compared to wild-type mice. Preference tests for lipid mixtures have been previously performed in other studies, but the specific contributions of the gustatory or olfactory systems to the behavioral responses could not be distinguished in these reported experiments19. Our study aimed to test specifically olfaction-driven instinctive behavioral responses. However, we cannot rule out the possibility that low concentrations of volatile fatty acids detected by CD36-taste cells contributed to the observed behavior. It is also possible that a generalized CD36-deficiency provokes specific impairment of lipid-elicited responses evaluated by preference tests, excluding an olfactory role for CD36. Additional experiments, including those involving OSN-specific Cd36 deficient mice, are necessary in order to support the hypothesis that CD36+-OSNs are true lipid sensors functionally coupled to a brain circuitry involved in lipid preference behavior. Nevertheless, our results raise the interesting possibility that CD36 expressing olfactory neurons are involved in the recognition of lipid odorants, mediating the attraction to this type of chemosensory cue, especially because long-chain fatty acids should be present at very low concentrations in the air-phase. In this case, lipid-associated signal transduction mechanisms in the MOE could influence important social and feeding behaviors. For instance, the fatty alcohol (Z)-5-tetradecen-1-ol from exocrine glands was recently identified as a compound that enhance male urine attractiveness to female mice36. Recently, a study performed with human subjects suggested that fat content of food can be detected via odors37, implying that lipid odorants may be more physiologically relevant than previously assumed. One may expect that fatty odors would be important to identify highly caloric food that should be strategically consumed, but this hypothetical behavior may function in the opposite direction in a different metabolic context.
Previous studies have implicated CD36 in neuronal functions. For instance, it was shown that a subpopulation of glucose-responsive hypothalamic neurons senses fatty acids through CD3638,39. In addition, different studies implicated CD36 in lipid intake, satiety and obesity40,41,42,43. Although there is insufficient evidence to conclude that olfactory CD36 mediates fatty acid detection, the identification of a novel population of olfactory neurons expressing CD36 provides a new perspective to neuronal signaling with relevant impact to olfactory functions. It is known that each subpopulation of OSNs makes distinct connections in the OB, indicating that they provide information from different chemosensory inputs27. It will be interesting to determine how the axonal projection from CD36 expressing neurons to the main OB is organized. The fact that, unlike other OSN subpopulations, the CD36+-neurons are widely distributed all over the olfactory epithelium, and not segregated in different expression zones, suggests that the projection pattern of these neurons to the OB is different. It is possible that these neurons relay information in non-conventional forms that still need to be clarified.
Importantly, the possibility that CD36 may not work as a receptor involved in odorant signal transduction cannot be excluded. For example, CD36 could function as a metabolic sensor that informs the neurons about nutritional status or even transport fatty acids through the cell membrane. In addition, as an innate immune signaling co-receptor, CD36 can also sense lesions or nasal flora imbalance. These possibilities will have to be carefully addressed in order to determine the roles played by CD36 in olfactory physiology. During the preparation of the revised version of this manuscript two independent groups44,45 also reported expression of CD36 in OSNs and supporting cells.
Methods
Animal experimentation
Cd36obl /Mmucd mice15 were acquired from MMRRC (UC Davis) in heterozygosis and were intercrossed to yield homozygotes mutants and wild-type mice; genotyping was performed by PCR followed by DNA sequencing. Wild-type C57BL/6J and Olfr17tm7Mom /MomJ (P2-IRES-tauGFP) mice were obtained from the Jackson Laboratories. Animal experimentation was approved by the local Committees (COBEA-USP and Comitê de Ética em Pesquisa – UNIFESP) and the methods were carried out in accordance with the approved guidelines.
Tissue dissection and total RNA extraction
Tissues were dissected from male mice. Dorsal tongue dissection was conducted by minimizing skeletal muscle contamination and prioritizing superficial epithelial tissue. Total RNA was extracted using Trizol® reagent (Invitrogen™) according to the manufacturer instructions. Prior to cDNA synthesis we carried out DNAse I treatment (Qiagen®) on the RNA, followed by purification with RNeasy® mini kit (Qiagen®).
cDNA synthesis and qPCR
cDNA was synthesized from 1 μg of purified total RNA as described previously46. Briefly, 1 μg of total RNA and 100 ng of oligo (dT) were incubated for 2 minutes at 70 °C followed by rapid chilling on ice. cDNA was synthesized in the presence of RNAse inhibitor (RNaseOUT™, Invitrogen™) and 200 U of Superscript II reverse transcriptase (Invitrogen™) in 1x Superscript II first strand buffer at 42 °C for 60 minutes. Cd36, Actb and Gapdh transcripts were quantified by real-time PCR using the ABI 7300 Real-Time PCR system (Applied Byosystems®). Primer sequences for Cd36 were GATGACGTGGCAAAGAACAG (forward) and TCCTCGGGGTCCTGAGTTAT (reverse)19. Primer sequences for Actb were GCAAGCAGGAGTACGATGAG (forward) and CCATGCCAATGTTGTCTCTT (reverse). Primer sequences for Gapdh were AAATGGTGAAGGTCGGTGTG (forward) and TGAAGGGGTCGTTGATGG (reverse). All reactions were performed by using a standard real time PCR protocol (1 cycle of 95 °C for 10 min, 40 cycles of 95 °C for 15 s, and 60 °C for 1 min). Data was normalized by using Gapdh and Actb as references. Relative gene expression across tissues was then calculated as 2−ΔΔCt with the respective standard deviation values, using the OE sample as calibrator, according to Livak et al.47. Each reaction was performed in triplicate and the standard deviation was inferior to 0.3. Actb was preferred to Hprt as normalizer because the former showed less variability among the different tissues. Nevertheless, both normalizers produced similar results (Fig. S1).
Tissue Preparation for Histology
Newborns were sacrificed by decapitation and heads were immediately fixed by immersion. The other mice were deeply anesthetized via an intraperitoneal injection of a mixture of ketamine hydrochloride and xylazine and then rapidly perfused transcardially with 0.9% saline, followed by 4% formaldehyde in Borax pH 9, at 4 °C. The noses were removed rapidly, postfixed and decalcified in 2% formaldehyde/0.25 M EDTA, and then placed in 20% sucrose solution overnight at 4 °C for cryoprotection. For fresh frozen sections the fixation step was skipped. The noses were embedded in Tissue-Tek® O.C.T.™ media and were cut on a Microm cryostat (Thermo Scientific™) into 16 μm coronal sections. The slices were collected onto silane-coated adhesive microscope slides and stored at −20 °C.
Immunofluorescence (IF)
The fresh frozen sections were fixed in cold methanol (−20 °C for 20 min), washed with PBS several times and incubated with blocking solution (PBS supplemented with 1% BSA and 4% donkey serum) followed by the addition anti-CD36 (EMD Millipore, #MAB1258) and anti-acetyl-α-tubulin (Lys40) (Cell Signaling Technologie®, # 5335S) antibodies. Subsequently, the sections were washed and primary antibody detection was conducted with compatible Alexa Fluor® -488 and -546 secondary antibodies (Molecular Probes®). Conventional immunofluorescence in fixed tissue was performed by incubating washed sections in blocking solution (PBS containing 4% donkey serum, 1% BSA, and 0.4% Triton™ X-100) for 30 min. Using the same buffer solution composition, the sections were incubated over night at 4 °C with primary antibody anti-OMP (WAKO, Ltd. Japan #544–10001) or anti-TUBB3 (Aviva System Biology, CA #ARP63784). After incubation with the primary antibody, OE sections were rinsed in PBS and incubated with compatible Alexa Fluor® -488 and -546 secondary antibodies for 120 min. Sections were then rinsed with PBS and counterstained with DAPI for nuclear labeling. Staining controls were performed to assure the specificity of CD36 labeling (Fig. S3).
cRNA probe synthesis
Cd36 sense and antisense riboprobes were synthesized as described before20, except that digoxigenin (DIG)-11-UTP (Roche) or DNP-11-UTP (PerkinElmer) were used instead of an isotopic labeled nucleotide. Gnal and Omp riboprobes were synthetized as described48, while Taar7b, Cyp2a4, Olfr78, Olfr124, Olfr287, Olfr569, Olfr638, Olfr646, Olfr691, Olfr692, Olfr1264, Olfr1372-ps1, Olfr1507, Olfr1509 and Olfr1512 DNA segments were amplified by PCR from OE cDNA. Primers used for Taar7b were CTACCCCACCAGGTTTACT (forward) and AGGAATTCTCCCTCAAGACT (reverse); primers for Cyp2a4 were AGAGATTTGCAGACCTGATCC (forward) and ATGCAGCCCAGGATCAACG (reverse); primers for Olfr1507 were TTTTAAATTGTCCTGACAAACTGG (forward) and TCTGATTCTCTCAGTCCCTTCA (reverse); primers for Olfr78 were GAGGAAGCTCACTTTTGGTTTGG (forward) and CAGCTTCAATGTCCTTGTCACAG (reverse); primers for Olfr124 were GGTAATATCTCCATTATCCTAGTTTCCC (forward) and TTGACCCAAAACTCCTTTGTTAGTG (reverse); primers for Olfr287 were GGGCTCTTGGGATAGGTAG (forward) and GGGCTGGCTTACAGGTTTAG (reverse); primers for Olfr569 were TAACAGCTCTTCCCATCCCCTGTTC (forward) and TAGGGTTGAGCATGGGAGGAACAAGC (reverse); primers for Olfr638 were AGAAGTAACTAACACCACTCATGGC (forward) and TTAGTGCACCTTTCTTTGCAAC (reverse); primers for Olfr646 were ATGAGCGAAAGTCTCCCAGTCACTC (forward) and AAGTGAAGCAGTTTTAGAAGCCGAC (reverse); primers for Olfr691 were TGGGTTGGAGGCTTATCATACCTG (forward) and AAGAACAACACAGAGTCTTGATGTC (reverse); primers for Olfr692 were AAACTTCATCCTTACAGAATGGCAG (forward) and ACTGGCTTTGGGACAGTGTGA (reverse); primers for Olfr1264 were TTTCTCAGAGGCCAGAGATCCAG (forward) and CTACAACTTCTGAGCGTGTGAAGAG (reverse); primers for Olfr1372-ps1 were CCCTAGGTCATTTAGCAGTTATTGTCAC (forward) and CAGATCTTGACTTGATCTTCAACACTG (reverse); primers for Olfr1509 were ATGGGAGCTCTAAATCAAACAAGAG (forward) and TAGAAAACCGATACCACCTTGTCG (reverse); primers for Olfr1512 were TACATCCTGACTCAGCTGGGGAACG (forward) and GGGCACATAGTACACAGTAACAATAGTC (reverse).
In situ hibridization, two-color fluorescence in situ hybridization (FISH) and double-labeling ISH/IF
Pre-hybridization/hybridization was conducted according to an adapted protocol from a previously described procedure49. Briefly, the OE sections were immersed in 4% formaldehyde, followed proteinase K treatment at 37 °C and acetylation in triethanolamine solution. Tissue sections were dehydrated through graded concentrations of ethanol and air-dried. Hybridization solution containing the probe was spotted onto the slides, which were coverslipped and incubated overnight at 60 °C in a slide moat incubator (Boekel; Feasterville, PA). Post-hybridization was carried out with several washes in decreasing concentrations of standard saline citrate (SSC) solution and RNAse treatment. Chromogenic DIG detection was performed according to manufacturer by employing alkaline phosphatese (AP)-conjugated antibodies against DIG and NBT/BCIP as substrate (Roche). Two-color FISH was performed adapting a previously described protocol50. DIG-labeled probes signals were developed with tyramide-Alexa Fluor® 488 (Tyramide signal amplification kit, Molecular Probes®), which was followed by HRP quenching with 3% H2O2, several washes and anti-rabbit-HRP (Jackson ImmunoResearch; West Grove, PA) incubation. DNP-labeled probes signals were developed with tyramide-Alexa Fluor® 555 (Tyramide signal amplification kit, Molecular Probes®). In some assays fluorophore labeling for each hapten detection was inverted, and we also made use of tyramide-Alexa Fluor® 546, and tyramide-biotin (PerkinElmer)/Avidin- Alexa Fluor® 488 reagents (Molecular Probes®) along with Avidin/Biotin Blocking kit (Vector Laboratories). For ISH/IF combination, IF was performed after ISH signal development using a shorter proteinase K treatment protocol.
Olfactory behavioral assays
Risk assessment behavior exhibited towards predator odors was used as a VNO-mediated behavior in our control investigation of the effect of CD36 genetic ablation on VNO function. Risk assessment behaviors have been consistently used as indicators of defensive behavior in many animals51. The experiments were performed as previously described29. Animals were habituated in the procedure room for two consecutive days, in the dark, for 90 min each time. On the following day, mice were assayed by exposing them to a control clean sterile and deodorized medical gauze, or to a medical gauze rubbed against the fur of a domestic cat. Each exposure session was filmed and the digitalized movies were scored blindly to annotate risk assessment episodes, which were quantified for the first 12 min of the assay.
The buried food test was carried out based on protocols described in previous studies52. Mice were placed in home cages (polysulfone, 46 cm L × 23.5 cm W × 20 cm H) with 5 cm of clean bedding and subjected to fasting starting on the afternoon on the day before the behavioral assays. Mice were manipulated and allocated into the experimental room for one hour habituation 48 and 24 h prior the test day, and were fasted for 24 hours before the test. The experiment was performed placing each mouse, individually, in the middle of a clean cage containing 5 cm deep of clean bedding and the stimulus (food pellet measuring about 1 cm) was hidden at the bottom of the cage, in a random corner of the cage. We considered that the animal found the food pellet when it started eating it, usually holding the pellet with its forepaws. Each experimental trial lasted for 5 minutes; therefore, the maximum score was 300 sec. After the test, mice returned to their home cage and food was replaced ad libitum.
The innate odor preference test was conducted as previously described53. In these experiments, mice were used only once in order to avoid adding the confounding factor of olfactory learning. Mice (n = 5–10) were individually caged (test cages were 50 L × 30 W × 25 H cm) two days prior to the onset of each experimental trial. They were then habituated for 30 min four times on two consecutive days by placing them on a clean cage identical to the home cage, but without bedding, food grid or water bottle. On the test day, mice were transferred to the empty test cage with a deodorized filter paper (2 × 2 cm) scented with PBS (control), amyl acetate (1 mM), or a chemically defined lipid concentrate (Gibco®; lipid composition in (mg/l): arachidonic acid (2.0), cholesterol (220.0), DL-alpha-tocopherol acetate (70.0), linoleic acid (10.0), linolenic acid (10.0), myristic acid (10.0), oleic acid (10.0), palmitic acid (10.0), palmitoleic acid (10.0), and stearic acid (10.0)). Similar results were obtained when the concentration of amyl acetate used was 100 μM (not shown). In each trial, the filter paper was placed inside a metallic wire ball (tea ball) to prevent the animals from directly contacting the paper or probing it with the tongue. Behaviors were recorded on digital video camera and investigation times were measured for a 5 min exposure session.
Image processing
Bright-field and epifluorescence photomicrographs (Nikon® or Carl Zeiss) or confocal acquisitions (Carl Zeiss LSM 780 and Leica SP8) were processed to enhance contrast and image quality using GIMP (http://www.gimp.org/) and were assembled using Inkscape (http://inkscape.org). Pictures representing different groups received equivalent image treatment. Orthogonal projections from confocal acquisitions were exported from BioImageXD free analysis software (www.bioimagexd.net).
Quantitative analysis
During postnatal development, the density of Cd36-positive cells in OSN layer were determined by counting the total number of completely stained nucleated cells per linear length of the OE (lamina basal as reference) in each field (region of interest), or per OE area. It is important to point out that robustly stained Cd36 cells in OSN layer, contrast with the weakly stained supporting cells, since signal development was controlled. Transcript coexpression in the same cells was carefully conducted both in widefield and confocal microscope acquisitions. We only considered for counting transcript-positive cells in the OSN layer with robust staining, defined nucleus, and restricting Cd36+-OSNs counting to the same OE zone where the ORs are expressed. In some few cases, coexpression could not be determined and was classified as “unsure”.
Statistical analysis
A student’s t-test variation, Welch’s test, was employed for comparison between two groups according to homogeneity of variances and normality tests. For more groups and two grouping categorical independent variables, comparison of means was performed using a two-way analysis of variance (ANOVA) followed by a Tukey’s HSD procedure as post hoc multiple comparisons test when pertinent. Frequencies were evaluated with binomial test (expected frequencies) or Fisher’s exact test (contingency tables). The analyses were carried out with R (cran.r-project.org; base, and packages RVAideMemoire, car and phia). Heatmap was generated using ordered rows imposed by dendrogram according to the function heatmap.2 (gplots; R). Co-localization probability was calculated with BioImageXD (www.bioimagexd.net) according to Costes methods54.
Additional Information
How to cite this article: Xavier, A. M. et al. CD36 is expressed in a defined subpopulation of neurons in the olfactory epithelium. Sci. Rep. 6, 25507; doi: 10.1038/srep25507 (2016).
Supplementary Material
Acknowledgments
This work was supported by research grants to I.G. from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP: 2007/53732-8) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq 484869/2012-4). I.G. is member of the CEPID Redoxoma (FAPESP 2013/07937-8). B.M. is supported by research grant from FAPESP (FAPESP11/51604-8). We are grateful to Darren Logan (University of Cambridge) for sharing unpublished data. We thank Umberto Crisafulli for assistance with OE cryosections, Daniela Gonzalez-Kristeller for providing the Olfr1507 plasmid, Thiago Seike Nakahara for providing several ORs plasmids, and Maria Christina W. Avellar for acquisitions of wide field microscope images. We also thank the technical support for confocal images acquisitions in Laboratórios Multiusuários do INFAR-UNIFESP, and the staff of the animal facilities for excellent animal care.
Footnotes
Author Contributions Conceived and designed the experiments: A.X., I.G., B.M. and F.P. Performed the experiments: A.X., H.W., R.L., M.N., T.R., M.H. and T.A. Analyzed the data: A.X., M.N., I.G., B.M. and F.P. Contributed reagents/materials/analysis tools: I.G., B.M. and F.P. Wrote the paper: A.X., I.G., B.M. and F.P.
References
- Su C. Y., Menuz K. & Carlson J. R. Olfactory Perception: Receptors, Cells, and Circuits. Cell 139, 45–59 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat. Rev. Neurosci. 5, 263–278 (2004). [DOI] [PubMed] [Google Scholar]
- Farbman A. I. & Margolis F. L. Olfactory marker protein during ontogeny: immunohistochemical localization. Dev. Biol. 74, 205–215 (1980). [DOI] [PubMed] [Google Scholar]
- Munger S. D., Leinders-Zufall T. & Zufall F. Subsystem organization of the mammalian sense of smell. Annu. Rev. Physiol. 71, 115–140 (2009). [DOI] [PubMed] [Google Scholar]
- Ihara S., Yoshikawa K. & Touhara K. Chemosensory signals and their receptors in the olfactory neural system. Neuroscience 254, 45–60 (2013). [DOI] [PubMed] [Google Scholar]
- Buck L. & Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65, 175–187 (1991). [DOI] [PubMed] [Google Scholar]
- Malnic B., Hirono J., Sato T. & Buck L. B. Combinatorial receptor codes for odors. Cell 96, 713–723 (1999). [DOI] [PubMed] [Google Scholar]
- Dewan A., Pacifico R., Zhan R., Rinberg D. & Bozza T. Non-redundant coding of aversive odours in the main olfactory pathway. Nature 497, 486–489 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberles S. D. & Buck L. B. A second class of chemosensory receptors in the olfactory epithelium. Nature 442, 645–650 (2006). [DOI] [PubMed] [Google Scholar]
- Zhang J., Pacifico R., Cawley D., Feinstein P. & Bozza T. Ultrasensitive detection of amines by a trace amine-associated receptor. J. Neurosci. 33, 3228–39 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinders-Zufall T. et al. Contribution of the receptor guanylyl cyclase GC-D to chemosensory function in the olfactory epithelium. Proc. Natl. Acad. Sci. USA 104, 14507–14512 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. et al. Detection of near-atmospheric concentrations of CO2 by an olfactory subsystem in the mouse. Science 317, 953–957 (2007). [DOI] [PubMed] [Google Scholar]
- Silverstein R. L. & Febbraio M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci. Signal. 2, re3 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abumrad N. A., el-Maghrabi M. R., Amri E. Z., Lopez E. & Grimaldi P. A. Cloning of a rat adipocyte membrane protein implicated in binding or transport of long-chain fatty acids that is induced during preadipocyte differentiation. Homology with human CD36. J. Biol. Chem. 268, 17665–17668 (1993). [PubMed] [Google Scholar]
- Hoebe K. et al. CD36 is a sensor of diacylglycerides. Nature 433, 523–527 (2005). [DOI] [PubMed] [Google Scholar]
- Benton R., Vannice K. S. & Vosshall L. B. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature 450, 289–293 (2007). [DOI] [PubMed] [Google Scholar]
- Li Z., Ni J. D., Huang J. & Montell C. Requirement for Drosophila SNMP1 for rapid activation and termination of pheromone-induced activity. PLoS Genet. 10, e1004600 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard D., Passilly-Degrace P. & Besnard P. Molecular mechanisms of fat preference and overeating. Ann. N. Y. Acad. Sci. 1141, 163–175 (2008). [DOI] [PubMed] [Google Scholar]
- Laugerette F. et al. CD36 involvement in orosensory detection of dietary lipids, spontaneous fat preference, and digestive secretions. J. Clin. Invest. 115, 3177–3184 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glezer I., Bittencourt J. C. & Rivest S. Neuronal expression of Cd36, Cd44 and Cd83 antigen transcripts maps to distinct and specific murine brain circuits. J. Comp. Neurol. 517, 906–924 (2009). [DOI] [PubMed] [Google Scholar]
- Boehm U., Zou Z. & Buck L. B. Feedback loops link odor and pheromone signaling with reproduction. Cell 123, 683–695 (2005). [DOI] [PubMed] [Google Scholar]
- Guillamon A. & Segovia S. Sex differences in the vomeronasal system. Brain Res. Bull. 44, 377–382 (1997). [DOI] [PubMed] [Google Scholar]
- Scalia F. & Winans S. S. The differential projections of the olfactory bulb and accessory olfactory bulb in mammals. J. Comp. Neurol. 161, 31–55 (1975). [DOI] [PubMed] [Google Scholar]
- Yoon H., Enquist L. W. & Dulac C. Olfactory inputs to hypothalamic neurons controlling reproduction and fertility. Cell 123, 669–682 (2005). [DOI] [PubMed] [Google Scholar]
- Tsuboi A. et al. Olfactory neurons expressing closely linked and homologous odorant receptor genes tend to project their axons to neighboring glomeruli on the olfactory bulb. J. Neurosci. 19, 8409–8418 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M., Vaes E. & Mombaerts P. Temporal patterns of odorant receptor gene expression in adult and aged mice. Mol. Cell. Neurosci. 57, 120–9 (2013). [DOI] [PubMed] [Google Scholar]
- Mombaerts P. et al. Visualizing an olfactory sensory map. Cell 87, 675–686 (1996). [DOI] [PubMed] [Google Scholar]
- Saraiva L. R. et al. Hierarchical deconstruction of mouse olfactory sensory neurons: from whole mucosa to single-cell RNA-seq. Sci. Rep. 5, 18178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papes F., Logan D. W. & Stowers L. The Vomeronasal Organ Mediates Interspecies Defensive Behaviors through Detection of Protein Pheromone Homologs. Cell 141, 692–703 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teboul L. et al. Structural and functional characterization of the mouse fatty acid translocase promoter: activation during adipose differentiation. Biochem. J. 360, 305–312 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard D. et al. The gustatory pathway is involved in CD36-mediated orosensory perception of long-chain fatty acids in the mouse. FASEB J. 22, 1458–1468 (2008). [DOI] [PubMed] [Google Scholar]
- Karunadasa D. K., Chapman C. & Bicknell R. J. Expression of pheromone receptor gene families during olfactory development in the mouse: Expression of a V1 receptor in the main olfactory epithelium. Eur. J. Neurosci. 23, 2563–2572 (2006). [DOI] [PubMed] [Google Scholar]
- Pascarella G. et al. NanoCAGE analysis of the mouse olfactory epithelium identifies the expression of vomeronasal receptors and of proximal LINE elements. Front. Cell. Neurosci. 8, 41 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Yassimi A., Hichami A., Besnard P. & Khan N. A. Linoleic acid induces calcium signaling, Src kinase phosphorylation, and neurotransmitter release in mouse CD36-positive gustatory cells. J. Biol. Chem. 283, 12949–12959 (2008). [DOI] [PubMed] [Google Scholar]
- Omura M. & Mombaerts P. Trpc2-Expressing Sensory Neurons in the Main Olfactory Epithelium of the Mouse. Cell Rep. 8, 583–595 (2014). [DOI] [PubMed] [Google Scholar]
- Yoshikawa K., Nakagawa H., Mori N., Watanabe H. & Touhara K. An unsaturated aliphatic alcohol as a natural ligand for a mouse odorant receptor. Nat. Chem. Biol. 9, 160–2 (2013). [DOI] [PubMed] [Google Scholar]
- Boesveldt S. & Lundström J. N. Detecting fat content of food from a distance: Olfactory-based fat discrimination in humans. PLoS One 9, 1–5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Foll C., Irani B. G., Magnan C., Dunn-Meynell A. A. & Levin B. E. Characteristics and mechanisms of hypothalamic neuronal fatty acid sensing. Am. J. Physiol. Regul. Integr. Comp. Physiol. 297, R655–R664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Foll C., Dunn-Meynell A., Musatov S., Magnan C. & Levin B. E. FAT/CD36: a major regulator of neuronal fatty acid sensing and energy homeostasis in rats and mice. Diabetes 62, 2709–2716 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdener M. H. et al. CD36- and GPR120-mediated Ca2+ signaling in human taste bud cells mediates differential responses to fatty acids and is altered in obese mice. Gastroenterology 146, 995–1005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. S. Y. et al. Preference for linoleic acid in obesity-prone and obesity-resistant rats is attenuated by the reduction of CD36 on the tongue. Am. J. Physiol. Integr. Comp. Physiol. 305, R1346–R1355 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller K. L. et al. Common Variants in the CD36 Gene Are Associated With Oral Fat Perception, Fat Preferences, and Obesity in African Americans. Obesity 20, 1066–1073 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepino M. Y., Love-Gregory L., Klein S. & Abumrad N. A. The fatty acid translocase gene CD36 and lingual lipase influence oral sensitivity to fat in obese subjects. The Journal of Lipid Research 53, 561–566 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. et al. Expression of CD36 by Olfactory Receptor Cells and Its Abundance on the Epithelial Surface in Mice. PLoS One 10, e0133412 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberland S. et al. CD36 is involved in oleic acid detection by the murine olfactory system. Front. Cell. Neurosci. 9, doi: 10.3389/fncel.2015.00366 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaloski J. S. et al. Common promoter elements in odorant and vomeronasal receptor genes. PLoS One 6, doi: 10.1371/journal.pone.0029065 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Von Dannecker L. E. C., Mercadante A. F. & Malnic B. Ric-8B, an olfactory putative GTP exchange factor, amplifies signal transduction through the olfactory-specific G-protein Galphaolf. J. Neurosci. 25, 3793–3800 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme N., Lacroix S. & Rivest S. An essential role of interleukin-1beta in mediating NF-kappaB activity and COX-2 transcription in cells of the blood-brain barrier in response to a systemic and localized inflammation but not during endotoxemia. J. Neurosci. 19, 10923–10930 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T., Omura M. & Mombaerts P. Protocols for two- and three-color fluorescent RNA in situ hybridization of the main and accessory olfactory epithelia in mouse. J. Neurocytol. 33, 657–669 (2005). [DOI] [PubMed] [Google Scholar]
- Blanchard R. J., Yang M., Li C. I., Gervacio A. & Blanchard D. C. Cue and context conditioning of defensive behaviors to cat odor stimuli. Neurosci. Biobehav. Rev. 25, 587–595 (2001). [DOI] [PubMed] [Google Scholar]
- Yang M. & Crawley J. N. Simple behavioral assessment of mouse olfaction. Curr. Protoc. Neurosci. doi: 10.1002/0471142301.ns0824s48 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayakawa K. et al. Innate versus learned odour processing in the mouse olfactory bulb. Nature 450, 503–508 (2007). [DOI] [PubMed] [Google Scholar]
- Costes S. V. et al. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 86, 3993–4003 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.